Abstract
Cigars and cigarillos are emerging as popular tobacco alternatives to cigarettes. However, these products may be equally harmful to human health than cigarettes and are associated with similar adverse health effects. We used 16S rRNA gene amplicon sequencing to extensively characterize the microbial diversity and investigate differences in microbial composition across 23 different products representing three different cigar product categories: filtered cigar, cigarillo, and large cigar. High throughput sequencing of the V4 hypervariable region of the 16 s rRNA gene revealed 2124 Operational Taxonomic Units (OTUs). Our findings showed that the three categories of cigars differed significantly in observed richness and Shannon diversity, with filtered cigars exhibiting lower diversity compared to large cigars and cigarillos. We also found a shared and unique microbiota among different product types. Firmicutes was the most abundant phylum in all product categories, followed by Actinobacteria. Among the 16 genera shared across all product types were Bacillus, Staphylococcus, Pseudomonas, and Pantoea. Nine genera were exclusively shared by large cigars and cigarillos and an additional thirteen genera were exclusive to filtered cigars. Analysis of individual cigar products showed consistent microbial composition across replicates for most large cigars and cigarillos while filtered cigars showed more inter-product variability. These findings provide important insights into the microbial diversity of the different cigar product types.
Keywords: Cigar, Cigarillo, Filtered cigar, 16S rRNA gene amplicon sequencing, Microbiome
Introduction
Cigars in all product categories are emerging as popular tobacco products due to multiple factors that include lower cost as a result of the lower taxation rate when compared to cigarettes and the addition of flavors to cigar products [1, 2]. In recent years, cigars have become the most reported combustible tobacco product used by youth [3, 4]. A study conducted in the United States in 2020 of almost one million middle and high school students that self-reported having smoked cigars in the past 30 days, showed that the largest proportion of students reported using cigarillos (44.1%), followed by traditional cigars (33.1%), and filtered cigars (22.6%) [5, 6]. In this study, we focus on three different types of cigars, namely large (traditional) cigars, filtered cigars, and cigarillos. Large cigars come in a range of sizes and are made up of tobacco wrapped in leaf tobacco or homogenized tobacco leaf (HTL) material. Cigarillos are similar to large cigars but may be thinner and longer than a normal cigarette, while filtered cigars resemble cigarettes but are wrapped with HTL [7]. Cigar products are manufactured using specific varieties of tobacco which typically undergo a fermentation process that impacts the microbial population [8]. Additionally, the commercially available products used in this study use homogenized tobacco leaves for wrapper and binders, where applicable, as not all products contain both wrapper and binder. The products were expected to have differences as a result of the blend and ratio of the variety of cigar tobacco used to make the filler, the region of tobacco growth impacts the quality, and manufacturing processes. Contrary to popular belief, cigars are not less toxic than cigarettes but are associated with the same adverse health effects, including addiction to nicotine, oral lesions, oral cancer, lung cancer, cardiovascular disease, and chronic obstructive pulmonary disease [9–12]. Some studies have suggested that cigar products may be more harmful to human health than cigarettes [13–15]. Tobacco-specific nitrosamines (TSNAs) are a class of harmful compounds known to be carcinogenic and the formation of TSNAs has been shown to correlate with microbial metabolic activities [16, 17]. By studying microbial communities in cigars, organisms and pathways relating to the formation and possible reduction of TSNAs in tobacco products may be unveiled [18].
In this study, we use 16S rRNA gene amplicon to characterize the bacterial communities of cigar products in multiple product categories. 16S rRNA gene amplicon study is useful when examining multiple taxonomic domains simultaneously, especially when some of the microbes are not visually different and cannot be cultured in media [19]. Similar culture-independent methods have been used to investigate the microbial diversity of tobacco products, which revealed increased diversity compared to traditional culture-based approaches [18, 20]. We utilized the MiSeq platform to sequence the V4 region of samples representative of multiple cigar product categories which then allowed us to compare the similarities and differences between product types and analyze the associated microbial communities. The objective of the study is to extensively characterize microbial populations in cigar products that will fill a research gap and provide data to assess the potential health impact of cigars, providing important insights for the development of more effective tobacco control policies and public health interventions.
Materials and Methods
Sample Collection and DNA Extraction
A total of 23 different cigar products were obtained from online vendors and commercial distributors, including four research cigars (1C1, 1C2, 1C3, and 1C4) from the Center for Tobacco Reference Products (CTRP) at Kentucky Tobacco Research and Development Center (University of Kentucky, USA). The commercially available products were selected based on market share and sales within the United States to provide a comprehensive overview of the microbial compositions. The products were categorized into three groups: large standard cigars, filtered cigars, and cigarillos, consisting of five, six, and eight brands, respectively (Table 1). Cigars were randomly selected from the commercial packaging. Filtered cigars were selected from three packs within a carton, and 3 filtered cigars were randomly selected from various locations within each pack. Cigarillos were selected from three foil packs within an upright containing 15 foil packs, the industrial standard for packaging of cigarillos, and only one of the two cigarillos was used for analysis. The large cigars were randomly selected from a box of cigars containing up to 50 individually wrapped cigars. For all products tested, all tobacco material was homogenized prior to DNA extraction. DNA extraction was performed on freshly opened packages of all products using the ZymoBIOMICS™ DNA miniprep D4300 kit (Zymo Research, Irvine, CA, USA) and following the manufacturer’s recommended protocol. The extracted DNA was quantified using QUBIT 4.0. To minimize variability, we performed three technical replicates for each biological replicate, and three biological replicates were taken. The technical replicates were combined prior to sequencing to ensure that the data presented in the figures represent the averaged result of the technical replicates, thereby reducing technical variability. To ensure sterility, each product was opened under sterile conditions, homogenized in sterile saline solution (0.85%) using a bag mixer, filtered, and processed according to the kit instructions.
Table 1.
Tabulation of the different cigar products used with their name and category
| Large cigars | Filtered cigars | Cigarillos |
|---|---|---|
| 1C1 | 1C2 | 1C3 |
| 1C4 | Phillies | Phillies Black |
| William Penn | Captain Black | Phillies Sweet |
| Garcia y Vega | King Edward | White Owl black |
| White Owl | Talon | White Owl silver |
| Swisher Sweets | Cheyenne | Black & Mild |
| Dutch Masters | Djarum | Pom Pom sweet |
| Dutch Masters | ||
| Swisher Sweets |
Sequencing and Analysis of 16S rRNA Gene Sequences
We shipped the DNA samples from cigar products to The University of Michigan, Microbial Systems Molecular Biology Laboratory core sequencing facility (http://microbe.med.umich.edu/services/microbial-community-analysis) for PCR amplification and sequencing of the V4 region of the 16S rRNA gene on the Illumina MiSeq platform (dual-barcoded, paired-end reads, 2 × 250 flow cell) according to Kozich, et al. [21]. Sequence data from MiSeq sequencing was processed using MOTHUR software (v1.48.5) following the MiSeq SOP (https://www.mothur.org/wiki/MiSeq SOP, accessed January 2023) [22]. We followed the analysis methodology previously published by Law et al., 2020, using the SILVA reference alignment of the MOTHUR-formatted version of the RDP training set (SSU Silva 138 v.18) for classification [23, 24]. The cigar data set was subsampled and normalized to 3436 sequences per sample, which resulted in 2124 OTUs (operational taxonomic units) after classification at the 0.03 cutoff level. Raw sequence reads for all samples in this study were uploaded to the NCBI Bio Project database under accession number PRJNA1073920.
Statistical analysis was performed using the built-in functions in MOTHUR [22]. We compared bacterial community structure using the analysis of similarities function (ANOSIM) [25]. Observed Richness and Shannon’s diversity were calculated in MOTHUR and imported into the R program to compare alpha diversity measures and abundance using the Kruskal–Wallis rank sum test, with any significant results further tested by Dunn’s test [26]. The package ggplot2 was used to plot the data generated from MOTHUR [27]. The Venn diagram was made using VENNY 2.1 [28].
Results
Sequencing Dataset and Diversity
In this study, we extracted DNA from 69 different samples of cigar and cigarillo products, resulting in a total of 2,308,008 raw sequencing reads. After filtering out PCR and sequencing errors, 2,147,392 reads remained, leaving us with an error rate of 0.95%. We used Good’s coverage estimator to confirm that all samples had sufficient sampling depth, with values greater than 98.84%. We then calculated alpha diversity, which is based on the number of species within a sample, and measures for richness and Shannon diversity. We then compared them across cigar types using the non-parametric Kruskal–Wallis rank sum test. Our results indicate that the three categories of cigars (large, filtered, cigarillos) differ significantly in observed richness and Shannon diversity. Specifically, filtered cigars showed lower observed richness and Shannon diversity compared to large cigars and cigarillos, with large cigars having the highest measure of alpha diversity (Fig. 1). The Kruskal–Wallis rank sum test revealed a chi-squared value of 10.02 with a p-value of 0.01 for Shannon diversity. Using the Dunn test with Holm correction, we found a significant reduction in Shannon diversity in filtered cigars compared to both large cigars and cigarillos. Additionally, we observed that the microbial populations of filtered cigars were statistically different from those of cigarillos (p = 0.0388) and large cigars (p = 0.0026), while the microbial populations of cigarillos and large cigars were not statistically different.
Fig. 1.
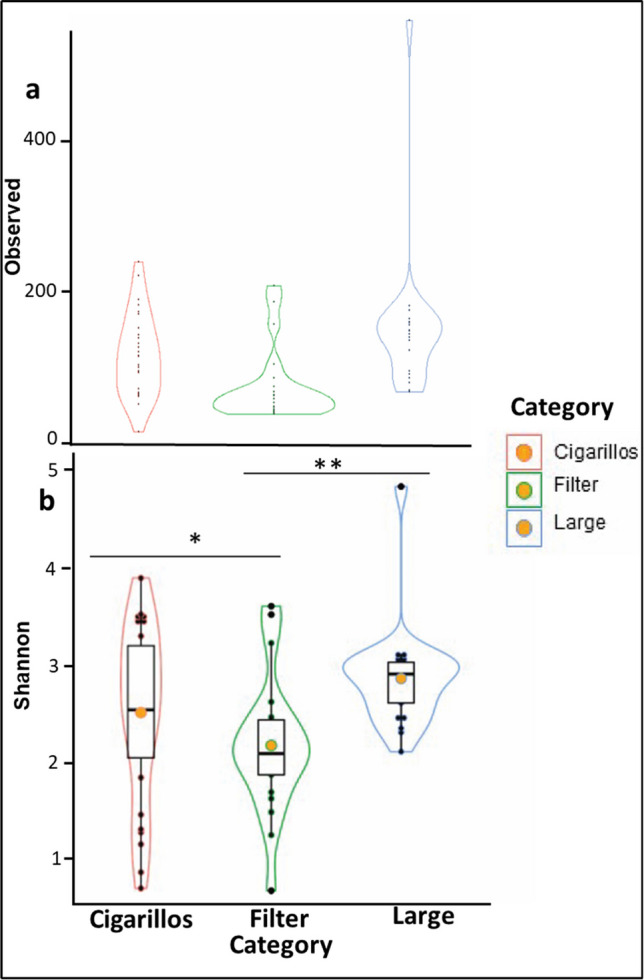
Alpha diversity (a) Observed richness (b) Shannon diversity by product category (Lines connecting box plots indicate significant difference using the Dunn test. (** P < 0.01; *P < 0.05). Filtered cigars showed lower observed richness and Shannon diversity compared to large cigars and cigarillos, with large cigars having the highest measure of alpha diversity
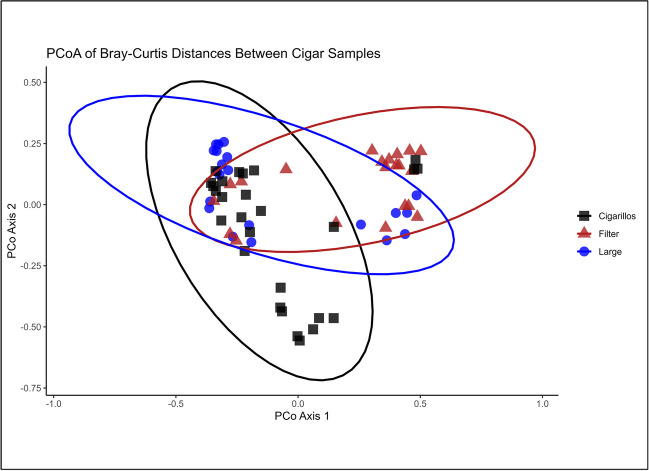
Furthermore, we employed Principal Coordinate Analysis (PCoA) to assess the dissimilarity of various cigar samples based on Bray–Curtis distances plotted as the results in Fig. 2. Subsequently, we subjected the data to the ANOSIM test, which yielded an R statistic value of 0.193, and the significance level was set at 0.001. These findings indicate the presence of significant beta diversity, which compares the diversity across samples, among the three cigar categories. Beta diversity analysis is crucial for comparing the diversity between different environments or groups, even when raw materials, formulations, and processes differ. It highlights differences in the presence or absence and abundance of microbial species among cigar types. This approach allows us to understand the distinct microbial communities associated with each cigar product category. When comparing filtered cigars with cigarillos, the computed R-value was 0.29 at a significance level of 0.01, indicating a substantial dissimilarity between the two groups. Similarly, the comparison between filtered cigars and large cigars also exhibited a significant level of dissimilarity. However, in contrast, the comparison between large cigars and cigarillos did not reach a level of statistical significance (Table 2).
Fig. 2.
Principal component analysis (PCoA) of Bray–Curtis distances between cigar samples of different product categories. The ellipse is drawn at a 90% confidence level. PCoA analysis indicated significant differences in beta diversity measures between the three cigar categories
Table 2.
Analysis of Similarities (ANOSIM) test with distance and category of product. Pairwise comparison using Mothur for each product type. Experiment-wise error rate: 0.05, pairwise error rate (Bonferroni): 0.0167. Microbial populations of filtered cigars are statistically different from those of cigarillos and large cigars
| Group Comparison | R-value | P-value |
|---|---|---|
| Cigarillos-Filter-Large | 0.19 | < 0.001* |
| Cigarillos-Filter | 0.291 | 0.001* |
| Cigarillos-Large | 0.044 | 0.087 |
| Filter-Large | 0.257 | < 0.001* |
Core and Shared Microbiomes Across Products
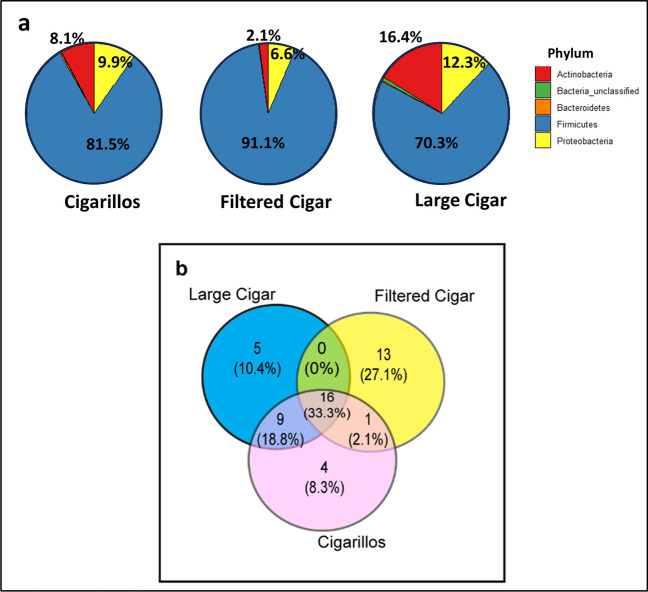
In our study, we clustered the overall sequences into 2124 Operational Taxonomic Units (OTUs) with 97% identity across all 69 cigar samples. The top five bacterial phyla identified across all cigar types were Firmicutes, Actinobacteria, Proteobacteria, Acidobacteria, and Bacteroidetes. The most abundant phyla in all product categories were Firmicutes, followed by Actinobacteria. Filtered cigars exhibited a slightly higher abundance of Firmicutes (91.12%) compared to large cigars (70.29%) and cigarillos (81.55%), while also showing a decrease in Actinobacteria (2.09%) compared to large cigars (16.42%) and cigarillos (8.05%) (Fig. 3a). When we compared the top 30 genera for the three cigar categories, 16 genera (33.3%) were shared among all product types, while nine genera were specifically shared only by large cigars and cigarillos. Apart from the core microbiome, there was no genus similarity between filtered cigars and large cigars (Fig. 3b). However, there were 13 exclusive genera only for filtered cigars. The list of shared and unique genera for each cigar type is provided in Table 3.
Fig. 3.
Core and shared microbiome in different categories in (a) Phylum level showing in each category in percentage (b) Genus level similarity and specific to each category in Venn diagram
Table 3.
List of genera shared and unique to specific cigar category types
| 16 common genera in all products | 9 common genera in “Large Cigar” and “Cigarillos” | 1 genus in “Filtered Cigar” and “Cigarillos” |
| Bacillus | Yaniella | Paenibacillaceae_1_unclassified |
| Acinetobacter | Lentibacillus | |
| Aerococcus | Brachybacterium | |
| Bacillaceae_2_unclassified | Gracilibacillus | |
| Bacillales_unclassified | Atopostipes | |
| Brevibacterium | Tetragenococcus | |
| Corynebacterium | Geomicrobium | |
| Enterobacteriaceae_unclassified | Aurantimonas | |
| Oceanobacillus | Rhodobacteraceae_unclassified | |
| Paenibacillus | ||
| Pantoea | ||
| Planococcaceae_unclassified | ||
| Pseudomonas | ||
| Sphingomonas | ||
| Staphylococcus | ||
| Terribacillus | ||
| 13 genera exclusively in “Filtered Cigar” only | 5 genera exclusively in “Large Cigar” only | 4 genera exclusively in “Cigarillos” only |
| Tissierella | Ralstonia | Sedimentibacter |
| Clostridium_XlVa | Bacteria_unclassified | Weissella |
| Planococcaceae_incertae_sedis | Desemzia | Thermoactinomyces |
| Sporomusa | Stenotrophomonas | Actinomycetales_unclassified |
| Enterococcus | Staphylococcaceae_unclassified | |
| Lachnospiraceae_unclassified | ||
| Veillonellaceae_unclassified | ||
| Firmicutes_unclassified | ||
| Garciella | ||
| Bacilli_unclassified | ||
| Clostridiales_unclassified | ||
| Clostridium_sensu_stricto | ||
| Dendrosporobacter |
Microbial Populations Within each Category
Total Genus Population Within Filtered Cigars
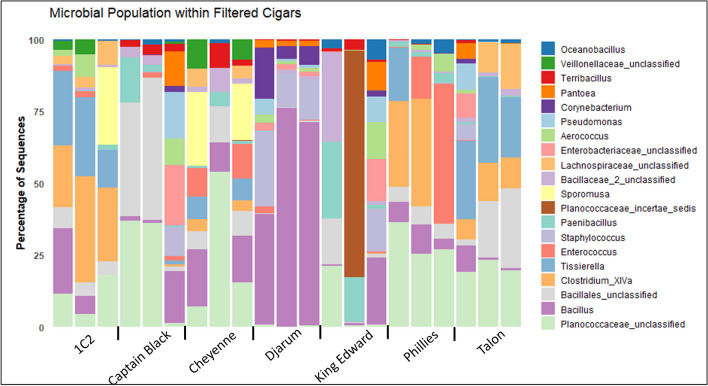
We analyzed seven different filtered cigars including one research filtered cigar (1C2). The top twenty genera of each product type were assessed based on the most numerous OTUs. The filtered cigars 1C2 and Phillies had uniform microbiome composition for each replicate (Fig. 4). Other products including Captain Black, Djarum, Talon, and Cheyenne have similar microbiomes for two of the three replicates. Djarum, the only filtered cigar product tested that included cloves with tobacco, had a higher percentage of Bacillus and Staphylococcus populations compared to other filtered cigars, whereas 1C2 and Talon had higher Tissierella genus. Of the seven products in the filtered cigar category, only the King Edward samples were significantly different for each replicate tested, making it difficult to determine the microbial population of this product.
Fig. 4.
Stacked bar chart showing the percentage of the dominant top 20 genera within the microbial population of each filtered cigar sample
Total Genus Population Within Cigarillos
In the case of cigarillos, we tested nine products. Cigarillo samples had a more consistent and uniform microbiome composition between the replicates. The research cigarillo (1C3), Black and Mild, White Owl Black, White Owl Silver, and Swisher Sweets have uniform microbiomes for each replicate (Fig. 5). For Black and mild, Pseudomonas represented more than 40% of the microbial population, and Pantoea represented 12% of the population which was the highest compared to other cigarillo types. White Owl Black and White Owl Silver had the highest percentage of Staphylococcus compared to the other products tested. Swisher Sweets and PomPom had a higher percentage of Terribacillus compared to other products, and they had similar microbiomes for two of the three replicates. Bacillus was the most common genus present in most samples. Two out of three replicates of Phillies Sweet showed higher levels of Lentibacillus. Dutch Masters and Phillies Black samples were different for each replicate.
Fig. 5.
Stacked bar chart showing the dominant top 20 genera of each cigarillos
Total Genus Population Within Large Cigars
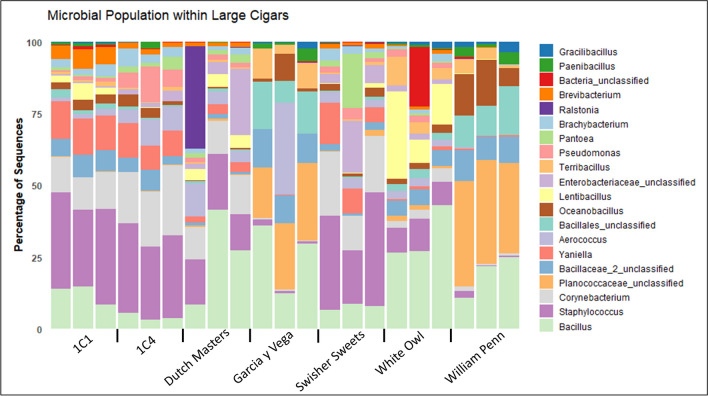
A total of seven products in the large cigar category were taken including two research cigar products (1C1 and 1C4). All products have a uniform microbiome for each replicate, except White Owl which had a consistent microbiome for two of the three replicates. All large cigar samples show a higher percentage (8.20–39.46%) of Staphylococcus genus except William Penn and Garcia y Vega (0.16–2.09%) (Fig. 6). Additionally, these products showed an increased percentage of Planococcaceae unclassified (17.62–36.70%) and Paenibacillus (0.72–4.3%) which was observed at low levels for the other products tested (0.01–0.08%, with one replicate on the 1C4 having 2.3%).
Fig. 6.
Stacked bar chart showing the dominant top 20 genera of each large cigar
Discussion
The use of 16S rRNA gene amplicon sequencing allowed for a comprehensive characterization of the microbial diversity of cigar and cigarillo products, revealing differences between product types and providing information on various microbial populations. This study explores the microbial diversity of large cigars, filtered cigars, and cigarillos, to provide insight into the composition of the microbial communities that are unique and shared in these products. Our study revealed a significant difference in microbial diversity among the three categories of cigars (large, filtered, cigarillos) based on alpha diversity measures for richness and Shannon diversity. Specifically, filtered cigars exhibited lower diversity compared to both large cigars and cigarillos. PCoA analysis with the ANOSIM test indicated significant differences in beta diversity measures between the three cigar categories, with the microbial populations of filtered cigars being statistically different from those of cigarillos and large cigars. Furthermore, when comparing the top 30 genera for the three cigar categories, the core microbiome analysis revealed that 16 genera were shared among all three categories of cigars, while nine genera were shared only by large cigars and cigarillos. These shared genera such as Oceanobacillus, Corynebacterium, Staphylococcus, Bacillus, and Pseudomonas are commonly found as the most predominant genera in tobacco products [29, 30]. The first three genera are shown to have a direct effect on the fermentation process [31–33]. Additionally, our study identified 13 exclusive genera only for filtered cigars, such as Tissierella, and Clostridium_XlVa. In contrast, there was no genus similarity between filtered cigars and large cigars (Table 3).
The presence of certain genera in all cigar types suggests the existence of a core microbiome in these products. However, there was no significant difference between the microbial populations of cigarillos and large cigars. The differences observed in the microbial composition of filtered cigars compared to large cigars and cigarillos could be due to the type of cigar tobacco (filler leaf, wrapper leaf), the region the tobacco was grown, the fermentation method during curing, or the manufacturing process. The most abundant phyla across all product categories were Firmicutes, followed by Actinobacteria, which agrees with similar studies to the one presented here [33–35]. Interestingly, filtered cigars had a slightly higher abundance of Firmicutes compared to large cigars and cigarillos, while also showing a decrease in Actinobacteria. A recent study showed that filtered cigar products have a unique bacterial signature and certain genera such as Enterobacteriaceae, Pantoea, Pseudomonas, and Staphylococcus were more abundant in the cigar tobacco used to produce the filtered cigar [1]. Microbial diversity studies conducted on cigarettes and various tobacco products, including smokeless forms, have uncovered a similar microbial population as found in the current study of cigar products [36]. Research on Burley and Flue-cured tobacco leaves has found Bacillus and Pseudomonas to be the predominant genera, while Proteobacteria is the dominant phylum, comprising over 90% of the operational taxonomic units (OTUs) in burley tobacco leaves [36, 37]. A recent study where sequencing was performed on the samples cultured from the mainstream smoke of unfiltered cigarettes showed Bacillus, Terribacillus, Paenibacillus, and Desulfotomaculum as their predominant genera [38]. Furthermore, bacterial community profiling in smokeless tobacco products revealed Firmicutes, Proteobacteria, Actinobacteria and Bacteroidetes as dominant phyla and Acinetobacter, Bacillus, Prevotella, Acetobacter, Lactobacillus were enriched bacterial genera. Additionally, microbial diversity and composition were determined to be variable across multiple smokeless tobacco product types and brands [18, 39–41].
We evaluated for inter-product variability within the microbiome composition of seven different filtered cigars, nine cigarillos, and seven large cigars. The filtered cigars 1C2 and Phillies had a uniform microbiome composition for each replicate, while other products showed some variability. Djarum had a higher percentage of Bacillus and Staphylococcus population, while 1C2 and Talon had a higher Tissierella genus. Cigarillo samples had a more consistent and uniform microbiome composition between the replicates, and Bacillus was the most common genus present in all samples. Compared to the other cigar categories, large cigars showed the highest percentage of Staphylococcus genus, except for William Penn and Gracia y Vega, which showed an increased percentage of Planococcaceae unclassified and Paenibacillus. There could be several factors causing the variation among the products, natural differences in the microbial populations between individual cigars as a result of the blend of tobacco used in manufacturing. The impact on the microbial population as a result of the manufacturer was not investigated for this study, as many manufacturers have production facilities in multiple locations, oftentimes in different countries. A study, conducted by Di Giacomo et al. (2007) on the microbial community in Italian Toscano cigar tobacco during fermentation, observed changes in the relative abundances of specific microbial groups over time [20]. They found that Bacillus species, including B. licheniformis and B. subtillis, could reduce NO3 without producing N2 gas, while Corynebacterium ammonia genes accumulated nitrite during subsequent stages of tobacco maturation. During the curing process of tobacco, changes in microbial communities were observed as nutrients were depleted, pH changed, moisture evaporated, and temperature increased. It also demonstrated a gradual decrease in tobacco pH, which was attributed to the metabolic byproducts of acid-producing species [20]. Various discrepancies in abundance and composition are not unexpected considering that brands are manufactured under different industrial conditions with proprietary tobacco blends. Another study showed that filtered cigar microbiome composition was dynamic and influenced by environmental factors such as temperature, humidity, and storage time [42].
Overall, our study provides new insights into the microbial diversity and composition of different categories of cigars, highlighting the impact of various cigar types and sizes on microbial populations. These findings hold significant implications for the regulation of tobacco products and the comprehension of potential health impacts arising from smoking diverse types of cigars. Prior research has demonstrated the abundance of human pathogens in tobacco products, particularly in cigarettes and smokeless tobacco, which could result in the development of chronic or infectious respiratory disease, as well as cancer [43–46]. Similarly, it is crucially important for further studies to elucidate the functional roles of these microorganisms in cigar products. In future studies, it may be possible to link pathogenicity to the microbial population as species with the genera Bacillus, Staphylococcus, and Pseudomonas have been identified as human pathogens. The use of MiSeq technology could be limited due to amplification bias, sequencing errors, and taxonomic resolutions. Utilizing advanced sequencing technologies and conducting experiments to identify metabolically active bacteria [34] could be one of the several approaches to dissecting the impact of microbes on human health. A recent study highlighted how viable bacteria could survive cigarette combustion and then be transferred to the upper respiratory system via mainstream smoke [38]. Additionally, future studies could focus on variation within microbiomes based on various production locations for specific manufacturers.
Author Contribution
SJ and RM contributed to the design and implementation of the research. SJ and KP contributed to the analysis of the sequences. SJ, KP, LM, and RM contributed to the writing and reviewing of the manuscript.
Funding
This work was supported by the Food and Drug Administration (grant UC2FD006890). The views expressed in written materials or publications and by speakers and moderators do not necessarily reflect the official policies of the Department of Health and Human Services nor does any mention of trade names, commercial practices, or organization imply endorsement by the United States Government.
Data Availability
Sequences can be found in the NCBI SRA database under the Bio project accession number PRJNA1073920.
Declarations
Competing Interests
The authors declare no competing interests.
References
- 1.Smyth EM, Chattopadhyay S, Babik K, Reid M, Chopyk J, Malayil L, Kulkarni P, Hittle LE, Clark PI, Sapkota AR (2019) The bacterial communities of little cigars and cigarillos are dynamic over time and varying storage conditions. Front Microbiol 10:2371 10.3389/fmicb.2019.02371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chattopadhyay S, Smyth EM, Kulkarni P, Babik KR, Reid M, Hittle LE, Clark PI, Mongodin EF, Sapkota AR (2019) Little cigars and cigarillos harbor diverse bacterial communities that differ between the tobacco and the wrapper. PLoS One 14:e0211705. 10.1371/journal.pone.0211705 10.1371/journal.pone.0211705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang TW, Gentzke AS, Creamer MR, Cullen KA, Holder-Hayes E, Sawdey MD, Anic GM, Portnoy DB, Hu S, Homa DM (2019) Tobacco product use and associated factors among middle and high school students—United States, 2019. MMWR Surveill Summ 68:1 10.15585/mmwr.ss6812a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phan L, McNeel TS, Chen-Sankey J, Niederdeppe J, Tan ASL, Choi K (2022) US trends in age of cigar smoking initiation by race/ethnicity and education. Am J Prev Med 63:624–629. 10.1016/j.amepre.2022.04.004 10.1016/j.amepre.2022.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gentzke AS, Wang TW, Cornelius M, Park-Lee E, Ren C, Sawdey MD, Cullen KA, Loretan C, Jamal A, Homa DM (2022) Tobacco product use and associated factors among middle and high school students—National Youth Tobacco Survey, United States, 2021. MMWR Surveill Summ 71:1 10.15585/mmwr.ss7105a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parms TA, Head SK, Sawdey MD, Rostron BL, Cullen KA (2022) Characteristics of past 30-day cigar smoking, U.S. Adolescents, 2020. Am J Prev Med 62:e39–e44. 10.1016/j.amepre.2021.06.011 10.1016/j.amepre.2021.06.011 [DOI] [PubMed] [Google Scholar]
- 7.Milam AJ, Bone L, Furr-Holden D, Coylewright M, Dachille K, Owings K, Clay E, Holmes W, Lambropoulos S, Stillman F (2012) Mobilizing for policy: using community-based participatory research to impose minimum packaging requirements on small cigars. Prog Community Health Partnersh 6:205–212. 10.1353/cpr.2012.0027 10.1353/cpr.2012.0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Xu Q, Yang M, Yang Y, Fu J, Miao C, Wang G, Hu L, Hu Z (2023) Analysis of differences in tobacco leaf microbial communities after redrying in Chinese provinces and from abroad. AMB Express 13:80. 10.1186/s13568-023-01580-5 10.1186/s13568-023-01580-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulin LM, Halenar MJ, Edwards KC, Lauten K, Stanton CA, Taylor K, Hatsukami D, Hyland A, MacKenzie T, Mahoney MC, Niaura R, Trinidad D, Blanco C, Compton WM, Gardner LD, Kimmel HL, Lauterstein D, Marshall D, Sargent JD (2022) Association of tobacco product use with chronic obstructive pulmonary disease (COPD) prevalence and incidence in Waves 1 through 5 (2013–2019) of the Population Assessment of Tobacco and Health (PATH) Study. Resp Res 23:273. 10.1186/s12931-022-02197-1 10.1186/s12931-022-02197-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker F, Ainsworth SR, Dye JT, Crammer C, Thun MJ, Hoffmann D, Repace JL, Henningfield JE, Slade J, Pinney J, Shanks T, Burns DM, Connolly GN, Shopland DR (2000) Health risks associated with cigar smoking. JAMA 284:735–740. 10.1001/jama.284.6.735 10.1001/jama.284.6.735 [DOI] [PubMed] [Google Scholar]
- 11.Chen-Sankey JC, Mead-Morse EL, Le D, Rose SW, Quisenberry AJ, Delnevo CD, Choi K (2021) Cigar-smoking patterns by race/ethnicity and cigar type: a nationally representative survey among US adults. Am J Prev Med 60:87–94 10.1016/j.amepre.2020.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang CM, Corey CG, Rostron BL, Apelberg BJ (2015) Systematic review of cigar smoking and all cause and smoking related mortality. BMC Public Health 15:1–20 10.1186/s12889-015-1617-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richardson A, Xiao HJ, Val DM (2012) Primary and dual users of cigars and cigarettes: profiles, tobacco use patterns, and relevance to policy. Nicotine Tob Res 14:927–932. 10.1093/ntr/ntr306 10.1093/ntr/ntr306 [DOI] [PubMed] [Google Scholar]
- 14.Symm B, Morgan MV, Blackshear Y, Tinsley S (2005) Cigar smoking: an ignored public health threat. J Prim Prev 26:363–375. 10.1007/s10935-005-5389-z 10.1007/s10935-005-5389-z [DOI] [PubMed] [Google Scholar]
- 15.Ross JC, Suerken CK, Reboussin BA, Denlinger-Apte RL, Spangler JG, Sutfin EL (2022) Cigar harm beliefs and associations with cigar use among young adults. Subst Use Misuse 57:1478–1485. 10.1080/10826084.2022.2092149 10.1080/10826084.2022.2092149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humans IWGotEoCRt (2007) Smokeless tobacco and some tobacco-specific N-nitrosamines. IARC Monogr Eval Carcinog Risks Hum 89:1–592 [PMC free article] [PubMed] [Google Scholar]
- 17.Law AD, Fisher C, Jack A, Moe LA (2016) Tobacco, microbes, and carcinogens: correlation between tobacco cure conditions, tobacco-specific nitrosamine content, and cured leaf microbial community. Microb Ecol 72:120–129. 10.1007/s00248-016-0754-4 10.1007/s00248-016-0754-4 [DOI] [PubMed] [Google Scholar]
- 18.Rivera AJ, Tyx RE (2021) Microbiology of the American Smokeless Tobacco. Appl Microbiol Biotechnol 105:4843–4853. 10.1007/s00253-021-11382-z 10.1007/s00253-021-11382-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tessler M, Cunningham SW, Ingala MR, Warring SD, Brugler MR (2023) An environmental DNA primer for microbial and restoration ecology. Microb Ecol. 10.1007/s00248-022-02168-5 10.1007/s00248-022-02168-5 [DOI] [PubMed] [Google Scholar]
- 20.Di Giacomo M, Paolino M, Silvestro D, Vigliotta G, Imperi F, Visca P, Alifano P, Parente D (2007) Microbial community structure and dynamics of dark fire-cured tobacco fermentation. Appl Environ Microbiol 73:825–837. 10.1128/Aem.02378-06 10.1128/Aem.02378-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM (2014) Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633-642. 10.1093/nar/gkt1244 10.1093/nar/gkt1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Law AD, McNees CR, Moe LA (2020) The microbiology of hemp retting in a controlled environment: steering the hemp microbiome towards more consistent fiber production. Agronomy 10:492 10.3390/agronomy10040492 [DOI] [Google Scholar]
- 25.Clarke KR (1993) Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143 10.1111/j.1442-9993.1993.tb00438.x [DOI] [Google Scholar]
- 26.Dinno A (2015) Nonparametric pairwise multiple comparisons in independent groups using Dunn’s test. Stand Genomic Sci 15:292–300 [Google Scholar]
- 27.Wickham H, Chang W, Henry L, Pendersen T, Takahashi K, Wilke C, Woo K, Yutani H, Dunnington D (2016) ggplot2: elegant graphics for data analysis.[Google Scholar]
- 28.Oliveros JC (2007) VENNY. An interactive tool for comparing lists with Venn Diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html. Accessed July 2023
- 29.Chattopadhyay S, Malayil L, Mongodin EF, Sapkota AR (2021) A roadmap from unknowns to knowns: advancing our understanding of the microbiomes of commercially available tobacco products. Appl Microbiol Biotechnol 105:2633–2645 10.1007/s00253-021-11183-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar RS, Mishra N, Kumar A (2022) Characterization of tobacco microbiome by metagenomics approach. Methods Mol Biol 2413:229–244. 10.1007/978-1-0716-1896-7_22 10.1007/978-1-0716-1896-7_22 [DOI] [PubMed] [Google Scholar]
- 31.Gao R, Sun-Waterhouse D, Xiang H, Cui C, Waterhouse GI (2022) The effect of the Corynebacterium glutamicum on the shortening of fermentation time, physicochemical and sensory properties of soy sauce. Int J Food Sci Technol 57:4316–4327 10.1111/ijfs.15758 [DOI] [Google Scholar]
- 32.Jia Y, Niu C-T, Xu X, Zheng F-Y, Liu C-F, Wang J-J, Lu Z-M, Xu Z-H, Li Q (2021) Metabolic potential of microbial community and distribution mechanism of Staphylococcus species during broad bean paste fermentation. Food Res Int 148:110533 10.1016/j.foodres.2021.110533 [DOI] [PubMed] [Google Scholar]
- 33.Liu T, Guo S, Wu C, Zhang R, Zhong Q, Shi H, Zhou R, Qin Y, Jin Y (2022) Phyllosphere microbial community of cigar tobacco and its corresponding metabolites. Front Microbiol 13:1025881. 10.3389/fmicb.2022.1025881 10.3389/fmicb.2022.1025881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chattopadhyay S, Ramachandran P, Malayil L, Mongodin EF, Sapkota AR (2023) Conventional tobacco products harbor unique and heterogenous microbiomes. Environ Res 220:115205. 10.1016/j.envres.2022.115205 10.1016/j.envres.2022.115205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Y, Wang H-C, Cai L-T, Li W, Pan D, Xiang L, Su X, Li Z, Adil MF, Shamsi IH (2021) Phyllospheric microbial composition and diversity of the tobacco leaves infected by Didymella segeticola. Front Microbiol 12:699699 10.3389/fmicb.2021.699699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pauly JL, Paszkiewicz G (2011) Cigarette smoke, bacteria, mold, microbial toxins, and chronic lung inflammation. J Oncol 2011:819129. 10.1155/2011/819129 10.1155/2011/819129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang J, Yang J, Duan Y, Gu W, Gong X, Zhe W, Su C, Zhang K-Q (2010) Bacterial diversities on unaged and aging flue-cured tobacco leaves estimated by 16S rRNA sequence analysis. Appl Microbiol Biotechnol 88:553–562. 10.1007/s00253-010-2763-4 10.1007/s00253-010-2763-4 [DOI] [PubMed] [Google Scholar]
- 38.Malayil L, Chattopadhyay S, Mongodin EF, Sapkota AR (2022) Bacterial communities of hookah tobacco products are diverse and differ across brands and flavors. Appl Microbiol Biotechnol 106:5785–5795. 10.1007/s00253-022-12079-7 10.1007/s00253-022-12079-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sajid M, Srivastava S, Kumar A, Kumar A, Singh H, Bharadwaj M (2021) Bacteriome of moist smokeless tobacco products consumed in India with emphasis on the predictive functional potential. Front Microbiol 12:784841 10.3389/fmicb.2021.784841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smyth EM, Kulkarni P, Claye E, Stanfill S, Tyx R, Maddox C, Mongodin EF, Sapkota AR (2017) Smokeless tobacco products harbor diverse bacterial microbiota that differ across products and brands. Appl Microbiol Biotechnol 101:5391–5403. 10.1007/s00253-017-8282-9 10.1007/s00253-017-8282-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vishwakarma A, Verma D (2020) 8 - Exploring the microbiome of smokeless tobacco. In: Chowdhary P, Raj A, Verma D, Akhter Y (eds.) Microorganisms for sustainable environment and health. Elsevier, pp 167-178. 10.1016/B978-0-12-819001-2.00008-5
- 42.Chopyk J, Chattopadhyay S, Kulkarni P, Smyth EM, Hittle LE, Paulson JN, Pop M, Buehler SS, Clark PI, Mongodin EF (2017) Temporal variations in cigarette tobacco bacterial community composition and tobacco-specific nitrosamine content are influenced by brand and storage conditions. Front Microbiol 8:358 10.3389/fmicb.2017.00358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simba H, Menya D, Mmbaga BT, Dzamalala C, Finch P, Mlombe Y, Mremi A, Narh CT, Schuz J, McCormack V (2023) The contribution of smoking and smokeless tobacco to oesophageal squamous cell carcinoma risk in the African oesophageal cancer corridor: results from the ESCCAPE multicentre case-control studies. Int J Cancer. 10.1002/ijc.34458 10.1002/ijc.34458 [DOI] [PubMed] [Google Scholar]
- 44.Sawant S, Dugad J, Parikh D, Srinivasan S, Singh H (2023) Oral microbial signatures of tobacco chewers and oral cancer patients in India. Pathogens 12. 10.3390/pathogens12010078 [DOI] [PMC free article] [PubMed]
- 45.Vishwakarma A, Srivastava A, Mishra S, Verma D (2022) Taxonomic and functional profiling of Indian smokeless tobacco bacteriome uncovers several bacterial-derived risks to human health. World J Microbiol Biotechnol 39:20. 10.1007/s11274-022-03461-8 10.1007/s11274-022-03461-8 [DOI] [PubMed] [Google Scholar]
- 46.Sapkota AR, Berger S, Vogel TM (2010) Human pathogens abundant in the bacterial metagenome of cigarettes. Environ Health Perspect 118:351–356. 10.1289/ehp.0901201 10.1289/ehp.0901201 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences can be found in the NCBI SRA database under the Bio project accession number PRJNA1073920.