Abstract
Background
Helicobacter pylori (H. pylori) is a significant global health concern, posing a high risk for gastric cancer. Conventional diagnostic and screening approaches are inaccessible, invasive, inaccurate, time-consuming, and expensive in primary clinics.
Objective
This study aims to apply machine learning (ML) models to detect H. pylori infection using limited laboratory parameters from routine blood tests and to investigate the association of these biomarkers with clinical outcomes in primary clinics.
Methods
A retrospective analysis with three ML and five ensemble models was conducted on 1409 adults from Hubei Provincial Hospital of Traditional Chinese Medicine. evaluating twenty-three blood test parameters and using the urea breath test as the gold standard for diagnosing H. pylori infection.
Results
In our comparative study employing three different feature selection strategies, Random Forest (RF) model exhibited superior performance over other ML and ensemble models. Multiple evaluation metrics underscored the optimal performance of the RF model (ROC = 0.951, sensitivity = 0.882, specificity = 0.906, F1 = 0.906, accuracy = 0.894, PPV = 0.908, NPV = 0.880) without feature selection. Key biomarkers identified through importance ranking and shapley additive Explanations (SHAP) analysis using the RF model without feature selection include White Blood Cell Count (WBC), Mean Platelet Volume (MPV), Hemoglobin (Hb), Red Blood Cell Count (RBC), Platelet Crit (PCT), and Platelet Count (PLC). These biomarkers were found to be significantly associated with the presence of H. pylori infection, reflecting the immune response and inflammation levels.
Conclusion
Abnormalities in key biomarkers could prompt clinical workers to consider H. pylori infection. The RF model effectively identifies H. pylori infection using routine blood tests, offering potential for clinical application in primary clinics. This ML approach can enhance diagnosis and screening, reducing medical burdens and reliance on invasive diagnostics.
Keywords: Helicobacter pylori, Machine learning, Routine blood test, Mass screening
1. Introduction
Helicobacter pylori (H. pylori), a prevalent bacterium infecting over half the global population [1], is increasingly recognized for pivotal risk factor to gastric cancer [2], cardiovascular disease [3], colorectal cancer [4], and Alzheimer's disease [5]. H. pylori infection is associated with to 89 % of noncardiac stomach tumors [6], accounting for 78 % of all cases of gastric cancer [2]. A sixfold increase in the risk of gastric cancer was observed in populations with 100 % H. pylori infection, as opposed to those without infection [7]. Chronic infection of H. pylori decreases the release of gastric acid, leading to genetic instability and promoting the growth of the microbiome, which converts food components into substances that might cause cancer. H. pylori-induced chronic inflammation facilitates the progression of gastric cancer, resulting in the emergence of precancerous diseases such atrophic gastritis and intestinal metaplasia [8].
Eliminating H. pylori is an efficient way to combat inflammation [9], stop the advancement of mucosal damage [10], prevent further DNA damage [11], improve gastric acid secretion [12], and restore a healthy microbiome [13]. Given the potential for H. pylori eradication through a brief course of antibiotic treatment [14], the identification and elimination of H. pylori infection emerge as a promising strategy to mitigate the substantial disease burden associated with gastric cancer [15,16]. However, current diagnostic methods for H. pylori, including breath tests [17], serological assays [18], stool antigen tests [19], gastroscopy with biopsy [20], and culture of biopsy samples [21], are essential yet often time-consuming, costly, inaccurate, and require extensive preparation and clinical guidance [[21], [22], [23], [24], [25]]. This highlights an urgent need for the development of more efficient and accessible screening and diagnostic techniques for H. pylori.
Routine blood tests are the most common and fundamental diagnostic procedures in primary healthcare systems, with most patients undergoing these examinations. However, few patients presenting with gastrointestinal symptoms are initially screened for H. pylori infection. Specifically, primary clinics lack accessibility for further experiments such as gastroscopy and histopathology. This poses a challenge in primary clinics: how to detect H. pylori infection at an early stage using only routine blood tests, without relying on additional laboratory parameters. Routine blood tests, notable for their cost-efficiency and swift outcomes [26], are indispensable in early disease identification [27], bespoke therapies [28], chronic illnesses managements [29], and medical research [30]. Recently, routine blood tests have emerged as a minimally invasive alternative to traditional biopsies, notably in lung cancer diagnostics [31]. Moreover, contemporary research highlights routine blood tests’ role in detecting and diagnosing brain tumors, underscoring the importance of early intervention [32]. Another study confirms the effectiveness of the Biochemistry and Hematology Outcome model, which is developed by Portsmouth Hospital NHS Trust researchers, in predicting patient mortality from routine blood tests for 9497 adults, emphasizing its adaptability across diverse hospital settings [33]. Various studies have assessed the efficacy of routine blood tests to differentiate between viral and bacterial infections [34] such as COVID-19 [35], tuberculosis [36], and Clostridium difficile [37]. Consequently, emerging evidence on using routine blood tests for H. pylori screening shows promising, unexplored potential.
Machine learning (ML) is transforming medical screening, augmenting routine blood tests with its advanced pattern recognition and predictive analytics capabilities, enabling the detection of nuanced biomarkers for early and non-invasive screening. Previous studies have confirmed the effectiveness of ML models, demonstrating outstanding sensitivity and precision in predicting diverse health outcomes [[38], [39], [40]]. To date, several studies have investigated that ML models have an excellent performance for predicting bacterial infection [[41], [42], [43]]. Furthermore, ML has been demonstrated the feasible approaches in identifying intricate biomarkers for complex relationships with datasets [[44], [45], [46]]. Therefore, integrating ML into routine blood tests for detecting H. pylori appears viable without further investigation.
Given the automation and regularity of routine blood tests, we aimed to apply ML models to detect H. pylori infection using routine blood tests and investigate the biomarkers’ association with outcomes. In this study, we propose a ML-based strategy for a cost-effective, and efficient diagnostic method, leveraging ML to identify subtle biomarkers indicative of H. pylori infection. Using our ML-based strategy, the detection rate of H. pylori infection could be increased in a cost-effective manner.
2. Materials and methods
2.1. Study design
A retrospective cross-sectional analysis was conducted on adult participants at the Hubei Provincial Hospital of Traditional Chinese Medicine in Wuhan from January 1, 2021, to June 30, 2023.
2.2. Participants and data collection
The study enrolled participants aged 18 and above who underwent the urea breath test at Hubei Provincial Hospital of Traditional Chinese Medicine, encompassing both inpatient and outpatient cases. Eligible participants had at least one recorded urea breath test and routine blood test. No exclusions were made based on gender, race, comorbidities, or illness severity. For this analysis, we considered only the first urea breath test and the first routine blood test from each unique patient encounter, with a maximum interval of 24 h between the two tests.
Data extraction was conducted by two independent reviewers who retrieved relevant data elements from the hospital systems for all eligible patients. The dataset, provided by the Department of Spleen and Gastroenterology at Hubei Provincial Hospital of Traditional Chinese Medicine, included 1409 cases. Data collected comprised participant age, gender, routine blood test parameters, and urea breath test results. Patients with known malignancies, chronic inflammatory diseases, or other infections that could influence blood test parameters were excluded after a thorough review of their medical histories and clinical records. H. pylori infection was confirmed using the urea breath test, a specific and reliable diagnostic tool for detecting active infection.
2.3. Measurements
2.3.1. Demographic characteristics
Participants reported sociodemographic information, including age and gender.
2.3.2. H. pylori infection
According to the Chinese clinical guideline for H. pylori infection in primary clinics [24], an active H. pylori infection can be diagnosed if any of the following three criteria are met as follows: a positive result from any one of the following tests on gastric mucosal tissue—rapid urease test, histological staining of tissue sections, or bacterial culture; positive results from a or -urea breath test; or positive results from HpSA testing (clinically validated monoclonal antibody method). Positive serum H. pylori antibody testing (using clinically validated, highly accurate reagents) indicates a past infection, and if the patient has never been treated, this can be considered an active infection.
Since the urea breath test is commonly used in Chinese primary clinics, it is often utilized as the gold standard diagnostic method for detecting H. pylori infection. This test is based on the principle that H. pylori secrete an enzyme called urease, which facilitates the hydrolysis of ingested urea, resulting in the production of carbon dioxide () and ammonia. By using -labelled urea, the subsequent exhalation of can be tracked as a reliable marker for H. pylori within the digestive tract. Thus, the diagnosis of whether the patient has an H. pylori infection is primarily based on the urea breath test.
2.3.3. Routine blood test
Our routine blood tests include a total of 23 parameters in ML models: white blood cell count (WBC), red blood cell count (RBC), hemoglobin (Hb), absolute lymphocyte count (ALC), absolute monocyte count (AMC), absolute neutrophil count (ANC), absolute eosinophil count (AEC), absolute basophil count (ABC), lymphocyte percentage (Lymph%), monocyte percentage (Mono%), neutrophil percentage (Neut%), eosinophil percentage (Eos%), and basophil percentage (Baso%), platelet distribution width (PDW), hematocrit (HCT), mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), red cell distribution width (RDW), platelet count (PLC), mean platelet volume (MPV), platelet crit (PCT), C-reactive protein (CRP). WBC may suggest H. pylori infection or immunodeficiency. A long-lasting H. pylori infection can make anemia or thirst worse, which can be shown by abnormal RBC. Because of stomach bleeds caused by H. pylori, Hb values may also show anemia or polycythemia. ALC and AMC indicate about how the immune system reacts to H. pylori. Infections with H. pylori bugs are one type of disease that ANC can show. Immune response markers (AEC, ABC, Lymph%, Mono%, Neut%, Eos%, and Baso%) may indicate inflammation caused by H. pylori infection. PDW alterations might suggest platelet abnormalities associated with H. pylori-related bleeding. Hct abnormalities may suggest anemia or polycythemia caused by a protracted H. pylori infection. MCV, MCH, and MCHC aid in the diagnosis of anemia and hemoglobinopathies caused by H. pylori's effect on nutritional absorption. RDW variations may indicate anemia from chronic inflammation or dietary deficiencies caused by H. pylori. PLC abnormalities might suggest bleeding or bone marrow concerns associated with severe H. pylori disorders. MPV might indicate the alteration of platelet production and function caused by chronic H. pylori infections. PCT evaluates platelet status, indicating changes owing to H. pylori issues. CRP is an inflammatory measure that might indicate the systemic response to H. pylori infection.
2.3.4. Feature selection
For feature selection, we employed three strategies: Lasso L1 regularization, t-test/Chi-square test, and an approach without selection. Lasso L1 regularization [47] is a feature selection method that enhances model accuracy by penalizing the absolute size of the regression coefficients, effectively shrinking less important feature coefficients to zero. The penalty coefficient used in our study was set to 1.0, which was determined based on cross-validation to balance model complexity and prediction accuracy. We implemented this method using the Scikit-learn library in Python. The t-test/Chi-square test is utilized for feature selection by statistically evaluating the independence between features and the target variable, helping to identify the most relevant predictors for the model. Features with p-values below 0.05 were considered statistically significant and retained for model training, while those above this threshold were excluded. This approach helps in identifying the most relevant predictors for the model. We performed these tests using the scipy.stats library in Python. As a baseline, we also trained models without any feature selection to compare the impact of feature selection methods on model performance. This approach provided a reference point for evaluating the effectiveness of the feature selection strategies employed.
2.3.5. ML models
We selected five ensemble models and three ML algorithms, each with established effectiveness in classification tasks. Logistic Regression (LR) [48] is a fundamental linear model used for binary classification tasks due to its simplicity and interpretability. Support Vector Machine (SVM) [49] excels in high-dimensional and complex settings, providing robust classification performance. Multilayer Perceptron (MLP) [50] is a neural network model that effectively identifies complex patterns through its layered structure.
Ensemble models enhance prediction performance by combining multiple models to mitigate overfitting and improve generalization. Random Forest (RF) [51] is esteemed for its high accuracy and ability to mitigate overfitting through ensemble learning, aggregating the predictions of decision trees via a majority vote to enhance predictive strength. LightGBM [52] is a highly efficient gradient boosting framework that uses a histogram-based method to bin continuous features, accelerating training speed, optimizing memory usage, and excelling in processing large-scale datasets with remarkable speed and effectiveness. AdaBoost [53] prioritizes difficult scenarios, enhancing classification precision by iteratively adjusting weights to improve the model. XGBoost [54], an advanced gradient boosting system developed by Chen, iteratively refines models by splitting tree nodes and fitting residuals, demonstrating exceptional scalability and superior performance across diverse applications. CatBoost [55], introduced in 2018, is a cutting-edge gradient boosting algorithm known for its exceptional handling of categorical features, reduced training times, and the use of a greedy strategy to pinpoint optimal tree splits, thereby enhancing prediction accuracy.
2.4. Statistical analysis
Statistical analyses were conducted using Python 3.11.5 within the Microsoft Visual Code environment. Samples with missing data were initially excluded. Categorical variables were encoded using scikit-learn's LabelEncoding, and string data were transformed into numeric types. Descriptive statistics, such as frequencies, analyzed categorical variables at each level, while chi-square tests validated the accuracy coefficients for each subscale. Continuous variables were described using means () and standard deviations (SD), supplemented by unpaired t-tests and chi-square tests where applicable. Data distributions were visualized using violin plots. To ensure uniformity, each feature was standardize by the formula and then normalized using the StandardScaler function from the Scikit-learn package in Python. To tackle the issue of imbalanced samples, the RandomOverSampler function was applied, which led to notable enhancements in the models' Receiver Operating Characteristic (ROC) and overall accuracy. Correlation heatmaps were then generated to depict relationships among variables.
Feature selection was implemented using three approaches: Lasso L1 regularization, t-test/Chi-square test, and no selection. Subsequently, the data were divided into training (80 %) and testing (20 %) sets using leave-one-out cross-validation. Eight conventional ML and ensemble models were trained: LR, SVM, MLP, RF, LightGBM, AdaBoost, XGBoost, and CatBoost. Their performances were assessed on the testing sets using metrics such as ROC, sensitivity, specificity, F1 Score, accuracy, positive predictive value (PPV), and negative predictive value (NPV).
Additionally, importance rankings and Shapley Additive Explanations (SHAP) values were utilized to determine each parameter's contribution to the models. Features were scored for their importance using version 0.42.1 of the SHAP Python package, with all features of each model selected for analysis. A two-sided p-value of less than 0.05 was set as the threshold for statistical significance. The Python environment was equipped with essential packages including pandas 2.1.4, numpy 1.24.3, scikit-learn 1.3.0, scipy 1.11.4, catboost 1.2, lightgbm 4.1.0, seaborn 0.12.2, SHAP 0.42.1, and matplotlib 3.8.0, enabling comprehensive data analysis.
3. Results
3.1. Characteristics of the dataset
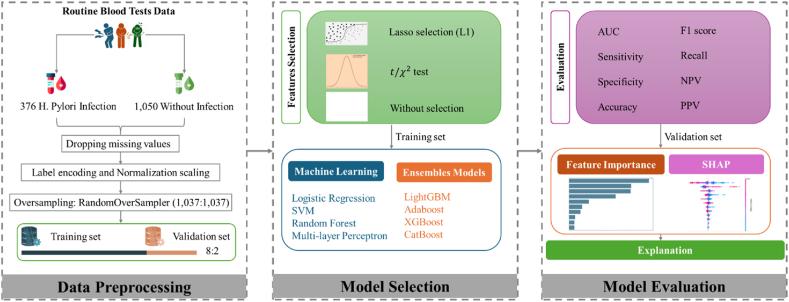
Fig. 1 presents a flowchart outlining the study's methodology, including data preprocessing, model selection, and evaluation. Table 1 complements this by detailing the input variables used in the study and their respective rates of missing data. The data show minimal missing values, with only four individuals (1.064 %) with H. pylori infection and thirteen individuals (1.238 %) without the infection having incomplete data. This indicates a high level of data integrity across both groups.
Fig. 1.
Overview of the study flowchart.
Table 1.
Input variables and their missing rates.
| Variables | Missing values |
% Missing |
||
|---|---|---|---|---|
| H. pylori Infection(n = 376) | Without H. pylori Infection(n = 1050) | H. pylori Infection(n = 376) | Without H. pylori Infection(n = 1050) | |
| Gender | 0 | 0 | 0.000 % | 0.000 % |
| Age | 0 | 0 | 0.000 % | 0.000 % |
| WBC | 2 | 1 | 0.532 % | 0.095 % |
| RBC | 2 | 1 | 0.532 % | 0.095 % |
| Hb | 2 | 1 | 0.532 % | 0.095 % |
| ALC | 2 | 1 | 0.532 % | 0.095 % |
| AMC | 2 | 1 | 0.532 % | 0.095 % |
| ANC | 2 | 1 | 0.532 % | 0.095 % |
| AEC | 2 | 1 | 0.532 % | 0.095 % |
| ABC | 2 | 1 | 0.532 % | 0.095 % |
| Baso% | 2 | 1 | 0.532 % | 0.095 % |
| Eos% | 2 | 1 | 0.532 % | 0.095 % |
| Lymph% | 2 | 1 | 0.532 % | 0.095 % |
| Mono% | 2 | 1 | 0.532 % | 0.095 % |
| Neut% | 2 | 1 | 0.532 % | 0.095 % |
| PDW | 2 | 1 | 0.532 % | 0.095 % |
| HCT | 2 | 1 | 0.532 % | 0.095 % |
| MCV | 2 | 1 | 0.532 % | 0.095 % |
| MCH | 2 | 1 | 0.532 % | 0.095 % |
| MCHC | 2 | 1 | 0.532 % | 0.095 % |
| RDW | 2 | 1 | 0.532 % | 0.095 % |
| PLC | 2 | 1 | 0.532 % | 0.095 % |
| MPV | 2 | 1 | 0.532 % | 0.095 % |
| PCT | 2 | 1 | 0.532 % | 0.095 % |
| CRP | 4 | 13 | 1.064 % | 1.238 % |
Table 2 in the study provides a comprehensive comparison of the dataset (N = 1409) between two groups: those with H. pylori infection (n = 372) and those without the infection (n = 1037). The study reports 222 females and 150 males with H. pylori infection, compared to 589 females and 448 males without the infection. The age average is similar in both groups, with no significant difference. Notably, significant differences ( < 0.05) are observed in several parameters such as WBC (), ALC (), AMC (, ANC (, and PDW (, suggesting significant differences between individuals with and without H. pylori infection. Other parameters, like RBC, Hb, and various percentage distributions of blood components (Baso%, Eos%, Lymph%, Mono%, Neut%), along with MCV, MCH, MCHC, RDW, PLC, MPV, PCT, and CRP, show no statistically significant difference between the two groups, as indicated by higher p-values.
Table 2.
Characteristics of the dataset (N = 1409).
| Parameters | H. pylori Infection(n = 372) | Without H. pylori Infection(n = 1037) | t/Chi-Square | -value | |
|---|---|---|---|---|---|
| Gender | Female | 222 | 589 | 0.815 | 0.367 |
| Male | 150 | 448 | |||
| Age | 54.204(12.168) | 54.943(22.933) | 0.777 | 0.438 | |
| WBC | 5.582(1.515) | 5.308(1.742) | −2.872 | 0.004 | |
| RBC | 4.358(0.454) | 4.398(0.508) | 1.418 | 0.157 | |
| Hb | 132.166(14.866) | 133.029(14.277) | 0.971 | 0.332 | |
| ALC | 1.533(0.523) | 1.455(0.492) | −2.524 | 0.012 | |
| AMC | 0.358(0.144) | 0.337(0.134) | −2.423 | 0.016 | |
| ANC | 3.553(1.299) | 3.384(1.515) | −2.067 | 0.039 | |
| AEC | 0.102(0.134) | 0.102(0.119) | −0.013 | 0.990 | |
| ABC | 0.019(0.037) | 0.020(0.038) | 0.611 | 0.541 | |
| Baso% | 0.565(0.409) | 0.591(0.426) | 1.046 | 0.296 | |
| Eos% | 1.907(2.181) | 1.953(1.855) | 0.363 | 0.717 | |
| Lymph% | 28.286(8.109) | 28.376(8.182) | 0.182 | 0.855 | |
| Mono% | 6.493(2.138) | 6.496(2.010) | 0.028 | 0.977 | |
| Neut% | 62.749(9.245) | 62.584(9.42) | −0.295 | 0.768 | |
| PDW | 16.609(0.613) | 16.684(0.622) | 2.005 | 0.045 | |
| Hct | 0.396(0.042) | 0.399(0.040) | 1.118 | 0.264 | |
| MCV | 91.096(5.531) | 91.049(5.587) | −0.139 | 0.889 | |
| MCH | 30.380(2.205) | 30.36(2.222) | −0.150 | 0.881 | |
| MCHC | 333.317(9.061) | 333.237(8.538) | −0.149 | 0.882 | |
| RDW | 13.097(1.083) | 13.082(0.956) | −0.228 | 0.820 | |
| PLC | 211.656(50.747) | 206.85(57.543) | −1.511 | 0.131 | |
| MPV | 8.786(1.044) | 8.913(1.165) | 1.950 | 0.052 | |
| PCT | 0.184(0.039) | 0.181(0.045) | −0.986 | 0.324 | |
| CRP | 1.754(4.926) | 1.475(6.081) | −0.879 | 0.380 | |
3.2. Heatmap analysis, distribution visualization, and feature selection in the dataset
Table 3 demonstrates that the use of random oversampling enhances model performance after preprocessing without feature selection. For classifiers not employing random oversampling, the highest performance metrics are achieved with the LR model (ROC = 0.577, Sensitivity = 0, Specificity = 1, F1 Score = 0, Accuracy = 0.791, NPV = 0.791). Upon applying RandomOversampler, the performance metrics improve, with the RF model attaining ROC = 0.951, Sensitivity = 0.882, Specificity = 0.906, F1 Score = 0.895, Accuracy = 0.894, and NPV = 0.880. Fig. 2 illustrates the diverse characteristics of all variables in the study. Fig. 2A shows violin plot distributions for each variable, facilitating a visual comparison between the control group (without H. pylori infection) and the experimental group (with H. pylori infection). Despite the overall distributions being broadly similar, significant differences are observed in the outliers between the two groups. Fig. 2B indicates a heatmap that reveals strong correlations between certain variables, highlighting the importance of feature selection for enhancing model interpretability and efficiency. Fig. 2C depicts the relationship between the coefficients and C (the inverse of regularization strength) using Lasso L1 regularization. This visualization indicates that many variables are non-essential and can be omitted to improve model performance or prevent overfitting. Table 4 supports these findings by listing the coefficients derived from Lasso L1 regularization, with variables such as WBC, Neut%, MCHC, and RDW showing zero coefficients, suggesting their minimal influence and potential for exclusion.
Table 3.
Performance metrics of classifiers using random oversampling preprocessing and no preprocessing without feature selection.
| Classifier | ROC | Sensitivity | Specificity | F1 Score | Accuracy | NPV | |
|---|---|---|---|---|---|---|---|
| Without RandomOversample | LR | 0.577 | 0 | 1 | 0 | 0.791 | 0.791 |
| SVM | 0.535 | 0 | 1 | 0 | 0.791 | 0.791 | |
| AdaBoost | 0.5 | 0 | 1 | 0 | 0.791 | 0.791 | |
| RF | 0.502 | 0.051 | 0.978 | 0.09 | 0.784 | 0.796 | |
| XGBoost | 0.514 | 0.102 | 0.897 | 0.136 | 0.73 | 0.791 | |
| CatBoost | 0.539 | 0.051 | 0.978 | 0.09 | 0.784 | 0.796 | |
| LightGBM | 0.53 | 0.119 | 0.901 | 0.159 | 0.738 | 0.794 | |
| MLP | 0.571 | 0 | 0.991 | 0 | 0.784 | 0.789 | |
| After RandomOversample | LR | 0.577 | 0.538 | 0.571 | 0.552 | 0.554 | 0.542 |
| SVM | 0.743 | 0.684 | 0.680 | 0.687 | 0.682 | 0.673 | |
| RF | 0.951 | 0.882 | 0.906 | 0.895 | 0.894 | 0.880 | |
| MLP | 0.797 | 0.741 | 0.700 | 0.730 | 0.720 | 0.721 | |
| LightGBM | 0.942 | 0.892 | 0.833 | 0.869 | 0.863 | 0.880 | |
| AdaBoost | 0.686 | 0.660 | 0.581 | 0.641 | 0.622 | 0.621 | |
| XGBoost | 0.943 | 0.901 | 0.818 | 0.868 | 0.860 | 0.888 | |
| CatBoost | 0.936 | 0.877 | 0.818 | 0.855 | 0.848 | 0.865 |
Fig. 2.
Characteristics of all variables. (A) We display violin plot distributions for each variable, providing a visual comparison of their distributions. (B) We illustrate a heatmap of the correlations between variables, highlighting interdependencies. (C) We present a graph depicting the relationship between coefficients and C (the inverse of regularization strength) using Lasso L1 regularization, offering insights into the influence of regularization on variable selection.
Table 4.
Coefficients derived from lasso l1 regularization.
| Variables | Coefficient |
|---|---|
| Gender | −0.097 |
| Age | −0.133 |
| WBC | 0.000 |
| RBC | −0.749 |
| Hb | 0.377 |
| ALC | 0.253 |
| AMC | 0.160 |
| ANC | −0.017 |
| AEC | −0.104 |
| ABC | −0.100 |
| Baso% | 0.101 |
| Eos% | 0.104 |
| Lymph% | −0.218 |
| Mono% | −0.067 |
| Neut% | 0.000 |
| PDW | −0.149 |
| HCT | 0.219 |
| MCV | −0.154 |
| MCH | −0.291 |
| MCHC | 0.000 |
| RDW | 0.000 |
| PLC | −0.453 |
| MPV | −0.230 |
| PCT | 0.350 |
| CRP | 0.029 |
3.3. Performance of various models with different feature selection strategies
Table 5 presents a comprehensive comparison of three ML models and five ensemble models, evaluated using performance metrics such as ROC, sensitivity, specificity, F1 score, accuracy, PPV, and NPV.
Table 5.
Performance metrics of various classifiers under different feature selection methods.
| Classifier | ROC | Sensitivity | Specificity | F1 Score | Accuracy | PPV | NPV | |
|---|---|---|---|---|---|---|---|---|
| Feature selection with Lasso | LR | 0.575 | 0.542 | 0.581 | 0.558 | 0.561 | 0.575 | 0.549 |
| SVM | 0.742 | 0.689 | 0.626 | 0.673 | 0.658 | 0.658 | 0.658 | |
| RF | 0.951 | 0.901 | 0.882 | 0.895 | 0.892 | 0.888 | 0.895 | |
| MLP | 0.763 | 0.717 | 0.680 | 0.709 | 0.699 | 0.700 | 0.697 | |
| LightGBM | 0.937 | 0.901 | 0.793 | 0.858 | 0.848 | 0.820 | 0.885 | |
| AdaBoost | 0.687 | 0.684 | 0.567 | 0.652 | 0.627 | 0.622 | 0.632 | |
| XGBoost | 0.944 | 0.906 | 0.828 | 0.875 | 0.867 | 0.846 | 0.894 | |
| CatBoost | 0.925 | 0.844 | 0.833 | 0.842 | 0.839 | 0.840 | 0.837 | |
| Feature selection with t/Chi-Square | LR | 0.575 | 0.462 | 0.586 | 0.497 | 0.523 | 0.538 | 0.511 |
| SVM | 0.600 | 0.519 | 0.586 | 0.542 | 0.552 | 0.567 | 0.538 | |
| RF | 0.951 | 0.925 | 0.818 | 0.881 | 0.872 | 0.841 | 0.912 | |
| MLP | 0.585 | 0.538 | 0.591 | 0.557 | 0.564 | 0.579 | 0.550 | |
| LightGBM | 0.871 | 0.854 | 0.700 | 0.797 | 0.778 | 0.748 | 0.821 | |
| AdaBoost | 0.685 | 0.722 | 0.542 | 0.668 | 0.634 | 0.622 | 0.651 | |
| XGBoost | 0.900 | 0.906 | 0.749 | 0.844 | 0.829 | 0.790 | 0.884 | |
| CatBoost | 0.846 | 0.830 | 0.724 | 0.793 | 0.778 | 0.759 | 0.803 | |
| Without Feature selection | LR | 0.577 | 0.538 | 0.571 | 0.552 | 0.554 | 0.567 | 0.542 |
| SVM | 0.743 | 0.684 | 0.680 | 0.687 | 0.682 | 0.690 | 0.673 | |
| RF | 0.951 | 0.882 | 0.906 | 0.895 | 0.894 | 0.908 | 0.880 | |
| MLP | 0.797 | 0.741 | 0.700 | 0.730 | 0.720 | 0.720 | 0.721 | |
| LightGBM | 0.942 | 0.892 | 0.833 | 0.869 | 0.863 | 0.848 | 0.880 | |
| AdaBoost | 0.686 | 0.660 | 0.581 | 0.641 | 0.622 | 0.622 | 0.621 | |
| XGBoost | 0.943 | 0.901 | 0.818 | 0.868 | 0.860 | 0.838 | 0.888 | |
| CatBoost | 0.936 | 0.877 | 0.818 | 0.855 | 0.848 | 0.834 | 0.865 |
The RF classifier stands out for its consistently high performance across all feature selection strategies, achieving an ROC of 0.951 and the highest accuracy of 0.894 without feature selection. The superior performance of RF can be attributed to its ensemble learning method, which constructs multiple decision trees and averages their outputs. This approach reduces overfitting and enhances robustness. Additionally, RF's flexibility allows it to effectively handle various data types and high-dimensional datasets.
When considering ROC values, most models without feature selection outperformed those with feature selection methods. The performance metrics for all models without feature selection are as follows: XGBoost (ROC = 0.943, Sensitivity = 0.901, Specificity = 0.818, F1 Score = 0.868, Accuracy = 0.860, PPV = 0.838, NPV = 0.888), LightGBM (ROC = 0.942, Sensitivity = 0.892, Specificity = 0.833, F1 Score = 0.869, Accuracy = 0.863, PPV = 0.848, NPV = 0.880), CatBoost (ROC = 0.936, Sensitivity = 0.877, Specificity = 0.818, F1 Score = 0.855, Accuracy = 0.848, PPV = 0.834, NPV = 0.865), MLP (ROC = 0.797, Sensitivity = 0.741, Specificity = 0.700, F1 Score = 0.730, Accuracy = 0.720, PPV = 0.720, NPV = 0.721), SVM (ROC = 0.743, Sensitivity = 0.684, Specificity = 0.680, F1 Score = 0.687, Accuracy = 0.682, PPV = 0.690, NPV = 0.673), AdaBoost (ROC = 0.686, Sensitivity = 0.660, Specificity = 0.581, F1 Score = 0.641, Accuracy = 0.622, PPV = 0.622, NPV = 0.621), and LR (ROC = 0.577, Sensitivity = 0.538, Specificity = 0.571, F1 Score = 0.552, Accuracy = 0.554, PPV = 0.567, NPV = 0.542).
LR performed poorly, with an ROC of 0.577 and an accuracy of 0.554. This underperformance is due to LR's assumption of linear relationships, struggles with multicollinearity, and sensitivity to outliers, which necessitate additional preprocessing. AdaBoost showed the worst performance among the ensemble algorithms, with an ROC of 0.686 and an accuracy of 0.622. Its sensitivity to noisy data and outliers, reliance on weak learners, and iterative re-weighting mechanism can lead to overfitting, adversely affecting its performance. Thus, the RF model demonstrated the highest robustness and accuracy, while LR and AdaBoost underperformed due to their inherent limitations and sensitivity to data characteristics.
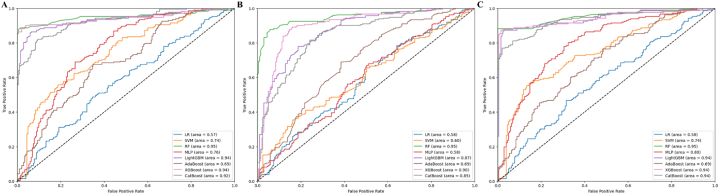
Fig. 3 illustrates the ROC curves for three different feature selection strategies. Fig. 3A shows the ROC curve using Lasso L1 feature selection, Fig. 3B presents the ROC curve employing t/Chi-Square feature selection, and Panel C depicts the ROC curve without any feature selection.
Fig. 3.
ROC curves for various feature selection strategies. (A) ROC curve using lasso l1 feature selection. (B) ROC curve employing t/chi-square feature selection. (C) ROC curve without feature selection.
3.4. Analysis of feature importance: importance ranking and SHAP values
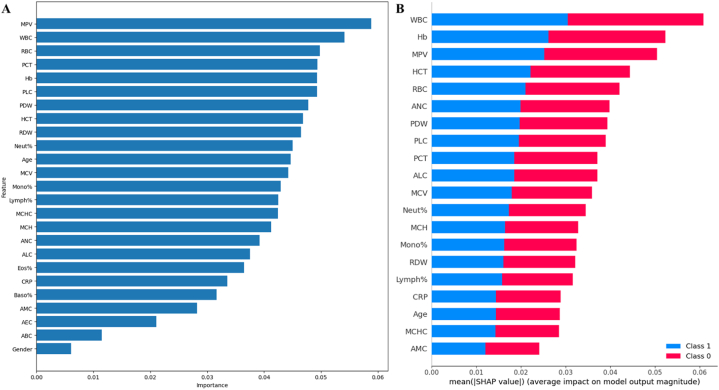
Fig. 4 presents an integrated analysis of routine blood tests using importance ranking and SHAP values, applied to the RF model. In Fig. 4A, the importance ranking of variables is as follows: MPV, WBC, RBC, PCT, Hb, PLC, PDW, Neut%, Age, MCV, Mono%, Lymph%, MCHC, MCH, ANC, ALC, Eos%, CRP, Baso%, AMC, AEC, ABC, and Gender. Notably, the SHAP analysis depicted in Fig. 4B shows a slightly different order of importance among these parameters, with WBC, Hb, MPV, HCT, RBC, ANC, PDW, PLC, PCT, ALC, MCV, Neut%, MCH, Mono%, RDW, Lymph%, CRP, Age, MCHC, and AMC ranking. Although the rankings are broadly similar, they exhibit some variations. This comparative ranking highlights the relative impact of each factor on the model's predictions, providing valuable insights into which features most influence outcomes.
Fig. 4.
Comprehensive analysis of feature importance. (A) Ranking of feature importance. (B) SHAP value-based feature importance bar chart.
The RF model ranks MPV as the most important feature, suggesting its crucial role in distinguishing between conditions in the dataset. However, SHAP values place MPV third in importance, indicating that while significant, its direct impact on the model's output may be slightly less than other top features. WBC is ranked second in importance but emerges as the most influential feature in SHAP analysis, with high SHAP values indicating a strong association between increased WBC count and H. pylori infection. RBC, ranked third in importance, is confirmed by SHAP analysis to be a significant predictor, where lower counts indicate potential infection due to chronic inflammation's effect on red blood cell production and lifespan.
Other key features include PCT, Hb, PLC, and PDW, which are consistently highlighted by both importance ranking and SHAP values. These features are significant predictors of infection, reflecting various aspects of the body's immune response and inflammation processes. Neut%, Age, MCV, Mono%, and Lymph% are also important, with SHAP analysis emphasizing their roles in predicting infection. Lower-ranked features such as MCHC, MCH, ANC, and others still contribute to the model, indicating diverse aspects of the immune response. This detailed interpretation of both importance ranking and SHAP values provides a comprehensive understanding of each feature's contribution to predicting H. pylori infection, offering valuable insights for clinical application and decision-making.
4. Discussion
H. pylori is a bacterium that infects over half the global population [1] and is the primary cause of various gastrointestinal issues, such as gastritis and peptic ulcers [56]. Therefore, there is a pressing need for fast, low-cost, accurate, and accessible digital screening tools to identify H. pylori infections. Integrating ML algorithms with routine blood tests provides a viable foundation for mass screening. Despite this potential, few studies have developed comprehensive clinical decision support systems using ML to aid in predicting, classifying, and treating illnesses. This research establishes a basis for advancing digital screening techniques to explore ML models for detecting H. pylori infection via routine blood tests. Specifically, our ML system can accurately identify H. pylori infection using only standard blood samples, with high precision regardless of disease severity.
This study is the first to investigate the predictive capabilities of routine blood tests for detecting H. pylori infection at all stages, facilitating early intervention and effective screening. Our findings demonstrate that basic blood indicators can effectively detect H. pylori, indicating a paradigm shift in diagnostic approaches. We propose using regular blood tests combined with ML models as a low-cost, accessible screening tool that avoids invasive procedures. This integration has significant implications for healthcare delivery, potentially improving the speed and accuracy of detecting and treating H. pylori infections. This approach can provide healthcare professionals with a valuable diagnostic tool and pave the way for substantial improvements in clinical screening practices.
Our research identified the RF model as optimal for detecting H. pylori infection using routine blood tests. Our findings showed remarkable performance, with an ROC of 0.951, sensitivity of 0.882, specificity of 0.906, F1 score of 0.895, accuracy of 0.894, PPV of 0.908, and NPV of 0.880. These metrics align closely with existing studies. For example, Ibrahim et al. [57] reported an accuracy of 0.924, sensitivity of 0.879, and specificity of 0.938 using deep learning on histopathological images. Li et al. [58] achieved an ROC of 0.973, sensitivity of 0.915, and specificity of 0.902 using deep learning techniques on endoscopic videos. Although these studies reported slightly superior performance metrics, they involved more invasive and complex methodologies compared to the non-invasive routine blood tests used in our study. This highlights the potential of our approach as a practical and efficient alternative for detecting H. pylori in primary clinical settings. Arai et al. [59] employed ML on endoscopic and histologic findings, reporting slightly lower sensitivity (0.832) and specificity (0.886) compared to our RF model. The comparable performance of our non-invasive method underscores its clinical significance. Differences in data types, model complexities, and population demographics can account for variations in performance metrics across studies. Nevertheless, our study demonstrates that routine blood tests combined with ML can effectively detect H. pylori infection, offering a valuable diagnostic tool for primary clinics by balancing accessibility and diagnostic accuracy.
The main biomarkers identified in our study are WBC, MPV, Hb, RBC, PCT, and PLC, which are well-known indicators of infection and inflammation. Our findings corroborate previous research. H. pylori infection contributes to atherothrombosis through chronic inflammation, direct vascular damage, and systemic inflammatory responses that promote prothrombotic changes in blood plasma factors and platelet activity [60]. Metabolic and inflammatory parameters, such as blood sugar, lipid profiles, insulin resistance, white blood cell count, and CRP levels, remain unchanged following H. pylori eradication treatment [61]. Infected individuals exhibit lower hemoglobin, serum iron, and serum ferritin levels, alongside higher total iron-binding capacity [62]. Chronic H. pylori infection correlates with increased neutrophil counts and platelet-to-lymphocyte ratios, and decreased neutrophil-to-lymphocyte ratios [63]. Mean platelet count is lower, while MPV is higher in infected patients, suggesting ongoing compensated platelet destruction-production [64]. The infection is marked by an inflammatory infiltrate predominantly of neutrophils and T cells [65], with associated monocyte phenotypic changes [66] and increased hepatoma-derived growth factor secretion promoting neutrophil infiltration [67]. Gastric mucosa in infected patients also shows eosinophil [68] and basophil infiltration [69], with intraepithelial lymphocyte counts predictive of infection [70]. Hemoglobin and hematocrit levels are significantly lower in H. pylori antibody-positive individuals, indicating an association with normocytic and normochromic anemia [71], particularly in elderly males with comorbidities [72]. Elevated leukocyte counts serve as markers of inflammation and infection in these patients [73]. Our study is the first to explore the relationship between these common blood biomarkers and H. pylori infection, indicating the feasibility of routine blood tests for predicting H. pylori infection.
The findings of our study also have significant implications for medical treatment. Firstly, this method has the potential to expedite early treatment for individuals with H. pylori infection. Evaluating the efficacy of preventive measures is challenging due to the varying stages of H. pylori infections within the population. However, our study lays a foundation for future randomized controlled trials that will further evaluate the effectiveness and potential preventive methods of using ML algorithms to forecast early H. pylori infection. Another significant contribution is towards the development of efficient universal screening procedures for H. pylori infection. The ongoing debate between universal and selective screening technologies is in diagnosing H. pylori infection is a primary focus. Currently, selective screening procedures are prevalent. However, our study indicates that predictive models have the capacity to identify low-risk individuals, offering valuable insights into the likelihood of H. pylori infection and associated illnesses. This aids physicians in effectively prioritizing patients and establishes screening protocols that are more efficient, focused, and cost-effective. Precise universal screening is particularly advantageous as it reduces the financial and temporal limitations for individuals requiring medical assessments. This innovative approach not only makes diagnostic processes painless and inexpensive, but also surpasses the limitations of common methods like endoscopy and biopsy. The results of this study are suitable in different healthcare situations, particularly those lacking access to modern diagnostic tools. By incorporating ML models into routine blood tests, the approach for healthcare systems becomes more streamlined and the necessity for invasive diagnostic procedures is diminished.
ML emerges as a valuable alternative for reducing reliance on demanding diagnostic procedures, enhancing patient comfort, and expending the diagnosis process, especially in areas lacking specialized healthcare infrastructure. Conventional H. pylori diagnostic techniques, such as the breath test for urea, serological testing, stool antigen tests, gastroscopy, and biopsy, are often linked with challenges such as invasiveness, lengthy procedures, and high costs. Recent studies revealing significant alterations in blood-related parameters among patients demonstrate the efficacy of ML models [[74], [75], [76], [77]]. Introducing this procedure can lead to significant cost savings, particularly in situations where advanced diagnostics are not accessible or where a less aggressive treatment is desired. Comprehending this concept helps in fostering a medical environment where a non-invasive and rapid method for assessing H. pylori infection will be employed. The laboratory-based ML diagnostic paradigm includes regular blood tests, early identification of key indicators, and quick result reporting. Data scientists and doctors need to become acquainted with fundamental ML techniques, receive standardized training, communicate this innovative approach to patients, and adhere to data protection and ethical guidelines. This can be achieved by implementing a combination of technologies designed to enhance the precision of diagnosing gastrointestinal diseases.
With the help of ML models, the regular blood tests can solve some of the major health problems, especially in the areas where healthcare is inadequate, when many patients suffer from digestive issues induced by H. pylori. Traditional techniques for diagnosing H. pylori are invasive, time-consuming, expensive, and demand specialized equipment, all of which puts impediment to the early detection and diagnosis in low resource areas in healthcare. These gadgets facilitate diagnosis and cut down on the use of costly and time-consuming techniques like endoscopy and biopsy to a considerable degree. ML tests are a valuable commodity in cases where conventional diagnostic tests fail for one reason or another because they are accessible and low-cost. ML applied to the medical diagnosis may result in the reduction of cost and improvement of precision. According to the study, ML could be applied to public health initiatives in order to be able to detect and treat digestive problems earlier, therefore reducing symptoms, and world health campaigns.
The screening methods of the H. pylori-related disorders done by medical practitioners is significant due to the relation of H. pylori and stomach abscesses such as stomach cancer and ulcers. H. pylori is also frequent in areas with high digestive system disease occurrence and their poor health access, particularly in developing nations [78]. The research points that using ML models would come up with non-invasive blood tests for H. pylori detection and make thus the diagnostic methods more unique. This diagnostic approach simplifies research methods and decreases reliance on expensive and resource-intensive procedures like endoscopy, which is especially advantageous in resource-limited settings. Implementing affordable and non-invasive screening methods can improve the timely detection and eradication of H. pylori infections, reducing associated illnesses. The results could impact healthcare plans in developing nations by highlighting the need of utilizing innovative technology. The proposed integration is anticipated to address gaps in symptom management, emphasize the significance of outcomes, and significantly enhance global efforts in the treatment or control of digestive illnesses. Heatmap examination of the map reveals consistent and significant connections between variables, highlighting their importance in predictive modelling. These links open a deeper awareness of the working principle of the model, therapeutic implication and possible disease mechanisms. The outcomes of our studies are of utmost importance in this time of sharpened misinformation. The paper points out the utility of different ML algorithms in precisely pinpointing the H. pylori infections and suggests that more widespread use of these methods can help overcome diverse clinical problems. Such a finding enhances opportunities for ML in medical diagnosis and influences the existing ways for identifying H pylori infection, which might upgrade the diagnostic precision and the process overall.
There are some limitations that must be considered. The strategy although a good starting point for research, might not be comprehensive enough to tackle the grandeur and the intricacy of the bigger sample. This restriction may affect the credibility of the data, especially when addressing the contribution of genetics, food, and environmental aspects to the high susceptibility to H. pylori among different populations. The data collected is from Wuhan and thus it is likely that the range of the study's findings may be different than for a different type of population. Genetic variations are the major contributing factor to the disparities in the reaction of people with diseases and it may lead to incomplete results in a single group. Food and environmental conditions are often key determinants of this condition, as they depend on the geographical location and lifestyle customs. Consequently, one must be careful while generalizing the results to populations with a gene pool, dietary habit and environments that are completely different. Future research should strive to replicate and validate these findings in diverse and representative populations to enhance the reliability and applicability of the proposed diagnostic technique. Furthermore, while the utilization of ML-based models proved effective, it may not be universally suitable. Examining diverse models or employing deep learning techniques could yield more accurate and precise results. Examining these models across multiple demographic groups, including individuals of different races, age and socioeconomic background, is vital to determine the accuracy and generalizability of the conclusion.
5. Conclusion
The RF model effectively identifies H. pylori infection using routine blood tests, offering a potential for clinical application. This ML approach can enhance diagnosis and screening, reducing medical burdens and reliance on invasive diagnostics.
Funding
This study did not receive financial support from public, commercial, or not-for-profit funding organizations.
Ethics approval and consent to participate
The ethical approval was obtained from the Ethics Committee of Hubei Provincial Hospital of Traditional Chinese Medicine in January 2024, which waived the requirement for informed patient consent. The analysis included patients who had undergone the urea breath test and routine blood test at Hubei Provincial Hospital of Traditional Chinese Medicine, providing the necessary clinical data from January 1, 2021, to June 30, 2023. The documented clinical characteristics included gender, age, and results from both the urea breath test and the routine blood test.
Consent for publication
Not applicable.
Data availability statement
Data associated with our study will be made available upon request. However, our code is available at https://github.com/BigbenZHU/Machine-Learning.
CRediT authorship contribution statement
Shiben Zhu: Writing – review & editing, Writing – original draft, Methodology, Formal analysis, Conceptualization. Xinyi Tan: Writing – original draft, Formal analysis, Data curation. He Huang: Formal analysis, Data curation. Yi Zhou: Formal analysis. Yang Liu: Supervision, Project administration, Methodology, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e35586.
Abbreviations
- ABC
Absolute basophil count
- AEC
Absolute eosinophil count
- ALC
Absolute lymphocyte count
- AMC
Absolute monocyte count
- ANC
Absolute neutrophil count
- Baso%
Basophil percentage
- CRP
C-reactive protein
- Eos%
Eosinophil percentage
- H. pylori
Helicobacter pylori
- Hb
Hemoglobin
- HCT
Hematocrit
- LR
Logistic regression
- Lymph%
Lymphocyte percentage
- MCH
Mean corpuscular hemoglobin
- MCHC
Mean corpuscular hemoglobin concentration
- MCV
Mean corpuscular volume
- ML
Machine learning
- MLP
Multilayer perceptron
- Mono%
Monocyte percentage
- MPV
Mean platelet volume
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Hooi J.K.Y., Lai W.Y., Ng W.K., Suen M.M.Y., Underwood F.E., Tanyingoh D., Malfertheiner P., Graham D.Y., Wong V.W.S., Wu J.C.Y., et al. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology. 2017;153:420–429. doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y.-C., Chiang T.-H., Chou C.-K., Tu Y.-K., Liao W.-C., Wu M.-S., Graham D.Y. Association between Helicobacter pylori eradication and gastric cancer incidence: a systematic review and meta-analysis. Gastroenterology. 2016;150:1113–1124.e1115. doi: 10.1053/j.gastro.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 3.Ridker P.M., Hennekens C.H., Buring J.E., Kundsin R., Shih J. Baseline IgG antibody titers to Chlamydia pneumoniae, Helicobacter pylori, herpes simplex virus, and cytomegalovirus and the risk for cardiovascular disease in women. Ann. Intern. Med. 1999;131:573–577. doi: 10.7326/0003-4819-131-8-199910190-00004. [DOI] [PubMed] [Google Scholar]
- 4.Butt J., Varga M.G., Blot W.J., Teras L., Visvanathan K., Le Marchand L., Haiman C., Chen Y., Bao Y., Sesso H.D., et al. Serologic response to Helicobacter pylori proteins associated with risk of colorectal cancer among diverse populations in the United States. Gastroenterology. 2019;156:175–186.e172. doi: 10.1053/j.gastro.2018.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butt J., Varga M.G., Blot W.J., Teras L., Visvanathan K., Le Marchand L., Haiman C., Chen Y., Bao Y., Sesso H.D., et al. Clinically apparent Helicobacter pylori infection and the risk of incident Alzheimer's disease: A population-based nested case-control study. Alzheimer's Dementia. 2023;20(3):1716–1724. doi: 10.1002/alz.13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garman K.S., Brown H., Alagesan P., McCall S.J., Patierno S., Wang Q., Wang F., Hyslop T., Epplein M. Helicobacter pylori testing prior to or at gastric cancer diagnosis and survival in a diverse US patient population. Gastric Cancer. 2023:1–8. doi: 10.1007/s10120-023-01448-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The E.S.G. An international association between Helicobacter pylori infection and gastric cancer. Lancet. 1993;341:1359–1363. doi: 10.1016/0140-6736(93)90938-D. [DOI] [PubMed] [Google Scholar]
- 8.Matysiak-Budnik T., Mégraud F. Helicobacter pylori infection and gastric cancer. Eur. J. Cancer. 2006;42:708–716. doi: 10.1016/j.ejca.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W., Zhou Y., Fan Y., Cao R., Xu Y., Weng Z., Ye J., He C., Zhu Y., Wang X. Metal–organic‐framework‐based hydrogen‐release platform for multieffective Helicobacter pylori targeting therapy and intestinal flora protective capabilities. Adv. Mater. 2022;34 doi: 10.1002/adma.202105738. [DOI] [PubMed] [Google Scholar]
- 10.Graham D.Y. Helicobacter pylori update: gastric cancer, reliable therapy, and possible benefits. Gastroenterology. 2015;148:719–731.e713. doi: 10.1053/j.gastro.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahm K.B., Lee K.J., Choi S.Y., Kim J.H., Cho S.W., Yim H., Park S.J., Chung M.H. Possibility of chemoprevention by the eradication of Helicobacter pylori: oxidative DNA damage and apoptosis in H. pylori infection. Am. J. Gastroenterol. 1997;92 [PubMed] [Google Scholar]
- 12.Yasunaga Y., Shinomura Y., Kanayama S., Yabu M., Nakanishi T., Miyazaki Y., Murayama Y., Bonilla-Palacios J., Matsuzawa Y. Improved fold width and increased acid secretion after eradication of the organism in Helicobacter pylori associated enlarged fold gastritis. Gut. 1994;35:1571. doi: 10.1136/gut.35.11.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liou J.-M., Chen C.-C., Chang C.-M., Fang Y.-J., Bair M.-J., Chen P.-Y., Chang C.-Y., Hsu Y.-C., Chen M.-J., Chen C.-C. Long-term changes of gut microbiota, antibiotic resistance, and metabolic parameters after Helicobacter pylori eradication: a multicentre, open-label, randomised trial. Lancet Infect. Dis. 2019;19:1109–1120. doi: 10.1016/S1473-3099(19)30272-5. [DOI] [PubMed] [Google Scholar]
- 14.Chiba N., Rao B.V., Rademaker J.W., Hunt R.H. Meta-analysis of the efficacy of antibiotic therapy in eradicating Helicobacter pylori. Am. J. Gastroenterol. 1992;87 [PubMed] [Google Scholar]
- 15.Towards the Eradication of Helicobacter pylori Infection. 2024. (IntechOpen). [Google Scholar]
- 16.Wang L., Li Z., Tay C.Y., Marshall B.J., Gu B., Tian Y., Dai X., Du H., Dai Q., Feng C., et al. Multicentre, cross-sectional surveillance of Helicobacter pylori prevalence and antibiotic resistance to clarithromycin and levofloxacin in urban China using the string test coupled with quantitative PCR. The Lancet Microbe. 2024;5:e512–e513. doi: 10.1016/S2666-5247(24)00027-2. [DOI] [PubMed] [Google Scholar]
- 17.Ferwana M., Abdulmajeed I., Alhajiahmed A., Madani W., Firwana B., Hasan R., Altayar O., Limburg P.J., Murad M.H., Knawy B. Accuracy of urea breath test in Helicobacter pylori infection: meta-analysis. World J. Gastroenterol. 2015;21:1305–1314. doi: 10.3748/wjg.v21.i4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miftahussurur M., Yamaoka Y. Diagnostic methods of Helicobacter pylori infection for epidemiological studies: critical importance of indirect test validation. BioMed Res. Int. 2016;2016 doi: 10.1155/2016/4819423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gisbert J.P., Pajares J.M. Stool antigen test for the diagnosis of Helicobacter pylori infection: a systematic review. Helicobacter. 2004;9:347–368. doi: 10.1111/j.1083-4389.2004.00235.x. [DOI] [PubMed] [Google Scholar]
- 20.Laheij R.J.F., de Boer W.A., Jansen J.B.M.J., van Lier H.J.J., Sneeberger P.M., Verbeek A.L.M. Diagnostic performance of biopsy-based methods for determination of Helicobacter pylori infection without a reference standard. J. Clin. Epidemiol. 2000;53:742–746. doi: 10.1016/S0895-4356(99)00222-X. [DOI] [PubMed] [Google Scholar]
- 21.Sabbagh P., Mohammadnia-Afrouzi M., Javanian M., Babazadeh A., Koppolu V., Vasigala V.R., Nouri H.R., Ebrahimpour S. Diagnostic methods for Helicobacter pylori infection: ideals, options, and limitations. Eur. J. Clin. Microbiol. Infect. Dis. 2019;38:55–66. doi: 10.1007/s10096-018-3414-4. [DOI] [PubMed] [Google Scholar]
- 22.Sousa C., Ferreira R., Santos S.B., Azevedo N.F., Melo L.D.R. Advances on diagnosis of Helicobacter pylori infections. Crit. Rev. Microbiol. 2023;49:671–692. doi: 10.1080/1040841X.2022.2125287. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y.K., Kuo F.C., Liu C.J., Wu M.C., Shih H.Y., Wang S.S., Wu J.Y., Kuo C.H., Huang Y.K., Wu D.C. Diagnosis of Helicobacter pylori infection: current options and developments. World J. Gastroenterol. 2015;21:11221–11235. doi: 10.3748/wjg.v21.i40.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chinese Medical Association, C.M.J.P.H., Chinese Society of General Practice, Helicobacter pylori Study Group of Chinese Digestive Diseases Society Editorial board of Chinese journal of general practitioners of Chinese medical association, expert group of guidelines for primary care of digestive disease, lyu nonghua, zhou liya (2019). Guideline for primary care of Helicobacter pylori infection: practice version. Chinese Journal of General Practitioners. 2019;19:403–407. doi: 10.3760/cma.j.cn114798-20200223-00159. [DOI] [Google Scholar]
- 25.Tang J.W., Li F., Liu X., Wang J.T., Xiong X.S., Lu X.Y., Zhang X.Y., Si Y.T., Umar Z., Tay A.C.Y., et al. Detection of Helicobacter pylori infection in human gastric fluid through surface-enhanced Raman spectroscopy coupled with machine learning algorithms. Lab. Invest. 2024;104 doi: 10.1016/j.labinv.2023.100310. [DOI] [PubMed] [Google Scholar]
- 26.Bodansky D.M.S., Lumley S.E., Chakraborty R., Mani D., Hodson J., Hallissey M.T., Tucker O.N. Potential cost savings by minimisation of blood sample delays on care decision making in urgent care services. Annals of Medicine and Surgery. 2017;20:37–40. doi: 10.1016/j.amsu.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrara G., Losi M., D'Amico R., Roversi P., Piro R., Meacci M., Meccugni B., Dori I.M., Andreani A., Bergamini B.M., et al. Use in routine clinical practice of two commercial blood tests for diagnosis of infection with Mycobacterium tuberculosis: a prospective study. Lancet. 2006;367:1328–1334. doi: 10.1016/S0140-6736(06)68579-6. [DOI] [PubMed] [Google Scholar]
- 28.Altman R.S., Altman L.J., Altman J.S. A proposed set of new guidelines for routine blood tests during isotretinoin therapy for acne vulgaris. Dermatology. 2002;204:232–235. doi: 10.1159/000057887. [DOI] [PubMed] [Google Scholar]
- 29.Liu M., Zhou J., Xi Q., Liang Y., Li H., Liang P., Guo Y., Liu M., Temuqile T., Yang L. A computational framework of routine test data for the cost-effective chronic disease prediction. Briefings Bioinf. 2023;24:bbad054. doi: 10.1093/bib/bbad054. [DOI] [PubMed] [Google Scholar]
- 30.Di N., He W., Zhang K., Cui J., Chen J., Cheng J., Chu B., Li S., Xie Y., Xiang H. Association of short-term air pollution with systemic inflammatory biomarkers in routine blood test: a longitudinal study. Environ. Res. Lett. 2021;16 [Google Scholar]
- 31.Morelli D., Cantarutti A., Valsecchi C., Sabia F., Rolli L., Leuzzi G., Bogani G., Pastorino U. Routine perioperative blood tests predict survival of resectable lung cancer. Sci. Rep. 2023;13 doi: 10.1038/s41598-023-44308-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Podnar S., Kukar M., Gunčar G., Notar M., Gošnjak N., Notar M. Diagnosing brain tumours by routine blood tests using machine learning. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-51147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faisal M., Howes R., Steyerberg E.W., Richardson D., Mohammed M.A. Using routine blood test results to predict the risk of death for emergency medical admissions to hospital: an external model validation study. QJM: An International Journal of Medicine. 2017;110:27–31. doi: 10.1093/qjmed/hcw110. [DOI] [PubMed] [Google Scholar]
- 34.Gunčar G., Kukar M., Smole T., Moškon S., Vovko T., Podnar S., Černelč P., Brvar M., Notar M., Köster M. Differentiating viral and bacterial infections: a machine learning model based on routine blood test values. Heliyon. 2024;10(8) doi: 10.1016/j.heliyon.2024.e29372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrari D., Motta A., Strollo M., Banfi G., Locatelli M. Routine blood tests as a potential diagnostic tool for COVID-19. Clin. Chem. Lab. Med. 2020;58:1095–1099. doi: 10.1515/cclm-2020-0398. [DOI] [PubMed] [Google Scholar]
- 36.Li K., Liu S.-X., Yang C.-Y., Jiang Z.-C., Liu J., Fan C.-Q., Li T., Dong X.-M., Wang J., Ran R.-Y. A routine blood test-associated predictive model and application for tuberculosis diagnosis: a retrospective cohort study from northwest China. J. Int. Med. Res. 2019;47:2993–3007. doi: 10.1177/0300060519851673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Planche T.D., Davies K.A., Coen P.G., Finney J.M., Monahan I.M., Morris K.A., O’Connor L., Oakley S.J., Pope C.F., Wren M.W., et al. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. Lancet Infect. Dis. 2013;13:936–945. doi: 10.1016/S1473-3099(13)70200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parikh R.B., Manz C., Chivers C., Regli S.H., Braun J., Draugelis M.E., Schuchter L.M., Shulman L.N., Navathe A.S., Patel M.S., O'Connor N.R. Machine learning approaches to predict 6-month mortality among patients with cancer. JAMA Netw. Open. 2019;2 doi: 10.1001/jamanetworkopen.2019.15997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall I.J., Noel-Storr A., Kuiper J., Thomas J., Wallace B.C. Machine learning for identifying Randomized Controlled Trials: an evaluation and practitioner's guide. Res. Synth. Methods. 2018;9:602–614. doi: 10.1002/jrsm.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garriga R., Mas J., Abraha S., Nolan J., Harrison O., Tadros G., Matic A. Machine learning model to predict mental health crises from electronic health records. Nat Med. 2022;28:1240–1248. doi: 10.1038/s41591-022-01811-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rawson T.M., Hernandez B., Wilson R.C., Ming D., Herrero P., Ranganathan N., Skolimowska K., Gilchrist M., Satta G., Georgiou P., Holmes A.H. Supervised machine learning to support the diagnosis of bacterial infection in the context of COVID-19. JAC Antimicrob Resist. 2021;3 doi: 10.1093/jacamr/dlab002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang F., Wang H., Liu L., Su T., Ji B. Machine learning model for the prediction of gram-positive and gram-negative bacterial bloodstream infection based on routine laboratory parameters. BMC Infect. Dis. 2023;23:675. doi: 10.1186/s12879-023-08602-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rawson T.M., Hernandez B., Moore L.S.P., Blandy O., Herrero P., Gilchrist M., Gordon A., Toumazou C., Sriskandan S., Georgiou P., Holmes A.H. Supervised machine learning for the prediction of infection on admission to hospital: a prospective observational cohort study. J. Antimicrob. Chemother. 2019;74:1108–1115. doi: 10.1093/jac/dky514. [DOI] [PubMed] [Google Scholar]
- 44.Xiong T., Lv X.S., Wu G.J., Guo Y.X., Liu C., Hou F.X., Wang J.K., Fu Y.F., Liu F.Q. Single-cell sequencing analysis and multiple machine learning methods identified G0S2 and HPSE as novel biomarkers for abdominal aortic aneurysm. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.907309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parmar C., Grossmann P., Bussink J., Lambin P., Aerts H.J. Machine learning methods for quantitative radiomic biomarkers. Sci. Rep. 2015;5 doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi K., Lin W., Zhao X.-M. Identifying molecular biomarkers for diseases with machine learning based on integrative omics. IEEE ACM Trans. Comput. Biol. Bioinf. 2020;18:2514–2525. doi: 10.1109/TCBB.2020.2986387. [DOI] [PubMed] [Google Scholar]
- 47.Fonti V., Belitser E. Feature selection using lasso. VU Amsterdam research paper in business analytics. 2017;30:1–25. [Google Scholar]
- 48.Nusinovici S., Tham Y.C., Yan M.Y.C., Ting D.S.W., Li J., Sabanayagam C., Wong T.Y., Cheng C.-Y. Logistic regression was as good as machine learning for predicting major chronic diseases. Journal of clinical epidemiology. 2020;122:56–69. doi: 10.1016/j.jclinepi.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 49.Suthaharan S., Suthaharan S. 2016. Support Vector Machine. Machine Learning Models and Algorithms for Big Data Classification: Thinking with Examples for Effective Learning; pp. 207–235. [Google Scholar]
- 50.Tang J., Deng C., Huang G.-B. Extreme learning machine for multilayer perceptron. IEEE Transact. Neural Networks Learn. Syst. 2015;27:809–821. doi: 10.1109/TNNLS.2015.2424995. [DOI] [PubMed] [Google Scholar]
- 51.Segal M.R. 2004. Machine Learning Benchmarks and Random Forest Regression. [Google Scholar]
- 52.Shehadeh A., Alshboul O., Al Mamlook R.E., Hamedat O. Machine learning models for predicting the residual value of heavy construction equipment: an evaluation of modified decision tree, LightGBM, and XGBoost regression. Autom. ConStruct. 2021;129 [Google Scholar]
- 53.Zhang P.B., Yang Z.X. A novel AdaBoost framework with robust threshold and structural optimization. IEEE Trans. Cybern. 2018;48:64–76. doi: 10.1109/tcyb.2016.2623900. [DOI] [PubMed] [Google Scholar]
- 54.Chen T., Guestrin C. 2016. Xgboost: A Scalable Tree Boosting System; pp. 785–794. [Google Scholar]
- 55.Hancock J.T., Khoshgoftaar T.M. CatBoost for big data: an interdisciplinary review. Journal of big data. 2020;7:1–45. doi: 10.1186/s40537-020-00369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peterson W.L. Helicobacter pylori and peptic ulcer disease. N. Engl. J. Med. 1991;324:1043–1048. doi: 10.1056/NEJM199104113241507. [DOI] [PubMed] [Google Scholar]
- 57.Ibrahim A.U., Dirilenoğlu F., Hacisalihoğlu U.P., Ilhan A., Mirzaei O. Classification of H. pylori infection from histopathological images using deep learning. J Imaging Inform Med. 2024;37:1177–1186. doi: 10.1007/s10278-024-01021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y.D., Wang H.G., Chen S.S., Yu J.P., Ruan R.W., Jin C.H., Chen M., Jin J.Y., Wang S. Assessment of Helicobacter pylori infection by deep learning based on endoscopic videos in real time. Dig. Liver Dis. 2023;55:649–654. doi: 10.1016/j.dld.2023.02.010. [DOI] [PubMed] [Google Scholar]
- 59.Arai J., Aoki T., Sato M., Niikura R., Suzuki N., Ishibashi R., Tsuji Y., Yamada A., Hirata Y., Ushiku T., et al. Machine learning-based personalized prediction of gastric cancer incidence using the endoscopic and histologic findings at the initial endoscopy. Gastrointest. Endosc. 2022;95:864–872. doi: 10.1016/j.gie.2021.12.033. [DOI] [PubMed] [Google Scholar]
- 60.Hofmann R., Bäck M. Time for routine Helicobacter pylori screening in coronary artery disease? Circulation. 2023;147:1731–1733. doi: 10.1161/CIRCULATIONAHA.123.064944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park S.H., Jeon W.K., Kim S.H., Kim H.J., Park D.I., Cho Y.K., Sung I.K., Sohn C.I., Kim B.I., Keum D.K. Helicobacter pylori eradication has no effect on metabolic and inflammatory parameters. J. Natl. Med. Assoc. 2005;97:508–513. [PMC free article] [PubMed] [Google Scholar]
- 62.Nashaat E.H., Mansour G.M. Helicobacter pylori and anemia with pregnancy. Arch. Gynecol. Obstet. 2014;289:1197–1202. doi: 10.1007/s00404-013-3138-8. [DOI] [PubMed] [Google Scholar]
- 63.Sağlam N., Civan H.A. Impact of chronic Helicobacter pylori infection on inflammatory markers and hematological parameters. Eur. Rev. Med. Pharmacol. Sci. 2023;27:969–979. doi: 10.26355/eurrev_202302_31190. [DOI] [PubMed] [Google Scholar]
- 64.Umit H., Umit E.G. Helicobacter pylori and mean platelet volume: a relation way before ımmune thrombocytopenia? Eur. Rev. Med. Pharmacol. Sci. 2015;19:2818–2823. [PubMed] [Google Scholar]
- 65.Amedei A., Munari F., Bella C.D., Niccolai E., Benagiano M., Bencini L., Cianchi F., Farsi M., Emmi G., Zanotti G., et al. Helicobacter pylori secreted peptidyl prolyl cis, trans-isomerase drives Th17 inflammation in gastric adenocarcinoma. Intern Emerg Med. 2014;9:303–309. doi: 10.1007/s11739-012-0867-9. [DOI] [PubMed] [Google Scholar]
- 66.Harris P.R., Mobley H.L., Perez-Perez G.I., Blaser M.J., Smith P.D. Helicobacter pylori urease is a potent stimulus of mononuclear phagocyte activation and inflammatory cytokine production. Gastroenterology. 1996;111:419–425. doi: 10.1053/gast.1996.v111.pm8690207. [DOI] [PubMed] [Google Scholar]
- 67.Chu T.H., Huang S.T., Yang S.F., Li C.J., Lin H.W., Weng B.C., Yang S.M., Huang S.C., Wu J.C., Chang Y.C., et al. Hepatoma-derived growth factor participates in Helicobacter pylori-induced neutrophils recruitment, gastritis and gastric carcinogenesis. Oncogene. 2019;38:6461–6477. doi: 10.1038/s41388-019-0886-3. [DOI] [PubMed] [Google Scholar]
- 68.Kim J.M., Kim J.S., Lee J.Y., Sim Y.S., Kim Y.J., Oh Y.K., Yoon H.J., Kang J.S., Youn J., Kim N., et al. Dual effects of Helicobacter pylori vacuolating cytotoxin on human eosinophil apoptosis in early and late periods of stimulation. Eur. J. Immunol. 2010;40:1651–1662. doi: 10.1002/eji.200939882. [DOI] [PubMed] [Google Scholar]
- 69.de Paulis A., Prevete N., Fiorentino I., Walls A.F., Curto M., Petraroli A., Castaldo V., Ceppa P., Fiocca R., Marone G. Basophils infiltrate human gastric mucosa at sites of Helicobacter pylori infection, and exhibit chemotaxis in response to H. pylori-derived peptide Hp(2-20) J. Immunol. 2004;172:7734–7743. doi: 10.4049/jimmunol.172.12.7734. [DOI] [PubMed] [Google Scholar]
- 70.Bosch D.E., Liu Y.J., Truong C.D., Lloyd K.A., Swanson P.E., Upton M.P., Yeh M.M. Duodenal intraepithelial lymphocytosis in Helicobacter pylori gastritis: comparison before and after treatment. Virchows Arch. 2021;478:805–809. doi: 10.1007/s00428-020-02941-2. [DOI] [PubMed] [Google Scholar]
- 71.Abe Y., Kusano C., Takano C., Morioka I., Gotoda T. Association between Helicobacter pylori antibody-positive status and extragastric diseases in Japanese junior high school students. Pediatr. Int. 2021;63:1087–1094. doi: 10.1111/ped.14585. [DOI] [PubMed] [Google Scholar]
- 72.Hou B., Zhang M., Liu M., Dai W., Lin Y., Li Y., Gong M., Wang G. Association of active Helicobacter pylori infection and anemia in elderly males. BMC Infect. Dis. 2019;19:228. doi: 10.1186/s12879-019-3849-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jafarzadeh A., Akbarpoor V., Nabizadeh M., Nemati M., Rezayati M.T. Total leukocyte counts and neutrophil-lymphocyte count ratios among Helicobacter pylori-infected patients with peptic ulcers: independent of bacterial CagA status. Southeast Asian J Trop Med Public Health. 2013;44:82–88. [PubMed] [Google Scholar]
- 74.Lancet D., Pecht I. Spectroscopic and immunochemical studies with nitrobenzoxadiazolealanine, a fluorescent dinitrophenyl analog. Biochemistry. 1977;16:5150–5157. doi: 10.1021/bi00642a031. [DOI] [PubMed] [Google Scholar]
- 75.Karthikeyan A., Garg A., Vinod P.K., Priyakumar U.D. Machine learning based clinical decision support system for early COVID-19 mortality prediction. Front. Public Health. 2021;9 doi: 10.3389/fpubh.2021.626697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li C., Liu Y.-c., Zhang D.-r., Han Y.-x., Chen B.-j., Long Y., Wu C. A machine learning model for distinguishing Kawasaki disease from sepsis. Sci. Rep. 2023;13 doi: 10.1038/s41598-023-39745-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bai Q., Su C., Tang W., Li Y. Machine learning to predict end stage kidney disease in chronic kidney disease. Sci. Rep. 2022;12:8377. doi: 10.1038/s41598-022-12316-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hooi J.K., Lai W.Y., Ng W.K., Suen M.M., Underwood F.E., Tanyingoh D., Malfertheiner P., Graham D.Y., Wong V.W., Wu J.C. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology. 2017;153:420–429. doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with our study will be made available upon request. However, our code is available at https://github.com/BigbenZHU/Machine-Learning.