Abstract
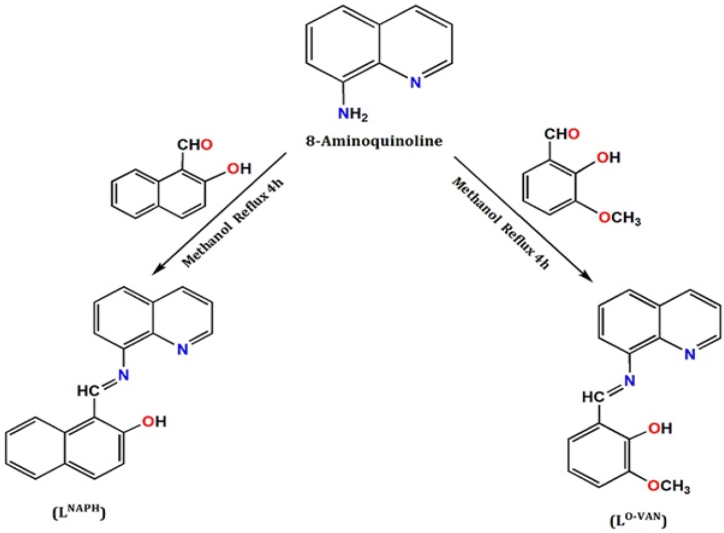
The current research divulges the synthesis of two new Schiff base (SB) (LNAPH/LO-VAN) derived from 8-aminoquinoline (8-AMQ) in the presence of 2-hydroxy naphthaldehyde (NAPH) and ortho-vanillin (O-VAN) in CH3OH solvent. They are structurally characterized by spectroscopic methods (IR/Raman/UV–vis/DRS/NMR) and SEM-EDX. SB compounds have a biologically active avenue of azomethine/imine group (H–C=N) that can donate N e's to Mn + ions, showing coordinating flexibility. The –OH and imine (H–C=N) groups are stable in air, light, and alkalis but undergo acidic environments hydrolysis, separating –NH2 and carbonyl compounds. Moreover, buffer solutions with a pH range of 4–6 release aldehyde. Molecular electrostatic potential (MEP), Frontier molecular orbitals (FMO), Fukui function, and Non-linear optical (NLO) were conducted to elucidate SBs chemical potency, optoelectronic significance, and corrosion inhibitor. Accordingly, the calculated ΔE of FMO for LNAPH and LO-VAN is 3.82 and 4.08 eV, ensuring potent biological function. DFT supported the experimental and theoretical IR spectral correlation to enrich better structural insights. NLO-based polarizability (α) and hyperpolarizability (β) factors successfully explore the potential optoelectronic significance. Molecular docking experiments were simulated against DNA, anti-COVID-19, and E. coli. The potential microbiological activity was screened against the bacterial strains E. coli, Klebsiella, Bacillus, and Pseudomonas sp. based on zone of inhibition and MIC values. These experiments also explored the fact that LNAPH and LO-VAN discourage microbial cell biofilms and corrosion. We extensively covered the as-prepared compounds' pH-dependent bacterial effects.
Keywords: Schiff base, DFT, Corrosion inhibitor, molecular docking, pH-dependent antibacterial
Graphical abstract
Highlights
-
•
LNAPH and LO-VAN ligands were synthesized from 8-aminoquinoline and characterized.
-
•
Molecular geometry was optimized using the B3LYP/6–311++G (d, p) level of theory.
-
•
FMO, MEP, Fukui function, corrosion, and NLO properties were analysed.
-
•
MD was performed against E. coli./anti-COVID-19/DNA to ensure antibacterial potency.
-
•
PH-dependent antibacterial was conducted in favour of LNAPH/LO-VAN.
1. Introduction
Contemporary research on coordination chemistry is hugely beneficial in modern times, particularly in understanding the multifaceted role of Schiff base (SB) [1,2]. Due to SB's modular synthesis and bridging capabilities, denticity, steric, and electronic properties are now called "privileged ligands" [3]. Hugo Schiff first developed the SB synthetic platforms in 1864 [1,2] and constituted a diverse group of substances containing an azomethine or imine group (H–C=N-) [4]. Generally, a nucleophilic (Nu) reaction involving a primary amine and an R-substituted carbonyl group characterizes the synthetic results of SB (Schem S1) [[5], [6], [7], [8]]. The R group improved the number of SB functional activities. Concerning the synthetic aspect, a few promising candidates for the novel synthesis of SBs are 8-aminoquinoline (8-AMQ), 2-hydroxy naphthaldehyde (NAPH), and ortho-vanillin (O-VAN). These blocking ligands are not encountered in today's literature [[9], [10], [11]]. Scientists' efforts on synthesis and outcomes related to SB have notably increased. Despite its limitless potential, research is thoroughly explored in numerous areas. The imine group (H–C=N-) is employed to ascertain the mechanism of racemization reaction transformation in biological systems [[12], [13], [14], [15]]. Due to the active imine functional group, certain bioactive SBs exhibit anticancer, antibacterial, antifungal, herbicidal, and other properties in biological systems. They are significant in medicinal chemistry [16] (Scheme S2). SBs exhibit thermochromic, photochromic, and thermochromic properties in the solid state [17]. The strong antiviral, antifungal, and antibacterial qualities make SB derivatives valuable medicinal products [16,18,19]. They are also commonly used as model substances to understand the structure-activity relationship (SAR) between pharmaceuticals and biomolecules [[20], [21], [22]]. Sriram et al. [23] have shown that antiviral prodrugs modified with SBs are incredibly effective against mouse hepatitis viruses at inadequate doses (EC50 of 1.6 μM). Several recent studies have investigated the function of SB bacteria. Some germs cause disease by damaging tissues, while others use toxins [17]. S. aureus is one of the most prevalent pathogens of infectious agents obtained from hospitals and the community [17]. However, researchers should pay little attention to SB chemistry in DFT investigations, including FMO, MEP, corrosion activities, NLO, Fukui function, and molecular docking against DNA binding, anti-COVID-19, and E. coli [2,24,25]. Research on experimental and theoretical correlation in the DFT has recently established SB's distinctive biological properties [[26], [27], [28], [29]]. Meanwhile over the past few years, DFT has offered valuable insights into the study of chemical and biological systems [[30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40]]. NLO materials can be used in telecom through intramolecular charge transfer, electron delocalisation, and optical communication technology [[41], [42], [43]]. A corrosion-fighting electroactive imine group helps SBs adsorb onto substrate metals. Aromaticity/multiple bonds and hetero atoms/functional groups can boost this effectiveness by enhancing electrical contact with Fe's d-orbital. Therefore, Schiff bases work as corrosion inhibitors [44].
Considering all the precedence significance of the SBs, our research attempts to comprehend compound (LNAPH/LO-VAN) DFT-based reactivity and establish antibacterial properties. Spectroscopic methods and SEM-EDX have characterized the synthesized compounds. The quantum chemical parameters were evaluated using DFT to determine the correlation between the compounds' biological activity and molecular structures. The ligand's reactivity was checked using FMO, MEP, Fukui function, and NLO properties based on α and β values. The correlation between experimental and theoretical IR provides improved structural insights for SB. Molecular docking investigation against DNA, E. coli, and anti-COVID-19 confirmed biological potency. Docking aimed to explore the potential broad-spectrum efficacy of SB's diverse biological targets. SB ligands inhibit biofilm development and microbial-induced corrosion. We also investigated pH-dependent bacterial impacts on the synthesized ligands.
2. Experimental
2.1. Materials and instrumentation
The chemicals used in the research, such as CH3OH, ortho-vanillin (O-VAN), 8-aminoquinoline (8-AMQ), and 2-hydroxy naphthaldehyde (NAPH), are of reagent grade (RG) quality. The PerkinElmer instrument measures the elemental C, H, and N. PerkinElmer Spectrum, specifically the RX 1 instrument, analyses IR spectra. The Bruker RFS 27 model obtains Raman spectra in KBr pellets (4000-50 cm−1). NMR spectra in DMSO‑d6 have been received using a Bruker FT-NMR spectrometer (75.45 MHz–400 MHz). The Oxford XMX N model collected EDX spectrum data, while the JEOL (JSM-6390LV) and BRUKER AXS visualized the SEM figures. U-3501 spectrophotometer model is used to analyse UV–visible absorption spectra. We used the PerkinElmer Lambda 365 model for diffuse reflectance spectroscopy (DRS), with a DRS integrating sphere (50 mm diameter, 8° angle).
2.2. Broth dilution: Bacterial cultures
The efficiency of LNAPH, LO-VAN, NAPH, 8-AMQ, and O-VAN against four clinical pathogens, namely E. coli, P. aeruginosa, Klebsiella sp., and Bacillus sp. was analysed based on the BDT (Broth dilution technique). All bacterial strains for research are maintained at REVA University's Department of Biotechnology in Bengaluru. Microbial cultures grown overnight and pathogenic were utilized for the experimental analysis. The broth dilution technique determines the antibacterial effect of the samples and optimizes their concentration. The above test would also help assess the samples' capacity to avoid MIC (microbial-induced corrosion). The pathogens were mixed with L-B M (Luria-Bertani medium) and suspended. It was then incubated with compounds under shaking conditions for 24 h. OD (optical density) was measured with an ELISA reader to check bacterial growth inhibition after incubation (Biotek-elx800). The inhibition of pathogenic strains by the components is correlated with the efficacy of avoiding microbial-induced corrosion. A bacterial suspension was kept without the addition of any antibacterial material as a control. The OD of the control was compared to that of the test samples to calculate the bacterial inhibition percentage (%). An antibacterial assay was done thrice (n = 3), and the mean and SD presented the reported experimental data [45]. The abovementioned experiment was conducted at pH 4, pH 7, and pH 9 to analyse the impact of pH on the study.
2.3. Synthesis of ligands
Schiff bases (LNAPH and LO-VAN) have been synthesized following the literature-based method (Scheme 1) [46]. For in a mortar, the solid 2-hydroxy naphthaldehyde (NAPH) (0.173 g,1 mmol) is thoroughly mixed with 8-aminoquinoline (8-AMQ) (0.144 g,1 mmol) after grinding. Then, the mixture was poured in 30 mL methanol. At this moment, the yellow colour of the solution is generated. Then, the overall mixture is refluxed for 4 h at 75 °C. The reaction's progress is tracked using thin-layer chromatography (TLC). After removing the solvent under a rotary vacuum, the solution was cooled to isolate a yellow solid product (LNAPH). We consider the synthesis of LO-VAN a similar experimental procedure by taking ortho-vanillin (0.152 g, 1 mmol) instead of 2-hydroxy naphthaldehyde. Here, the ligand solution colour is deep red. The SB products are light and air-stable, and both compounds are collected in a tight air-dried desiccator. LNAPH: Yield: 91 % Anal. Calc. for C20H14NO: C, 84.48; H, 4.96; N, 4.93 % Found: C, 85.01; H, 5.00; N, 4.59 %. IR (KBr cm−1) selected bands: ν(C꞊N), 1629 s, ν(C-Ophenolic) 1215 s, ν(O–H) 3456 s, ν(C꞊C), 1469 s, ν(C–H), 3058 s, Raman (cm−1) bands: ν(C꞊N), 1601 s, ν(C-Ophenolic) 1237 m, ν(O–H) 3076 s, ν(C꞊C), 1421–1500 s, 1H NMR (DMSO‑d6, 400 MHz): δ (ppm): 6.73–7.87 (m, H), 8.39–8.41 (m, 1H), 15.87 (m,1H), 13C NMR (DMSO‑d6, 75.45 MHz): δ (ppm):108.67–147.76 (Arom-C),150.69 (C–OH), 181.92 (CH=N), UV–Vis (λmax CH3OH): 243, and 475 nm, DRS: 283, 369, and 720 nm. LO-VAN: Yield: 89 % Anal. Calc. for C17H14N2O2: C, 73.37; H, 5.07; N, 10.07 Found: C, 74.00; H, 5.10; N, 10.08 %. IR (KBr cm−1) selected bands: ν(C꞊N), 1615 vs, ν(C-Ophenolic) 1255 s, ν(O–H) 3620 s, ν(C꞊C), 1465 m, ν(C–H), 3065 s, ν(O–CH3), 2962 m, Raman (cm−1) bands: ν(C꞊N), 1617 s, ν(C-Ophenolic) 1210 s, ν(O–H) 2835–3019 m, ν(C꞊C), 1462–1542 s, ν(C–H), 3089 s, 1H NMR (DMSO‑d6, 400 MHz): δ (ppm): 3.84 (s, H), 6.87–7.12 (m, 1H), 9.84 (m, H), 12.91 (m, H),13C NMR (DMSO‑d6, 75.45 MHz): δ (ppm): 56.20 (O–CH3),114.66–151.50 (Arom-C),163.56 (C–OH), 196.69 (CH=N), UV–Vis (λmax CH3OH): 219, 264, and 345 nm, DRS:278, 367, and 758 nm.
Scheme 1.
The synthetic outline for LNAPH and LO-VAN.
2.4. DFT methodology
The DFT method optimized synthesized compounds' geometry (LNAPH/LO-VAN) at the B3LYP functional level (gas phase). The Gaussian 09W program [47] and Gauss View 6.0 performed DFT calculations on ground-state geometries. C, H, N, and O have been optimized using a 6–311++g (d, p) basis set. Regarding their purpose, (LNAPH/LO-VAN) structures had the absolute minimum of necessary components. Concurrent frequency calculations and geometry optimization were carried out to generate theoretical IR spectra. Different quantum properties, like FMO (HOMO-LUMO), MEP, and quantum chemical reactivity parameters, were studied using DFT. Based on molecular docking, both compounds were simulated for potential DNA binding and antimicrobial against E. coli. Furthermore, research on combating anti-COVID-19 also involves conducting molecular docking experiments. Tools like Auto Dock 4.2 [48] and Auto Dock Tools 1.5.6 [49] were used to find the best binding energy between compounds and macromolecules. The DNA 3D structure (PDB ID: 453D) and SARS-CoV-2 virus receptor protein (PDB ID: 6LU7) were received from the website www.rcsb.org/pdb (a Protein Data Bank) for molecular docking purposes. The synthesized compounds' optimized structure was converted to pdb format and used as input in Auto Dock Tools software for docking simulation. Gasteiger and Kollman charges were added to the primary structures of DNA and the receptor protein of SARS-CoV-2, respectively. The resolution of 453D [50], 2VF5 [51], and 6LU7 [51] structures was 1.80, 2.90, and 2.16 Å, respectively. Polar hydrogens were then added to these structures. The crystal structures were purged of water molecules and other ligands. Utilizing the Auto Dock Tools 1.5.6 package, we define binding sites with grid centers of X = 15.117, Y = 20.789, and Z = 8.718 (for PDB ID: 453D), X = 26.638, Y = 22.694, and Z = 8.036 (for PDB ID: 2VF5), and X = −26.283, Y = 12.599, and Z = 58.965 (for PDB ID: 6LU7). Furthermore, the grid size for 453D, 2VF5, and 6LU7 targets was selected at 50 × 40 × 60, 80 × 80 × 80, and 90 × 80 × 90 Å, respectively, with 0.375 Å spacing. The Lamarckian genetic algorithm (LGA) and default docking parameters initiated the docking simulation [52]. Docking interactions and binding affinities of compounds were computed using Discovery Studio Visualizer 4.1 software. The corrosion inhibitor, Fukui function, and NLO calculations were done at DFT's B3LYP, HF, and M06-2X levels of theory [[53], [54], [55]]. Fukui function calculations determine the electrophilic (E) and nucleophilic (Nu) attacking areas of SB compounds.
3. Results and discussion
3.1. Research perspective
Ligands were obtained using the literature-based method [46]. After conducting multiple synthetic experiments, we are unsuccessful to synthesize them into crystal products. The ligands are available in crystalline powder and highly stable towards air and light. The products obtained through these practices yield satisfactory results, producing yellow and red amorphous crystalline powders. Both compounds are soluble in CH3OH, including all the common organic solvents (Scheme 1). They are structurally characterized using various spectroscopic (IR/Raman/NMR) and elemental techniques, including SEM-EDX and DFT-conducted theoretical IR. The experimental and theoretical IR study provides conclusive evidence for aromatic C/imines C-atoms (H–C=N) and other significant C-atoms, including the –OH group in their structural framework. An extensive literature survey on these ligands has been unveiled, especially for DFT studies, corrosion inhibitors, antibacterial/pH-dependent antibacterial studies, and molecular docking research with DNA binding, anti-COVID-19, and E. coli. However, ligands' versatile complexing activities are explored in the literature [9,46]. We are divulging ligand activities based on DFT, antibacterial/pH-dependent antibacterial studies, and molecular docking experiments for the first time. Our article's exciting research potential includes global reactivity parameters, theoretical corrosion inhibitor, FMO (HOMO-LUMO) reactivity, Fukui function, NLO, and MEP, docking experiments with DNA binding, anti-COVID-19, and E. coli, and finally, antibacterial/pH-dependent properties.
3.2. Characterization
3.2.1. Experimental and theoretical IR
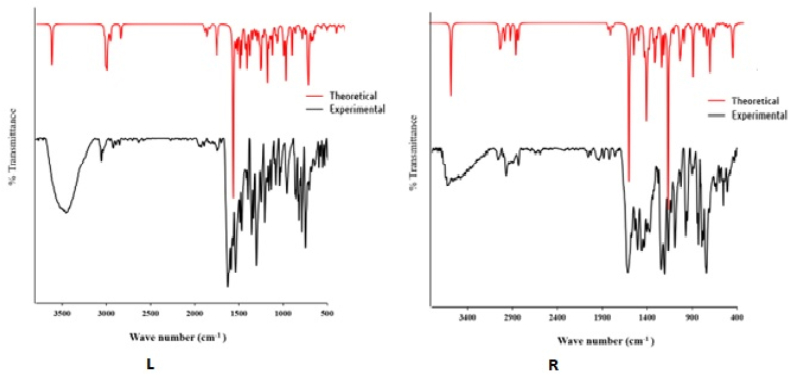
IR spectroscopy uses molecular vibrations to identify functional groups and predict the molecular structure of synthesized compounds (LNAPH/LO-VAN). Here, the structural frameworks highlighted that two SBs encompass functional groups, notably imines ν(C=N), ν(C-Ophenolic), free ν(O–H), ν(C=C), and ν(C–H) (Scheme 1,Fig.S1-Fig.S2). Therefore, the IR study identifies ν(C=N), ν(C-Ophenolic), free ν(O–H), ν(C=C), and ν(C–H) stretching values (cm−1) in the ligand's structural frameworks. The IR stretching values are 1629, 1215, 3456, 1469, and 3058 cm−1 (LNAPH), and 1615, 1255, 3620, 1465, and 3065 cm−1 (LO-VAN), respectively [56,57]. Apart from that, in LO-VAN, the O–CH3 IR peak value was observed at 2962 cm−1 [57]. Concerning the analysis, the experimental IR spectral values are correlated to the theoretical IR evaluated by DFT. The B3LYP/6–311++g (d, p) method (gas phase) computed the theoretical IR (cm−1). The significant experimental and theoretical IR bands for LNAPH/LO-VAN correlate with Fig. 1(L–R)/Table S1 [24]. Herein, the following theoretical IR peaks were observed and assigned and correlated with experimental values (cm−1): ν(C=N), ν(C-Ophenolic), free ν(O–H), ν(C=C), and ν(C–H). The group ν(C=N) IR was experimentally observed in both compounds at 1629-1615 cm−1, deviating from the theoretical value range of 1690–1700 cm−1. The free-OH IR stretching fell within the theoretical range of 3445–3550 cm−1, while the experimental value is 3456–3620 cm−1. The C-Ophenolic stretching was identified at 1215 and 1255 cm−1, while the DFT computed values are 1258–1260 cm−1. Further, the experimental ν(C=C) and ν(C–H) values are 1469-1465 cm−1, and 3058-3065 cm−1 whereas theoretical values are correlated with 1421–1580 cm−1 and 3055-3070 cm−1. The ligands' structure can be confirmed by comparing the experimental and theoretical IR spectra.
Fig. 1.
(L–R). Experimental and DFT computed FT-IR plot of LNAPH (L) and LO-VAN (R).
3.2.2. Raman spectra
We have also structurally characterized the as-prepared compounds based on the Raman spectral study. Raman's study assigned the stretching (cm−1) mainly ν(C=N), ν(C–O), free ν(O–H), ν(C=C), and ν(C–H) in the ligand's structural frameworks. The reference peak values are 1601, 1237, 3076, and 1421-1500 cm−1 for LNAPH. In contrast, the observed peak values for LO-VAN are assigned 1617, 1210, 2835–3019, 1462–1542, and 3089 cm−1, respectively (Fig.S3-Fig.S4).
3.2.3. UV–visible, and DRS
The absorption spectra of the prepared ligands were analysed using a CH3OH solvent. The assigned UV absorption band values were 226 nm, 475 nm, (LNAPH) 219 nm, 264 nm, and 345 nm (LO-VAN) (Fig.S5-Fig.S6). The provided data validates that the ligands display optical shifts of the π→π*/n→π* nature, which occur within the aromatic rings [24]. The spectral data mentioned are unequivocally identical to the previously reported ligands [58,59]. We also investigated diffuse reflectance spectroscopy (DRS) for ligands to check for any shoulder peak. In this observation, slightly shifted bands are seen along with a shoulder. The assigned bands for the ligands are 283 and 369 nm, and 278 and 367 nm correspond to the intra-ligand π→π* or n→ π* transitions. A completely new shoulder peak is identified at 720 and 758 nm (LNAPH/LO-VAN), respectively (Fig.S7-Fig.S8).
3.2.4. NMR study
The two SB ligands are formed when 2-hydroxy naphthaldehyde and ortho-vanillin react with 8-aminoquinoline. NMR study ensures the ligand's aromatic ring structure, imine (H–C=N), and free-OH groups. 1H NMR identifies all protons' environments, while 13C characterizes the C-atoms in the ligand structure (Fig.S9-Fig.S12). In LNAPH, the aromatic, imine (H–C=N) and –OH proton NMR peak appears at δ 6.73–7.87, δ 8.39–8.41, and δ 15.87 ppm, while for LO-VAN, such protons appear at δ 6.87–7.12, δ 9.84 and δ 12.91 ppm, respectively. The 1H NMR study has effectively verified the imine bond formation and the existence of an unbound –OH group. Notably, the O–CH3 proton appears at δ 3.84 ppm in LO-VAN. Further, a13C NMR study in both ligands confirmed the C-nature in Arom-C, C–OH, and CH=N. Therefore, the 13C NMR study δ (ppm) value for LNAPH appears at 108.67–147.76, 150.69, and 181.92 ppm, while for LO-VAN, 114.66–151.50, 114.66–151.50, and 114.66–151.50 ppm respectively. The methoxy C-atoms δ (ppm) in LO-VAN is 56.20 ppm [24]. Thus, 13C NMR confirms that both SBs included Aro-C, C–OH, and imine-C, and the LO-VAN structure includes an O–CH3 group.
3.2.5. EDX-SEM
EDX analysis is becoming increasingly influential in synthetic chemistry. It provides accurate insight into the elemental composition of the synthesized compounds (LNAPH/LO-VAN), making it a valuable tool for researchers. Using the EDX tool, we identify the primary elements in the ligands as C, N, and O. This information is valuable and informative (Table S2/Fig. S13). In addition, the scanning electron microscope (SEM) technique is utilized to examine the surface morphological characteristics of the current ligands. Our solid crystalline powder compounds underwent an SEM experiment (Fig. S14). SEM micrographs indicate that the morphology is beautifully distributed in ice-type sheets, resembling other Schiff base compounds.
4. DFT profiles
4.1. Optimized geometric parameters
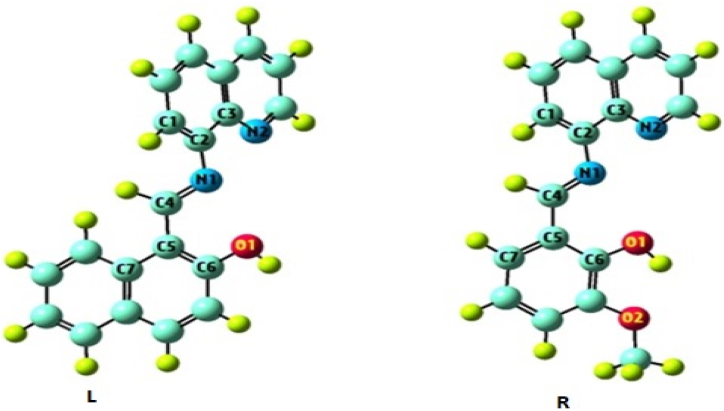
The DFT concept will give an overview of geometry for LNAPH/LO-VAN. The compounds' structural parameters, which include BL (bond lengths) (Å) and BA (bond angles (°), were seen using the Gauss View 6.0 visualization package. The optimized geometries of the compounds are demonstrated in Fig. 2(L–R). In the geometry of LNAPH, the estimated bond lengths of N1–C2, N1–C4, N2–C3, and O1–C6 are found to be 1.393, 1.275, 1.358, and 1.354 Å, respectively. In contrast, these parameters for LO-VAN geometry are observed at 1.393, 1.273, 1.358, and 1.347 Å. The calculated bond angles of N2–C3–C2, C3–C2–N1, C2–N1–C4, and O1–C6–C5 for LNAPH are 118.618, 118.921, 119.798, and 119.755°, respectively, whereas these values for LO-VAN are determined at 118.611, 119.054, 119.914, and 121.717°, respectively. Furthermore, the most stable geometries of LNAPH and LO-VAN are illustrated by the minimum energies of −955.507 and −916.391 Hartree, respectively.
Fig. 2.
(L–R). DFT optimized geometry of the compounds LNAPH (L) and LO-VAN (R).
4.2. MEP surface
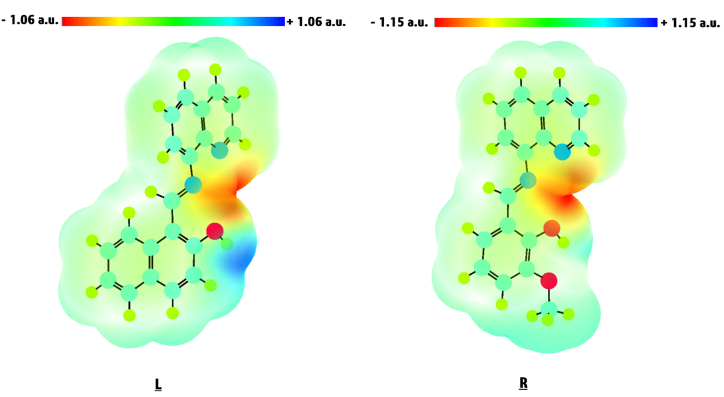
The MEP surface visualizes the reactive sites and H bonding forces in a compound's structure [60]. The regions with electrophilic (E) and nucleophilic (Nu) tendencies can be identified by examining the molecular electrostatic potential (MEP) surface. The molecule's reactive sites can be identified by analysing areas with the highest negative (-ve) and positive (+ve) electrostatic potential. Furthermore, electrostatic potential surfaces reflect molecular shape and size, making them a beneficial tool for investigating the correlation between the molecular geometry of bioactive compounds and their physiochemical capabilities [61]. The MEP surface with the highest negative electrostatic potential, or red-coloured area, shows the relative abundance of electrons and is fit for the electrophilic attack. The MEP surface with the highest positive electrostatic potential, or blue-coloured area, demonstrates the relative absence of electrons and is ideal for nucleophilic attack [62]. On the other hand, the green-coloured area is related to the neutral zone with zero electrostatic potential. The predicted MEP surfaces of LNAPH and LO-VAN compounds are displayed in Fig. 3(L–R). MEP findings for LNAPH and LO-VAN indicate that red-coloured surfaces appear around the N1, N2, and O1 atoms, which suggests these areas are ideal targets for electrophilic attack. In the MEP surface of LNAPH, hydrogen attached to the O1 atom possesses the highest positive electrostatic potential, illustrating that this atom is suitable for nucleophilic attack.
Fig. 3.
(L–R). MEP surface of the compounds LNAPH (L) and LO-VAN (R). The isodensity values for LNAPH(L) and LO-VAN(R) are in the range of −1.06 a.u. (red) to +1.06 a.u. (blue) and −1.15 a.u. (red) to +1.15 a.u. (blue), respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4.3. FMO analysis
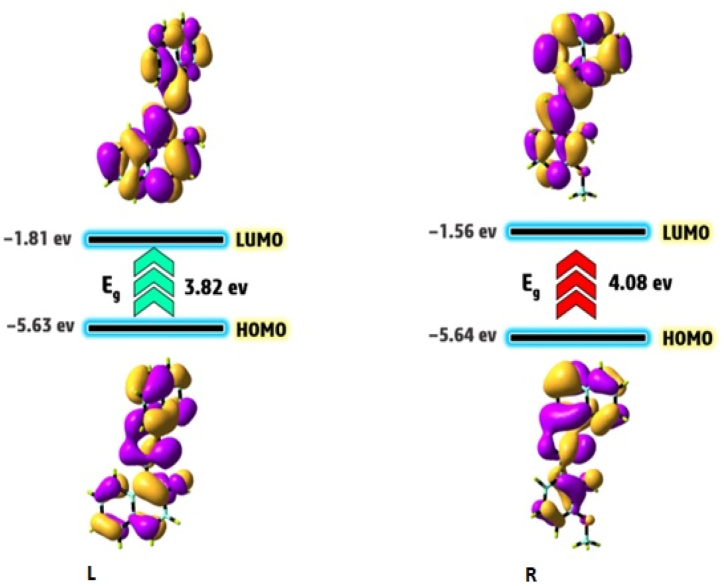
The HOMO-LUMO energies for the compounds (LNAPH/LO-VAN) were determined using the DFT/B3LYP/6–311++g (d, p) level method. The FMO parameters and its energy gap (Eg = ELUMO-EHOMO) are popular quantum mechanical parameters that play a crucial role in the compounds' biological activity, stability, and chemical reactivity [63]. The smaller Eg is related to the high biological activity, high chemical reactivity, and lower stability. In contrast, the high energy gap reveals low biological activity, chemical reactivity, and stability [64]. Fig. 4(L–R) exhibits the HOMO, LUMO, and Eg of the compounds. We observed the HOMO and LUMO electronic clouds of LNAPH and LO-VAN are centered over the whole molecule. The Eg parameter for the compounds LNAPH and LO-VAN is calculated at 3.82 and 4.08 eV. These findings indicate that LNAPH possesses a smaller energy gap than LO-VAN, which is associated with higher biological activity and chemical reactivity of this compound compared to LO-VAN [65].
Fig. 4.
(L–R). HOMO and LUMO contour plots for LNAPH (L) and LO-VAN (R).
Additional quantum reactivity factors were developed to expand on the reactivity of the compounds (Table 1). These parameters consist of chemical potential (Pi), global electrophilicity (ω), absolute hardness (η), absolute electronegativity (χ), and absolute softness (σ), which are obtained from the following equations (1), (2), (3), (4), (5)) for the synthesized compounds at the same level of theory [66].
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
Table 1.
Calculated quantum chemical reactivity parameters.
| Quantum chemical parameter | LNAPH | LO−VAN |
|---|---|---|
| EHOMO | −5.63 | −5.64 |
| ELUMO | −1.81 | −1.56 |
| ΔE(LUMO-HOMO) | 3.82 | 4.08 |
| χ | 3.72 | 3.60 |
| η | 1.91 | 2.04 |
| σ | 0.52 | 0.49 |
| Pi | −3.72 | −3.60 |
| ω | 3.62 | 3.17 |
It is clear from Table 1 that.
-
[1]
LO-VAN has a higher hardness value (η = 2.04 eV), indicating that this compound is more rigid with a lower softness than LNAPH.
-
[2]
The electrophilicity (ω) values, 3.62 eV for LNAPH and 3.17 eV for LO-VAN highlight the robust electrophilic (E) capability of LNAPH, making it the more reactive compound in this context.
-
[3]
Based on the negative values of Pi, both compounds are stable and do not undergo decomposition into the primary elements [67].
-
[4]
LNAPH interacts with macromolecules like protein or DNA more effectively than LO-VAN, based on ω values [68].
4.4. Corrosion inhibitor: A true theoretical approach
We have explored the corrosion inhibitor function of the compounds (LNAPH/LO-VAN) purely from a theoretical background. Based on the HOMO-LUMO parameters calculated using different levels of DFT basis sets in the gas phase (Table S3-Table S4), the corrosion inhibitor activities of the prepared compounds at protonated w. r. to –OH functional group & non-protonated concepts can be explained. It is more appropriate to determine the better corrosion inhibitor based on these calculations [69]. Notably, DFT calculations were more realistic for an acidic environment; further calculations were made by protonating the most negative atom of the compound (Table S4). Based on the HOMO parameter, calculations indicate that LNAPH is a better inhibitor. Therefore, LNAPH is a promising inhibitor based on its impressive LUMO parameter value.
4.5. Fukui function
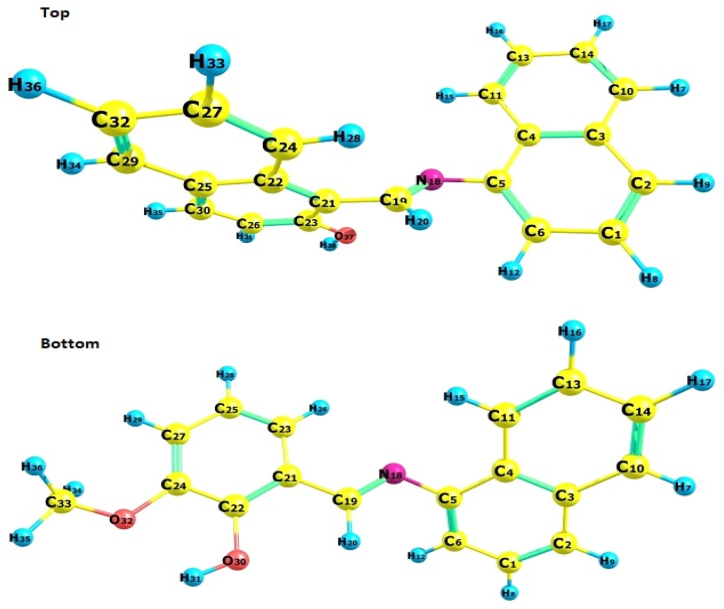
Fukui indices reveal a molecule's propensity to give or take electrons. It also provides crucial information regarding the molecule's atom susceptible to nucleophilic (Nu) or electrophilic (E) attacks. The Fukui functions (condensed/atomic) corresponding to the jth atomic domain can be defined using the formulas presented in ESI (Equation S1). The formulas (ESI) represent an E attack (r) or a Nu attack (r) on the synthesized molecule's atoms (C/H). The symbol qj (Mulliken population analysis, electrostatic derived charge) represents the N, N+1, or N–1 atomic charge. The ESI equations provide a binary descriptor, Δf(r), distinguishing E and Nu Fukui functions [70,71]. Generally, an area is chosen for a Nu attack zone if the parameter Δf(r) > 0, and for an E attack if the Δf(r) value < 0. Δf(r) defines E and Nu assault at a given site concerning the binary identifier. Consequently, they offer a +ve value for Nu attack-prone atoms and a -ve value for E attack-prone atoms [72]. We successfully have computed the Fukui function for LNAPH and LO-VAN based on the atom numbering in Fig. 5(Top-Bottom). For the Fukui function calculations in LNAPH, the Nu zone is 5C if (Δf(r) > 0), where the maximum is +0.12. Similarly, the E zone is 4C if(Δf(r) < 0), where the lowest is −0.44. In LO-VAN, the Nu zone (Δf(r) > 0) is 30C, and, here, the maximum is +1.54 atoms. Similarly, the E region (Δf(r) < 0) is 5C and the lowest is −0.12 atoms. Table S5 provides all calculated Fukui function values. The behavior of a molecule during an E and Nu attack is determined by its local behavior. The Fukui function analysis's numerical values and the MEP computations' outcomes were in excellent agreement [73].
Fig. 5.
(Top-Bottom). Fukui atom representation in LNAPH (Top) and LO-VAN (Bottom).
4.6. NLO characteristics
The NLO activities of LNAPH and LO-VAN were evaluated and presented in Table 2. For NLO purposes, versatile parameters have been computed. The NLO operations and other advanced optical technologies are essential for optical modulation/memory. Each calculated parameters measure yields necessary information about the as-prepared compounds. Non-linear optical (NLO) activities of a newly synthesized material can be explored through experimental parameters polarizability (α) and hyperpolarizability (β). Two essential terms, α and β, comprise the molecular dipole moment (μ). The NLO properties can be calculated using the following ESI (Equation S2). Herein, the hyperpolarizability and polarizability values of the compounds have been calculated. The conversion of estimated parameters into electrostatic units (esu) involves multiplication by constant values of the atomic unit (a.u) (Equation S3). The characteristics of NLOs increase with the magnitude of the parameters such as μ, α, and β.
Table 2.
NLO calculated parameters.
| LNAPH |
LO−VAN |
LNAPH |
LO−VAN |
||||||
|---|---|---|---|---|---|---|---|---|---|
| a.u. | esu | a.u. | esu | a.u. | esu | a.u. | esu | ||
| 296.817 | 4.39 10−23 | 287.735 | 4.25 10−23 | 31.240 | 2.70 10−28 | −1084.909 | −9.37 10−27 | ||
| 4.238 | 6.27 10−25 | −6.090 | −9.01 10−25 | −118.841 | −1.02 10−27 | 79.401 | 6.86 10−28 | ||
| 193.133 | 2.85 10−23 | 170.419 | 2.52 10−23 | 142.375 | 1.23 10−27 | 27.677 | 2.39 10−28 | ||
| −15.026 | −2.22 10−24 | 24.955 | 3.69 10−24 | −61.914 | −5.35 10−28 | 136.6594 | −1.18 10−27 | ||
| 4.226 | 6.25 10−25 | −4.441 | −6.57 10−25 | 90.195 | 7.79 10−28 | −141.186 | −1.22 10−27 | ||
| 161.745 | 2.39 10−23 | 122.892 | 1.81 10−23 | −53.110 | −4.58 10−28 | 2.259 | 1.95 10−29 | ||
| 296.817 | 4.39 10−23 | 150.688 | 2.23 10−23 | 2.377 | 2.05 10−29 | 6.141 | 5.30 10−29 | ||
| 1.240 | 1.83 10−25 | 2.157 | 3.19 10−25 | −0.829 | −7.16 10−30 | −47.743 | −4.12 10−28 | ||
| 0.077 | 1.14 10−26 | −0.444 | −6.57 10−26 | 4.065 | 3.51 10−29 | −2.179 | −1.88 10−29 | ||
| 0.219 | 3.24 10−26 | −0.838 | −1.24 10−25 | 101.882 | 8.80 10−28 | 1220.668 | 1.05 10−26 | ||
| 1.261 | 2.356 | ||||||||
5. Molecular docking
Molecular docking simulations can be used to evaluate the synthesized compounds (LNAPH and LO-VAN) for their potential to bind to DNA and inhibit the activity of E. coli and COVID-19. We explain all the docking simulations as follows.
5.1. DNA binding
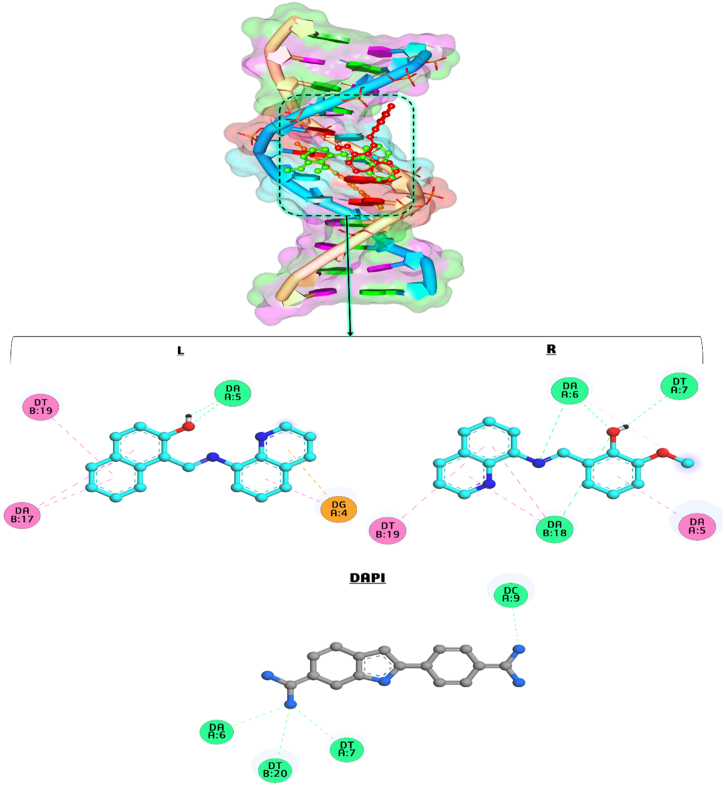
The docking simulation of synthesized compounds and DAPI (4′,6-diamidino-2-phenylindole, as standard DNA-groove binder) against DNA (PDB ID: 453D) was first conducted to investigate the best binding poses and interactions involved in these binding systems. Fig. 6 (L-R) exhibits the docking viewpoint of LNAPH, LO-VAN, and DAPI with DNA and their interactions. The best compound-DNA binding affinity for LNAPH, LO-VAN, and DAPI is calculated at −5.07, −4.29, and −4.18 kcal/mol, respectively. This result suggested that LNAPH has a more robust interaction with DNA compared with LO-VAN and DAPI. On the other hand, the synthesized compounds interact with DNA through groove binding (Fig. 6(L–R)). The binding of LNAPH with DNA involves three hydrophobic interactions with DT B:19 and DA B:17, two H bonds with DA A:5, and two electrostatic attractions with DG A:4 base pair. Furthermore, LO-VAN creates five hydrophobic interactions with DT B:19, DA B:18, DA A:5, and DA A:6, and four hydrogen bonds with base pairs of DA B:18, DA A:6, and DT A:7. The binding of DAPI with DNA involves four H bonds with DA A:6, DT B:20, DT A:7, and DC A:9 base pairs. As a result, DNA docking simulation indicates that LNAPH and LO-VAN bind to DNA through groove binding, mainly by H bonding and hydrophobic interactions. Docking protocol was validated by re-docking (Fig. S15) LNAPH and LO-VAN against DNA and calculating root mean square deviation (RMSD). RMSD values of 0.6 (for LNAPH) and 0.8 Å (for LO-VAN) between the docked conformations of each compound depicted the accuracy of the docking protocol. The re-docking process indicated that the new conformers bind to the same binding site as the previous conformers on DNA.
Fig. 6.
(L–R). Docking of LNAPH (L), LO-VAN (R), and DAPI (as standard DNA-groove binder) during the interaction with DNA (PDB ID: 453D).
5.1.1. Anti-COVID-19
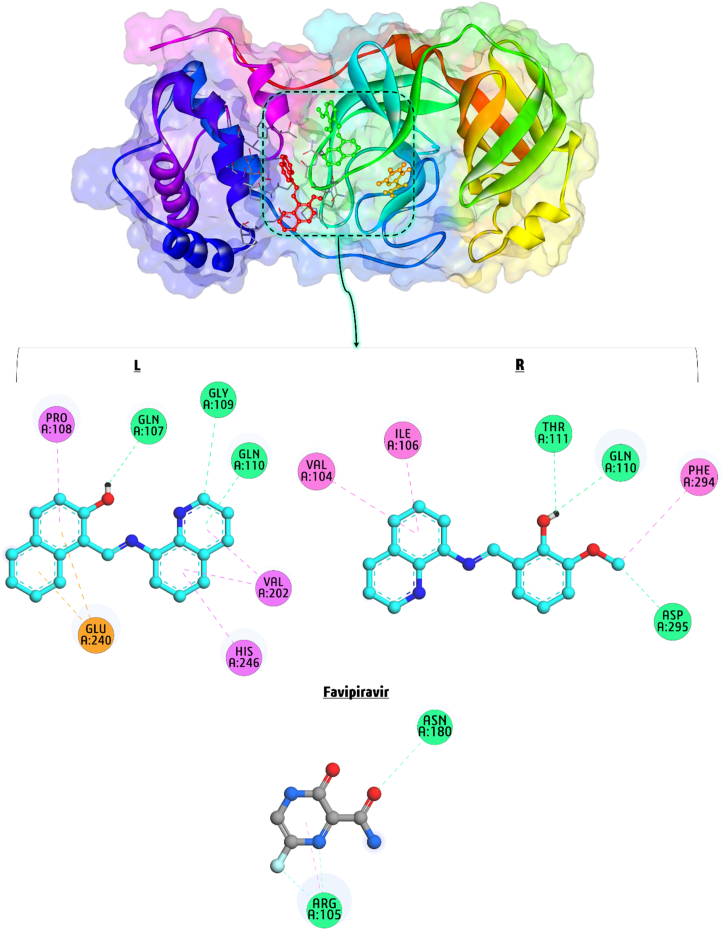
The compounds LNAPH and LO-VAN were tested for anti-COVID-19 activity against the virus receptor protein specified as SARS-CoV-2 (PDB ID: 6LU7). The overall docking findings are presented in Fig. 7(L–R). The best binding poses of interaction between LNAPH, LO-VAN, and Favipiravir (as standard anti-COVID-19 drug) and SARS-CoV-2 virus receptor protein have a binding affinity of −5.22, −4.94, and −3.69 kcal/mol, respectively, indicating LNAPH possesses a more substantial effect on the active site of this protein compared to LO-VAN and Favipiravir. Furthermore, LNAPH and LO-VAN demonstrated higher binding affinity to the active site of the protein target than Favipiravir. LNAPH establishes four hydrophobic interactions with PRO A:108, HIS A:246, and VAL A:202, three H bonds with GLN A:107, GLY A:109, and GLN A:110, and two electrostatic attractions with GLU A:240 amino acid residue. LO-VAN demonstrates robust interaction with the amino acid residues in protein structure PDB ID 6LU7. It forms three HI (hydrophobic interactions) with VAL A:104, ILE A:106, and PHE A:294 and three H bonds with THR A:111, GLN A:110, and ASP A:295. Favipiravir establishes one hydrophobic interaction with ARG A:105, two H bonds with ASN A:180 and ARG A:105, and one halogen interaction with ARG A:105 amino acid residue. H bonding and HI (hydrophobic interactions) are essential in interacting with the title compounds for docking receptor proteins. Docking simulation of LNAPH and LO-VAN versus SARS-CoV-2 virus makes them potential candidates for future coronavirus therapeutic research. Molecular docking simulations demonstrate that compound LNAPH interacts effectively with DNA and SARS-CoV-2 receptor proteins. The docking result is consistent with the DFT data, precisely the global electrophilicity values.
Fig. 7.
(L–R). Docking of LNAPH (L), LO-VAN (R), and Favipiravir (as standard anti-COVID-19 drug) during the interaction with SARS-CoV-2 virus receptor protein (PDB ID: 6LU7).
5.1.2. E. coli
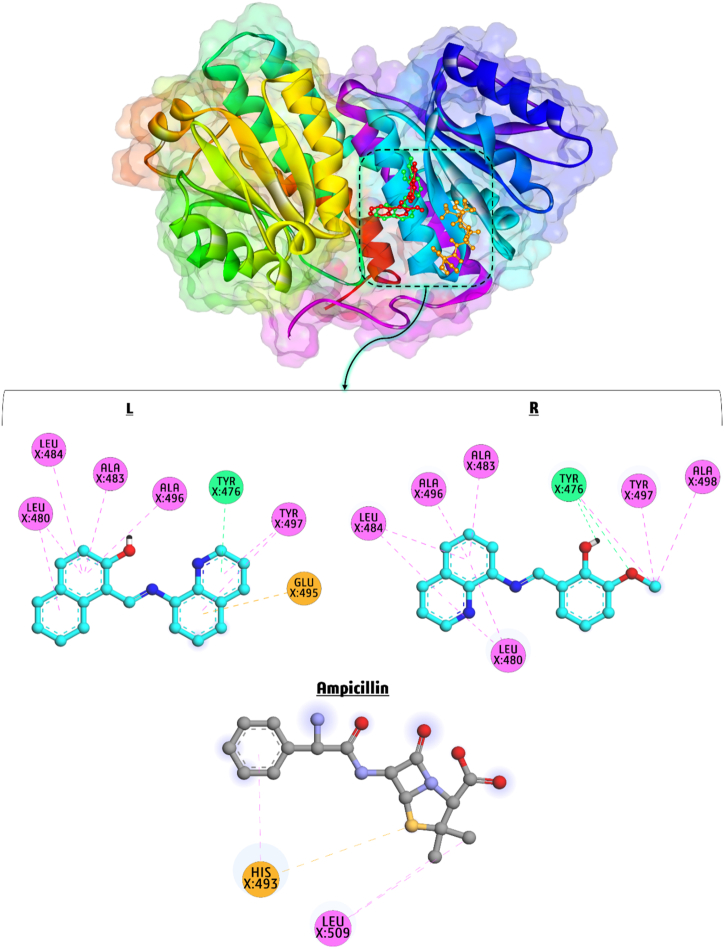
Based on the in-vitro antimicrobial activity results of LNAPH and LO-VAN, we concluded that they significantly impact E. coli's protein. We used molecular docking simulations to verify the antimicrobial activity and highlight the crucial interaction in these binding systems. Docking simulations of LNAPH, LO-VAN, and Ampicillin (as standard antibacterial drug) were performed on the active site of E. coli (PDB ID:2VF5), and the findings are represented in Fig. 8(L–R). The binding affinity of LNAPH, LO-VAN, and Ampicillin against E. coli target is determined at −6.30, −5.72, and −5.57 kcal/mol, respectively. LNAPH demonstrates stronger binding affinity to E. coli receptors than LO-VAN and Ampicillin. LNAPH is stabilized by seven hydrophobic interactions with residues of LEU X:480, LEU X:484, ALA X:483, ALA X:496 and TYR X:497, one H bond with TYR X:476, and one electrostatic attraction with GLU X:495. LO-VAN develops two H bonds with TYR X:476 and nine hydrophobic interactions with residues of LEU X:484, ALA X:496, ALA X:483, LEU X:480, TYR X:497, and ALA X:498. Ampicillin forms three hydrophobic interactions with residues of LEU X:509 and HIS X:493, and one electrostatic attraction with HIS X:493. As represented via the docking simulation, H bonding and hydrophobic interactions significantly affect the active site of E. coli (PDB ID: 2VF5). So, docking simulations validate the antimicrobial activity of the synthesized compounds (LNAPH and LO-VAN) by demonstrating high binding affinities and many interactions.
Fig. 8.
(L–R). Docking of LNAPH (L), LO-VAN (R), and Ampicillin (as standard antibacterial drug) during the interaction E. coli (PDB ID: 2VF5).
5.1.3. Docking objectives
The primary objective of our research was to explore the potential broad-spectrum efficacy of the studied compounds against diverse biological targets, including COVID-19 activity, DNA binding, and antibacterial activity. Here, we provide a detailed explanation for selecting the specific proteins and DNA for docking.
-
[1]
E. coli Protein: Escherichia coli serves as a model organism for understanding bacterial infections and developing antibacterial agents. By targeting E. coli proteins, we aim to investigate the potential of our compounds to inhibit bacterial growth and function. This choice is particularly relevant given the rise of antibiotic-resistant strains of bacteria, making the discovery of new antibacterial agents a critical area of research.
-
[2]
COVID-19 Protein: The COVID-19 pandemic has underscored the urgent need for effective antiviral therapies. By including SARS-CoV-2 proteins (such as the main protease) in our docking simulations, we aim to identify compounds that may interfere with the virus's ability to infect host cells or replicate. This dual-target approach helps in assessing whether our compounds have antiviral properties that could contribute to the fight against COVID-19.
-
[3]
DNA Receptor: Investigating the interaction of compounds with DNA receptors is crucial for understanding their potential effects on genetic material. Such interactions can provide insights into the compound's ability to act as antitumor agents or modulators of gene expression. This aspect of our study helps in assessing the broader pharmacological potential of the compounds, including their ability to bind to and possibly modulate DNA function.
By examining the binding affinities of our compounds to these varied targets, we aim to demonstrate their potential multifunctionality, which is highly desirable in drug development. This comprehensive approach not only broadens the scope of potential therapeutic applications but also provides a foundation for future studies on the mechanism of action and optimization of these compounds.
6. Antibacterial study
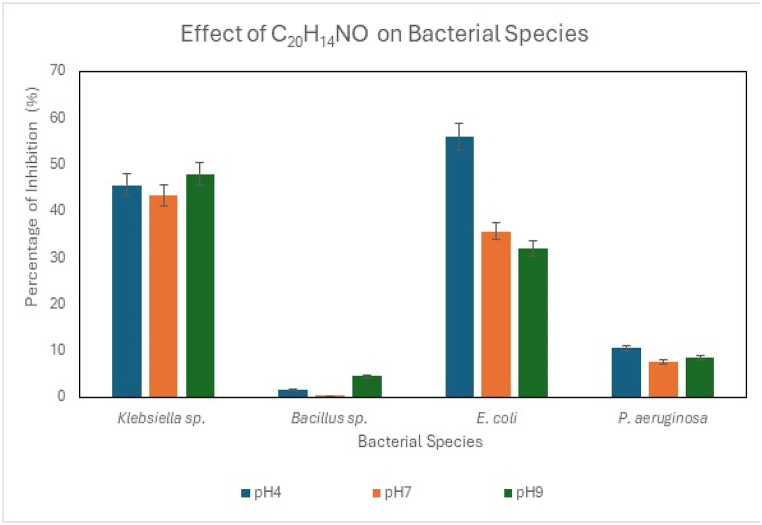
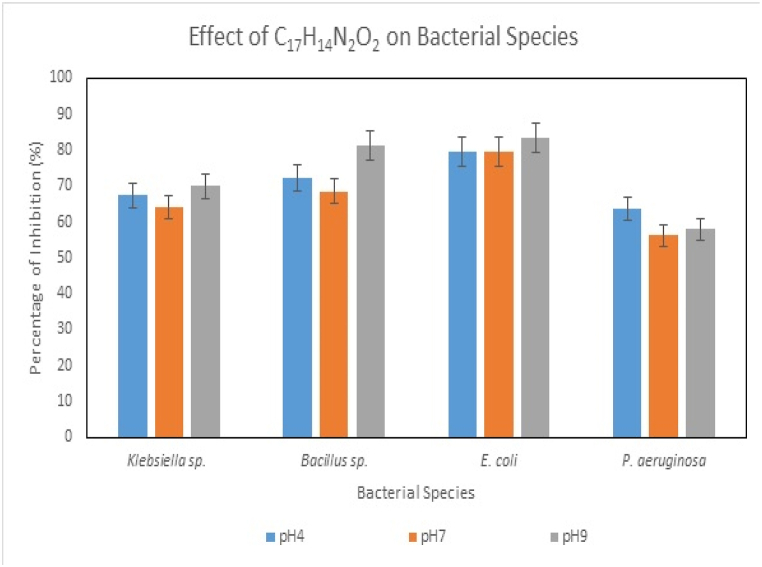
LNAPH (C20H14NO), LO-VAN (C17H14N2O2), NAPH (2-hydroxy naphthaldehyde), O-VAN (ortho vanillin) and 8-AMQ (8-aminoquinoline) were exposed to antibacterial study against E. coli, Klebsiella, Bacillus and Pseudomonas sp. The last three compounds belong to synthetic components. The experimental trials were conducted and observed in triplicates to document the statistically significant results, as represented in Fig. 9, Fig. 10 and Fig. S16. The compounds' antibacterial activity was studied with increasing concentrations (0–1 mg/L). The MIC (minimum inhibitory concentrations) of the compounds against the bacterial species are tabulated in Table 3. MIC indicates the least concentration of any antimicrobial compound at which the complete prevention of microbial growth is observed [74]. In this study, the range of MIC of the compounds against individual bacterial strains varied within the range of 0.2–2.5 mg/mL. The values exhibited the minimum concentration at which the growth of E. coli, Klebsiella, Bacillus, and Pseudomonas sp. can be hindered. The presence of carbon in the LNAPH and LO-VAN helps exhibit antimicrobial activity, as carbon can cause mechanical destruction of the cell wall of the bacterial cells. Mechanical damage further leads to the efflux of cytoplasmic constituents, thus resulting in cell death [75]. O-VAN showed the highest inhibition rate against Klebsiella sp., 67.5 %, followed by LO-VAN, which showed 64.15 % inhibition against the strain. Both compounds efficiently controlled the growth of Pseudomonas, Bacillus, and E. coli. It can be claimed from the antibacterial analyses that these compounds restrict the formation of biofilms of the mentioned microbial cells and thus restrict microbial-induced corrosion [76,77].
Fig. 9.
Effect of bacterial species for LNAPH.
Fig. 10.
Effect of bacterial species for LO−VAN.
Table 3.
Details effect of the compounds on Bacterial Strains.
| Sample Code | Name of the Bacterial Strain | pH | OD Value |
Percentage of Inhibition (%) | MIC (mg/ml) | MIC (μM) | |
|---|---|---|---|---|---|---|---|
| Control | Test | ||||||
| C20H14NO (LNAPH) | Klebsiella sp. | 4 | 0.366 | 0.199 | 45.6284153 | 0.303 | 0.086 |
| 7 | 0.385 | 0.218 | 43.37662338 | 0.304 | 0.086 | ||
| 9 | 0.304 | 0.158 | 48.02631579 | 0.297 | 0.084 | ||
| Pseudomonas aeruginosa | 4 | 0.581 | 0.52 | 10.49913941 | 0.49 | 0.139 | |
| 7 | 0.582 | 0.538 | 7.560137457 | 0.569 | 0.16 | ||
| 9 | 0.564 | 0.516 | 8.510638298 | 0.537 | 0.15 | ||
| Bacillus sp. | 4 | 0.315 | 0.31 | 1.587301587 | 2.5 | 0.71 | |
| 7 | 0.558 | 0.556 | 0.36 | 1.136 | 0.32 | ||
| 9 | 0.487 | 0.465 | 4.517453799 | 0.764 | 0.22 | ||
| Escherichia coli | 4 | 0.492 | 0.216 | 56.09756098 | 0.285 | 0.080 | |
| 7 | 0.518 | 0.333 | 35.71428571 | 0.322 | 0.09 | ||
| 9 | 0.418 | 0.284 | 32.05741627 | 0.33 | 0.094 | ||
| C17H14N2O2(LO-VAN) | Klebsiella sp. | 4 | 0.366 | 0.119 | 67.4863388 | 0.27 | 0.075 |
| 7 | 0.385 | 0.138 | 64.15584416 | 0.276 | 0.077 | ||
| 9 | 0.304 | 0.091 | 70.06578947 | 0.271 | 0.075 | ||
| Pseudomonas aeruginosa | 4 | 0.581 | 0.21 | 63.85542169 | 0.277 | 0.077 | |
| 7 | 0.582 | 0.255 | 56.18556701 | 0.285 | 0.08 | ||
| 9 | 0.564 | 0.237 | 57.9787234 | 0.284 | 0.079 | ||
| Bacillus sp. | 4 | 0.315 | 0.087 | 72.38095238 | 0.268 | 0.074 | |
| 7 | 0.558 | 0.176 | 68.45878136 | 0.272 | 0.076 | ||
| 9 | 0.487 | 0.092 | 81.10882957 | 0.262 | 0.073 | ||
| Escherichia coli | 4 | 0.492 | 0.1 | 79.67479675 | 0.263 | 0.073 | |
| 7 | 0.518 | 0.106 | 79.53667954 | 0.263 | 0.073 | ||
| 9 | 0.418 | 0.069 | 83.49282297 | 0.26 | 0.072 | ||
| NAPH | Klebsiella sp. | 4 | 0.366 | 0.347 | 5.191256831 | 0.699 | 4.06 |
| 7 | 0.385 | 0.353 | 8.311688312 | 0.544 | 3.16 | ||
| 9 | 0.304 | 0.268 | 11.84210526 | 0.467 | 2.71 | ||
| Pseudomonas aeruginosa | 4 | 0.581 | 0.564 | 2.925989673 | 1.07 | 6.21 | |
| 7 | 0.582 | 0.579 | 0.515463918 | 1.742 | 10.12 | ||
| 9 | 0.564 | 0.541 | 4.078014184 | 0.819 | 4.76 | ||
| Bacillus sp. | 4 | 0.315 | 0.295 | 6.349206349 | 0.623 | 3.62 | |
| 7 | 0.558 | 0.509 | 8.781362007 | 0.53 | 3.08 | ||
| 9 | 0.487 | 0.319 | 34.49691992 | 0.325 | 1.89 | ||
| Escherichia coli | 4 | 0.492 | 0.395 | 19.71544715 | 0.386 | 2.24 | |
| 7 | 0.518 | 0.403 | 22.2007722 | 0.371 | 2.15 | ||
| 9 | 0.418 | 0.318 | 23.92344498 | 0.362 | 2.10 | ||
| O-VAN | Klebsiella sp. | 4 | 0.366 | 0.116 | 68.30601093 | 0.273 | 1.79 |
| 7 | 0.385 | 0.125 | 67.53246753 | 0.273 | 1.79 | ||
| 9 | 0.304 | 0.08 | 73.68421053 | 0.268 | 1.76 | ||
| Pseudomonas aeruginosa | 4 | 0.581 | 0.202 | 65.232358 | 0.276 | 1.81 | |
| 7 | 0.582 | 0.232 | 60.13745704 | 0.281 | 1.84 | ||
| 9 | 0.564 | 0.219 | 61.17021277 | 0.279 | 1.83 | ||
| Bacillus sp. | 4 | 0.315 | 0.099 | 68.57142857 | 0.272 | 1.78 | |
| 7 | 0.558 | 0.133 | 76.16487455 | 0.266 | 1.74 | ||
| 9 | 0.487 | 0.128 | 73.71663244 | 0.267 | 1.75 | ||
| Escherichia coli | 4 | 0.492 | 0.085 | 82.72357724 | 0.261 | 1.71 | |
| 7 | 0.518 | 0.091 | 82.43243243 | 0.261 | 1.71 | ||
| 9 | 0.418 | 0.064 | 84.68899522 | 0.259 | 1.70 | ||
| 8-AMQ | Klebsiella sp. | 4 | 0.366 | 0.277 | 24.31693989 | 0.36 | 2.5 |
| 7 | 0.385 | 0.29 | 24.67532468 | 0.359 | 2.49 | ||
| 9 | 0.304 | 0.19 | 37.5 | 0.317 | 2.19 | ||
| Pseudomonas aeruginosa | 4 | 0.581 | 0.253 | 56.45438898 | 0.286 | 1.98 | |
| 7 | 0.582 | 0.288 | 50.51546392 | 0.294 | 2.03 | ||
| 9 | 0.564 | 0.256 | 54.60992908 | 0.29 | 2.01 | ||
| Bacillus sp. | 4 | 0.315 | 0.288 | 8.571428571 | 0.536 | 3.71 | |
| 7 | 0.558 | 0.5 | 10.39426523 | 0.492 | 3.41 | ||
| 9 | 0.487 | 0.41 | 15.8110883 | 0.417 | 2.89 | ||
| Escherichia coli | 4 | 0.492 | 0.454 | 7.723577236 | 0.563 | 3.90 | |
| 7 | 0.518 | 0.481 | 7.142857143 | 0.586 | 4.06 | ||
| 9 | 0.418 | 0.398 | 4.784688995 | 0.736 | 5.10 | ||
6.1. pH-dependent antibacterial
pH-dependent studies revealed that the compounds are most effective when exposed to bacterial species at an alkaline pH of 9 (Fig. 9, Fig. 10) and Fig. S15. The pH of the medium was also observed to increase after the initial phase of the interaction. The gradual increase of the pH affects the kinetics of the redox reaction of the bacterial metabolism and further interferes with the microbial structures. The high pH weakens the plasma membrane and thereby bursts the bacteria's membrane, killing the cells. The proteins, lipids, and enzymes associated with the bacterial cytoplasmic membrane are highly solubilized in the alkaline environment. The denaturation of these microbial components leads to bacterial cell death in the medium [75,78].
7. Conclusion
We synthesized two SBs (LNAPH/LO-VAN) with satisfactory yields, characterized using various spectroscopic tools and EDX-SEM. The DFT/B3LYP/6–311++g (d, p) basis set was used to optimize the molecular geometry in the gas phase. FMO, MEP, corrosion inhibitors, Fukui function, NLO, and global reactivity parameters have been calculated to explain the versatile properties of the ligands. The NLO computed α and β parameters suggest that the compounds may have significant optoelectronic applications. Molecular docking was conducted to test the biological potency of anti-COVID-19 agents, DNA, and E. coli. Accordingly, results proved the antimicrobial effect of compounds by binding to E. coli proteins. The antibacterial was screened against E. coli, Klebsiella, Bacillus, and Pseudomonas sp. with increasing concentrations (0–1 mg/L). The investigation relied on the zone of inhibition and MIC values. MIC of the compounds against individual bacterial strains varied within the 0.2–2.5 mg/mL range. O-VAN exhibited the highest inhibition rate of 67.5 % against Klebsiella sp., while LO-VAN showed an inhibition rate of 64.15 %. The ligand's impact on bacteria was tested at pH levels (4–9). The research provides a new avenue for developing antimicrobial agents and holds the potential to serve as a model for developing new drugs. The antibacterial findings are exciting and significant for antimicrobial treatments. The quantum chemical DFT investigation explores novel research findings, especially for theoretical community chemists. The NLO study finds ligands industrial applications like optical filters, switches, photoelectronic devices, frequency mixing, electro-optic modulators, and photorefractive media. The investigation of SB's corrosion inhibitors further emphasizes its diverse industrial applications.
Research funding
This research has not received funding from any public, commercial, or non-profit agencies.
Conflicts of interest
The authors declared no conflicts of interest or personal relationships that could have influenced the research work.
Data availability statement
The data that has been used is confidential.
CRediT authorship contribution statement
Dhrubajyoti Majumdar: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Ankita Chatterjee: Writing – review & editing, Visualization, Validation, Software, Formal analysis, Data curation. Mehran Feizi-Dehnayebi: Writing – review & editing, Visualization, Validation, Supervision, Software, Formal analysis, Data curation. Neelakanta Sarvashiva Kiran: Visualization, Validation, Supervision, Software, Resources, Formal analysis, Data curation. Burak Tuzun: Visualization, Validation, Supervision, Software, Formal analysis, Data curation. Dipankar Mishra: Visualization, Validation, Supervision, Software, Investigation, Formal analysis, Data curation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
All the authors are thankful to the Central Laboratory of Tamralipta Mahavidyalaya, Purba Medinipur, Tamluk-721636, West Bengal, India. We acknowledge the Raman spectra, DRS, and SEM-EDX analysis conducted by SAIF, IIT Madras, Chennai-600036, and STIC, Cochin, Chennai-600036.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e35591.
Contributor Information
Dhrubajyoti Majumdar, Email: dmajumdar30@gmail.com.
Dipankar Mishra, Email: dmishra@tmv.ac.in.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Schiff Hugo. third ed. 1970-1979. The Great Soviet Encyclopaedia. [Google Scholar]
- 2.Majumdar D.J., Philip J.E., Dubey A., Tufail A., Roy S. Heliyon. 2023 doi: 10.1016/j.heliyon.2023.e16103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Salihovi M., Pazalja M., Halilovi S.Š., Veljovi E., Mahmutovic-Dizdarevi I., Sunčica Roca S., Novakovi I., Trifunovi S. J. Mol. Struct. 2021;1241 [Google Scholar]
- 4.El-Attar M.S., Ahmed F.M., Sadeek S.A., Mohamed S.F., Zordok W.A., El-Shiny W.H. Appl. Organomet. Chem. 2022;36 [Google Scholar]
- 5.Siddappa K., Mayana N.S. Bioinorgan. Chem. Appl. 2014;2014 doi: 10.1155/2014/483282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wozniczka M., Sutradhar M., Pombeiro A., Swiatek M., Pajak M., Gadek J., Chmiela M., Gonciarz W., Pasternak B., Kufelnicki A. Molecules. 2020;25:3462. doi: 10.3390/molecules25153462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ibrahim F., Jassim A.H., Muhyedeen B.R.J. Academic Publishing; 2018. LAP LAMBERT; p. 116. [Google Scholar]
- 8.Gavalyan V.B. Carbohydr. Polym. 2016;145:37–47. doi: 10.1016/j.carbpol.2016.02.076. [DOI] [PubMed] [Google Scholar]
- 9.Ghorai P., Brandao P., Bauza A., Frontera A., Saha A. Inorg. Chim. Acta. 2018;469:189–196. [Google Scholar]
- 10.Shyamal M., Mandal T.K., Panja A., Saha A. RSC Adv. 2014;4:53520–53530. [Google Scholar]
- 11.Ghorai P., Chakraborty A., Panja A., Mandal T.K., Saha A. RSC Adv. 2016;6:36020–36030. [Google Scholar]
- 12.Keskioglu E., Gunduzalp A.B., Cete S., Hamurcu F., Erk B. Spectrochim. Acta Mol. Biomol. Spectrosc. 2008;70(3):634–640. doi: 10.1016/j.saa.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 13.Abu-Hussen, Azza A.A. J. Coord. Chem. 2006;59(2):157–176. [Google Scholar]
- 14.Singh K., Barwa M.S., Tyagi P. Eur. J. Med. Chem. 2006;41(1):147–153. doi: 10.1016/j.ejmech.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Panneerselvam P., Nair R.R., Vijayalakshmi G., Subramanian E.H., Sridhar S.K. Eur. J. Med. Chem. 2005;40(2):225–229. doi: 10.1016/j.ejmech.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Elmac G., Duyar H., Aydıner B., Yahaya I., Seferoglu N., Sahin E., Çelik S.P., Açık L., Seferoglu Z. J. Mol. Struct. 2019;1184:271–280. [Google Scholar]
- 17.Süleymanoglu N., Demir E.E., Direkel S., Ünver Y. J. Mol. Struct. 2020;1218 [Google Scholar]
- 18.Abo-Aly M., Salem A., Sayed M., Aziz A.A. Spectrochim. Acta Mol. Biomol. Spectrosc. 2015;136:993–1000. doi: 10.1016/j.saa.2014.09.122. [DOI] [PubMed] [Google Scholar]
- 19.Shebl M. Spectrochim. Acta Mol. Biomol. Spectrosc. 2014;117:127–137. doi: 10.1016/j.saa.2013.07.107. [DOI] [PubMed] [Google Scholar]
- 20.Abd-Elzaher M.M., Labib A.A., Mousa H.A., Moustafa S.A., Ali M.M., El- Rashedy A.A. University Journal of Basic and Applied Sciences. 2016;5:85–96. [Google Scholar]
- 21.Duan J.-R., Liu H.-B., Jeyakkumar P., Gopala L., Li S., Geng R.-X., Zhou C.-H. MedChemComm. 2017;8:907–916. doi: 10.1039/c6md00688d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zehra S., Shavez Khan M., Ahmad I., Arjmand F. J. Biomol. Struct. Dyn. 2018:1–17. doi: 10.1080/07391102.2018.1467794. [DOI] [PubMed] [Google Scholar]
- 23.Sriram D., Yogeeswari P., Myneedu N.S. Bioorg. Med. Chem. Lett. 2006;16:2127–2129. doi: 10.1016/j.bmcl.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 24.Majumdar D.J., Dubey A., Tufail A., Sutradhar D., Roy S. Heliyon. 2023 doi: 10.1016/j.heliyon.2023.e16057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majumdar D.J., Philip J.E., Roy S., Tuzun B. Results in Chemistry. 2022;4 [Google Scholar]
- 26.Beegum S., Sheena Mary Y., Varghese H.T., Panicker C.Y., Armaković S., Armaković S.J., Zitko J., Dolezal M., Alsenoy C.V. J. Mol. Struct. 2017;1131:1–15. [Google Scholar]
- 27.Abraham C.S., Muthu S., Prasana J.C., Rizwana B.F., Armaković S., Armaković S.J. J. Mol. Struct. 2018;1171:733–746. [Google Scholar]
- 28.Abraham C.S., Muthu S., Prasana J.C., Armaković S.J., Armaković S., F. R. B, B. G. A. S CB (Curr. Biol.) 2018;77:131–145. doi: 10.1016/j.compbiolchem.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Armaković S.J., Abramović B.F. J. Mol. Model. 2016;22:240. doi: 10.1007/s00894-016-3101-2. [DOI] [PubMed] [Google Scholar]
- 30.Houas N., Kitouni S., Chafai N., Ghedjati S., Djenane M., Tounsi A. J. Mol. Struct. 2023;1284 doi: 10.1016/j.molstruc.2023.135356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elkolli M., Chafai N., Chafaa S., Kadi I., Bensouici C., Hellal A. J. Mol. Struct. 2022;1268 doi: 10.1016/j.molstruc.2022.134005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.kerkour R., Chafai N., Moumeni O., Chafaa S. J. Mol. Struct. 2023;1272 doi: 10.1016/j.molstruc.2022.134196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tabbiche A., Bouchama A., Chafai N., Zaidi F., Chiter C., Yahiaoui M., Abiza A. J. Mol. Struct. 2022;1261 doi: 10.1016/j.molstruc.2022.132865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adjissi L., Chafai N., Benbouguerra K., Kirouani I., Hellal A., Layaida H., Elkolli M., Bensouici C., Chafaa S. J. Mol. Struct. 2022;1270 doi: 10.1016/j.molstruc.2022.134005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tlidjane H., Chafai N., Chafaa S., Bensouici C., Benbouguerra K. J. Mol. Struct. 2022;1250 doi: 10.1016/j.molstruc.2021.131853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benbouguerra K., Chafai N., Chafaa S., Touahria Y.I., Tlidjane H. J. Mol. Struct. 2021;1239 doi: 10.1016/j.molstruc.2021.130480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chafai N., Chafaa S., Benbouguerra K., Hellal A., Mehri M. J. Mol. Struct. 2019;1181:83–92. [Google Scholar]
- 38.Djenane M., Chafaa S., Chafai N., Kerkour R., Hellal A. J. Mol. Struct. 2019;1175:398–413. [Google Scholar]
- 39.Benbouguerra K., Chafaa S., Chafai N., Mehri M., Moumeni O., Hellal A. J. Mol. Struct. 2018;1157:165–176. [Google Scholar]
- 40.Chafai N., Chafaa S., Benbouguerra K., Daoud D., Hellal A., Mehri M. J. Taiwan Inst. Chem. Eng. 2017;70:331–344. [Google Scholar]
- 41.Basri R., Khalid M., Shafiq Z., Tahir M.S., Khan M.U., Tahir M.N., Naseer M.M., Braga A.A.C. ACS Omega. 2020;5:30176–30188. doi: 10.1021/acsomega.0c04653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chalkha M., Nour H., Chebbac K., Nakkabi A., Bahsis L., Bakhouch M., Akhazzane M., Bourass M., Chtita S., Bin Jardan Y.A., Augustyniak M., Bourhia M., Aboul-Soud M.A.M., El Yazidi M. ACS Omega. 2022;7:46731–46744. doi: 10.1021/acsomega.2c05788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imran M., Khalid M., Jawaria R., Ali A., Asghar M.A., Shafiq Z., Assiri M.A., Lodhi H.M., Braga A.C. ACS Omega. 2021;6:33914–33922. doi: 10.1021/acsomega.1c04984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narayanswamy A., Ramakrishna D., Raja Shekar P.V., Rajendrachari S., Sudhakar R. ACS Omega. 2024;9(11):13262–13273. doi: 10.1021/acsomega.3c10048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choudhary R., Chatterjee A., Venkatraman S.K., Koppala S., Abraham J., Swamiappan S. S. Bioactive Materials. 2018;3(3):218–224. doi: 10.1016/j.bioactmat.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nayak S., Gamez P., Kozlevčar B., Pevec A., Roubeau O., Dehnen S., Reedijk J. Polyhedron. 2010;29:2291–2296. [Google Scholar]
- 47.Frisch M., Trucks G., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G. Gaussian, Inc.; Wallingford CT: 2009. Gaussian 09, Revision D. 01; p. 201. [Google Scholar]
- 48.Garrett M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MGL Tools. 1.5. 6 (ADT)/MGL Tools 1. 6. The Scripps Research Institute: La Jolla; CA, USA: 2016. [Google Scholar]
- 50.Neidle S., Rayner E.L., Simpson I.J., Smith N.J., Mann J., Baron A., Opoku-Boahen Y., Fox K.R., Hartley J.A., KellaND L.R. Chem. Commun. 1999;10:929–930. [Google Scholar]
- 51.(a) Mouilleron S., Badet-Denisot M.-A., Golinelli-Pimpaneau B. JMB (J. Mol. Biol.) 2008;377:1174–1185. doi: 10.1016/j.jmb.2008.01.077. [DOI] [PubMed] [Google Scholar]; (b) Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 52.Milusheva M., Todorova M., Gledacheva V., Stefanova I., Feizi-Dehnayebi M., Pencheva M., Nedialkov P., Tumbarski Y., Yanakieva V., Tsoneva S. Pharmaceuticals. 2023;16(12):1660. doi: 10.3390/ph16121660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Becke A.D. J. Chem. Phys. 1992;96(3):2155–2160. [Google Scholar]
- 54.Vautherin D., Brink D.T. Phys. Rev. C. 1972;5(3):626. [Google Scholar]
- 55.Hohenstein E.G., Chill S.T., Sherrill C.D. J. Chem. Theor. Comput. 2008;4(12):1996–2000. doi: 10.1021/ct800308k. [DOI] [PubMed] [Google Scholar]
- 56.Shakir M., Hanif S., Asif Sherwani Mohd, Mohammad O., Al-Resayes S.I. J. Mol. Struct. 2015;1092:143–159. [Google Scholar]
- 57.Fugu M.B., Ndahi N.P., Paul B.B., Mustapha A.N. J. Chem. Pharmaceut. Res. 2013;5(4):22–28. [Google Scholar]
- 58.Hohenstein E.G., Chill S.T., Sherrill C.D. J. Chem. Theor. Comput. 2008;4(12):1996–2000. doi: 10.1021/ct800308k. [DOI] [PubMed] [Google Scholar]
- 59.Sadhukhan D., Ray A., Rosair G., Charbonnière L., Mitra S. BCSJ. 2011;84:211–217. [Google Scholar]
- 60.Akbari Z., Stagno C., Iraci N., Efferth T., Omer E.A., Piperno A., Montazerozohori M., Feizi-Dehnayebi M., Micale N. J. Mol. Struct. 2024;1301 [Google Scholar]
- 61.Šponer Jiří, Hobza P. Int. J. Quant. Chem. 1996;57(5):959–970. [Google Scholar]
- 62.Zinatloo-Ajabshir S., Rakhshani S., Mehrabadi Z., Farsadrooh M., Feizi-Dehnayebi M., Rakhshani S., Dušek M., Eigner V., Rtimi S., Aminabhavi T.M. J. Environ. Manag. 2024;350 doi: 10.1016/j.jenvman.2023.119545. [DOI] [PubMed] [Google Scholar]
- 63.Elangovan N., Thomas R., Sowrirajan S. J. Mol. Struct. 2022;1250 [Google Scholar]
- 64.Abu‐Dief A.M., El‐Khatib R.M., El‐Dabea T., Feizi‐Dehnayebi M., Barnawi I.O., Alsehli A.H., Al‐Ghamdi K., El‐Remaily M.A.E.A.A.A. Appl. Organomet. Chem. 2024:e7358. [Google Scholar]
- 65.Kurbanova M., Ashfaq M., Sadigova A., Feizi-Dehnayebi M., Maharramov A., Tahir M. J. Struct. Chem. 2024;65(1):92–106. [Google Scholar]
- 66.Parr R.G., Pearson R.G. J. Am. Chem. Soc. 1983;105:7512–7516. [Google Scholar]
- 67.Alieva Qudrat M., Tahir M.N., Munawar K.S., Feizi-Dehnayebi M., Ashfaq M., Hasanova Saadat S., Hasanova Ulviyya M., Kerimova Tahira Q., Iskenderova Simuzer A., Alieva Shebnem C., Muhammed M.E. J. Mol. Struct. 2023;1297 [Google Scholar]
- 68.Milusheva M., Gledacheva V., Stefanova I., Feizi-Dehnayebi M., Mihaylova R., Nedialkov P., Cherneva E., Tumbarski Y., Tsoneva S., Todorova M. Int. J. Mol. Sci. 2023;24(18) doi: 10.3390/ijms241813855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chalkha M., Chebbac K., Nour H., Nakkabi A., El Moussaoui A., Tüzün B., El Yazidi M. Arab. J. Chem. 2024;17(1) [Google Scholar]
- 70.Saber A.L., Tuzun B., Alessa H., Althakafy J.T. Curr. Anal. Chem. 2023;19(3):262–271. [Google Scholar]
- 71.Sakr M.A., Sherbiny F.F., El-Etrawy A.A.S. J. Fluoresc. 2022;32(5):1857–1871. doi: 10.1007/s10895-022-03000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vennila M., Rathikha R., Muthu S., Jeelani A., Irfan A., A J. Mol. Liq. 2022;359(2022) [Google Scholar]
- 73.Selvakumari S., Potla K.M., Shanthi D., Irfan A., A, Muthu S., S J. Mol. Liq. 2023;382(2023) [Google Scholar]
- 74.Kowalska-Krochmal B., Dudek-Wicher R. Pathogens. 2021;10(2):165. doi: 10.3390/pathogens10020165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yan Y., Li Y., Zhang Z., Wang X., Niu Y., Zhang S., Xu W., Ren C. Colloids Surf., B: Bio interfaces. 2021;202 doi: 10.1016/j.colsurfb.2021.111682. [DOI] [PubMed] [Google Scholar]
- 76.Choudhary R., Venkatraman S.K., Chatterjee A., Vecstaudza J., Yáñez-Gascón M.J., Perez-Sanchez H., Locs J., Abraham J., Swamiappan S. Adv. Powder Technol. 2019;30(9):1950–1964. [Google Scholar]
- 77.Dagur A.H., Kartha A.A., Subodh M.A., Vishnu C., Arun D., Kumar M.G., Abraham W.S., Chatterjee A., Abraham J. J. Manuf. Process. 2017;30:27–40. [Google Scholar]
- 78.Sampathkumar B., Khachatourians G.G., Korber D.R. Appl. Environ. Microbiol. 2003;69(1):122–129. doi: 10.1128/AEM.69.1.122-129.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that has been used is confidential.