Abstract
“An impregnable stronghold where one or more warrior clans can evade enemy attacks" may serve as a description of bacterial biofilm on a smaller level than human conflicts. Consider this hypothetical conflict: who would emerge victorious? The occupants of secure trenches or those carrying out relentless assault? Either faction has the potential for triumph; the defenders will prevail if they can fortify the trench with unwavering resolve, while the assailants will succeed if they can devise innovative means to breach the trench. Hence, bacterial biofilms pose a significant challenge and are formidable adversaries for medical professionals, often leading to the failure of antibiotic treatments in numerous hospital infections. Phage engineering has become the foundation for the targeted enhancement of various phage properties, facilitating the eradication of biofilms. Researchers across the globe have studied the impact of engineered phages and phage-derived enzymes on biofilms formed by difficult-to-treat bacteria. These novel biological agents have shown promising results in addressing biofilm-related challenges. The compilation of research findings highlights the impressive capabilities of engineered phages in combating antibiotic-resistant bacteria, superbugs, and challenging infections. Specifically, these engineered phages exhibit enhanced biofilm destruction, penetration, and prevention capabilities compared to their natural counterparts. Additionally, the engineered enzymes derived from phages demonstrate improved effectiveness in addressing bacterial biofilms. As a result, these novel solutions, which demonstrate high penetration, destruction, and inhibition of biofilms, can be regarded as a viable option for addressing infectious biofilms in the near future.
Keywords: Biofilms, Engineered phages, Engineered endolysin, Superbugs, Antibiotic-resistant bacteria
1. Introduction
Antibiotic-resistant bacteria present a significant challenge in the realm of infectious diseases. These microscopic pathogens pose a formidable threat, often evading detection and causing harm without clear evidence. A key tactic employed by these pathogens is the formation of protective polysaccharide biofilm coatings, enabling them to conceal themselves, resist antibiotics, evade the host's immune response, and complicate their identification [[1], [2], [3], [4], [5]]. When bacteria reside within biofilms, their ability to resist antibiotics is heightened as these medications struggle to penetrate the protective barrier. Moreover, the exchange of resistance genes among different bacterial species serves as a fundamental mechanism driving the escalation of antibiotic resistance. The presence of biofilms and antibiotic resistance presents a formidable obstacle for healthcare providers in managing infections, often resulting in treatment challenges, extended hospital stays, elevated healthcare expenses, and heightened mortality rates among patients [[6], [7], [8], [9]]. Hence, experts across diverse disciplines have united their efforts to establish a formidable defense against the concealed threat posed by antibiotic-resistant bacteria dwelling within protective biofilm havens. This interdisciplinary collaboration has furnished the defense with cutting-edge tools to efficiently tackle biofilms and antibiotic resistance [1,10]. Phages have emerged as powerful biological tools with strong antimicrobial capabilities, demonstrating notable successes in combating antibiotic-resistant bacteria across various research environments, including laboratories, animal studies, and clinical trials. Natural phages and their enzymes can target antibiotic-resistant biofilms through three key mechanisms and can disrupt various cellular components of the planktonic, antibiotic-resistant bacteria, leading to their death (Table 1). While their effectiveness has been evident, researchers have engaged in discussions regarding the constraints associated with natural phages, including bacterial resistance and their restricted efficacy against certain biofilms [9,11,12].
Table 1.
The primary mechanisms through which natural phages and enzymes derived from them serve as the foundation for eliminating bacteria in both planktonic and biofilm forms involve.
| The mode of action of natural phage and their enzymes | |
|---|---|
| Planktonic and antibiotic-resistant bacteria | Antibiotic-resistant biofilms |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
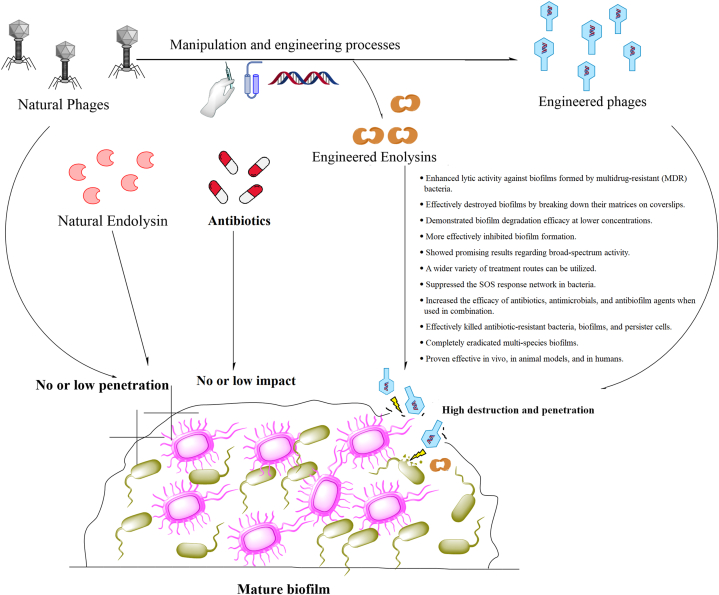
Phage therapy presents various challenges, as highlighted in multiple studies. These challenges encompass the risk of transferring antibiotic-resistance genes between bacteria, the absence of well-defined official protocols for administering phage cocktails, the potential for the patient's immune system to swiftly recognize and eliminate phages as foreign entities, the limited knowledge regarding the genomes of many phages, the rapid bacterial lysis leading to the release of endotoxins and superantigens triggering an inflammatory response, the specificity of phages to particular hosts complicating their preparation for diverse bacterial strains, the necessity for the target bacteria to be present for phage replication, the lack of widespread acceptance of phages as medicinal products, and the absence of coverage for phage therapy under public health insurance schemes [13,14]. In response to the challenges posed by phage therapy constraints, scientists have innovated novel antimicrobial agents through the modification of phages and endolysins. These tailored substances are designed to specifically target bacteria and eradicate biofilms, presenting a fresh strategy in the fight against antibiotic-resistant infections and biofilms (Fig. 1). The efficacy of engineered phages and endolysins has been evidenced in laboratory settings (in vitro) as well as in living organisms (in vivo), showcasing their impact on biofilms and antibiotic-resistant infections [13,14]. The objective of this research is to explore the clinical obstacles posed by biofilms and their interaction with the immune system, elucidate the role of engineered phages and endolysins, and assess the impact of these modified agents on biofilms formed by antibiotic-resistant bacteria.
Fig. 1.
Intricate communities of antibiotic-resistant bacteria reside within biofilms, which can be challenging to penetrate and eradicate due to their low permeability or lack of response to antibiotics, endolysins, and natural phages. To overcome these challenges, researchers have explored the engineering or targeted manipulation of phages and endolysins to enhance their abilities to penetrate and destroy biofilms, as well as eliminate antibiotic-resistant bacteria.
2. Biofilm
Bacterial biofilms are intricate and organized communities enclosed within their self-generated extracellular polymeric substances (EPSs). These biofilms comprise cells existing in diverse physiological and morphological conditions, firmly adhered to either living or non-living surfaces [1,10,15]. While the composition of adhesive fibers, proteins, nucleic acids, and exopolysaccharides in biofilms may vary, the fundamental stages of biofilm formation exhibit remarkable similarity among diverse bacteria. These stages typically encompass: A) initial reversible attachment to a surface or aggregation in a liquid environment, B) the development of microcolonies embedded in a shared extracellular polymeric substance (EPS), C) the progression and maturation of the biofilm, and D) the dispersal of planktonic cells for colonization at new locations [16]. Within bacterial biofilms, microorganisms typically constitute a mere 10 % of the dry weight, with over 90 % attributed to extracellular polymeric substances (EPSs). EPSs exhibit a hydrated, gel-like, three-dimensional structure and encompass an array of extracellular polysaccharides, proteins, lipids, nucleic acids (including extracellular DNA and RNA), and other biological molecules. These elements play a crucial role in upholding the mechanical integrity of biofilms, shielding bacteria from diverse environmental challenges like metal toxicity, UV radiation, salinity, desiccation, antimicrobial agents, and phagocytosis [17,18]. The composition and arrangement of bacterial biofilms can exhibit significant diversity influenced by factors such as the specific bacterial species, nutrient levels, and environmental surroundings. However, a prevalent feature across various bacterial biofilms is the presence of extracellular polysaccharides and proteins, serving as fundamental structural elements in these complex microbial communities [19]. Extracellular polysaccharides exhibit robust binding capabilities and complexation properties, engaging with divalent cations like calcium, magnesium, and zinc. This interaction enhances microbial adhesion to surfaces and reinforces the cohesion of biofilms [20,21]. Proteins are recognized as vital elements within biofilms, serving pivotal functions in triggering inflammation and sustaining biofilm integrity. Moreover, proteins can act as extracellular enzymes, contributing to biofilm breakdown by facilitating the liberation of bacteria residing within the biofilm and the initiation of fresh biofilm formations [22,23]. Lipids, although constituting a minor fraction within biofilms, have the capacity to bind with proteins, creating lipoproteins that are crucial for preserving cellular integrity, initiating infections, and facilitating the development of biofilms [24]. Moreover, lipids play a crucial role in conferring a vital characteristic to EPS known as hydrophobicity [19]. Extracellular DNA (eDNA) is a newly identified element that is prevalent in biofilms, potentially originating from the controlled disintegration of specific cells within the biofilm. Its purpose remained enigmatic for an extended period. Nevertheless, disrupting early-stage biofilms with DNase I results in destabilization and premature dispersal, underscoring the structural role of DNA within the three-dimensional matrix. Additionally, alternative functions have been proposed, including acting as a nutrient reservoir for neighboring colonies and functioning as a reservoir for horizontal gene transfer among the biofilm population [25,26]. The eDNA plays a significant role in the maturation of biofilms by interacting with various molecular constituents like exopolysaccharides, lipoproteins, and amyloidogenic peptides, contributing to the organization and reinforcement of biofilm structures. While eDNA's involvement is well-established, the presence of additional cellular components within biofilms suggests the need for further exploration to elucidate their contributions to the formation and progression of biofilms [19]. Each polymer constituent within the EPS is associated with specific enzymes like proteases, saccharolytic enzymes, lipases, and DNases, which play a role in their degradation. This enzymatic activity is crucial for processes like nutrient acquisition during biofilm development and restructuring, as well as for promoting biofilm dispersal and the liberation of planktonic bacteria [16]. Moreover, these enzymes are pivotal not only in the context of biofilms but also in host infection and colonization. They can effectively neutralize or break down host molecules, aiding bacteria in evading the host immune response and facilitating their dissemination. Proteases exhibit a dual role: directly influencing biofilms and playing significant functions in host infections as crucial virulence factors. Additionally, proteases are instrumental in breaking down host proteins within the extracellular matrix to impede host invasion. They also have a regular role in deactivating and targeting host proteases, immunoglobulins, and components of the complement system, ensuring prolonged undetected infection [16].
2.1. Biofilm role in antibiotics resistant
Bacteria residing in biofilms exhibit antibiotic tolerance levels that are notably higher, ranging from approximately 10 to 1000 times greater than individual sessile cells [27]. Moreover, biofilms enhance nutrient utilization, metabolic exchange, genetic material transfer, and intercellular communication. The capacity and propensity to form biofilms are recognized as pivotal virulence factors for numerous microorganisms, amplifying their pathogenicity and treatment resilience in contrast to planktonic bacteria. The development of biofilms on medical implants and devices, coupled with antibiotic resistance, poses a substantial medical challenge [16]. Alterations in the morphology of bacteria that form biofilms enhance characteristics like hydrophobicity, surface adhesion, autoaggregation, and limited motility. These changes play a crucial role in bolstering the resilience of biofilms in demanding environmental conditions [28,29]. Biofilm-related infections pose significant challenges in clinical environments, frequently necessitating the use of multiple antibiotics for treatment, thereby elevating the potential for antibiotic toxicity [30]. The utilization of combined antibiotic therapy may contribute to the development of antibiotic tolerance or resistance, complicating the treatment of infections. Moreover, the absence of established diagnostic protocols in healthcare settings can lead to delayed or inaccurate diagnoses, further challenging effective patient care [31,32]. Antibiotic tolerance in biofilm-forming bacteria arises from two key features: (A) The structural changes induced by matrix formation in the bacteria, and (B) the elevated numbers of bacterial persister cells, which are characterized by reduced metabolic activity [33]. Resistance mechanisms in bacteria, particularly in biofilm-associated and resistant strains, encompass various strategies such as efflux pumps, alterations in membrane protein expression (porins), decreased cellular membrane permeability, and limited diffusion of small inhibitory molecules. These mechanisms exhibit similarities across Gram-negative and Gram-positive bacteria, involving actions like efflux pump upregulation, enzymatic degradation of antibiotics (e.g., β-lactamase activity), and mutations in target sites. Additionally, bacteria employ diverse tactics to resist antibiotics, including covalent modification of antibiotics, hindering antibiotic permeability, shielding antibiotic targets with proteins, excessive production of penicillin-binding proteins (PBPs), and reducing the affinity for antibiotic binding [33]. Diagnosing infections caused by biofilms often involves intricate and costly processes. Antibiotic sensitivity tests typically assess bacterial susceptibility in planktonic states rather than in biofilm structures, potentially resulting in the prescription of incorrect dosages or antibiotics [32]. Creating antibiofilm medications alongside precise, swift, and cost-effective diagnostic methods could significantly enhance the treatment of biofilm-related infections. Researchers are investigating streamlined and rapid techniques to assess the vulnerability of bacterial biofilms, aiming to move beyond solely relying on the susceptibility of planktonic cells [34,35]. Pseudomonas aeruginosa, Acinetobacter baumannii, and Staphylococcus aureus are recognized as significant pathogens in healthcare environments. Their capacity to exhibit diverse virulence factors, including the formation of biofilms, plays a crucial role in the pathogenesis of the diseases they induce [36,37].
2.2. Host immune system function against biofilm
Certain components of planktonic bacteria, such as flagella and lipopolysaccharide (LPS), are identified by the innate immune system's pathogen recognition receptors (PRRs) through interactions with pathogen-associated molecular patterns (PAMPs). These PAMPs are recognized via Toll-like receptors 5 and 4, respectively [38]. Likewise, bacteria thriving within biofilms trigger the immune system via identical pathways as individual planktonic bacteria. However, biofilm-forming bacteria are enclosed by extracellular polymeric substances, which reduces the exposure of PAMPs to the immune system. Consequently, the extracellular matrix components of biofilms play a significant role in inducing an immune response for biofilm infections [[39], [40], [41]]. The innate immune response identifies PAMPs and Biofilm-Associated Molecular Patterns (BAMPs) present within the biofilm matrix. PRRs mediate the detection of BAMPs and PAMPs by PMNs and macrophages. When BAMPs and PAMPs bind to PRRs, they stimulate PMNs and macrophages, leading to the consumption of O2 for the release of tissue-toxic reactive oxygen species (ROS) and nitric oxide (NO). PMNs also secrete proteases that can cause tissue lesions, while macrophages can intensify inflammation by releasing pro-inflammatory cytokines including IL-6, IL-8, TNF-α, IL-1, and IL-12 [42]. T-cells and B-cells are effector cells of the adaptive immune response located in secondary lymphoid organs, while plasma cells, which are primarily found in the bone marrow, are another type of effector cell. The innate immune response to P. aeruginosa biofilm involves the activation of NK cells, macrophages, neutrophils, dendritic cells, and the complement system. The most convincing evidence of the function of innate immune responses in bacterial biofilm has been obtained by founding human neutrophils and macrophages to P. aeruginosa biofilms that lack planktonic bacteria [[43], [44], [45], [46]]. Similarly, research on mouse lungs exposed to biofilms has demonstrated that the innate immune response involves a significant accumulation of activated neutrophils in the airways. This phenomenon is also observed in experimentally infected chronic wounds in mice, where neutrophils accumulate early at the site of biofilm infection [[46], [47], [48]]. The activation of adaptive immune system is facilitated by dendritic cells (DC), which are necessary for sufficient adequate activation during the initial encounter with a pathogen, and by macrophages. Immature DCs in the peripheral tissue are efficient in capturing antigens and are particularly abundant in regions exposed to pathogens, such as mucosal surfaces and secondary lymphoid tissue [42]. Following antigen uptake and the impact of inflammatory cytokines, DCs mature into dedicated antigen-processing and presenting mature DCs. As a result, DCs play an essential role in connecting the innate and adaptive immune systems and have the unique ability to prime naive T-cells for subsequent Th1, Th2, or Th17 cell responses [[49], [50], [51]]. Activated T-cells can induce cytokines that further enhance inflammation by stimulating the activation and accumulation of PMNs as well as the generation of IgG. The enhanced aggregation of activated PMNs contributes to local inflammation, which is further accelerated by the attachment of antigens to IgG. This leads to the activation of the classical complement pathway and the immune complex-related induction of the PMNs [42]. An imbalanced oral microbial biofilm has been demonstrated to induce systemic inflammation, as evidenced by elevated levels of oxidative stress markers (e.g., 8-Hydroxyguanosine (8-OHdG), malondialdehyde (MDA)), prostaglandins (PG) (e.g., PGE2), acute phase proteins (e.g., C-reactive protein (CRP)), and cytokines such as interleukins (IL) (e.g., IL-1β, IL-1α, IL-6, IL-8), and tumor necrosis factors (TNF) (e.g., TNF-α) [52]. Furthermore, systemic inflammation activates neutrophils, which are part of the innate immune defense and are responsible for identifying and eliminating microorganisms, to release their enzymes, such as neutrophil elastase (NE), myeloperoxidase (MPO), and matrix metalloproteinases (MMPs) [53,54]. Consequently, microorganisms and their biofilm components in the oral cavity, along with inflammatory mediators, may spread throughout the body. They can attack the fetal-placental unit, leading to an inflammatory response, or spread to other organs, thereby enhancing systemic inflammation via acute phase protein responses, which could later affect the fetal-placental unit [55,56]. This phenomenon may lead to adverse pregnancy outcomes such as pregnancy-induced hypertension (PIH), preterm birth, low birth weight (LBW), early pregnancy loss (EPL), or preeclampsia (PE). In this context, biofilm-induced inflammatory disease in periodontitis is a polymicrobial condition and has previously been linked to adverse outcomes [[57], [58], [59]]. Therefore, the inflammatory condition induced by biofilms uncommonly involves the activation of both the adaptive and innate immune systems due to the chronic nature of biofilm-related infections. Neither of these immune responses is capable of destroying biofilms, but they do lead to extensive secondary damage [42]. Intestinal microorganisms can exist in three distinct states: fully embedded in biofilms, fully planktonic, or recently dispersed from biofilms. This latter state, characterized by a distinct phenotype, naturally occurs between biofilms and the planktonic lifestyle. Several factors, including fatty acid signaling, oxygen levels, nutrient availability, nitric oxide, iron, and proteases, can induce the dispersion of biofilm bacteria [[60], [61], [62]]. Key questions to consider when examining gut microorganisms on intestinal surfaces are whether there is a specific host response to biofilms and whether biofilm-dispersed bacteria exhibit a different phenotype compared to biofilms or purely planktonic bacteria. Although not yet investigated for gut microbiota, examples of host responses to biofilms versus planktonic cells include reduced oxidative burst and neutrophil extracellular trap responses in vitro [[63], [64], [65]]. The biofilm-dispersed phenotype has strikingly different characteristics, including increased antimicrobial resistance, iron intake capacity, and overall virulence. This observation is crucial for understanding human gut diseases associated with altered commensal biofilms. Therefore, identifying factors that induce bacterial dispersion from biofilms, whether from the host (e.g., immune cells, neuronal cells, fibroblasts, or enterocytes) or the environment (e.g., diet, pollutants, or invading pathogens), is essential. These factors can be associated with pathologies but also released in health [66]. Even in regions with poor taxonomic diversity (e.g., the stomach and upper gastrointestinal tract), biofilms can be heterogeneous, comprising cells with different phenotypes, multiple genotypic variants of a strain, or different strains of the same species. This heterogeneity, combined with the complex spatial structure of biofilms, leads to cell-cell interactions and the emergence of social behaviors such as cooperation, competition, and cheating, which are important for understanding microbiota-associated health and disease [67]. The host tissues and their mucosal biofilms have a complex symbiotic relationship, with numerous factors and pathways involved. The host can directly and persistently influence its mucosal biofilm through mechanisms such as mucin secretion, membrane vesicle release, antimicrobial peptide production, immunoglobulin secretion, hydrogen sulfide generation, and protease activity. Conversely, the mucosal biofilm can secrete various components, including proteins, polysaccharides, proteases, hydrogen sulfide, secondary metabolites, membrane vesicles, and nucleic acids, which may in turn trigger host defense mechanisms [66]. The human immune system may be unable to complete eliminate many pathogenic biofilms, and due to the high antibiotic resistance of bacteria in biofilms, new approaches such as phage therapy have been proposed. While phage therapy has demonstrated some effectiveness in eradicating biofilms, natural phages have limitations, including low penetration in biofilms [68,69]. As a result, the use of advanced methods such as engineered phages and engineered endolysins has been recommended.
3. Engineered phages and engineered endolysin
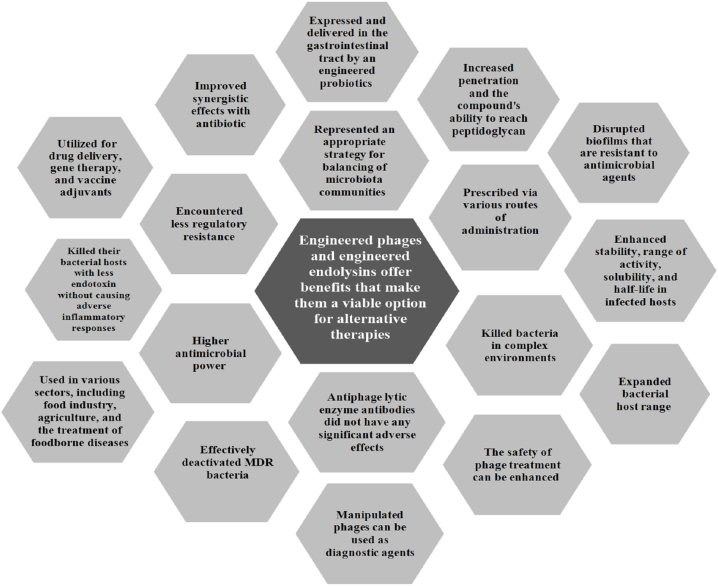
Engineered phages and modified endolysins have demonstrated the ability to address numerous challenges posed by natural phages and conventional antibiotics when combating multidrug-resistant (MDR), extensively drug-resistant (XDR), and biofilm-forming bacteria [9,12]. The development and application of modified enzymes and tailored phages necessitate interdisciplinary cooperation spanning biotechnology, microbiology, medicine, genetics, and biology. This intricate procedure encompasses genetic manipulation techniques like CRISPR-Cas-mediated genome editing, homologous recombination, traditional methods reliant on homologous recombination, yeast-mediated assembly of phage genetic material, cell-free transcription-translation systems, bacteriophage recombination of introduced DNA via electroporation, in vivo genetic recombination, and in vitro manipulation of phage genetic material. These methodologies are pivotal in the creation of genetically engineered phages and endolysins [12]. Genetic engineering has significantly improved the utilization of phages and endolysins in pathogen treatment, leading to enhanced production and organic synthesis of lysins. This advancement has proven successful across diverse fields such as the food industry, agriculture, and the management of foodborne illnesses. As a result, the European Medicine's Agency (EMA) has advocated for heightened innovation in phage-based therapies [12,70]. Synthetic biology is currently offering resources for producing, prototyping, and virtually testing innovative engineered phages. As molecular biology and sequencing technology continue to evolve, the creation of modular designer-phages is anticipated to become a standard procedure. These modular designer-phages will broaden the spectrum of innovative strategies for efficiently neutralizing MDR bacteria [71]. The detrimental effects of disrupted communities within the body's microbiota on human health are widely recognized. Conversely, engineered phages offer a suitable approach for restoring balance within microbiota communities [[72], [73], [74], [75]]. Bioengineering surpasses the constraints of natural phages in challenging conditions. Improving the safety of phage therapy involves developing phage variants devoid of lysins, the lytic enzymes, to prevent the release of bacterial toxins upon cell destruction [71]. Through immunomodulation and genetic manipulation of phages by deleting holin-endolysin system genes, it is feasible to generate nonlytic phage variations that effectively eliminate bacterial hosts with reduced endotoxin production, mitigating adverse inflammatory responses [[76], [77], [78]]. Engineered phages with an extended bacterial host range have proven the efficacy of customized phage mixtures. Notably, these modified phages are anticipated to face reduced regulatory hurdles, increasing their likelihood of acceptance for therapeutic purposes [79]. Engineered phages have the potential to serve as diagnostic tools. For instance, the incorporation of luciferases into phage genetic material enables the sensitive, quick, and direct identification of bacteria [80]. Phage bioengineering has been employed to replace tail-fiber and tail genes of phages, expanding the repertoire of encoded proteins to target a particular bacterial host at the species level. These genetic substitutions allow for the modification of phages to infect and differentiate a new bacterial host, surmounting species boundaries [81]. Reprogramming via genetic engineering can result in the creation of new phages that trigger different immune responses at the site of infection, offering potential applications in drug delivery, gene therapy, and vaccine adjuvants [[76], [77], [78]]. Engineered phages have the capability to dismantle biofilms that exhibit resistance to antimicrobial agents [12]. While certain phages may not naturally produce depolymerase, the creation of depolymerase-producing phages is feasible through genetic manipulation. These modified phages, with the ability to break down biofilms and capsule polysaccharides, have demonstrated effectiveness in combating bacterial biofilms [82,83]. To be efficacious in medical settings, an engineered endolysin must possess the capability to eliminate bacteria in intricate surroundings like mucosal membranes, animal tissue, bodily fluids, and blood. Beyond enhancing its bactericidal efficacy, modifications to phage lytic enzymes can boost various characteristics such as stability, activity range, solubility, and persistence within infected hosts. While phage lytic enzymes are anticipated to prompt an immune reaction due to their protein nature and bacterial lysis induction, it is noteworthy that antibodies against these enzymes did not yield notable adverse effects [84]. These modified lytic enzymes can be administered through different methods to address infections, including oral, intratracheal, intramammary, intramuscular, intravenous, intranasal, topical, subcutaneous, and intraperitoneal routes [9]. Different modified enzymes and endolysins have undergone examination for their improved functional characteristics. The enhancement of endolysin or enzyme properties has been achieved through chemical alterations, involving fusion with diverse substances like silver nanoparticles, AIEgens, pheophorbide A, cellulose membrane, and indium tin oxide. These innovative compounds have demonstrated effectiveness in fighting infections and exhibit diverse mechanisms to combat bacteria, posing challenges for the development of resistance mechanisms [85]. For example, the fusion of phage T4 lysozyme with pesticin led to enhanced penetration and improved capability of the compound to access peptidoglycan [86]. A new endolysin, CP25L, has been effectively produced and transported into the gastrointestinal tract using a genetically modified probiotic strain [87]. Moreover, Artilysin®, a fusion of an additional endolysin with a peptide, has exhibited increased antimicrobial potency and enhanced penetration abilities [88]. Two original lysins have been merged to produce chimeolysins ClyR and ClyF, which demonstrate resistance to heat-inactivated human serum, mouse serum, human serum, and rabbit serum. Additionally, they display strong lytic activity and an expanded spectrum of streptococcal hosts [[89], [90], [91]]. PM-477, a compound created via domain shuffling, exhibits heightened bactericidal efficacy and biofilm removal capabilities while preserving the vaginal microbiome unaffected [92]. A new compound, LNT113, was developed through the fusion of cecropin A with EC340 endolysin, leading to enhanced outer membrane permeability, heightened activity, and synergistic interactions with antibiotics [93]. The advantages of engineered phages and enzymes highlighted in this section indicate that these antimicrobial agents can function as practical substitutes for conventional antibiotic therapies. Furthermore, they present encouraging remedies for addressing extensively antibiotic-resistant bacteria and biofilms, which natural phages and antibiotics struggle to combat (Fig. 2).
Fig. 2.
The advantages of engineered phages and engineered endolysins have been highlighted, demonstrating how they overcome the limitations of natural phages through genetic and non-genetic manipulations.
4. The effect of engineered phages and phage-derived enzymes to destroy biofilms
In a research study, scientists developed a hybrid endolysin by replacing an enzymatically active domain endolysin with a cell wall-binding endolysin. ClyC, a novel chimeric endolysin, exhibited enhanced lytic activity against MDR S. aureus compared to its original endolysin form. Additionally, ClyC effectively destroyed biofilms of clinical MDR bacteria, including methicillin-resistant Staphylococcus epidermidis, and methicillin-resistant S. aureus. ClyC demonstrated a 2-fold increase in biofilm degradation efficacy at a low concentration (50 nM), and it exhibited more than 4-fold enhanced efficacy at 100 nM. It also demonstrated effectiveness against methicillin-resistant S. aureus without inducing any harmful effects in an in vivo animal model [94]. In a 2022 study, researchers created an engineered form of endolysin by combining cationic carbosilane (CBS) dendrimers with an endolysin (KP27). They investigated the antimicrobial function of this combination for killing P. aeruginosa infections in both biofilm and planktonic forms. The protective outer membrane shield of Gram-negative bacteria can lead to resistance to endolysin alone, but the recombinant KP27-CBS dendrimer combination exhibits a synergistic effect that allows this combined form to penetrate the membrane by destroying the peptidoglycan layer [95]. Son et al. developed a novel chimeric endolysin, Lys109, by swapping engineered endolysins from a random domain of four endolysins isolated from S. aureus phages. Lys109 demonstrated remarkable staphylolytic activity against staphylococcal biofilms and planktonic cells. Compared to its parental endolysins, Lys109 exhibited over a 3-fold enhanced efficacy in removing biofilms, leading to improved removal of S. aureus from steel and milk surfaces [96]. A research group developed an engineered phage (ɸlexA3) that targeted overexpressed proteins, attack gene networks, and non-SOS gene networks. The phage remarkably enhanced the survival of infected mice with E. coli by suppressing the SOS network in the bacterium and increasing quinolone-induced killing. Additionally, ɸlexA3 demonstrated killing functionality against antibiotic-resistant bacteria, biofilms, and persister cells. The phage not only significantly reduced the number of antibiotic-resistant bacteria but also had an excellent adjuvant role for aminoglycosides and β-lactams [97]. Three new artilysins, namely AL-3AA, AL-9AA, and AL-15AA, have been developed by fusing the antimicrobial peptide SMAP29 at the N-terminus of LysPA26. The findings demonstrate that AL-3AA exhibits high bactericidal activity, with a mere 0.05 mg/mL of AL-3AA capable of reducing P. aeruginosa by 5.81 log units. This artilysin rapidly and dose-dependently eradicates P. aeruginosa through cell lysis. Additionally, AL-3AA hinders P. aeruginosa biofilm formation and significantly reduces mature P. aeruginosa biofilm. The new artilysins have demonstrated promising results in terms of broad-spectrum activity against various susceptible Gram-negative bacteria found in hospitals, such as K. pneumoniae and E. coli [98]. The C-terminal amino acids of phage lysin PlyF307, referred to as P307, demonstrated the ability to kill MDR A. baumannii. In comparison to their parental forms, more than three logs of endolysins showed an increased lytic effect against MDR S. aureus. Both P307 and the engineered form P307SQ-8C displayed high in vitro activity against A. baumannii biofilms. Biofilms were exposed to 250 μg/mL of P307 or P307SQ-8C for 2 or 24 h. After 2 h, approximately 3- and 4-log reductions in CFU/ml were observed with P307 and P307SQ-8C, respectively. After 24 h, P307 resulted in an additional approximately 1.3-log reduction, while no further decrease was observed with P307SQ-8C. Furthermore, these peptides disrupted the bacterial membrane without affecting human red blood cells or B cells and reduced the MDR A. baumannii burden in mouse skin infections [99]. The endolysin of phage vB_AbaP_D2 was applied in an attempt to kill the biofilm of antibiotic-resistant bacteria. An engineered endolysin, namely Abtn-4, with an amphipathic helix, represented killing function against MDR infection caused by Gram-negative bacteria. On one side, Abtn-4 (5 μM) destroyed A. baumannii in 2 h. On the other side, it showed broad killing properties against Gram-negative and Gram-positive bacteria, including K. pneumoniae, S. aureus, P. aeruginosa, Enterococcus, and Salmonella. In addition to Abtn-4's potential to decrease biofilm formation, it also demonstrated a bactericidal role in coping with phage-resistant bacterial mutants. The biofilm matrices on coverslips were broken down, and only a few bacteria remained in the Abtn-4 treatment groups [100]. The PlyF307 lysin demonstrated the highest activity among 21 lysins with varying activities, effectively eliminating all tested MDR A. baumannii isolates (>5 log-unit reduction). In addition to eradicating lethal A. baumannii bacteremia in mice, PlyF307 significantly decreased both planktonic and biofilm A. baumannii in vitro and in vivo. Biofilms were established on catheter sections by incubating them with A. baumannii for 3 days in vitro, followed by treatment with PlyF307 after washing. After 2 h, the remaining biofilm bacteria were removed and counted in buffer. Scanning electron microscopy confirmed a significant reduction in total biofilm biomass on the catheters [101]. In a subsequent investigation, the impact of treating S. pneumoniae P046 24-h biofilms with the engineered endolysin Cpl-711 at a concentration of 1 μg/mL for 2 h was examined. The results showed that this treatment effectively killed approximately 4 logs of the bacterial population. Additionally, Cpl-1 and Cpl-7 (parental phage lysozymes) reduced biofilm cell counts by around 1.5 logarithmic units. Mice were administered an intraperitoneal injection of Cpl-711 1 h after being infected with pneumococcal strain. This treatment resulted in approximately 50 % greater protection compared to the use of Cpl-1 alone [102]. The engineered chimeric lysin Cpl-711 is notable for its powerful bactericidal effects. There is evidence demonstrating the synergistic collaboration of the catalytically diverse lysins Cpl-711 and PL3 in various tests, such as purified cell wall enzymatic degradation, in vitro bacterial cell growth inhibition, and the eradication of both planktonic and biofilm-cultivated cells. The synergy between Cpl-711 and PL3 has been shown to decrease the amount of enzyme required to inhibit growth in checkerboard assays, while also significantly increasing the bactericidal effect by ≥ 2 logs compared to the sum of the activities of individual Cpl-711 or PL3 treatments. On planktonic cells of S. pneumoniae, the combination of 0.5 × MIC of each enzyme causes a 2.4-log increase in killing activity, while on biofilm cells, the synergistic effect increases by as much as 3.6 logs with respect to the sum of the activities of the individual Cpl-711 or PL3 treatments [103]. Arroyo-Moreno et al. demonstrated the effectiveness of CHAPk, a truncated derivative of staphylococcal phage K endolysin (LysK), in inhibiting and disrupting MRSA biofilms. In this study, they combined a 165-amino acid fragment containing CHAPk with a 136-amino acid fragment containing the cell-binding domain of the bacteriocin lysostaphin to create a chimeric enzyme designated CHAPk-SH3blys. For biofilm prevention, a concentration of 1 μg/mL of the chimeric enzyme proved effective, while CHAPk required 125 μg/mL for the same purpose. Furthermore, the chimeric enzyme exhibited complete biofilm disruption when 5 μg/mL was employed in 4-h assays, whereas CHAPk could only partially disrupt the biofilms at this concentration [104]. PM-477 is a novel investigational candidate that is a genetically engineered endolysin with specificity for bacteria of the genus Gardnerella. The minimum inhibitory concentration (MIC) for PM-477 was determined, and it was found that all Gardnerella isolates were highly susceptible to the endolysin. There was no evidence of cross-resistance formation, as metronidazole-resistant Gardnerella strains remained highly susceptible to PM-477, both in suspension and in preformed biofilms. Strains resistant to metronidazole in suspension were also tolerant to metronidazole at 2048 mg/mL when growing as biofilms. All strains were susceptible to PM-477 when grown as preformed biofilms, at minimum biofilm eradication concentrations in the range of 1–4 mg/mL [105]. The investigation explored the effectiveness of a genetically engineered phage, PM-477, in eliminating dual-species biofilms consisting of Gardnerella vaginalis and Fannyhessea vaginae or G. vaginalis and Prevotella bivia. The study found that PM-477 was highly effective in disrupting and reducing the culturability of G. vaginalis biofilms. Although PM-477 demonstrated lower efficiency in dual-species biofilms, it still managed to selectively and significantly eliminate G. vaginalis. These promising results aim to completely eradicate multi-species biofilms associated with bacterial vaginosis [106]. The bacteriophage BG3P encodes a glycosyl-hydrolase domain (lysozyme) and a SH3 domain in its orf28, which produces the endolysin LysCP28. The endolysin LysCP28 (38.8 kDa) exhibits broad lytic activity against a significant percentage (80.21 %) of Clostridium perfringens strains, encompassing types A, B, C, and D, sourced from diverse origins. LysCP28 (18.7 μg/mL) was effective in removing biofilms and inhibiting their formation. To test LysCP28's efficacy in a food matrix, duck meat contaminated with C. perfringens was treated with endolysin (100 μg/mL and 50 μg/mL), which reduced viable bacteria by 3.2 and 3.08 units-log, respectively, in 48 h at 4 °C [107]. A phage qdsa002 with high virulence against MRSA was discovered, and its endolysin (Lys84) and domains were expressed and purified. Lys84 consists of two enzymatic domains, CHAP and Amidase_2, along with a cell-binding domain, SH3b. This endolysin has a potent lytic activity against S. aureus and a broader bactericidal spectrum than qdsa002. Additionally, Lys84 effectively removed around 90 % of S. aureus biofilms at concentrations exceeding 10 μM. The CHAP and Amidase_2 domains retained 61.20 % and 59.46 % of the lytic activity and 84.31 % and 70.11 % of the anti-biofilm activity of Lys84, respectively. CHAP and Amidase_2 The combination of CHAP and Amidase_2 domains exhibited lytic and anti-biofilm activities close to 90 % of those of Lys84 [108]. Landlinger et al. produced several modified endolysins through domain shuffling, which exhibited a tenfold increase in bactericidal activity compared to any natural enzyme. When tested on Gardnerella strains, the most powerful endolysin, referred to as PM-477, exhibited minimum inhibitory concentrations spanning from 0.13 to 8 μg/mL. PM-477 had no impact on beneficial lactobacilli or other types of vaginal bacteria. It eradicated the Gardnerella bacteria and disintegrated the biofilms without affecting the rest of the vaginal microbiome. The exceptional selectivity and efficacy in eradicating Gardnerella, both in isolated strain cultures and in naturally occurring polymicrobial biofilms from clinical samples, position PM-477 as a promising substitute for antibiotics in the treatment of bacterial vaginosis, particularly in patients prone to frequent recurrence [92]. In 2023, two modified enzybiotics were assessed either alone or in combination against a dual biofilm created by S. aureus and E. faecalis on an inert glass surface. An engineered lysin BP404 and a hybrid protein P16-17/100 were produced by fusing 141 amino acids (catalytic domain) from the endolysin P16 of the bacteriophage ϕ44AHJD with a 100-amino-acid cell wall binding domain from the minor tail protein P17 of phage ϕ44AHJD. The combination of proteins exhibited an additive effect in rapidly disrupting the preformed dual biofilm, compared to monotherapy. The biofilms treated with the cocktail were dispersed by over 90 % within 3 h of treatment. In addition to biofilm disruption, bacterial cells embedded in the biofilm matrix were also significantly reduced by more than 90 % within 3 h of treatment [109]. The engineered phage-derived enzybiotic Vplys60, expressed in Pichia pastoris X33 as a whole cell biocatalyst, was evaluated for its efficacy in controlling Vibrio parahaemolyticus in aquaculture. Vplys60 exhibited stable activity across a wide range of pH (6–10), temperatures (37–75 °C), and salinity (100–600 mM NaCl). Additionally, Vplys60 demonstrated robust amidase activity by cleaving the peptidoglycan of V. parahaemolyticus. The data also indicated that Vplys60 (75 μg/mL) significantly suppressed biofilm formation (91.6 %) and reduced the bacterial population [110]. The genetically modified phage phagefEf11/fFL1C(D36)PnisA was evaluated to disrupt biofilms of two Enterococcus faecalis strains: JH2-2 (vancomycin-sensitive) and V583 (vancomycin-resistant). The results demonstrated a 10-100-fold decrease in viable cells (CFU/biofilm) after phage treatment, and the biomass of both vancomycin-sensitive and vancomycin-resistant E. faecalis biofilms was significantly reduced following infection by bacteriophage fEf11/fFL1C(D36)PnisA [111]. In 2023, an engineered endolysin, NC5, was developed with in vitro intracellular activity against streptococcal pathogens causing bovine mastitis. A leading candidate was identified, demonstrating a 4.0 log reduction in Streptococcus uberis and exhibiting similar efficacy against Streptococcus agalactiae and Streptococcus dysgalactiae. This top candidate eliminated S. uberis biofilm and displayed intracellular activity in two bovine mammary epithelial cell lines, as confirmed by confocal microscopy. Furthermore, it was observed that this top candidate engineered endolysin had a potentiating effect on cloxacillin, a beta-lactam penicillin used for intramammary treatment of bovine Gram-positive mastitis, in raw cow's milk from (sub)clinically infected udders [112]. Because biofilms are a significant concern in food safety, agriculture, and medicine, the study measured the degrading efficacy of engineered phages and engineered endolysins against a comprehensive panel of clinical isolates (Table 2). As a result of these studies, engineered phages and engineered endolysins have demonstrated improved antimicrobial properties, increased penetration of cell walls, and the ability to destroy biofilms.
Table 2.
A summary of studies using engineered phages and engineered endolysins against biofilms of antibiotic-resistant bacteria and other challenging bacteria.
| Authors | Year | Name of engineered phage or enzymes | Targeted bacterial biofilm | Summary of result | Reference |
|---|---|---|---|---|---|
| Lu et al. | 2009 | ɸlexA3 | E. coli | ɸlexA3 demonstrated killing functionality against antibiotic-resistant bacteria, biofilms, and persister cells. | [97] |
| Díez-Martínez et al. | 2015 | Cpl-711 | S. pneumoniae | The results showed that this treatment effectively killed approximately 4 logs of the bacterial population. | [102] |
| Thandar et al. | 2016 | P307SQ-8C | MDR S. aureus and A. baumannii | After 2 h, approximately 3- and 4-log reductions in CFU/ml were observed with P307 and P307SQ-8C, respectively. After 24 h, P307 resulted in an additional approximately 1.3-log reduction, while no further decrease was observed with P307SQ-8C. | [99] |
| Tinoco et al. | 2016 | fEf11/fFL1C(D36)PnisA | vancomycin-sensitive and vancomycin-resistant E. faecalis | The biomass of biofilms was significantly reduced following infection by bacteriophage. | [111] |
| Vázquez et al. | 2019 | Cpl-711 | S. pneumoniae | On planktonic cells of S. pneumoniae, the combination of 0.5 × MIC of each enzyme causes a 2.4-log increase in killing activity, while on biofilm cells, the synergistic effect increases by as much as 3.6 logs. | [103] |
| Srinivasan et al. | 2020 | Vplys60 | V. parahaemolyticus | Vplys60 demonstrated robust amidase activity by cleaving the peptidoglycan. The data also indicated that Vplys60 (75 μg/mL) significantly suppressed biofilm formation (91.6 %) and reduced the bacterial population. | [110] |
| Son et al. | 2021 | Lys109 | S. aureus | Lys109 exhibited over a 3-fold enhanced efficacy in removing biofilms. | [96] |
| Yuan et al. | 2021 | Abtn-4 | A. baumannii, K. pneumoniae, S. aureus, P. aeruginosa, Enterococcus, and Salmonella | The biofilm matrices on coverslips were broken down, and only a few bacteria remained in the Abtn-4 treatment groups. | [100] |
| Lee | 2021 | ClyC | MDR methicillin-resistant Staphylococcus epidermidis, and methicillin-resistant S. aureus | ClyC effectively destroyed biofilms of clinical MDR bacteria and demonstrated a 2-fold increase in biofilm degradation efficacy at a low concentration (50 nM), and it exhibited more than 4-fold enhanced efficacy at 100 nM. | [94] |
| Wang et al. | 2021 | AL-3AA, AL-9AA, and AL-15AA | P. aeruginosa, K. pneumoniae, and E. coli | Additionally, AL-3AA hinders P. aeruginosa biofilm formation and significantly reduces mature P. aeruginosa biofilm. | [98] |
| Landlinger et al. | 2021 | PM-477 | Gardnerella | PM-477 eradicated the Gardnerella bacteria and disintegrated the biofilms without affecting the rest of the vaginal microbiome. | [92] |
| Arroyo-Moreno et al. | 2021 | CHAPk-SH3blys | MRSA | In terms of biofilm prevention, a concentration of 1 μg/mL of the chimeric enzyme was sufficient, whereas for CHAPk, 125 μg/mL was needed. | [104] |
| Ning et al. | 2021 | CHAP and Amidase_2 | MRSA | The combination of CHAP and Amidase_2 domains exhibited lytic and anti-biofilm activities close to 90 % of those of Lys84. | [108] |
| Quintana-Sanchez et al. | 2022 | KP27-CBS | P. aeruginosa | KP27-CBS dendrimer combination exhibits a synergistic effect against biofilm that allows this combined form to penetrate the membrane by destroying the peptidoglycan layer. | [95] |
| Castro et al. | 2022 | PM-477 | G. vaginalis, F. vaginae, and P. bivia | Although PM-477 demonstrated lower efficiency in dual-species biofilms, it still managed to selectively and significantly eliminate G. vaginalis. | [106] |
| Landlinger et al. | 2022 | PM-477 | Gardnerella | All strains were susceptible to PM-477 when grown as preformed biofilms, at minimum biofilm eradication concentrations in the range of 1–4 mg/mL. | [105] |
| Lu et al. | 2023 | LysCP28 | C. perfringens | LysCP28 (18.7 μg/mL) was effective in removing biofilms and inhibiting their formation. | [107] |
| Manoharadas et al. | 2023 | BP404 and P16-17/100 | S. aureus and E. faecalis | In addition to biofilm disruption, bacterial cells embedded in the biofilm matrix were also significantly reduced by more than 90 % within 3 h of treatment. | [109] |
| Vander Elst et al. | 2023 | NC5 | Streptococcus uberis, Streptococcus agalactiae, and Streptococcus dysgalactiae | This top candidate eliminated S. uberis biofilm and displayed intracellular activity in two bovine mammary epithelial cell lines, as confirmed by confocal microscopy. | [112] |
5. Perspectives and future of phage and engineered phages therapy
Therapy based on engineered phages and enzymes, acting as living therapeutic agents, offers a promising approach in modern medicine. This alternative method provides a way to combat resistant and biofilm-producing pathogens. However, before it can be adopted as a routine treatment globally, extensive scientific and clinical research should be conducted. Additionally, the misconception of phages as potential causes of human infections must be addressed and dispelled. Implementing this treatment on a global scale could have significant benefits for improving the health status of millions of people infected with MDR, XDR, and pan-drug resistance (PDR) bacteria each year [13,113]. Zoonotic pathogens that enter the human food chain pose a significant challenge for phage therapy, representing a major medical problem worldwide. The increasing demand for meat necessitates the development of alternative methods to prevent and treat bacterial infections, which are often resistant to multiple antibiotics. Genetic engineering has significantly enhanced the use of phages in combating zoonotic and foodborne pathogens by boosting the production of lysins. This breakthrough has led to the widespread adoption of phage therapy in agriculture, the food industry, and the management of foodborne illnesses. In response to these advancements, the European Medicines Agency (EMA) has encouraged further innovation in bacteriophage-based treatments. Unfortunately, in the EU, there is no legal framework for introducing phages into water or feed, for example, as part of washing procedures for farm animals or in modified packaging in slaughterhouses. Regulations limiting the release of phages in the environment should also be considered. Therefore, it is crucial to design phage interventions and engineered phages in the future with all safety standards to prevent the release of phages outside livestock farms [70,114]. The therapeutic use of engineered phages and enzymes involves identifying and cultivating the patient's clinical bacterial isolate responsible for a specific infection and then testing their sensitivity to engineered phages and enzymes. This knowledge requires interdisciplinary collaboration among doctors, microbiologists, geneticists, and biotechnologists, as several phages targeting a specific strain of bacteria must be engineered to increase their effect on a wide range of bacterial hosts. Treatment by engineered phages and enzymes requires the existence of a large bank containing these biologically produced compounds, which can be prepared in the form of a medicinal product intended for a specific patient [115]. Due to the continuous growth of bacterial resistance to antibiotics, there is a real need to continue the search for engineered phages and enzymes and their adaptation as potential therapeutic agents. However, the commercialization of engineered phage products and engineered enzymes by pharmaceutical companies is not high due to the high financial costs involved. Additionally, there are problems with patenting phage-based products. The lack of regulatory approval for such a large-scale treatment (e.g., FDA) is also a significant problem, close to the absence of flexible approval mechanisms. Therefore, more efforts are needed to overcome regulatory hurdles and achieve therapeutic guidelines based on engineered phages and enzymes [115,116]. In the future, significant technological progress may solve the problem of patenting phage preparations, which can increase the specificity and availability of phage therapy. Phages can also be modified using genome editing techniques (such as sequencing, CRISPR/Cas-based phage engineering, homologous recombination, assembly of phage genomic DNA) to obtain preparations capable of killing only antibiotic-resistant bacteria without affecting the commensal microflora. Such unique phages or phage cocktails can be easily patented and commercialized. The need for available phage products can be met by phage libraries created by various research teams and phage therapy centers around the world. The production of safe engineered phage drugs and engineered enzymes, whether on a small scale for personal therapy or on a larger scale, requires valid quality control measures. In any case, sterility, stability, and the absence of endo- and exotoxin or other harmful impurities in the therapeutic preparation should be considered [116]. Regardless of the fate of engineered phages and enzymes as living medicines in the future, most researchers and experienced experts believe that this form of therapy will never replace antibiotics. A therapeutic strategy based on engineered phages and enzymes can be used in combination with antibiotics or as a last line of defense for patients suffering from infections that have not been effectively treated by any other available treatment. Due to the rapid increase in the number of life-threatening MDR, XDR, and PDR infections in recent years, there is a need to review the potential role of engineered phages and enzymes and other alternative therapies to antibiotics in combating these strains. There is resistance to antibiotics. Therapy based on engineered phages and enzymes certainly deserves further investigation in the clinic [113,115,116].
6. Conclusion
Our advanced research on the impact of engineered phages on biofilms has demonstrated that these enhanced antimicrobial compounds possess a strong ability to destroy and inhibit biofilms in various bacteria, including antibiotic-resistant bacteria and superbugs. Although these studies are still in their early stages, it is evident that these enhanced antimicrobial compounds hold potential for further investigation towards commercialization and clinical application. Consequently, we propose to conduct additional research on engineered phages and engineered endolysins in laboratory animals to better understand their effects on bacterial biofilms. Uncovering the precise mechanisms of action of these enhanced compounds on biofilms will bring us one step closer to the clinical application of these compounds. Because engineered phages and engineered endolysins have demonstrated the ability to not only overcome the limitations of natural phages but also serve as a viable alternative to traditional antibiotics.
Funding
No financial support was provided relevant to this article.
Data availability statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
CRediT authorship contribution statement
Fatemeh Eghbalpoor: Writing – original draft. Mahdieh Gorji: Writing – original draft. Maryam Zamani Alavigeh: Writing – original draft. Majid Taati Moghadam: Writing – review & editing, Writing – original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
None.
References
- 1.Moghadam M.T., Chegini Z., Khoshbayan A., Farahani I., Shariati A. Helicobacter pylori biofilm and new strategies to combat it. Curr. Mol. Med. 2021;21(7):549–561. doi: 10.2174/1566524020666201203165649. [DOI] [PubMed] [Google Scholar]
- 2.Moradi M., Norouzi A. Prevalence of bla-CTX-M, bla-SHV, and bla-TEM genes and comparison of antibiotic resistance pattern in extended-spectrum β-lactamase producing and non-producing groups of Klebsiella pneumoniae isolated from clinical samples in kerman hospitals. Journal of Advanced Biomedical Sciences. 2016;6(1):120–128. [Google Scholar]
- 3.Mohebi S., Hossieni Nave H., Norouzi A., Kandehkar Gharaman M., Taati Moghadam M. Detection of extended spectrum beta lactamases on class I integron in Escherichia coli isolated from clinical samples. Journal of Mazandaran University of Medical Sciences. 2016;26(138):66–76. [Google Scholar]
- 4.Taati Moghadam M., Hossieni Nave H., Mohebi S., Norouzi A. The evaluation of connection between integrons class I and II and ESBL-producing and Non-ESBL klebsiella pneumoniae isolated from clinical samples, Kerman. Iran. J. Med. Microbiol. 2016;10(4):1–9. [Google Scholar]
- 5.Khoshbayan A., Golmoradi Zadeh R., Taati Moghadam M., Mirkalantari S., Darbandi A. Molecular determination of O25b/ST131 clone type among extended spectrum β-lactamases production Escherichia coli recovering from urinary tract infection isolates. Ann. Clin. Microbiol. Antimicrob. 2022;21(1):35. doi: 10.1186/s12941-022-00526-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asgharzadeh S., Golmoradi Zadeh R., Taati Moghadam M., Farahani Eraghiye H., Sadeghi Kalani B., Masjedian Jazi F., Mirkalantari S. Distribution and expression of virulence genes (hlyA, sat) and genotyping of Escherichia coli O25b/ST131 by multi-locus variable number tandem repeat analysis in Tehran, Iran. Acta Microbiol. Immunol. Hung. 2022;69(4):314–322. doi: 10.1556/030.2022.01826. [DOI] [PubMed] [Google Scholar]
- 7.Hosseini M., Ahmed Hamad M., Mohseni G., Salamy S., Dehghan Tarzjani S., Taati Moghadam M. Prediction of tsunami of resistance to some antibiotics is not far‐fetched which used during COVID‐19 pandemic. J. Clin. Lab. Anal. 2023;37(15–16) doi: 10.1002/jcla.24959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hosseininasab S.S., Gorji M., Hosseini S.S., Moghadam M.T. The use of last-line antibiotics for the treatment of COVID-19 is a risk to disarm humanity against future antibiotic-resistant infectious diseases: suggestions for prevention. Infect. Dis. Clin. Pract. 2023;31(4):1–2. [Google Scholar]
- 9.Hassannia M., Naderifar M., Salamy S., Akbarizadeh M.R., Mohebi S., Moghadam M.T. Engineered phage enzymes against drug-resistant pathogens: a review on advances and applications. Bioproc. Biosyst. Eng. 2023:1–12. doi: 10.1007/s00449-023-02938-6. [DOI] [PubMed] [Google Scholar]
- 10.Moghadam M.T., Chegini Z., Norouzi A., Dousari A.S., Shariati A. Three-decade failure to the eradication of refractory Helicobacter pylori infection and recent efforts to eradicate the infection. Curr. Pharmaceut. Biotechnol. 2021;22(7):945–959. doi: 10.2174/1389201021666200807110849. [DOI] [PubMed] [Google Scholar]
- 11.Taati Moghadam M., Khoshbayan A., Chegini Z., Farahani I., Shariati A. Bacteriophages, a new therapeutic solution for inhibiting multidrug-resistant bacteria causing wound infection: lesson from animal models and clinical trials. Drug Des. Dev. Ther. 2020:1867–1883. doi: 10.2147/DDDT.S251171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boroujeni M.B., Mohebi S., Malekian A., Shahraeini S.S., Gharagheizi Z., Shahkolahi S., et al. The therapeutic effect of engineered phage, derived protein and enzymes against superbug bacteria. Biotechnol. Bioeng. 2024;121(1):82–99. doi: 10.1002/bit.28581. [DOI] [PubMed] [Google Scholar]
- 13.Taati Moghadam M., Amirmozafari N., Shariati A., Hallajzadeh M., Mirkalantari S., Khoshbayan A., Masjedian Jazi F. How phages overcome the challenges of drug resistant bacteria in clinical infections. Infect. Drug Resist. 2020:45–61. doi: 10.2147/IDR.S234353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J., Du F., Long M., Li P. Limitations of phage therapy and corresponding optimization strategies: a review. Molecules. 2022;27(6):1857. doi: 10.3390/molecules27061857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chegini Z., Khoshbayan A., Taati Moghadam M., Farahani I., Jazireian P., Shariati A. Bacteriophage therapy against Pseudomonas aeruginosa biofilms: a review. Ann. Clin. Microbiol. Antimicrob. 2020;19:1–17. doi: 10.1186/s12941-020-00389-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramírez-Larrota J.S., Eckhard U. An introduction to bacterial biofilms and their proteases, and their roles in host infection and immune evasion. Biomolecules. 2022;12(2):306. doi: 10.3390/biom12020306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S., Lu H., Zhang S., Shi Y., Chen Q. Phages against pathogenic bacterial biofilms and biofilm-based infections: a review. Pharmaceutics. 2022;14(2):427. doi: 10.3390/pharmaceutics14020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.González J.F., Hahn M.M., Gunn J.S. Chronic biofilm-based infections: skewing of the immune response. Pathogens and disease. 2018;76(3) doi: 10.1093/femspd/fty023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siddik A., Satheesh S. Characterization and assessment of barnacle larval settlement-inducing activity of extracellular polymeric substances isolated from marine biofilm bacteria. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-54294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Low K.E., Howell P.L. Gram-negative synthase-dependent exopolysaccharide biosynthetic machines. Curr. Opin. Struct. Biol. 2018;53:32–44. doi: 10.1016/j.sbi.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Steiger E.L., Muelli J.R., Braissant O., Waltimo T., Astasov-Frauenhoffer M. Effect of divalent ions on cariogenic biofilm formation. BMC Microbiol. 2020;20(1):1–11. doi: 10.1186/s12866-020-01973-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devaraj A., Novotny L.A., Robledo-Avila F.H., Buzzo J.R., Mashburn-Warren L., Jurcisek J.A., et al. The extracellular innate-immune effector HMGB1 limits pathogenic bacterial biofilm proliferation. J. Clin. Investig. 2021;131(16) doi: 10.1172/JCI140527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paharik A.E., Horswill A.R. The staphylococcal biofilm: adhesins, regulation, and host response. Virulence mechanisms of bacterial pathogens. 2016:529–566. doi: 10.1128/microbiolspec.VMBF-0022-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaplan E., Greene N.P., Crow A., Koronakis V. Insights into bacterial lipoprotein trafficking from a structure of LolA bound to the LolC periplasmic domain. Proc. Natl. Acad. Sci. USA. 2018;115(31):E7389–E7397. doi: 10.1073/pnas.1806822115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arciola C.R., Visai L., Ravaioli S., Cangini I., Campoccia D., Montanaro L. Extracellular DNA in biofilms. Int. J. Artif. Organs. 2011;34(9):824–831. doi: 10.5301/ijao.5000051. [DOI] [PubMed] [Google Scholar]
- 26.Whitchurch C.B., Tolker-Nielsen T., Ragas P.C., Mattick J.S. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295(5559):1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 27.Uruén C., Chopo-Escuin G., Tommassen J., Mainar-Jaime R., Arenas J. Biofilms as promoters of bacterial antibiotic resistance and tolerance. Antibiotics. 2021;10:3. doi: 10.3390/antibiotics10010003. New Insights on Biofilm Antimicrobial Strategies. 2021:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Toole G., Kaplan H.B., Kolter R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000;54(1):49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 29.Banerjee D., Shivapriya P., Gautam P.K., Misra K., Sahoo A.K., Samanta S.K. A review on basic biology of bacterial biofilm infections and their treatments by nanotechnology-based approaches. Proc. Natl. Acad. Sci. India B Biol. Sci. 2020;90:243–259. [Google Scholar]
- 30.Lin Y.-K., Yang S.-C., Hsu C.-Y., Sung J.-T., Fang J.-Y. The antibiofilm nanosystems for improved infection inhibition of microbes in skin. Molecules. 2021;26(21):6392. doi: 10.3390/molecules26216392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh A., Amod A., Pandey P., Bose P., Pingali M.S., Shivalkar S., et al. Bacterial biofilm infections, their resistance to antibiotics therapy and current treatment strategies. Biomedical Materials. 2022;17(2) doi: 10.1088/1748-605X/ac50f6. [DOI] [PubMed] [Google Scholar]
- 32.Silva N., Marques L., Röder D. Diagnosis of biofilm infections: current methods used, challenges and perspectives for the future. J. Appl. Microbiol. 2021;131(5):2148–2160. doi: 10.1111/jam.15049. [DOI] [PubMed] [Google Scholar]
- 33.Mohamad F., Alzahrani R.R., Alsaadi A., Alrfaei B.M., Yassin A.E.B., Alkhulaifi M.M., Halwani M. An explorative review on advanced approaches to overcome bacterial resistance by curbing bacterial biofilm formation. Infect. Drug Resist. 2023:19–49. doi: 10.2147/IDR.S380883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wannigama D.L., Hurst C., Pearson L., Saethang T., Singkham-In U., Luk-In S., et al. Simple fluorometric-based assay of antibiotic effectiveness for Acinetobacter baumannii biofilms. Sci. Rep. 2019;9(1):6300. doi: 10.1038/s41598-019-42353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wannigama D.L., Hurst C., Hongsing P., Pearson L., Saethang T., Chantaravisoot N., et al. A rapid and simple method for routine determination of antibiotic sensitivity to biofilm populations of Pseudomonas aeruginosa. Ann. Clin. Microbiol. Antimicrob. 2020;19:1–8. doi: 10.1186/s12941-020-00350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sedarat Z., Taylor-Robinson A.W. Biofilm formation by pathogenic bacteria: applying a staphylococcus aureus model to appraise potential targets for therapeutic intervention. Pathogens. 2022;11(4):388. doi: 10.3390/pathogens11040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gedefie A., Demsis W., Ashagrie M., Kassa Y., Tesfaye M., Tilahun M., et al. Acinetobacter baumannii biofilm formation and its role in disease pathogenesis: a review. Infect. Drug Resist. 2021:3711–3719. doi: 10.2147/IDR.S332051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raoust E., Balloy V., Garcia-Verdugo I., Touqui L., Ramphal R., Chignard M. Pseudomonas aeruginosa LPS or flagellin are sufficient to activate TLR-dependent signaling in murine alveolar macrophages and airway epithelial cells. PLoS One. 2009;4(10) doi: 10.1371/journal.pone.0007259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moser C., Pedersen H.T., Lerche C.J., Kolpen M., Line L., Thomsen K., et al. Biofilms and host response–helpful or harmful. Apmis. 2017;125(4):320–338. doi: 10.1111/apm.12674. [DOI] [PubMed] [Google Scholar]
- 40.Jensen P.Ø., Givskov M., Bjarnsholt T., Moser C. The immune system vs. Pseudomonas aeruginosa biofilms. FEMS Immunol. Med. Microbiol. 2010;59(3):292–305. doi: 10.1111/j.1574-695X.2010.00706.x. [DOI] [PubMed] [Google Scholar]
- 41.Rybtke M., Jensen P.Ø., Nielsen C.H., Tolker-Nielsen T. The extracellular polysaccharide matrix of Pseudomonas aeruginosa biofilms is a determinant of polymorphonuclear leukocyte responses. Infect. Immun. 2020;89(1) doi: 10.1128/iai.00631-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moser C., Jensen P.Ø., Thomsen K., Kolpen M., Rybtke M., Lauland A.S., et al. Immune responses to Pseudomonas aeruginosa biofilm infections. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.625597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jesaitis A.J., Franklin M.J., Berglund D., Sasaki M., Lord C.I., Bleazard J.B., et al. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J. Immunol. 2003;171(8):4329–4339. doi: 10.4049/jimmunol.171.8.4329. [DOI] [PubMed] [Google Scholar]
- 44.Bjarnsholt T., Jensen P.Ø., Burmølle M., Hentzer M., Haagensen J.A., Hougen H.P., et al. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology. 2005;151(2):373–383. doi: 10.1099/mic.0.27463-0. [DOI] [PubMed] [Google Scholar]
- 45.Leid J.G., Willson C.J., Shirtliff M.E., Hassett D.J., Parsek M.R., Jeffers A.K. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-γ-mediated macrophage killing. The journal of immunology. 2005;175(11):7512–7518. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- 46.Jensen P.Ø., Bjarnsholt T., Phipps R., Rasmussen T.B., Calum H., Christoffersen L., et al. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology. 2007;153(5):1329–1338. doi: 10.1099/mic.0.2006/003863-0. [DOI] [PubMed] [Google Scholar]
- 47.Trøstrup H., Thomsen K., Christophersen L.J., Hougen H.P., Bjarnsholt T., Jensen P.Ø., et al. P seudomonas aeruginosa biofilm aggravates skin inflammatory response in BALB/c mice in a novel chronic wound model. Wound Repair Regen. 2013;21(2):292–299. doi: 10.1111/wrr.12016. [DOI] [PubMed] [Google Scholar]
- 48.Alhede M., Bjarnsholt T., Jensen P.Ø., Phipps R.K., Moser C., Christophersen L., et al. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology. 2009;155(11):3500–3508. doi: 10.1099/mic.0.031443-0. [DOI] [PubMed] [Google Scholar]
- 49.Kaliński P., Hilkens C.M., Wierenga E.A., Kapsenberg M.L. T-cell priming by type-1and type-2 polarized dendritic cells: the concept of a third signal. Immunol. today. 1999;20(12):561–567. doi: 10.1016/s0167-5699(99)01547-9. [DOI] [PubMed] [Google Scholar]
- 50.Lai L., Alaverdi N., Maltais L., Morse H.C. Mouse cell surface antigens: nomenclature and immunophenotyping. J. Immunol. 1998;160(8):3861–3868. [PubMed] [Google Scholar]
- 51.Banchereau J., Briere F., Caux C., Davoust J., Lebecque S., Liu Y.-J., et al. Immunobiology of dendritic cells. Annu. Rev. Immunol. 2000;18(1):767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 52.Lieske B., Makarova N., Jagemann B., Walther C., Ebinghaus M., Zyriax B.-C., Aarabi G. Inflammatory response in oral biofilm during pregnancy: a systematic review. Nutrients. 2022;14(22):4894. doi: 10.3390/nu14224894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi N. Oral microbiome metabolism: from “who are they?” to “what are they doing?”. Journal of dental research. 2015;94(12):1628–1637. doi: 10.1177/0022034515606045. [DOI] [PubMed] [Google Scholar]
- 54.Jang H., Patoine A., Wu T.T., Castillo D.A., Xiao J. Oral microflora and pregnancy: a systematic review and meta-analysis. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-96495-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanz M., Kornman K., Workshop W. Periodontitis and adverse pregnancy outcomes: consensus report of the joint EFP/AAP workshop on periodontitis and systemic diseases. J. Periodontol. 2013;84:S164–S169. doi: 10.1902/jop.2013.1340016. [DOI] [PubMed] [Google Scholar]
- 56.Mor G., Aldo P., Alvero A.B. The unique immunological and microbial aspects of pregnancy. Nat. Rev. Immunol. 2017;17(8):469–482. doi: 10.1038/nri.2017.64. [DOI] [PubMed] [Google Scholar]
- 57.Saadaoui M., Singh P., Al Khodor S. Oral microbiome and pregnancy: a bidirectional relationship. J. Reprod. Immunol. 2021;145 doi: 10.1016/j.jri.2021.103293. [DOI] [PubMed] [Google Scholar]
- 58.Corbella S., Taschieri S., Del Fabbro M., Francetti L., Weinstein R., Ferrazzi E. Adverse pregnancy outcomes and periodontitis: a systematic review and meta-analysis exploring potential association. Quintessence international. 2016;47(3) doi: 10.3290/j.qi.a34980. [DOI] [PubMed] [Google Scholar]
- 59.Daalderop L., Wieland B., Tomsin K., Reyes L., Kramer B., Vanterpool S., Been J. Periodontal disease and pregnancy outcomes: overview of systematic reviews. JDR Clinical & Translational Research. 2018;3(1):10–27. doi: 10.1177/2380084417731097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chua S.L., Liu Y., Yam J.K.H., Chen Y., Vejborg R.M., Tan B.G.C., et al. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat. Commun. 2014;5(1):4462. doi: 10.1038/ncomms5462. [DOI] [PubMed] [Google Scholar]
- 61.Guilhen C., Charbonnel N., Parisot N., Gueguen N., Iltis A., Forestier C., Balestrino D. Transcriptional profiling of Klebsiella pneumoniae defines signatures for planktonic, sessile and biofilm-dispersed cells. BMC Genom. 2016;17:1–15. doi: 10.1186/s12864-016-2557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rumbaugh K.P., Sauer K. Biofilm dispersion. Nat. Rev. Microbiol. 2020;18(10):571–586. doi: 10.1038/s41579-020-0385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jensen E.T., Kharazmi A., Høiby N., Costerton J.W. Some bacterial parameters influencing the neutrophil oxidative burst response to Pseudomonas aeruginosa biofilms. Apmis. 1992;100(7‐12):727–733. [PubMed] [Google Scholar]
- 64.Lockhart J.S., Buret A.G., Ceri H., Storey D.G., Anderson S.J., Morck D.W. Mixed species biofilms of Fusobacterium necrophorum and Porphyromonas levii impair the oxidative response of bovine neutrophils in vitro. Anaerobe. 2017;47:157–164. doi: 10.1016/j.anaerobe.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 65.Kernien J.F., Johnson C.J., Nett J.E. Conserved inhibition of neutrophil extracellular trap release by clinical Candida albicans biofilms. Journal of Fungi. 2017;3(3):49. doi: 10.3390/jof3030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Motta J.-P., Wallace J.L., Buret A.G., Deraison C., Vergnolle N. Gastrointestinal biofilms in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2021;18(5):314–334. doi: 10.1038/s41575-020-00397-y. [DOI] [PubMed] [Google Scholar]
- 67.Nadell C.D., Drescher K., Foster K.R. Spatial structure, cooperation and competition in biofilms. Nat. Rev. Microbiol. 2016;14(9):589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 68.Byun K.-H., Han S.H., Choi M.W., Kim B.-H., Park S.H., Ha S.-D. Biofilm eradication ability of phage cocktail against Listeria monocytogenes biofilms formed on food contact materials and effect on virulence-related genes and biofilm structure. Food Res. Int. 2022;157 doi: 10.1016/j.foodres.2022.111367. [DOI] [PubMed] [Google Scholar]
- 69.Pires D.P., Melo L.D., Boas D.V., Sillankorva S., Azeredo J. Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Curr. Opin. Microbiol. 2017;39:48–56. doi: 10.1016/j.mib.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 70.São-José C. Engineering of phage-derived lytic enzymes: improving their potential as antimicrobials. Antibiotics. 2018;7(2):29. doi: 10.3390/antibiotics7020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kilcher S., Loessner M.J. Engineering bacteriophages as versatile biologics. Trends Microbiol. 2019;27(4):355–367. doi: 10.1016/j.tim.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 72.Hsu B.B., Gibson T.E., Yeliseyev V., Liu Q., Lyon L., Bry L., et al. Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell host & microbe. 2019;25(6):803–814. e5. doi: 10.1016/j.chom.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gibb B., Hyman P., Schneider C.L. The many applications of engineered bacteriophages—an overview. Pharmaceuticals. 2021;14(7):634. doi: 10.3390/ph14070634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taati Moghadam M., Amirmozafari N., Mojtahedi A., Bakhshayesh B., Shariati A., Masjedian Jazi F. Association of perturbation of oral bacterial with incident of Alzheimer's disease: a pilot study. J. Clin. Lab. Anal. 2022;36(7) doi: 10.1002/jcla.24483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moghadam M.T., Bakhshayesh B., Babakhani S., Ajorloo P., Shariati A., Mirzaei M., et al. The effect of bacterial composition shifts in the oral microbiota on alzheimer’s disease. Curr. Mol. Med. 2024;24(15):167–181. doi: 10.2174/1566524023666220819140748. [DOI] [PubMed] [Google Scholar]
- 76.Paul V.D., Sundarrajan S., Rajagopalan S.S., Hariharan S., Kempashanaiah N., Padmanabhan S., et al. Lysis-deficient phages as novel therapeutic agents for controlling bacterial infection. BMC Microbiol. 2011;11(1):1–9. doi: 10.1186/1471-2180-11-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsuda T., Freeman T.A., Hilbert D.W., Duff M., Fuortes M., Stapleton P.P., Daly J.M. Lysis-deficient bacteriophage therapy decreases endotoxin and inflammatory mediator release and improves survival in a murine peritonitis model. Surgery. 2005;137(6):639–646. doi: 10.1016/j.surg.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 78.Meile S., Du J., Dunne M., Kilcher S., Loessner M.J. Engineering therapeutic phages for enhanced antibacterial efficacy. Current opinion in virology. 2022;52:182–191. doi: 10.1016/j.coviro.2021.12.003. [DOI] [PubMed] [Google Scholar]
- 79.Yehl K., Lemire S., Yang A.C., Ando H., Mimee M., Torres M.D.T., et al. Engineering phage host-range and suppressing bacterial resistance through phage tail fiber mutagenesis. Cell. 2019;179(2):459–469. e9. doi: 10.1016/j.cell.2019.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meile S., Kilcher S., Loessner M.J., Dunne M. Reporter phage-based detection of bacterial pathogens: design guidelines and recent developments. Viruses. 2020;12(9):944. doi: 10.3390/v12090944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goren M.G., Yosef I., Qimron U. Programming bacteriophages by swapping their specificity determinants. Trends Microbiol. 2015;23(12):744–746. doi: 10.1016/j.tim.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 82.Lu T.K., Collins J.J. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. USA. 2007;104(27):11197–11202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pei R., Lamas-Samanamud G.R. Inhibition of biofilm formation by T7 bacteriophages producing quorum-quenching enzymes. Appl. Environ. Microbiol. 2014;80(17):5340–5348. doi: 10.1128/AEM.01434-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gerstmans H., Criel B., Briers Y. Synthetic biology of modular endolysins. Biotechnol. Adv. 2018;36(3):624–640. doi: 10.1016/j.biotechadv.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 85.Guo D., Chen J., Zhao X., Luo Y., Jin M., Fan F., et al. Genetic and chemical engineering of phages for controlling multidrug-resistant bacteria. Antibiotics. 2021;10(2):202. doi: 10.3390/antibiotics10020202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lukacik P., Barnard T.J., Keller P.W., Chaturvedi K.S., Seddiki N., Fairman J.W., et al. Structural engineering of a phage lysin that targets Gram-negative pathogens. Proc. Natl. Acad. Sci. USA. 2012;109(25):9857–9862. doi: 10.1073/pnas.1203472109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gervasi T., Horn N., Wegmann U., Dugo G., Narbad A., Mayer M.J. Expression and delivery of an endolysin to combat Clostridium perfringens. Appl. Microbiol. Biotechnol. 2014;98:2495–2505. doi: 10.1007/s00253-013-5128-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gerstmans H., Rodríguez-Rubio L., Lavigne R., Briers Y. From endolysins to Artilysin® s: novel enzyme-based approaches to kill drug-resistant bacteria. Biochem. Soc. Trans. 2016;44(1):123–128. doi: 10.1042/BST20150192. [DOI] [PubMed] [Google Scholar]
- 89.Yang H., Bi Y., Shang X., Wang M., Linden S.B., Li Y., et al. Antibiofilm activities of a novel chimeolysin against Streptococcus mutans under physiological and cariogenic conditions. Antimicrobial agents and chemotherapy. 2016;60(12):7436–7443. doi: 10.1128/AAC.01872-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jia M., Zhou W., Luo D., Xue H., Hu F., Zhang X., et al. Calcium-binding motif-mediated binding of redundant calcium offers a chimeolysin enhanced bactericidal activity and extended host range under physiological conditions. J. Antimicrob. Chemother. 2023;78(5):1182–1190. doi: 10.1093/jac/dkad059. [DOI] [PubMed] [Google Scholar]
- 91.Yang H., Linden S.B., Wang J., Yu J., Nelson D.C., Wei H. A chimeolysin with extended-spectrum streptococcal host range found by an induced lysis-based rapid screening method. Sci. Rep. 2015;5(1) doi: 10.1038/srep17257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Landlinger C., Tisakova L., Oberbauer V., Schwebs T., Muhammad A., Latka A., et al. Engineered phage endolysin eliminates Gardnerella biofilm without damaging beneficial bacteria in bacterial vaginosis ex vivo. Pathogens. 2021;10(1):54. doi: 10.3390/pathogens10010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hong H.-W., Kim Y.D., Jang J., Kim M.S., Song M., Myung H. Combination effect of engineered endolysin EC340 with antibiotics. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.821936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee C., Kim J., Son B., Ryu S. Development of advanced chimeric endolysin to control multidrug-resistant Staphylococcus aureus through domain shuffling. ACS Infect. Dis. 2021;7(8):2081–2092. doi: 10.1021/acsinfecdis.0c00812. [DOI] [PubMed] [Google Scholar]
- 95.Quintana-Sanchez S., Gómez-Casanova N., Sánchez-Nieves J., Gómez R., Rachuna J., Wąsik S., et al. The antibacterial effect of PEGylated carbosilane dendrimers on P. aeruginosa alone and in combination with phage-derived endolysin. Int. J. Mol. Sci. 2022;23(3):1873. doi: 10.3390/ijms23031873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Son B., Kong M., Lee Y., Ryu S. Development of a novel chimeric endolysin, Lys109 with enhanced lytic activity against Staphylococcus aureus. Front. Microbiol. 2021;11 doi: 10.3389/fmicb.2020.615887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu T.K., Collins J.J. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc. Natl. Acad. Sci. USA. 2009;106(12):4629–4634. doi: 10.1073/pnas.0800442106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang T., Zheng Y., Dai J., Zhou J., Yu R., Zhang C. Design SMAP29-LysPA26 as a highly efficient Artilysin against Pseudomonas aeruginosa with bactericidal and antibiofilm activity. Microbiol. Spectr. 2021;9(3) doi: 10.1128/Spectrum.00546-21. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thandar M., Lood R., Winer B.Y., Deutsch D.R., Euler C.W., Fischetti V.A. Novel engineered peptides of a phage lysin as effective antimicrobials against multidrug-resistant Acinetobacter baumannii. Antimicrobial agents and chemotherapy. 2016;60(5):2671–2679. doi: 10.1128/AAC.02972-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yuan Y., Li X., Wang L., Li G., Cong C., Li R., et al. The endolysin of the Acinetobacter baumannii phage vB_AbaP_D2 shows broad antibacterial activity. Microb. Biotechnol. 2021;14(2):403–418. doi: 10.1111/1751-7915.13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lood R., Winer B.Y., Pelzek A.J., Diez-Martinez R., Thandar M., Euler C.W., et al. Novel phage lysin capable of killing the multidrug-resistant gram-negative bacterium Acinetobacter baumannii in a mouse bacteremia model. Antimicrobial agents and chemotherapy. 2015;59(4):1983–1991. doi: 10.1128/AAC.04641-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Díez-Martínez R., De Paz H.D., García-Fernández E., Bustamante N., Euler C.W., Fischetti V.A., et al. A novel chimeric phage lysin with high in vitro and in vivo bactericidal activity against Streptococcus pneumoniae. J. Antimicrob. Chemother. 2015;70(6):1763–1773. doi: 10.1093/jac/dkv038. [DOI] [PubMed] [Google Scholar]
- 103.Vázquez R., García P. Synergy between two chimeric lysins to kill Streptococcus pneumoniae. Front. Microbiol. 2019;10:1251. doi: 10.3389/fmicb.2019.01251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arroyo-Moreno S., Begley M., Dembicka K., Coffey A. Engineering of the CHAPk staphylococcal phage endolysin to enhance antibacterial activity against stationary-phase cells. Antibiotics. 2021;10(6):722. doi: 10.3390/antibiotics10060722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Landlinger C., Oberbauer V., Podpera Tisakova L., Schwebs T., Berdaguer R., Van Simaey L., et al. Preclinical data on the gardnerella-specific endolysin PM-477 indicate its potential to improve the treatment of bacterial vaginosis through enhanced biofilm removal and avoidance of resistance. Antimicrob. Agents Chemother. 2022;66(5) doi: 10.1128/aac.02319-21. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Castro J., Sousa L.G., França Â., Podpera Tisakova L., Corsini L., Cerca N. Exploiting the anti-biofilm effect of the engineered phage Endolysin PM-477 to disrupt in vitro single-and dual-species biofilms of vaginal pathogens associated with bacterial vaginosis. Antibiotics. 2022;11(5):558. doi: 10.3390/antibiotics11050558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lu R., Liu B., Wu L., Bao H., García P., Wang Y., et al. A broad-spectrum phage endolysin (LysCP28) able to remove biofilms and inactivate Clostridium perfringens strains. Foods. 2023;12(2):411. doi: 10.3390/foods12020411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ning H., Lin H., Wang J., He X., Lv X., Ju L. Characterizations of the endolysin Lys84 and its domains from phage qdsa002 with high activities against Staphylococcus aureus and its biofilms. Enzym. Microb. Technol. 2021;148 doi: 10.1016/j.enzmictec.2021.109809. [DOI] [PubMed] [Google Scholar]
- 109.Manoharadas S., Ahmad N., Altaf M., Alrefaei A.F., Al-Rayes B.F. An enzybiotic cocktail effectively disrupts preformed dual biofilm of Staphylococcus aureus and Enterococcus faecalis. Pharmaceuticals. 2023;16(4):564. doi: 10.3390/ph16040564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Srinivasan R., Chaitanyakumar A., Subramanian P., Mageswari A., Gomathi A., Aswini V., et al. Recombinant engineered phage-derived enzybiotic in Pichia pastoris X-33 as whole cell biocatalyst for effective biocontrol of Vibrio parahaemolyticus in aquaculture. Int. J. Biol. Macromol. 2020;154:1576–1585. doi: 10.1016/j.ijbiomac.2019.11.042. [DOI] [PubMed] [Google Scholar]
- 111.Tinoco J.M., Buttaro B., Zhang H., Liss N., Sassone L., Stevens R. Effect of a genetically engineered bacteriophage on Enterococcus faecalis biofilms. Arch. Oral Biol. 2016;71:80–86. doi: 10.1016/j.archoralbio.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vander Elst N., Bert J., Favoreel H., Lavigne R., Meyer E., Briers Y. Development of engineered endolysins with in vitro intracellular activity against streptococcal bovine mastitis‐causing pathogens. Microb. Biotechnol. 2023;16(12):2367–2386. doi: 10.1111/1751-7915.14339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hassannia M., Naderifar M., Salamy S., Akbarizadeh M.R., Mohebi S., Moghadam M.T. Engineered phage enzymes against drug-resistant pathogens: a review on advances and applications. Bioproc. Biosyst. Eng. 2024;47(3):301–312. doi: 10.1007/s00449-023-02938-6. [DOI] [PubMed] [Google Scholar]
- 114.Huang Z., Zhang Z., Tong J., Malakar P.K., Chen L., Liu H., et al. Phages and their lysins: toolkits in the battle against foodborne pathogens in the postantibiotic era. Compr. Rev. Food Sci. Food Saf. 2021;20(4):3319–3343. doi: 10.1111/1541-4337.12757. [DOI] [PubMed] [Google Scholar]
- 115.Boroujeni M.B., Mohebi S., Malekian A., Shahraeini S.S., Gharagheizi Z., Shahkolahi S., et al. The therapeutic effect of engineered phage, derived protein and enzymes against superbug bacteria. Biotechnol. Bioeng. 2024;121(1):82–99. doi: 10.1002/bit.28581. [DOI] [PubMed] [Google Scholar]
- 116.Zalewska-Piątek B. Phage therapy—challenges, opportunities and future prospects. Pharmaceuticals. 2023;16(12):1638. doi: 10.3390/ph16121638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.