Abstract
Biobanks are valuable service units that ensure the usage of high-quality biological samples. They contribute to translational research, and their support may improve future therapeutic approaches. They store biological samples that can be used to examine circulation biomarkers, immune cells, and immunohistochemistry aspects of illnesses and further in-depth examinations using NGS techniques. The IRCCS Synlab SDN Biobank has about 70,000 well-preserved cryopreserved human samples from various diseases, primarily oncological but also neurological and cardiovascular. These biospecimens were taken from 25,000 participants underwent imaging with a contrast agent. The goal is to propose quality control assays that meet the requirements of the international standard ISO 9001:2015 and ISO 20387:2020 accreditation. PBMCs viability was determined, and immune subset cells were analyzed by flow cytometry. Furthermore, the expression of ubiquitous miRNAs was used to assess plasma sample integrity. The quality controls demonstrated that the biological samples were correctly cryopreserved; the preservation of human biological samples did not affect the quality of the biological samples tested. Indeed, the cryopreserved PBMCs had a vitality of more than 80%, and the lymphocyte subsets could be selected for future immune cell investigations. Furthermore, miRNA expression was highest in thawed plasma samples compared to the positive and negative controls. We evaluated the quality of our randomly selected biobank-thawed human samples. Both PBMCs and plasma samples fulfill the high-quality standards needed for biomedical research, assuring their long-term preservation. However, further research is needed in the biobanking field to establish globally accepted procedures to confirm the quality of biological samples.
Keywords: Biobanking, Quality controls, Quality management system, Biobanks
Subject terms: Biomarkers, Health care, Medical research
Introduction
Several papers highlight the mismatch between financial investment and outcomes in biomedical research due to the non-reproducibility of preclinical research data, which remains a critical issue for possible clinical applications1–3. Human bioresources are typically gathered from pathology, genetics, and other research facilities that store biological sample collections, as well as from established infrastructure such as biobanks4.
All biobanks are required to establish a Quality Management System (QMS) that defines procedures and responsibilities to help them comply with international standards and guidelines such as ISO 9001:2015 (Quality Management Systems), ISO 17025:2017 (General Requirements for the Competence of Testing and Calibration Laboratories) and finally ISO 203987:2020 (ISO 20387:2020—Biotechnology—Biobanking)5,6. The biobanking community is aware that quality control (QC) is essential to ensure the quality and integrity of biological samples throughout the entire workflow, from collection to storage. However, specific, and unambiguously recognized quality controls are lacking. The use of QC during workflows not only ensures that biological samples are of high quality, but it also improves preclinical research by eliminating difficulties related to data non-reproducibility.
The IRCCS Synlab SDN Biobank is a service unit dedicated to the processing, storage, and sharing of high-quality human biological samples and clinical data7. It has been developed according to international biobanking standards in terms of staffing, establishment, governance, and standard operating procedures (SOPs) for biospecimen management8. It is a partner of the Pan-European Biobanking and Biomolecular Resources Research Infrastructure Consortium (BBMRI-ERIC) and operates according to ethics, quality, and international guidelines9,10. In addition, the biological sample workflow is well defined and strictly defined in compliance with ISO9001:2015 certification, with a future goal of ISO 20387:2020 accreditation6,11. The biological samples consist of human bioresources, mainly human peripheral blood collected from participants undergoing imaging. The biobank team actively participates in working groups organized by the BBMRI-ERIC network and in the activities of the Italian Ministry of Health activities to deepen and explore the open issues of biobanking10.
It is noteworthy that one of the most discussed topics has been focused on the QC of biological samples12; in this scenario, it is essential to underline that each biobank has its specific workflow and can manage different types of biological samples, identifying the appropriate QC. To address this critical open question, we describe our biobanking experience and our QC procedures related to the management of biological samples, focusing on peripheral blood management.
Materials and methods—quality management system
A set of standardized QMS documents is available, including a quality manual with a dedicated section on procedural documents, SOPs, forms for storage temperature registration, and instrument checks displayed in Table 1; these documents have been introduced to cover all biobank workflows to ensure a homogenous and controlled biobank workflow.
Table 1.
Enumeration of documents (I-VIII) associated with the biobank QMS.
| QMS documents | |
|---|---|
| I. BBMRI partnership | Partner charter: The IRCCS Synlab Biobank is a partner of the European BBMRI-ERIC network; the partnership guarantees continuous updates on legal, ethical, and other issues relating to the quality of biological samples. It also allows for a continuous exchange of information on biological samples that can potentially be shared within the network at national and international levels. The BBMRI-ERIC network also regularly proposes activities aimed at biobank development, training, and public health improvement |
| II. ISO 9001:2015 | Our biobank is periodically evaluated with a dedicated audit aimed at proper biospecimen management |
| III. Biobank reagents quality certification check | All reagents used are rigorously controlled. The certificate of analysis is verified for the reagents to guarantee the quality of biological samples |
| IV. Biobank workflow procedures | The management of biological samples in terms of collection, processing, and storage is described in the dedicated documentation. In addition, checklist modules for the monitoring of the instruments (mechanical freezers and nitrogen tanks) and the sharing of biological samples are used to verify traceability |
| V. Instruments calibration | The instruments are periodically checked to provide certificates of temperature calibration. All instruments in the biobank are rigorously checked to ensure the correct storage temperature |
| VI. Quality checks: Peripheral blood mononuclear cells (PBMCs) samples and Serum/Plasma | The PBMCs are evaluated in terms of cell viability after thawing; the serum and plasma samples are checked to evaluate the publication of scientific manuscripts obtained with biological sample analyses; also, circulating microRNA expression is tested to check proper sample storage |
| VII. Biobank personnel training | Staff are trained in the ethical, legal, privacy, and quality issues associated with biobanking |
| VIII. Science popularization activities | Biobank staff periodically organizes science outreach events for public schools and citizens to promote the importance of biobanks and the high-quality preservation of biosamples |
Participants and biological samples
The storage of human biological samples has been approved by the medical ethics committee of the Istituto Nazionale per lo studio e la Cura dei Tumori “Fondazione Giovanni Pascale-Naples, Italy” (Protocol number 4/15 approved, July 22, 2015). Research activities were performed according to the ethical principles introduced in 1964 by the Declaration of Helsinki13.
Bioresources description
The IRCCS Synlab SDN Biobank stores human biological samples such as whole blood, formalin-fixed paraffin-embedded (FFPE) blocks, freshly obtained breast tissue samples, bone marrow mononuclear cells (BMMCs), and immortalized cell lines. Peripheral blood biosamples contribute to approximately 90% of all specimens handled; they are collected from individuals who sign an informed consent form and undergo contrast medium imaging. The remaining biological samples are collected through collaborations with affiliated health facilities, and all stored biological samples undergo QC. Tissue samples are frozen in nitrogen vapor using portable canisters to maintain the cold chain, and the sample is then safely moved to the long storage facility. Furthermore, the characteristics of the human tissue samples are verified by the information acquired from the pathology laboratory that handled the specific component for the diagnostic activity. BMMCs are obtained from medical facilities as part of joint research activities, with the same quality control procedures used for PBMCs samples. Finally, for immortalized cell lines, controls based on Short Tandem Repeats (STRs) analysis and mycoplasma identification are carried out. In the context of the several workflows related with the preservation of biological samples, we will examine in detail peripheral blood, as this typology is the most abundant and represents the first type of biomaterial that we want to accredit under ISO20387:2020.
Peripheral blood: biobanking workflow, standard operatives procedures, and quality control
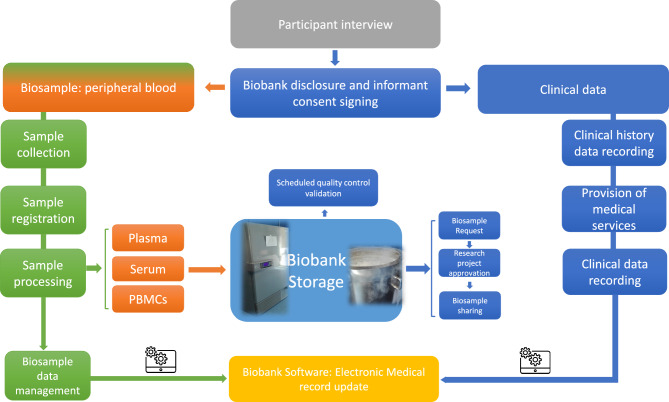
Figure 1 displays the biobank workflow as a flowchart; after participants provide informed consent, the collected peripheral blood is processed (flow depicted in green on the left) to yield plasma, serum, and PBMCs (orange rectangles). Clinical and imaging data are collected simultaneously (flow displayed in blue on the right). All information about biological samples and clinical data is recorded in dedicated software (shown in yellow below); additionally, human biosamples are selected for (i) sharing following approval of a research project proposal, and (ii) QC to ensure the integrity/quality of the human biological sample. The main steps of the biobank workflow are as follows:
(i) The biobank unit nurse interviews the possible participants during the diagnostic procedure in the nuclear medicine, CT, or MR unit; the subjects willing to share a peripheral blood sample sign a disclosure and informed consent.
(ii) The IRCCS Synlab SDN Biobank sample processing unit follows the established SOPs; dedicated biologists and technicians are responsible for receiving, recording, storing, and managing the medical data; a General Data Protection Regulation (GDPR) compliant biobank management software, Olohealth (Olomedia Palermo, Italy), is used for the digital collection, including vial location, sample information (type, hemolysis, volume/µl), and clinical data. The human biological samples are stored at low temperatures (−80 ℃ mechanical freezer for plasma and serum samples; −140 ℃ vapor nitrogen phase; and −196 ℃ liquid nitrogen phase for PBMCs samples) to preserve biological quality.
(iii) The sharing of human biological samples is highly regulated: the project’s proposing researcher schedules and submits a request to the biobank curator via a specific form, stating the type of human biomaterial, volume, and number of biosamples required. Requests to the IRCCS Synlab SDN Biobank are delivered in two main ways: the first is represented by the presence of the biobank unit on the Synlab IRCCS SDN official website in the research section (https://sdn.synlab.it/facilities/) with references to be able to contact the scientific direction (https://sdn.synlab.it/contatti-ricerca/); The second refers to the Directory of the European BBMRI network website (https://directory.bbmri-eric.eu/#/collection/bbmri-eric:ID:IT_1463060660514422:collection:1478245289805104); it can be consulted with all the biobanks associated with the network, with references associated with the individual Biobanks (name, addresses, contact details), types of biological samples that can be requested (plasma, serum, PBMCs, tissues, saliva, feces, sputum, etc.). However, in this scenario, only the BBMRI-ERIC affiliated biobank could request biological samples through a login negotiator website area (https://negotiator.bbmri-eric.eu). Finally, the IRCCS scientific director evaluates the research project and decides whether to allow the sharing of human biological samples.
Figure 1.
Biobank workflow: The workflow model for collection, storage, distribution, and peripheral blood quality control evaluation.
The main goal of the biobank workflow is to ensure the integrity of biological samples; to this aim, QC analysis is an essential procedure for ensuring the integrity of biological samples during processing and storage.
Some key aspects that can affect QC include:
- biological specimen collection: this phase focuses on the proper collection and handling of biological samples to ensure that they are not contaminated or degraded during the biological specimen collection procedures;
- sample processing: sample processing and storage SOPs must be followed to maintain sample quality and integrity;
- storage condition monitoring: regular monitoring of storage conditions, such as temperature and power, is essential to prevent sample degradation;
- quality audits: quality assurance audits are essential for ensuring regulatory compliance and best practices.
- document sample information, including metadata, for traceability.
- provide ongoing education and training for sample handling professionals to guarantee proper sample management.
The QC study is the critical step in determining biobank efficiency; in this case, we examined biological sample quality at random from 2018 to 2023, examining two biological samples per year (January–June).
PBMCs QC analysis
The cell viability was assessed by staining PBMC with 7-AAD (Beckman Coulter, # IM3422); dead cells were calculated as the percentage of 7-AAD positive events using the 7-AAD vs. SSC dot plot. The negative control corresponds to 30% DMSO-treated PBMCs for 3 h, while the positive control corresponds to fresh recovered PBMCs.
The CytoFLEX (Beckman Coulter) flow cytometer was used to analyze lymphocyte subpopulations after labeling with CD45FITC, CD56PE, CD19ECD, and CD3PC5 antibody mix (Beckman Coulter # 6607073) according to the gating technique described in Supplementary Fig. 1.
Plasma QC analysis
To examine the quality of plasma samples, microRNA was extracted, using an aliquot thawed for the first time in an ultra-freezer at − 80 ℃, and its integrity was evaluated using PCR analysis. The miRNA extraction was carried out using the miRNeasy Serum/Plasma Advantage kit (217204; Qiagen). For the initial step, 200 μl of plasma was lysed with 60 μl of QIAzol Buffer RPL and 1 μl of spike-in mix (reconstituted as per manufacturer’s instructions) (339,390, RNA spike-in kit; Qiagen). Afterward, the samples were purified following the manufacturer’s guidelines. The RNA was eluted in 14 μl RNase-free water.
The cDNA was then generated using the miRCURY LNA RT kit (339340; Qiagen). 0.5 µl of UniSp6 and cel-miR-39-3p spike-in mix (339390, RNA spike-in kit; Qiagen) were added to the RT reaction mix. The reaction volume was increased to 10 µl by adding a predetermined volume of 1 μl of input RNA eluate from each sample. Reverse transcription-quantitative PCR (RT-qPCR) was carried out in a 384-well plate format utilizing a Bio-Rad CFX384 real-time PCR equipment (Bio-Rad Laboratories, Ltd.) and the miRCURY LNA SYBR Green PCR Kit (339346; Qiagen). A Bio-Rad CFX384, Real-time C1000 Touch Thermal Cycler (Bio-Rad Laboratories, Ltd.) was used to perform a 2-step cycling qPCR procedure (95 ℃ for 2 min, followed by 40 cycles of 95 ℃ for 10 s and 56 ℃ for 60 s). RT-qPCR involved no-template controls (no RNA template). Furthermore, as negative control we used plasma stored at 4 ℃ for 1 month14,15, and as positive control we used a fresh unfrozen plasma sample. Overall, five locked nucleic acid (LNA) PCR assays for miRNA detection were used: 1) the RNA isolation spike-in controls added at the beginning of each isolation procedure (UniSp2), 2) the cDNA synthesis spike-in controls (UniSp6), and 4) three target miRNAs: miR-16-5p, miR-21-5p and miR-150-5p.
Results
Peripheral blood and derivatives: quality controls to verify the correct use of standard operating procedures
To assess the quality of our biobank specimens, we randomly thawed human biological samples from 2018 to 2023 and tested PBMC and plasma samples (2 tests per year, one in January and one in June). For the PBMCs samples, we thawed cells, carried out a test to evaluate cell viability, and studied the cellular subsets using flow cytometry. As shown in Fig. 2 (panel A) the percentage of dead cells after thawing was not > 15% of the total. Furthermore, the study of lymphocyte subsets highlighted the high quality of the biological samples for possible studies focused on immune cell characterization (Fig. 2, panel B).
Figure 2.
PBMCs analyses: (A) Evaluation of the percentage of dead cells stored in IRCCS Synlab SDN Biobank PBMCs from 2018 to 2023. Negative control corresponds to 30% DMSO-treated PBMCs for 3 h. Positive control corresponds to fresh recovered PBMCs. Error bars represent the standard deviation of 2 independent samples. (B) Exemplified representation of cytofluorimetric lymphocytes typing of PBMCs stored in IRCCS Synlab SDN Biobank (sample of 2018); the symbol % represents the percentage of gated cells.
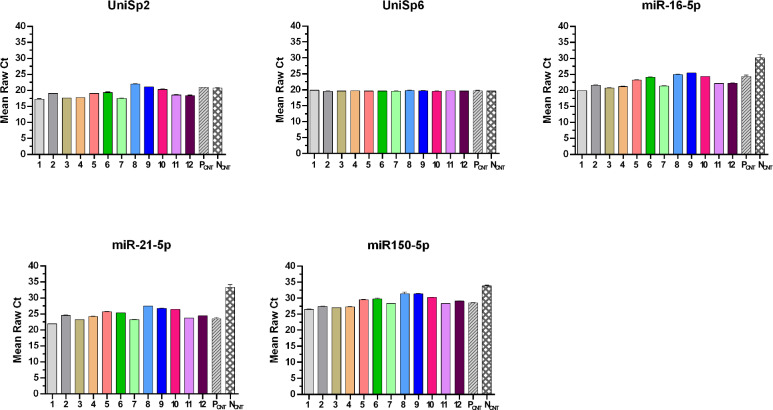
In addition, we investigated the stability of miRNAs isolated from the plasma samples and evaluated the abundance and solidity of miRNAs subjected to a long period of freezing. These samples were compared with a fresh sample, collected and immediately processed without undergoing any conservation steps (positive control), and with a degraded sample (negative control). For degraded conditions, we left the sample at 4 ℃ for one month. We applied these conditions based on previous published results14, in which was demonstrated that endogenous circulating miRNA levels were unstable when plasma specimens were stored at 4 ℃ after 24 h, 48 h, 1 month, and 4 months with an increase in Cq values in all cases. Figure 3 depicts the expression of the analyzed exogenous and endogenous miRNAs. Exogenous miRNAs, such as UniSp2 and UniSp6, were utilized to monitor extraction and retro-transcription processes, respectively. Because both exogenous miRNA templates were added to outside samples, the expression pattern of UniSp2 and UniSp6 overlapped with evaluated plasma samples stored in the biobank and controls, as expected. UniSp2 expression was more variable among the tested samples compared to UniSP6. Because it is added before extraction, it undergoes more experimental steps and suffers more from potential interferences related to the nature of the sample itself. Conversely, the exogenous UniSp2 miRNA was added to the already extracted miRNA and utilized as a control for the retro-transcription reaction.
Figure 3.
Stability study of circulating miRNAs in plasma samples at different storage times. Stability study of circulating miRNAs in plasma samples at different storage times. Has-miR-16-5p, has-miR-21-5p, and has-miR-150-5p are used to measure each sample miRNA signal. UniSp2 and UniSp6 were used as internal controls. Results are represented as differences in raw Cq values. Data represent the means ± SD of three independent experiments. 1–2 biological samples refer to January-June 2018; 3–4 refer to 2019; 5–6 refer to January-June 2020; 7–8 refer to January-June 2021; 9–10 refer to January-June 2022; 11–12 refer to January-June 2023.
As shown in Table 2, miR-16-5p, miR-21-5p, and miR-150-5p had mean raw Cq values of 22.63, 24.49, and 28.71 in plasma samples (1–12). The miRNAs expression appeared stable at − 80 ℃ for the last 5 years in all kept samples, with a Cq trend that overlapped with the fresh sample but differed from the degraded control (Fig. 3). About endogenous miRNAs, Cq values differed by a few cycles between samples, which did not appear to be connected to dating. Because the samples were chosen at random, regardless of health status, it cannot be excluded that discrepancies across samples can be due to the participants’ condition at the time of sample collection.
Table 2.
Cq values of exogenous and endogenous miRNAs in plasma samples retrieved from the IRCCS Synlab SDN biobank dating from 2018–2023 were compared to a positive control (fresh plasma without any freezing steps) and a negative control (a sample degraded and kept at 4 ℃ for one month).
| miRNAs | Sample 1–12 mean ± ds | Positive control mean ± ds | Negative control mean ± ds |
|---|---|---|---|
| UniSp2 | 19.24 ± 0.55 | 19.5 ± 0.67 | 19.79 ± 0.06 |
| UniSP6 | 19.63 ± 0.08 | 19.65 ± 0.23 | 19.60 ± 0.11 |
| hsa-miR-16-5p | 22.63 ± 1.78 | 24.43 ± 0.31 | 30.20 ± 0.95 |
| hsa-miR-21-5p | 24.49 ± 1.31 | 23.49 ± 0.45 | 33.27 ± 0.88 |
| hsa-miR-150-5p | 28.71 ± 1.39 | 28.54 ± 1.28 | 33.85 ± 0.23 |
Scientific manuscripts realized utilizing the biological samples from the IRCCS Synlab SDN biobank
To provide additional evidence of biological sample quality, we classified all published publications that used biological samples from the IRCCS Synlab SDN Biobank using the SOPs (for more details see the materials and methods section). Table 3 displays all accepted publications that employed IRCCS Synlab Biobank samples, together with information on the biobank sample, title, size, analyte determined, year, journal, and associated reference.
Table 3.
List of publications produced 2019–2023 with bioresources obtained from the IRCCS Synlab SDN.
| Biobank sample | Title | Size | Analyte determined | Year | Journal | Ref |
|---|---|---|---|---|---|---|
| PBMCs and cell lines | Influence of breast cancer extracellular vesicles on immune cell activation: a pilot study | 5 | extracellular vesicles and immune cells | 2023 | Biology | 16 |
| BMMCs | A comprehensive analysis of the expression profiles of KCTD proteins in acute lymphoblastic leukemia: evidence of selective expression of KCTD1 in T-ALL | 34 | lncRNA | 2023 | Journal of clinical medicine | 17 |
| Cell lines | The oncosuppressive properties of KCTD1: its role in cell growth and mobility | 2 | Proteins and genes expression | 2023 | Biology | 18 |
| Plasma | A peripheral signature of Alzheimer’s disease featuring microbiota-gut-brain axis markers | 84 | Cytokines and ultrasensitive neuroproteins detection | 2023 | Alzheimer’s research and therapy | 19 |
| Cell lines | Caveolin-mediated internalization of Fmoc-FF nanogels in breast cancer cell lines | 6 | Protein expression | 2023 | Pharmaceutics | 20 |
| Serum | Stratification of patients with coronary artery disease by circulating cytokines profile: a pilot study | 75 | Cytokines | 2023 | Journal of clinical medicine | 21 |
| BMMCs | Specific lncRNA signatures discriminate childhood acute leukaemias: a pilot study | 34 | lncRNA | 2022 | Cancer cell international | 22 |
| Plasma | An innovative approach for the evaluation of prolonged disorders of consciousness using NF-L and GFAP biomarkers: a pivotal study | 16 | Proteins | 2022 | Scientific reports | 23 |
| Serum | SARS-CoV-2 antibody responses before and after a third dose of the BNT162b2 vaccine in Italian healthcare workers aged ≤ 60 years: one year of surveillance | 186 | Proteins | 2022 | Frontiers in immunology | 24 |
| Breast tissues | Identification of immune cell components in breast tissues by a multiparametric flow cytometry approach | 29 | Proteins | 2022 | Cancers | 25 |
| Breast tissues and cell lines | KCTD15 Is Overexpressed in her2 + Positive Breast Cancer Patients and Its Silencing Attenuates Proliferation in SKBR3 CELL LINE | 34 | Proteins | 2022 | Diagnostics | 26 |
| BMMCs and cell lines | KCTD15 deregulation is associated with alterations of the NF-κβ signaling in both pathological and physiological model systems | 4 | Proteins | 2021 | Scientific Reports | 27 |
| BMMCs | The lncRNA TEX41 is upregulated in pediatric B-Cells Acute Lymphoblastic Leukemia and it is necessary for leukemic cell growth | 10 | lncRNA | 2021 | Biomarker Research | 28 |
| Plasma and serum | Impact of breast tumor onset on blood count, carcinoembryonic antigen, cancer antigen 15–3, and lymphoid subpopulations supported by automatic classification approach: a pilot study | 127 | Blood count, CA 15–3, and lymphocytes subsets | 2021 | Cancer Control | 29 |
| Breast cancer cells | Peptide-based hydrogels and nanogels for delivery of doxorubicin | 1 | hydrogels (HGs) and nanogels (NGs) study | 2021 | International Journal of Nanomedicine | 30 |
| Plasma, breast tissues, cell lines | miR-622 is a novel potential biomarker of breast carcinoma and impairs motility of breast cancer cells through targeting NUAK1 kinase | 98 | miR-622 | 2020 | British Journal of Cancer | 31 |
| PBMCs and cell lines | KCTD15 protein expression in peripheral blood and acute myeloid Leukemia | 17 | Proteins | 2020 | Diagnostics | 32 |
| Plasma and breast tissues | Circulating miRNAs in untreated breast cancer: an exploratory multimodality morpho-functional study | 177 | miRNA | 2019 | Cancers | 33 |
| Whole blood, PBMCs, cell lines | Purification of viable peripheral blood mononuclear cells for biobanking using a robotized liquid handling workstation | 120 | PBMCs isolations | 2019 | Journal of translational Medicine | 34 |
From 2019 to 2023, a total of nineteen publications were achieved using high-quality biological samples stored in our biobank. These publications show that the unit has preserved the quality and integrity of biological samples, serving as an important resource for the scientific activities of our institute.
Discussion
Today, using of high-quality resources is essential to ensure a high level of biomedical research. A biobank unit must be focused on the research topic purposes (disease-oriented, population-based, genetic, rare diseases, etc.)35 and its QC system must be designed according to the needs of the individual biobank36. Currently, although researchers and clinicians receive recognition for the scientific activities produced within the research community, biobanks receive very sporadic recognition for the preservation and provision of bioresources37. Furthermore, although biomedical research requires the use of high-quality bioresources, there is currently no homogeneous system capable of clearly tracing the bioresources used; additionally, the clear recognition through traceability of bioresources obtained from biobanks could trigger a mechanism whereby only the best-performing service units could obtain “biobank status”38.
In this way, evaluation rankings could be generated that could provide a rationale for determining the distribution of research funding. This could address the issue of sustainability of biobanks, an open question that has never been adequately addressed worldwide35. Each biobank manages different types of biological samples, has a different governance (private company, university, non-profit company, etc.), a different logistical organization from the others in terms of areas dedicated to biobanking and personnel skills (biologists, laboratory technicians, nurses, doctors, chemists, etc.); therefore, it is essential to identify appropriate QC strategies that cover all stages of biological samples, from handling and collection to storage. For this purpose, the accreditation ISO 20387:2020, specific for biobanks, which focuses on the examination of ethical, legal, privacy and quality management aspects, is of fundamental importance10; the evaluation of the experts who carry out the audit is fundamental to evaluate and possibly implement the activities of the biobank examined, all for the benefit of biological samples quality used for research purposes; nevertheless, it is important to underline that the QC to be adopted and the time controls are not specifically defined.
The description of our biobank workflow and the QC performed represents our context, which can be partially replicated in other scenarios. In the evaluation of the PBMCs biosamples (2018–2023), we evaluated the lymphocyte subsets that proved to be preserved for possible future studies; we also estimated plasma integrity using ubiquitously expressed miRNAs. Nevertheless, we cannot exclude that proteins, cytokines, or other molecules very sensitive to the thawing phase may have undergone a certain degree of degradation not reported by our selected QC39. However, we preferred to assess miRNA because many projects that we have been and are currently involved in concern the study of miRNA. It is important to define the downstream activities that will be performed and to ensure that the selected QC does not neglect the health of the molecules to be studied. Indeed, biobanks identify the appropriate QC depending on the biological matrix stored and the scientific goals of the facility12,40–42. Another open issue, closely associated with the QC analyses is the traceability of biobank bioresources citation; although the scientific community is much more up-to-date on the role of biobanks, researchers should be provided with the tools to cite them in a homogenous and easily traceable manner43. Indeed, the contribution of a biobank unit is cited heterogeneously, making its traceability difficult. In this context, scientific journals should only accept manuscripts that focus on certified bioresources, as all published results are strictly related to the correct preservation of the biological sample. There are several publications describing the potential biases introduced by variables associated with the incorrect preservation of the biomaterial44. Therefore, the traceability of bioresources is closely related to the use of QC to validate the biobank workflow. It should be homogeneous depending on the biological matrix managed and universally accepted. This would allow a system to assess the quality of bioresources, provide a “proof” to enable the sharing of biological data, and lay the foundations for large-scale assessments in the era of big data.
Conclusions
Today, although the concept of QC is mentioned in the ISO 20387:2020 standard, the techniques to be used to validate the bioresource are not defined. Finally, QC analysis and traceability of bioresources would guarantee greater reproducibility of biomedical data, favoring the possibility of easier clinical translation and reducing the associated costs. We have reported our experience facing this open issue; we hope it can represent a starting point for comparing and improving the world’s biobanks.
Supplementary Information
Acknowledgements
We want to thank all the patients involved in the contribution of research biobanks.
Abbreviations
- BBMRI-ERIC
Biobanking and Biomolecular Resources Research Infrastructure Consortium
- BMMCs
Bone marrow mononuclear cells
- FFPE
Formalin-fixed paraffin-embedded
- GDPR
Data protection regulation
- PBMCs
Peripheral blood mononuclear cells
- QC
Quality control
- QMS
Quality management system
- SOPs
Standard operating procedures
- STRs
Short tandem repeat analysis
Author contributions
L.C. designed and conceptualized the study; designed and performed the experiments; analyzed the data and drafted the manuscript for intellectual content; A.M.G. performed the experiments, analyzed the data, and drafted the manuscript for intellectual content. S.D.F.M and G.S. performed the experiments; G.S. performed the experiments, analyzed the data, and drafted the manuscript for intellectual content; M.S. revised intellectual content; supervised the project, and gave the final approval. All authors reviewed the manuscript.
Funding
This work is supported by the Ministero della Salute “Ricerca Corrente” project.
Data availability
The biological data used and analyzed during the study are available from the corresponding author on a reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-70263-3.
References
- 1.Freedman, L. P., Cockburn, I. M. & Simcoe, T. S. The economics of reproducibility in preclinical research. PloS Biol.10.1371/journal.pbio.1002165 (2015). 10.1371/journal.pbio.1002165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poldrack, R. The costs of reproducibility. Neuron101, 11–14 (2019). 10.1016/j.neuron.2018.11.030 [DOI] [PubMed] [Google Scholar]
- 3.Browne, D., Miller, C. & Doolan, D. Technical pitfalls when collecting, cryopreserving, thawing, and stimulating human T-cells. Front. Immunol.10.3389/fimmu.2024.1382192 (2024). 10.3389/fimmu.2024.1382192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castillo-Pelayo, T., Babinszky, S., LeBlanc, J. & Watson, P. H. The importance of biobanking in cancer research. Biopreservation Biobanking13, 172–177. 10.1089/bio.2014.0061 (2015). 10.1089/bio.2014.0061 [DOI] [PubMed] [Google Scholar]
- 5.Ohn, H. M. Internal audit techniques for testing laboratories: ISO/IEC 17025:2017 perspective. Accredit. Qual. Assur.29, 263–266. 10.1007/s00769-024-01592-z (2024). 10.1007/s00769-024-01592-z [DOI] [Google Scholar]
- 6.Tarling, T. et al. Comparison and analysis of two internationally recognized biobanking standards. Biopreservation Biobanking18, 82–89. 10.1089/bio.2019.0126 (2020). 10.1089/bio.2019.0126 [DOI] [PubMed] [Google Scholar]
- 7.Mirabelli, P. et al. SDN biobank: Bioresource of human samples associated with functional and/or morphological bioimaging results for the study of oncological, cardiological, neurological, and metabolic diseases. Open J. Bioresour.4, 2–2 (2017). 10.5334/ojb.26 [DOI] [Google Scholar]
- 8.Schenk, M. et al. Biobanking of different body fluids within the frame of IVF-a standard operating procedure to improve reproductive biology research. J. Assist. Reprod. Genet.34, 283–290. 10.1007/s10815-016-0847-5 (2017). 10.1007/s10815-016-0847-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Litton, J. E. Launch of an infrastructure for Health Research: BBMRI-ERIC. Biopreservation Biobanking16, 233–241. 10.1089/bio.2018.0027 (2018). 10.1089/bio.2018.0027 [DOI] [PubMed] [Google Scholar]
- 10.Linsen, L. et al. Biobank quality management in the BBMRI be network. Front. Med.10.3389/fmed.2019.00141 (2019). 10.3389/fmed.2019.00141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.López-Púa, Y. et al. Implementation of a quality management system in a liver transplant programme. BMJ Open Qual.10.1136/bmjoq-2023-002440 (2023). 10.1136/bmjoq-2023-002440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ornskov, D., Waldstrom, M., Thomsen, L. T., Munk, C. & Kjaer, S. K. Quality control of biospecimens in a Danish clinical cytology biobank. Biopreservation Biobanking21, 184–190. 10.1089/bio.2021.0162 (2023). 10.1089/bio.2021.0162 [DOI] [PubMed] [Google Scholar]
- 13.World Medical Association. World Medical Association Declaration of Helsinki Ethical Principles for Medical Research Involving Human Subjects. JAMA J. Am. Med. Assoc.310, 2191–2194. 10.1001/jama.2013.281053 (2013). 10.1001/jama.2013.281053 [DOI] [PubMed] [Google Scholar]
- 14.Sourvinou, I., Markou, A. & Lianidou, E. Quantification of circulating miRNAs in plasma. J. Mol. Diagn.15, 827–834 (2013). 10.1016/j.jmoldx.2013.07.005 [DOI] [PubMed] [Google Scholar]
- 15.Blondal, T. et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods59, S1–S6. 10.1016/j.ymeth.2012.09.015 (2013). 10.1016/j.ymeth.2012.09.015 [DOI] [PubMed] [Google Scholar]
- 16.Santoro, J. et al. Influence of breast cancer extracellular vesicles on immune cell activation: A pilot study. Biol. Basel10.3390/biology12121531 (2023). 10.3390/biology12121531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buono, L. et al. A comprehensive analysis of the expression profiles of KCTD proteins in acute lymphoblastic leukemia: Evidence of selective expression of KCTD1 in T-ALL. J. Clin. Med.10.3390/jcm12113669 (2023). 10.3390/jcm12113669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smaldone, G. et al. The oncosuppressive properties of KCTD1: Its role in cell growth and mobility. Biol. Basel10.3390/biology12030481 (2023). 10.3390/biology12030481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marizzoni, M. et al. A peripheral signature of Alzheimer’s disease featuring microbiota-gut-brain axis markers. Alzheimers Res. Ther.10.1186/s13195-023-01218-5 (2023). 10.1186/s13195-023-01218-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smaldone, G. et al. Caveolin-mediated internalization of Fmoc-FF nanogels in breast cancer cell lines. Pharmaceutics10.3390/pharmaceutics15031026 (2023). 10.3390/pharmaceutics15031026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iside, C. et al. Stratification of patients with coronary artery disease by circulating cytokines profile: A pilot study. J. Clin. Med.10.3390/jcm12206649 (2023). 10.3390/jcm12206649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buono, L. et al. Specific lncRNA signatures discriminate childhood acute leukaemias: A pilot study. Cancer Cell Int.10.1186/s12935-022-02789-3 (2022). 10.1186/s12935-022-02789-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coppola, L. et al. An innovative approach for the evaluation of prolonged disorders of consciousness using NF-L and GFAP biomarkers: A pivotal study. Sci. Rep.10.1038/s41598-022-21930-w (2022). 10.1038/s41598-022-21930-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franzese, M. et al. SARS-CoV-2 antibody responses before and after a third dose of the BNT162b2 vaccine in Italian healthcare workers aged ≤60 years: One year of surveillance. Front. Immunol.10.3389/fimmu.2022.947187 (2022). 10.3389/fimmu.2022.947187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coppola, L. et al. Identification of immune cell components in breast tissues by a multiparametric flow cytometry approach. Cancers10.3390/cancers14163869 (2022). 10.3390/cancers14163869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coppola, L. et al. KCTD15 Is overexpressed in her2+ positive breast cancer patients and its silencing attenuates proliferation in SKBR3 cell line. Diagnostics10.3390/diagnostics12030591 (2022). 10.3390/diagnostics12030591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smaldone, G. et al. KCTD15 deregulation is associated with alterations of the NF-κB signaling in both pathological and physiological model systems. Sci. Rep.10.1038/s41598-021-97775-6 (2021). 10.1038/s41598-021-97775-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orlandella, F. M. et al. The lncRNA TEX41 is upregulated in pediatric B-cells acute lymphoblastic leukemia and it is necessary for leukemic cell growth. Biomark. Res.10.1186/s40364-021-00307-7 (2021). 10.1186/s40364-021-00307-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baselice, S. et al. Impact of breast tumor onset on blood count, carcinoembryonic antigen, cancer antigen 15–3 and lymphoid subpopulations supported by automatic classification approach: A pilot study. Cancer Control10.1177/10732748211048612 (2021). 10.1177/10732748211048612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diaferia, C., Rosa, E., Accardo, A. & Morelli, G. Peptide-based hydrogels as delivery systems for doxorubicin. J. Pept. Sci.10.1002/psc.3301 (2022). 10.1002/psc.3301 [DOI] [PubMed] [Google Scholar]
- 31.Orlandella, F. M. et al. miR-622 is a novel potential biomarker of breast carcinoma and impairs motility of breast cancer cells through targeting NUAK1 kinase. Br. J. Cancer123, 426–437. 10.1038/s41416-020-0884-9 (2020). 10.1038/s41416-020-0884-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smaldone, G. et al. KCTD15 protein expression in peripheral blood and acute myeloid leukemia. Diagnostics10, 11. 10.3390/diagnostics10060371 (2020). 10.3390/diagnostics10060371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Incoronato, M. et al. Circulating miRNAs in untreated breast cancer: An exploratory multimodality morpho-functional study. Cancers10.3390/cancers11060876 (2019). 10.3390/cancers11060876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coppola, L. et al. Purification of viable peripheral blood mononuclear cells for biobanking using a robotized liquid handling workstation. J. Transl. Med.10.1186/s12967-019-2125-7 (2019). 10.1186/s12967-019-2125-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coppola, L. et al. Biobanking in health care: Evolution and future directions. J. Transl. Med.10.1186/s12967-019-1922-3 (2019). 10.1186/s12967-019-1922-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Servais, M. D. et al. Addressing the quality challenge of a human biospecimen biobank through the creation of a quality management system. PloS ONE10.1371/journal.pone.0278780 (2022). 10.1371/journal.pone.0278780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Napolitani, F. et al. Biobankers: Treat the poison of invisibility with CoBRA, a systematic way of citing bioresources in journal articles. Biopreservation Biobanking14, 350–352. 10.1089/bio.2015.0105 (2016). 10.1089/bio.2015.0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howard, H. C. et al. How to responsibly acknowledge research work in the era of big data and biobanks: Ethical aspects of the bioresource research impact factor (BRIF). J. Community Genet.9, 169–176. 10.1007/s12687-017-0332-6 (2018). 10.1007/s12687-017-0332-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jain, K., Salamat-Miller, N. & Taylor, K. Freeze-thaw characterization process to minimize aggregation and enable drug product manufacturing of protein based therapeutics. Sci. Rep.10.1038/s41598-021-90772-9 (2021). 10.1038/s41598-021-90772-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, Y. H. et al. Quality control system in an obstetrics and gynecology disease biobank. Biopreservation Biobanking17, 27–38. 10.1089/bio.2018.0056 (2019). 10.1089/bio.2018.0056 [DOI] [PubMed] [Google Scholar]
- 41.Verberk, I. M. W., Nossent, E. J., Bontkes, H. J. & Teunissen, C. E. Pre-analytical sample handling effects on blood cytokine levels: Quality control of a COVID-19 biobank. Biomark. Med.15, 987–997. 10.2217/bmm-2020-0770 (2021). 10.2217/bmm-2020-0770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhat, B. V. & Adhisivam, B. Human milk banking and challenges in quality control. Indian J. Pediatr.85, 255–256. 10.1007/s12098-018-2635-y (2018). 10.1007/s12098-018-2635-y [DOI] [PubMed] [Google Scholar]
- 43.Bravo, E. et al. Developing a guideline to standardize the citation of bioresources in journal articles (CoBRA). BMC Med.10.1186/s12916-015-0266-y (2015). 10.1186/s12916-015-0266-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Batheja, D. et al. Understanding the value of biobank attributes to researchers using a conjoint experiment. Sci. Rep.10.1038/s41598-023-49394-6 (2023). 10.1038/s41598-023-49394-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The biological data used and analyzed during the study are available from the corresponding author on a reasonable request.