Abstract
Porcine Circovirus type (PCV) 2 is an important pathogen that has been circulating worldwide and has cuased serious economic loss in pig industry. However, both PCV3 and PCV4 are newly emerging viruses. In Taiwan, PCV2 has been one of the critical pathogens in pig frams and PCV3 has been detected since 2016; however, the epidemiolog of PCV3 in Taiwan remains unclear and PCV4 has yet to be identified. Therefore, in order to detect the positive rate of PCV2, to investigate the epidemiolog of PCV3 in the pig farms, and to examine whether pigs were infected with PCV4 in Taiwan, a total of 128 samples from 46 clinical cases of pigs were collected from September 2020 to December 2021. The case detection rates were 54.3 % for PCV2, 43.5 % for PCV3, and 2.2 % for PCV4. The results suggested that the positivity rates for both PCV2 and PCV3 were still high in Taiwan. In addition, PCV3 was detected among cases from all 7 sampled counties and in 11 of the 16 sampling months, suggesting that PCV3 may lead to endemic pig disease in Taiwan. Surprisingly, the PCV4 was also detected, suggesting the first PCV4 case in Taiwan. The complete genomes derived from the identified PCV3 and PCV4 strains were subsequently sequenced followed by phylogenetic analysis. The results suggested that the 17 identified PCV3 strains could be divided into Taiwanese-like and Japanese-like strains. In addition, the amino acid residues at positions 27, 80, and 212 in the identified PCV4 cap protein were asparagine, isoleucine, and methionine, respectively, and thus the identified PCV4 was catalorized into clade PCV4b. Consequently, it is concluded that (i) the prevalence of PCV2 and PCV3 is still high in Taiwanese pigs, (ii) PCV3 has may be an endemic infection in Taiwan and can be classified into Japanese-like and Taiwanese-like strains, (iii) PCV4 was detected for the first time in Taiwanese pigs and can be classified into PCV4b. It remains unclear how PCV2, PCV3, and PCV4 were introduced to Taiwan, and thus continuous investigation of emerging pathogens in pigs is needed.
Keywords: Porcine circovirus (PCV), PCV3, PCV4, Emerging disease, phylogenetic analysis
1. Introduction
Porcine circoviruses (PCVs) are circular single-stranded DNA viruses with a diameter of 17 nm and a nonenveloped icosahedral virion. PCVs belong to the family Circoviridae and the genus Circovirus [1]. The PCV genome encodes two main open reading frames (ORFs): ORF1 encodes a replication-related protein, and ORF2 encodes a structural capsid protein [1,2]. Four species of pig circoviruses, PCV1, PCV2, PCV3, and PCV4, have been identified. PCV1 can be grown in pig kidney cell lines [2,3]. PCV2 is the primary pathogen for porcine circovirus–associated diseases, including postweaning multisystemic wasting syndrome (PMWS), porcine dermatitis, and nephropathy syndrome (PDNS), and is associated with reproductive failure in sows [[4], [5], [6]]. PCV3 was first reported by Palinski et al. in the United States in 2017 [7]. This report described that PCV3 was isolated in 2015 from samples originating from sows with chronic reproductive failure and clinical signs of PDNS that were kept on a pig farm in North Carolina. On the histopathological examination, acute necrotizing dermatitis and epidermis were observed. In addition, cortical tubular dilation, lymphocytes in the cortical interstitium and glomerulus, and macrophage infiltration were also observed in the kidneys [7].
After PCV3 was first reported in the United States in 2017 [7], it was detected in Asia, Europe, South America, and Africa. In Asia, the positive rates of PCV3-positive pig samples in China in 2017 and 2021 were 34.7 % and 36.36 %, respectively [8,9]; in South Korea, 360 pig samples were collected from 73 pig farms in 2017, and the percentage of PCV3-positive saliva samples was 44.2 % [10]; in Thailand, PCV3 viral nucleic acid was detected in the lungs and lymph nodes of porcine respiratory disease complex (PRDC) specimens, with a positive rate of 62.5 % [11]; and in India, PCV3 viral nucleic acid was also confirmed in nasal swabs and serum of aborted fetuses and sows [12]. On the other hand, in European countries, PCV3 nucleic acid was detected in pig specimens, including aborted fetuses, serum, tissue specimens, and feces, in Poland (with a herd prevalence of 85.7 % (12/14)) [13], Italy (with a seroprevalence positivity rate of 18 % (6/33)) [14,15], Denmark (with a seroprevalence positivity rate of 30 % (6/20)) [14], Spain (with a herd prevalence of 50 % (7/14)) [14], Germany (with a herd prevalence of 75 % (40/53)) [16], the United Kingdom (with a prevalence of 5 0.5–100 % in fecal samples between 2002 and 2017) [17], Hungary [18], Ukraine [19], and Serbia (with a herd prevalence of 100 % (10/10)) [20]. In addition, in a retrospective study in Sweden, pig lymph node specimens were collected from 1993 to 2003, and the earliest PCV3-positive specimen in Sweden could be traced back to 1993 [21]. Another retrospective study in Brazil detected PCV3 viral nucleic acid in specimens in 1967, which is the earliest known evidence of PCV3 [22]. In Central and South American countries, the presence of PCV3 has also been detected in Brazil [23], Colombia [24], and Argentina [25].
PCV4 has been identified in pigs in China [26,27] and South Korea [28] since 2019. A document published in 2023 also revealed that PCV4 was detected in Thailand in three pig samples during 2019–2020 [29]. Although Italy and Spain were thought to be PCV4-free countries between 2015 and 2018 [30], most recent research has revealed that Spanish wild boar and Iberian pigs in mid-southwestern Spain were PCV4 positive, as determined by quantitative polymerase chain reaction (qPCR) [31]. In 2023, a molecular prevalence of 4.08 % obtained from 49 lung tissue samples by conventional PCR for the detection of the Cap gene of PCV4 was reported in Peninsular Malaysia [32]. PCV4 can be classified into PCV4a and PCV4b genotypes based on the open reading frame (ORF) 1 (Rep gene) and ORF2 (Cap gene) sequences and complete genome sequences [33,34]. PCV4 was initially detected in pigs with severe respiratory and gastrointestinal symptoms and PDNS symptoms [26]. In subsequent studies, PCV4 was also detected in pigs with neurological symptoms, abortions [28], and PRDC [29]. More studies are needed to clarify the pathogenesis and transmission of PCV4.
PCV2 has been one of the important pathogens in pig frams of Taiwan. In addition, PCV3 and PCV4 are known to be emerging viruses and have been identified in several countries. In Taiwan, however, there has only been one report on the positivity rate for PCV3, which was found to be 10.6 % in 2016 and increased markedly to 34.78 % in 2019 [35]. In addition, although PCV4 has been identified in East Asia (China, South Korea, and Thailand), Malaysia and Spain, there are no studies on the identification of PCV4 in Taiwan. Thus, the epidemiology of PCV3 and PCV4 in pig farms remains unclear. Although it remains unknown how PCV2 and PCV3 were introduced to Taiwan, continuous investigations of newly emerging infectious diseases in pigs are needed. The aim of this study is therefore to explore the recent status of PCV2, PCV3 and PCV4 infections in Taiwan using molecular diagnostic technology and pathological diagnosis based on clinical cases submitted to the Animal Disease Diagnostic Center of National Chung Hsing University from 2020 to 2021. Based on the analysis results, including the positivity rate, major clinical symptoms, and distribution, the positivity rate of PCV2, the epidemiology of PCV3 and the existence of PCV4 in Taiwan can be revealed.
2. Materials and methods
2.1. Sample collection and preparation
Because the ADDC at National Chung Hsing University provides disease diagnosis services for pigs submitted by pig farmers and veterinarians, the samples in this passive epidemiological study were selected from the submitted pigs with PCV-like syndromes, including abortion, PDNS, or PMWS.
Clinical cases from pig farms located in northern and central Taiwan (New Taipei City, Hsinchu County, Miaoli County, Taichung City, Changhua County, Nantou County, and Yunlin County) between September 2020 and December 2021 were selected for analysis. Each case was considered independent because all cases were submitted to the ADDC for diagnosis at different times. The specimens were collected during rigorous autopsies by veterinary pathologists, and cross-contamination between pigs was avoided based on standard procedures for specimen sampling. Each pig specimen was collected independently.
The collected samples, which included tonsils, lungs, spleens, hilar lymph nodes, and mesenteric lymph nodes, were sent to the responsible laboratory independently, stored individually in a freezer, and homogenized into emulsions, followed by the extraction of nucleic acids and the application of PCR (and/or qPCR).
2.2. Nucleic acid extraction
To ensure the reliability of the molecular diagnosis, the extraction of nucleic acids from each pig specimen was performed independently. The extraction was performed by authorized laboratory members who had undergone rigorous training. DNA was extracted from mixed tissue samples (tonsils, lungs, spleens, and lymph nodes) using a viral nucleic acid kit (High Pure Viral Nucleic Acid Kit, Roche Diagnostics, Mannheim, Germany). Briefly, 200-μL tissue emulsions were gently mixed with 200 μL of binding buffer, 50 μL of proteinase K, and 4 μL of poly (A) and incubated at 72 °C for 10 min. Subsequently, 100 μL of binding buffer was added, and the solution was mixed gently. The solution was then poured into a spin column and centrifuged at 8000×g for 1 min. Subsequently, 500 μL of inhibitor removal buffer and 450 μL of washing buffer were sequentially poured into the spin column, and the mixture was centrifuged at 8000×g for 1 min. The spin column was attached to a 1.5-mL Eppendorf tube, and 50 μL of elution buffer was added. The mixture was centrifuged at 8000 g for 1 min. The DNA samples were stored at −20 °C.
2.3. Molecular detection of PCV2, PCV3, and PCV4
PCV2 and PCV3 DNA were detected from samples using qPCR protocols [7,36], and PCV4 DNA was detected from samples using only the recently reported PCR protocol [27]. For the detection of PCV2 and PCV3 by qPCR, 10-fold serial dilutions of plasmids containing amplicons were used to establish the standard curves. The linear range of 2 x 102 to 2 x 108 copies/μL of PCV2 and 5.74 x 102 to 5.74 x 106 copies/μL of PCV3 for standard curves were used in this study. The Ct values less than 30.1 and 35 indicated positive detection for PCV2 and PCV3, respectively.
PCV2 examination was conducted based on a previously described qPCR protocol [36]. qPCR reactions were carried out to detect the partial ORF2 sequence in PCV2 using primers for PCV2 q3F and PCV2 q3R (Table 1) together with a reaction mixture (PerfeCTa SYBR Green FastMix, Blossom Biotechnologies, Taipei, Taiwan). The components of the mixture were as follows: 5 μL of reaction mixture, 0.4 μL of PCV2 q3F primer (10 μM), 0.4 μL of PCV2 q3R primer (10 μM), 3.2 μL of water treated with diethylpyrocarbonate (DEPC) and 1 μL of DNA template. The samples were placed in a 96-well plate in triplicate. The qPCR assay was performed using a polymerase chain reaction (PCR) platform (LightCycler 480 II, Roche Diagnostics, Mannheim, Germany) under the following thermocycling conditions: 3 min at 95 °C, followed by 40 cycles of denaturation at 95 °C for 5 s and annealing and extension at 60 °C for 20 s, as previously described [36].
Table 1.
Primers used for PCV2, PCV3, and PCV4 detection.
| Primer | Nucleotide sequence | Amplicons (bp) | Reference |
|---|---|---|---|
| PCV2 q3F | 5′-ATAACCCAGCCCTTCTCCTACC-3′ | 145 | [36] |
| PCV2 q3R | 5′-GGCCTACGTGGTCTACATTTCC-3′ | ||
| PCV3 1137F | 5′-TTGGGGTGGGGGTATTTATT-3′ | 425 | [16] |
| PCV3 1561R | 5′-ACACAGCCGTTACTTCAC-3′ | ||
| PCV3 1427F | 5′-AGTGCTCCCCATTGAACG-3′ | 135 | [16] |
| PCV3 1561R | 5′-ACACAGCCGTTACTTCAC-3′ | ||
| PCV3 39F | 5′-GGGAGAAAAAGTGGTATCCC-3′ | 1106 | This study |
| PCV3 1144R | 5′-CACCCCAACGCAATAATTGTA-3′ | [16] | |
| PCV3 910F | 5′-CTGTTAAGGGTGGGTTTG-3′ | 652 | [16] |
| PCV3 1561R | 5′-ACACAGCCGTTACTTCAC-3′ | ||
| PCV3 1427F | 5′-AGTGCTCCCCATTGAACG-3′ | 612 | [16] |
| PCV3 38R | 5′-TACAGACCTCCGTGGATCCG-3′ | This study | |
| PCV4 1347F | 5′-GTTTTTCCCTTCCCCCACATAG-3′ | 391 | [27] |
| PCV4 1737R | 5′-ACAGATGCCAATCAGATCTAGGTAC-3′ | ||
| PCV4 312F | 5′-CCCGCGGVACTGATTGTGAT-3′ | 932 | This study |
| PCV4 1243R | 5′-AAACCCACACCCTCCACTTC-3′ | ||
| PCV4 1047F | 5′-TTATCCCTGTTTGGGGTAG-3′ | 687 | [26] |
| PCV4 1733R | 5′-ATGCCAATCAGATCTAGGTAC-3′ | ||
| PCV4 464F | 5′-CCGTGAGTTCCCGTCTGTAT-3′ | 1274 | [28] |
| PCV4 1737R | 5′-ACAGATGCCAATCAGATCTAGGTAC-3′ | [27] | |
| PCV4 1347F | 5′-GTTTTTCCCTTCCCCCACATAG-3′ | 879 | [27] |
| PCV4 469R | 5′-TCACGGGCCACTTCACTCAT-3′ | [37] |
PCV3 was examined using a modification of the qPCR protocol published by Palinski et al. [7], and the ORF2 gene sequence (nt position 1427–1561) was used as the target for detection using primers for PCV3 1427F and PCV3 1561R (Table 1) and the PerfeCTa SYBR Green SuperMix reaction mix (Blossom Biotechnologies). The samples were prepared by combining the following: 5 μL of reaction mixture, 0.2 μL of PCV3 1427F primer (10 μM), 0.2 μL of PCV3 1561R primer (10 μM), 3.6 μL of DEPC-treated water, and 1 μL of positive standard or sample nucleic acid. The samples were placed in a 96-well plate in triplicate. The qPCR assay was performed using a PCR platform (LightCycler 480 II, Roche Diagnostics) under the following thermocycling conditions: 2 min at 95 °C, followed by 45 cycles of denaturation at 95 °C for 10 s and annealing and extension at 60 °C for 20 s, as described previously [7].
PCV4 was examined using modified PCR protocols published previously [27]. The expected PCR product was 391 bp amplified from nt 1347–1737 and primers for PCV4 1347F and PCV4 1737R were used (Table 1). The PCR mixture was prepared as follows: 12.5 μL of 2 × Taq Plus Master Mix, 1 μL of PCV4 1347F primer (10 μM), 1 μL of PCV4 1737R primer (10 μM), 5 μL of DNA (50–500 ng), and 5.5 μL of double distilled water (DDW), which were added to a 0.2-mL PCR tube to a total reaction volume of 25 μL. PCR was performed using a thermal cycler (Applied Biosystems 2720 Thermal Cycler, ThermoFisher Scientific, USA) under the following thermocycling conditions: 4 min at 95 °C, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, elongation at 72 °C for 30 s, and final elongation at 72 °C for 7 min.
In addition, positive and negative control samples were subjected to parallel PCR and qPCR. The positive control samples were used to ensure the accuracy and sensitivity of each examination, and the negative control samples were used to ensure that these tested samples were not contaminated. Note that the laboratory member who performed the PCR experiments was authorized to perform these examinations after sufficient training, the equipment and instruments were regularly calibrated, and the environment where the examinations were performed was monitored to ensure the validity of all tests.
2.4. Genome sequencing of PCV3 and PCV4
The entire length of the PCV3 genome was amplified from the PCV3 qPCR-positive samples via PCR. The total length of the PCV3 genome was identified using modified reaction conditions published by Fux et al., in 2018 [16]. The complete sequence of the PCV3 genome was assembled by aligning three sequences derived from PCR. Three pairs of primers (PCV3 39F–1144R, PCV3 910F–1561R, and PCV3 1427F–38R; Table 1) were used to amplify three PCV3 fragments, and the expected sizes were 1106, 652, and 612 bp, respectively. PCR was performed using a thermal cycler (Applied Biosystems 2720 Thermal Cycler, ThermoFisher Scientific). The 25-μL final reaction mixture consisted of 2.5 μL of PCR buffer (10X AccuPrime PCR Buffer II, ThermoFisher Scientific), 0.2 μL of DNA polymerase (5 U/μL; AccuPrime Taq DNA Polymerase High Fidelity, ThermoFisher Scientific), 1 μL of PCV3 front primer (10 μM), 1 μL of PCV3 reverse primer (10 μM), 5 μL of DNA (50–500 ng), and 15.3 μL of DDW. The thermocycling conditions were as follows: 2 min at 94 °C, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, elongation at 68 °C for 1 min, and a final elongation at 68 °C for 7 min. The PCR products were subjected to direct Sanger sequencing using a Sanger sequencing kit (Tri-I Biotech, Taipei, Taiwan).
The complete sequence of the PCV4 genome was assembled by aligning four sequences derived from PCR. Four pairs of primers (PCV4 312F–1243R, PCV4 1047F–1733R, PCV4 464F–1737R, and PCV4 1347F–469R; Table 1) were used to amplify four PCV4 fragments with sizes of 932, 687, 1274, and 879 bp, respectively. PCR was performed using a thermal cycler (Applied Biosystems 2720 Thermal Cycler, ThermoFisher Scientific). The 25-μL final mixture consisted of 2.5 μL of PCR buffer (10X AccuPrime PCR Buffer II, ThermoFisher Scientific), 0.2 μL of DNA polymerase (5 U/μL; AccuPrime Taq DNA Polymerase High Fidelity, ThermoFisher Scientific), 1 μL of PCV3 front primer (10 μM), 1 μL of PCV3 reverse primer (10 μM), 5 μL of DNA (50–500 ng), and 15.3 μL of DDW. The thermocycling conditions were as follows: 2 min at 94 °C, followed by 38–40 cycles of denaturation at 94 °C for 30 s; annealing for 30 s at 55 °C (primer pair PCV4 312F–PCV4 1243R), 50 °C (primer pair PCV4 1047F–PCV4 1733R), 55 °C (primer pair PCV4 464F–PCV4 1737R), and 56 °C (primer pair PCV4 1347F–PCV4 469R); elongation at 72 °C for 1 min; and a final elongation at 72 °C for 8 min. The PCR products were also subjected to direct Sanger sequencing, which was performed using a Sanger sequencing kit (Tri-I Biotech Inc., New Taipei City, Taiwan).
2.5. Sequence alignment and phylogenetic analysis
The complete genome sequences of PCV3 and PCV4 were edited and assembled using the EditSeq program (DNASTAR, Madison, WI, USA). Multiple aligned sequences were created using Molecular Evolutionary Genetics Analysis (MEGA) software version 11.0.13 [38]. In MEGA11, phylogenetic trees were produced using the maximum likelihood method with a bootstrap analysis of 1000 replicates [38,39].
3. Results
3.1. Molecular detection and epidemiology of PCV2, PCV3, and PCV4 in Taiwan
The recent status of PCV2, PCV3 and PCV4 and their prevalence in Taiwan remained unclear. To explore the recent positivity rates of PCV2 and the infection status of PCV3 and PCV4, a total of 128 tissue samples were collected from 46 clinical cases from pig farms in northern and central Taiwan between September 2020 and December 2021. Of the collected sample, 13 were from suckling piglets, 94 were from nursery pigs, 8 were from finishing pigs, 3 were from sows, and 10 were from aborted stillborn piglets. Note that live animals were not included in this study. As shown in Table 2, it was suggested that among the 46 clinical cases, 25 (54.3 %), 20 (43.5 %), and 1 (2.2 %) had positive results for PCV2, PCV3, and PCV4, respectively. Furthermore, among the 128 specimens, 47 (36.7 %), 36 (28.1 %), and 1 (0.8 %) were positive for PCV2, PCV3 and PCV4, respectively (Table 2). The results suggested that even though the tissue samples collected from pigs with PCV-like symptoms were selected for detection, the positive rates of PCV2-PCV3 cases and specimens were 54-43 % and 36-28 %, respectively. In addition, the positive rates of PCV4 cases and specimens were even lower and were only 2 % and 0.8 %, respectively. Consequently, more studies are needed to be done in order to evaluate the correlation between PCV positive rates and PCV-like clinical symptoms. The geographic distribution of the 46 clinical cases across the counties and cities where the samples were collected is shown in Fig. 1. Positive clinical cases for PCV2 and PCV3 were identified in two sampled cities and five sampled counties in northern and central Taiwan. A newly emerged case for PCV4 (Fig. 1, solid triangle) was detected from sick pig samples collected from a pig farm in Changhua County (CH).
Table 2.
Detection of PCV2, PCV3, and PCV4 through PCR in cases and samples collected in Taiwan from September 2020 to December 2021.
| Item | Number of samples collecteda | Positive (%) |
||
|---|---|---|---|---|
| PCV2 | PCV3 | PCV4 | ||
| Cases | 46 | 25 (54.3) | 20 (43.5) | 1 (2.2) |
| Specimens | 128 | 47 (36.7) | 36 (28.1) | 1 (0.8) |
The tissue samples were collected from pigs with PCV-like syndromes based on the prevous study, including abortion, PDNS, or PMWS.
Fig. 1.
Geographic distribution of Porcine circovirus infection by city and county. The bold text indicates the cities and counties in Taiwan. The numbers in parentheses indicate the number of positive PCV2, PCV3, and PCV4 cases. The number after the slash indicates the number of collected cases. The black triangle represents the location of the pig farm where the first PCV4 case was identified. NTC: New Taipei City, HC: Hsinchu County, ML: Miaoli County, TC: Taichung City, CH: Changhua County, YL: Yunlin County, NT: Nantou County. Solid square (■): Animal Disease Diagnostic Center at National Chung Hsing University; solid triangle (▲): position of the pig farm (CH01) where PCV4 was first detected. The corresponding author drew the map, and thus, there are no copyright issues.
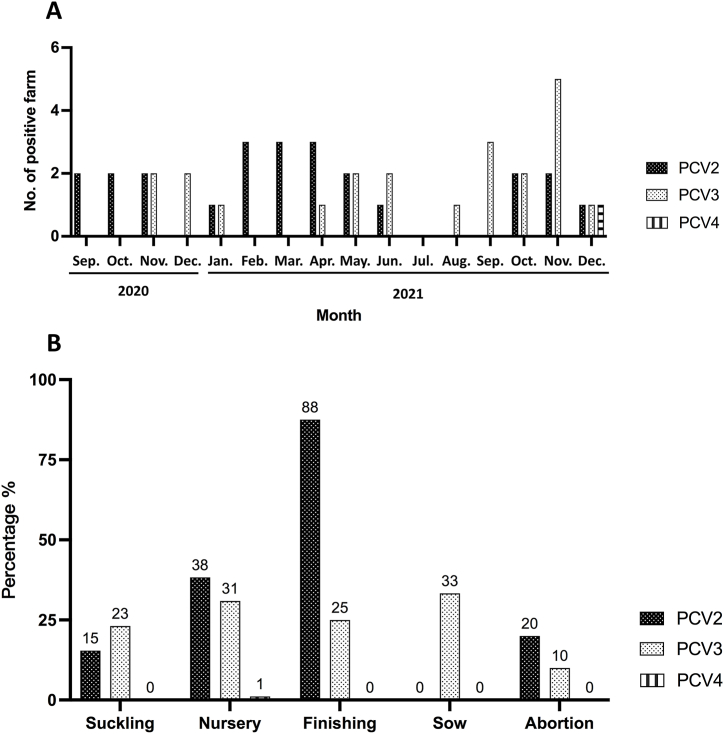
PCV2-positive clinical cases were detected between September and December 2020 and between January and December 2021 (Fig. 2A). Based on the results, it is suggested that (i) the number of PCV2-positive cases was similar across all months, except for January, July, August, and December in 2021, (ii) PCV3-positive cases occurred in 11 of the 16 months, indicating that PCV3 may be an endemic swine disease in Taiwan and (iii) a PCV4-positive specimen was collected in December 2021, suggesting the first PCV4 case was identified in Taiwan (Fig. 2A).
Fig. 2.
Monthly positive cases of Porcine circovirus infection and positive percentage of Porcine circovirus infections by pig growth stage. (A) Number of PCV2-, PCV3-, and PCV4-positive cases per month from September 2020 to December 2021. (B) Percentage of PCV2-, PCV3-, and PCV4-positive specimens from suckling piglets (N = 13), nursery pigs (N = 94), finishing pigs (N = 8), sows (N = 3), and aborted fetuses, placentas, or stillborn pigs (N = 10).
Regarding the growth phases of pigs that were PCV2-, PCV3-, and PCV4-positive (Fig. 2B), the PCV2-infection detection rate was the highest in fattening pigs (88 %). The positive detection rates of PCV3 were similar across pigs at different growth stages, ranging from 23 % to 31 %. The positive detection rate in sows was 33 %, which was higher than the abortion fetus detection rate of 10 %. On the other hand, the first PCV4 case was detected in a nursery pig (Fig. 2B).
Of the 46 clinical cases, six (13 %) coinfections, including 5 (10.8 %) with coinfections with PCV2 and PCV3 and 1 (2 %) with coinfections with PCV2 and PCV4 (Table 3). Of the six clinical cases with detected coinfections, five were from nursery pigs, and 1 was from a growing pig, indicating that nursery pigs are still the main target for PCV infection. In addition, based on the detection results (Table 3), five of the six clinical cases occurred between October and December, and only one of the six cases occurred in June, a month with higher temperatures in Taiwan, indicating that lower ambient temperatures may contribute to the transmission and infection of PCV2 and PCV3. Among the co-infection cases, respiratory signs, swollen inguinal lymph nodes, and interstitial pneumonia were noticed (IP2101098, IP2106103, IP2111121, IP2111122 in Table 4), although these lesions also appear in PCV2- or PCV3-infected pigs. Consequently, no specific lesions or clinical signs among these co-infection pigs were observed.
Table 3.
The cases with co-infection of PCV2, PCV3 or PCV4.
| Sample No. | Farm No. | Sampling pig growth stage | Sampling Month | PCV2 | PCV3 | PCV4 |
|---|---|---|---|---|---|---|
| IP2110098 | NT01 | Growing | Oct. | + | + | N* |
| IP2106103 | ML01 | Nursery | Jun. | + | + | N |
| IP2111121 | YL02 | Nursery | Nov. | + | + | N |
| IP2111122 | YL02 | Nursery | Nov. | + | + | N |
| IP2110112 | CH07 | Nursery | Oct. | + | + | N |
| IP2112138 | CH01 | Nursery | Dec. | + | N | + |
*N: not detected.
Table 4.
Clinical signs and lesions in PCV3-positive cases.
|
Sample No.a |
bFarm No. |
Growth phase | Clinical signs |
Pathological lesions |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Respiratory syndrome | Macule | Neurological sign | Dermatitis | Swollen ILN% | Interstitial pneumonia | Myocarditis | Vasculitis | Lymphoid depletion | Interstitial nephritis | Periarteritis in the spleen | Splenitis | ||||
| IP2005116 | TC01 | Suckling | – | + | – | – | + | + | – | – | – | – | – | – | |
| IP2101098 | NT01 | Growing | – | – | – | – | + | + | – | – | – | – | – | – | |
|
IP2105099 IP2105100 IP2105101 |
CH02 | Nursery | + | – | – | – | + | + | + | – | – | – | – | – | |
| IP2105128 | NT02 | Nursery | + | + | – | – | + | + | + | – | – | – | – | + | |
| IP2106102 | HC01 | Nursery | + | + | – | + | + | + | + | + | – | + | – | – | |
| IP2106103 | ML01 | Nursery | – | – | – | – | + | + | + | – | – | + | – | – | |
|
IP2108118 IP2108119 IP2108120 |
CH03 | Nursery | + | – | – | – | + | + | + | – | + | – | – | + | |
| IP2109104 | NTC01 | Nursery | + | – | + | – | + | – | – | – | – | – | – | – | |
|
IP2109105 IP2109106 |
CH04 | Nursery | – | + | – | – | + | + | – | – | – | – | – | – | |
|
IP2109108 IP2109109 |
YL01 | Nursery | + | – | + | – | + | – | – | – | – | – | + | – | |
|
IP2110129 IP2110130 IP2110131 |
CH01 | Nursery | – | – | – | + | – | + | – | – | – | – | – | – | |
| IP2111132 | CH01 | Nursery | – | – | – | – | + | + | – | – | – | – | – | – | |
|
IP2111133 IP2111134 |
CH08 | Piglet | – | + | – | – | – | – | – | – | – | – | – | – | |
|
IP2111121 IP2111122 |
YL02 | Nursery | + | – | – | – | + | + | – | – | – | + | – | – | |
|
IP2111135 IP2111136 |
NT02 | Nursery | + | – | + | – | + | + | – | – | – | + | – | – | |
|
IP2112123 IP2112124 IP2112125 IP2112126 |
CH05 | Nursery | – | – | – | + | + | + | + | + | – | + | – | – | |
| IP2111115 | TC02 | Sow | – | – | – | – | – | – | – | – | – | – | – | – | |
| No. of “+” | 8 | 5 | 3 | 3 | 14 | 13 | 6 | 2 | 1 | 5 | 1 | 2 | |||
| “+” % | 47.1 | 29.4 | 17.6 | 17.6 | 82.3 | 76.5 | 35.3 | 11.8 | 5.9 | 29.4 | 5.9 | 11.8 | |||
% swollen inguinal lymph nodes.
Because the owners of the pig farm only sent the specimens for IP201110, IP2104111, IP2010112, IP2101113 (Nursery pigs from Farm CH07) and IP2011114 (Finishing pigs from Farm YL03), information on the clinical signs and pathological lesions in the pigs is lacking.
NTC: New Taipei City, HC: Hsinchu County, ML: Miaoli County, TC: Taichung City, CH: Changhua County, YL: Yunlin County, NT: Nantou County.
3.2. Clinical and pathological profile of PCV3 cases
Since PCV3 was continuously detected in Taiwan, it is important to define the clinical and pathological profile of pigs with PCV3 infection. For this aim, the medical records of the PCV3-positive cases, including clinical and pathological characteristics of PCV3-infected pigs, were applied. Of the 20 PCV3-positive cases, three cases only requested a specimen examination and thus did not have medical records. Consequently, we analyzed the clinical signs and pathological lesions of the other 17 PCV3-positive cases. Most of the PCV3-positive cases were identified in nursery pigs (13 cases), and 1 case occurred in suckling piglets, piglets, growing pigs, or sows (Table 4). PCV3-positive pigs, respiratory syndrome (47.1 %) and macules (29.4 %) were the main clinical signs found in nursery pigs. Neurological signs and dermatitis were also observed in nursery pigs and piglets. The main pathological lesions included swollen inguinal lymph nodes (82.3 %), interstitial pneumonia (76.5 %), myocarditis (35.3 %), and interstitial nephritis (29.4 %) were noticed. Vasculitis, lymphoid depletion, periarteritis in the spleen, and splenitis were also identified. Notably, although these clinical symptoms may not all be caused by PCV3 infection, the PCV3-positive pigs had symptoms similar to those of pigs with respiratory infectious disease. It is still suggested that the attention on the main clinical symptoms and pathological lesions should be particularly paid during the diagnosis of PCV3 cases in the future.
3.3. Clinical and pathological profile of the first PCV4 infection case in Taiwan
Although PCV4 has been identified in East Asia (China, South Korea, and Thailand), Malaysia and Spain, there are no studies on the identification of PCV4 in Taiwan. Based on the results described above, the first PCV4 infection case was identified in Taiwan as detected by PCR (Supplementary Fig. S1). Consequently, the clinical and pathological profile of the first PCV4 infection case was investigated, and the results were as follows. In December 2021, a farmer at farm CH01 (Fig. 1, solid triangle) reported that a 6-week-old nursery pig had mushy yellow diarrhea, and respiratory symptoms appeared the following week. The sick pig from the farm CH01 was sent for pathological examination (sample no. IP2112138). The sick pig was thin, with sticky secretions in the nasal cavity. Necropsy examination revealed considerable fibrin in the chest and abdominal cavity (Fig. 3A), consolidation of the cranial and middle lobes of the lungs (Fig. 3B), and thickening of the pericardium with white effusion in the pericardial sac (Fig. 3C). Histopathological examination revealed considerable neutrophil and lymphocyte infiltration and accumulation of necrotic cell debris in the alveoli and bronchial lumen of the cranial and middle lobes (Fig. 3D). The number of macrophages in the tonsillar lymphoid follicles was increased (Fig. 3E). A small amount of fibrous exudate was accumulated in the interstitium of the hilar lymph node and tracheobronchial lymph node and was accompanied by neutrophil infiltration (Fig. 3F). Microbiological examination revealed Glaesserella parasuis and Streptococcus suis infection, and molecular diagnostic tests were positive for Mycoplasma hyorhinis, PCV2, and PCV4 (Supplementary Fig. S1) but negative for porcine reproductive and respiratory syndrome virus (PRRSV). This case was diagnosed with PRDC infection and was the first PCV4 infection case detected by PCR in Taiwan.
Fig. 3.
Clinical signs, gross findings, and histopathology (hematoxylin & eosin stain) of a pig with PCV4 infection, which was the first PCV4-positive pig in Taiwan (sample no. IP2112138, farm CH01). (A) Polyserositis was found in the pleural and abdominal cavities. (B) The lungs were mottled and firm upon palpation. (C) The epicardium was covered with fibrin. (D) Type II pneumocyte hyperplasia with neutrophil, lymphocyte, plasma cell, and histiocyte infiltration of the lung interstitium. (E) Macrophage infiltration of lymphoid follicles observed in a tonsil. (F) Lymphoid depletion was found in the tracheobronchial lymph node with abundant neutrophil infiltration.
In May 2022, two 10-week-old sick pigs from the same farm (CH01) were sent to ADDC for pathological examination (sample no. IP2205150). This case was also diagnosed with PRDC infection (Supplementary Fig. S2) and was PCV4-positive as determined by PCR again.
3.4. Characterization of the PCV3 and PCV4 genomes
Although PCV3 and PCV4 were identified in Taiwan, the characteristics of their genomes remain unclear. To characterize the genomes, PCV3-positive samples were used to amplify and sequence the complete PCV3 genome. Among the 46 PCV3-positive specimens with higher amounts of viral DNA (lower Ct values), as determined by qPCR, were selected for sequencing. In addition, at least one positive specimen from each pig farm was selected for sequencing. Consequently, seventeen complete full-length PCV3 sequences were obtained for subsequent analysis. The amplified PCV3 sequences were assembled into 2000-bp genomes. The sequences of the YH108, YH109, and YH110 strains had a deletion at position 1224; thus, the length of the genome size was 1999 bp. Three strains, YH110, YH114, and YH116, were detected in 2020, and the remaining 14 PCV3 strains were detected in 2021 (Supplementary Table S1).
The sequences of the 17 PCV3 isolates (accession no. OQ557960–OQ557976) in this study were closely related (less than 1.1 % difference; Supplementary Table S2, lower part). The results of sequence analysis revealed that (i) YH099 and YH101 were isolated from farm CH02 in January 2021 and exhibited a sequence divergence of 0.8 %, (ii) YH108 and YH109 were isolated from farm YL01 in September 2021, and had completely identical gene sequences, and (iii) YH118 and YH119 were isolated from farm CH03 in August 2021 and exhibited a sequence divergence of 0.1 %. Furthermore, YH110, YH111, and YH113 were isolated from farm CH07 in November 2020, April 2021, and January 2021, respectively, and their sequence divergences were only 0.6 % and 0.7 % (Supplementary Table S2). These results indicate that the divergence of PCV3 sequences isolated at the same or different times on the same farm was less than 0.8 % in the current study.
We obtained two PCV4 isolates, YH138C4 (accession no. OQ557977) and YH150 (accession nos. OQ557978 and OQ939948). The first Taiwanese isolate, YH138C4, was only partially sequenced (1376 bp) due to an insufficient specimen. This 1376-bp sequence of the YH138C4 strain was identical to that of the YH150 strain obtained from the follow-up samples from the same pig farm (CH01) (Supplementary Table S1). Thus, the two strains, YH138C4 and YH150, were considered the same PCV4 strain. The Taiwanese strain YH150 was closely related to the South Korean KU-02011 strain (accession no. MW712667) that was collected in November 2020 because YH150 and KU-02011 strain shared 98.7 % (1748/1770) of sequence identity. (Supplementary Table S3).
3.5. Phylogenetic analysis of the PCV3 and PCV4 full-length sequences
Since PCV3 and PCV4 were identified in Taiwan, the relationship of these isolates with others identified in other countries remained to be clarified. To this aim, a phylogenetic tree was generated based on the 17 PCV3 Taiwanese isolate sequences and 36 published full-length PCV3 sequences (Fig. 4, Supplementary Fig. S3). In Supplementary Fig. S3, the additional sequences of PCV1 and PCV2 were used as outgroups to indicate that the PCV3 sequence cluster was independent of that in PCV1 and PCV2 based on the result of a long genetic distance. Phylogenetic analysis of the PCV3 sequences demonstrated that most of the Taiwanese isolates in this study were closely related to previously published Taiwanese isolates (accession nos. MN510446 and MN510467) that belonged to clade 1 (Fig. 4, Group 1) [35]. However, the YH118 and YH119 isolates were grouped in another cluster, most closely related to a 2016 Japanese isolate (accession number LC383840) (Fig. 4, Group 2). Therefore, in this study, two groups of PCV3, including Taiwanese strain-like and Japanese strain-like, were identified.
Fig. 4.
Phylogenetic analysis of PCV3 isolates. The phylogenetic tree was constructed based on the genomes of 53 PCV3 isolates, 17 Taiwanese isolates (black solid circles), and 36 PCV3 isolates obtained from GenBank. The phylogenetic analysis of PCV3 isolates was performed using Molecular Evolutionary Genetics Analysis (MEGA) software version 11.0.13. The phylogenetic tree was established using the maximum likelihood method by bootstrap resampling with 1000 replicates. The scale bar indicates nucleotide substitutions per site.
According to medical records from pig farms, YH098 from farm NT01 and YH099 from farm CH02 were 100 % identical, the distance between the two farms was more than 40 km. In addition, although farms CH07, YL03, TC01, and YL02 were in different locations in Taiwan, strain YH110 from farm CH07 and strain YH114 from farm YL03 shared 99.6 % sequence identity and thus were grouped in the same group. Strain YH116 from farm TC01 and strain YH121 from farm YL02 shared 99.8 % identity and were also grouped in the same group (Fig. 4). Consequently, the results indicated that the same virus strain can potentially be isolated from different farms that are a certain distance from each other.
In contrast, strains YH110 (1999 bp), YH111 (2000 bp), and YH113 (2000 bp) were all isolated from farm CH07 in November 2020, April 2021, and January 2021, respectively. They were distributed in three branches of the phylogenetic tree, showing that different strains of PCV3 may have been circulating on farm CH07 (Fig. 4).
The phylogenetic tree for the PCV4 sequences, including 54 full-length sequences from GenBank (48 Chinese strains, 3 South Korean strains, and 3 Thai strains) and 2 Taiwanese sequences (Fig. 5), was similar to that of the most recently reported study in 2023 [29]. It was noticed that the bootstrap values resulting from the analysis were low. The lower bootstrap values were also observed in recent studies regarding the phylogenetic analysis of PCV4 sequences [29,32]. It is speculated that the lower bootstrap values may be attributed to (i) the current number of PCV4 sequences in Genbank is not sufficient to obtain a reliable analysis and (ii) the sequences between the PCV4 isolates are highly similar. Consequently, it is expected that, with the increase of PCV studies, the issue could possibly be solved. These 2 Taiwanese isolates were closely related to previously published isolates from South Korea (accession no. MW712667) and were grouped in the same clade. All of these Taiwanese, South Korean, and Thai strains have been published, and some of the Chinese strains belong to the PCV4b clade [29].
Fig. 5.
Phylogenetic analysis of PCV4 isolates. The phylogenetic tree was constructed based on the 54 PCV4 sequences derived from 56 PCV4 isolates obtained from GenBank and 2 Taiwanese sequences (black solid circles) with the accession number, country/zone, and collection year. The phylogenetic analysis of PCV4 isolates was performed using Molecular Evolutionary Genetics Analysis (MEGA) software version 11.0.13. The phylogenetic tree was established using the maximum likelihood method by bootstrap resampling with 1000 replicates. The scale bar indicates nucleotide substitutions per site.
4. Discussion
PCV2 has been circulating in Taiwan and PCV3 and PCV4 are known to be emerging viruses in several countries [8,[10], [11], [12], [13], [14],[16], [17], [18], [19], [20],[23], [24], [25]]. However, most of these studies have focused mainly on the detection of PCV2 and PCV3 in different pig specimens; thus, the epidemiology of PCVs in pig farms in Taiwan remains unclear. In addition, to eradicate foot and mouth disease (FMD) in pigs, Taiwan has imported only breeding gilts and semen from countries free from FMD, classical swine fever (CSF), and African swine fever (ASF) in the past 20 years. Because Taiwan is an island, opportunities for the introduction of newly emerging infectious diseases for pigs across borders may be limited. Although it remains unknown how PCV2, PCV3, and PCV4 were introduced to Taiwan, continuous investigations of newly emerging infectious diseases in pigs are needed. In this study, we investigated pig cases submitted to the Animal Disease Diagnostic Center of National Chung Hsing University from 2020 to 2021. A total of 128 pig samples from 46 pig farms in northern and central Taiwan were collected from September 2020 to December 2021. The collected pig samples were used to explore the rates of infection with PCV2, PCV3, and PCV4 in Taiwan using molecular diagnostic technology and pathological diagnosis. Based on the analysis results, including the positivity rate, major clinical symptoms, and distribution, we attempted to explain the epidemiology of PCV2 and PCV3 and to provide evidence for the existence of PCV4 in Taiwan as follows.
Based on the results shown in Tables 2 and it is suggested that PCV2 (54 % of the cases detected) is still widespread in Taiwan. Compared with that of PCV3, the PCV2 detection rate was greater in nursery and fattening pigs, consistent with a previous study in Taiwan [40]; however, the PCV3 detection rate was greater in nursery pigs and sows. A study in Spain indicated that PCV3 infection is not age-specific [41]. The detection rates in pigs of different growth periods suggest that PCV3 is more easily detected in nursery pigs (31 %), similar to PCV2 (38 % in nursery pigs). Although vaccines against PCV2 infection have been used on pig farms in Taiwan, the high positivity rate of PCV2 and subclinical PCV2 infection suggests that PCV2 infection is still a long-term problem on pig farms. Whether insufficient protection from the PCV2 vaccine is correlated with coinfection of PCV3 and other pathogens, including PCV4, and whether infection with PCV2 and PCV3 is age-specific remains to be elucidated by continuous surveillance.
The percentages of PCV3-positive specimens in Taiwan were 10.6 %, 12.64 %, 33.3 %, and 34.78 % in 2016, 2018, 2018, and 2019, respectively, in a previous study [35]. In the present study, the positivity rate for PCV3 was 28.1 % during 2020–2021, suggesting that the positivity rate did not increase with time. This study also showed that PCV3-positive cases (43.5 %) were identified in all seven counties and in 11 of the 16 months of the sampling periods (September 2020 to December 2021). Consequently, the results suggest that PCV3 has been spreading on pig farms across Taiwan and that PCV3 may lead to an endemic disease. In addition, the rate (10.8 %, 5/46) (Table 3) of coinfection with PCV2 and PCV3 in the current study was lower than that (12.5 %–70 %) in the previous studies [25].
Among 17 identified cases of PCV3 infection, the clinical symptoms and pathological findings were similar to those of PDNS in pigs [7,42,43] and included respiratory symptoms [11,12,44], and multisystemic inflammatory responses [42,45,46]. However, PRDC-induced porcine respiratory syndrome is one of the most severe pig diseases in Taiwan, and many pathogens, such as PRRSV, PCV2, and Mycoplasma hyopneumoniae, can also cause interstitial pneumonia. Therefore, it cannot be ruled out that pathogenic lesions caused by PRDC may potentially be the result of PCV3. Furthermore, PCV3 nucleic acid can also be detected in healthy pigs [43,47,48], indicating that PCV3 may be a subclinical infection and thus explaining the high PCV3 detection rate in pig farms in Taiwan. Finally, the differences between the 17 PCV3 virus sequences obtained in this study ranged from 0 to 1.1 % (Supplementary Table S2). These results indicate that it may take at least 468 years to accumulate a 1.1 % difference in the complete PCV3 sequences based on a study in which the molecular evolutionary rate of PCV3 was approximately 2.35 × 10−5 substitutions/site/year according to a molecular clock [49]. Consequently, it is possible that PCV3 may have been circulating on pig farms in Taiwan but was not identified until recently.
To date, PCV4 has been identified in only 5 countries, namely, China, South Korea, Thailand, Malaysia, and Spain. In the present study, PCV4 was detected for the first time in pigs in Taiwan (YH 138C4 strain, accession number OQ557977). This is also the first case in which PCV4 was detected on an island. The follow-up detection of PCV4 at the same farm (YH150 strain, accession number OQ557978 and OQ939948) further demonstrated the existence of PCV4 in Taiwan. A study published in 2022 indicated that the amino acid position 27 of the PCV4 Cap gene was a critical site for separating PCV4a (serine; S) from PCV4b (asparagine; N) based on PCV4 sequences [34]. However, another study in 2023 revealed that the amino acid at position 27 in PCV4a is serine (S), but PCV4b can contain both serine and asparagine (N). In addition, amino acid position 212 of the PCV4 Cap protein could play a critical role in differentiating PCV4a (leucine; L) from PCV4b (methionine; M) [29]. For the PCV4 strain YH150 in Taiwan, the amino acid residues at positions 27, 80, and 212 on the Cap protein are asparagine (N), isoleucine (I), and methionine (M), respectively. As a result, the amino acid sequences of these three sites in Taiwanese strains are identical to those of the South Korean KU-02011 strain (accession no. MW712667) [29]. The phylogenetic tree analysis in this study showed that PCV4 strains from Taiwan, South Korea, and Thailand were clustered in the same PCV4b branch, which is also supported by the conclusion of a study in 2023 [29].
In South Korea, China, and Thailand, PCV4 can be transmitted through susceptible animals carrying PCV4. Due to the occurrence of FMD, CSF, and ASF in These countries, Taiwan has not imported pigs or pork products from these countries for decades. The pathway of PCV4 introduction remains unclear; thus, additional studies on the etiology, epidemiology, immunology, impact on animals, and transboundary transmission of PCV4 infection in pig populations are urgently needed.
5. Conclusions
In this study, a total of 128 pig samples from 46 pig farms in northern and central Taiwan were collected from September 2020 to December 2021 to identify the PCV2, PCV3 and PCV4 infection rates in Taiwan using molecular diagnostic technology and pathological diagnosis. Based on the analysis results, including the positivity rate, major clinical symptoms, time and distribution, it can be concluded that (i) the positive rates of PCV2 and PCV3 are still high in Taiwanese pigs, (ii) PCV3 has spreaded on the pig farms across Taiwan, and PCV3 may lead to an endemic infection in Taiwan in terms of epidemiology, (iii) the identified PCV3 strains can be classified into Japanese-like and Taiwanese-like strains based on the sequence analysis, (iv) PCV4 was detected for the first time in Taiwanese pigs based on DNA sequence detection, and (v) the first identified PCV4 strain was closely related to the South Korean KU-02011 strain. It remains unknown how PCV2, PCV3, and PCV4 were introduced to Taiwan, and thus continuous investigation of newly emerging infectious diseases in pigs is needed.
Funding
This work was supported by the Endowment Fund from National Chung Hsing University (108GP04376).
Ethics statement
The protocols for animal sample collection and agreements for research purposes were conducted according to the “Council of Agriculture Executive Yuan Guideline for the Care and Use of Laboratory Animals”. These measures were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of National Chung Hsing University (Affidavit of Approval of Animal Use Protocol, IACUC No.110–113). The biomaterial examination protocol was carried out according to the “Chung Hsing University Infectious Biomaterials Management Regulations” and reviewed and approved by the Biological Safety Committee (no. 110GR085) of the National Chung Hsing University.
Data availability statement
The datasets generated and analyzed during the current study (accession numbers: OQ557960 - OQ557978, and OQ939948) are available in the GenBank repository. The data supporting this article's conclusions are included in the publication.
CRediT authorship contribution statement
Yu Fan Hung: Writing – original draft, Visualization, Validation, Methodology, Data curation. Po-Chen Liu: Visualization, Validation, Methodology, Data curation. Ching-Hung Lin: Validation, Methodology, Investigation, Data curation. Chao-Nan Lin: Validation, Methodology, Data curation. Hung-Yi Wu: Writing – review & editing, Writing – original draft, Methodology, Conceptualization. Ming-Tang Chiou: Writing – review & editing, Validation, Methodology, Conceptualization. Hung-Jen Liu: Writing – review & editing, Resources, Conceptualization. Cheng-Yao Yang: Writing – review & editing, Writing – original draft, Methodology, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank D.V.M. Yao-Hsuan Liu for supporting the clinical diagnosis and Chung-Ping Huang, Mr. Yi-Fan Chao, and Mrs. Tsung-Ting Chuang for supporting the molecular diagnostic examinations.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e35579.
List of abbreviations
- ASF
African swine fever
- CSF
Classical swine fever
- FMD
foot and mouth disease
- MEGA
Molecular Evolutionary Genetics Analysis
- ORFs
open reading frames
- PCR
polymerase chain reaction
- PCV
porcine circovirus
- PCV1
porcine circovirus type 1
- PCV2
porcine circovirus type 2
- PCV3
porcine circovirus type 3
- PCV4
porcine circovirus type 4
- PCVs
porcine circoviruses
- PDNS
porcine dermatitis and nephropathy syndrome
- PMWS
postweaning multisystemic wasting syndrome
- PRDC
porcine respiratory disease complex
- PRRS
porcine reproductive and respiratory syndrome
- qPCR
quantitative polymerase chain reaction
Appendix A. Supplementary data
The following is the supplementary data to this article.
References
- 1.Rosario K., Breitbart M., Harrach B., Segalés J., Delwart E., Biagini P., et al. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch. Virol. 2017;162(5):1447–1463. doi: 10.1007/s00705-017-3247-y. [DOI] [PubMed] [Google Scholar]
- 2.Allan G.M., Ellis J.A. Porcine circoviruses: a review. J. Vet. Diagn. Invest. 2000;12(1):3–14. doi: 10.1177/104063870001200102. [DOI] [PubMed] [Google Scholar]
- 3.Allan G.M., McNeilly F., Cassidy J.P., Reilly G.A., Adair B., Ellis W.A., et al. Pathogenesis of porcine circovirus; experimental infections of colostrum deprived piglets and examination of pig foetal material. Vet. Microbiol. 1995;44(1):49–64. doi: 10.1016/0378-1135(94)00136-k. [DOI] [PubMed] [Google Scholar]
- 4.Grasland B., Loizel C., Blanchard P., Oger A., Nignol A.C., Bigarré L., et al. Reproduction of PMWS in immunostimulated SPF piglets transfected with infectious cloned genomic DNA of type 2 porcine circovirus. Vet Res. 2005;36(5–6):685–697. doi: 10.1051/vetres:2005024. [DOI] [PubMed] [Google Scholar]
- 5.Segalés J. Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Res. 2012;164(1–2):10–19. doi: 10.1016/j.virusres.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Ellis J., Hassard L., Clark E., Harding J., Allan G., Willson P., et al. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can. Vet. J. 1998;39(1):44–51. [PMC free article] [PubMed] [Google Scholar]
- 7.Palinski R., Piñeyro P., Shang P., Yuan F., Guo R., Fang Y., et al. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J. Virol. 2017;91(1) doi: 10.1128/JVI.01879-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ku X., Chen F., Li P., Wang Y., Yu X., Fan S., et al. Identification and genetic characterization of porcine circovirus type 3 in China. Transbound Emerg Dis. 2017;64(3):703–708. doi: 10.1111/tbed.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu T., Zhang Y.-H., Tian R.-B., Hou C.-Y., Li X.-S., Zheng L.-L., et al. Prevalence and genetic analysis of porcine circovirus type 2 (PCV2) and type 3 (PCV3) between 2018 and 2020 in central China. Infect. Genet. Evol. 2021;94 doi: 10.1016/j.meegid.2021.105016. [DOI] [PubMed] [Google Scholar]
- 10.Kwon T., Yoo S.J., Park C.K., Lyoo Y.S. Prevalence of novel porcine circovirus 3 in Korean pig populations. Vet. Microbiol. 2017;207:178–180. doi: 10.1016/j.vetmic.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Kedkovid R., Woonwong Y., Arunorat J., Sirisereewan C., Sangpratum N., Lumyai M., et al. Porcine circovirus type 3 (PCV3) infection in grower pigs from a Thai farm suffering from porcine respiratory disease complex (PRDC) Vet. Microbiol. 2018;215:71–76. doi: 10.1016/j.vetmic.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Bera B.C., Choudhary M., Anand T., Virmani N., Sundaram K., Choudhary B., et al. Detection and genetic characterization of porcine circovirus 3 (PCV3) in pigs in India. Transbound Emerg Dis. 2020;67(3):1062–1067. doi: 10.1111/tbed.13463. [DOI] [PubMed] [Google Scholar]
- 13.Stadejek T., Woźniak A., Miłek D., Biernacka K. First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transbound Emerg Dis. 2017;64(5):1350–1353. doi: 10.1111/tbed.12672. [DOI] [PubMed] [Google Scholar]
- 14.Franzo G., Legnardi M., Hjulsager C.K., Klaumann F., Larsen L.E., Segales J., et al. Full-genome sequencing of porcine circovirus 3 field strains from Denmark, Italy and Spain demonstrates a high within-Europe genetic heterogeneity. Transbound Emerg Dis. 2018;65(3):602–606. doi: 10.1111/tbed.12836. [DOI] [PubMed] [Google Scholar]
- 15.Faccini S., Barbieri I., Gilioli A., Sala G., Gibelli L.R., Moreno A., et al. Detection and genetic characterization of Porcine circovirus type 3 in Italy. Transbound Emerg Dis. 2017;64(6):1661–1664. doi: 10.1111/tbed.12714. [DOI] [PubMed] [Google Scholar]
- 16.Fux R., Söckler C., Link E.K., Renken C., Krejci R., Sutter G., et al. Full genome characterization of porcine circovirus type 3 isolates reveals the existence of two distinct groups of virus strains. Virol. J. 2018;15(1):25. doi: 10.1186/s12985-018-0929-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins P.J., McKillen J., Allan G. Porcine circovirus type 3 in the UK. Vet. Rec. 2017;181(22):599. doi: 10.1136/vr.j5505. [DOI] [PubMed] [Google Scholar]
- 18.Deim Z., Dencső L., Erdélyi I., Valappil S.K., Varga C., Pósa A., et al. Porcine circovirus type 3 detection in a Hungarian pig farm experiencing reproductive failures. Vet. Rec. 2019;185(3):84. doi: 10.1136/vr.104784. [DOI] [PubMed] [Google Scholar]
- 19.Rudova N., Lymanska O., Stegniy B., Bolotin V., Solodiankin O., Gerilovych A. First detection of porcine circovirus type 3 in Ukraine. Agricultural Science and Practice. 2021;8(2):16–23. [Google Scholar]
- 20.Savic B., Milicevic V., Radanovic O., Zdravkovic N., Stevancevic O., Kureljusic B., et al. Identification and genetic characterization of porcine circovirus 3 on pig farms in Serbia. Arch. Virol. 2020;165(1):193–199. doi: 10.1007/s00705-019-04455-y. [DOI] [PubMed] [Google Scholar]
- 21.Ye X., Berg M., Fossum C., Wallgren P., Blomström A.L. Detection and genetic characterisation of porcine circovirus 3 from pigs in Sweden. Virus Gene. 2018;54(3):466–469. doi: 10.1007/s11262-018-1553-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodrigues I.L.F., Cruz A.C.M., Souza A.E., Knackfuss F.B., Costa C.H.C., Silveira R.L., et al. Retrospective study of porcine circovirus 3 (PCV3) in swine tissue from Brazil (1967-2018) Braz. J. Microbiol. 2020;51(3):1391–1397. doi: 10.1007/s42770-020-00281-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tochetto C., Lima D.A., Varela A.P.M., Loiko M.R., Paim W.P., Scheffer C.M., et al. Full-Genome Sequence of Porcine Circovirus type 3 recovered from serum of sows with stillbirths in Brazil. Transbound Emerg Dis. 2018;65(1):5–9. doi: 10.1111/tbed.12735. [DOI] [PubMed] [Google Scholar]
- 24.Vargas-Bermudez D.S., Campos F.S., Bonil L., Mogollon D., Jaime J. First detection of porcine circovirus type 3 in Colombia and the complete genome sequence demonstrates the circulation of PCV3a1 and PCV3a2. Vet. Med. Sci. 2019;5(2):182–188. doi: 10.1002/vms3.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serena M.S., Cappuccio J.A., Barrales H., Metz G.E., Aspitia C.G., Lozada I., et al. First detection and genetic characterization of porcine circovirus type 3 (PCV3) in Argentina and its association with reproductive failure. Transbound Emerg Dis. 2021;68(4):1761–1766. doi: 10.1111/tbed.13893. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H.H., Hu W.Q., Li J.Y., Liu T.N., Zhou J.Y., Opriessnig T., et al. Novel circovirus species identified in farmed pigs designated as Porcine circovirus 4, Hunan province, China. Transbound Emerg Dis. 2020;67(3):1057–1061. doi: 10.1111/tbed.13446. [DOI] [PubMed] [Google Scholar]
- 27.Tian R.B., Zhao Y., Cui J.T., Zheng H.H., Xu T., Hou C.Y., et al. Molecular detection and phylogenetic analysis of porcine circovirus 4 in henan and shanxi provinces of China. Transbound Emerg Dis. 2021;68(2):276–282. doi: 10.1111/tbed.13714. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen V.G., Do H.Q., Huynh T.M., Park Y.H., Park B.K., Chung H.C. Molecular-based detection, genetic characterization and phylogenetic analysis of porcine circovirus 4 from Korean domestic swine farms. Transbound Emerg Dis. 2022;69(2):538–548. doi: 10.1111/tbed.14017. [DOI] [PubMed] [Google Scholar]
- 29.Sirisereewan C., Nguyen T.C., Piewbang C., Jittimanee S., Kedkovid R., Thanawongnuwech R. Molecular detection and genetic characterization of porcine circovirus 4 (PCV4) in Thailand during 2019-2020. Sci. Rep. 2023;13(1):5168. doi: 10.1038/s41598-023-32382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franzo G., Ruiz A., Grassi L., Sibila M., Drigo M., Segalés J. Lack of porcine circovirus 4 genome detection in pig samples from Italy and Spain. Pathogens. 2020;9(6) doi: 10.3390/pathogens9060433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holgado-Martín R., Arnal J.L., Sibila M., Franzo G., Martín-Jurado D., Risco D., et al. First detection of porcine circovirus 4 (PCV-4) in Europe. Virol. J. 2023;20(1):230. doi: 10.1186/s12985-023-02181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan C.Y., Thanawongnuwech R., Arshad S.S., Hassan L., Lee C.Y., Low S.E., et al. First molecular detection of porcine circovirus type 4 (PCV4) in Malaysia. Trop. Biomed. 2023;40(3):301–306. doi: 10.47665/tb.40.3.005. [DOI] [PubMed] [Google Scholar]
- 33.Xu T., Hou C.Y., Zhang Y.H., Li H.X., Chen X.M., Pan J.J., et al. Simultaneous detection and genetic characterization of porcine circovirus 2 and 4 in Henan province of China. Gene. 2022;808 doi: 10.1016/j.gene.2021.145991. [DOI] [PubMed] [Google Scholar]
- 34.Wang D., Mai J., Yang Y., Xiao C.-T., Wang N. Current knowledge on epidemiology and evolution of novel porcine circovirus 4. Veterinary Research. 2022;53(1):38. doi: 10.1186/s13567-022-01053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang C.C., Wu C.W., Chang Y.C., Wu C.Y., Chien M.S., Huang C. Detection and phylogenetic analysis of porcine circovirus type 3 in Taiwan. Arch. Virol. 2021;166(1):259–263. doi: 10.1007/s00705-020-04870-6. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z.Z., Habib M., Shuai J.B., Fang W.H. Detection of PCV2 DNA by SYBR green I-based quantitative PCR. J. Zhejiang Univ. - Sci. B. 2007;8(3):162–169. doi: 10.1631/jzus.2007.B0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Yan S., Ji Y., Yang Y., Rui P., Ma Z., et al. First identification and phylogenetic analysis of porcine circovirus type 4 in Fur animals in hebei, China. Animals (Basel) 2022;12(23) doi: 10.3390/ani12233325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38(7):3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanderson M.J. Confidence limits on phylogenies: the bootstrap revisited. Cladistics. 1989;5(2):113–129. doi: 10.1111/j.1096-0031.1989.tb00559.x. [DOI] [PubMed] [Google Scholar]
- 40.Tsai G.T., Lin Y.C., Lin W.H., Lin J.H., Chiou M.T., Liu H.F., et al. Phylogeographic and genetic characterization of porcine circovirus type 2 in Taiwan from 2001-2017. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-47209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klaumann F., Franzo G., Sohrmann M., Correa-Fiz F., Drigo M., Núñez J.I., et al. Retrospective detection of Porcine circovirus 3 (PCV-3) in pig serum samples from Spain. Transbound Emerg Dis. 2018;65(5):1290–1296. doi: 10.1111/tbed.12876. [DOI] [PubMed] [Google Scholar]
- 42.Arruda B., Piñeyro P., Derscheid R., Hause B., Byers E., Dion K., et al. PCV3-associated disease in the United States swine herd. Emerg Microbes Infect. 2019;8(1):684–698. doi: 10.1080/22221751.2019.1613176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhai S.L., Zhou X., Zhang H., Hause B.M., Lin T., Liu R., et al. Comparative epidemiology of porcine circovirus type 3 in pigs with different clinical presentations. Virol. J. 2017;14(1):222. doi: 10.1186/s12985-017-0892-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S.H., Park J.Y., Jung J.Y., Kim H.Y., Park Y.R., Lee K.K., et al. Detection and genetic characterization of porcine circovirus 3 from aborted fetuses and pigs with respiratory disease in Korea. J. Vet. Sci. 2018;19(5):721–724. doi: 10.4142/jvs.2018.19.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayashi S., Ohshima Y., Furuya Y., Nagao A., Oroku K., Tsutsumi N., et al. First detection of porcine circovirus type 3 in Japan. J. Vet. Med. Sci. 2018;80(9):1468–1472. doi: 10.1292/jvms.18-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen G.H., Mai K.J., Zhou L., Wu R.T., Tang X.Y., Wu J.L., et al. Detection and genome sequencing of porcine circovirus 3 in neonatal pigs with congenital tremors in South China. Transbound Emerg Dis. 2017;64(6):1650–1654. doi: 10.1111/tbed.12702. [DOI] [PubMed] [Google Scholar]
- 47.Kim S.C., Nazki S., Kwon S., Juhng J.H., Mun K.H., Jeon D.Y., et al. The prevalence and genetic characteristics of porcine circovirus type 2 and 3 in Korea. BMC Vet. Res. 2018;14(1):294. doi: 10.1186/s12917-018-1614-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saporiti V., Martorell S., Cruz T.F., Klaumann F., Correa-Fiz F., Balasch M., et al. Frequency of detection and phylogenetic analysis of porcine circovirus 3 (PCV-3) in healthy primiparous and multiparous sows and their mummified fetuses and stillborn. Pathogens. 2020;9(7) doi: 10.3390/pathogens9070533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Franzo G., He W., Correa-Fiz F., Li G., Legnardi M., Su S., et al. A shift in porcine circovirus 3 (PCV-3) history paradigm: phylodynamic analyses reveal an ancient origin and prolonged undetected circulation in the worldwide swine population. Adv. Sci. 2019;6(22) doi: 10.1002/advs.201901004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study (accession numbers: OQ557960 - OQ557978, and OQ939948) are available in the GenBank repository. The data supporting this article's conclusions are included in the publication.