Abstract
Background
The significant rebound of influenza A (H1N1) virus activity, particularly among children, with rapidly growing number of hospitalized cases is of major concern in the post-COVID-19 era. The present study was performed to establish a prediction model of severe case in pediatric patients hospitalized with H1N1 infection during the post-COVID-19 era.
Methods
This is a multicenter retrospective study across nine public tertiary hospitals in Yunnan, China, recruiting pediatric H1N1 inpatients hospitalized at five of these centers between February 1 and July 1, 2023, into the development dataset. Screening of 40 variables including demographic information, clinical features, and laboratory parameters were performed utilizing Least Absolute Shrinkage and Selection Operator (LASSO) regression and logistic regression to determine independent risk factors of severe H1N1 infection, thus constructing a prediction nomogram. Receiver operating characteristic (ROC) curve, calibration curve, as well as decision curve analysis (DCA) were employed to evaluate the model's performance. Data from four independent cohorts comprised of pediatric H1N1 inpatients from another four hospitals between July 25 and October 31, 2023, were utilized to externally validate this nomogram.
Results
The development dataset included 527 subjects, 122 (23.1 %) of whom developed severe H1N1 infection. The external validation dataset included 352 subjects, 72 (20.5 %) of whom were eventually confirmed as severe H1N1 infection. The LASSO regression identified 19 candidate predictors, with logistic regression further narrowing down to 11 independent risk factors, including underlying conditions, prematurity, fever duration, wheezing, poor appetite, leukocyte count, neutrophil-lymphocyte ratio (NLR), erythrocyte sedimentation rate (ESR), lactate dehydrogenase (LDH), interleukin-10 (IL-10), and tumor necrosis factor-α (TNF-α). By integrating these 11 factors, a predictive nomogram was established. In terms of prediction of severe H1N1 infection, excellent discriminative capacity, favorable accuracy, and satisfactory clinical usefulness of this model were internally and externally validated via ROC curve, calibration curve, and DCA, respectively.
Conclusion
Our study successfully established and validated a novel nomogram model integrating underlying conditions, prematurity, fever duration, wheezing, poor appetite, leukocyte count, NLR, ESR, LDH, IL-10, and TNF-α. This nomogram can effectively predict the occurrence of serious case in pediatric H1N1 inpatients during the post-COVID-19 era, facilitating the early recognition and more efficient clinical management of such patients.
Keywords: Influenza A (H1N1) virus, Children, Nomogram, Post-COVID-19 era, Machine learning
Abbreviations
- ALT
alanine transaminase
- AST
aspartate transaminase
- AUC
area under the curve
- CNS
central nervous system
- Cr
creatinine
- CRP
c-reactive protein
- DCA
decision curve analysis
- ESR
erythrocyte sedimentation rate
- hs-TnT
high-sensitivity troponin T
- IFN-γ
interferon-γ
- IL6
interleukin 6
- IQR
interquartile range
- LASSO
least absolute shrinkage and selection operator
- LDH
lactate dehydrogenase
- NLR
neutrophil-lymphocyte ratio
- NPIs
non-pharmaceutical interventions
- PCCM
pediatric pulmonary and critical care medicine
- PCT
procalcitonin
- ROC
receiver operating characteristic
- TNF-α
tumor necrosis factor-α
1. Introduction
Influenza is a highly infectious virus that was responsible for 3 to 5 million cases of severe illness and 290,000–650,000 respiratory deaths globally every year [1]. Despite the population-wide susceptibility to the virus, children suffer from a disproportionate burden of influenza infection, which is well known to be a common cause of acute lower respiratory infection (ALRI) during childhood [2]. It was estimated that 109.5 million influenza virus episodes occurred among children under 5 years in 2018 resulting in 34,800 influenza-associated ALRI deaths, accounting for 4 % of ALRI-associated deaths among this age group [3]. Since early 2020, incidence of influenza infection has been significantly low globally and this reduction has been attributed mainly to the strict non-pharmaceutical interventions (NPIs) against the spread of SARS-CoV-2. However, concerns are increasing regarding the potential for more serious influenza epidemics during the post-COVID-19 era because of the so-called immune debt, a term proposed to characterize the decrease of protective immunity caused by prolonged low exposure to a given pathogen, generating a larger proportion of susceptible population and posing great hidden dangers to the whole population [4]. In fact, a recent study documented a more dramatic influenza pandemic with an elevated number of severe influenza infections and influenza-related hospitalizations among children in the post-COVID-19 era compared to the influenza seasons before and during the COVID-19 pandemic in Nicaragua [5]. More worryingly, as one of the countries experiencing the longest duration of COVID‐19‐associated restrictions, China may suffer from a stronger impact of immunity debt.
In addition, prior infection of SARS-CoV-2 may introduce a negative clinical impact for the severity of influenza infection during the post-pandemic period. A previous report indicated that over 82 % of the Chinese populace contracted SARS-CoV-2 in the most recent surge of the SARS-CoV-2 Omicron variant in China (December 2022–February 2023) [6]. After recovery from COVID-19, a considerable number of individuals have experienced SARS-CoV-2-associated enduring symptoms, which are theorized to stem from the presence of residual virus in diverse tissues, dysregulation of immune system, and autoimmune reactions triggered by the cross-reactivity of SARS-CoV-2-specific antibodies with the host's own proteins [[7], [8], [9], [10]]. According to Peluso et al. [11], a reduction in frequency of degranulating virus-specific CD8+ T cells was demonstrated in a certain proportion of individuals who had recovered from the acute phase of SARS-CoV-2 infection for over eight months, suggesting either a decline in the functionality of these cells or an issue with the immune system's response to viral infection. Consequently, a history of SARS-CoV-2 infection might be a significant consideration that could potentially worsen the clinical outcomes for children afflicted with influenza during the period after the end of COVID-19 pandemic.
Besides, despite the surge of COVID-19 cases at the initial stage after lifting NPIs, COVID-19 pandemic in China tended to stabilize and maintained at an extremely low level since the end of January 2023 (https://www.chinacdc.cn/). Subsequently, however, a swift resurgence of influenza A (H1N1) activity was noted, with a sharp increase in the hospitalization of children due to H1N1 infection from February 2023 onwards. Up to July 2023, daily reports of new infections continue to emerge.
Considering the soaring hospitalizations of children infected by H1N1 and the increased clinical severity of H1N1 cases, early recognition of severe pediatric H1N1 infection in the post-COVID-19 period is essential for clinical management of such patients, which may aid in delivering proper medical care and optimizing use of medical resources. Although characteristics of H1N1 infection among children during the pre-COVID-19 period were clearly elucidated and analyzed [[12], [13], [14]], whereas those in the post-COVID-19 era have changed significantly compared to traditional H1N1 infection in the pre-pandemic period [5,[15], [16], [17]]. Limited data are available regarding the risk prediction of severe pediatric H1N1 infection in the post-COVID-19 era. Hence, in this study, we aimed to develop and validate a fast, simple, accurate, and generalizable nomogram based on pediatric H1N1 inpatients from multiple centers to help predict risk of severe H1N1 infection at the time of hospital admission.
2. Materials and methods
2.1. Study population
This was a multicenter retrospective study conducted in Yunnan, China. Medical records from hospitalized cases with laboratory-confirmed H1N1 infection in five public tertiary hospitals in Yunnan between February 1, 2023, and July 1, 2023, were retrospectively collected as development dataset in this study. The inclusion criteria were clearly defined as follows: 1) children who fulfilled the diagnostic standards of influenza A (H1N1) infection [18,19]; 2) patients who were ≤14 years old at the time of their initial diagnosis (almost all children's hospitals or pediatric departments in general hospitals in China receive patients aged ≤14 years, while all age groups among these children are susceptible to influenza infections); and 3) the duration from onset of symptom to hospital admission did not exceed 48 h. Patients with incomplete information were excluded.
Ethical clearance of this study was received from the Ethics Committee of Kunming Children's Hospital Affiliated to Kunming Medical University (approval number: 2023-05-121-K11). This study was performed strictly adhering to the 1964 Declaration of Helsinki and local regulations. Given the retrospective design of the present study, the requirement for written informed consent was waived.
2.2. Specimen collection and processing
Collection of venous blood samples of patients was performed within 24h after admission and these sample were delivered immediately to the clinical laboratory department. Etiological examination was performed using the multiple pathogens IgM-capture ELISA kit (Antu Biotechnology, Zhengzhou, China; or Zhuochenghuisheng, Beijing, China), which included influenza virus A (H1N1 and H3N2), influenza virus B, respiratory syncytial virus (RSV), adenovirus (ADV), parainfluenza virus (PIV), Bordetella pertussis (BP), Mycoplasma pneumoniae (MP), and Chlamydia pneumoniae (CP). Blood routine examination was performed using the automated hematology analyzer (Mindray, Shenzhen, China; or Beckman Coulter, Fullerton, USA); lymphocyte subpopulation analysis was conducted on automated flow cytometer (Beckman Coulter, Fullerton, USA; or NanoFCM, Xiamen, China); liver and renal function was assessed using the automated biochemical analyzer (Mindray, Shenzhen, China; or Roche Cobas, Basel, Switzerland); cytokines, c-reactive protein (CRP) and procalcitonin (PCT) were measured using automated chemiluminescence immunoassay analyzer (Antu Biotechnology, Zhengzhou, China; or Roche Cobas, Basel, Switzerland); fibrinogen and D-dimer levels were measured using the automatic coagulation analyzer (Sysmex Corporation, Kobe, Japan). All equipment and reagents required for the above examinations were used in accordance with the manufacturers’ instructions.
2.3. Potential predictive variables
Potential predictive variables included the following 40 characteristics at patient admission, including demographic characteristics, medical history, clinical symptoms, and laboratory findings. Specifically, demographic variables included age, gender, body mass index (BMI), ethnicity, as well as history of parental consanguinity. The vaccination data encompassed records of both H1N1 and COVID-19 immunization, where individuals who had received a minimum of one vaccination dose were classified as having been vaccinated. Medical history included underlying conditions and prematurity. Clinical symptoms included fever, fever duration, fever peak, cough, rhinorrhea, wheezing, sore throat, headache, myalgia, seizure, poor appetite, and abdominal pain. Laboratory findings included leukocyte count, neutrophil-lymphocyte ratio (NLR), CD4+/CD8+ T cell ratio, hemoglobin, erythrocyte sedimentation rate (ESR), CRP, PCT, alanine transaminase (ALT), aspartate transaminase (AST), lactate dehydrogenase (LDH), creatinine (Cr), high-sensitivity troponin T (hs-TnT), D-dimer, interleukin 6 (IL-6), IL-10, tumor necrosis factor-α (TNF-α), interferon-γ (IFN-γ), fibrinogen and the presence or absence of co-infection, which was defined as the confirmation of one or more other pathogens concurrently with H1N1 infection. All data underwent a thorough review and cross-check conducted by two experienced clinicians (Cong-Yun Liu, Min Wang) and two trained Ph.D. students (Ming-Ze Sui, Jia-Wu Yang).
2.4. Outcomes
The severity of H1N1 infection (severe and general) was classified according to the WHO guidelines for influenza A (H1N1) given the wide acceptance of this guideline [20], which has already been described in detail and is easily available online at https://www.ncbi.nlm.nih.gov/books/NBK138515. Based on this guideline, our subjects were eventually assigned to the severe and general groups, respectively.
2.5. Statistical analysis
Statistical analyses were conducted using R software version 3.5.1 (R Foundation, Vienna, Austria). Statistical significance was inferred from a two-sided P value < 0.05.
2.5.1. Descriptive statistics of all subjects
Continuous variables were presented utilizing medians with interquartile range (IQR), while categorical variables were summarized using frequencies and percentages. Comparison between severe and general groups was calculated by Pearson's chi-square or Fisher's exact test for categorical variables, and Mann-Whitney U test for continuous variables due to their skewed distribution which had been checked by Shapiro-Wilk test. It should be pointed out that the aim of the above univariate analyses was to show the characteristics of patients and their differences between severe and general groups, rather than to screen candidate predictors.
2.5.2. Selection of candidate predictor variables
Least Absolute Shrinkage and Selection Operator (LASSO) regression, a machine learning variable selection algorithm, was adopted to minimize potential multicollinearity and overfitting as well as capture interaction effects between variables, thus selecting candidate predictor variables more effectively. This is a penalized regression model with L1-penalized regression method, which controls the model's complexity by shrinking the value of regression coefficients based on the value of lambda. Before the LASSO regression, the continuous variables were standardized using Z-score by subtracting the mean and dividing by the standard deviation of each continuous variable. To avoid omission of potentially vital predictors, all 40 variables were included into the LASSO model. The function “cv.glmnet” from the R package “glmnet” was applied to tune lambda by a ten-fold cross-validation, which was utilized to reduce the error generated by the randomization of the samples, ensuring the stability of variable selection and the generalization ability of the final model. To maintain both precision and parsimoniousness of this model, a lambda within one standard error of the minimum criterion (lambda.1se) was selected as the optimal value of lambda for preliminary determining the candidate predictor variables.
2.5.3. Establishment and internal validation of the prediction model
The covariates acquired from LASSO regression were added into logistic regression, identifying the independent influencing factors of severe H1N1 infection as well as obtaining regression coefficients (βi) and intercept. The predicted risk probability was calculated according to the logistic regression defined by the following equations:
The R package “rms” was utilized to plot the nomogram based on the results of logistic regression analysis. We randomly split the subjects in the development dataset into a training set and an internal validation set with a 7:3 ratio employing the R package “caret”, then receiver operating characteristic curve (ROC), calibration curve, and decision curve analysis (DCA) were subsequently performed utilizing the R packages “pROC”, “rms”, and “rmda”, respectively, to assess the discriminative capability, accuracy, and clinical benefit of the model in both training and internal validation sets.
2.5.4. External validation
To validate the generalizability of the prediction model, we retrospectively constructed an external validation dataset, which was comprised of pediatric H1N1 inpatients from four other public tertiary hospitals between July 25, 2023, and October 31, 2023, that were not included in the development dataset. These four hospitals were located in Wenshan, Baoshan, Lincang, and Dali, respectively, with different characteristics of ethnic composition and varied levels of healthcare resources. The data required for risk prediction from the validation dataset were still reviewed and cross-checked by the four investigators mentioned earlier. The final external validation was performed based on the area under the ROC curve (AUC), calibration curve, and DCA.
3. Results
3.1. Characteristics of subjects in the development dataset
In the development dataset (Fig. 1 and Table 1), we included 527 pediatric H1N1 inpatients with a median age of 5.2 (3.5–6.4) years and a median BMI of 17.3 (16.1–19.2) kg/m2. Of these, 300 (56.9 %) were males and 227 (43.1 %) were females. The majority of subjects were Han Chinese (507 [96.2 %]), with a minority belonging to Chinese ethnic minorities (20 [3.8 %]). Out of the total subjects, 6 (1.1 %) had parents who were consanguineous, 15 (2.8 %) received the H1N1 vaccine, 499 (94.7 %) received the COVID-19 vaccine, 29 (5.5 %) had underlying diseases, and 50 (9.5 %) had a history of prematurity. Overall, 122 (23.1 %) patients eventually developed severe H1N1 infection (severe group), while the remaining 405 (76.9 %) were considered to be general (general group).
Fig. 1.
Flow chart of the study design.
Table 1.
Characteristics of subjects in the development dataset.
| Characteristics | Total (n = 527) | Severe cases (n = 122) | General cases (n = 405) | P value |
|---|---|---|---|---|
| Age, y, median (IQR) | 5.2 (3.5–6.4) | 4.2 (2.4–5.9) | 5.3 (4.0–6.6) | <0.001 |
| Gender, n (%) | 0.247 | |||
| Male | 300 (56.9) | 75 (61.5) | 225 (55.6) | |
| Female | 227 (43.1) | 47 (38.5) | 180 (44.4) | |
| BMI, kg/m2, median (IQR) | 17.3 (16.1–19.2) | 17.5 (16.3–19.5) | 17.3 (16.0–19.0) | 0.284 |
| Ethnicity, n (%) | 0.841 | |||
| Chinese Han | 507 (96.2) | 117 (95.9) | 390 (96.3) | |
| Chinese ethnic minorities | 20 (3.8) | 5 (4.1) | 15 (3.7) | |
| Consanguineous family, n (%) | 6 (1.1) | 2 (1.6) | 4 (1.0) | 0.626 |
| Vaccination status, n (%) | ||||
| H1N1 vaccination | 15 (2.8) | 3 (2.5) | 12 (3.0) | 0.769 |
| COVID-19 vaccination | 499 (94.7) | 112 (91.8) | 387 (95.6) | 0.105 |
| Underlying disease, n (%) | 29 (5.5) | 19 (15.6) | 10 (2.5) | <0.001 |
| Prematurity, n (%) | 50 (9.5) | 30 (24.6) | 20 (4.9) | <0.001 |
| Symptoms, n (%) | ||||
| Fever | 516 (97.9) | 122 (100.0) | 394 (97.3) | 0.066 |
| Duration of fever, d, median (IQR) | 3.0 (2.0–5.0) | 5.0 (4.0–7.0) | 3.0 (2.0–4.0) | <0.001 |
| Fever peak, °C, median (IQR) | 39.0 (38.7–39.4) | 39.1 (38.8–39.5) | 39.0 (38.7–39.4) | 0.003 |
| Cough | 479 (90.9) | 113 (92.6) | 366 (90.4) | 0.448 |
| Rhinorrhea | 188 (35.7) | 43 (35.2) | 145 (35.8) | 0.910 |
| Wheezing | 155 (29.4) | 63 (51.6) | 92 (22.7) | <0.001 |
| Sore throat | 99 (18.8) | 32 (26.2) | 67 (16.5) | 0.016 |
| Headache | 66 (12.5) | 17 (13.9) | 49 (12.1) | 0.591 |
| Myalgia | 148 (28.1) | 32 (26.2) | 116 (28.6) | 0.603 |
| Seizure | 119 (22.6) | 34 (27.9) | 85 (21.0) | 0.111 |
| Poor appetite | 189 (35.9) | 76 (62.3) | 113 (27.9) | <0.001 |
| Abdominal pain | 40 (7.6) | 20 (16.4) | 20 (4.9) | <0.001 |
| Laboratory findings, median (IQR) | ||||
| Leukocyte count, × 109/L | 11.8 (9.8–14.4) | 14.8 (11.8–17.9) | 11.1 (9.6–12.9) | <0.001 |
| NLR | 4.2 (3.6–5.8) | 9.1 (5.7–11.0) | 3.9 (3.5–4.8) | <0.001 |
| CD4+/CD8+ T cell ratio | 1.3 (1.0–1.8) | 1.1 (0.8–1.5) | 1.4 (1.0–1.9) | <0.001 |
| Hemoglobin, g/L | 124.0 (114.0–133.0) | 119.0 (109.0–129.0) | 125.0 (116.0–134.0) | <0.001 |
| ESR, mm/H | 21.0 (17.0–33.0) | 38.0 (31.0–49.0) | 19.0 (16.0–25.0) | <0.001 |
| CRP, mg/L | 9.8 (6.5–17.1) | 17.5 (16.9–19.1) | 8.7 (4.2–11.3) | <0.001 |
| PCT, ng/mL | 0.3 (0.2–0.6) | 0.5 (0.3–1.3) | 0.3 (0.2–0.5) | <0.001 |
| ALT, U/L | 24.0 (20.0–31.0) | 23.0 (19.0–32.0) | 23.0 (21.0–30.0) | 0.146 |
| AST, U/L | 26.0 (19.0–37.0) | 27.0 (19.0–37.0) | 26.0 (19.0–36.0) | 0.406 |
| LDH, U/L | 392.5 (342.7–483.4) | 451.8 (385.4–655.1) | 381.9 (339.2–451.5) | <0.001 |
| Cr, umol/L | 29.5 (23.9–37.6) | 30.2 (24.5–37.8) | 29.3 (23.8–37.5) | 0.213 |
| hs-TnT, pg/mL | 15.6 (13.9–17.8) | 18.2 (15.1–24.1) | 15.1 (13.5–16.6) | <0.001 |
| D-dimer, ug/mL | 0.6 (0.4–0.8) | 0.6 (0.4–0.9) | 0.5 (0.4–0.8) | 0.006 |
| IL-6, pg/mL | 14.2 (9.9–24.1) | 20.4 (12.7–42.9) | 12.4 (9.4–21.3) | <0.001 |
| IL-10, pg/mL | 15.4 (11.6–21.3) | 21.6 (15.8–27.1) | 13.8 (10.9–18.6) | <0.001 |
| TNF-α, pg/mL | 20.1 (15.8–25.0) | 24.2 (20.7–29.7) | 18.3 (15.3–23.6) | <0.001 |
| IFN-γ, pg/mL | 11.1 (9.0–12.5) | 11.3 (8.9–12.8) | 11.0 (9.1–12.4) | 0.617 |
| Fibrinogen, g/L | 6.8 (6.4–7.6) | 7.1 (6.7–8.0) | 6.8 (6.3–7.5) | <0.001 |
| Co-infection, n (%) | 97 (18.4) | 25 (20.5) | 72 (17.8) | 0.498 |
ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; Cr, creatinine; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; hs-TnT, high-sensitivity troponin-T; IFN-γ, interferon-γ; IL-6, interleukin 6; IL-10, interleukin 10; IQR, interquartile ranges; LDH, lactate dehydrogenase; NLR, neutrophil-lymphocyte ratio; PCT, procalcitonin; TNF-α, tumor necrosis factor-α.
Comparison between the two groups was performed based on demographic, clinical, and laboratory characteristics. Compared to the general group, the severe group had a younger median age (4.2 vs. 5.3 years, P < 0.001), and showed greater proportions of underlying conditions (15.6 % vs. 2.5 %, P < 0.001) as well as prematurity history (24.6 % vs. 4.9 %, P < 0.001). In terms of clinical symptoms, significantly longer fever duration (5.0 vs. 3.0 days, P < 0.001), greater fever peak (39.1 vs. 39.0 °C, P = 0.003), and higher incidences of wheezing (51.6 % vs 22.7 %, P < 0.001), sore throat (26.2 % vs. 16.5 %, P = 0.016), poor appetite (62.3 % vs. 27.9 %, P < 0.001), abdominal pain (16.4 % vs. 4.9 %, P < 0.001) were observed in the severe group than the general group. Besides, marked differences regarding laboratory parameters were also identified among the two groups, including elevated leukocyte count, NLR, ESR, CRP, PCT, LDH, hs-TnT, D-dimer, IL-6, IL-10, TNF-α, fibrinogen, and declined CD4+/CD8+ T cell ratio as well as hemoglobin in the severe group (all P < 0.05, Table 1).
3.2. Identification of candidate variables
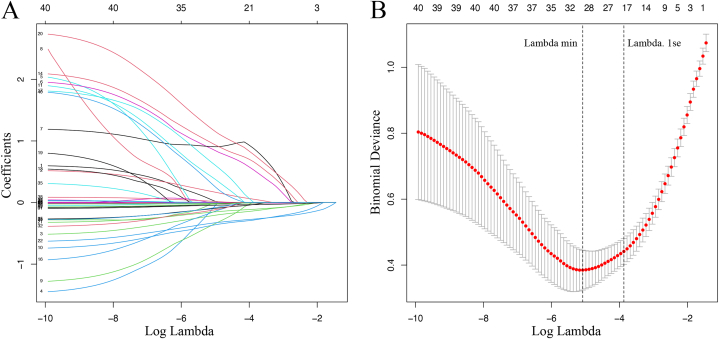
Forty variables (Table 1) were entered into the LASSO regression. After LASSO regression based on lambda.1se, 19 variables with non-zero coefficients were chosen, including age, BMI, underlying conditions, prematurity, duration of fever, wheezing, poor appetite, abdominal pain, leukocyte count, NLR, hemoglobin, ESR, CRP, PCT, AST, LDH, hs-TnT, IL-10, and TNF-α (Fig. 2A and B).
Fig. 2.
Selection of candidate predictors by LASSO regression. (A) LASSO coefficient profiles of these 40 potential predictive variables. (B) Tuning parameter (lambda) selection in the LASSO model using ten-fold cross-validation via one standard error of the minimum criteria (lambda.1se). Nineteen candidate variables with non-zero coefficients were finally outputted. LASSO, Least Absolute Shrinkage and Selection Operator.
3.3. Development of the prediction nomogram
The inclusion of these above 19 candidate variables into a logistic regression model outputted 11 variables which were independently associated with severe H1N1 infection. Specifically, these 11 risk factors were underlying conditions (OR = 5.231, 95%CI: 1.443–18.965), prematurity (OR = 4.087, 95%CI: 1.199–13.936), fever duration (OR = 1.786, 95%CI: 1.358–2.348), wheezing (OR = 7.157, 95%CI: 2.512–20.393), poor appetite (OR = 14.517, 95%CI: 4.808–43.831), leukocyte count (OR = 1.310, 95%CI: 1.148–1.496), NLR (OR = 1.588, 95%CI:1.361–1.853), ESR (OR = 1.072, 95%CI: 1.042–1.103), LDH (OR = 1.005, 95%CI: 1.003–1.006), IL-10 (OR = 1.056, 95%CI: 1.031–1.081), and TNF-α (OR = 1.069, 95%CI: 1.036–1.104), respectively (Fig. 3A). A prediction model was established by integrating the above 11 independent predictors and visualized as a nomogram (Fig. 3B). The equation of this model was generated as follows:
Fig. 3.
Construction and visualization of prediction model. (A) Eleven independent risk factors for severe H1N1 infection were identified by the logistic regression. (B) A prediction model integrating these 11 independent predictors was constructed and visualized as a nomogram.
Logit (P) = −15.625 + 1.655 × Underlying conditions (yes) + 1.408 × Prematurity (yes) + 0.58 × Fever duration (d) + 1.968 × Wheezing (yes) + 2.675 × Poor appetite (yes) + 0.27 × Leukocyte count ( × 109/L) + 0.463 × NLR + 0.069 × ESR (mm/H) + 0.005 × LDH (U/L) + 0.054 × IL-10 (pg/mL) + 0.067 × TNF-α (pg/mL).
3.4. Evaluation of the nomogram
The ROC curves demonstrated that this nomogram achieved an AUC of 0.965 (specificity: 93.8 %; sensitivity: 91.4 %) in the training set (Fig. 4A) as well as an AUC of 0.981 (specificity: 93.1 %; sensitivity: 96.6 %) in the internal validation set (Fig. 4B), indicating an excellent discrimination ability. Meanwhile, in the calibration curves of the nomogram in both training (Fig. 4C) and internal validation sets (Fig. 4D), the bias-corrected curves were found to be quite close to the 45° diagonal (Hosmer-Lemeshow test P = 0.731 for training set and P = 0.545 for validation set), indicating satisfactory consistency between the model-predicted probability and observed probability of severe H1N1 infection. Besides, DCA was performed to evaluate the clinical usefulness of this nomogram. As shown in the decision curves, threshold probability was denoted by the abscissa, with the net benefit as the ordinate. When the threshold probability was above 0.07 in the training set (Fig. 4E) and above 0.03 in the internal validation set (Fig. 4F), the nomogram provided a higher net benefit than the “all” and “none” schemes, demonstrating that the nomogram was clinically useful.
Fig. 4.
Performance evaluation of the prediction nomogram. (A) and (B) ROC curves of the nomogram in training set and internal validation set, respectively. (C) and (D) Calibration curves of the nomogram in training and internal validation sets, respectively. (E) and (F) DCA of the nomogram in training and internal validation sets, respectively. ROC, receiver operating characteristic; DCA, decision curve analysis.
3.5. External validation of the nomogram
The overall external validation dataset (Table 2), including four subsets (Wenshan, Baoshan, Lincang, Dali), consisted of 352 patients, 29 (8.2 %) of whom were accompanied by at least one underlying condition, 36 (10.2 %) had history of prematurity, 104 (29.5 %) had wheezing, and 122 (34.7 %) had poor appetite. The median (IQR) fever duration, leukocyte count, NLR, ESR, LDH, IL-10, as well as TNF-α of the external validation dataset were 3.0 (2.0–4.0) days, 11.2 (9.5–14.2) × 109/L, 4.2 (3.0–7.3), 19.0 (15.0–25.8) mm/H, 399.4 (363.8–448.8) U/L, 16.4 (11.6–21.7) pg/mL, and 19.5 (15.6–25.2) pg/mL, respectively. Severe H1N1 infection was eventually confirmed in 72 (20.5 %) of these patients. The nomogram was assessed externally based on the external validation dataset and its subsets. The AUC values of the nomogram were 0.976 in Wenshan subset, 0.870 in Baoshan subset, 0.929 in Lincang subset, 0.979 in Dali subset, and 0.936 in the overall validation dataset (Fig. 5A), respectively, revealing excellent discrimination. The calibration curves of the nomogram for the overall validation dataset and its four subsets showed favorable consistency between the predicted and observed outcomes (all Hosmer-Lemeshow P > 0.05) (Fig. 5B). Furthermore, based on the DCA conducted in the validation dataset and its subsets, the net benefits acquired from the use of this model across a broad range of threshold probabilities were found to be superior to those derived from “all” and “none” schemes (Fig. 5C), suggesting that the nomogram could offer important clinical value in the prediction of severe H1N1 infection.
Table 2.
Characteristics of subjects in the external validation dataset.
| Characteristics | Total (Overall dataset) (n = 352) | Wenshan subset (n = 82) | Baoshan subset (n = 150) | Lincang subset (n = 61) | Dali subset (n = 59) |
|---|---|---|---|---|---|
| Severe case, n (%) | 72 (20.5) | 21 (25.6) | 26 (17.3) | 13 (21.3) | 12 (20.3) |
| Underlying disease, n (%) | 29 (8.2) | 9 (11.0) | 8 (5.3) | 5 (8.2) | 7 (11.9) |
| Prematurity, n (%) | 36 (10.2) | 9 (11.0) | 15 (10.0) | 6 (9.8) | 6 (10.2) |
| Duration of fever, d, median (IQR) | 3.0 (2.0–4.0) | 3.0 (2.0–5.0) | 3.0 (2.0–3.0) | 3.0 (2.0–4.0) | 2.0 (2.0–4.0) |
| Wheezing, n (%) | 104 (29.5) | 24 (29.3) | 52 (34.7) | 15 (24.6) | 13 (22.0) |
| Poor appetite, n (%) | 122 (34.7) | 21 (25.6) | 57 (38.0) | 23 (37.7) | 21 (35.6) |
| Leukocyte count, × 109/L, median (IQR) | 11.2 (9.5–14.2) | 12.5 (9.8–17.1) | 10.9 (8.8–13.3) | 11.7 (10.1–14.1) | 10.6 (9.5–14.2) |
| NLR, median (IQR) | 4.2 (3.0–7.3) | 4.2 (2.9–7.6) | 4.4 (3.3–7.5) | 3.8 (2.8–6.2) | 4.0 (2.9–7.0) |
| ESR, mm/H, median (IQR) | 19.0 (15.0–25.8) | 21.0 (17.0–29.5) | 17.0 (13.0–25.0) | 18.0 (16.0–24.0) | 19.0 (15.0–27.0) |
| LDH, U/L, median (IQR) | 399.4 (363.8–448.8) | 400.3 (372.0–429.6) | 410.8 (366.2–479.2) | 395.3 (366.7–414.7) | 395.0 (331.6–419.5) |
| IL-10, pg/mL, median (IQR) | 16.4 (11.6–21.7) | 15.8 (10.3–21.5) | 16.9 (10.4–24.3) | 14.5 (10.4–17.9) | 17.0 (14.1–21.5) |
| TNF-α, pg/mL, median (IQR) | 19.5 (15.6–25.2) | 20.4 (15.0–26.6) | 20.1 (16.4–25.4) | 16.7 (12.5–20.3) | 19.5 (15.6–23.4) |
ESR, erythrocyte sedimentation rate; IL-10, interleukin 10; IQR, interquartile ranges; LDH, lactate dehydrogenase; NLR, neutrophil-lymphocyte ratio; TNF-α, tumor necrosis factor-α.
Fig. 5.
External validation of the nomogram. ROC curves (A), calibration curves (B), and DCA (C) of the nomogram in the Wenshan subset, Baoshan subset, Lincang subset, Dali subset, as well as the overall external validation dataset, respectively. ROC, receiver operating characteristic; DCA, decision curve analysis.
4. Discussion
This research established and validated a straightforward yet potent nomogram (including 11 common variables) for predicting the risk of occurrence of severe pediatric H1N1 infection during the post-COVID-19 period. This model showed great potentials for clinical application due to its satisfactory performance confirmed by comprehensive assessments.
As described earlier, underlying conditions, history of prematurity, fever duration, wheezing, poor appetite, leukocyte count, NLR, ESR, LDH, IL-10, and TNF-α were integrated into our prediction nomogram. Several of these variables have been previously reported to be the risk factors for severe cases linked with classical influenza infection in the past. It is widely acknowledged that children with a prior history of preterm birth and/or certain underlying diseases are deemed to be at an increased risk for severe progression of influenza infection, irrespective of additional physiological and pathophysiological conditions [14,[21], [22], [23], [24]]. Meanwhile, there is already evidence that poor appetite can further aggravate clinical outcomes by weakening the body's immune system, and persistent high fever are closely associated to neurological damage as well as worse outcomes in individuals with severe viral infection [[25], [26], [27]]. Raised leukocyte count, LDH, and inflammatory markers (ESR, IL-10, TNF-α, etc.) were also commonly used as the outcome measures representing the severity of infectious disorders, including influenza infection [[28], [29], [30]].
Noteworthy, differed from other common indicators in some previous prediction models for classical influenza infection in the pre-COVID-19 era, wheezing and NLR were identified as valuable predictors and were introduced into our risk model. As we described earlier, the latest wave of Omicron variant in China, spanning from December 2022 to February 2023, resulted in a huge proportion of the Chinese population (approximately 82 %) having a history of SARS-CoV-2 infection [6]. Substantial evidence has suggested a higher risk of long-term abnormalities in the respiratory tract in patients who recovered from acute SARS-CoV-2 infection compared to the general population [9,31,32]. The ultra-high rate of prior SARS-CoV-2 infection in the population and its persistent pathophysiological impact on the respiratory tract perhaps partially explain why wheezing has become a more prominent clinical feature in the progression of H1N1 infection during the post-pandemic era of COVID-19 than the classical seasonal H1N1 infection in the past [33]. As for NLR, it is a relatively emerging hematological biomarker of systemic inflammation and stress with a short history of invention. Unlike the total white blood cell count or the counts of its various subsets, all of which presented limited specificity and sensitivity in assessing inflammation and the severity of disease, NLR is gaining increasing recognition as a simple, potent, and dependable indicator of immune-inflammatory response's intensity [34]. It is progressively being incorporated as a key metric for gauging the severity of infections. In the present study, NLR was also identified as one of the most vital components of our nomogram and played an important role in predicting serious pediatric H1N1 infection during the post-COVID-19 era.
Additionally, age had been consistently acknowledged as a pivotal factor influencing the progress of influenza infection. For traditional influenza in the pre-COVID-19 period, pediatric cases within the first two years of life are more prone to progress to severe illnesses, while the hospitalization rate due to influenza infection for children aged ≥5 years does not exceed 50 per 100,000 and gradually decreases with advancing age [35,36]. However, according to our calculation, age did not appear to be an independent risk factor for severe H1N1 infection in this study. The median age of severe and general groups as well as the total population in our development dataset was 4.2, 5.3, and 5.2 years, respectively, all of which were around 5 years, suggesting a trend of increasing age among the pediatric H1N1 patients in the post-COVID-19 era compared to the pre-COVID-19 era [28]. In the post-COVID-19 era, the age structure of various respiratory tract infections in children seems to be undergoing changes. For instance, the median age of children hospitalized with RSV infection in Western Australia increased from 8.1 months in the pre-COVID-19 era to 16.4 months in the post-pandemic period [37]. This phenomenon may be partially attributed to the different extent of impact of immune debt on children in different age groups, with older children appearing to be more profoundly affected. For example, a recent study confirmed that influenced by immune debt, the levels of RSV antibodies in children in post-COVID-19 period have prominently decreased compared to that in pre-pandemic period, especially for older children (>3 years old), where the decline was most significant [38]. A similar mechanism may also exist in the pediatric H1N1 infection. This suggests that previous feature of age and its relationship with disease severity in children with classical H1N1 infection in the past might be not fully applicable to the pediatric H1N1 cases during the post-COVID-19 era. Clinicians should pay heightened attention to this significant change, avoiding missed diagnosis and misdiagnosis.
In this study, 11 variables, including underlying conditions, prematurity, fever duration, wheezing, poor appetite, leukocyte count, NLR, ESR, LDH, IL-10, and TNF-α were identified as the influencing factors independently associated with the occurrence of severe H1N1 infection in post-COVID-19 era, and we further integrated these 11 factors into a nomogram, where each independent risk factor was transformed into a scoring system, thus providing an intuitive way to interpret our prediction model. An important and attractive feature of the nomogram is that this model was composed of highly common clinical and laboratory characteristics, and all 11 variables are relatively easy to obtain in many healthcare institutions. Meanwhile, we conducted internal and external validations to verify the performance and clinical value of this prediction nomogram. Encouragingly, the eventual results suggested great discrimination, accuracy, and clinical usefulness of the model. In the post-COVID-19 era, this innovative predictive model may offer substantial benefits for the clinical handling of pediatric patients with H1N1 infection, particularly considering the heightened severity and a surge in the incidence of such patients compared to classical influenza infection prior to the COVID-19 pandemic [5]. The threat of H1N1 infection to children's health during the post-COVID-19 period deserves special clinical attention, and the early identification and prevention of severe cases are crucial for improving prognosis of such patients. As the first department of Pediatric Pulmonary and Critical Care Medicine (PCCM) in China, our team has been committed to combination of pediatric respiratory medicine and critical care medicine, both in aspects of clinical management and scientific research, working for the best interests of patients and their family. Based on our nomogram, early identification of potential severe cases among children hospitalized for H1N1 infection is achievable, aiding clinicians in decision-making. For patients estimated to be at high risk, clinicians may choose aggressive therapy or transfer them to intensive care unit (ICU), while others with non‐high risk might be administered with conventionally supportive treatment and monitoring. This can facilitate the efficient allocation of medical resources based on local or regional conditions during the H1N1 surge, particularly in areas with limited resources and/or a high prevalence of H1N1, where clinicians can allocate limited resources to severe patients with more emergent needs, maximizing the availability and value of ICU beds and ventilators.
Several potential limitations of this study warrant consideration. With the retrospective design of the study, there is likely recording, and selection bias associated with data collection. Meanwhile, two of predictors (IL-10 and TNF-α) in our nomogram might not be routinely determined in some primary hospitals. Besides, despite the multicenter nature, the sample size was relatively small, and subjects were entirely from Yunnan, which might limit the generalizability of the prediction model in other parts of China. Future studies with greater closeness to clinical practice in hospitals of all levels should include larger cohorts from areas outside Yunnan using a prospective multicenter design.
5. Conclusions
A novel nomogram integrating underlying conditions, prematurity, fever duration, wheezing, poor appetite, leukocyte count, NLR, ESR, LDH, IL-10, and TNF-α was developed to predict the occurrence of severe case among children hospitalized with H1N1 infection in the post-COVID-19 era. The internal and external validations demonstrated the good predictive performance of this nomogram, which may have great potential for application in clinical work in the future.
Data availability statement
Data will be made available on request.
CRediT authorship contribution statement
Hai-Feng Liu: Writing – original draft, Software, Project administration, Methodology, Funding acquisition, Conceptualization. Xiao-Zhong Hu: Writing – original draft, Validation, Investigation, Conceptualization. Cong-Yun Liu: Validation, Methodology, Investigation, Formal analysis. Zheng-Hong Guo: Methodology, Investigation, Formal analysis. Rui Lu: Validation, Investigation, Formal analysis. Mei Xiang: Visualization, Investigation, Formal analysis. Ya-Yu Wang: Validation, Investigation, Formal analysis. Zhao-Qing Yin: Investigation, Formal analysis, Data curation. Min Wang: Visualization, Investigation, Formal analysis. Ming-Ze Sui: Software, Data curation. Jia-Wu Yang: Investigation, Formal analysis. Hong-Min Fu: Writing – review & editing, Supervision, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.World Health Organization Vaccines against influenza WHO position paper – november 2012. Wkly. Epidemiol. Rec. 2012;87(47):461–476. [PubMed] [Google Scholar]
- 2.Shen K.L., Namazova-Baranova L., Yang Y.H., Wong G.W.K., Rosenwasser L.J., Rodewald L.E., et al. Global Pediatric Pulmonology Alliance recommendation to strengthen prevention of pediatric seasonal influenza under COVID-19 pandemic. World J Pediatr. 2020;16(5):433–437. doi: 10.1007/s12519-020-00389-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X., Li Y., O'Brien K.L., Madhi S.A., Widdowson M.A., Byass P., et al. Global burden of respiratory infections associated with seasonal influenza in children under 5 years in 2018: a systematic review and modelling study. Lancet Global Health. 2020;8(4):e497–e510. doi: 10.1016/s2214-109x(19)30545-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen R., Pettoello-Mantovani M., Somekh E., Levy C. European pediatric societies call for an implementation of regular vaccination programs to contrast the immunity debt associated to coronavirus disease-2019 pandemic in children. J. Pediatr. 2022;242 doi: 10.1016/j.jpeds.2021.11.061. 260-1.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoy G., Maier H.E., Kuan G., Sánchez N., López R., Meyers A., et al. Increased influenza severity in children in the wake of SARS-CoV-2. Influenza Other Respir Viruses. 2023;17(7) doi: 10.1111/irv.13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu D., He G., Li H., Tan H., Ji X., Lin Z., et al. Effectiveness of COVID-19 vaccination against SARS-CoV-2 omicron variant infection and symptoms - China, December 2022-February 2023. China CDC Wkly. 2023;5(17):369–373. doi: 10.46234/ccdcw2023.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wulf Hanson S., Abbafati C., Aerts J.G., Al-Aly Z., Ashbaugh C., Ballouz T., et al. Estimated global proportions of individuals with persistent fatigue, cognitive, and respiratory symptom clusters following symptomatic COVID-19 in 2020 and 2021. JAMA. 2022;328(16):1604–1615. doi: 10.1001/jama.2022.18931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao X.H., He Z.C., Li T.Y., Zhang H.R., Wang Y., Mou H., et al. Pathological evidence for residual SARS-CoV-2 in pulmonary tissues of a ready-for-discharge patient. Cell Res. 2020;30(6):541–543. doi: 10.1038/s41422-020-0318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carfì A., Bernabei R., Landi F. Persistent symptoms in patients after acute COVID-19. JAMA. 2020;324(6):603–605. doi: 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu X., Wang S., Zhang W., Li C., Guo L., Wang Z., et al. Probing long COVID through a proteomic lens: a comprehensive two-year longitudinal cohort study of hospitalised survivors. EBioMedicine. 2023;98 doi: 10.1016/j.ebiom.2023.104851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peluso M.J., Deitchman A.N., Torres L., Iyer N.S., Munter S.E., Nixon C.C., et al. Long-term SARS-CoV-2-specific immune and inflammatory responses in individuals recovering from COVID-19 with and without post-acute symptoms. Cell Rep. 2021;36(6) doi: 10.1016/j.celrep.2021.109518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halasa N.B. Update on the 2009 pandemic influenza A H1N1 in children. Curr. Opin. Pediatr. 2010;22(1):83–87. doi: 10.1097/MOP.0b013e3283350317. [DOI] [PubMed] [Google Scholar]
- 13.Hernández-Bou S., Novell C.B., Alins J.G., García-García J.J. Hospitalized children with influenza A H1N1 (2009) infection: a Spanish multicenter study. Pediatr. Emerg. Care. 2013;29(1):49–52. doi: 10.1097/PEC.0b013e31827b528f. [DOI] [PubMed] [Google Scholar]
- 14.Morgan C.I., Hobson M.J., Seger B., Rice M.A., Staat M.A., Wheeler D.S. 2009 pandemic influenza A (H1N1) in critically ill children in Cincinnati, Ohio. Pediatr. Crit. Care Med. 2012;13(3):e140–e144. doi: 10.1097/PCC.0b013e318228845f. [DOI] [PubMed] [Google Scholar]
- 15.Fossum E., Rohringer A., Aune T., Rydland K.M., Bragstad K., Hungnes O. Antigenic drift and immunity gap explain reduction in protective responses against influenza A(H1N1)pdm09 and A(H3N2) viruses during the COVID-19 pandemic: a cross-sectional study of human sera collected in 2019, 2021, 2022, and 2023. Virol. J. 2024;21(1):57. doi: 10.1186/s12985-024-02326-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain A., Mahesh S., Prakash O., Khan D.N., Verma A.K., Rastogi Y. Effect of COVID-19 pandemic on influenza; observation of a tertiary level virology laboratory. Virusdisease. 2024;35(1):27–33. doi: 10.1007/s13337-024-00860-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin F., Chen M.T., Zhang L., Wu M., Xie H., Guan Z.X., et al. Resurgence of influenza A after SARS-CoV-2 omicron wave and comparative analysis of hospitalized children with COVID-19 and influenza A virus infection. Front. Med. 2024;10 doi: 10.3389/fmed.2023.1289487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar V. Influenza in children. Indian J. Pediatr. 2017;84(2):139–143. doi: 10.1007/s12098-016-2232-x. [DOI] [PubMed] [Google Scholar]
- 19.Gaitonde D.Y., Moore F.C., Morgan M.K. Influenza: diagnosis and treatment. Am. Fam. Physician. 2019;100(12):751–758. [PubMed] [Google Scholar]
- 20.World Health Organization . World Health Organization; Geneva: 2010. WHO Guidelines Approved by the Guidelines Review Committee. WHO Guidelines for Pharmacological Management of Pandemic Influenza A(H1N1) 2009 and Other Influenza Viruses. [PubMed] [Google Scholar]
- 21.Mikić D., Nozić D., Kojić M., Popović S., Hristović D., Dimitrijević R.R., et al. Clinical manifestations, therapy and outcome of pandemic influenza A (H1N1) 2009 in hospitalized patients. Vojnosanit. Pregl. 2011;68(3):248–256. doi: 10.2298/vsp1103248m. [DOI] [PubMed] [Google Scholar]
- 22.Gill P.J., Ashdown H.F., Wang K., Heneghan C., Roberts N.W., Harnden A., et al. Identification of children at risk of influenza-related complications in primary and ambulatory care: a systematic review and meta-analysis. Lancet Respir. Med. 2015;3(2):139–149. doi: 10.1016/s2213-2600(14)70252-8. [DOI] [PubMed] [Google Scholar]
- 23.Principi N., Esposito S. Severe influenza in children: incidence and risk factors. Expert Rev. Anti Infect. Ther. 2016;14(10):961–968. doi: 10.1080/14787210.2016.1227701. [DOI] [PubMed] [Google Scholar]
- 24.Centers for Disease Control and Prevention (CDC) Update: influenza-associated deaths reported among children aged <18 years--United States, 2003-04 influenza season. MMWR Morb. Mortal. Wkly. Rep. 2004;52(53):1286–1288. [PubMed] [Google Scholar]
- 25.Walsh T.S., Salisbury L.G., Boyd J., Ramsay P., Merriweather J., Huby G., et al. A randomised controlled trial evaluating a rehabilitation complex intervention for patients following intensive care discharge: the RECOVER study. BMJ Open. 2012;2(4) doi: 10.1136/bmjopen-2012-001475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polderman K.H. Induced hypothermia and fever control for prevention and treatment of neurological injuries. Lancet. 2008;371(9628):1955–1969. doi: 10.1016/s0140-6736(08)60837-5. [DOI] [PubMed] [Google Scholar]
- 27.Launey Y., Nesseler N., Mallédant Y., Seguin P. Clinical review: fever in septic ICU patients--friend or foe? Crit. Care. 2011;15(3):222. doi: 10.1186/cc10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y., Chen W., Zeng M., Shen G., Sun C., Liu G., et al. Clinical features and risk factors for severe influenza in children: a study from multiple hospitals in Shanghai. Pediatr Neonatol. 2021;62(4):428–436. doi: 10.1016/j.pedneo.2021.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Hofto M.E., Schmit E.O., Sharma M., Samuy N. Acute appendicitis associated with multisystem inflammatory syndrome in children. Cureus. 2021;13(6) doi: 10.7759/cureus.15893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joli J., Buck P., Zipfel S., Stengel A. Post-COVID-19 fatigue: a systematic review. Front. Psychiatr. 2022;13 doi: 10.3389/fpsyt.2022.947973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lippi G., Sanchis-Gomar F., Henry B.M. COVID-19 and its long-term sequelae: what do we know in 2023? Pol. Arch. Intern. Med. 2023;133(4) doi: 10.20452/pamw.16402. [DOI] [PubMed] [Google Scholar]
- 33.Jiang M.Y., Duan Y.P., Tong X.L., Huang Q.R., Jia M.M., Yang W.Z., et al. Clinical manifestations of respiratory syncytial virus infection and the risk of wheezing and recurrent wheezing illness: a systematic review and meta-analysis. World J Pediatr. 2023;19(11):1030–1040. doi: 10.1007/s12519-023-00743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zahorec R. Neutrophil-to-lymphocyte ratio, past, present and future perspectives. Bratisl. Lek. Listy. 2021;122(7):474–488. doi: 10.4149/bll_2021_078. [DOI] [PubMed] [Google Scholar]
- 35.Silvennoinen H., Peltola V., Vainionpää R., Ruuskanen O., Heikkinen T. Incidence of influenza-related hospitalizations in different age groups of children in Finland: a 16-year study. Pediatr. Infect. Dis. J. 2011;30(2):e24–e28. doi: 10.1097/inf.0b013e3181fe37c8. [DOI] [PubMed] [Google Scholar]
- 36.Thompson W.W., Shay D.K., Weintraub E., Brammer L., Cox N., Anderson L.J., et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289(2):179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 37.Foley D.A., Phuong L.K., Peplinski J., Lim S.M., Lee W.H., Farhat A., et al. Examining the interseasonal resurgence of respiratory syncytial virus in Western Australia. Arch. Dis. Child. 2022;107(3) doi: 10.1136/archdischild-2021-322507. [DOI] [PubMed] [Google Scholar]
- 38.Jiang W., Xu L., Wang Y., Hao C. Exploring immunity debt: dynamic alterations in RSV antibody levels in children under 5 years during the COVID-19 pandemic. J. Infect. 2024;88(1):53–56. doi: 10.1016/j.jinf.2023.10.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.