Abstract
Although the E3 ligase Mdm2 and its homologue and binding partner MdmX are the major regulators of the p53 tumor suppressor protein, it is now evident that Mdm2 and MdmX have multiple functions that do not involve p53. As one example, it is known that Mdm2 can regulate cell migration, although mechanistic insight into this function is still lacking. Here we show in cells lacking p53 expression that knockdown of Mdm2 or MdmX, as well as pharmacological inhibition of the Mdm2/MdmX complex, not only reduces cell migration and invasion, but also impairs cell spreading and focal adhesion formation. In addition, Mdm2 knockdown decreases metastasis in vivo. Interestingly, Mdm2 downregulates the expression of Sprouty4, which is required for the Mdm2 mediated effects on cell migration, focal adhesion formation and metastasis. Further, our findings indicate that Mdm2 dampening of Sprouty4 is a prerequisite for maintaining RhoA levels in the cancer cells that we have studied. Taken together we describe a molecular mechanism whereby the Mdm2/MdmX complex through Sprouty4 regulates cellular processes leading to increase metastatic capability independently of p53.
Subject terms: Metastasis, Focal adhesion, Focal adhesion, Cell signalling, Sarcoma
The molecular mechanism of how Mdm2 regulates cell migration remains to be elucidated. Here the authors report the role of Mdm2-x complexes, independent of p53, modulates the expression of Sprouty4 and regulates the cellular processes by decreasing the metastatic capability of cancer cells.
Introduction
Mouse Double Minute 2 (Mdm2) was first described as a potential oncogene in mouse fibroblasts1. Shortly thereafter Mdm2 was discovered to bind the p53 tumor suppressor protein, which was followed by studies validating it as the most critical and obligate regulator of p532–5. Mdm2 is an E3 ligase that can function either alone or with its homolog MdmX (aka Mdm4) in a complex to ubiquitinate, SUMOylate or NEDDylate proteins, most notably p533,6.
While TP53 is found mutated in more than 50% of all human cancers7, Mdm2 is rarely lost or deleted in cancer. Instead, the Mdm2 gene is found amplified in a number of tumors, notably sarcomas, but also in melanomas, glioblastomas and breast cancers8. MdmX is also over-expressed in several cancers, both where Mdm2 is also amplified as well as others, such as retinoblastomas and hematological cancers, where MdmX alone is amplified9,10.
Although the great majority of published studies have focused on the relationship between Mdm2, MdmX, and p53, there is substantial literature that describes p53-independent activities of Mdm2 and MdmX6,11. Mdm2 has been shown to regulate many cellular processes involved in tumorigenesis, such as regulation of chromatin state12 and DNA repair11, maintenance of cell survival and growth13,14, modulation of cell migration15,16 and promotion of metastasis17,18. In aggregate, the abovementioned findings have reintroduced Mdm2 as a potential oncogene in its own right, as first suggested by Fakharzadeh in 1991.
Upstream regulation of Mdm2 levels, implicating both post-translational modifications and microRNAs, have been shown to modulate cell migration16,19–21, but the downstream underlying mechanisms remain to be elucidated. Most reported experiments testing migration/invasion were performed in “2D” settings (i.e., cells attached to and growing on plastic culture dishes) as opposed to the “3D” model where cells are placed on or within a substratum that resembles the extracellular matrix (ECM) and form clusters that more closely recapitulate the bona fide tissue architecture. Further, previous studies describing changes in cell migration have only explored the impact of altering Mdm2 protein levels and did not address whether the E3-ligase activity of Mdm2 or the Mdm2/MdmX complex are required for this function. The same can be said for studies examining the potential roles of Mdm2 in metastasis17,18,22,23.
One additional confounding issue of previous studies characterizing pro-oncogenic activities of Mdm2 has been that, for the most part, researchers have used cell lines that either harbor wild-type or mutant versions of p53 to make their observations. While in such cases, modulation of Mdm2 levels in different cell lines leads to changes in cell migration15,20,21,24–27, the presence of either form of p53 might impact cellular responses, however, and thereby complicate the interpretation of data.
Here, by both altering Mdm2 levels and inhibiting the E3 ligase activity of the Mdm2/MdmX complex in cells that lack any version of the p53 protein, we delve into the fully p53-independent mechanism(s) by which Mdm2 regulates migration and invasion in vitro and metastasis in vivo. We show that the presence of a functional Mdm2/MdmX complex is needed for the migratory and metastatic potential of the cells that we have examined. We also demonstrate that the Mdm2/MdmX complex produces such effects by interfering with reciprocal cell-ECM interactions through impairment of the formation or stabilization of focal adhesions. Finally, we show that Mdm2 acts by regulating the levels and subcellular localization of Sprouty4, which acting through a non-canonical pathway, affects migration by modulation of Rho signaling.
Results
Mdm2 regulates cell migration, cell attachment to extracellular matrix components, and cell spreading independently of p53
To eliminate possible complications in data interpretation stemming from the presence of p53 protein, we used both a CRISPR-derived human HT1080 fibrosarcoma p53 knock-out cell line (HT1080 p53KO; Supplementary Fig. 1A) that does not express p53 and the H1299 lung carcinoma-derived cell line that is endogenously p53-null. While it was previously reported that lowering Mdm2 levels, typically with either si- or shRNAs, interferes with cell migration23,28,29, to our knowledge, the importance of the E3-ligase activity of Mdm2 and the role of the Mdm2/MdmX complex in this process has not been reported. To test this, together with Mdm2 or MdmX silencing using siRNAs, we have used a small molecule inhibitor of Mdm2, MEL23, that was previously shown to inhibit the function of the Mdm2/MdmX complex, leading to the accumulation of Mdm2 and MdmX proteins30.
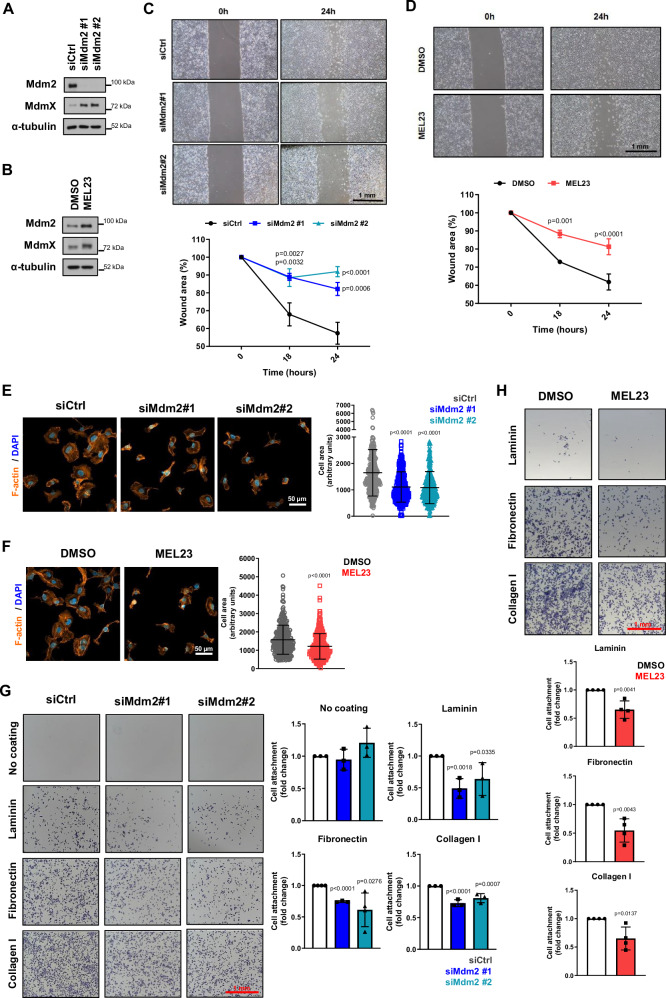
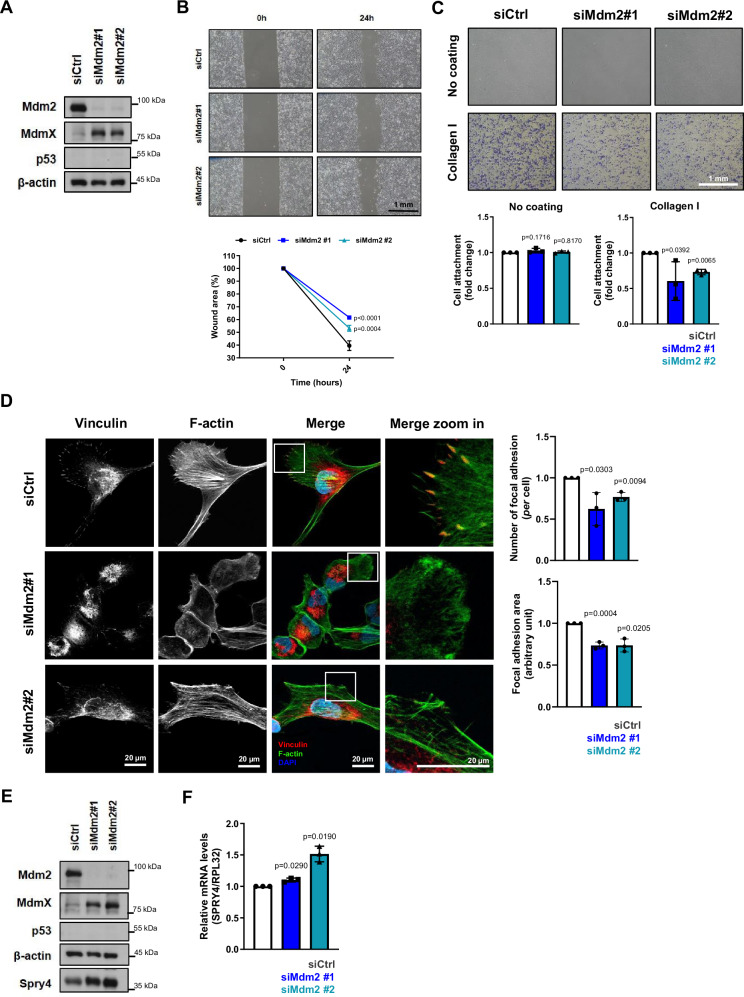
We treated HT1080 p53KO cells either with Mdm2 siRNAs (Fig. 1A) or with MEL23, which, as expected, led to increased Mdm2 and MdmX protein levels (Fig. 1B). Cells were also treated with MdmX siRNAs (Supplementary Fig. 1B). Either siRNA-mediated knockdown of Mdm2 (Fig. 1C), MEL23 treatment (Fig. 1D) or MdmX siRNAs (Supplementary Fig. 1C) led to reduced cell migration using the wound scratch assay.
Fig. 1. Loss of Mdm2 or functional impairment of the Mdm2/MdmX complex decreases migration, impairs cell spreading and cell attachment to ECM in a p53-independent manner.
HT1080 p53KO cells were transfected with siRNAs against Mdm2 and siCtrl or treated with the Mdm2 inhibitor, MEL23. A, B Protein levels of Mdm2 and MdmX after (A) transfection using siRNAs (n = 6 samples) as indicated or (B) treatment with MEL23 (7 µM) for 24 h (n = 4 samples). α-tubulin was used as a loading control. C, D Cell migration assay. Representative images (top) and quantification (bottom) of wound scratch migration assay with cells (C) silenced for Mdm2 or (D) treated with MEL23 for 24 h, n = 3 groups. E, F Representative images of the morphology of cells attached to collagen-coated coverslips and quantification of cell area after (E) silencing of Mdm2 or (F) treatment with MEL23 for 24 h, n = 3 groups. Cells stained for actin (orange) and DNA via DAPI (blue). The cell area was quantified and plotted. Graphs show all three independent experimental replicates combined, for each independent experimental replicate a minimum of 100 cells were quantified summing a total of at least 300 cells/condition. G, H Quantification and representative micrographs showing attachment to different ECM components of cells (G) silenced for Mdm2, n = 3 samples or (H) treated with MEL23 for 24 h, n = 4 samples. The graphs shown represent the mean ± SD of independent experimental replicates. More details about the statistical tests used can be found in the Source Data file.
We also analyzed the morphology of cells growing in 2D in response to those stimuli and found that the area of the attached cells was significantly reduced (Fig. 1E, F and Supplementary Fig. 1D). Interestingly, cells in solution (unattached) did not display any difference in size after such treatments (Supplementary Fig. 1E) demonstrating that the changes in area upon Mdm2 knockdown require processes involved in cell attachment.
Since the ability of cells to attach and spread are correlated directly to interaction and binding to molecules in the extracellular environment, we performed an attachment assay using the three main extracellular matrix (ECM) components, collagen (represented by collagen I), fibronectin, and laminin. When Mdm2 was silenced by siRNA or functionally inhibited by MEL23, the attachment capability of the HT1080 p53KO cells to all three ECM components was significantly reduced to similar extents (Fig. 1G, H and Supplementary Fig. 1F). It should be pointed out that, although Mdm2 modulation has been associated with epithelial-mesenchymal transition/mesenchymal-epithelial transition (EMT/MET) in some cells17,23,31,32, there were no changes associated with different EMT markers (E-cadherin, N-cadherin, vimentin and Zeb1) in response to silencing Mdm2 as shown by immunoblot analysis (Supplementary Fig. 1G) or when a panel of markers was analyzed by mass spectrometry (Supplementary Table 1).
Taken together, our results support the likelihood that the active Mdm2/MdmX complex is required for efficient migration of cells growing in 2D culture conditions, which likely results from impaired ability to interact with or respond to the extracellular environment.
Functional impairment of the Mdm2/MdmX complex decreases individual and collective invasion in a 3D in vitro model of cancer cell dissemination
Experimental results related to cell migration often differ substantially between 2D and 3D models33,34. To gain insights on the impact of manipulation of Mdm2 levels or activity in a 3D setting, we examined individual and collective invasion of HT1080 KO cells using a physiologically relevant 3D tumor spheroid model35. Here, spheroid invasion was used to assess individual cancer cell invasion in a low-density collagen I matrix, since this matrix is more permissive, while invasion in a more dense composite matrix (collagen I/BME, Basement Membrane Extract) was used to assess collective invasion36.
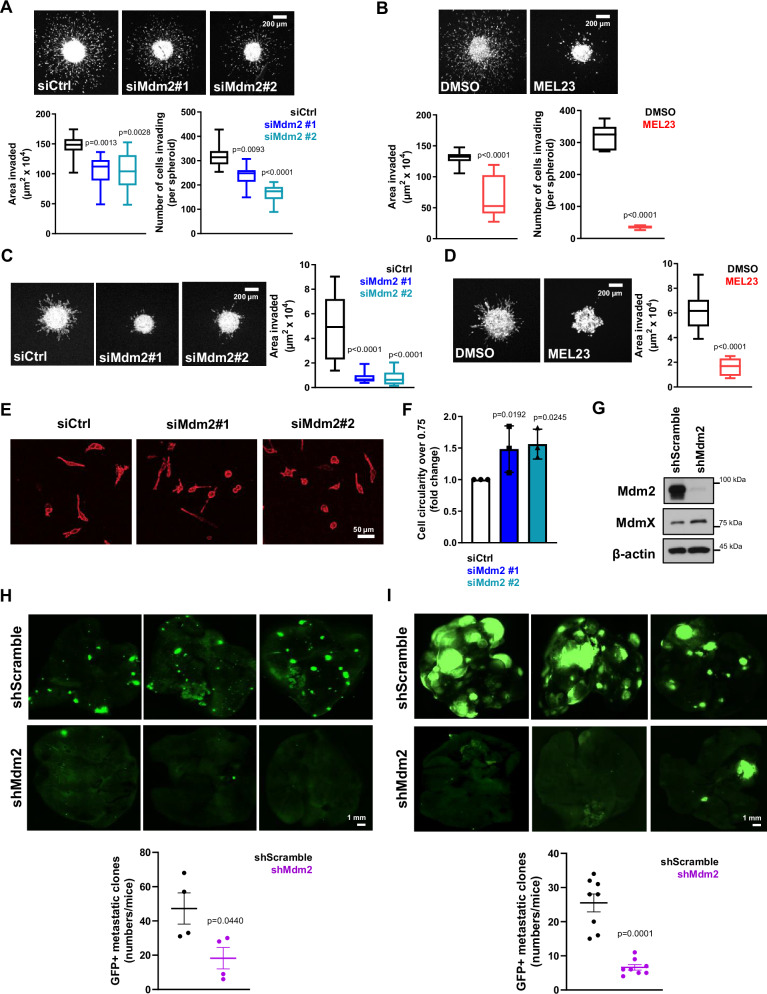
Silencing of Mdm2, as well as treatment with MEL23, led to a significant decrease in individual 3D invasion, which is typically determined by measuring the area invaded over a given time period and by counting the number of cells invading from the spheroid into the surrounding ECM (Fig. 2A, B). When using the biomechanically more challenging composite matrix (collagen/BME), a matrix of a much tighter mesh forming smaller pores for locomotion, we observed that both Mdm2 siRNA and MEL23 treatment significantly decreased collective invasion as well (Fig. 2C, D). We also embedded dispersed cells in collagen I matrices to investigate if cell morphology in this 3D setting was affected by Mdm2 knockdown, as we had observed in 2D cultures. Again, silencing of Mdm2 led to altered cell morphology; specifically, there was a statistically significant shift towards a rounder cell morphology (cells with higher circularity) in 3D, when compared to the control group (Fig. 2E, F). Note that Mdm2 knockdown did not affect spheroid proliferation for the duration of the invasion assay (Supplementary Fig. 2A).
Fig. 2. Loss of Mdm2 function decreases individual and collective invasion in 3D and metastatic burden in vivo.
A, B Tumor spheroid invasion in collagen matrix after (A) transfection of HT1080 p53KO cells with siRNAs against Mdm2 or (B) treatment with MEL23. Representative images above and quantification below of the area invaded and the number of cells invading 24 h after implantation. C, D Tumor spheroid invasion in the collagen-BME matrix after (C) transfection of HT1080 p53KO cells with siRNAs against Mdm2 or (D) treatment with MEL23. Representative images and quantification of the area invaded 24 h after implantation. Graphs show three pooled independent experimental replicates, the total number of spheroids in each condition is: siCtrl = 10, siMdm2#1 = 15, siMdm2#2 = 12 in panel (A); DMSO = 25, MEL23 = 24 in panel (B); siCtrl = 12, siMdm2#1 = 14, siMdm2#2 = 15 in panel (C), and DMSO = 8, MEL23 = 9 in panel (D). E Representative images of cell morphology of HT1080 p53KO cells transfected with siRNAs against Mdm2 or siCtrl in collagen matrix, 6 h after implantation. F Quantification of more circular cells (circularity of 0.75 or higher) in each condition shown in (E), n = 3 groups. The graph represents the average fold change of more circular morphology in Mdm2 silenced cells compared to control in three independent experimental replicates. More details about the quantification can be found in the method’s session. G–I HT1080 p53KO cells stably expressing shRNA scramble (shScramble) or a pool of Mdm2 shRNAs (shMdm2) were used to analyze metastatic burden using mouse models. G Protein levels of Mdm2 and MdmX in HT1080 shScramble and shMdm2 stable cell lines. β-actin was used as a loading control. H, I Representative images above and quantification below of metastatic foci in the lungs after implantation of shScramble or shMdm2 cells using (H) orthotropic model, n = 4 mice/group or (I) tail-vein model, n = 8 mice/group. In all box and whisker plots the boxes extend from 25 to 75 percentiles and whiskers show min and max values, The line in the center of each box represents the median. Graphs shown in panels (F, H, I) represent the mean ± SD of independent experimental replicates. More details about the statistical tests used can be found in the Source Data file.
These findings indicating that a functional Mdm2/MdmX complex is needed for the regulation of cell migration and invasion by Mdm2 provided the impetus for us to examine whether Mdm2 might play a role in processes associated with metastasis in vivo.
Mdm2 knockdown decreases metastatic burden in vivo
To evaluate if decreased Mdm2 levels can impact metastasis, we performed experiments using both orthotopic and tail-vein injections in athymic nude mice. We first established GFP-expressing cell lines derived from HT1080 p53KO cells that stably express either normal (shScramble cells) or much lower Mdm2 levels (shMdm2, cells that harbored four different shRNAs against Mdm2) (Fig. 2G). Assessment of cell motility (Supplementary Fig. 2B), cell area in 2D (Supplementary Fig. 2C), and invasion in 3D culture conditions (Supplementary Fig. 2D) demonstrated that the HT1080 cell line with stable Mdm2 knockdown (shMdm2) displayed the same characteristics observed in cells transiently transfected with siRNA or in cells treated with MEL23, when compared to the shScramble cells.
Using these GFP+ HT1080 p53KO cell lines, we performed an analysis of metastatic burden in the lungs first using an orthotopic model in vivo where cells were introduced into the quadriceps femoris muscle of mice, and cells from tumors formed in situ were allowed to metastasize to the lungs. After 6 weeks (counting from the injection of tumor cells) mice were euthanized and the number of metastatic foci in the lungs were analyzed. The mice injected with shMdm2 cells displayed a dramatic decrease in the number of metastatic foci in comparison to the control shScramble group (Fig. 2H). Measurement of tumor weight in our orthotopic model showed that tumors with Mdm2 knocked down were significantly smaller than in the control group at the time of harvest (Supplementary Fig. 2E). While these findings revealed that reducing Mdm2 levels impacted metastasis, we could not rule out the possibility that at least some of the effects seen were due to the initial smaller sized shMdm2 tumors that formed over the course of these experiments.
To better understand if the changes in metastatic burden observed in mice were due to decreased metastatic ability or changes in primary tumor growth between the two groups, we performed lateral-tail-vein injections into athymic nude mice. This model encompasses the stages of metastatic cascade after the tumor cells enter the bloodstream and is not influenced by differences in initial tumor size, since the same number of cells for the two experimental groups are injected into the mice tail-veins. The analysis of metastatic burden in the lungs of mice injected with the GFP+ HT1080 p53KO cells using this model demonstrated once again that cells expressing less Mdm2 had markedly reduced metastatic capability in comparison to the control group in vivo (Fig. 2I). These data indicate that defects in cell attachment and motility of cancer cells lacking full levels of Mdm2 may lead to a decreased ability of cancer cells to metastasize into distant organs.
Mdm2 depletion or inhibition decreases the number and size of focal adhesions
Our findings showing that knockdown or pharmacological inhibition of Mdm2 decreased cell spreading and cell attachment to the ECM accompanied by reduced migration, invasion, and metastasis, suggested that this might be due to inhibition of one or more signaling processes responsible for intermediating cellular contacts with the extracellular environment and/or cellular movement. It is well established that the main proteins that mediate ECM attachment are integrins, that each have specific sets of ECM substrates37. Yet, since the ECM substrates that we tested were affected to similar extents, we speculated that integrins are not the likely step in the process of ECM attachment that is affected by Mdm2 downregulation. In concert with this, when we measured the levels of those integrins by mass spectrometry (which is described in the next section) or the levels of some of the most expressed integrins in HT1080 cells (integrin beta1, integrin alpha-2, and integrin alpha-3) by immunoblot we did not identify any significant differences upon Mdm2 knockdown or inhibition (Supplementary Table 2 and Supplementary Fig. 3A). Further, silencing of Mdm2 did not affect the presentation of integrins in the membrane either, as determined by measuring integrins present in the cell surface (Supplementary Fig. 3B).
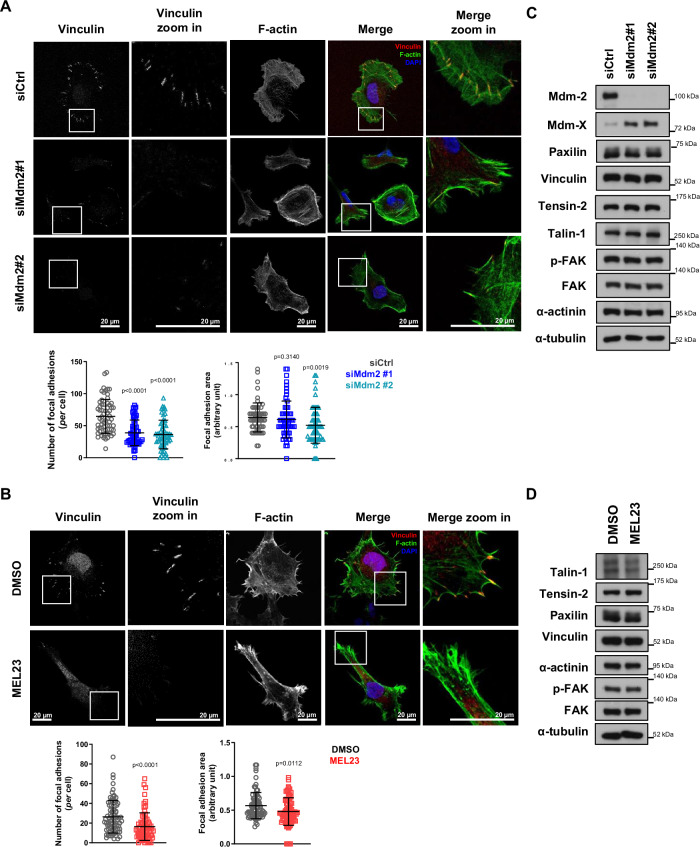
Activation of integrins leads to the recruitment of a set of proteins that form structures known as focal adhesions (FA), which are responsible for integrin activation via linking cells to the ECM with an ensuing reorganization of the cytoskeleton in order to promote anchorage and support motility38,39. Strikingly, when HT1080 cells were transfected with Mdm2 siRNAs (Fig. 3A), treated with MEL23 (Fig. 3B), or transfected with MdmX siRNAs (Supplementary Fig. 3C), the number and size of FA foci decreased significantly. Notably, as well, the stable cell line expressing shMdm2 used in the metastasis experiments described above also displayed reduced FAs compared to control cells (Supplementary Fig. 3D). Thus, inhibition of the Mdm2/MdmX complex either by silencing of Mdm2 or MdmX or by pharmacological inhibition leads to less FA formation. That the stable shMdm2 cell line also displayed lower FA’s indicates that this impairment can be stably maintained.
Fig. 3. Loss of Mdm2 or functional impairment of Mdm2/MdmX complex decreases FA formation in a p53-independent manner.
HT1080 p53KO cells were (A) silenced for Mdm2 using siRNAs or (B) treated with 7 µM MEL23 for 24 h. A, B Immunofluorescence showing FA foci by vinculin staining (red), cell surface was outlined by phalloidin staining (green), and nuclei (blue) detected by DAPI staining of DNA. Representative images are shown above, with quantification of FA parameters below. The graphs shown represent the mean ± SD of three independent experimental replicates. For each independent experimental replicate all parameters were quantified in at least 20 events/condition for a total of at least 60 events/condition. C, D Protein levels of FA-related proteins in HT1080 p53KO cells (C) silenced for Mdm2 or (D) treated with MEL23 for 24 h. α-tubulin was used as a loading control, n = 3 samples. More details about the statistical tests used can be found in the Source Data file.
To understand if the process impacted by Mdm2/MdmX complex inhibition is the formation or recycling of the FA we performed an FA disassembly assay that indicates whether the recycling of FAs by the microtubule network in cells is being altered. Since control cells or cells silenced for Mdm2 responded similarly to treatment with nocodazole, an inhibitor of FA recycling, the disassembly of FAs was likely not affected by Mdm2 modulation (Supplementary Fig. 3E).
Although we had linked Mdm2 functionally with FA formation, when we probed the levels of several proteins known to be involved in the formation of FAs, somewhat confoundingly none of those proteins tested were significantly altered in response to Mdm2 knockdown or MEL23 treatment (Fig. 3C, D). Having failed to identify any candidates by focusing on FA components, we broadened our search by performing an untargeted proteomic analysis of cells treated with MEL23 or transfected with siRNAs against Mdm2.
Modulation of Mdm2 levels or activity leads to changes in the cell proteome associated with cell migration and cell-ECM interaction
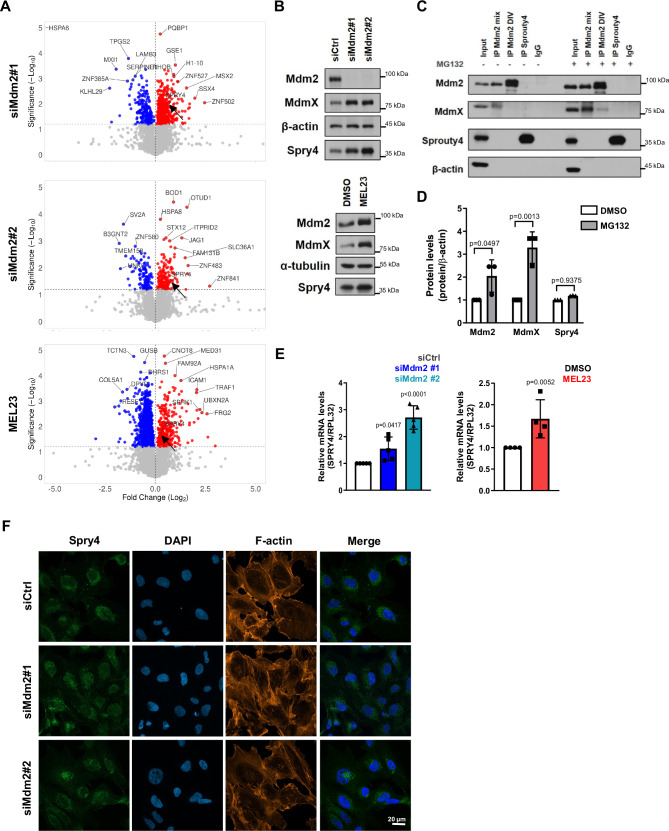
We treated HT1080 cells with two separate siRNAs or with MEL23 prior to subjecting them to the proteomics protocol. Mass spectrometry analysis revealed that levels of many proteins were significantly up- or downregulated after silencing of Mdm2 or treatment with the inhibitor. The volcano plots show all identified proteins and their significance based on statistical analysis (Fig. 4A). Validation of the conditions tested was reflected by levels of Mdm2 that were in accordance with both the immunoblot analysis of proteomics samples and the results of the proteomics analysis itself (Supplementary Fig. 4A, B).
Fig. 4. The Mdm2/MdmX complex regulates the expression and subcellular localization of Sprouty4.
HT1080 p53KO cells were transfected with siRNAs against Mdm2 or siCtrl for 24 h; or treated with 7 µM MEL23 or DMSO (vehicle) for 24 h. A Volcano plots show all proteins identified by mass spectrometry. Colored dots represent significantly differentially expressed proteins that were downregulated (blue dots) and upregulated (red dots) in each condition shown at the left of the plot. Gray dots represent non-significant changes. MEL23-treated cells were compared to DMSO-treated cells. Cells transfected with siMdm2#1 or #2 were compared to siCtrl-transfected cells, n = 3 samples. Black arrows point to the location of Spry4 in each Volcano plot. B Spry4 expression in HT1080 p53KO cells in response to Mdm2 knockdown or treatment with MEL23 for 24 h by immunoblotting. β-actin and α-tubulin were used as loading control for immunoblot, n = 6 samples. C Co-immunoprecipitation of Mdm2 and Spry4 in the presence or absence of MG132. β-actin was used as a loading control. Mdm2 was pulled down by using either a mix of antibodies against Mdm2 (4B11, 3G5, and 2A9) that recognize different domains within the protein, this condition was called “mix”, or by using a single monoclonal antibody D1V2Z, this condition was called “DIV”, n = 3 samples. D Quantification of Spry4, Mdm2, and MdmX protein levels after treatment with MG132 (or vehicle, DMSO) for 4 h, n = 3 samples. E Spry4 mRNA levels in response to Mdm2 knockdown (n = 5 samples) or treatment with MEL23 for 24 h (n = 4 samples). RPL32 was used as housekeeping. F Localization of Spry4 in HT1080 p53KO cells in response to Mdm2 knockdown. Immunofluorescence showed Spry4 staining (green), the cell surface was outlined by phalloidin staining (orange), and nuclei (blue) were detected by DAPI staining of DNA, n = 3 groups. Graphs shown in (D, E) represent the mean ± SD of independent experimental replicates. More details about the statistical tests used can be found in the Source Data file.
Comparison of significantly differentially expressed proteins between the different conditions in the two groups (siRNA and drug) showed that in the siRNA group (siRNA#1 and siRNA#2 vs. siCtrl), 799 proteins were differentially expressed, while in the drug group (MEL23 vs. DMSO) 1854 proteins were significantly altered (see heatmaps in Supplementary Fig. 4C).
Surprisingly, analysis of the false discovery rate of the datasets revealed that only eight proteins were modulated significantly by all three conditions (Supplementary Fig. 4D). Further, for five of these, the fold changes were inverse between drug and siRNA, i.e. one protein was significantly upregulated by MEL23 treatment while the same protein was found significantly downregulated by silencing of Mdm2 or vice versa (Supplementary Fig. 4E, pink and green rows). This would not reflect a functional consequence of inhibition of the Mdm2/MdmX complex. Only three proteins were commonly upregulated by all three conditions (Supplementary Fig. 4E, blue rows), and none were found significantly downregulated by all conditions.
Because of the low number of proteins regulated by all three conditions, we performed pathway analysis examining proteins commonly regulated by at least two out of the three conditions (a total of 119 proteins). Reassuringly, two different pathway databases revealed focal adhesions as one of the top pathways associated with the changes in the proteome of HT1080 cells in response to Mdm2 inhibition (Supplementary Fig. 4F).
The Mdm2/MdmX complex regulates Sprouty4 expression and localization
The three proteins that were significantly similarly regulated by all conditions were: the lysine demethylase KDM3A, the deubiquitinase OTUD1, and the RTK signaling antagonist protein Sprouty4. Among those three hits, Sprouty4 (Spry4) emerged as the most promising candidate for further investigation since one of the main functions of this protein (and other members of the Sprouty/Spred family) in cancer cells is the regulation of cell motility and migration40. Spry4 is one of four members of the Sprouty family (which includes Spry 1, 2, 3, and 4, as well as Spred1 and 2) that have been shown to be key negative regulators of Ras/ERK signaling. The expression and cancer relevance of different Sprouty proteins varies amongst different tumor types40,41. This was supported by our analysis of TCGA and GTEX databases which showed that expression of the different Sprouty family members varies in different tissues, as well as when tumors are compared to normal tissue (Supplementary Fig. 5), suggesting that they play different roles in tumorigenesis depending on the context. The same is seen for their potential value as prognostic markers40,41.
In accordance with the differences seen in the proteomics analysis (Supplementary Fig. 6A), Spry4 levels were significantly increased (~1.5 to 2-fold) in HT1080 p53KO cells that were treated with either siMdm2 or MEL23 measured by immunoblot (Fig. 4B and Supplementary Fig. 6B). Furthermore, our stable HT1080 shMdm2 cells also displayed increased Spry4 levels upon Mdm2 downregulation (Supplementary Fig. 6C). Although HT1080 cells express other Sprouty family members, including Spry2 as well as Spred1 and 2, these were not differentially expressed in response to Mdm2 siRNAs or MEL23 treatment as determined by mass spectrometry (Supplementary Fig. 6D).
We continued to focus on Spry4 and asked whether Mdm2 or the Mdm2/MdmX complex directly regulates Spry4 protein stability. For that we performed a co-immunoprecipitation and, as expected, MdmX was co-immunoprecipitated with Mdm2, however, Spry4 was not pulled down with Mdm2, and the reverse co-immunoprecipitation provided the same negative result (Fig. 4C). Further, cells treated with the proteasome inhibitor MG132 revealed no change in Spry4 protein levels in comparison to non-treated cells (Fig. 4C), which we confirmed by densitometry in all independent experimental replicates (Fig. 4D). By contrast, as expected, Mdm2 and MdmX, both known targets of the proteasome, accumulated in the presence of MG132 (Fig. 4C, D). This showed that Spry4 not only is not a direct binding-partner of Mdm2, but also that the protein levels of Spry4 are not sensitive to proteasomal inhibition.
We then determined that there was a significant increase in Spry4 mRNA in cells harboring siMdm2 or treated with MEL23 (Fig. 4E). The stable shMdm2 cell line also showed a significant increase in Spry4 mRNA levels (Supplementary Fig. 6E). To understand which aspect of Spry4 mRNA expression was being regulated by Mdm2 we tested both the Spry4 promoter’s activation and Spry4 mRNA stability. Surprisingly, both mRNA stability, as measured using two different transcription inhibitors (Supplementary Fig. 6F, G), and Spry4 transcription, as determined by the reporter assay (Supplementary Fig. 6H), were increased upon Mdm2 inhibition.
In addition to the effects of Mdm2 inhibition on Spry4 mRNA expression, we found that the cellular localization of Spry4 protein was altered in response to Mdm2 knockdown. While in control conditions, Spry4 was localized preferentially to the perinuclear region, depletion of Mdm2 led to a more diffused cytosolic distribution of Spry4 both in the transient (Mdm2 siRNA treated) and stable (shMdm2) cells (Fig. 4F and Supplementary Fig. 6I).
Finally, to confirm that the results are not unique to HT1080 cells, we found that knockdown of Mdm2 in H1299 cells (Fig. 5A) led to decreased cell migration (Fig. 5B), decreased attachment to collagen (Fig. 5C) and impaired FA formation (Fig. 5D). As seen in HT1080 cells, Mdm2 silencing induced Spry4 expression at both protein and mRNA levels (Fig. 5E, F) in H1299 cells, thereby extending our results to a cell line which had evolved naturally to lack expression of p53. Further physiological relevance of these observations was derived from analysis of patient samples from TCGA and GTEX databases which revealed that Spry4 expression is decreased in lung cancers (both squamous and adenocarcinoma) compared to normal lung tissues (Supplementary Fig. 6J).
Fig. 5. Mdm2 knockdown decreases cell migration, attachment to the ECM, and FA formation while induces expression of Sprouty4 in naturally p53 null lung adenocarcinoma cell line.
H1299 cells were transfected with siRNAs against Mdm2 and siCtrl. A Protein levels of Mdm2, MdmX, and p53 after. β-actin was used as a loading control, n = 4 samples. B Cell migration assay. Representative images (top) and quantification (bottom) of wound scratch migration assay, n = 3 groups. C Quantification and representative micrographs showing attachment to ECM component, collagen I, n = 3 samples. D Immunofluorescence showing FA foci by vinculin staining (red), cell surface was outlined by phalloidin staining (green), and nuclei (blue) detected by DAPI staining of DNA, n = 3 groups. Representative images are shown on the left, and the quantification of FA parameters is shown on the right. E Protein levels of Spry4 as well as Mdm2, MdmX, and p53 after Mdm2 silencing using siRNAs. β-actin was used as a loading control, n = 4 samples. F mRNA levels of Spry4 after Mdm2 silencing using siRNAs, n = 3 samples. RPL32 was used as housekeeping. Graphs shown represent the mean ± SD of independent experimental replicates. More details about the statistical tests used can be found in the Source Data file.
Sprouty4 regulation is required for Mdm2 to promote FA formation and metastasis
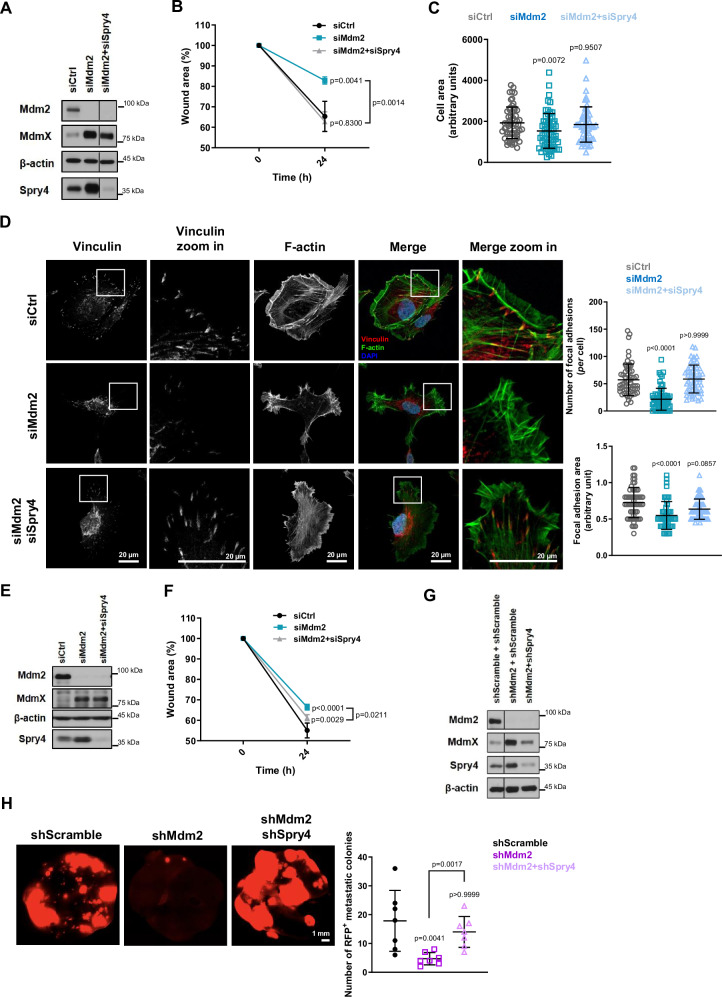
Overexpression of Spry4 has been reported to decrease migration of tumor cells42–45. In order to understand if the noted increase in Spry4 protein levels was linked to changes in cell migration induced by Mdm2 inhibition, we tested the impact of reducing Spry4 under these experimental conditions. Immunoblotting showed that in the double knockdown (siMdm2 + siSpry4) both Mdm2 and Spry4 levels were greatly diminished (Fig. 6A), although cell viability was unaffected by depletion of either Mdm2 and Spry4 individually or together (Supplementary Fig. 7A). Analysis of cell migration under the same conditions revealed that co-silencing of Mdm2 and Spry4 completely rescued the reduction in cell migration triggered by Mdm2 silencing alone (Fig. 6B). Spry4 knockdown also partially rescued migration in MEL23-treated HT1080 p53KO cells (Supplementary Fig. 7B). Co-depletion of Mdm2 and Spry4 also rescued the impaired cell spreading seen when Mdm2 alone was silenced, as shown by the restoration of HT1080 p53KO cells’ area (Fig. 6C). Importantly, the double knockdown (siMdm2+siSpry4) led to the same FA pattern that was observed in the control condition (siCtrl), showing that ablation of Spry4 significantly rescued the FA impairment induced by siMdm2 (Fig. 6D). As confirmation we performed the migration rescue experiment in H1299 cells and here too found that silencing of Spry4 was also able to rescue migration (Fig. 6E, F).
Fig. 6. Knockdown of Sprouty4 rescues changes in cell migration, FA formation, and metastasis resulting from loss of Mdm2.
A–D HT1080 p53KO cells were transfected with siRNA against Mdm2 alone or both siRNA against Mdm2 and a pool of Spry4 siRNAs. A Protein levels of Mdm2, MdmX, and Spry4 after transfection with indicated siRNAs, n = 3 samples. β-actin was used as a loading control. B Quantification of wound scratch migration assay in cells treated with the indicated siRNAs as in Fig. 1C, n = 3 samples. C Quantification of cell area after attachment to collagen-coated coverslips as in Fig. 1E, n = 3 samples. D Immunofluorescence showing FA foci by vinculin staining (red), cell surface was outlined by phalloidin staining (green), and nuclei (blue) as detected by DAPI staining, n = 3 groups. Representative images are shown on the left, and the quantification of FA parameters is shown on the right. In (C, D) the graphs shown represent mean ± SD of independent experimental replicates and in each replicate all parameters were quantified in at least 20 events/condition for total of at least 60 events/condition. E, F H1299 cells were transfected with siRNA against Mdm2 alone or both siRNA against Mdm2 and a pool of Spry4 siRNAs. E Protein levels of Mdm2, MdmX, and Spry4 after transfection with indicated siRNAs. β-actin was used as a loading control, n = 3 samples. F Quantification of wound scratch migration assay comparing migration into wound scratches in cells treated with the indicated siRNAs as in Fig. 1C, n = 3 samples. G, H HT1080 p53KO cell lines were established stably expressing a pool of shRNAs against Mdm2 alone or Mdm2 and Spry4 together. G Protein levels of Mdm2, MdmX, and Spry4 in stable cell lines. β-actin was used as a loading control, n = 3 samples. H Analysis of metastatic burden in vivo using tail-vein injection model as in Fig. 2H, I. Representative images above and quantification below of metastatic foci in the lungs after 8 weeks of injection, n = 7 mice/group. The graphs shown represent the mean ± SD of independent experimental replicates. More details about the statistical tests used can be found in the Source Data file.
We then turned to determine whether the regulation of Spry4 expression by Mdm2 might be involved in the role of Mdm2 in metastasis in vivo. For this, we generated cells stably co-expressing shRNAs against Mdm2 and Spry4 which allowed us to compare outcomes with the cells we had generated expressing shRNAs vs. Mdm2 alone. Confirmation of the silencing of Spry4 and Mdm2 in these stable cell lines was measured by immunoblotting (Fig. 6G) and a migration assay confirmed that the stable cells behaved similarly to transiently transfected cells in that the double knockdown of Mdm2 and Spry4 was able to rescue migration decreased by Mdm2 silencing alone (Supplementary Fig. 7C). These experiments served as a prelude to the lateral-tail-vein injection protocol that was performed as described in Fig. 2I. Remarkably, using the same assay our results showed that in vivo the silencing of Spry4 was able to completely rescue the effects of Mdm2 loss in metastasis formation in vivo (Fig. 6H). Controls with siRNAs and stable shRNAs against Spry4 alone were performed and are shown in Supplementary Fig. 8 and reviewed in the Discussion section.
Sprouty4 regulates migration through a non-canonical mechanism by downregulating RhoA in Mdm2 silenced cells
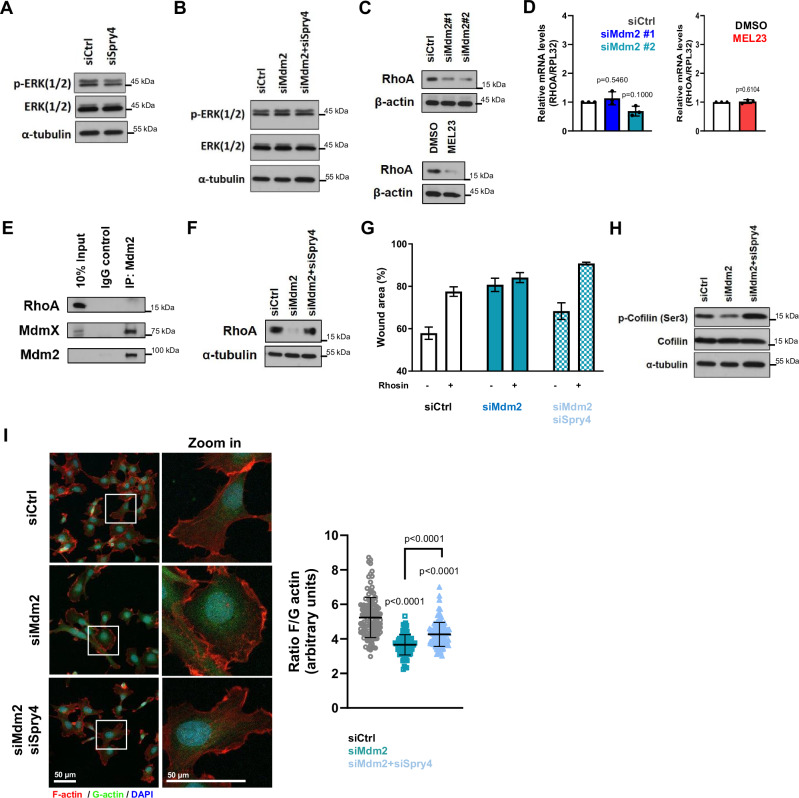
As mentioned above, Sprouty family members are most well characterized as negative regulators of Ras/ERK signaling40, which led us to measure ERK phosphorylation in HT1080 p53KO cells under our experimental conditions. Surprisingly, Spry4 knockdown did not affect ERK phosphorylation in HT1080 p53KO cells (Fig. 7A). We investigated whether the increase in Spry4 levels induced by modulation of Mdm2 affected this signaling pathway and observed that neither Mdm2 silencing nor the double knockdown (siMdm2 + siSpry4) changed phosphorylation of ERK (Fig. 7B). As additional evidence, we probed the phosphorylation of c-Myc at serine 62, a target of phosphorylated (active) ERK kinase46, and did not detect changed phosphorylation of this residue upon Mdm2 silencing (Supplementary Fig. 7D). This indicates that Spry4 might act through a non-canonical pathway to induce the changes in cell migration in response to Mdm2 inhibition.
Fig. 7. Sprouty4 acts through a non-canonical pathway, by regulating RhoA levels, to induce the effects of Mdm2 knockdown in cell migration.
A, B Effect of Spry4 modulation in the phosphorylation of ERKs in HT1080 p53KO cells. Protein levels of phospho-ERK and total ERK levels (A) after Spry4 silencing using a pool of siRNAs and (B) in response to silencing of Mdm2 alone or Mdm2 and Spry4 together. C, D RhoA modulation in HT1080 p53KO cells transfected with siRNAs against Mdm2 or treated with MEL23. C Protein levels and (D) mRNA levels of RhoA after transfection with indicated siRNAs (n = 3 samples) or treatment with MEL23 (n = 3 samples) for 24 h. β-actin was used as a loading control for immunoblots. RPL32 was used as a housekeeping control for qPCR. E Immunoprecipitation of Mdm2 in the presence of MG132. Lysates were probed for the presence of RhoA, and MdmX was used as a positive control. Mdm2 was pulled down by using a mix of antibodies against Mdm2 that recognize different domains within the protein. F RhoA protein levels in HT1080 p53KO cells transfected with siRNAs against Mdm2 alone or against Mdm2 and Spry4. α-tubulin was used as a loading control. G Quantification of wound scratch migration assay comparing cells transfected with siRNAs against Mdm2 alone or against Mdm2 and a pool of Spry4 siRNAs in the presence or absence of the RhoA inhibitor Rhosin (50 µM) for 24 h. The graph represents mean ± SD, n = 3 technical replicates. The graph for the other two independent experimental replicates can be found in the supplementary material. H Immunoblot of levels of total and phospho-cofilin-1(Ser3) in HT1080 p53KO cells silenced for Mdm2 alone or with double KD of Mdm2 and Spry4. I Immunofluorescence showing F- (red) and G-actin (green) staining. Nuclei (blue) as detected by DAPI staining, n = 3 groups. Representative images (left) and quantification of F/G ratio (right). Graph represents mean ± SD of independent experimental replicates, in each replicate the F/G-actin ratio was quantified in at least 30 events/condition for a total of at least 90 events/condition. More details about the statistical tests used can be found in the Source Data file.
Other members of the Sprouty family, Spred1 and Sprouty2, were reported to regulate migration in an ERK-independent manner47. Specifically, the authors showed that cell motility was regulated by Spred1 through reorganization of the actin cytoskeleton by the regulation of RhoA, a member of the Rho family. As RhoA is known to be associated with migration and focal adhesion density48, we hypothesized that Sprouty4 may regulate migration through RhoA and actin reorganization.
Supporting this possibility was our observation that upon Mdm2 inhibition, either by using siRNA or MEL23, there was a significant decrease in RhoA protein levels (Fig. 7C and Supplementary Fig. 7E) as well as less detectable alpha-smooth muscle actin (α-SMA), a downstream target of RhoA (Supplementary Fig. 7F). The level of RhoA mRNA, however, was unchanged (Fig. 7D and Supplementary Fig. 7G). To understand the sequence of events connecting Mdm2, Spry4, and RhoA, we checked if RhoA and Mdm2 were interacting directly. While, as expected, Mdm2 and MdmX could be co-immunoprecipitated, we were unable to show such an association between RhoA and Mdm2 (Fig. 7E). Strikingly, however, Spry4 upregulation was required for the decrease in RhoA levels upon Mdm2 knockdown, as co-depletion of Mdm2 and Spry4 together fully restored RhoA levels found in the control condition (Fig. 7F).
These observations led us to ask if RhoA is involved in the effects of Mdm2 in migration, and for this, we used a RhoA inhibitor, Rhosin. Treatment with Rhosin reduced migration under control conditions, mimicking what happens when Mdm2 is silenced. When cells were depleted of Mdm2 the addition of this inhibitor did not affect migration (which was already decreased by Mdm2 knockdown). Remarkably, while co-knockdown of Mdm2 and Spry4 was able to rescue the effects of Mdm2 silencing, when RhoA was also inhibited, the co-knockdown of Mdm2 and Spry4 failed to rescue migration (Fig. 7G and Supplementary Fig. 7H). Taken together, our findings strongly suggest that RhoA is involved in the effects of Mdm2 on migration through Spry4.
The Rho family regulates actin dynamics through cofilin phosphorylation49,50 and Spry4 has been reported previously to regulate phosphorylation of Cofilin-151, an important event in actin filament dynamics that is involved in FA stability52. Indeed, we observed that Mdm2 knockdown significantly decreased Cofilin-1 phosphorylation, and this was rescued by the co-knockdown of Mdm2 and Spry4 in HT1080 p53KO cells (Fig. 7H) and H1299 cells (Supplementary Fig. 7I). Staining for filamented (F-actin) and monomeric (G-actin) actin showed that silencing of Mdm2 favored monomeric actin, which was partially but significantly reversed in the double knockdown condition (Fig. 7I), supporting our other observations of altered cofilin phosphorylation.
Taken together, our findings point to the regulation of Spry4 by Mdm2 as a critical molecular mechanism by which Mdm2 knockdown leads to impaired cell attachment, cell migration, FA formation, and development of metastasis in vivo in HT1080 cells. Further, Spry4 involvement in the regulation of RhoA, and not ERK activation, is at least one of the mechanisms by which Mdm2 regulates metastasis in our experimental model.
Discussion
Mdm2 regulates several activities involved in tumorigenesis, one of which is migration of cancer cells. We bring several new findings concerning the mechanism by which Mdm2 regulates migration and metastasis in cells that do not express p53.
First, our data strongly support the likelihood that the E3 ligase activity of the Mdm2/MdmX complex is involved in the cellular processes we have studied. Second, Mdm2 is required for robust FA formation in order to maintain the attachment of cells to the ECM and this likely does not involve regulation of EMT. Third, we find that Mdm2 suppression of Spry4 in the cells that we have studied is critical for Mdm2 regulation of migration, FA formation, and metastasis. Fourth, our findings implicate RhoA signaling as a key target of Spry4 that needs to be counteracted by Mdm2.
We discuss the ramifications of these conclusions below.
The E3 ligase activity of the Mdm2/MdmX complex is required for Mdm2 to promote migration and invasion. To date, those studies reporting regulation of Mdm2 altering cell migration have modulated Mdm2 protein levels, either by overexpression of exogenous Mdm2, by silencing of the protein using short hairpin or small interference RNAs or by using inhibitors of Mdm2 such as SP14153 and InulanolideA54, that reduce Mdm2 protein levels. Although it is clear that modulation of Mdm2 protein levels leads to changes in cell migration, information as to whether the E3-ligase function of the Mdm2/MdmX complex is required for this process is lacking. MEL23, an inhibitor of the Mdm2/MdmX complex E3 ligase activity, which leads to the accumulation of inactive Mdm2 and MdmX30, recapitulates all the effects observed in response to Mdm2 silencing.
Further, MdmX silencing reproduces the effects of Mdm2 downregulation; the effects of MdmX silencing on Mdm2 levels, however, vary from cell line to cell line55, and silencing of MdmX in HT1080 cells leads to downregulation of Mdm256. This needs to be taken into consideration when analyzing the role of MdmX in the phenotype. Because MdmX levels also decreased Mdm2 protein, we cannot exclude the possibility that part of the effect observed in siMdmX conditions is due to Mdm2 decrease and not only because of MdmX knockdown. Nevertheless, our results together strongly point to the involvement of the E3-ligase activity in the regulation of cell migration and related phenotypes induced by Mdm2.
The proteomics analysis of significantly differently regulated proteins in response to Mdm2 knockdown using siRNA or MEL23 treatment provided us with potentially interesting insights into the complex-dependent and –independent functions of Mdm2. While a few proteins were found to be regulated in the same manner by MEL23 and Mdm2 siRNAs, showing a correlation with the function of the Mdm2-X complex, other proteins were inversely modulated by MEL23 treatment or Mdm2 knockdown. The latter case may suggest regulation associated with Mdm2 levels but not its E3-ligase function, since MEL23 treatment leads to increased Mdm2 levels while lower Mdm2 levels are seen upon Mdm2 silencing by siRNAs. Further investigation of those proteins could reveal interesting roles of Mdm2 independent of its E3-ligase activity as has been described by other groups57.
Mdm2 is required for focal adhesion formation in HT1080 cells. Focal adhesions are essential structures involved in cell anchorage that connect the cell cytoskeleton with membrane receptors which are in contact with the extracellular environment. As such, their function is intrinsic to cell invasion and metastasis. Formation of FAs has been shown to regulate metastasis in different models58–61 where activation of the pathway is associated with increased invasion. In our work, we show that the regulation of this process by Mdm2 is needed for cell attachment, migration, and metastasis. Loss or inhibition of Mdm2 sets up a unique condition where Spry4 is needed to prevent FA formation.
Based on the lack of change in the expression of relevant EMT markers, as well as the fact that Spry4 silencing fully rescued the impact of Mdm2 inhibition, EMT/MET does not seem to be required for Mdm2 regulation of migration, invasion, and metastasis in our experimental conditions. That said, we do not discard the possibility that in other cell types EMT/MET may be a significant contributing factor. The same applies to FAs, where, although we demonstrate this regulation happens in two different cell types, more studies are needed to determine the generalizability of our findings.
Methodological insights derived from untargeted proteomics. We were able to identify Sprouty4 as a protein involved in the regulation of migration by Mdm2 based on our exploratory proteomics analysis. Although for our purposes, this approach was sufficient, as we were able to identify hits commonly differentially expressed in cells inhibited for Mdm2 by different strategies, we acknowledge that based on the statistical power of our experimental design, using three independent experimental replicate,s it is possible we overlooked true positives in this experiment.
While the proteomics analysis revealed a small number of differentially expressed proteins as a result of Mdm2/MdmX complex inhibition using different strategies, the number of common hits was lower than anticipated. First, off-target effects of siRNAs have been well described in the literature62,63 and can influence the panel of significantly modulated molecules. Indeed, when comparing the two siRNAs, we saw far fewer common differentially expressed proteins (DEPs) than expected, given that both siRNAs targeted the same protein with apparently similar efficiency. Second, knockdown induced by siRNAs does not fully obliterate Mdm2 expression and the residual protein might be sufficient to prevent reaching significance for some proteins in the siRNA conditions, while a pharmacological inhibitor might reach every Mdm2 molecule. Indeed, MEL23 treatment elicited a much greater number of differentially changed proteins than either siRNA (Supplementary Fig. 4).
Considerations on the regulation and modulation of Sprouty4 in migration and metastasis. We establish in this work an association between Mdm2 and the Sprouty protein family, specifically with Spry4. Although we show evidence that Spry4 expression is caused by changes in mRNA being the focus of Mdm2 regulation, we could not rule out as well an effect of Mdm2 on Spry4 protein turnover. It is difficult to assess Spry4 protein stability using cyclohexamide in our experimental conditions due to the very short half-life of Mdm2 (~2 h). However, our experiments with the proteasome inhibitor MG132 suggest that regulation of Spry4 protein levels do not require proteasomal activity. Furthermore, consistent with our findings, iron depletion reportedly increases the expression of Spry4 by increasing both transcription and stability of Spry4 mRNA64.
Overexpression of Spry4 leads to decreased cell migration in the majority of cell models tested to date42–45,65–68 with a few reports showing no significant difference in migration after overexpression69,70. By contrast, migration in response to Spry4 knockdown was found to have widely varying effects in different studies, ranging from increasing cell migration65,71,72, to having no effect on the process73 or even having the opposite effect, where silencing of Spry4 led to decreased migration74,75. This reinforces the context dependence of Sprouty family members and, more specifically, Spry4. In fact, we found that silencing of Spry4 alone led to decreased migration in HT1080 cells (Supplementary Fig. 8A, B), which was unexpected when considering that when silenced together with Mdm2 complete migration was restored. Breast cancer cells depleted of Spry4 were reported to increase the metastatic burden in mice71, while we found that shSpry4 stable cell lines barely metastasized to the lungs using our fibrosarcoma model (Supplementary Fig. 8C, D). It is notable that while cell migration was decreased in siSpry4-silenced cells, cell area or FA formation were not altered (Supplementary Fig. 8E, F), thereby indicating that while siMdm2 alone and siSpry4 alone each lead to reduced migration, they are likely working through different pathways.
Although Sprouty family members are most well understood as negative regulators of receptor tyrosine kinases (RTKs), through Ras/Raf/ERK activation40,72,76,77, these proteins have been shown to play non-canonical functions in cells as well. Spry4, in particular, has been shown to regulate different kinases/kinase-driven pathways, including PI3K/Akt75, PLC/PIP278, c-Src43, and TESK151 independently of their activity on the Ras/Raf/ERK pathway. We demonstrate here that in HT1080 cells, changes in Spry4 do not lead to modulation of ERK phosphorylation (Fig. 7A, B), pointing to a non-canonical Spry4 function in regulating migration in this context.
Previously, Gao et al. found a link between Spry4 and Mdm267, although the authors of that study described the opposite regulation shown by us, where Spry4 can regulate the expression of Mdm2 in colorectal cancer cells. Although their data, together with ours might suggest the existence of a feedback loop between Mdm2 and Spry4, in our model while transient silencing of Spry4 does slightly increase the expression of Mdm2 (Supplementary Fig. 8A), this difference was not observed in the stable shMdm2 cell line (Supplementary Fig. 8C).
Mdm2 and promotion of tumor metastasis. Mdm2’s role in primary tumor growth kinetics is still somewhat controversial, with reports of Mdm2 inhibition leading to decreased tumor mass in one study18 and data showing no difference in tumor volume after Mdm2 silencing in another22. Hauck and colleagues showed that silencing of Mdm2 leads to decreased tumor volume in MDA-MB-231 cells, but the tumor growth using an MDA-MB-231-derived cell line, TMD231, is not impacted by Mdm2 knockdown17. Its role in metastasis, however, is more consistent with reports of decreased circulating tumor cells as well as decreased metastatic burden when Mdm2 is decreased17,18,22,23.
We found that Mdm2 knockdown affects tumor growth in vivo (Supplementary Fig. 2E, F), which might make it difficult to uncouple the roles of Mdm2 in cell migration, attachment, and metastasis alone. However, cell cycle analysis of HT1080 p53KO stable cell lines showed that while silencing of Mdm2 leads to cell cycle arrest, the double silencing of Mdm2 and Spry4 does not rescue this effect (Supplementary Fig. 7J) despite their co-depletion being able to rescue cell migration and metastasis in vivo (Fig. 6F, H). This result indicates that Spry4 is not involved in the growth-related effects of Mdm2 ablation and that the rescue of metastasis formation seen upon Spry4 co-depletion is independent of the cell cycle arrest induced by inhibition of the Mdm2/MdmX complex. This conclusion is supported by our in vitro results showing that cell migration is affected by Mdm2 inhibition even in non-proliferating cells (due to treatment with mitomycin-C) (Figs. 1C, D, 5B, 6B, F, 7G and Supplementary Figs. 1C, 2B, 7B, C, H). This is also supported by Gao and colleagues, who reported differences in metastatic burden when modulating Mdm2 while tumor growth is unaffected in their model22.
Our work poses several questions. How does Sprouty4 regulate RhoA levels? Does Mdm2 regulate the expression of other Spred-Sprouty family members and is such regulation required for Mdm2 roles in migration and metastasis in different contexts? Finally, how can our findings be exploited for clinical benefit? Our findings reveal the potential of targeting Mdm2 for cancer treatment with a special gain for some patients in advanced stages of the disease. While most inhibitors of Mdm2 in clinical trials target tumors with wild-type p53 with drugs that dissociate Mdm2 and p53, we suggest that blocking the E3 ligase function of Mdm2 might have more general therapeutic benefits that could positively affect even patients with mutated p53.
Methods
Ethical statement
All animal studies were approved by the Columbia University Institutional Animal Care and Use Committee.
Chemicals and reagents
Antibodies: Primary and secondary antibodies used for immunoblotting were at 1:1000 and 1:2000 dilution, respectively. Antibodies anti-Mdm2 (cat.#86934), anti-N-cadherin (cat.#13116S), anti-tensin-2 (cat.#11990), anti-talin-1 (cat.#4021), anti-vinculin (cat.#4650), anti-paxillin (cat.#12065), anti-alpha-actinin (cat.#6487), anti-focal adhesion kinase (FAK) (cat.#3285), anti-integrin beta1 (cat#9699), anti-cofilin-1 (cat.#5175T), and anti-p-cofilin-1 (Ser3) (cat.#3313T) were purchased from Cell Signaling Technology. Anti-β-actin (A2066 and A2228), anti-fibronectin (cat.#F3648), anti-mouse peroxidase (cat.#A4416), and anti-rabbit peroxidase (cat.#A6154) were purchased from Sigma-Aldrich. Anti-E-cadherin (cat.#sc-8426) and anti-RhoA (cat.#sc-418) were purchased from Santa Cruz Biotechnology. Anti-p-FAK(Tyr397) (cat.#05-1140) was purchased from Millipore. Anti-laminin (cat.#AHP2491) was purchased from Bio-Rad. Anti-α-tubulin (cat.#AA4.3) was purchased from DSHB. Anti-sprouty4 (cat.#A04343-2), anti-integrin alpha-3 (cat#A02902), and anti-integrin alpha-2 (cat#A01933-2) were purchased from Boster Biological Technology. Anti-vinculin AlexaFluor647 (cat.#ab196579) and anti-Ki67 (cat.#ab16667) were purchased from Abcam. Phalloidin Alexa Fluor Plus 555 (cat.#A30106) was purchased from Thermo Fisher Scientific. Goat anti-rabbit IgG (H + L) biotinylated secondary antibody (cat.# BA-1000-1.5) was purchased from Vector Laboratories. FITC anti-CD51/61 (cat.#304403), PE anti-CD49e (cat.#328009), APC/Cyanine7 anti-human CD18 (cat.# 302133), APC anti-CD29 (cat.# 303007), and FITC anti-CD49b (cat.# 359305) were purchased from BioLegend. Anti-p53 DO-I and 1801 were purified from hybridomas produced in-house. Anti-MdmX mAb 8C6 was produced in Dr. Jiandong Chen’s lab and kindly gifted to our group.
Constructs: For Mdm2 transient knockdown cells were transfected with Silencer Select siRNA s8630 (siMdm2#1) or s224037 (siMdm2#2). For MdmX knockdown cells were transfected with FlexiTube siRNA Mdm4 (cat.#SI00037163, siMdmX#4) from QIAGEN or costume siRNA against MdmX (siMdmX#1, sequence: 5’-AGAUUCAGCUGGUUAUUAA-3’) from Sigma-Aldrich. For Spry4 knockdown cells were transfected with a pool of 3 siRNAs against Spry4 (s37824, s37825, and s37826). Silencer Select Negative Control siRNA #1 (cat.#4390843) was used as transfection control. All siRNAs but the ones against MdmX were purchased from Ambion and transfection was done using Lipofectamine RNAiMax transfection reagent (cat.#13778150, Thermo Scientific). Stable cell lines were established transducing cells with a set of 4 shRNAs against Mdm2 (Origene, cat.#TL311529) or shRNA negative control (Origene, cat.#TR30021) and using the adequate drug for cell selection. For shSpry4 stable cell lines, cells were transduced with a set of four shRNAs against Spry4 (OriGene, cat.#HC108594) or shRNA negative control (OriGene, cat.#TR30033), and using the appropriate drug for cell selection.
Drugs and other reagents: MEL23 (Sigma-Aldrich, cat#373227), MG132 (Selleck Chemicals, cat.#S2619), Accutase (Thermo Fisher Scientific, cat.#ICN1000449), Actinomycin D (Cayman Chemichals, cat#11421), DRB (Sigma-Aldrich, cat#D1916), CellTracker dye (Thermo Fisher Scientific, cat.#C34551), pepsin-treated (PT) bovine type I collagen (Advanced BioMatrix, cat.#5010), growth factor-reduced Phenol Red-free BME/Matrigel (8.9-10 mg/ml solution from BD Biosciences), DMEM solution (10×), NaOH (1 N) and sodium bicarbonate solution (7.5%) (Sigma-Aldrich), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (1 M) (Invitrogen, cat.#15630080), CellMask membrane dye (Thermo Fisher Scientific, cat.#C10045), Alexa Fluor Plus 555 Phalloidin (Thermo Fisher Scientific, cat.#A30106), Molecular Probes Deoxyribonuclease I Alexa Fluor 488 Conjugate (Thermo Fisher Scientific, cat.#D12371).
Cell culture
The human fibrosarcoma cell line HT1080 was obtained from Dr. Brent Stoclwell and CRISPR-engineered to produce the isogenic HT1080 p53 knock-out (KO) cells.56. These cells were grown in Dulbecco’s modified Eagle’s Medium (DMEM) (cat.#12100-061, Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (cat.#900108H, Gemini), 100 units of penicillin, and 100 mg/ml streptomycin (cat.#15140122, Thermo Scientific). The human lung adenocarcinoma H1299 was purchased from ATCC (cat.#CRL-5803). H1299 cells were grown in RPMI 1640 medium (cat.#11875119, Thermo Scientific) supplemented with 10% heat-inactivated FBS, 100 units of penicillin, and 100 mg/ml streptomycin. Cells were maintained at 37 °C with 5% CO2 and were sub-cultured using trypsin-EDTA (cat.#15090-046, Gibco) every 2 or 3 days. All cells used in this work were previously tested for mycoplasma contamination and found to be negative.
Mdm2 and Mdm2/Spry4 knockdown stable cell lines
To perform in vivo experiments, we established an Mdm2 knockdown and an Mdm2/Sprouty4 double knockdown stable cell line, as well as control cell lines expressing shScramble contructs. For the single Mdm2 knockdown, HT1080 p53KO cells were transduced with lentivirus carrying a pool of shRNAs against Mdm2 (4 different shRNAs purchased from OriGene, cat.#TL311529) or shScramble sequence as control (OriGene, cat.#TR30021). Constructs co-expressed GFP. Stable cell lines were generated by selecting transduced cells with 1 µg/ml puromycin for 72 h. Cells were kept on media with a lower concentration (0.5 µg/ml) of puromycin after selection for 1 week, then challenged with 1 µg/ml puromycin every 4 weeks in culture. Newly established cell lines were sorted for GFP-positive cells.
For the double knock-down, cells expressing shMdm2 or shScramble were transduced with lentivirus carrying a pool of shRNAs against Sprouty4 (four different shRNAs purchased from OriGene, cat.#HC108594) or shScramble sequence as control (OriGene, cat.#TR30033). Constructs co-expressed RFP. Stable cell lines were generated by selecting transduced cells with 5 µg/ml blasticidin for 96 h. Cells were kept on media with a lower concentration (2 µg/ml) of blasticidin after selection for one week then challenged with 5 µg/ml blasticidin every 4 weeks in culture. Newly established cell lines were sorted for GFP- and RFP-positive cells.
Cell viability
To ensure the treatments used in this work did not interfere with cell viability, cells were stained with Trypan Blue (Bio-Rad, cat.#1450011) 48 h after transfections with siRNA or after 24 h of treatment with MEL23 or vehicle. Cell viability in 3D was determined using CellTiter Glo (Promega, cat.#G9681) according to the manufacturer’s instructions. Luminescence was measured by a BioTek SynergyH1 Hybrid Reader.
Immunoblotting
After the desired treatments, cells were harvested with trypsin, pelleted, and lysed in RIPA buffer (50 mM Tris, pH 8, 0.5% deoxycholate, 0.1% SDS, 1% Nonidet P-40, 5 mM EDTA, and 150 mM NaCl) along with protease and phosphatase inhibitors. Samples were loaded in equal protein amounts and polypeptides were separated by SDS–PAGE. Proteins were transferred to a PVDF membrane (IPVH00010, Immobilon, Millipore) and blocked for 1 h with Tris-buffered saline containing 0.05% Tween 20 and 3% bovine serum albumin. Membranes were incubated overnight at 4 °C with primary antibody followed by three washes with Tris-buffered saline, 0.05% Tween 20. A secondary HRP-conjugated antibody was added and incubated for 1 h at room temperature (RT). After washes, blots were visualized by chemiluminescence detection using chemiluminescent horseradish peroxidase reagents (Thermo Fisher, Pierce, cat# 32106 or EMD Millipore, Immobilon, cat# WBKLS0050) according to the manufacturer’s instructions. Quantification of protein expression was performed using ImageJ software. Molecular weight markers are indicated on the right side of each blot shown in the figures. All uncropped and unprocessed scans of all blots in Figures can be found in the Source Data file.
Co-immunoprecipitation
Cells were seeded in 10 cm dishes, and after desired treatments, MG132 was added for four hours, followed by lysing samples in NP-40 lysis buffer. Lysates (400 µg of protein) were then incubated with primary antibody or IgG control antibody for 2 h at 4 °C with constant rotation, followed by 1 h incubation with Protein G agarose beads (Millipore, catalog no. 16-266) at 4 °C with constant rotation. Beads were washed four times with lysis buffer or PBS then protein was eluted from beads with Laemmli buffer and boiled for 5 min. Samples were analyzed by immunoblotting. All uncropped and unprocessed scans of all blots in Figures can be found in the Source Data file.
Quantitative real-time PCR (qPCR)
Total RNA was isolated using a Qiagen RNeasy Minikit (Qiagen, cat.#74106) according to the manufacturer’s instructions. A total of 1 μg of RNA was used for reverse transcription (RT) using QuantiTect Reverse Transcription Kit (cat.#205311). The cDNA products were diluted (1:10) with nuclease-free water and analyzed by qPCR using Power SYBR Green PCR Master Mix (Applied Biosystems, cat.#4368708) according to the manufacturer’s instructions. Quantitative PCR analysis was performed using the following protocol: polymerase activation and DNA denaturation for 30 s at 95 °C; amplification denaturation for 5 s at 95 °C and annealing for 30 s at 60 or 62 °C with 40 cycles; and melt curve 65–95 °C with 0.5 °C increment 5 s/step. The quantification cycle (Cq) value was recorded by StepOne software. Relative changes in cDNA levels were calculated using the comparative Ct method (ΔΔCT method). Transcripts were normalized to RPL32. Primers for SPRY4: forward 5’-AGAAGTGTACTGAAGGGACTGGAG-3’; reverse 5’-GTGTGTAGACCACCAAGATCACC-3’. Primers for MDM2: forward 5’-TTGGCGTGCCAAGCTTCTCT-3’, reverse 5’-TACCTGAGTCCGATGATTCC-3’. Primers for ARG1: forward 5’-GTCTGTGGGAAAAGCAAGCG-3’, reverse 5’-CACCAGGCTGATTCTTCCGT-3’. Primers for RPL32: forward 5’-TTCCTGGTCCACAACGTCAAG-3’; reverse 5’-TGTGAGCGATCTCGGCAC-3’.
Immunofluorescence
Cells were seeded in six-well plates and, after desired treatments, were detached using PBS-EDTA 5 mM and reseeded onto coverslips. For cell spreading analysis and focal adhesion staining, cells were fixed 6 h after seeded in collagen I coated (Advanced BioMatrix, cat.#5056) coverslips. Once ready to be processed, cells were fixed and permeabilized in one step with paraformaldehyde (PFA) 4% - 0.2% Triton X-100 for 10 min at room temperature. After washes with PBS, cells were blocked in PBS-1%-BSA for 30 min and incubated with anti-vinculin antibody overnight in a humid chamber. Coverslips were then washed with PBS and incubated with fluorescent phalloidin for 1 h RT, washed with PBS, and incubated with DAPI (5 µM) for 10 min. Coverslips were mounted and micrographs were acquired using ZEISS LSM800. Quantifications were performed using Fiji software. Vinculin foci were used for the quantification of FAs after the exclusion of the nuclear area. The number of vinculin foci and their area in individual cells were quantified in an automated manner using Fiji software. To prevent the influence of background, FA were quantified using an exclusion criterion of a minimum size of 0.2 micron2.
Focal adhesion disassembly assay and FA quantification
The disassembly of FAs was assessed using the protocol described by Ezratty et al.79. Briefly, cells were seeded in 6-well plates, after desired treatments cells were detached using PBS-EDTA 5 mM, washed with FBS-free media and reseeded in coverslips. Cells in complete media were used as a positive control for the presence of FAs (control condition). The day after reseeding, nocodazole (10 µM) was added (nocodazole condition) or not (serum starved condition) for 4 h then washed out with FBS-free media, and cells were incubated in FBS-free media for another 30 min at 37 °C with 5% CO2 (washout condition). Cells were then fixed and permeabilized for immunofluorescence as described above.
Quantification of images considered “cells presenting FA” as cells showing 15 or more FA foci. The graph shows cells “presenting FA” as a percentage of the total number of cells counted. To prevent the influence of the background, FA were quantified using an exclusion criterion of a minimum size of 0.2 micron2. Quantifications were performed using Fiji software.
F/G-actin staining
Filamented actin (F-actin) was stained using fluorescently-conjugated phalloidin while monomeric actin (G-actin) was probed using fluorescently-labeled DNase I. Cells were fixed in PFA 4% for 10 min, washed with PBS and permeabilized with PBS-0.12% Triton X-100 for 5 min RT. After washes with PBS, cells were incubated with a fluorescent phalloidin and DNAse I in the manufacturer’s suggested concentrations for 1 h RT, washed with PBS, and incubated with DAPI for 10 min. Coverslips were mounted and micrographs were acquired using ZEISS LSM800. Quantifications were performed using Fiji software.
Cell motility/migration assay
Cell migration was measured by the wound scratch assay. Briefly, cells were seeded in six-well plates and allowed to reach 100% confluence. The confluent layers were each subjected to a scratch with a 200 µl tip, then washed and media added back. HT1080 cells were serum starved and treated with 2 µg/ml mitomycin-C, while H1299 cells were kept in 2% FBS-RPMI with 2 µg/ml mitomycin-C for the entire duration of the experiment. At the time of scratching, 0 h micrographs were taken and after the indicated time intervals, micrographs of the same points in the wound were taken for comparison. Quantification of wound area was performed using Fiji software. At 0 h, the images were considered to have 100% wound areas.
Determination of cell size in solution and expression of integrins in the membrane by flow cytometry
To analyze cell size in solution, cells (2.5 × 105) were seeded in 6-well plates. After desired treatments cells were detached using PBS-EDTA and cell size was analyzed by forward scatter (FSC) by flow cytometry in a FACS Celesta, using FACS diva software (BD).
For quantification of integrin expression at the cell surface, cells (2.5 × 105) were seeded in six-well plates. After 48 h treatment with siRNAs against Mdm2 cells were detached and stained using fluorescent-conjugated antibodies against integrins alpha-2, alpha5, alphaVbeta3, beta2, and beta1. Briefly, cells were detached using PBS-EDTA 5 mM and washed with FACS buffer (HBSS 1%BSA 0.1% sodium azide). Samples were then blocked using 10% Normal Goat Serum (Invitrogen, cat# 50062Z), followed by incubation with conjugated antibodies. Next, samples were washed twice with FACS buffer and resuspended in the same buffer for analysis by flow cytometry in a FACS Celesta, using FACS diva software (BD).
Cell cycle analysis
Analysis of the cell cycle was performed by quantification of cell DNA content by staining with Propidium Iodide (PI). Briefly, HT1080 p53KO stable cells were detached from plates and fixed in PBS-50% ethanol for 48 h. Cells were then washed with PBS and resuspended in a buffer containing PI. Cells were incubated for 15 min at 37 °C. The PI solution was then washed out and cells were resuspended in PBS for acquisition and analysis by flow cytometry in a FACS Celesta, using FACS diva software (BD).
ECM attachment assay
Cells were seeded in six-well plates, after desired treatments cells were detached using PBS-EDTA 5 mM and collected. After washing and resuspension in FBS-free media, 105 cells were reseeded on Laminin- (Corning, cat.#354412), Fibronectin- (Corning, cat.#356271), Collagen I-coated (Corning, cat.#356271) 24-well, or on uncoated 24-well plates, all of which had been pre-blocked with PBS-1% BSA. Plates were kept at 37 °C with 5% CO2 for 1 h. After incubation, the culture dish wells were washed three times with FBS-free media to remove unattached cells. The plates were then kept in complete media for 1 h at 37 °C with 5% CO2 for cell recovery, and then the remaining cells were stained using 0.2% crystal violet and 10% ethanol. Micrographs of crystal violet-stained wells were taken, followed by dissolving material in the wells in 2% SDS for quantification. Absorbance was measured at 540 nm by a BioTek SynergyH1 Hybrid Reader and was normalized to control conditions.
Analysis of mRNA stability
mRNA stability was assessed by treatment with two transcription inhibitors: actinomycin D (actD) as described previously80 or 5, 6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB)81. Briefly, cells were seeded in 12-well plates and treated with MEL23 for 24 h. Then, act D (10 µg/ml) or DRB (100 µM) were added to wells for indicated time intervals. Cells were harvested, RNA was isolated, and cDNA was generated as described above. mRNA levels were analyzed by qPCR. Data from actD- or DRB-treated cells were normalized to cells treated with vehicle in each condition (DMSO or MEL23 treatment).
Luciferase assay
A Spry4 Firefly luciferase reporter plasmid containing the promoter region of Spry4 (kindly provided by Dr. Meredith Tennis, University of Colorado, USA)44 or negative control were used in co-transfection assays with Renilla-luciferase-expressing plasmids, as internal controls. Briefly, cells were seeded in 12-well plates and transfected the next day with siRNA directed against Mdm2. After 24 h, cells were co-transfected with 3.9 µg of a Firefly luciferase-expressing construct (Spry4 promoter or mock) and 0.1 µg of the Renilla-luciferase-expressing plasmid. Firefly and Renilla signals were measured 24 h after transfection using the Dual-Glo Luciferase Assay System (Promega, cat#E2920) according to the manufacturer’s instructions. Luminescence was measured by BioTek SynergyH1 Hybrid Reader, and the Firefly signal was normalized to the Renilla signal.
Untargeted proteomics
Cells were transfected using siRNA control or two different siRNAs against Mdm2 for 24 h for the siRNA sample set; or treated with MEL23 (7 µM) or DMSO for 24 h for the drug sample set. Three independent experimental replicates were prepared for each of the six groups (untreated, DMSO treated, MEL23 treated, siRNA Ctrl, siMdm2#1, and siMdm2#2) for a total of 18 samples analyzed by LC-MS/MS.
Proteins were isolated from the cell samples using a phenol-guanidinium isothiocyanate (P/GTC) reagent according to the manufacturer’s protocol82,83. After RNA was precipitated from the aqueous phase with isopropanol, proteins were precipitated from the phenol/ethanol phase by the addition of acetone. The protein precipitate was redissolved in 2% SDS and 100-mM Tris-HCl (pH 8.5) using a sonicator (BIORUPTOR II, CosmoBio, Tokyo, Japan). The protein content was measured using a BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) and adjusted to 1 mg/ml with 2% SDS and 100 mM Tris-HCl, pH 8.5. Then 20 µL of protein extract was treated with 10 mM dithiothreitol at 50 °C for 30 min and subjected to alkylation with 35 mM iodoacetamide in the dark at room temperature for 30 min while being protected from light. The alkylated sample was subjected to clean up and digestion with SP3 method84,85 with minor modifications. Briefly, two types of Sera-Mag SpeedBead carboxylate-modified magnetic particles (hydrophilic particles, cat# 45152105050250; hydrophobic particles, cat# 65152105050250; Cytiva, Marlborough, MA, USA) were used. These beads were combined at a 1:1 (v/v) ratio, washed twice with distilled water, and reconstituted in distilled water at a concentration of 10 μg solids/μL. About 20 μL of reconstituted beads were then added to the alkylated protein sample, followed by 99.5% ethanol to bring the final concentration to 75% (v/v), with mixing for 10 min. The supernatant was discarded, and the beads were washed twice with 80% ethanol, then resuspended in 100 μL of 50 mM Tris-HCl (pH 8.0) with 1 μg of trypsin/Lys-C Mix (cat# V5072, Promega, Madison, WI, USA) and mixed gently at 37 °C for 15 h to digest proteins. The digested sample was acidified with 20 μL of 5% trifluoroacetic acid (TFA) and then sonicated using a Bioruptor II. The sample was desalted using an SDB-STAGE tip (cat# 7820-11200, GL Sciences Inc., Tokyo, Japan) according to the manufacturer’s protocol, followed by drying in a centrifugal evaporator (miVac Duo concentrator, Genevac Ltd., Ipswich, UK) and redissolving in 2% ACN in 0.1% TFA. The peptide concentration in the redissolved sample was determined using a Lunatic instrument (Unchained Labs, Pleasanton, CA, USA) and transferred to a hydrophilic MS vial (cat# 11-19-1021-10; AMR Inc., Tokyo, Japan).
The redissolved peptides were directly injected onto an Aurora column (C18, 75 μm ID, 25 cm length, 1.6 μm beads, IonOpticks, Fitzroy, Australia) at 50 °C and then separated with a 120-min gradient (A = 0.1% formic acid in the water, B = 0.1% formic acid in 80% ACN) consisting of 0 min 5% B, 114 min 35% B, 117 min 60% B, 120 min 60% B at a flow rate of 200 nL/min using an UltiMate 3000 RSLCnano LC system. The peptides eluted from the column were analyzed using an Orbitrap Exploris 480 with an InSpIon system86 for overlapping window DIA-MS87,88. MS1 spectra were collected in the range of 495–745 m/z at a 15,000 resolution to set AGC targets of 3 × 106 and a maximum injection time of “Auto”. MS2 spectra were collected at m/z 200–1800 at a 30,000 resolution to set AGC targets of 3 × 106, a maximum injection time of “Auto”, and stepped normalized collision energies of 22, 26, and 30%. The isolation width for MS2 was set to 4 Da, and overlapping window patterns at m/z 500–740 were used for window placements optimized via Scaffold DIA v3.0.1 (Proteome Software, Inc., Portland, OR, USA).
The MS files were searched against an in silico human spectral library using Scaffold DIA v3.0.1. The human spectral library was generated from the human protein sequence database (UniProt, proteome ID UP000005640, reviewed, canonical, downloaded on March 31, 2021) by Prosit (http://www.proteomicsdb.org/prosit)89,90. The search parameters on the Scaffold DIA were as follows: experimental data search enzyme, trypsin; maximum missed cleavage sites, 1; precursor mass tolerance, 10 ppm; fragment mass tolerance, 10 ppm; static modification; and cysteine carbamidomethylation. The threshold for protein identification was set such that both protein and peptide false discovery rates (FDRs) were <1%. Peptide quantification was calculated by EncyclopeDIA algorithm v 1.2.291. Protein quantification was estimated from the summed unique peptide quantification. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the jPOST partner repository (http://jpostdb.org) with the dataset identifier PXD033789. Files containing the full analysis with fold changes and statistical parameters can be found in the supplementary material (Supplementary Data 1).
For the proteomics analysis, volcano plots were generated using VolcaNoseR92. Heatmaps showing significantly differentially expressed proteins were generated using Heatmapper93. Heatmaps were created using Pearson’s correlation to measure the distance between samples, clustering was done using average linkage. Pathway analysis was performed using Enrichr25,94,95.
Database analysis of gene expression
Analysis of Spry4 gene expression in normal tissue and tumors was performed using GEPIA96, grouping patient samples from the TCGA and GTEx projects.
Invasion assay and 3D morphology in hydrogels
Generation and treatment of multicellular tumor spheroids: Cells were seeded in 6-well plates and either silenced for Mdm2 using different siRNAs or treated with MEL23 (7 µM) or DMSO for 24 h. After the desired treatments, cells were detached using Accutase. Spheroids were formed in an appropriate growth medium supplemented with 0.25 mg/ml BME (Basement Membrane Extract) using a centrifugation method described previously97. Spheroids were allowed to form for 24 h at 37 °C under 5% carbon dioxide. For MEL23 treatment, the same concentration of inhibitor was added to the biomatrix and the overlay media to maintain continuous treatment during the invasion assay. Preparation of collagen solutions, collagen (1 mg/ml) and collagen/BME matrices, were described in Guzman et al., Biomaterials 201435. For the preparation of cell-embedded gels loaded with a single spheroid, the collagen solution was added to a chamber, and the spheroid was added to the liquid collagen, which was then transferred to a 37 °C incubator for gelation. For collagen gels loaded with dispersed cells, the collagen solution was prepared, omitting 50% of the water, and neutralized. The water volume was substituted with a growth medium containing the desired number of cells (2 × 104–4 × 104 cells/ml). Subsequently, cell-loaded collagen was added to the chamber and gelled at 37 °C. For staining the actin cytoskeleton in collagen-embedded cells, samples were fixed with 4% PFA for 15 min at RT, permeabilized with 0.2% Triton X/H2O for 10 min at RT, and incubated with fluorescently-labeled phalloidin dye for 16–20 h at 4 °C. For invasion analysis, spheroids were imaged at 1 and 18 h after implantation. For spheroids invading in pure collagen I gels, magnified fluorescence images were used to determine the invaded area, which was defined as the difference between the area of the spheroid at t = 1 h and an ellipse that circumscribes 90% of the invasive cells at t = 18 h. For spheroids invading collagen/BME composite matrices, maximum intensity projections were thresholded and used to generate masks from which the non-invading spherical core of the spheroid was removed. The remaining invasive structures were analyzed. The total area of invasive structures per spheroid was used to assess invasion efficiency.
Regarding the quantification of cell circularity in 3D, a minimum of 100 cells in each independent experimental replicate was measured. For each independent experimental replicate, the percentage of cells with circularity equal to or higher than 0.75 among the total number of events was analyzed. Using this percentage for each independent experimental replicate, we generated the fold change of this more circular population relative to the control condition.
Metastasis assays: lateral-tail-vein injections and orthotopic implantation
All animal studies were approved by the Institutional Animal Care and Use Committee at Columbia University, and all experiments were conducted in compliance with the NIH guidelines for animal research. To establish the lung metastasis model, an equal number of 5-week-old male and female athymic nude mice were purchased (NCr-Foxn1nu, n = 19, Taconic). Mice were housed under controlled environmental conditions (22–24 °C, 45–60% humidity) in a 12-h dark–light cycle with ad libitum access to food and tap water.
At the age of 7 weeks, 1.0 × 106 (tail-vein) and 5.0 × 105 (orthotopics) cells were resuspended in 100 μL of cold Dulbecco’s phosphate-buffered saline (DPBS) and injected into the distal end of the lateral-tail-vein (tail-vein) or right quadriceps femoris muscle (orthotopics) with a 28- gauge 0.5 in needle. After injection, mice were monitored daily for signs of pain or discomfort and weight loss. The maximal tumor size/burden set by IACUC (>20 mm in any direction) was not exceeded in any experiments (under approved IACUC protocols AC-AABB4504, AC-AABS7612, and AC-AABV0653). To detect the formation of lung metastatic lesions, mice were euthanized at 8 weeks (tail-vein) or 6 weeks (orthotopics) time points by CO2 and cervical dislocation. To identify lung metastases, the mouse lungs were flushed with 20 mL Heparin-PBS to deplete red blood cells and were imaged with a Keyence fluorescent microscope at 2X magnification.
Immunohistochemistry
Harvested tissues were fixed in buffered zinc formalin (Thermo Fisher, 22-050-259) for 16 h at 4 °C, washed in PBS for 30 min, and placed in 70% ethanol until paraffin embedding. Antigen retrieval was performed on 4 µm dewaxed and rehydrated FFPE sections using 10 mM boiling citric acid buffer at pH 6. Endogenous peroxidases were quenched with 3% H2O2 (Sigma-Aldrich, H1009), followed by blocking with Avidin (Sigma-Aldrich, A9275), Biotin (Sigma-Aldrich, B4501), and Starting Block T20 buffer (Thermo Fisher, 37543) for 15 min each at room temperature. The tissue sections were incubated with Ki67 primary antibody overnight at 4 °C, and followed by incubation with biotinylated secondary antibody for 30 min at 37 °C. The primary and secondary antibodies were diluted in PBS + 1% Bovine serum albumin+0.3% Triton X buffer. After the application of the ABC reagent (Vector Laboratories, PK-6100) for 30 min at 37 °C, the slides were treated with a DAB substrate kit (Vector Laboratories, SK4100), counterstained with Hematoxylin, dehydrated, and mounted for imaging. Bright-field images were taken with Keyence BZ-X800 (Keyence).
Statistical analysis and reproducibility
Data reported in this paper were expressed as the mean ± S.D, except for Supplementary Fig. 7H (bottom graph), which shows mean ± SEM, and Fig. 2A–D, which shows the median, from at least three independent experiments. The sample size for each experiment (n) is indicated in the figure legends. A significant difference from the respective control for each experimental condition or between experimental conditions was assessed by one or two-tailed analysis of variance (or non-parametric equivalent) with respective post-tests, or Student’s t-test using GraphPad Prism 10 software. Further details on which tests were used for each comparison can be found in the available Source Data file. Values of p < 0.05 were considered statistically significant. Exact p values are indicated in the panels and refer to comparison with the respective control for a particular condition, unless shown otherwise by the use of brackets. For proteomics analysis, the false discovery rate cutoff was set as FDR <0.2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We are grateful to members of the Prives laboratory for helpful suggestions and discussion. This study was supported by grants CA87497 and CA220526 to C.P., as well by grants P30 CA013696, PO1 CA098101 to A.K.R. and F31 CA275369 to G.E. and shared resources: Confocal and Specialized Microscopy, Molecular Pathology, Cancer Biostatistics, and Genetically Modified Mouse Models at Columbia University. This work was also supported by grants from the Japan Society for the Promotion of Science KAKENHI [Grants-in-Aid for Scientific Research (B) #19H03708, #21H02974, (C) #18K07439, (Challenging Exploratory Research) #21K19398] to T.T. This work was partly supported by the Uehara Memorial Foundation, Naito Foundation, Princes Takamatsu Cancer Research Fund, Takeda Science Foundation, Yamaguchi Endocrine Research Foundation, Novartis Foundation (Japan) for Promotion of Science, Kose Cosmetology Research Foundation, and Medical Institute of Bioregulation Kyushu University Cooperative Research Project Program to T.T.
Author contributions
R.M.d.Q. conceived the idea for the project. R.M.d.Q., A.G., G.E., A.K.R. and C.P. designed the experiments. R.M.d.Q., A.G., G.E., N.H. and Y.K. conducted the experiments. R.M.d.Q., G.E., A.G. and T.T., analyzed the results. T.T. deposited proteomics database. R.M.d.Q. wrote the paper with the support of C.P., A.G., G.E., N.H., T.T.; and C.P. contributed to manuscript preparation. All authors reviewed the results and approved the final version of the manuscript.
Peer review
Peer review information
Nature Communications thanks Adam Byron, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the jPOST partner repository (http://jpostdb.org) with the dataset identifier PXD033789. Files containing full analysis with fold changes and statistical parameters can be found in the supplemental material (Supplementary Data 1). Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51488-2.
References
- 1.Fakharzadeh, S. S., Trusko, S. P. & George, D. L. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J.10, 1565–1569 (1991). 10.1002/j.1460-2075.1991.tb07676.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karni-Schmidt, O., Lokshin, M. & Prives, C. The roles of MDM2 and MDMX in cancer. Annu. Rev. Pathol.11, 617–644 (2016). 10.1146/annurev-pathol-012414-040349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwakuma, T. & Lozano, G. MDM2, an introduction. Mol. Cancer Res.1, 993–1000 (2003). [PubMed] [Google Scholar]
- 4.Manfredi, J. J. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev.24, 1580–1589 (2010). 10.1101/gad.1941710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tackmann, N. R. & Zhang, Y. Mouse modelling of the MDM2/MDMX-p53 signalling axis. J. Mol. Cell Biol.9, 34–44 (2017). 10.1093/jmcb/mjx006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein, A. M., de Queiroz, R. M., Venkatesh, D. & Prives, C. The roles and regulation of MDM2 and MDMX: it is not just about p53. Genes Dev.35, 575–601 (2021). 10.1101/gad.347872.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donehower, L. A. & Lozano, G. 20 years studying p53 functions in genetically engineered mice. Nat. Rev. Cancer9, 831–841 (2009). 10.1038/nrc2731 [DOI] [PubMed] [Google Scholar]
- 8.Bagci, O. & Kurtgoz, S. Amplification of cellular oncogenes in solid tumors. N. Am. J. Med. Sci.7, 341–346 (2015). 10.4103/1947-2714.163641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berberich, S. J. Mdm2 and MdmX involvement in human cancer. Subcell. Biochem.85, 263–280 (2014). 10.1007/978-94-017-9211-0_15 [DOI] [PubMed] [Google Scholar]
- 10.Hou, H., Sun, D. & Zhang, X. The role of MDM2 amplification and overexpression in therapeutic resistance of malignant tumors. Cancer Cell Int.19, 216 (2019). 10.1186/s12935-019-0937-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eischen, C. M. Role of Mdm2 and Mdmx in DNA repair. J. Mol. Cell Biol.9, 69–73 (2017). 10.1093/jmcb/mjw052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wienken, M., Moll, U. M. & Dobbelstein, M. Mdm2 as a chromatin modifier. J. Mol. Cell Biol.9, 74–80 (2017). 10.1093/jmcb/mjw046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feeley, K. P., Adams, C. M., Mitra, R. & Eischen, C. M. Mdm2 is required for survival and growth of p53-deficient cancer cells. Cancer Res.77, 3823–3833 (2017). 10.1158/0008-5472.CAN-17-0809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein, A. M. et al. MDM2, MDMX, and p73 regulate cell-cycle progression in the absence of wild-type p53. Proc. Natl Acad. Sci. USA118, e2102420118 (2021). [DOI] [PMC free article] [PubMed]
- 15.Miao, Z., Liu, S., Xiao, X. & Li, D. LINC00342 regulates cell proliferation, apoptosis, migration and invasion in colon adenocarcinoma via miR-545-5p/MDM2 axis. Gene743, 144604 (2020). 10.1016/j.gene.2020.144604 [DOI] [PubMed] [Google Scholar]
- 16.Sha, M. X., Huang, X. W. & Yin, Q. MiR-548b-3p inhibits proliferation and migration of breast cancer cells by targeting MDM2. Eur. Rev. Med. Pharmacol. Sci.24, 3105–3112 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Hauck, P. M. et al. Early-stage metastasis requires Mdm2 and not p53 gain of function. Mol. Cancer Res.15, 1598–1607 (2017). 10.1158/1541-7786.MCR-17-0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, W. et al. A novel inhibitor of MDM2 oncogene blocks metastasis of hepatocellular carcinoma and overcomes chemoresistance. Genes Dis.6, 419–430 (2019). 10.1016/j.gendis.2019.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jana, A. et al. NFkB is essential for activin-induced colorectal cancer migration via upregulation of PI3K-MDM2 pathway. Oncotarget8, 37377–37393 (2017). 10.18632/oncotarget.16343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, D. et al. MiR-758-3p suppresses proliferation, migration and invasion of hepatocellular carcinoma cells via targeting MDM2 and mTOR. Biomed. Pharmacother.96, 535–544 (2017). 10.1016/j.biopha.2017.10.004 [DOI] [PubMed] [Google Scholar]
- 21.Qiu, W., Xia, X., Qiu, Z., Guo, M. & Yang, Z. RasGRP3 controls cell proliferation and migration in papillary thyroid cancer by regulating the Akt-MDM2 pathway. Gene633, 35–41 (2017). 10.1016/j.gene.2017.08.024 [DOI] [PubMed] [Google Scholar]
- 22.Gao, C. et al. Context-dependent roles of MDMX (MDM4) and MDM2 in breast cancer proliferation and circulating tumor cells. Breast Cancer Res.21, 5 (2019). 10.1186/s13058-018-1094-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen, Y. et al. MDM2 promotes epithelial-mesenchymal transition and metastasis of ovarian cancer SKOV3 cells. Br. J. Cancer117, 1192–1201 (2017). 10.1038/bjc.2017.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradbury, R., Jiang, W. G. & Cui, Y. X. MDM2 and PSMA play inhibitory roles in metastatic breast cancer cells through regulation of matrix metalloproteinases. Anticancer Res.36, 1143–1151 (2016). [PubMed] [Google Scholar]
- 25.Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics14, 128 (2013). 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan, Y., Peng, Y., Ou, Y. & Jiang, Y. MicroRNA-610 is downregulated in glioma cells, and inhibits proliferation and motility by directly targeting MDM2. Mol. Med. Rep.14, 2657–2664 (2016). 10.3892/mmr.2016.5559 [DOI] [PubMed] [Google Scholar]
- 27.Wu, D. et al. MicroRNA-379-5p plays a tumor-suppressive role in human bladder cancer growth and metastasis by directly targeting MDM2. Oncol. Rep.37, 3502–3508 (2017). 10.3892/or.2017.5607 [DOI] [PubMed] [Google Scholar]
- 28.Liu, L. et al. CP31398 attenuates endometrial cancer cell invasion, metastasis and resistance to apoptosis by downregulating MDM2 expression. Int. J. Oncol.54, 942–954 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polanski, R. et al. MDM2 interacts with NME2 (non-metastatic cells 2, protein) and suppresses the ability of NME2 to negatively regulate cell motility. Carcinogenesis32, 1133–1142 (2011). 10.1093/carcin/bgr070 [DOI] [PubMed] [Google Scholar]
- 30.Herman, A. G. et al. Discovery of Mdm2-MdmX E3 ligase inhibitors using a cell-based ubiquitination assay. Cancer Discov.1, 312–325 (2011). 10.1158/2159-8290.CD-11-0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiang, P., Shao, Y., Sun, Y. P., Zhang, J. & Chen, L. J. Metformin inhibits proliferation and migration of endometrial cancer cells through regulating PI3K/AKT/MDM2 pathway. Eur. Rev. Med. Pharmacol. Sci.23, 1778–1785 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Yang, J. Y. et al. MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. Mol. Cell Biol.26, 7269–7282 (2006). 10.1128/MCB.00172-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galarza, S., Kim, H., Atay, N., Peyton, S. R. & Munson, J. M. 2D or 3D? How cell motility measurements are conserved across dimensions in vitro and translate in vivo. Bioeng. Transl. Med.5, e10148 (2020). 10.1002/btm2.10148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baskaran, J. P. et al. Cell shape, and not 2D migration, predicts extracellular matrix-driven 3D cell invasion in breast cancer. APL Bioeng.4, 026105 (2020). 10.1063/1.5143779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guzman, A., Ziperstein, M. J. & Kaufman, L. J. The effect of fibrillar matrix architecture on tumor cell invasion of physically challenging environments. Biomaterials35, 6954–6963 (2014). 10.1016/j.biomaterials.2014.04.086 [DOI] [PubMed] [Google Scholar]
- 36.Poincloux, R., Lizarraga, F. & Chavrier, P. Matrix invasion by tumour cells: a focus on MT1-MMP trafficking to invadopodia. J. Cell Sci. 122, 3015–3024 (2009). 10.1242/jcs.034561 [DOI] [PubMed] [Google Scholar]
- 37.Janiszewska, M., Primi, M. C. & Izard, T. Cell adhesion in cancer: beyond the migration of single cells. J. Biol. Chem.295, 2495–2505 (2020). 10.1074/jbc.REV119.007759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rikitake, Y. & Takai, Y. Directional cell migration regulation by small G proteins, nectin-like molecule-5, and afadin. Int. Rev. Cell Mol. Biol.287, 97–143 (2011). 10.1016/B978-0-12-386043-9.00003-7 [DOI] [PubMed] [Google Scholar]
- 39.Burridge, K. & Guilluy, C. Focal adhesions, stress fibers and mechanical tension. Exp. Cell Res.343, 14–20 (2016). 10.1016/j.yexcr.2015.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawazoe, T. & Taniguchi, K. The Sprouty/Spred family as tumor suppressors: coming of age. Cancer Sci.110, 1525–1535 (2019). 10.1111/cas.13999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masoumi-Moghaddam, S., Amini, A. & Morris, D. L. The developing story of Sprouty and cancer. Cancer Metastasis Rev.33, 695–720 (2014). 10.1007/s10555-014-9497-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Celik-Selvi, B. E. et al. Sprouty3 and Sprouty4, two members of a family known to inhibit FGF-mediated signaling, exert opposing roles on proliferation and migration of glioblastoma-derived cells. Cells8, 808 (2019). [DOI] [PMC free article] [PubMed]
- 43.Gong, Y. et al. Sprouty4 regulates endothelial cell migration via modulating integrin beta3 stability through c-Src. Angiogenesis16, 861–875 (2013). 10.1007/s10456-013-9361-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tennis, M. A. et al. Sprouty-4 inhibits transformed cell growth, migration and invasion, and epithelial-mesenchymal transition, and is regulated by Wnt7A through PPARgamma in non-small cell lung cancer. Mol. Cancer Res.8, 833–843 (2010). 10.1158/1541-7786.MCR-09-0400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaggi, F., Cabrita, M. A., Perl, A. K. & Christofori, G. Modulation of endocrine pancreas development but not beta-cell carcinogenesis by Sprouty4. Mol. Cancer Res.6, 468–482 (2008). 10.1158/1541-7786.MCR-07-0255 [DOI] [PubMed] [Google Scholar]
- 46.Benassi, B. et al. c-Myc phosphorylation is required for cellular response to oxidative stress. Mol. Cell21, 509–519 (2006). 10.1016/j.molcel.2006.01.009 [DOI] [PubMed] [Google Scholar]
- 47.Miyoshi, K. et al. The Sprouty-related protein, Spred, inhibits cell motility, metastasis, and Rho-mediated actin reorganization. Oncogene23, 5567–5576 (2004). 10.1038/sj.onc.1207759 [DOI] [PubMed] [Google Scholar]
- 48.Tuntithavornwat, S. et al. Giant obscurin regulates migration and metastasis via RhoA-dependent cytoskeletal remodeling in pancreatic cancer. Cancer Lett.526, 155–167 (2022). 10.1016/j.canlet.2021.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sumi, T., Matsumoto, K., Takai, Y. & Nakamura, T. Cofilin phosphorylation and actin cytoskeletal dynamics regulated by rho- and Cdc42-activated LIM-kinase 2. J. Cell Biol.147, 1519–1532 (1999). 10.1083/jcb.147.7.1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, W., Halasz, E. & Townes-Anderson, E. Actin dynamics, regulated by RhoA-LIMK-Cofilin signaling, mediates rod photoreceptor axonal retraction after retinal injury. Invest. Ophthalmol. Vis. Sci.60, 2274–2285 (2019). 10.1167/iovs.18-26077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsumura, Y., Toshima, J., Leeksma, O. C., Ohashi, K. & Mizuno, K. Sprouty-4 negatively regulates cell spreading by inhibiting the kinase activity of testicular protein kinase. Biochem. J.387, 627–637 (2005). 10.1042/BJ20041181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tribollet, V. et al. ERRalpha coordinates actin and focal adhesion dynamics. Cancer Gene Ther. 29, 1429–1438 (2022). [DOI] [PubMed]
- 53.Wang, W. et al. Identification of a new class of MDM2 inhibitor that inhibits growth of orthotopic pancreatic tumors in mice. Gastroenterology147, 893–902.e892 (2014). 10.1053/j.gastro.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qin, J. J. et al. Inulanolide A as a new dual inhibitor of NFAT1-MDM2 pathway for breast cancer therapy. Oncotarget7, 32566–32578 (2016). 10.18632/oncotarget.8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu, B., Gilkes, D. M., Farooqi, B., Sebti, S. M. & Chen, J. MDMX overexpression prevents p53 activation by the MDM2 inhibitor Nutlin. J. Biol. Chem.281, 33030–33035 (2006). 10.1074/jbc.C600147200 [DOI] [PubMed] [Google Scholar]
- 56.Venkatesh, D. et al. MDM2 and MDMX promote ferroptosis by PPARalpha-mediated lipid remodeling. Genes Dev.34, 526–543 (2020). 10.1101/gad.334219.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riscal, R. et al. Chromatin-bound MDM2 regulates serine metabolism and redox homeostasis independently of p53. Mol. Cell62, 890–902 (2016). 10.1016/j.molcel.2016.04.033 [DOI] [PubMed] [Google Scholar]
- 58.Zhang, P. et al. CPNE8 promotes gastric cancer metastasis by modulating focal adhesion pathway and tumor microenvironment. Int. J. Biol. Sci.18, 4932–4949 (2022). 10.7150/ijbs.76425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun, T. et al. G9a promotes invasion and metastasis of non-small cell lung cancer through enhancing focal adhesion kinase activation via NF-kappaB signaling pathway. Mol. Cancer Res.19, 429–440 (2021). 10.1158/1541-7786.MCR-20-0557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong, Y. et al. MYH9-dependent polarization of ATG9B promotes colorectal cancer metastasis by accelerating focal adhesion assembly. Cell Death Differ28, 3251–3269 (2021). 10.1038/s41418-021-00813-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen, J. et al. Hippo component YAP promotes focal adhesion and tumour aggressiveness via transcriptionally activating THBS1/FAK signalling in breast cancer. J. Exp. Clin. Cancer Res.37, 175 (2018). 10.1186/s13046-018-0850-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neumeier, J. & Meister, G. siRNA specificity: RNAi mechanisms and strategies to reduce off-target effects. Front. Plant Sci.11, 526455 (2020). 10.3389/fpls.2020.526455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jackson, A. L. & Linsley, P. S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov.9, 57–67 (2010). 10.1038/nrd3010 [DOI] [PubMed] [Google Scholar]
- 64.Haigl, B., Mayer, C. E., Siegwart, G. & Sutterluty, H. Sprouty4 levels are increased under hypoxic conditions by enhanced mRNA stability and transcription. Biol. Chem.391, 813–821 (2010). 10.1515/bc.2010.082 [DOI] [PubMed] [Google Scholar]
- 65.Vanas, V., Muhlbacher, E., Kral, R. & Sutterluty-Fall, H. Sprouty4 interferes with cell proliferation and migration of breast cancer-derived cell lines. Tumour Biol.35, 4447–4456 (2014). 10.1007/s13277-013-1587-0 [DOI] [PubMed] [Google Scholar]
- 66.Wang, J., Thompson, B., Ren, C., Ittmann, M. & Kwabi-Addo, B. Sprouty4, a suppressor of tumor cell motility, is down regulated by DNA methylation in human prostate cancer. Prostate66, 613–624 (2006). 10.1002/pros.20353 [DOI] [PubMed] [Google Scholar]
- 67.Guo, J. et al. SPRY4 suppresses proliferation and induces apoptosis of colorectal cancer cells by repressing oncogene EZH2. Aging13, 11665–11677 (2021). 10.18632/aging.202859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, E. et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res.45, 3086–3101 (2017). 10.1093/nar/gkw1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rathmanner, N. et al. Sprouty2 but not Sprouty4 is a potent inhibitor of cell proliferation and migration of osteosarcoma cells. FEBS Lett.587, 2597–2605 (2013). 10.1016/j.febslet.2013.06.040 [DOI] [PubMed] [Google Scholar]
- 70.Stutz, A., Kamptner, A. Z. M. & Sutterluty, H. A Sprouty4 mutation identified in Kallmann syndrome increases the inhibitory potency of the protein towards FGF and connected processes. Int. J. Mol. Sci. 22, 2145 (2021). [DOI] [PMC free article] [PubMed]
- 71.Jing, H. et al. Suppression of Spry4 enhances cancer stem cell properties of human MDA-MB-231 breast carcinoma cells. Cancer Cell Int.16, 19 (2016). 10.1186/s12935-016-0292-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qiu, B. et al. Sprouty4 correlates with favorable prognosis in perihilar cholangiocarcinoma by blocking the FGFR-ERK signaling pathway and arresting the cell cycle. EBioMedicine50, 166–177 (2019). 10.1016/j.ebiom.2019.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.So, W. K. et al. Sprouty4 mediates amphiregulin-induced down-regulation of E-cadherin and cell invasion in human ovarian cancer cells. Tumour Biol.37, 9197–9207 (2016). 10.1007/s13277-016-4790-y [DOI] [PubMed] [Google Scholar]
- 74.Pan, Y. et al. LINC00675 suppresses cell proliferation and migration via downregulating the H3K4me2 level at the SPRY4 promoter in gastric cancer. Mol. Ther. Nucleic Acids22, 766–778 (2020). 10.1016/j.omtn.2020.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Das, M. K., Furu, K., Evensen, H. F., Haugen, O. P. & Haugen, T. B. Knockdown of SPRY4 and SPRY4-IT1 inhibits cell growth and phosphorylation of Akt in human testicular germ cell tumours. Sci. Rep.8, 2462 (2018). 10.1038/s41598-018-20846-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brock, E. J. et al. Sprouty4 negatively regulates ERK/MAPK signaling and the transition from in situ to invasive breast ductal carcinoma. PLoS ONE16, e0252314 (2021). 10.1371/journal.pone.0252314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sasaki, A. et al. Mammalian Sprouty4 suppresses Ras-independent ERK activation by binding to Raf1. Nat. Cell Biol.5, 427–432 (2003). 10.1038/ncb978 [DOI] [PubMed] [Google Scholar]
- 78.Ayada, T. et al. Sprouty4 negatively regulates protein kinase C activation by inhibiting phosphatidylinositol 4,5-biphosphate hydrolysis. Oncogene28, 1076–1088 (2009). 10.1038/onc.2008.464 [DOI] [PubMed] [Google Scholar]
- 79.Ezratty, E. J., Partridge, M. A. & Gundersen, G. G. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat. Cell Biol.7, 581–590 (2005). 10.1038/ncb1262 [DOI] [PubMed] [Google Scholar]
- 80.Ratnadiwakara, M. & Anko, M. L. mRNA stability assay using transcription inhibition by actinomycin D in mouse pluripotent stem cells. Bio Protoc.8, e3072 (2018). 10.21769/BioProtoc.3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Radha, K. S. et al. Iron-mediated stability of PAI-1 mRNA in adenocarcinoma cells-involvement of a mRNA-binding nuclear protein. Thromb. Res.116, 255–263 (2005). 10.1016/j.thromres.2004.12.006 [DOI] [PubMed] [Google Scholar]
- 82.Kawashima, Y. et al. Proteogenomic analyses of cellular lysates using a phenol-guanidinium thiocyanate reagent. J. Proteome Res.18, 301–308 (2019). [DOI] [PubMed] [Google Scholar]
- 83.Ochiiwa, H. et al. TAS4464, a NEDD8-activating enzyme inhibitor, activates both intrinsic and extrinsic apoptotic pathways via c-Myc-mediated regulation in acute myeloid leukemia. Oncogene40, 1217–1230 (2021). 10.1038/s41388-020-01586-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hughes, C. S. et al. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol.10, 757 (2014). 10.15252/msb.20145625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hughes, C. S. et al. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc.14, 68–85 (2019). 10.1038/s41596-018-0082-x [DOI] [PubMed] [Google Scholar]
- 86.Kawashima, Y., Ishikawa, M., Konno, R., Nakajima, D. & Ohara, O. Development of a simple and stable NanoESI spray system using suction wind from the MS inlet. J. Proteome Res.22, 1564–1569 (2023). 10.1021/acs.jproteome.3c00146 [DOI] [PubMed] [Google Scholar]
- 87.Kawashima, Y. et al. Single-shot 10K proteome approach: over 10,000 protein identifications by data-independent acquisition-based single-shot proteomics with ion mobility spectrometry. J. Proteome Res.21, 1418–1427 (2022). 10.1021/acs.jproteome.2c00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Amodei, D. et al. Improving precursor selectivity in data-independent acquisition using overlapping windows. J. Am. Soc. Mass Spectrom.30, 669–684 (2019). 10.1007/s13361-018-2122-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gessulat, S. et al. Prosit: proteome-wide prediction of peptide tandem mass spectra by deep learning. Nat. Methods16, 509–518 (2019). 10.1038/s41592-019-0426-7 [DOI] [PubMed] [Google Scholar]
- 90.Searle, B. C. et al. Generating high quality libraries for DIA MS with empirically corrected peptide predictions. Nat. Commun11, 1548 (2020). 10.1038/s41467-020-15346-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Searle, B. C. et al. Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nat. Commun9, 5128 (2018). 10.1038/s41467-018-07454-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goedhart, J. & Luijsterburg, M. S. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci. Rep.10, 20560 (2020). 10.1038/s41598-020-76603-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Babicki, S. et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res.44, W147–W153 (2016). 10.1093/nar/gkw419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xie, Z. et al. Gene set knowledge discovery with Enrichr. Curr. Protoc.1, e90 (2021). 10.1002/cpz1.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res.44, W90–W97 (2016). 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tang, Z. et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res.45, W98–W102 (2017). 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ivascu, A. & Kubbies, M. Rapid generation of single-tumor spheroids for high-throughput cell function and toxicity analysis. J. Biomol. Screen.11, 922–932 (2006). 10.1177/1087057106292763 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the jPOST partner repository (http://jpostdb.org) with the dataset identifier PXD033789. Files containing full analysis with fold changes and statistical parameters can be found in the supplemental material (Supplementary Data 1). Source data are provided with this paper.