Abstract
Vespula vulgaris (V. vulgaris), commonly known as the common wasp, poses a significant health threat due to its venom-induced allergic reactions. This research focused on the exploration of bioactive compounds from Bryophyllum pinnatum as potential inhibitors for V. vulgaris allergen proteins, including Phospholipase A1 (Ves V1), Hyaluronoglucosaminidase (Ves V2), and Antigen 5 (Ves V5). Through a multidisciplinary approach involving literature reviews, molecular docking analyses, ADMET assessments and Molecular Dynamics Simulations (MDS) of 100ns we identified two promising drug candidates from four bioactive compounds- Bryophyllin A, Bryophyllin B, Bryotoxin A, and Bryotoxin B of Bryophyllum pinnatum. Molecular docking results revealed strong binding interactions, with Bryophyllin B and Bryotoxin A consistently exhibiting the highest affinity (−9.6 kcal/mol and −10.0 kcal/mol) across the allergen proteins. ADMET analyses highlighted Bryophyllin B as a favorable candidate, showing high absorption (HIA: 92.1 %), minimal metabolic interactions (CYP1A2: No), and a low toxicity profile (LD50 (rat): 2.431). MDS analysis revealed Bryophyllin B and Bryotoxin A as promising drug inhibitors, exhibiting the highest binding stability with the allergen proteins of V. vulgaris, as indicated by the lowest Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), Radius of Gyration (RG) values and highest protein-ligand contacts. Our study provides valuable insights into the therapeutic potential of Bryophyllum pinnatum compounds as inhibitors for V. vulgaris allergen proteins having two promising candidates- Bryophyllin B and Bryotoxin A.
1. Introduction
Vespula vulgaris (V. vulgaris), commonly known as the common wasp, poses a significant health threat due to its venom-induced allergic reactions in humans. The venom of V. vulgaris contains a complex mixture of bioactive compounds, including allergen proteins such as Phospholipase A1 (Ves V1), Hyaluronoglucosaminidase (Ves V2), and Antigen 5 (Ves V5) [1,2], which are known to trigger allergic responses in susceptible individuals [[3], [4], [5], [6]]. These allergen proteins elicit a cascade of immunological reactions upon exposure, leading to various adverse effects ranging from localized swelling and itching to potentially life-threatening systemic reactions like anaphylactic shock [1,7,8].
The mechanism of allergic sensitization to wasp venom involves the recognition of these allergens by the immune system, leading to the production of specific immunoglobulin E (IgE) antibodies and subsequent activation of mast cells and basophils, resulting in the release of inflammatory mediators such as histamine and leukotrienes [[9], [10], [11], [12]]. As the incidence of wasp venom allergies continues to rise globally, there is an urgent need to explore innovative therapeutic strategies to mitigate allergic reactions induced by V. vulgaris venom [13]. Traditional treatment options for wasp venom allergies include allergen-specific immunotherapy (desensitization) and symptomatic management with antihistamines, corticosteroids, and epinephrine for severe reactions [10,[14], [15], [16],16]. However, these approaches have limitations, including the need for long-term treatment and potential adverse effects [[17], [18], [19], [20]].
In this context, the exploration of natural compounds with potential inhibitory effects on V. vulgaris allergen proteins presents a promising avenue for novel therapeutic interventions. Bryophyllum pinnatum, a succulent plant with a rich history in traditional medicine, has emerged as a source of bioactive compounds with pharmacological properties, including anti-inflammatory and immunomodulatory effects. Through a comprehensive literature review, four bioactive compounds- Bryophyllin A, Bryophyllin B, Bryotoxin A, and Bryotoxin B - have been identified within Bryophyllum pinnatum, showing promising inhibitory effects against different allergen proteins.
This study aimed to delve into the molecular interactions between the identified bioactive compounds and V. vulgaris allergen proteins. Through molecular docking analyses and structural examinations, we sought to elucidate the binding mechanisms and potential inhibitory effects of Bryophyllum pinnatum compounds on the allergen proteins. The insights gained from this exploration could pave the way for the development of novel therapeutics to alleviate the allergic burden imposed by V. vulgaris venom. As we embarked on this investigation, it is important to underscore the urgency of finding effective interventions for wasp venom allergies and the potential of natural compounds from Bryophyllum pinnatum as promising candidates.
2. Materials and methods
2.1. Data retrieval
The allergen proteins of V. vulgaris were identified through a comprehensive literature review. Subsequently, their three-dimensional structures were obtained from the RCSB PDB server [21]. The active compounds from Bryophyllum pinnatum were selected based on literature highlighting their potential as inhibitors, particularly in the context of COVID-19 cytokine storm therapy [[22], [23], [24]]. The chemical structures of these compounds were retrieved in sdf format from the PubChem server (https://pubchem.ncbi.nlm.nih.gov/). This approach ensured a thorough and well-informed selection of both the target proteins and the active compounds, laying the foundation for subsequent molecular docking analyses to explore potential inhibitory interactions between Bryophyllum pinnatum compounds and V. vulgaris allergen proteins.
2.2. Molecular docking analysis
The allergen proteins of V. vulgaris (1QNX - Ves V5, 2ATM - Ves V2, and 4QNN - Ves V1) were prepared using the Discovery Studio tool, involving tasks such as energy minimization, removal of water molecules, and assignment of appropriate charges. Additionally, drug binding cavities of the allergen proteins were identified utilizing the CBDOCK2 online server. For the active compounds from Bryophyllum pinnatum (Bryophyllin A, Bryophyllin B, Bryotoxin A, and Bryotoxin B), similar preparations were performed using the Open Babel tool, involving energy minimization and structural refinement. The molecular docking analysis was executed using PyRx software, wherein the prepared compounds and allergen proteins were docked to predict their binding interactions [25]. The site-specific docking was performed, using the coordinates and dimensions of the cavities detected by the CBDOCK2 online server [26]. The molecular docking results were subsequently analyzed to assess the binding affinities and key interactions between the active compounds and the allergen proteins, providing valuable insights into the potential inhibitory effects of Bryophyllum pinnatum compounds on V. vulgaris allergens.
2.3. Molecular properties and ADMET analysis of the bioactive compounds
ADMET properties refer to the pharmacokinetic characteristics of a drug candidate, encompassing Absorption, Distribution, Metabolism, Excretion and Toxicity. These properties play a crucial role in determining the bioavailability, efficacy, and safety of a drug [[27], [28], [29], [30]]. The molecular properties and ADMET analysis of the active compounds from Bryophyllum pinnatum, three online servers-pkCSM (Predicting the Activity and Specificity of Kinases using Molecular Signatures), SwissADME and Protox-II were employed. For molecular properties, the pkCSM and SwissADME servers were utilized to predict key physicochemical characteristics such as molecular weight, LogP, rotatable bonds, hydrogen bond acceptors and donors, surface area, Lipinski Rule of Five violations and ADME properties [[31], [32], [33], [34]]. pkCSM provided insights into absorption (Caco-2 permeability and human intestinal absorption) [35], distribution (blood-brain barrier and central nervous system permeability), metabolism (CYP1A2 inhibition) [36], and excretion (total clearance). On the other hand, Protox-II contributed toxicity predictions including hepatotoxicity, mutagenicity, cytoxicity, carcinogenicity, and immunotoxicity [37,38]. The combined utilization of these servers enabled a comprehensive evaluation of the molecular properties and pharmacokinetic parameters of the compounds, offering valuable information for further assessment of their drug-likeness and safety profiles.
2.4. Molecular dynamics (MD) simulation of the protein-ligand complexes
Molecular dynamics (MD) simulations are commonly utilized to assess the stability of candidate drug compounds bound to target proteins [[39], [40], [41]]. In this study, we conducted MD simulations lasting 100 ns using the Desmond software package by Schrödinger LLC. Prior to simulation, preprocessing steps were applied to the protein-ligand complexes, including optimization and minimization, using either the Protein Preparation Wizard in Maestro software or the System Builder tool. The simulations were set up in an orthorhombic simulation box with TIP3P solvent model to mimic realistic environmental conditions [42]. OPLS_2005 force field was employed, and counter ions were added as necessary to neutralize the system [41]. To mimic physiological conditions, a 0.15 M salt solution (NaCl) was introduced. Throughout the simulation, equilibrium was maintained using NVT and NPT ensembles, with temperature and pressure held constant at 300 K and 1 atm, respectively, to conserve moles (N), pressure (P), and temperature (T). Pre-simulation relaxation procedures were conducted, and parameters such as radius of gyration (RG), solvent-accessible surface area (SASA), root mean square deviation (RMSD) [43], root mean square fluctuations (RMSF), and torsion angles [44] were computed to evaluate simulation stability for both the control and top four selected complexes.
3. Results and discussion
3.1. Data retrieval
Three allergen proteins from V. vulgaris were identified through a literature review: Phospholipase A1 (Ves V1), Hyaluronoglucosaminidase (Ves V2), and Antigen 5 (Ves V5). The 3D structures of these proteins were obtained from the RCSB PDB server, with PDB IDs 4QNN, 2ATM, and 1QNX, respectively. Specifically, 4QNN (Ves V1) consists of four chains (chain A, B, C, and D) and has a length of 300 amino acids. 2ATM (Ves V2) comprises a single chain (chain A) with a length of 331 amino acids, while 1QNX (Ves V5) consists of a single chain (chain A) and comprises 209 amino acid residues.
In addition, we identified four bioactive compounds within Bryophyllum pinnatum from literature review. They are Bryophyllin A, Bryophyllin B, Bryotoxin A, and Bryotoxin B [23]. The molecular structures of these compounds were retrieved from the PubChem server [45,46], with PubChem IDs 5488801, 44575928, 441848, and 5489391, respectively (Table 01).
Table 01.
Four bioactive compounds from Bryophyllum pinnatum.
| Compounds | Pubchem ID | Formula | Canonical Smile |
|---|---|---|---|
| Bryophyllin A | 5488801 | C26H32O8 | CC12CC(C3C(C1(CCC2C4 COC(=O)C C4)O)CCC56C3(C7CC(C5)OC(O7)(O6)C)C O)O |
| Bryophyllin B | 44575928 | C26H34O9 | CC(=O)OC1CC(CC2(C13C4C(CC2)C5(CCC(C5(CC4OC3O)C)C6 COC(=O)C C6)O)O)O |
| Bryotoxin A | 441848 | C32H42O12 | CC1CC(C(O1)OC2CCC3(C4C(CCC3(C2)O)C5(CCC(C5(C(=O)C4O)C)C6 COC(=O)C C6)O)C O)O)OC(=O)C |
| Bryotoxin B | 5489391 | C26H32O9 | CC12C(CCC1(C3CCC45CC6CC(C4(C3C(C2 O)O)CO)OC(O6)(O5)C)O)C7 COC(=O)C C7 |
3.2. Molecular docking analysis
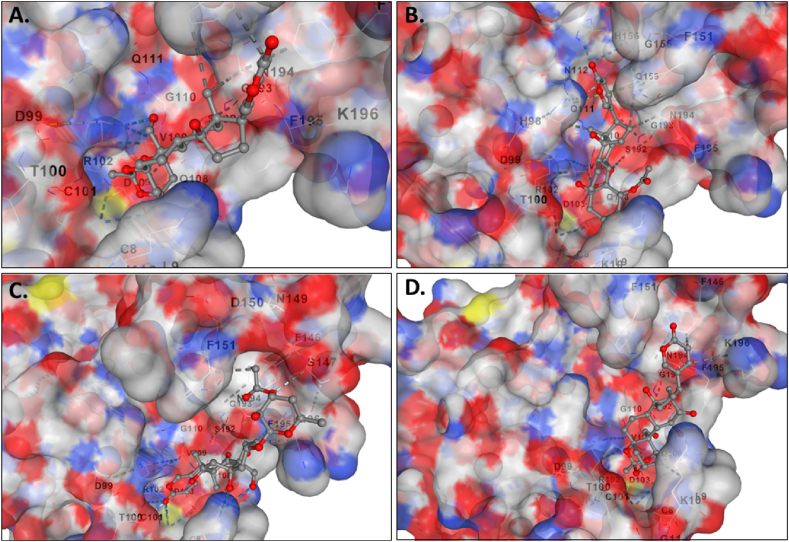
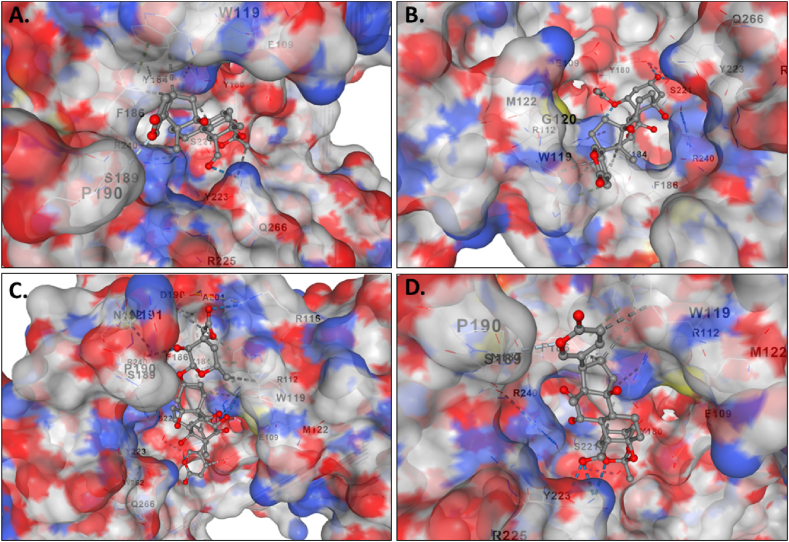
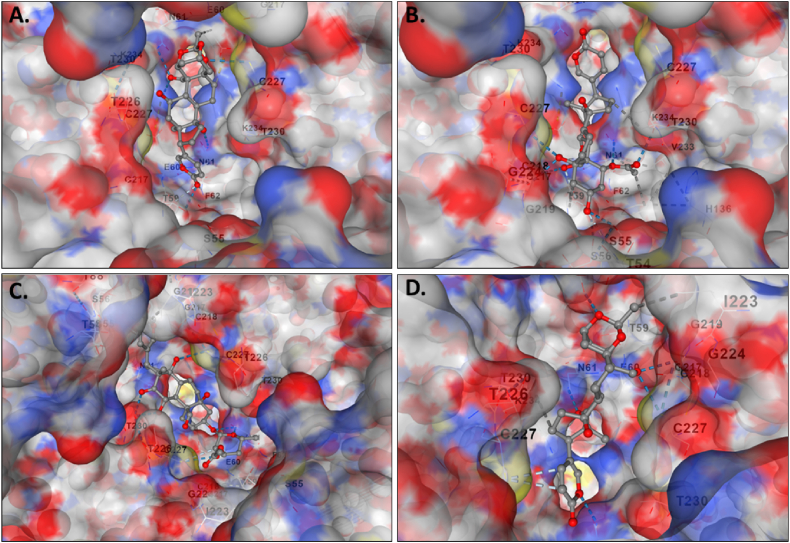
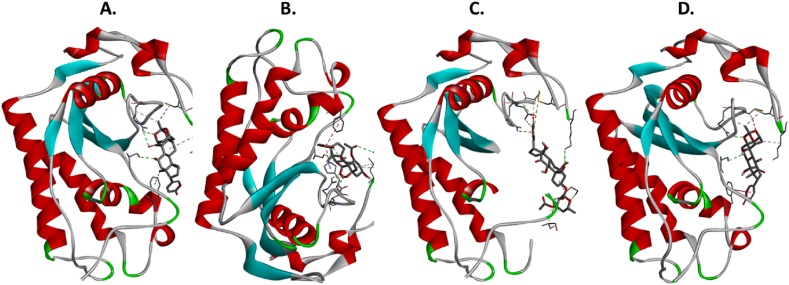
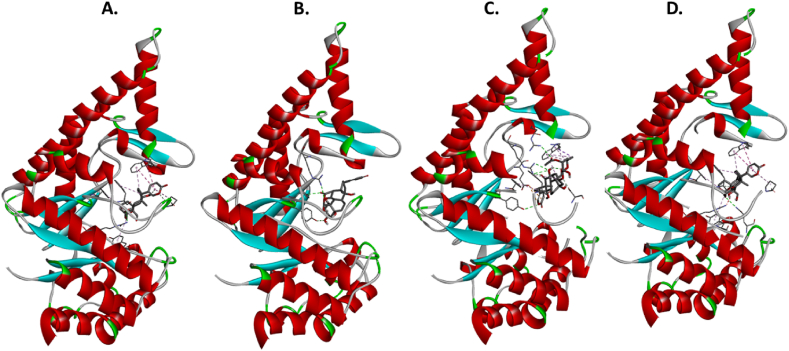
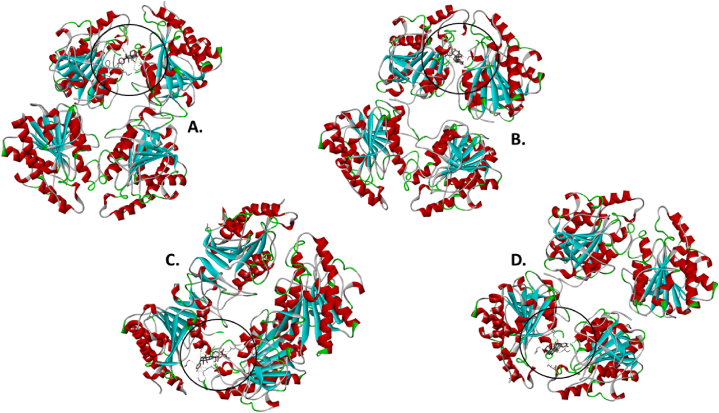
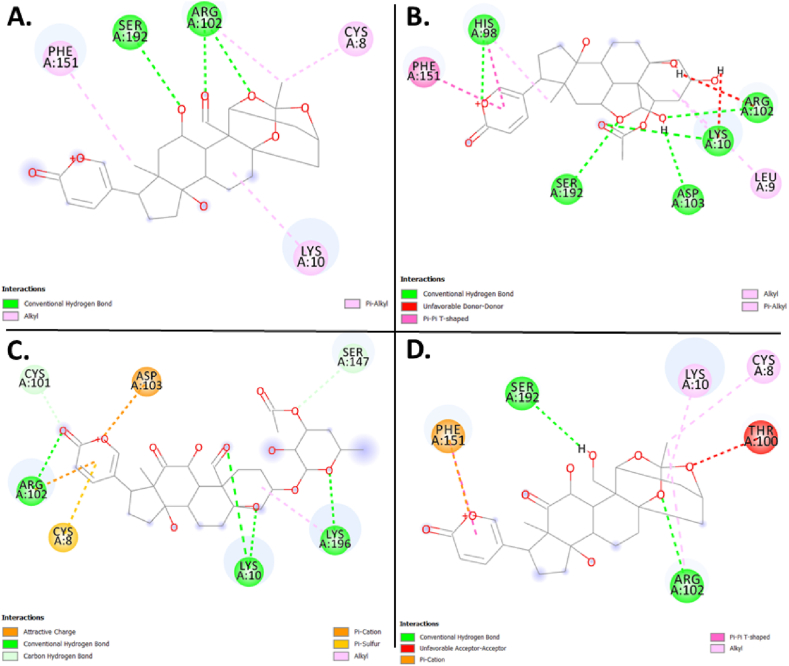
The molecular docking results for the active compounds (Bryophyllin A, Bryophyllin B, Bryotoxin A, and Bryotoxin B) of Bryophyllum pinnatum with the allergic proteins of V. vulgaris demonstrated robust binding interactions [47]. Across proteins 1QNX (Ves V5), 2ATM (Ves V2), and 4QNN (Ves V1), all compounds consistently exhibited low docking scores, indicating strong binding affinities (Table 02). For 1QNX, Bryophyllin B displayed the highest affinity with a score of −9.6 kcal/mol, followed closely by Bryotoxin B with a score of −8.3 kcal/mol. In 2ATM, Bryotoxin A exhibited the highest affinity with a score of −9.1 kcal/mol. In 4QNN, Bryotoxin A again demonstrated the strongest binding with an impressive score of −10.0 kcal/mol. The identified binding pockets (Table S1), such as C2 in 1QNX, C1 in 2ATM, and C1 in 4QNN, were consistent among different proteins, indicating potential shared interaction sites (Fig. 1(A–D), Fig. 2(A–D) and Fig. 3 (A–D)). Notably, Bryotoxin A consistently stood out as the compound with the highest binding fitness across all three proteins, suggesting its strong potential for interacting with the allergic proteins of V. vulgaris. The binding poses of protein-ligand complexes are demonstrated in Fig. 4(A–D), Fig. 5(A–D) and Fig. 6(A–D). The interacted amino acid residues (Table 03) and bond types are depicted in Fig. 7(A–D), Fig. 8(A–D) and Fig. 9 (A–D).
Table 02.
Molecular Docking analysis.
| Protein | Compound | Docking Score | CurPocket ID | Cavity Volume(Å3) | Center (x, y, z) | Docking size (x, y, z) |
|---|---|---|---|---|---|---|
| 1QNX (Ves V5) | Bryophyllin A | −8.4 | C2 | 187 | 21, 31, 36 | 23, 23, 23 |
| Bryophyllin B | −9.6 | C2 | 187 | 21, 31, 36 | 23, 23, 23 | |
| Bryotoxin A | −8.3 | C3 | 184 | 15, 25, 31 | 26, 26, 26 | |
| Bryotoxin B | −8.3 | C2 | 187 | 21, 31, 36 | 23, 23, 23 | |
| 2ATM (Ves V2) | Bryophyllin A | −7.7 | C1 | 504 | 19, 29, 8 | 23, 23, 23 |
| Bryophyllin B | −8.2 | C1 | 504 | 19, 29, 8 | 23, 23, 23 | |
| Bryotoxin A | −9.1 | C1 | 504 | 19, 29, 8 | 26, 26, 26 | |
| Bryotoxin B | −8.0 | C1 | 504 | 19, 29, 8 | 23, 23, 23 | |
| 4QNN (Ves V1) | Bryophyllin A | −9.9 | C1 | 9257 | −7, −7, 13 | 35, 33, 35 |
| Bryophyllin B | −9.4 | C1 | 9257 | −7, −7, 13 | 35, 33, 35 | |
| Bryotoxin A | −10.0 | C2 | 3809 | −3, 15, −20 | 32, 26, 26 | |
| Bryotoxin B | −9.0 | C2 | 3809 | −3, 15, −20 | 32, 23, 30 |
Fig. 1.
The binding of compounds to the cavities of the allergen protein Antigen 5 (Ves V5) of Vespula vulgaris. Specifically, A. Bryophyllin A, B. Bryophyllin B, C. Bryotoxin A, and D. Bryotoxin B are shown binding in cavities C2, C2, C3, and C1, respectively.
Fig. 2.
The binding of compounds to the cavities of the allergen protein Hyaluronoglucosaminidase (Ves V2) of Vespula vulgaris. Specifically, A. Bryophyllin A, B. Bryophyllin B, C. Bryotoxin A, and D. Bryotoxin B are all shown binding in cavities C1.
Fig. 3.
The binding of compounds to the cavities of the allergen protein Phospholipase A1 (Ves V1) of Vespula vulgaris. Specifically, A. Bryophyllin A, B. Bryophyllin B, C. Bryotoxin A, and D. Bryotoxin B are shown binding in cavities C1, C1, C2 and C2 respectively.
Fig. 4.
Molecular docking: Binding of the four bioactive compounds- A. Bryophyllin A, B. Bryophyllin B, C. Bryotoxin A, and D. Bryotoxin B from Bryophyllum pinnatum with the allergen protein Antigen 5 (Ves V5) of Vespula vulgaris.
Fig. 5.
Molecular docking: Binding of the four bioactive compounds- A. Bryophyllin A, B. Bryophyllin B, C. Bryotoxin A, and D. Bryotoxin B from Bryophyllum pinnatum with the allergen protein Hyaluronoglucosaminidase (Ves V2) of Vespula vulgaris.
Fig. 6.
Molecular docking: Binding of the four bioactive compounds- A. Bryophyllin A, B. Bryophyllin B, C. Bryotoxin A, and D. Bryotoxin B from Bryophyllum pinnatum with the allergen protein Phospholipase A1 (Ves V1) of Vespula vulgaris.
Table 03.
Interacted amino acid residues of allergen proteins of V. vulgaris with bioactive compounds of Bryophyllum pinnatum.
| Protein | Compound | Contact Residues |
|---|---|---|
| 1QNX (Ves V5) | Bryophyllin A | Chain A: CYS8, LYS10, ARG102, PHE151, SER192 |
| Bryophyllin B | Chain A: LEU9, LYS10, HIS98, ARG102, ASP103, PHE151, SER192 | |
| Bryotoxin A | Chain A: CYS8, LYS10, CYS101, ARG102, ASP103, SER147, LYS196 | |
| Bryotoxin B | Chain A: CYS8, LYS10, THR100, ARG102, PHE151, SER192 | |
| 2ATM (Ves V2) | Bryophyllin A | Chain A: TRP119, TYR184, PHE186, PRO190, TYR223, ARG240 |
| Bryophyllin B | Chain A: ARG112, TYR180, TYR184 | |
| Bryotoxin A | Chain A: GLU109, ARG112, ARG116, TRP119, TYR180, TYR184, PHE186, SER189 | |
| Bryotoxin B | Chain A: TRP119, TYR184, PHE186, PRO190, TYR223, ARG240, GLN266 | |
| 4QNN (Ves V1) | Bryophyllin A |
Chain A: ASN61, LYS234, CYS227 Chain C: GLU60, ASN61, PHE62, CYS227 |
| Bryophyllin B |
Chain A: ASN61, GLY217 Chain C: SER55, ASN61, CYS227, THR230 |
|
| Bryotoxin A |
Chain B: ASN61, CYS218, GLY219, ILE223, CYS227 Chain D: SER55, GLY224, CYS227, THR230 |
|
| Bryotoxin B |
Chain B: ASN61, CYS218, ILE223 Chain D: SER55, ASN61, CYS227 |
Fig. 7.
Molecular docking: Binding interactions (Bond types) of the four bioactive compounds- A. Bryophyllin A, B. Bryophyllin B, C. Bryotoxin A, and D. Bryotoxin B from Bryophyllum pinnatum with the allergen protein Antigen 5 (Ves V5) of Vespula vulgaris.
Fig. 8.
Molecular docking: Binding interactions (Bond types) of the four bioactive compounds- A. Bryophyllin A, B. Bryophyllin B, C. Bryotoxin A, and D. Bryotoxin B from Bryophyllum pinnatum with the allergen protein Hyaluronoglucosaminidase (Ves V2) of Vespula vulgaris.
Fig. 9.
Molecular docking: Binding interactions (Bond types) of the four bioactive compounds- A. Bryophyllin A, B. Bryophyllin B, C. Bryotoxin A, and D. Bryotoxin B from Bryophyllum pinnatum with the allergen protein Phospholipase A1 (Ves V1) of Vespula vulgaris.
3.3. Molecular properties analysis
The molecular properties of the active compounds from Bryophyllum pinnatum, including Bryophyllin A, Bryophyllin B, Bryotoxin A, and Bryotoxin B, were analyzed to gauge their drug-likeness. Bryophyllin A and Bryophyllin B, with molecular weights of 472.534 g/mol and 490.549 g/mol, respectively, demonstrated moderate sizes within the desirable range. Both compounds exhibited favorable logP values around 1, indicative of a balanced hydrophilic-lipophilic profile for efficient drug absorption and distribution. With two rotatable bonds, Bryophyllin A and Bryophyllin B showcased reasonable flexibility. Bryotoxin A, characterized by a higher molecular weight of 618.676 and five rotatable bonds, and Bryotoxin B, with three, indicated a greater degree of molecular flexibility. The number of hydrogen bond acceptors (HBA) and donors (HBD) varied, with Bryotoxin A possessing higher values, suggesting potential for increased hydrogen bonding interactions. While all compounds adhered to Lipinski's rule of five, Bryotoxin A's two violations indicate a departure from conventional drug-like criteria. These molecular property assessments provide valuable insights into the compounds' pharmacokinetic characteristics, aiding in the evaluation of their potential as drug candidates (Table 04).
Table 04.
Molecule properties of the four bioactive compounds from Bryophyllum pinnatum.
| Compounds | MW | LogP | RB | HBA | HBD | Surface Area | LOF Violations |
|---|---|---|---|---|---|---|---|
| Bryophyllin A | 472.534 | 2.2512 | 2 | 8 | 2 | 197.117 | 0 |
| Bryophyllin B | 490.549 | 1.2056 | 2 | 9 | 4 | 202.598 | 0 |
| Bryotoxin A | 618.676 | 1.1374 | 5 | 12 | 4 | 254.224 | 2 |
| Bryotoxin B | 488.533 | 1.2236 | 2 | 9 | 3 | 201.911 | 0 |
Here, LOF: Lipinski rule of five.
3.4. ADMET analysis
The ADMET analysis of the active compounds from Bryophyllum pinnatum provides valuable insights into their pharmacokinetic properties and potential therapeutic suitability [[48], [49], [50]]. Bryophyllin A and Bryophyllin B exhibited favorable absorption, as indicated by Caco-2 permeability (C2P) values of 1.351 and 0.57, respectively. Moreover, both compounds demonstrated high human intestinal absorption (HIA) percentages of 85.237 % and 92.164 %, respectively. Notably, Bryophyllin B showed the highest absorption, suggesting its potential as an orally bioavailable drug. The compounds exhibited negligible P-glycoprotein inhibition (P-gpI), indicating minimal impact on drug efflux. While Bryotoxin A and Bryotoxin B showed lower HIA percentages, Bryotoxin B displays excellent Caco-2 permeability. None of the compounds appeared to inhibit CYP1A2, reducing the likelihood of metabolic interactions. The blood-brain barrier (BBB) and central nervous system (CNS) permeability values suggested limited brain penetration for all compounds. Importantly, no hepatotoxicity was indicated, and the oral rat acute toxicity (LD50) values fallen within an acceptable range, with Bryotoxin B having the highest LD50 (Table 05).
Table 05.
ADME analysis of the four bioactive compounds from Bryophyllum pinnatum.
| Compounds | Absorption |
Distribution |
Metabolism |
Excretion |
|||
|---|---|---|---|---|---|---|---|
| C2P | HIA (%) | P-gpI | BBB |
CNS |
CYP1A2 | TC | |
| (Permeability) | |||||||
| Bryophyllin A | 1.351 | 85.237 | No | −0.635 | −3.077 | No | 0.254 |
| Bryophyllin B | 0.57 | 92.164 | No | −0.973 | −3.271 | No | 0.375 |
| Bryotoxin A | 0.616 | 56.276 | Yes | −1.318 | −3.32 | No | 0.373 |
| Bryotoxin B | 1.116 | 100 | No | −0.896 | −3.356 | No | 0.343 |
Here, TC = total clearance and measured in log mL/min/kg; I = inhibitor; Caco-2 permeability (C2P) (log Papp in 10−6 cm/s); human intestinal absorption (HIA) (% absorbed), and P-glycoprotein inhibitor (P-gpI); The blood-brain barrier (BBB) (log BB) and central nervous system (CNS) permeability (log PS).
The toxicity analysis provides crucial insights into their safety profiles and potential adverse effects. Bryophyllin A, Bryophyllin B, and Bryotoxin A shared a toxicity class of 2, with predicted LD50 values of 31 mg/kg, indicating moderate toxicity. In contrast, Bryophyllin B was classified in toxicity class 3, with a higher predicted LD50 of 144 mg/kg, suggesting a comparatively lower toxicity. Importantly, all compounds exhibited inactive hepatotoxicity, mutagenicity, and immunotoxicity, highlighting their favorable safety profiles in these aspects. However, cytoxicity was predicted to be active for Bryophyllin A, Bryophyllin B, and Bryotoxin A, suggesting a potential impact on cell viability. Carcinogenicity was predicted to be active for Bryophyllin A and Bryotoxin A, emphasizing the need for further investigation into their long-term effects (Table 06).
Table 06.
Toxicity analysis of the four bioactive compounds from Bryophyllum pinnatum.
| Compounds | Oral Toxicity |
Organ Toxicity |
Toxicity Endpoints Prediction |
||||
|---|---|---|---|---|---|---|---|
| Toxicity Class | Predicted LD50 (mg/kg) | Hepatotoxicity | Mutagenicity | Cyto Toxicity | Carcinogenicity | Immunotoxicity | |
| Bryophyllin A | 2 | 31 | Inactive | Inactive | Active | Inactive | Active |
| Bryophyllin B | 3 | 144 | Inactive | Inactive | Active | Inactive | Active |
| Bryotoxin A | 2 | 31 | Inactive | Inactive | Active | Inactive | Active |
| Bryotoxin B | 2 | 31 | Inactive | Inactive | Inactive | Inactive | Active |
Overall, Bryophyllin B emerged as a promising candidate with high absorption, minimal metabolic interactions, and low toxicity profile, marked by lower predicted oral toxicity and the absence of predicted carcinogenicity.
3.5. Analysis of molecular dynamics simulations (MDS): RMSD, RMSF, RG, SASA, MolSA, PSA and torsion angels
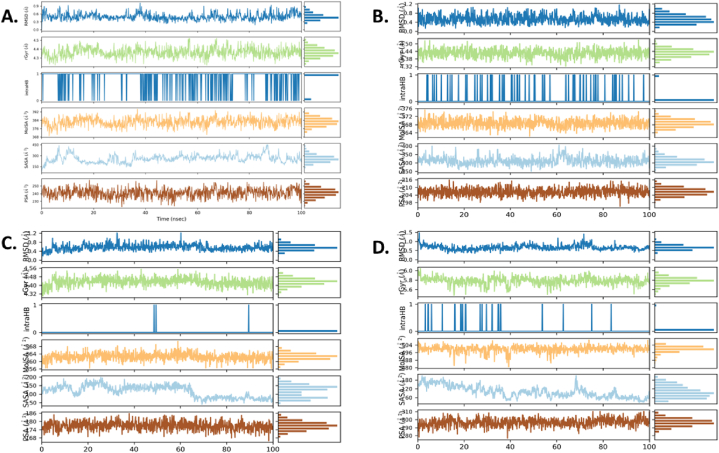
The molecular dynamics simulations provided insights into the dynamic behavior and stability of the protein-ligand complexes formed by the four bioactive compounds derived from Bryophyllum pinnatum. The RMSD values for the protein-ligand complexes ranged from 1.530682 Å to 2.111674 Å and individually for the ligands are ranges from 0.46445 Å to 0.699878 Å, indicating minimal structural deviation during the 100 ns simulation period (Fig. 10(A–D) and Fig. 11(A–D)). Notably, 4QNN-Bryophyllin A complex exhibited slightly higher RMSD value (2.111674 Å) compared to the other protein-ligand complexes, suggesting some degree of structural fluctuation. In terms of ligand stability, Bryophyllin A and B demonstrated the lowest RMSD value of 0.46445 Å and 0.475028 Å, indicating excellent structural preservation within the protein-ligand complex. Conversely, Bryotoxin A exhibited slightly higher RMSD value of 0.699878 Å, indicating relatively more significant structural deviations during the simulation.
Fig. 10.
Molecular Dynamics Simulations: Properties (PSA, SASA, MolSA, IntraHB, rGyr and RMSD) of the four bioactive compounds of Bryophyllum pinnatum: (A) Bryophyllin B, (B) Bryotoxin B, (C) Bryophyllin A, and (D) Bryotoxin A.
Fig. 11.
Root Mean Square Deviation (RMSD) of protein-ligand complexes: (A) 1QNX-Bryophyllin B, (B) 2ATM-Bryotoxin B, (C) 4QNN-Bryophyllin A, and (D) 4QNN-Bryotoxin A.
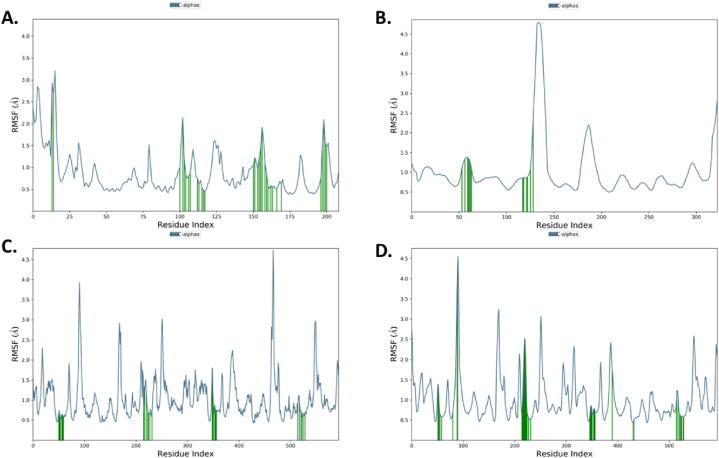
The RMSF analysis provided insights into the flexibility of individual atoms within the ligands. Bryophyllin A displayed the lowest RMSF value of 0.48435 Å, indicating minimal atom-level fluctuation, while Bryophyllin B exhibited the highest RMSF value of 3.63705 Å, suggesting greater flexibility. Bryotoxin A and Bryotoxin B displayed intermediate RMSF values, indicating moderate flexibility of atoms within the ligands (Fig. 12(A–D)).
Fig. 12.
Root Mean Square Fluctuation (RMSF) of the bioactive compounds- (A) Bryophyllin B, (B) Bryotoxin B, (C) Bryophyllin A, and (D) Bryotoxin A.
Regarding the radius of gyration (RG), Bryotoxin A exhibited the highest value of 5.792775 Å, indicating a larger spread of atoms from the center of mass, potentially reflecting a more extended conformation (Fig. 10D). In contrast, Bryophyllin B demonstrated the lowest RG value of 4.348928 Å, suggesting a more compact structure within the protein-ligand complex (Fig. 10A).
The solvent-accessible surface area (SASA) values provided insights into the accessibility of solvent molecules to the ligand surface. Bryophyllin A exhibited the lowest SASA value of 87.32623 Å2, indicating relatively lower solvent exposure, while Bryotoxin B showed the highest SASA value of 310.6630 Å2, indicating greater solvent accessibility (Fig. 10B and C).
Furthermore, the molecular surface area (MolSA) and polar surface area (PSA) values reflected the total surface area and the area occupied by polar atoms, respectively. Bryotoxin A exhibited the highest MolSA and PSA values, indicating a larger molecular surface area and a greater presence of polar atoms compared to the other compounds (Fig. 10(A–D)). These dynamic properties of the four bioactive compounds are summarized in Table 07 which actually a short form of Table S2.
Table 07.
Molecular Dynamics Simulations: The Summary of Properties of the top four compounds.
| Ligand | Protein- ligand complex |
Ligand |
|||||
|---|---|---|---|---|---|---|---|
| RMSD (Å) | RMSD (Å) | RMSF (Å) | RG (Å) | SASA (Å2) | MolSA (Å2) | PSA (Å2) | |
| Bryophyllin A | 2.111674 | 0.46445 | 0.48435 | 4.41730 | 123.3923 | 361.1234 | 176.326 |
| Bryophyllin B | 1.613691 | 0.475028 | 3.63705 | 4.348928 | 201.2910 | 377.2109 | 243.1312 |
| Bryotoxin A | 1.617276 | 0.699878 | 0.5681 | 5.792775 | 87.32623 | 498.5362 | 294.2162 |
| Bryotoxin B | 1.530682 | 0.557712 | 1.8984 | 4.209431 | 310.6630 | 269.8755 | 206.4726 |
Understanding the dynamic behavior of amino acids and pinpointing specific modification sites within a protein are critical for interpreting functional dynamics during molecular dynamics simulations. The root mean square fluctuation (RMSF) analysis offers valuable insights into the variability of individual amino acids over the simulation trajectory. The average RMSF values for Bryophyllin B, Bryotoxin B, Bryophyllin A, and Bryotoxin A were determined for three allergen proteins: Antigen 5 (Ves V5, PDB: 1QNX), Hyaluronoglucosaminidase (Ves V2, PDB: 2ATM), and Phospholipase A1 (Ves V1, PDB: 4QNN). Notable fluctuations (1.4–3.8 Å) were observed in specific residues of Antigen 5, such as LYS_5, ILE_6, LYS_7, CYS_8, LEU_9, LYS_10, GLY_11, ASN_194, PHE_195, LYS_196, ASN_197, GLU_198, and GLU_199, particularly with Bryophyllin B (Fig. 13A). Here LEU_9 and LYS_10 formed alkyl bond and conventional hydrogen bond respectively with Bryophyllin B and these two amino acids showed moderate fluctuation of 3.706 Å and 2.548 Å respectively which denotes the flexibility the interactions.
Fig. 13.
Molecular Dynamics Simulation: RMSF plotting of allergen proteins- Antigen 5 (Ves V5, PDB: 1QNX), Hyaluronoglucosaminidase (Ves V2, PDB: 2ATM), and Phospholipase A1 (Ves V1, PDB: 4QNN) with (A) Bryophyllin B, (B) Bryotoxin B, (C) Bryophyllin A, and (D) Bryotoxin A respectively.
For the allergen protein Hyaluronoglucosaminidase, residues including ASN_135, GLU_136, HIS_137, PRO_138, THR_139, TRP_140, ASN_141, LYS_142, and LYS_143 exhibited maximum fluctuations ranging from 2.1 to 9.8 Å (Fig. 13B). Similarly, for the allergen protein Phospholipase A1, residues like MET_91, SER_92, GLY_93, ILE_94, GLN_95, LEU_96, PRO_169, SER_170, PHE_171, LYS_172, SER_173, and ASN_174 showed fluctuations ranging from 2.5 to 7.5 Å with Bryotoxin B (Fig. 13C). Additionally, amino acid residues including ARG_18, GLU_19, ASN_20, LYS_21, LYS_22, HIS_23, ASP_24, GLU_89, GLU_90, MET_91, and others exhibited similar fluctuations with Bryotoxin A (Fig. 13D). Importantly, none of these highly fluctuating amino acid residues were directly involved in protein-ligand contacts. Despite the observed fluctuations, the protein-ligand complexes for all four compounds remained stable throughout the simulation. Notably, the allergen proteins Antigen 5, Hyaluronoglucosaminidase, and Phospholipase A1 demonstrated enhanced stability, indicating robust interactions with the ligands.
The compounds are visually represented in a 2D schematic diagram, showcasing color-coded rotatable bonds (Fig. 14(A–D)). To provide a clearer understanding of these rotatable torsional bonds, a radial plot and corresponding color bar plots were introduced. The radial plot depicts the evolution of torsion angles over the simulation period, emanating from the center and extending outward with time. Bar plots accompanying the radial diagram summarize the probability density of torsion angles, with data expressed in kcal/mol on the Y-axis. This combined visualization method elucidates the relationship between torsion potential and conformational strain of the compounds while preserving their protein-bound conformation.
Fig. 14.
Torsions of the four bioactive compounds- (A) Bryophyllin B, (B) Bryotoxin B, (C) Bryophyllin A, and (D) Bryotoxin A.
In summary, molecular dynamics simulations revealed distinct properties of the four bioactive compounds from Bryophyllum pinnatum. All compounds showed stable interactions within the protein-ligand complex, but differences in structural stability, flexibility, and surface properties were observed. These findings provide valuable insights for further investigation and development of these compounds as potential therapeutic agents.
3.5.1. Analysis of MDS: protein-ligand contacts
Based on the analysis of protein-ligand interactions during the 100ns molecular dynamics simulations, the interactions between the four bioactive compounds from Bryophyllum pinnatum and the respective allergen proteins of V. vulgaris were assessed, focusing on different types of interactions, including water bridges, hydrogen bonds, ionic interactions, and hydrophobic interactions.
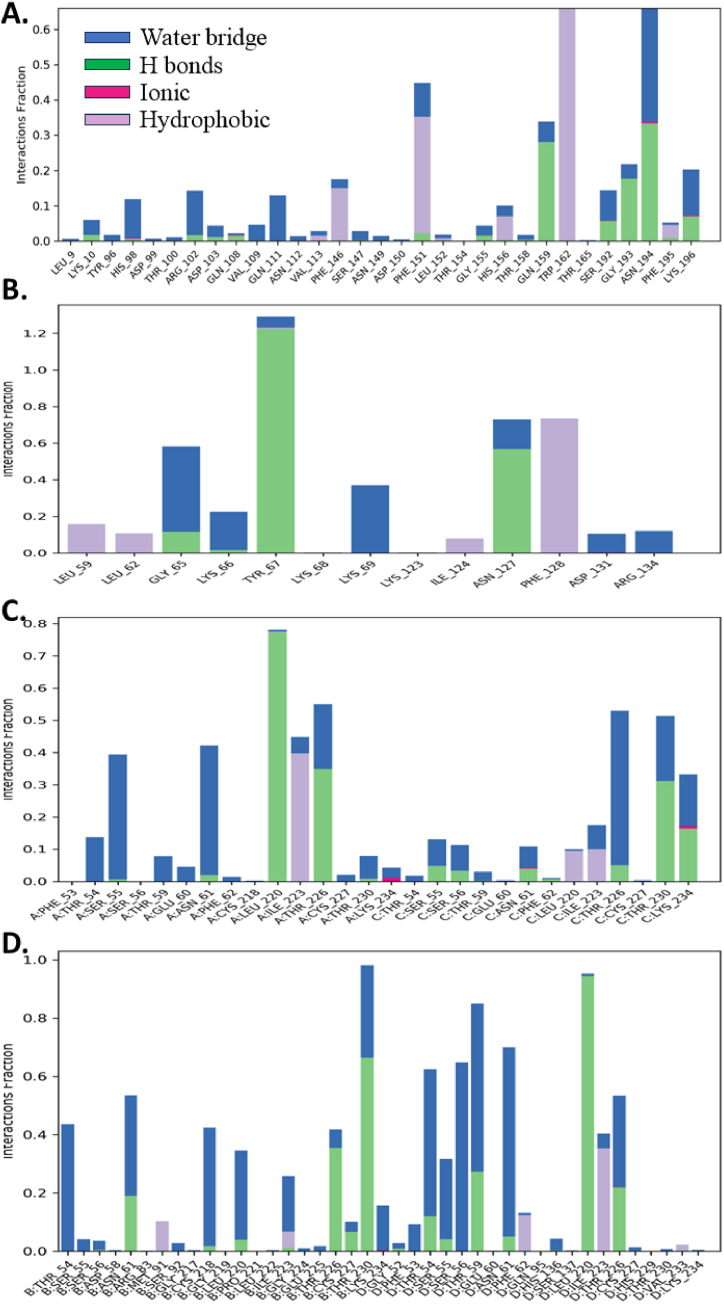
For the protein-ligand complex Antigen 5 (Ves V5, PDB: 1QNX) with Bryophyllin B, a substantial number of interactions were observed, primarily dominated by water bridges (24 amino acid residues) and hydrogen bonds (10 amino acid residues). These interactions persisted for an average duration of 47 % and 8 % of the simulation period, respectively, indicating a notable engagement between Bryophyllin B and the Antigen 5 (Ves V5) protein (Fig. 15A). Among them, TYR_162 demonstrated hydrophobic interaction throughout the whole simulation period and PHE_151 for 40 % time period. Again, ASN_194 showed both H bond and water bridge interaction for 50 % time of simulation period. These long-time interactions denote the compact nature and strong stability of the protein-ligand contacts.
Fig. 15.
Plot (stacked bar charts) of the bioactive compounds- (A) Bryophyllin B, (B) Bryotoxin B, (C) Bryophyllin A, and (D) Bryotoxin A interactions with the allergen proteins- Antigen 5 (Ves V5, PDB: 1QNX), Hyaluronoglucosaminidase (Ves V2, PDB: 2ATM), and Phospholipase A1 (Ves V1, PDB: 4QNN) of Vespula vulgaris supervised throughout the simulation period of 100ns.
In the case of the protein-ligand complex Hyaluronoglucosaminidase (Ves V2, PDB: 2ATM) with Bryotoxin B, fewer interactions were observed compared to 1QNX-Bryophyllin B. However, hydrogen bonds were the predominant interaction type, involving 4 amino acid residues and persisting for 60 % of the simulation time. Water bridges and hydrophobic interactions were also present, albeit to a lesser extent (Fig. 15B). The amino acid residues TYR_67 and ASN_127 showed H bond interactions for 120 % and 60 % time of simulation time periods. PHE_128 showed hydrophobic interaction for 70 % time period which denote the strong binding stability with Bryotoxin B.
Moving to the protein-ligand complex Phospholipase A1 (Ves V1, PDB: 4QNN), both Bryophyllin A and Bryotoxin A exhibited significant interactions. Bryophyllin A formed water bridges with 24 amino acid residues, hydrogen bonds with 10 amino acid residues, and ionic interactions with 3 amino acid residues. These interactions persisted for 65 %, 34 %, and 2 % of the simulation time, respectively. Similarly, Bryotoxin A formed water bridges with 27 amino acid residues and hydrogen bonds with 12 amino acid residues, with durations of 70 % and 87 % of the simulation time, respectively (Fig. 15C and D). Both the amino acid residue B:THR_230 and D:LEU_220 exhibited H bond interaction for 65 % and 98 % of simulation time period. Again, B:THR_54, B:CYS_218, D:THR_54, D:SER_56, D:THR_59 and D:ASN_61 all exhibited water bridges interactions with Bryotoxin A for more than 40 % time of the simulation time period. A timeline representation of these protein-ligand contacts was supplemented on Figs. S1(A–D).
Overall, the interactions between the compounds and the allergen proteins varied in terms of types and durations. Bryophyllin B and Bryotoxin A exhibited extensive interactions with the Phospholipase A1 (Ves V1) protein, while Bryophyllin B displayed notable engagement with the Antigen 5 (Ves V5) protein. These findings suggest distinct binding modes and potential binding affinities of the compounds with their respective target proteins, providing valuable insights for further drug development efforts.
4. Discussion
Different analysis in this study provides valuable insights into the potential therapeutic efficacy and safety profiles of bioactive compounds derived from Bryophyllum pinnatum against allergen proteins from V. vulgaris. Through a comprehensive analysis encompassing molecular docking, molecular properties assessment, and ADMET profiling, we have shed light on the promising candidates for inhibiting allergic reactions induced by wasp venom.
Firstly, the molecular docking analysis revealed robust binding interactions between the bioactive compounds and the allergen proteins, indicating strong potential for therapeutic intervention. Across all three proteins (Ves V1, Ves V2, and Ves V5), Bryophyllin B consistently exhibited high affinity (−9.6 kcal/mol), closely followed by Bryotoxin A (−10.0 kcal/mol), suggesting their strong binding capabilities with allergen proteins from V. vulgaris. The identified binding pockets were consistent among different proteins, suggesting potential shared interaction sites and highlighting the specificity of the compounds towards the target proteins.
Furthermore, the molecular properties analysis provided insights into the drug-likeness and pharmacokinetic characteristics of the compounds [[51], [52], [53], [54]]. Bryophyllin A and Bryophyllin B demonstrated moderate molecular weights and favorable logP values, indicative of balanced hydrophilic-lipophilic profiles essential for efficient drug absorption and distribution. While Bryotoxin A and Bryotoxin B exhibited higher molecular weights and greater molecular flexibility, they still adhered to Lipinski's rule of five, albeit with a few violations, suggesting their potential as drug candidates.
The ADMET analysis further elucidated the pharmacokinetic properties and safety profiles of the compounds [48]. Bryophyllin B emerged as a promising candidate with high absorption, minimal metabolic interactions, and a low toxicity profile, making it a favorable choice for further development as a therapeutic agent. Importantly, all compounds exhibited favorable safety profiles in terms of hepatotoxicity, mutagenicity, and immunotoxicity, although cytotoxicity and carcinogenicity predictions warrant further investigation.
The analysis of molecular dynamics simulations (MDS) offers a comprehensive understanding of the dynamic behavior and stability of protein-ligand complexes formed by four bioactive compounds derived from Bryophyllum pinnatum. These simulations provide valuable insights into various parameters such as RMSD, RMSF, RG, SASA, MolSA, PSA, and torsion angles, shedding light on the structural dynamics and interactions within the complexes.
The RMSD values, indicative of structural deviation during the simulation period, ranged from 1.530682 Å to 2.111674 Å for the protein-ligand complexes, suggesting minimal structural changes. Notably, Bryophyllin A and B exhibited the lowest RMSD values for individual ligands, indicating excellent structural preservation within the complexes. Conversely, Bryotoxin A displayed slightly higher RMSD values, suggesting relatively more significant structural deviations [[55], [56], [57]].
The RMSF analysis provided insights into the flexibility of individual atoms within the ligands [58]. Bryophyllin A displayed minimal atom-level fluctuation, while Bryophyllin B exhibited greater flexibility. Bryotoxin A and B displayed intermediate flexibility, indicating moderate fluctuation of atoms within the ligands.
Regarding the radius of gyration (RG) [59], Bryotoxin A exhibited the highest value, suggesting a more extended conformation, while Bryophyllin B demonstrated the lowest RG value, indicating a more compact structure within the complexes.
Solvent-accessible surface area (SASA) values reflected the accessibility of solvent molecules to the ligand surface, with Bryophyllin A exhibiting relatively lower solvent exposure and Bryotoxin B showing greater solvent accessibility. Additionally, MolSA and PSA values indicated differences in molecular surface area and the presence of polar atoms among the compounds.
The analysis of torsion angles provided further insights into the conformational dynamics of the compounds over the simulation period, elucidating their potential strain and stability within the protein-bound conformation.
Moreover, the analysis of protein-ligand contacts revealed extensive interactions between the compounds and allergen proteins, including water bridges, hydrogen bonds, and hydrophobic interactions. Each compound exhibited unique interaction profiles with the target proteins, indicating distinct binding modes and potential binding affinities.
Despite the observed fluctuations in dynamic properties, all protein-ligand complexes remained stable throughout the simulations, underscoring the robustness of their interactions. Notably, the allergen proteins demonstrated enhanced stability, further supporting the strong and stable binding affinity of the compounds.
In conclusion, this study highlights the potential of bioactive compounds from Bryophyllum pinnatum as inhibitors of V. vulgaris allergen proteins. Bryophyllin B, in particular, emerges as a most promising candidate with high binding affinity, favorable pharmacokinetic properties, a low toxicity profile and strong stability in protein-ligand complex. These findings pave the way for further research and development of novel therapeutics to mitigate the allergic burden imposed by wasp venom, emphasizing the importance of natural compounds in drug discovery and development.
5. Conclusion
In conclusion, this study unveiled Bryophyllum pinnatum compounds, particularly Bryophyllin B and Bryotoxin A as promising inhibitors for V. vulgaris allergen proteins. The strong binding affinities and favorable ADMET profiles make these compounds attractive candidates for further preclinical and clinical studies. The potential of Bryophyllin B as a lead compound emphasizes the importance of thorough drug development efforts. As we advance toward the goal of mitigating wasp venom allergies, these findings contribute valuable insights to the field, offering potential solutions to address the increasing prevalence of allergic reactions induced by V. vulgaris venom.
Data availability statement
Data included in article/supp. material/referenced in article.
CRediT authorship contribution statement
Sirajul Islam: Writing – original draft, Methodology, Data curation, Conceptualization. Abu Zaffar Shibly: Writing – review & editing, Visualization, Supervision, Data curation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e34713.
Contributor Information
Sirajul Islam, Email: sirajulislamsiru255@gmail.com.
Abu Zaffar Shibly, Email: zaffarshibly@mbstu.ac.bd.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Henriksen A., et al. Major venom allergen of yellow jackets, ves v 5: structural characterization of a pathogenesis-related protein superfamily. Proteins: Structure, Function and Genetics. Dec. 2001;45(4):438–448. doi: 10.1002/PROT.1160. [DOI] [PubMed] [Google Scholar]
- 2.Henriksen A., et al. Major venom allergen of yellow jackets, ves v 5: structural characterization of a pathogenesis-related protein superfamily. Proteins: Structure, Function and Genetics. Dec. 2001;45(4):438–448. doi: 10.1002/PROT.1160. [DOI] [PubMed] [Google Scholar]
- 3.Bohle B., Zwölfer B., G. F.-… E. Allergy, and undefined . Wiley Online Library; 2005. Characterization of the Human T Cell Response to Antigen 5 from Vespula Vulgaris (Ves V 5)https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1365-2222.2005.02180.x [Online]. Available: [DOI] [PubMed] [Google Scholar]
- 4.Kolarich D., et al. A proteomic study of the major allergens from yellow jacket venoms. Proteomics. May 2007;7(10):1615–1623. doi: 10.1002/PMIC.200600800. [DOI] [PubMed] [Google Scholar]
- 5.Bohle B., et al. Characterization of the human T cell response to antigen 5 from Vespula vulgaris (Ves v 5) Clin. Exp. Allergy. Mar. 2005;35(3):367–373. doi: 10.1111/J.1365-2222.2005.02180.X. [DOI] [PubMed] [Google Scholar]
- 6.Suck R., Weber B., Kahlert H., S. H.-… archives of allergy and, and undefined Purification and immunobiochemical characterization of folding variants of the recombinant major wasp allergen Ves v 5 (antigen 5) 2000. karger.comhttps://karger.com/iaa/article/121/4/284/164496 [Online]. Available: [DOI] [PubMed]
- 7.Kolarich D., Loos A., Léonard R., L. M.-, and undefined . Wiley Online Library; Accessed: 2007. A Proteomic Study of the Major Allergens from Yellow Jacket Venoms.https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/abs/10.1002/pmic.200600800 [Online]. Available: [DOI] [PubMed] [Google Scholar]
- 8.Henriksen A., King T., O. M.-P. S., and undefined . Wiley Online Library; 2001. Major Venom Allergen of Yellow Jackets, Ves V 5: Structural Characterization of a Pathogenesis‐related Protein Superfamily.https://onlinelibrary.wiley.com/doi/abs/10.1002/prot.1160 [Online]. Available: [DOI] [PubMed] [Google Scholar]
- 9.Hofmann S., Pfender N., Weckesser S., Huss-Marp J., Jakob T. Added value of IgE detection to rApi m 1 and rVes v 5 in patients with hymenoptera venom allergy. J. Allergy Clin. Immunol. 2011;127 doi: 10.1016/j.jaci.2010.06.042. [DOI] [PubMed] [Google Scholar]
- 10.Blank S., et al. Vitellogenins are new high molecular weight components and allergens (Api m 12 and Ves v 6) of apis mellifera and vespula vulgaris venom. PLoS One. Apr. 2013;8(4) doi: 10.1371/JOURNAL.PONE.0062009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korosec P., Valenta R., Mittermann I., Celesnik N., Silar M. High sensitivity of CAP-FEIA rVes v 5 and rVes v 1 for diagnosis of vespula venom allergy. J. Allergy Clin. Immunol. 2012;129 doi: 10.1016/j.jaci.2011.12.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffman S., Pfender N., Weckesser S., Blank S., Huss-Marp J. Detection of IgE to a panel of species specific allergens further improves discrimination of bee and wasp venom allergy (Reply) J. Allergy Clin. Immunol. 2011;128 [Google Scholar]
- 13.Schiener M., Graessel A., Ollert M., Schmidt-Weber C.B., Blank S. Allergen-specific immunotherapy of Hymenoptera venom allergy–also a matter of diagnosis. Hum Vaccin Immunother. Oct. 2017;13(10):2467–2481. doi: 10.1080/21645515.2017.1334745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Przybilla B., Ruëff F. Hymenoptera venom allergy. JDDG J. der Deutschen Dermatol. Gesellschaft. Feb. 2010;8(2):114–129. doi: 10.1111/J.1610-0387.2009.07125.X. [DOI] [PubMed] [Google Scholar]
- 15.Bilò B.M., Bonifazi F. Hymenoptera venom immunotherapy. Immunotherapy. Feb. 2011;3(2):229–246. doi: 10.2217/IMT.10.88. [DOI] [PubMed] [Google Scholar]
- 16.Sturm G.J., et al. EAACI guidelines on allergen immunotherapy: hymenoptera venom allergy. Allergy: European Journal of Allergy and Clinical Immunology. Apr. 2018;73(4):744–764. doi: 10.1111/ALL.13262. [DOI] [PubMed] [Google Scholar]
- 17.Ansotegui I.J., Sánchez-Borges M., Cardona V. Current trends in prevalence and mortality of anaphylaxis. Curr Treat Options Allergy. Sep. 2016;3(3):205–211. doi: 10.1007/S40521-016-0094-0. [DOI] [Google Scholar]
- 18.Cardona V., et al. World allergy organization anaphylaxis guidance 2020. World Allergy Organization Journal. Oct. 2020;13(10) doi: 10.1016/J.WAOJOU.2020.100472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner P.J., Campbell D.E., Motosue M.S., Campbell R.L. Global trends in anaphylaxis epidemiology and clinical implications. J. Allergy Clin. Immunol. Pract. Apr. 2020;8(4):1169–1176. doi: 10.1016/J.JAIP.2019.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tejedor Alonso M.A., Moro Moro M., Múgica García M.V. Epidemiology of anaphylaxis. Clin. Exp. Allergy. Jun. 2015;45(6):1027–1039. doi: 10.1111/CEA.12418. [DOI] [PubMed] [Google Scholar]
- 21.Bank P.D. Rcsb Pdb; 2019. RCSB PDB: Homepage. [Google Scholar]
- 22.Souza M., H. S.-… of A. in, and undefined An in silico analysis study of the chemical compounds from the crassulaceous plant Bryophyllum pinnatum (Lam.) oken against the SARS-COV-2 proteases. 2023. article.researchpromo.comhttp://article.researchpromo.com/id/eprint/1771/ [Online]. Available:
- 23.Rahman P., Syaban M., S. A.-… M. J. of, and undefined Molecular docking analysis from Bryophyllum pinnatum Compound as A COVID-19 cytokine storm therapy. 2022. https://oamjms.eu/index.php/mjms/article/view/8412 [Online]. Available:
- 24.Agrawal P., G. B.-N. P. Communications, and undefined . journals.sagepub.com; 2023. Phytochemicals against SARS-COV-2 Infection.https://journals.sagepub.com/doi/abs/10.1177/1934578X231152168 [Online]. Available: [Google Scholar]
- 25.Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 26.Kitchen D., Decornez H., J. F.-N. reviews D., and undefined . nature.comDB Kitchen, H Decornez, JR Furr, J BajorathNature Reviews Drug Discovery, 2004•nature.Com. 2004. Docking and scoring in virtual screening for drug discovery: methods and applications. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y., et al. In silico ADME/T modelling for rational drug design. Q. Rev. Biophys. Jul. 2015;48(4):488–515. doi: 10.1017/S0033583515000190. [DOI] [PubMed] [Google Scholar]
- 28.Rimmington F. Pharmacokinetics and pharmacodynamics. South. Afr. J. Anaesth. Analg. 2020;26(6) doi: 10.36303/SAJAA.2020.26.6.S3.2562. [DOI] [Google Scholar]
- 29.Egan W.J., Merz K.M., Baldwin J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. Oct. 2000;43(21):3867–3877. doi: 10.1021/JM000292E. [DOI] [PubMed] [Google Scholar]
- 30.Muegge I. Selection criteria for drug-like compounds. Med. Res. Rev. May 2003;23(3):302–321. doi: 10.1002/MED.10041. [DOI] [PubMed] [Google Scholar]
- 31.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. Mar. 2017;7 doi: 10.1038/SREP42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian S., Wang J., Li Y., Li D., Xu L., Hou T. The application of in silico drug-likeness predictions in pharmaceutical research. Adv. Drug Deliv. Rev. 2015;86 doi: 10.1016/j.addr.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 33.Lee K., Jang J., Seo S., Lim J., Kim W.Y. Drug-likeness scoring based on unsupervised learning. Chem. Sci. 2022;13(2) doi: 10.1039/d1sc05248a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia C.Y., Li J.Y., Hao G.F., Yang G.F. A drug-likeness toolbox facilitates ADMET study in drug discovery. Drug Discov. Today. 2020;25(1) doi: 10.1016/j.drudis.2019.10.014. [DOI] [PubMed] [Google Scholar]
- 35.Lee J.B., et al. Quantitative analysis of lab-to-lab variability in Caco-2 permeability assays. Eur. J. Pharm. Biopharm. 2017;114 doi: 10.1016/j.ejpb.2016.12.027. [DOI] [PubMed] [Google Scholar]
- 36.Li Y., et al. In vivo assessment of the effect of CYP1A2 inhibition and induction on pomalidomide pharmacokinetics in healthy subjects. J. Clin. Pharmacol. 2018;58(10) doi: 10.1002/jcph.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banerjee P., Eckert A.O., Schrey A.K., Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. Jul. 2018;46(W1):W257–W263. doi: 10.1093/NAR/GKY318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al Wasidi A.S., Hassan A.S., Naglah A.M. In vitro cytotoxicity and druglikeness of pyrazolines and pyridines bearing benzofuran moiety. J Appl Pharm Sci. 2020;10(4) doi: 10.7324/JAPS.2020.104018. [DOI] [Google Scholar]
- 39.Durrant J.D., McCammon J.A. Molecular dynamics simulations and drug discovery. BMC Biol. 2011;9 doi: 10.1186/1741-7007-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borhani D.W., Shaw D.E. The future of molecular dynamics simulations in drug discovery. J. Comput. Aided Mol. Des. 2012;26(1) doi: 10.1007/s10822-011-9517-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stalker M.R., Grant J., Yong C.W., Ohene-Yeboah L.A., Mays T.J., Parker S.C. Molecular simulation of hydrogen storage and transport in cellulose. Mol Simul. 2021;47(2–3) doi: 10.1080/08927022.2019.1593975. [DOI] [Google Scholar]
- 42.Kiss P.T., Baranyai A. Sources of the deficiencies in the popular SPCE and TIP3P models of water. J. Chem. Phys. 2011;134(5) doi: 10.1063/1.3548869. [DOI] [PubMed] [Google Scholar]
- 43.Sargsyan K., Grauffel C., Lim C. How molecular size impacts RMSD applications in molecular dynamics simulations. J Chem Theory Comput. Apr. 2017;13(4):1518–1524. doi: 10.1021/acs.jctc.7b00028. 10.1021/ACS.JCTC.7B00028/SUPPL_FILE/CT7B00028_SI_001.PDF. [DOI] [PubMed] [Google Scholar]
- 44.Hyberts S.G., Märki W., Wagner G. Stereospecific assignments of side‐chain protons and characterization of torsion angles in Eglin c. Eur. J. Biochem. 1987;164(3) doi: 10.1111/j.1432-1033.1987.tb11173.x. [DOI] [PubMed] [Google Scholar]
- 45.S. K.-C. protocols and undefined Exploring chemical information in PubChem. Wiley Online Library. Aug. 2021;1(8) doi: 10.1002/cpz1.217. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S., Thiessen P., Bolton E.…J. C.-N. acids, and undefined PubChem substance and compound databases. 2016. https://academic.oup.com/nar/article-abstract/44/D1/D1202/2503131 [Online]. Available: [DOI] [PMC free article] [PubMed]
- 47.Halayal R., Bagewadi Z., N. A.-S. P., and undefined . Elsevier; 2024. Phytocompounds from Kalanchoe Pinnata in the Treatment of Diabetes Mellitus by Integrating Network Pharmacology, Molecular Docking and Simulation ….https://www.sciencedirect.com/science/article/pii/S1319016424000768 [Online]. Available: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cruciani G. Molecular interaction fields: applications in drug discovery and ADME prediction. Molecular Interaction Fields: Applications in Drug Discovery and ADME Prediction. May 2006;27:1–303. doi: 10.1002/3527607676. [DOI] [Google Scholar]
- 49.Patel V., Lalani R., Bardoliwala D., Ghosh S., Misra A. Lipid-based oral Formulation strategies for lipophilic drugs. AAPS PharmSciTech. Nov. 2018;19(8):3609–3630. doi: 10.1208/S12249-018-1188-8. [DOI] [PubMed] [Google Scholar]
- 50.Patel V., Lalani R., Bardoliwala D., Ghosh S., Pharmscitech A.M.-A., undefined . Springer; 2018. Lipid-based Oral Formulation Strategies for Lipophilic Drugs.https://link.springer.com/article/10.1208/s12249-018-1188-8 [Online]. Available: [DOI] [PubMed] [Google Scholar]
- 51.Ghatole A., Gaidhane M., K. L.-… : B. J. of, and undefined Pharmacokinetics, drug-likeness, medicinal properties, molecular docking analysis of substituted β-lactams synthesized via -catalyzed. ceeol.com. 2020 https://www.ceeol.com/search/article-detail?id=854964 [Online]. Available: [Google Scholar]
- 52.Udugade S., Doijad R., B. U.-J. of Advanced, and undefined 2019 In silico evaluation of pharmacokinetics, drug-likeness and medicinal chemistry friendliness of momordicin1: an active chemical constituent of momordica charantia. sciensage.info. 2019;10(3):222–229. http://www.sciensage.info/index.php/JASR/article/view/369 [Online]. Available: [Google Scholar]
- 53.Alam M., Ahmed J., Chemistry D.L.-A.B. Springer; and undefined 2016. Biological Features, Drug-Likeness, Pharmacokinetic Properties, and Docking of 2-Arylidenehydrazinyl-4-Arylthiazole Analogues.https://link.springer.com/article/10.1007/s13765-016-0148-9 [Online]. Available: [Google Scholar]
- 54.Alam M.S., Ahmed J.U., Lee D.U. Biological features, drug-likeness, pharmacokinetic properties, and docking of 2-arylidenehydrazinyl-4-arylthiazole analogues. Appl Biol Chem. Apr. 2016;59(2):181–192. doi: 10.1007/S13765-016-0148-9. [DOI] [Google Scholar]
- 55.Kirchmair J., Markt P., Distinto S., Wolber G., Langer T. Evaluation of the performance of 3D virtual screening protocols: RMSD comparisons, enrichment assessments, and decoy selection - what can we learn from earlier mistakes? J. Comput. Aided Mol. Des. 2008;22(3–4):213–228. doi: 10.1007/S10822-007-9163-6. [DOI] [PubMed] [Google Scholar]
- 56.Brüschweiler R. Efficient RMSD measures for the comparison of two molecular ensembles. Proteins: Structure, Function and Genetics. Jan. 2003;50(1):26–34. doi: 10.1002/PROT.10250. [DOI] [PubMed] [Google Scholar]
- 57.Sargsyan K., Grauffel C., Lim C. How molecular size impacts RMSD applications in molecular dynamics simulations. J Chem Theory Comput. Apr. 2017;13(4):1518–1524. doi: 10.1021/ACS.JCTC.7B00028. [DOI] [PubMed] [Google Scholar]
- 58.Fonseca A.M., Caluaco B., Martinho J., Madureira C. Springer; 2023. Potential Inhibitors Targeting the Main Protease Structure of SARS-CoV-2 via Molecular Docking, and Approach with Molecular Dynamics, RMSD, RMSF, H-Bond, SASA …. [DOI] [PubMed] [Google Scholar]
- 59.Tamai Y., Konishi T., Einaga Y., Fujii M., Yamakawa H. Mean-square radius of gyration of oligo- and poly(methyl methacrylate)s in dilute solutions. Macromolecules. 1990;23(18):4067–4075. doi: 10.1021/MA00220A007. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.