Abstract
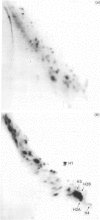
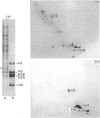
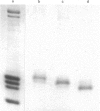
This study examines the histones pools in the nucleosol and cytosol of proliferating Friend cells. By using the conventional approach, detectable amounts of these molecules were found in both compartments; however, only H3 and H2B were identified in nucleosol, and H3, H2B and H4 in cytosol. The authenticity of each of these histones was verified by two independent methods, migration in SDS/polyacrylamide gels and peptide mapping. When the sensitivity of the approach was increased by radiolabelling with 125I, two additional proteins, migrating as H2A and H4, were observed in nucleosol. Even by this approach, however, H1 was not detected. Direct quantitative measurements of the histones in both compartments indicated that these pools are uneven and small. This was found also in experiments involving inhibition of protein synthesis by cycloheximide. Considered together, our data do not support the idea of the existence of preformed histone heterocomplexes or octamers. Instead the assembly of nucleosomes during replication occurs by a successive deposition of individual core histones.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson L. E., McClure W. O. An improved scintillation cocktail of high-solubilizing power. Anal Biochem. 1973 Jan;51(1):173–179. doi: 10.1016/0003-2697(73)90465-x. [DOI] [PubMed] [Google Scholar]
- Burlingame R. W., Love W. E., Wang B. C., Hamlin R., Nguyen H. X., Moudrianakis E. N. Crystallographic structure of the octameric histone core of the nucleosome at a resolution of 3.3 A. Science. 1985 May 3;228(4699):546–553. doi: 10.1126/science.3983639. [DOI] [PubMed] [Google Scholar]
- D'Anna J. A., Jr, Isenberg I. A complex of histones IIb2 and IV. Biochemistry. 1973 Mar 13;12(6):1035–1043. doi: 10.1021/bi00730a003. [DOI] [PubMed] [Google Scholar]
- D'Anna J. A., Jr, Isenberg I. A histone cross-complexing pattern. Biochemistry. 1974 Nov 19;13(24):4992–4997. doi: 10.1021/bi00721a019. [DOI] [PubMed] [Google Scholar]
- Djondjurov L. P., Yancheva N. Y., Ivanova E. C. Histones of terminally differentiated cells undergo continuous turnover. Biochemistry. 1983 Aug 16;22(17):4095–4102. doi: 10.1021/bi00286a016. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Glikin G. C., Ruberti I., Worcel A. Chromatin assembly in Xenopus oocytes: in vitro studies. Cell. 1984 May;37(1):33–41. doi: 10.1016/0092-8674(84)90298-8. [DOI] [PubMed] [Google Scholar]
- Jackson V., Chalkley R. A new method for the isolation of replicative chromatin: selective deposition of histone on both new and old DNA. Cell. 1981 Jan;23(1):121–134. doi: 10.1016/0092-8674(81)90277-4. [DOI] [PubMed] [Google Scholar]
- Jackson V., Chalkley R. A reevaluation of new histone deposition on replicating chromatin. J Biol Chem. 1981 May 25;256(10):5095–5103. [PubMed] [Google Scholar]
- Kleinschmidt J. A., Fortkamp E., Krohne G., Zentgraf H., Franke W. W. Co-existence of two different types of soluble histone complexes in nuclei of Xenopus laevis oocytes. J Biol Chem. 1985 Jan 25;260(2):1166–1176. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leffak I. M. Conservative segregation of nucleosome core histones. Nature. 1984 Jan 5;307(5946):82–85. doi: 10.1038/307082a0. [DOI] [PubMed] [Google Scholar]
- Leffak I. M., Grainger R., Weintraub H. Conservative assembly and segregation of nucleosomal histones. Cell. 1977 Nov;12(3):837–845. doi: 10.1016/0092-8674(77)90282-3. [DOI] [PubMed] [Google Scholar]
- Leffak I. M. Stability of the conservative mode of nucleosome assembly. Nucleic Acids Res. 1983 May 11;11(9):2717–2732. doi: 10.1093/nar/11.9.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louters L., Chalkley R. Exchange of histones H1, H2A, and H2B in vivo. Biochemistry. 1985 Jun 18;24(13):3080–3085. doi: 10.1021/bi00334a002. [DOI] [PubMed] [Google Scholar]
- Ostertag W., Melderis H., Steinheider G., Kluge N., Dube S. Synthesis of mouse haemoglobin and globin mRNA in leukaemic cell cultures. Nat New Biol. 1972 Oct 25;239(95):231–234. doi: 10.1038/newbio239231a0. [DOI] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969 Mar;130(1):337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- Ruberti I., Worcel A. Mechanism of chromatin assembly in Xenopus oocytes. J Mol Biol. 1986 Jun 5;189(3):457–476. doi: 10.1016/0022-2836(86)90317-7. [DOI] [PubMed] [Google Scholar]
- Sealy L., Cotten M., Chalkley R. Novobiocin inhibits passive chromatin assembly in vitro. EMBO J. 1986 Dec 1;5(12):3305–3311. doi: 10.1002/j.1460-2075.1986.tb04644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sealy L., Cotten M., Chalkley R. Xenopus nucleoplasmin: egg vs. oocyte. Biochemistry. 1986 May 20;25(10):3064–3072. doi: 10.1021/bi00358a049. [DOI] [PubMed] [Google Scholar]
- Senshu T., Akiyama K., Ohsawa T., Takahashi K. Immunochemical measurement of histone H3 in non-nucleosomal compartments of cultured mammalian cells. Eur J Biochem. 1985 Jan 15;146(2):261–266. doi: 10.1111/j.1432-1033.1985.tb08648.x. [DOI] [PubMed] [Google Scholar]
- Senshu T., Yamasu K., Ohsawa T. Fractionation of newly replicated nucleosomes by density labeling and rate zonal centrifugation for the analysis of the deposition sites of newly synthesized nucleosomal core histones. Eur J Biochem. 1985 Aug 1;150(3):575–580. doi: 10.1111/j.1432-1033.1985.tb09059.x. [DOI] [PubMed] [Google Scholar]
- Stein A., Bina-Stein M., Simpson R. T. Crosslinked histone octamer as a model of the nucleosome core. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2780–2784. doi: 10.1073/pnas.74.7.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A., Whitlock J. P., Jr, Bina M. Acidic polypeptides can assemble both histones and chromatin in vitro at physiological ionic strength. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5000–5004. doi: 10.1073/pnas.76.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov S., Ivanova E., Djondjurov L. Metabolic behaviors of the core histones in proliferating Friend cells. Exp Cell Res. 1989 Jan;180(1):94–105. doi: 10.1016/0014-4827(89)90215-2. [DOI] [PubMed] [Google Scholar]
- Weintraub H. The assembly of newly replicated DNA into chromatin. Cold Spring Harb Symp Quant Biol. 1974;38:247–256. doi: 10.1101/sqb.1974.038.01.028. [DOI] [PubMed] [Google Scholar]
- Worcel A., Han S., Wong M. L. Assembly of newly replicated chromatin. Cell. 1978 Nov;15(3):969–977. doi: 10.1016/0092-8674(78)90280-5. [DOI] [PubMed] [Google Scholar]