Abstract
In the antibiotic resistance era, utilizing understudied sources for novel antimicrobials or antivirulence agents can provide new advances against antimicrobial resistant pathogens. In this study, we aimed to investigate antibacterial and antibiofilm activities of Posidonia oceanica (L.) Delile against Enterococcus faecalis ATCC 29212, Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC 700603 and several S. aureus clinical isolates obtained from medical devices, including patient urinary catheters and breast implant infections, with varying antibiotic recalcitrance profiles. The ethanolic and methanolic extracts from P. oceanica rhizome exhibited significant antibacterial activity against E. faecalis and S. aureus, as well as drug resistant S. aureus clinical isolates. Furthermore, significant antibiofilm activity was observed against S. aureus and E. faecalis treated with ER, MR1, and MR2. P. oceanica extracts also exhibited synergistic antimicrobial activity with ciprofloxacin against E. faecalis, sensitizing E. faecalis to a lower ciprofloxacin concentration. Collectively, our data demonstrate the selective antibacterial and antibiofilm activity of the extracts of P. oceanica against Gram-positive bacteria and clinical isolates along with potentiation of current antibiotics, which suggests that P. oceanica can be further investigated as a potential source for novel therapeutic options in the treatment of drug resistant bacterial infections.
1. Introduction
Antimicrobial resistance (AMR) is a growing global health crisis and a serious public health threat, particularly spreading among pathogenic bacteria in nosocomial settings. The emergence and spread of new AMR among bacterial pathogens limit the efficacy of antibiotics, thus, negatively impacting the treatment of bacterial infections worldwide [1]. A fundamental mechanism by which pathogenic microorganisms negatively impact healthcare settings is by colonizing and forming biofilms on surfaces, prosthetics, or other implanted medical devices. Microbial biofilms, communities of bacteria enclosed in an exopolysaccharide matrix with a reduced metabolism, provide an increased level of recalcitrance to antimicrobial agents and many other environmental stressors, including the host immune system [2,3]. If new antibiotics aren't discovered and implemented, death by AMR pathogens is predicted to exceed 10 million each year and outnumber cancer deaths by 2050 [4] highlighting an unprecedented need for novel therapeutic approaches. One promising strategy to combat these infections is to identify and target essential mechanisms of virulence. Another strategy has been to potentiate current antibiotics that have been rendered ineffective against pathogens by AMR.
Marine organisms produce various bioactive compounds and secondary metabolites with the potential to combat pathogenic microorganisms [5,6]. Seagrasses, important members of marine ecosystems, are unique vascular plants that are tremendous sources of bioactive compounds which can grow only in sub-marine environments. Posidonia oceanica (L.) Delile, a member of the Posidoniaceae family, is an endemic seagrass to the Mediterranean Sea [7]. P. oceanica has a significant role in retaining marine ecosystems by providing shelter and nutrition sources for a wide range of marine life, while its rhizomes help stabilize the seafloor [8]. Several studies have shown that the plant organs of P. oceanica consist of various compounds, including phenols, carbohydrates, flavonoids and carboxylic acids with a variety of biological activities, including anticancer and antioxidant effects [[7], [8], [9], [10]]. For example, the anticancer activity of rhizome and leaf extracts of P. oceanica against HepG2 hepatocarcinoma cells were evaluated and the results revealed dose-dependent cytotoxicity [11]. Additionally, antidiabetic, antioxidant, and vasoprotective effects of P. oceanica lyophilized leaves were previously shown in vivo using rat models, where safety of aqueous preparation in rat models at 50, 150, and 250 mg/kg was demonstrated [12].
Similarly, molecules obtained from sea grasses such as linoleic acid were shown to inhibit Staphylococcus aureus growth via increasing membrane permeability [13]. It was previously shown that a novel sesquiterpene, posidonizol, obtained from the chloroformic extractions of P. oceanica exhibited promising antibacterial activity against S. aureus, Staphylococcus epidermidis, and Micrococcus luteus [14]. Furthermore, a polypeptide identified from aqueous extracts of P. oceanica was reported to exhibit antibacterial activity and displayed significant antibiofilm activity against well characterized S. aureus and Enterococcus faecalis [11]. However, the interaction of P. oceanica with commonly used antimicrobials or its impact on drug resistant/recalcitrant clinical isolates derived from patients’ medical device associated infections or their biofilm forming capabilities remained unknown.
In this study, we demonstrate the antibacterial and antibiofilm activities of P. oceanica extracts obtained from leaves and rhizomes collected from the northern shores of Cyprus, against selected Gram-positive and Gram-negative bacteria, including drug resistant clinical isolates, while also demonstrating synergistic activity of P. oceanica extracts with commonly prescribed antibiotics. We also demonstrate that P. oceanica has inhibitory function against medical device associated clinical isolates obtained from urinary catheters or breast implant infections with varying antimicrobial resistance profiles.
2. Results and discussion
2.1. Yield of different solvent extracts from leaves and rhizomes of P. oceanica
P. oceanica extracts were generated utilizing various organic solvents including acetone, chloroform, methanol, ethanol, and dichloromethane. The methanolic extraction protocol exhibited the highest yield whereas the dichloromethane extraction exhibited the lowest yield for both the leaves and rhizomes compared to other tested solvents (Table 1). The methanolic extraction of the rhizome revealed two layers of crude extract. Thus, they were separated into two parts as: Viscous (MR1) and Granulated (MR2). Ethanolic and methanolic extraction protocols led to higher yields in rhizome extracts. Methanol and ethanol solvents were shown to be good solvents for the extraction of bioactive compounds with antibacterial potencies from various marine organisms [15,16]. Solvents with lower polarities such as acetone, dichloromethane, and chloroform revealed higher yields in leaf extracts (Table 1) suggesting that the rhizome, subterranean organ of P. oceanica, possess higher amounts of polar compounds, whereas, the leaves, which arise from orthotropic rhizomes, possess higher amounts of nonpolar compounds.
Table 1.
Yield obtained from 10 g of each sample of leaves and rhizomes using selected solvents.
| Solvent(s) | Leaf |
Rhizome |
||
|---|---|---|---|---|
| Total (g) | Percentage (%) | Total (g) | Percentage (%) | |
| Methanol | 1.644 | 16.44 | 3.949 | 39.49 |
| Ethanol | 0.974 | 9.74 | 2.641 | 26.41 |
| Acetone | 0.390 | 3.90 | 0.192 | 1.92 |
| Dichloromethane | 0.109 | 1.09 | 0.089 | 0.89 |
| Chloroform | 0.193 | 1.93 | 0.144 | 1.44 |
2.2. Antimicrobial activities of the extracts against selected Gram-positive and Gram-negative bacteria
In order to assess the antimicrobial activities associated with P. oceanica extracts, a microdilution method was used to assess minimum inhibitory concentrations (MIC) against Enterococcus faecalis ATCC 29212, Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922, and Klebsiella pneumoniae ATCC 700603, as well as various antimicrobial resistant S. aureus clinical isolates. MIC assays revealed significant antibacterial activity of the rhizome (MR1, MR2, and ER) extracts of P. oceanica against S. aureus ATCC 25923 and E. faecalis ATCC 29212 at MIC of 1, 1, and 0.5 mg/mL, respectively (Table 2). Minimum bactericidal concentration (MBC) of ER against S. aureus ATCC 25923 and E. faecalis ATCC 29212 were >4 and 1 mg/mL, respectively, whereas that of MR1, and MR2 was >4 and 2 mg/mL, respectively. However, the leaf extracts did not exhibit antibacterial activity against S. aureus ATCC 25923 or E. faecalis ATCC 29212 (Supplemental Table 1). Additionally, none of the extracts had antibacterial activity against the tested Gram-negative bacteria (Table 2 and Supplemental Table 1), which may be due to the presence of an outer membrane in Gram-negative bacteria that can limit entry of bioactive molecules [17]. This is in line with previous findings from other marine plants where extracts of brown seaweeds were found to be more active against Gram-positive bacteria when compared with Gram-negative bacteria [18]. In addition to brown seaweeds, the same specific targeting of Gram-positive bacteria rather than Gram-negative bacteria was also observed with red seaweed extracts [19,20]. Interestingly, ER was found to have a slightly higher antibacterial activity compared to MR1, and MR2 likely due to the differences in the bioactive compound composition. Due to efficacy of rhizome extracts in comparison to the leaves, we further investigated the antibacterial and antibiofilm potency of rhizome extracts.
Table 2.
MIC values of the crude extracts obtained from different extractions rhizomes of Posidonia oceanica against Gram-positive and Gram-negative bacteria. Data represented as the standard error of mean (±S.E.M). MR1-viscous part from the methanolic crude extract of rhizome. MR2-granulated part from the methanolic crude extract of rhizome. ER-ethanolic extract of the rhizome. NC-no change.
| Extract | Gram- positive bacteria |
Gram-negative bacteria |
|||
|---|---|---|---|---|---|
| S. aureus ATCC 25923 | E. faecalis ATCC 29212 | E. coli ATCC 25922 | K. pneumoniae ATCC 700603 | ||
| Rhizome (mg/mL) | MR1 | 1 ± 0 | 1 ± 0.33 | NC | NC |
| MR2 | 1 ± 0 | 1 ± 0.17 | NC | NC | |
| ER | 0.5 ± 0 | 0.5 ± 0.17 | NC | NC | |
| Acetone | NC | NC | NC | NC | |
| Dichloromethane | NC | NC | NC | NC | |
| Chloroform | NC | NC | NC | NC | |
| Control (mg/L) | Ciprofloxacin | 0.125 ± 0 | 0.5 ± 0.083 | 0.008 ± 0 | 0.125 ± 0.021 |
In addition to the classical ATCC strains, we tested the P. oceanica ethanolic extract on the well-characterized JE2 strain, an MRSA (Methicillin Resistant S. aureus) skin infection isolate [21], as well as various S. aureus clinically derived isolates obtained from patients. These clinical isolates were obtained from patient urinary catheters [termed HUC (human urinary catheter)] [22] or from breast implant infections [termed BISA (breast implant S. aureus)] that occurred in women following reconstructive surgery post-mastectomy [23]. These isolates have varying antimicrobial resistance profiles (Table 3). To get insight towards the mechanisms of antimicrobial resistance, PCR was utilized to assess methicillin sensitivity of the clinical isolates via mecA gene amplification, which is known to confer methicillin resistance among S. aureus clinical isolates. Amplicon corresponding to 162 bp band indicated that the mecA gene is encoded only by the established MRSA strain, JE2 (Supplemental Fig. 1). Our previous study indicated that the MSSA (Methicillin Sensitive S. aureus) BISA isolates, obtained from two different breast implant infection patients are resistant to the antibiotic bacitracin, but susceptible to the antibiotics gentamicin and cefazolin, which are commonly prescribed clinically to prevent and treat breast implant infections (Table 3) [23]. However, during biofilm formation, these isolates are recalcitrant to more than four times the MIC of cefazolin and gentamicin, potentially explaining why these strains caused infection despite standard prophylactic cefazolin administration [23]. Here, we show the MSSA urinary catheter isolates HUC 86-07c, 95, 111-01c are sensitive to gentamicin, cefazolin, ciprofloxacin, and nitrofurantoin, which are commonly prescribed antibiotics to treat S. aureus urinary tract infections (Table 3). In contrast, HUC 97-02 displayed resistance to ciprofloxacin, but remained susceptible to cefazolin, gentamicin, and nitrofurantoin (Table 3). MIC studies carried out on these isolates with P. oceanica ethanolic extract revealed MIC values ranging from 0.2 to 0.4 mg/mL (Table 4). Specifically, while the BISA isolates were inhibited by the ethanolic extract at 0.2 mg/mL, the MRSA skin isolate JE2 was inhibited at 0.4 mg/mL (Table 4). The inhibitory concentration of the HUC isolates also ranged from 0.2 to 0.4 mg/mL (Table 4). Notably, the MSSA catheter isolate HUC 97-02 was inhibited at a relatively low concentration of 0.2 mg/mL, despite its antimicrobial resistance against ciprofloxacin. These clinical isolates occupy unique niches and have differential virulence factors presence/expression in order to optimally adapt to the site of infection. Therefore, the differential expression and niche profile of isolates may be a determining factor in the slight sensitivity differences. Future investigations will focus on understanding the determinant, including structural elements or virulence factors, that dictates bacterial sensitivity towards P. oceanica bioactive molecules.
Table 3.
Staphylococcus aureus clinical isolates obtained from patient urinary catheters or breast implants with varying antibiotic resistance/recalcitrance profiles. MRSA-Methicillin Resistant S. aureus. MSSA-Methicillin sensitive S. aureus. Gent-Gentamicin. Cef-Cefazolin. Bac-Bacitracin. Cipro-Ciprofloxacin. Nitro-Nitrofurantoin. BISA-breast implant S. aureus. HUC-human urinary catheter. S-susceptible. R-resistant. N/A-not applicable.
| Strains | Isolation site | Details | Antimicrobial Susceptibilities |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Gent | Cef | Bac | Cipro | Nitro | MethicillinR/mecA | Reference | |||
| JE2 | Skin | Well-characterized | S | S | R | R | S | MRSA/YES | This study and [24] |
| BISA 117 | Breast Implant | Clinical | S | S | R | N/A | N/A | MSSA/NO | 23 |
| BISA 158 | Breast Implant | Clinical | S | S | R | N/A | N/A | MSSA/NO | 23 |
| HUC 86–07c | Urinary Catheter | Clinical | S | S | N/A | S | S | MSSA/NO | This study and [25] |
| HUC 95 | Urinary Catheter | Clinical | S | S | N/A | S | S | MSSA/NO | This study and [25] |
| HUC 97–02 | Urinary Catheter | Clinical | S | S | N/A | R | S | MSSA/NO | This study and [25] |
| HUC 111–01c | Urinary Catheter | Clinical | S | S | N/A | S | S | MSSA/NO | This study and [25] |
Table 4.
MIC (mg/mL) values of the ethanolic rhizome extraction (ER) of Posidonia oceanica against clinical isolates of Staphylococcus aureus.
|
Extract |
Clinical Isolates of S. aureus |
||||||
|---|---|---|---|---|---|---|---|
| JE2 | BISA 117 | BISA 158 | HUC86-07c | HUC95 | HUC97-02 | HUC111-01c | |
| ER | 0.4 | 0.2 | 0.2 | 0.4 | 0.4 | 0.2 | 0.4 |
2.3. Biofilm inhibition potencies of MR1, MR2, and ER extracts on Gram-positive bacteria
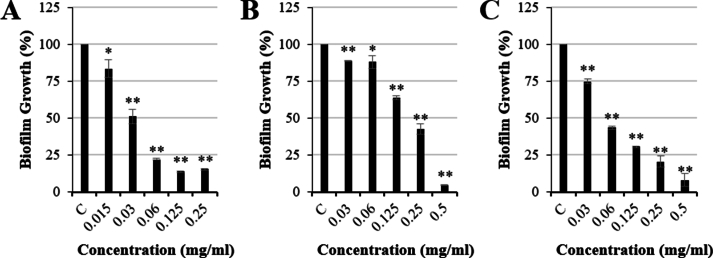
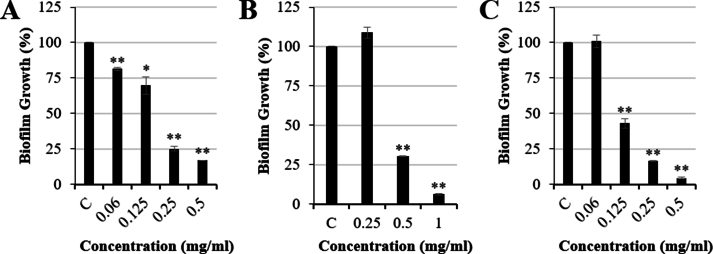
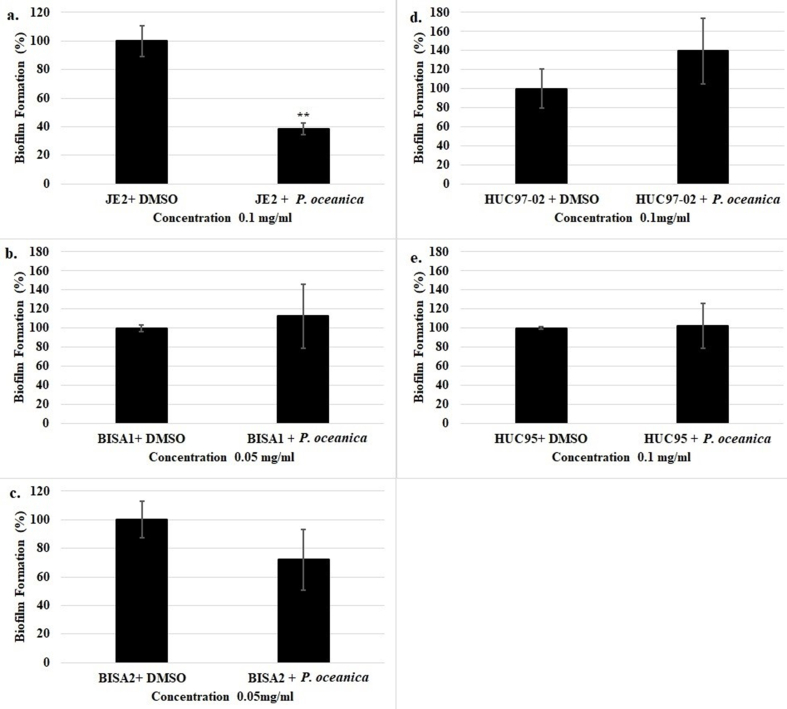
Next, we evaluated the antibiofilm activity associated with P. oceanica extracts. Ethanolic and methanolic extracts of P. oceanica revealed significant antibiofilm activity against S. aureus and E. faecalis. ER significantly reduced biofilm formation by 84.72 % (15.28 ± 0.58 % biofilm formation vs control, p < 0.01) at 0.25 mg/mL for S. aureus ATCC 25923 (Fig. 1A) and by 83.21 % (16.79 ± 0.17 % biofilm formation vs control, p < 0.01) at 0.5 mg/mL for E. faecalis ATCC 29212 (Fig. 2A). Furthermore, MR1 significantly inhibited the biofilm formed by S. aureus ATCC 25923 by 95.39 % (4.61 ± 0.16 % biofilm formation vs control, p < 0.01) at 0.5 mg/mL (Fig. 1B) and by 93.53 % (6.47 ± 0.52 % biofilm formation vs control, p < 0.01) at 1 mg/mL for E. faecalis ATCC 29212 (Fig. 2B). Moreover, MR2 displayed a concentration dependent biofilm inhibition activity for both E. faecalis ATCC 29212 and S. aureus ATCC 25923. The biofilm formation was reduced by 91.89 % (8.11 ± 4.38 % biofilm formation vs control, p < 0.01) and 95.69 % (4.31 ± 0.54 % biofilm formation vs control, p < 0.01) for S. aureus ATCC 25923 (Fig. 1C) and E. faecalis ATCC 29212 (Fig. 2C), respectively, at 0.5 mg/mL. In addition to the ATCC strains of S. aureus and E. faecalis, clinical isolates were assessed in the biofilm inhibition assays using P. oceanica ethanolic extract. P. oceanica ethanolic extract did not exhibit any significant biofilm inhibitory activity against the tested clinical isolates (Supplemental Fig. 2). However, a significant reduction in the biofilm formed by the MRSA JE2 isolate was observed using only ER at the sub-MIC concentration of 0.1 mg/mL (Supplemental Fig. 2). The specific biofilm inhibition of the JE2 MRSA skin isolate by P. oceanica but not the clinical catheter/breast implant MSSA strains indicate that bioactive molecules within these extracts may be developed into therapies that specifically target (i) drug resistant (i.e. MRSA) strains or (ii) niche specific biofilm factors required for the skin infection rather than device infection.
Fig. 1.
Antibiofilm activity of Posidonia oceanica rhizome extracts against Staphylococcus aureus ATCC 25923 treated with (A) ethanolic extract (ER), (B) viscous methanolic (MR1) and (C) granulated methanolic (MR2) extract in comparison to controls (C). Data represented as mean ± S.E.M (n = 3; * = p < 0.05 and ** = p < 0.01).
Fig. 2.
Antibiofilm activity of Posidonia oceanica rhizome extracts against Enterococcus faecalis ATCC 29212 (A) ethanolic extract (ER), (B) viscous methanolic (MR1) and (C) granulated methanolic (MR2) extract in comparison to controls (C). Data represented as mean ± S.E.M (n = 3; * = p < 0.05 and ** = p < 0.01).
2.4. Effect of MR1, MR2, and ER extracts on pre-formed biofilms of Gram-positive bacteria
To assess any effect of the extracts against a pre-formed biofilm, established biofilms were treated with P. oceanica extracts and MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide) assays were carried out. Since the MTT assay measures cellular metabolic activity, this assay is used as an indicator of cell viability and proliferation. This method was selected to be able to differentiate between the metabolically active and dead bacterial populations, since Crystal Violet stain detects dead cells/extracellular matrix in addition to the viable cells within the biofilm complex, in a non-discriminatory fashion [26,27]. When pre-formed biofilms were treated with P. oceanica extracts, there was a significant reduction in the viability of both S. aureus and E. faecalis treated with ER, MR1, and MR2 at high concentrations despite no change in biofilm mass (Table 5). Furthermore, at 1 mg/mL, only ER displayed an inhibitory effect against E. faecalis ATCC 29212 (64.46 % antimicrobial activity compared to the control). In contrast, all extracts inhibited S. aureus ATCC 25923 at 1 mg/mL (Table 5). At both 2 mg/mL and 4 mg/mL, ER, MR1, and MR2 exerted antimicrobial activity against E. faecalis ATCC 29212 and S. aureus ATCC 25923 in their pre-formed biofilm state. For example, at 4 mg/mL, ER, MR1, and MR2 antimicrobial activity values compared to controls were determined as 52.65 %, 54.00 % and 60.11 % for S. aureus and 72.93 %, 58.33 % and 52.48 % for E. faecalis (Table 5).
Table 5.
Percent eradication of Staphylococcus aureus ATCC 25923 and Enterococcus faecalis ATCC 29212 pre-formed biofilms treated with viscous methanolic (MR1), granulated methanolic (MR2) and ethanolic extractions (ER) in comparison to controls. Data represented as the standard error of mean (±S.E.M). NC-no change.
| Extracts (%) |
S. aureus ATCC 25923 |
E. faecalis ATCC 29212 |
||||
|---|---|---|---|---|---|---|
| 1 mg/mL | 2 mg/mL | 4 mg/mL | 1 mg/mL | 2 mg/mL | 4 mg/mL | |
| MR1 | 53.45 ± 2.66 | 50.95 ± 5.94 | 54.00 ± 7.52 | NC | 36.28 ± 5.26 | 52.48 ± 1.77 |
| MR2 | 40.38 ± 4.53 | 57.23 ± 2.40 | 60.11 ± 1.08 | NC | 29.42 ± 8.22 | 58.33 ± 5.89 |
| ER | 28.21 ± 4.92 | 44.79 ± 9.69 | 52.65 ± 3.32 | 62.46 ± 7.51 | 58.14 ± 6.54 | 72.93 ± 1.57 |
2.5. Synergism of MR1, MR2, and ER extracts with ciprofloxacin against Gram-positive bacteria
In order to investigate the synergistic activity of P. oceanica with commonly used antibiotics, we have established an OD based fractional inhibitory concentration (FIC) index assay with ciprofloxacin using E. faecalis ATCC 29212 and S. aureus ATCC 25923. When different concentrations of ciprofloxacin and ER were used against S. aureus, no additive difference was observed (Table 6). However, a combination of 0.16 mg/mL of ER and 0.002 mg/L ciprofloxacin was found to have an additive antimicrobial effect on E. faecalis. Furthermore, the combination of 0.08 mg/mL of both methanolic extracts (MR1 and MR2) with 0.25 mg/L of ciprofloxacin also revealed a synergistic effect against E. faecalis. Although, future studies will detail the mechanism behind the synergistic interactions of plant extract and antibiotic combinations, the bioactive compound complexity of plant extracts could be a contributing reason that enhances the therapeutic potentials of the antibiotics [[26], [27], [28], [29]]. For instance, the ER extract may have an influence on the cell wall integrity of E. faecalis or S. aureus which enhances the permeability of ciprofloxacin. Furthermore, with the increasing incidence of drug resistant enterococci, including VRE (Vancomycin Resistant E. faecalis) [30], the ability of these extracts to resensitize Enterococcus to antibiotics like ciprofloxacin could provide strategies for extending the efficacy of antibiotics. While indifference effect was observed against S. aureus, none of the combinations showed any antagonistic activity against any of the tested bacteria (Table 6).
Table 6.
Fractional inhibitory concentration (FIC) Index of ciprofloxacin in combination with bioactive extracts of Posidonia oceanica against Staphylococcus aureus ATCC 25923 and Enterococcus faecalis ATCC 29212. MR1-viscous part from the methanolic crude extract of rhizome. MR2-granulated part from the methanolic crude extract of rhizome. ER-ethanolic extract of the rhizome. A-additive. I-indifference. S-synergy.
| Species/Samples | Optimal Combination |
FIC Index |
||
|---|---|---|---|---|
| Ciprofloxacin (mg/L) | ER (mg/mL) | <0.5 | >0.5 | |
| S. aureus | 0.001 | 0.64 | 1.0 (I) | |
| E. faecalis | 0.002 | 0.16 | 0.5 (A) | |
| Ciprofloxacin (mg/L) | MR1(mg/mL) | < 0.5 | > 0.5 | |
| S. aureus | 0.001 | 1.28 | 1.0 (I) | |
| E. faecalis | 0.25 | 0.08 | 0.4 (S) | |
| Ciprofloxacin (mg/L) | MR2(mg/mL) | < 0.5 | > 0.5 | |
| S. aureus | 0.001 | 1.28 | 1.0 (I) | |
| E. faecalis | 0.25 | 0.08 | 0.4 (S) | |
2.6. Gas chromatography–mass spectrometry (GC-MS) study of bioactive extracts
P. oceanica, a common marine plant endemic in the Mediterranean Sea, is a promising resource for natural bioactive compounds. Previous studies have shown that the molecular composition of plant organs consisted of diverse compounds including, phenols, carbohydrates, flavonoids and carboxylic acids [[7], [8], [9], [10]]. Several studies have shown that plant extracts are capable of withstanding various physical factors including exposure to heat or sunlight, during and post extraction processes and long-term storage [31,32]. Additionally, plant extracts that are subjected to such physical factors were shown to have no change on their bioactivity [31,32]. Regardless, our extracts have been kept at 4 °C and in lightproof containers to avoid any potential degradation of bioactive molecules.
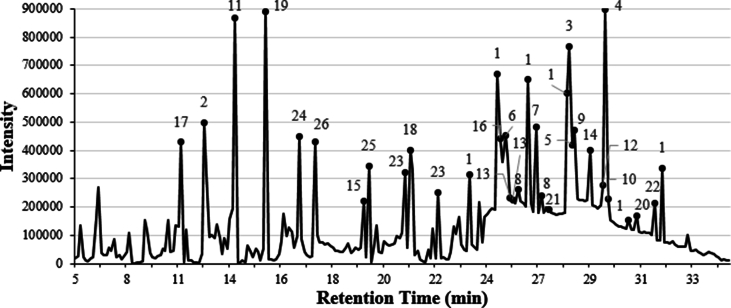
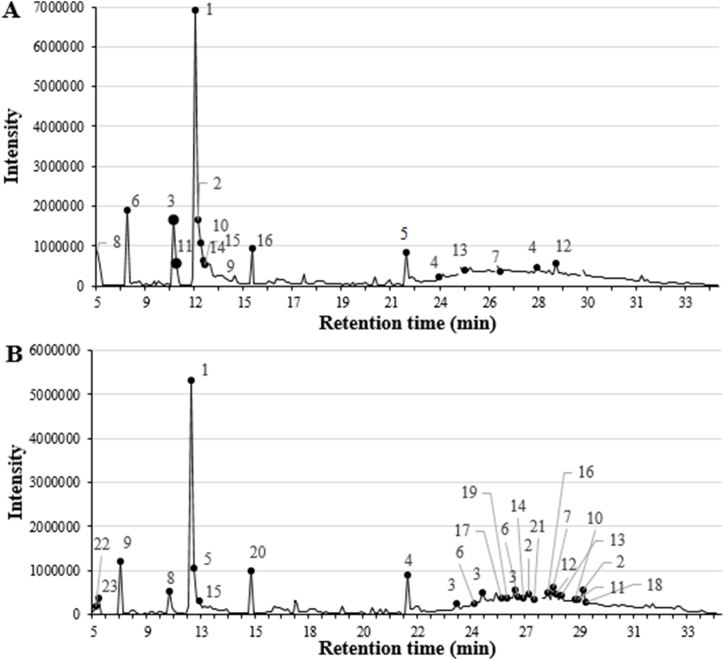
We have analyzed the bioactive compounds from the P. oceanica ER, MR1, and MR2 extracts used in our study via GC-MS. Our GC-MS analysis revealed that several bioactive compounds with various biological activities, including many with predicted or determined antimicrobial activities, were present (Table 7, Table 8, Table 9). The GC-MS analysis of the ER extract revealed the presence of heptasiloxane (7.70 %), 2-furancarboxaldehyde, 5-(hydroxymethyl)- (5.78 %), 1,3,5-trisilacyclohexane (4.35 %), iron, monocarbonyl-(1,3-butadiene-1,4-dicarbonic acid, diethyl ester) (4.29 %) and docosanedioic acid (3.13 %) as the main, prevailing compounds (Fig. 3 and Table 7). Since the ER extract showed lower MIC against S. aureus (Table 2) and lower MBC against E. faecalis in comparison to MR1, and MR2 extracts, we expected the ER to be rich in bioactive molecules with antimicrobial activity. As expected, the ER included tetradecane [33,34], heptadecane [35], nonadecane [33], docosane [36], dodecane [37,38] and docosanedioic acid [39], which were previously demonstrated to exhibit antimicrobial activities, are absent in both methanolic rhizome extracts MR1, and MR2. In contrast, the MR1 analysis revealed the presence of various bioactive compounds such as 2-furancarboxaldehyde, 5-(hydroxymethyl)- (24.30 %), 2,3-diacetoxy-1-2(2-methoxyphenoxy)propane (5.21 %), 4H-Pyran-4-one, 2,3-dihydro-3,5,-dihydroxy-6-methyl- (3.74 %), iron, monocarbonyl-(1,3-butadiene-1,4-dicarbonic acid, diethyl ester) (2.90 %) and 2-furancarboxaldehyde, 5,5’-[oxybis(methylene)]bis- (2.84 %) (Fig. 4A and Table 8), while the MR2 analysis demonstrated the presence of 2-furancarboxaldehyde, 5-(hydroxymethyl)- (22.01 %), iron, monocarbonyl-(1,3-butadiene-1,4-dicarbonic acid, diethyl ester) (9.17 %), heptasiloxane, hexadecamethyl- (3.75 %), 2-furancarboxaldehyde, 5,5’-[oxybis(methylene)]bis- (3.33 %) and cyclohexasiloxane, dodecamethyl- (2.91 %) as the main components (Fig. 4B and Table 9). Additionally, 2-furancarboxaldehyde, 5-(hydroxymethyl)- [40], tetracosanoic acid, methyl ester [41] and iron, monocarbonyl-(1,3-butadiene-1,4-dicarbonic acid, diethyl ester) a,a'-dipyridyl [42] are the only compounds that are present in both ethanolic and methanolic rhizome extracts of P. oceanica, which are shown to possess antioxidant, anti-inflammatory and antifungal activities, respectively [[40], [41], [42]]. Overall, ER was found to have a slightly higher antibacterial activity compared to MR1, and MR2 likely due to the differences in the bioactive compound composition, such as dodecane and docosanedioic acid. Dodecane [37,38] and docosanedioic acid [39] were previously reported to have antibacterial activity, and these compounds are notably absent in MR1 and MR2, which may be one of the factors leading to MR1 and MR2 having a relatively reduced antimicrobial activity. Furthermore, the leaf extracts which were revealed to have no antibacterial activity lack dodecane, docosanedioic acid and 2-furancarboxaldehyde, along with having an overall reduced concentration of each bioactive compound likely leading to the leaf extracts' collective inability to inhibit pathogenic bacteria.
Table 7.
Compounds determined through GC-MS analysis of Posidonia oceanica (L.) Delile ethanolic extract (ER) and their bioactivities.
| # | Compound Name | Compound (%) | Molecular Formula | Biological Activity |
|---|---|---|---|---|
| 1 | Heptasiloxane | 7.70 | O6Si7 | – |
| 2 | 2-Furancarboxaldehyde, 5-(hydroxymethyl)- | 5.78 | C6H6O3 | Antioxidant [40] |
| 3 | 1,3,5-Trisilacyclohexane | 4.35 | C3H6Si3 | – |
| 4 | Iron, monocarbonyl-(1,3-butadiene-1,4-dicarbonic acid, diethyl ester) a,a'-dipyridyl | 4.29 | C10H14O4 | Antifungal [42] |
| 5 | Docosanedioic acid | 3.13 | C22H42O4 | Antibacterial [39] |
| 6 | BIS [3-(3,5-di-tert-butyl-4-hydroxyphenyl)propyl]maleate | 3.02 | C38H56O6 | – |
| 7 | 1,2-Benzenedicarboxylic acid, bis(2-ethylhexyl) ester | 1.87 | C20H30O4 | Antibacterial and antifungal [43] |
| 8 | Docosane | 1.81 | C22H46 | Antibacterial [36] |
| 9 | Tetracosanoic acid, methyl ester | 1.75 | C25H50O2 | Antioxidant [41] |
| 10 | Tetratriacontane | 1.73 | C34H70 | Antibacterial [34] |
| 11 | Tetradecane | 1.54 | C14H30 | Antibacterial [33,34] |
| 12 | Pentadecanoic acid, ethyl ester | 1.52 | C17H34O2 | – |
| 13 | Methanone [2-(1-methylethyl)phenyl] | 1.47 | C16H16O | – |
| 14 | 1,2-Benzenedicarboxylic acid, dioctyl ester | 1.31 | C34H58O4 | Antibacterial [44] |
| 15 | Nonadecane | 1.30 | C19H40 | Antibacterial [33] |
| 16 | Hexanedioic acid, bis(2-ethylhexyl) ester | 1.27 | C22H42O4 | Antifungal [45] |
| 17 | Dodecane | 1.02 | C12H26 | Antibacterial and antifungal [37,38] |
| 18 | Pentadecanoic acid | 1.02 | C15H30O2 | Antimicrobial and anti-inflammatory [46] |
| 19 | Tetradecamethylcycloheptasiloxane | 0.99 | C14H42O7Si7 | – |
| 20 | Nonacosane | 0.95 | C29H60 | – |
| 21 | Heptacosanoic acid, methyl ester | 0.94 | C28H56O2 | – |
| 22 | Triacontanoic acid, methyl ester | 0.82 | C31H62O2 | – |
| 23 | Hexadecamethyl-octasiloxane | 0.78 | C16H48O7Si8 | – |
| 24 | Heptadecane | 0.67 | C17H36 | Antibacterial [35] |
| 25 | Tetradecamethylheptasiloxane | 0.66 | C14H42O6Si7 | – |
| 26 | Hexadecamethylcyclooctasiloxane | 0.59 | C16H48O8Si8 | – |
Table 8.
Compounds determined through GC-MS analysis of Posidonia oceanica (L.) Delile viscous methanolic extract (MR1) and their bioactivities.
| # | Compound Name | Compound (%) | Molecular Formula | Biological Activity |
|---|---|---|---|---|
| 1 | 2-Furancarboxaldehyde, 5-(hydroxymethyl)- | 24.30 | C6H6O3 | Antioxidant [40] |
| 2 | 2,3-diacetoxy-1-(2-methoxyphenoxy)propane | 5.21 | C10H12O3 | – |
| 3 | 4H-Pyran-4-one, 2,3-dihydro-3,5-dihydroxy-6-methyl- | 3.74 | C6H8O4 | – |
| 4 | Iron, monocarbonyl-(1,3-butadiene-1,4-dicarbonic acid, diethyl ester) a,a'-dipyridyl | 2.90 | C10H14O4 | Antifungal [42] |
| 5 | 2-Furancarboxaldehyde, 5,5'-[oxybis(methylene)]bis- | 2.84 | C12H10O5 | – |
| 6 | 4-oxo-5-methoxy-2-penten-5-olide | 2.71 | C6H6O4 | Antiproliferative [47] |
| 7 | Octadecane | 1.45 | C18H38 | Antibacterial [48] |
| 8 | 2-Furancarboxaldehyde | 1.40 | C5H4O2 | Antibacterial [49] |
| 9 | 1-Cyclohexene-1-ethanol, 2,6,6-trimethyl- | 1.31 | C11H20O | – |
| 10 | Cyclohexasiloxane, dodecamethyl- | 1.27 | C12H36O6Si6 | – |
| 11 | 2(1H)-Pyridinone, 5-methyl- | 1.19 | C6H7NO | – |
| 12 | Tetracosanoic acid, methyl ester | 1.14 | C25H50O2 | Antioxidant [41] |
| 13 | 14b-Pregnan | 1.12 | C21H36 | – |
| 14 | 1,3-Dioxolane, 4,5-diethenyl-2,2-dimethyl- | 1.09 | C9H14O2 | Antibacterial and antifungal [50] |
| 15 | 5-Formyl-2-furfurylmethanoate | 0.84 | C7H8O3 | – |
| 16 | Tetradecamethylcycloheptasiloxane | 0.55 | C14H42O7Si7 | – |
Table 9.
Compounds determined through GC-MS analysis of Posidonia oceanica (L.) Delile granulated methanolic extract (MR2) and their bioactivities.
| # | Compound Name | Compound (%) | Molecular Formula | Biological Activity |
|---|---|---|---|---|
| 1 | 2-Furancarboxaldehyde, 5-(hydroxymethyl)- | 22.01 | C6H6O3 | Antioxidant [40] |
| 2 | Iron, monocarbonyl-(1,3-butadiene-1,4-dicarbonic acid, diethyl ester) a,a'-dipyridyl | 9.17 | C10H14O4 | Antifungal [42] |
| 3 | Heptasiloxane, hexadecamethyl- | 3.75 | C16H48O6Si7 | – |
| 4 | 2-Furancarboxaldehyde, 5,5'-[oxybis(methylene)]bis- | 3.33 | C12H10O5 | – |
| 5 | Cyclohexasiloxane, dodecamethyl- | 2.91 | C12H36O6Si6 | – |
| 6 | Octadecane, 3-ethyl-5-(2-ethylbutyl)- | 2.57 | C26H54 | – |
| 7 | 1,3,5-Trisilacyclohexane | 2.51 | C3H6Si3 | – |
| 8 | 4H-Pyran-4-one, 2,3-dihydro-3,5-dihydroxy-6-methyl- | 2.20 | C6H8O4 | – |
| 9 | 4-oxo-5-methoxy-2-penten-5-olide | 2.05 | C6H6O4 | Antiproliferative [46] |
| 10 | 1,2-Benzenedicarboxylic acid, dioctyl ester | 1.42 | C34H58O4 | Antibacterial [43] |
| 11 | Octadecanoic acid, 2-hydroxy-1,3-propanediyl ester | 1.39 | C21H42O5 | – |
| 12 | Hexadecanoic acid, 2-hydroxy-1-(hydroxymethyl)ethyl ester | 1.38 | C19H38O4 | – |
| 13 | Tetracosanoic acid, methyl ester | 1.22 | C25H50O2 | Antioxidant [41] |
| 14 | Lucenin-2 | 1.10 | C27H30O16 | – |
| 15 | 2,6,6-Trimethylcyclohexene-1-carboxylic acid | 1.04 | C10H16O2 | – |
| 16 | Octadecamethylcyclononasiloxane | 1.03 | C18H54O9Si9 | – |
| 17 | Tricyclo[4.2.1.0(2,5)]nona-3,7-diene, 9-methoxy-1-phenyl- | 0.96 | C16H16O | – |
| 18 | Tetratriacontane | 0.83 | C34H70 | Antibacterial [34] |
| 19 | Tetradecamethylcycloheptasiloxane | 0.67 | C14H42O7Si7 | – |
| 20 | Octadecanoic acid | 0.61 | C18H36O2 | – |
| 21 | 2-Furancarboxaldehyde | 0.55 | C5H4O2 | Antibacterial [49] |
| 22 | Tetramethyldimethoxydisiloxane | 0.53 | C6H18O3Si2 | – |
| 23 | Hexadecamethylcyclooctasiloxane | 0.39 | C16H48O8Si8 | – |
Fig. 3.
Chromatogram of gas chromatography–mass spectroscopy analyses. Bioactive compounds obtained from ethanolic extraction. The numbers on each peak correspond to the compounds in Table 7.
Fig. 4.
Chromatogram of gas chromatography–mass spectroscopy analyses. Bioactive compounds obtained from (A) viscous methanolic (MR1) and (B) granulated methanolic (MR2) extractions. The numbers on each peak correspond to the compounds in Table 8, Table 9 respectively.
2.7. In silico pharmacokinetic studies
In silico pharmacokinetic studies done to determine pharmacokinetic parameters of major bioactive compounds identified in the ER, MR1, and MR2 extracts, as well as those specifically identified in the ER extract, were presented in Supplemental Table 2 and Supplemental Table 3, respectively. Major compounds (2-furancarboxaldehyde, 5-(hydroxymethyl)-1,3,5-trisilacyclohexane, tetracosanoic acid, methyl ester, and iron, monocarbonyl-(1,3-butadiene-1,4-dicarbonic acid, diethyl ester) a,a'-dipyridyl) present in all of the extracts exhibited good water solubility and gastrointestinal absorption, whereas, majority of the molecules possessed no blood-brain permeability (Supplemental Table 2). On the other hand, bioactive compounds (tetradecane [33,34], docosane [36], and docosanedioic acid [39]) that were present specifically in ER extracts possessed poor solubility in water, and exhibited low to none gastrointestinal absorption and blood-barrier permeability (Supplemental Table 3). Additionally, majority of the major molecules identified in the ER, MR1, and MR2 possessed favorable bioavailability scores (0.55) implying that these molecules possess promising pharmacokinetic properties (Supplemental Table 2 and Supplemental Table 3) [51].
Overall, in addition to detailing the molecular composition of P. oceanica, our results demonstrate significant antibacterial and antibiofilm activity of its ethanolic and methanolic rhizome extracts against E. faecalis and S. aureus as well as their synergistic activity with ciprofloxacin. Importantly, we demonstrate, for the first time, that P. oceanica inhibits drug resistant clinical S. aureus isolates, which suggests that bioactive molecules obtained from the rhizomes of the P. oceanica, an endemic Mediterranean Sea grass, may be further developed as potential resources for the treatment of drug resistant bacterial infections to help fight antimicrobial resistance.
3. Material and methods
3.1. Plant materials
The study complies with relevant institutional, national, and international guidelines and legislation. The fresh plant materials of P. oceanica were collected between the depths of 3–5 m of seawater of southern shores via free-diving in Northern Cyprus in June 2021. Specimens were kept in seawater on ice and brought to the laboratory within the day of collection. Identification of the plant samples were performed by Prof. Dr. F. Neriman Ozhatay, Chair of Pharmaceutical Botany Department, Eastern Mediterranean University, Faculty of Pharmacy. The voucher specimens were stored in the Cyprus International University (CIU) Public Herbarium (Voucher specimen ID: CIU-40).
3.2. Extraction methods
The leaves and rhizomes of the P. oceanica were separated, and washed with tap water to remove any debris, epiphytes or other marine organisms, followed by washing with distilled water. The samples were dried at room temperature overnight followed by a drying process at 45 °C in a drying oven for an additional 24h. A grinder was used to powder the dried plant samples and the powdered samples were stored at 4 °C until the extraction process.
The leaf and rhizome samples were extracted via maceration method using dichloromethane, chloroform, methanol, ethanol and acetone with 1:10 (w/v) ratio for three consecutive days [5,52]. All of the extracts were filtered via Whatman Noo 1 filter paper and stored at 4 °C till the evaporation process.
Solvents of the leaf and rhizome extracts were evaporated via rotary-evaporator (LabTech EV311) at 45 °C. The crude extracts were stored at 4 °C until further processes. The yields of the extractions of leaves and rhizomes using different solvents was calculated.
3.3. Preparation of the extracts
The stock solutions of the crude extracts were prepared using pure dimethyl sulfoxide (DMSO). The concentration of DMSO of the extracts was adjusted to 3 % with sterile dH2O for antibacterial and antibiofilm experiments.
Antibacterial activity assays of solvent extracts from leaves and rhizomes of P. oceanica.
3.4. Inoculum preparation
Enterococcus faecalis ATCC 29212, Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922, and Klebsiella pneumoniae ATCC 700603 were sub-cultured on Mueller-Hinton Agar (MHA). The media were incubated at 37oC. Upon incubations, strains from individual colonies were inoculated into Mueller-Hinton broth (MHB) and the turbidity was adjusted to 0.5 McFarland standard for each bacterial strain.
Additionally, the cultures of clinical strains of S. aureus within MHB were put into a thermal shaker at 37oC at 275 p.m. until OD600 of 0.4 ± 0.01. Upon desired OD600, all of the strains were prepared to obtain 1 × 106 cfu/mL for each S. aureus clinical strain.
3.5. Microdilution method
All antimicrobial susceptibility testing of clinical isolates was carried out as described in CLSI [[53], [54], [55]]. The MIC of the extracts was investigated by broth microdilution method [53]. Inocula of each bacterial strain was adjusted to 1 × 106 cfu/mL using MHB. The final concentrations of the extracts were serially diluted to range from 0.125 to 4 mg/mL for ATCC strains, and 0.0625–2 mg/mL for clinical isolates of S. aureus (Table 3). Ciprofloxacin was used as the positive control and the highest concentration of each extract in MHB was used as negative control for all tests. The microplates were incubated at 37oC for 18h.
The MIC was regarded as the minimum concentration of the extract that inhibited bacterial growth. Additionally, the microdilution experiments were conducted using different microplates with and without 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), a dye which is reduced by cellular enzymes of its barely colored tetrazolium salt into its formazan of visible purple color that acts as an indicator of cell viability of planktonic bacteria [26], to compare and further confirm the MIC of bacteria.
3.6. Minimum bactericidal concentration (MBC) determination
For the determination of MBCs [56] of the extracts, 10 μL of the sample was taken from the wells of each of the concentrations of the MICs above, and inoculated onto MHA. The media were incubated at 37 °C for 18h. The MBC was regarded as the minimum concentration of the extract that prevented bacterial growth on MHA.
3.7. Fractional inhibitory concentration (FIC) index assay of P. oceanica with ciprofloxacin
The synergistic effect assay [57] was performed for the bioactive extracts with ciprofloxacin on Gram-positive ATCC strains (Table 2). Inocula of each clinical isolate were adjusted to 1 × 106 cfu/mL using MHB. The final concentrations of the bioactive extracts ranged from 0.04 to 2.56 mg/mL, whereas ciprofloxacin ranged from 0.001 to 1 mg/L. Ciprofloxacin only and each extract in MHB was used as controls. The microplates were incubated at 37 °C for 18h.
To obtain the interaction of the tested antimicrobial agents in a combination, fractional inhibitory concentration (FIC) index calculation (FIC INDEX = A/MICA + B/MICB) was used where ‘A’ and ‘B’ are the MIC of each antimicrobial agent in combination within a single well plate; and MICA and MICB are the MIC of each drug individually. The interaction is considered to be synergistic at < 0.5; additive interaction is considered between 0.5 and 0.9; a value between 1 and 4 is considered to reveal indifference; and a value of >4 considered as an antagonistic interaction.
4. Antibiofilm activity
4.1. Biofilm inhibition assays
The biofilm inhibition assay was performed for the extracts that have exhibited antibacterial activity for the Gram-positive bacteria, as previously described [58]. ATCC strain bacterial species were incubated with 2 mL MHB containing 1 % glucose at 37 °C for 24h. The wells were mixed with serial dilutions of sub-MIC concentrations of each extract and the bacterial cultures. Inoculum of each bacterial species was used as the biofilm control, whereas serial dilutions of sub-MIC concentrations of each extract was used as negative controls. The microplates were incubated at 37 °C for 18h.
After incubation, the wells were washed with sterile dH2O to remove planktonic cells and loaded with 0.1 % crystal violet (CV) solution for 30 min followed by the application of 70 % ethanol solution. Absorbance readings were recorded upon readings via spectrophotometer (Thermo Scientific Varioskan Flash) at OD595. The biofilm inhibition experiments of the clinical isolates were modified from Ref. [22] such that the extract addition and isolate inoculation were carried out simultaneously without any use of antibiotics.
4.2. Biofilm eradication assays
The biofilm eradication assay was performed for the extracts that have exhibited significant antibacterial activity for further evaluation of antibiofilm activity [58]. Two different sets of experiments were used for the biofilm eradication assays. CV was used for determining the biofilm eradication and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), which is a useful tool for the evaluation of cellular metabolic activity [27,28], was used to quantify the metabolism of biofilm forming bacteria.
The bacterial strains were incubated within 2 mL MHB containing 1 % glucose at 37 °C for 24h. The wells were treated with the bacterial cultures and incubated at 37 °C for 18h. Inoculum of each bacterial species was used as the biofilm formation control, whereas extracts were used as negative controls for both microplates. After incubation, all of the wells were washed with sterile dH2O to remove planktonic cells.
The wells of the MTT plate were treated with serial dilutions of ER, MR1, and MR2. The wells were then immediately treated with MTT solution and the plate was incubated at 37 °C for 18h. After incubation, the solution was removed from each well. Subsequently, the wells were treated with DMSO for 30 min to dissolve the formazan product. The MTT was read via spectrophotometer (Thermo Scientific Varioskan Flash) at OD560.
The wells of CV plate were treated serial dilutions of ER, MR1 and MR2 and the plate was incubated at 37 °C for 18h. After incubation, the wells were washed with sterile dH2O. Subsequently, the wells were treated with 0.1 % CV solution for CV plate for 30 min followed by the application of 70 % ethanol solution. The CV plate was read via spectrophotometer (Thermo Scientific Varioskan Flash) at OD595.
4.3. Gas chromatography – mass spectrometry (GC-MS)
GC-MS analyses were performed using the GCMS-QP2010 PlusSystem as described by Isbilen et al. [59]. Helium was used as the carrier gas. The initial temperature of the column was 50 °C and it was maintained at the same temperature for 2 min. The temperature was increased by 10 °C until 280 °C and it was maintained for 9 min. The injection temperature was at 250 °C. The helium flow rate was used as 1 mL/min and the ion source temperature was ensured at 250 °C. The spectra were analyzed and the compounds were identified by using the MS data library WILEY7.LIB.
4.4. mecA carriage among S. aureus isolates
MRSA designation was confirmed by the carriage of the mecA gene using PCR, as the mecA gene is known to confer methicillin resistance. Genomic DNA was extracted as previously described [20]. Briefly, extractions were performed utilizing the Promega Miniprep Kit (Promega; cat # A1330) according to the manufacturer's guidelines with the exception that cells were resuspended in P1 buffer supplemented with lysostaphin for 1 h at 37 °C. PCR was then performed using the P4 (TCCAGATTACAACTTCACCAGG) and P7 (CCACTTCATATCTTGTAACG) primers as previously described [60] to amplify mecA if present, with the exception that the annealing temperature was 54 °C and time was 20 s. PCR products were then run on a 1 % agarose gel and imaged with a 162 bp band confirming mecA presence.
4.5. In silico pharmacokinetic studies
In silico pharmacokinetic studies were done for the major compounds (2-furancarboxaldehyde, 5-(hydroxymethyl)-1,3,5-trisilacyclohexane, tetracosanoic acid, methyl ester, and iron, monocarbonyl-(1,3-butadiene-1,4-dicarbonic acid, diethyl ester) a,a'-dipyridyl) which are present in ER, MR1, and MR2 extracts and several bioactive compounds (tetradecane [33,34], docosane [36], and docosanedioic acid [39]), which were previously shown to be a promising antibacterial agents, that are present specifically in ER extracts were evaluated with the aid of SwissADME software (http://www.swissadme.ch/). Several parameters such as, water solubility, bioavailability, blood-brain barrier permeation, and gastrointestinal absorption were considered to predict the pharmacokinetics and bioavailability of the bioactive extracts.
4.6. Statistical analyses
All of the experiments were performed in triplicates with at least two biological replicates, and the data were examined as means ± standard error of mean (S.E.M). Students t-test was carried out to determine the statistical significance using Excel and Prism version 8.4.3.
Data availability statement
The data used in this study are available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Ertugrul Ozbil: Writing – review & editing, Writing – original draft, Methodology, Investigation, Conceptualization. Mehmet Ilktac: Writing – review & editing, Resources, Methodology, Investigation. Sultan Ogmen: Methodology, Investigation. Ovgu Isbilen: Writing – review & editing, Validation, Methodology, Investigation, Conceptualization. Jesus M. Duran Ramirez: Methodology, Investigation. Jana Gomez: Methodology, Investigation. Jennifer N. Walker: Writing – review & editing, Writing – original draft, Validation, Resources, Methodology, Investigation, Conceptualization. Ender Volkan: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Authors would like to extend their acknowledgement to Prof. Dr. F. Neriman Ozhatay for her guidance in marine plant identification and preservation processes. We would also like to thank Scott J. Hultgren for providing the HUC isolates. Ender Volkan acknowledges the Fulbright Visiting Scholar Award for funding her visit to carry out part of the project.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e35592.
Contributor Information
Ertugrul Ozbil, Email: ertugrul.ozbil@emu.edu.tr.
Mehmet Ilktac, Email: mehmet.ilktac@emu.edu.tr.
Sultan Ogmen, Email: sultan.ogmen@emu.edu.tr.
Ovgu Isbilen, Email: oisbilen@ciu.edu.tr.
Jesus M. Duran Ramirez, Email: jesus.m.duranramirez@uth.tmc.edu.
Jana Gomez, Email: jana.gomez@uth.tmc.edu.
Jennifer N. Walker, Email: jennifer.n.walker@uth.tmc.edu.
Ender Volkan, Email: evolkan@ciu.edu.tr.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
References
- 1.Davies J., Davies D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roy R., Tiwari M., Donelli G., Tiwari V. Strategies for combating bacterial biofilms: a focus on antibiofilm agents and their mechanisms of action. Virulence. 2018;9:522–554. doi: 10.1080/21505594.2017.1313372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rutherford S.T., Bassler B.L. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraker M., Stewardson A.J., Harbarth S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016;13 doi: 10.1371/journal.pmed.1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pérez M.J., Falqué E., Domínguez H. Antimicrobial action of compounds from marine seaweed. Mar. Drugs. 2016;14:52. doi: 10.3390/md14030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zidorn C. Secondary metabolites of seagrasses (Alismatales and Potamogetonales; Alismatidae): chemical diversity, bio-activity, and ecological function. Phytochemistry. 2016;124:5–28. doi: 10.1016/j.phytochem.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Vassari M., De Biasi A.M., Barletta E., Pretti C., Degl'Innocenti D. An overview of new insights into the benefits of the seagrass Posidonia oceanica for human health. Mar. Drugs. 2021;19:476. doi: 10.3390/md19090476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larkum A.W.D., McComb A.J., Shephard S.A. Biology of seagrasses: a treatise on the biology of seagrasses with special reference to the Australian region. J. Appl. Psychol. 1989;1:105–112. [Google Scholar]
- 9.Benito-González I., et al. In-depth characterization of bioactive extracts from Posidonia oceanica waste biomass. Mar. Drugs. 2019;17:409. doi: 10.3390/md17070409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaal J., et al. Molecular composition of plant parts and sediment organic matter in a Mediterranean seagrass (Posidonia oceanica) mat. Aquat. Bot. 2016;133:50–61. [Google Scholar]
- 11.Punginelli D., et al. New bioactive peptides from the Mediterranean seagrass Posidonia oceanica (L.) Delile and their impact on antimicrobial activity and apoptosis of human cancer cells. Int. J. Mol. Sci. 2023;24:5650. doi: 10.3390/ijms24065650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gokce G., Haznedaroglu M.Z. Evaluation of antidiabetic, antioxidant and vasoprotective effects of Posidonia oceanica extract. J. Ethnopharmacol. 2008;115:122–130. doi: 10.1016/j.jep.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 13.Greenway D.L., Dyke K.G. Mechanism of the inhibitory action of linoleic acid on the growth of Staphylococcus aureus. J. Gen. Microbiol. 1979;115:233–245. doi: 10.1099/00221287-115-1-233. [DOI] [PubMed] [Google Scholar]
- 14.Hammami S., et al. A novel methylated sesquiterpene from seagrasses Posidonia oceanica (L.) Delile. Nat. Prod. Res. 2013;27:1265–1270. doi: 10.1080/14786419.2012.725401. [DOI] [PubMed] [Google Scholar]
- 15.Cox S., Abhu-Ghannam N., Gupta S. An assessment of the antioxidant and antimicrobial activity of six species of edible Irish seaweeds. Int. Food Res. J. 2010;17:205–220. [Google Scholar]
- 16.Rangaiah S.G., Lakshmi P., Manjula E. Antimicrobial activity of seaweeds Gracillaria, Padina and Sargassum sps. on clinical and phytopathogens. Int. J. Chem. Anal. Sci. 2010;1:114–117. [Google Scholar]
- 17.Zgurskaya H.I., Rybenkov V.V. Permeability barriers of Gram-negative pathogens. Ann. N. Y. Acad. Sci. 2019;1459:5–18. doi: 10.1111/nyas.14134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moubayed N.M.S., Al Houri H.J., Al Khulaifi M.M., Al Farraj D.A. Antimicrobial, antioxidant properties and chemical composition of seaweeds collected from Saudi Arabia (Red Sea and Arabian Gulf) Saudi J. Biol. Sci. 2017;24:162–169. doi: 10.1016/j.sjbs.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freitas M.V., et al. Antioxidant and antimicrobial properties of selected red seaweeds from central Portugal. Appl. Sci. 2023;13:157. [Google Scholar]
- 20.Cmiková N., et al. Determination of antioxidant, antimicrobial activity, heavy metals and elements content of seaweed extracts. Plants. 2022;11:1493. doi: 10.3390/plants11111493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fey P.D., et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. Am. Soc. Microbiol. News J. 2013;4 doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramirez J.M.D., Gomez J., Obernuefemann C.L.P., Gualberto N.C., Walker J.N. Semi-quantitative assay to measure urease activity by urinary catheter-associated uropathogens. Front. Cell. Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.859093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez J.M.D., Gomez J., Hanson B.M., Isa T., Myckatyn T.M., Walker J.N. Staphylococcus aureus breast implant infection isolates display recalcitrance to antibiotic pocket irrigants. Microbiol. Spectr. 2023;11 doi: 10.1128/spectrum.02884-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy A.D., et al. Complete nucleotide sequence analysis of plasmids in strains of Staphylococcus aureus clone USA300 reveals a high level of identity among isolates with closely related core genome sequences. J. Clin. Microbiol. 2010;48:4504–4511. doi: 10.1128/JCM.01050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nye T.M., et al. Microbial co-occurrences on catheters from long-term catheterized patients. Nat. Commun. 2024;15:61. doi: 10.1038/s41467-023-44095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh Y.J., Hong J. Application of the MTT-based colorimetric method for evaluating bacterial growth using different solvent systems. Lebensm. Wiss. Technol. 2022;153 [Google Scholar]
- 27.Rodríguez-Lázaro D., et al. Characterization of biofilms formed by foodborne methicillin-resistant Staphylococcus aureus. Front. Microbiol. 2018;9:3004. doi: 10.3389/fmicb.2018.03004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hemaiswarya S., Kruthiventi A.K., Doble M. Synergism between natural products and antibiotics against infectious diseases. Phytomedicine. 2008;15:639–652. doi: 10.1016/j.phymed.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Maheshwari M., et al. Bioactive extracts of Carum copticum L. enhances efficacy of ciprofloxacin against MDR enteric bacteria. Saudi J. Biol. Sci. 2019;26:1848–1855. doi: 10.1016/j.sjbs.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller W.R., Murray B.E., Rice L.B., Arias C.A. Resistance in vancomycin-resistant enterococci. Infect. Dis. Clin. 2020;34:751–771. doi: 10.1016/j.idc.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meena B.R., Chittora D., Meena S., Jain T., Sharma K. Effect of different physical factors on efficacy of Thevetia peruviana leaf extract and bio-formulations. Biochem Biophys Rep. 2022;30 doi: 10.1016/j.bbrep.2022.101271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meena S., Gehlot P., Meena B.R., Jain T., Sharma K. Impact of physical factors on bio-control potential of Lawsonia inermis leaf extract and bio-formulations as fungicides. Biochem Biophys Rep. 2022;32 doi: 10.1016/j.bbrep.2022.101361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naeim H., et al. Antibacterial activity of Centaurea pumilio L. root and aerial part extracts against some multidrug resistant bacteria. BMC Complement. Med. Ther. 2020;20:79. doi: 10.1186/s12906-020-2876-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Begum I.F., Mohankumar R., Jeevan M., Ramani K. GC–MS analysis of bioactive molecules derived from Paracoccus pantotrophus FMR19 and the antimicrobial activity against bacterial pathogens and MDROs. Indian J. Microbiol. 2016;56:426–432. doi: 10.1007/s12088-016-0609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumari N., Menghani E., Mithal R. Bioactive compounds characterization and antibacterial potentials of actinomycetes isolated from rhizospheric soil. J. Sci. Ind. Res. 2019;78:793–798. [Google Scholar]
- 36.Abdali E., Javadi S., Akhgari M., Hosseini S., Dastan D. Chemical composition and biological properties of Satureja avromanica Maroofi. J. Food Sci. Technol. 2017;54:727–734. doi: 10.1007/s13197-017-2512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Padma M., et al. Phytochemical screening and GC–MS analysis of bioactive compounds present in ethanolic leaves extract of Silybum marianum (L) J. Drug Deliv. Therapeut. 2019;9:85–89. [Google Scholar]
- 38.Stopiglia C.D.O., et al. Antimicrobial activity of [2-(methacryloyloxy)ethyl] trimethylammonium chloride against Candida spp. Rev. Iberoam. De. Micol. 2012;29:20–23. doi: 10.1016/j.riam.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Akbar N., Siddiqui R., Iqbal M., Khan A. Antibacterial activities of selected pure compounds isolated from gut bacteria of animals living in polluted environments. Antibiotics. 2020;9:190. doi: 10.3390/antibiotics9040190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao L., et al. In vitro antioxidant and antiproliferative activities of 5-hydroxymethylfurfural. J. Agric. Food Chem. 2013;61:10604–10611. doi: 10.1021/jf403098y. [DOI] [PubMed] [Google Scholar]
- 41.Gidik B. Antioxidant, antimicrobial activities and fatty acid compositions of wild Berberis spp. by different techniques combined with chemometrics (PCA and HCA) Molecules. 2021;76:7448. doi: 10.3390/molecules26247448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Derbalah A.S., Dewir Y.H., El-Sayed A.E.B. Antifungal activity of some plant extracts against sugar beet damping-off caused by Sclerotium rolfsii. Ann. Microbiol. 2012;62:1021–1029. [Google Scholar]
- 43.Ibrahim N.B.B.R., Puchooa D., Govinden-Soulange J., Facknath S. Chemical profiling and biological activity of Cassia abbreviata Oliv. South Afr. J. Bot. 2022;146:325–339. [Google Scholar]
- 44.Ajoke F.L., Kaita H., Ilyas M. Antibacterial activity of 1,2-benzenedicarboxylic acid, dioctyl ester isolated from the ethyl acetate soluble sub-portion of the unripe fruits of Nauclea latifolia. Int. J. Pure. App. Biosci. 2014;2:223–230. [Google Scholar]
- 45.Hernández-Ceja A., et al. In vitro antifungal activity of plant extracts on pathogenic fungi of blueberry (Vaccinium sp.) Plants. 2021;10:852. doi: 10.3390/plants10050852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berwal K.M., et al. The bioactive compounds and fatty acid profile of bitter apple seed oil obtained in hot, arid environments. Horticulturae. 2022;8:259. [Google Scholar]
- 47.Syazana M.S.N., Halim A.S., Gan S.H., Shamsuddin S. Antiproliferative effect of methanolic extraction of tualang honey on human keloid fibroblasts. BMC Compl. Alternative Med. 2011;11:82. doi: 10.1186/1472-6882-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rouis-Soussi L.S., et al. Chemical composition and antibacterial activity of essential oils from the Tunisian Allium nigrum L. EXCLI J. 2014;13:526–535. [PMC free article] [PubMed] [Google Scholar]
- 49.Chai W.M., et al. Antityrosinase and antimicrobial activities of furfuryl alcohol, furfural and furoic acid. Int. J. Biol. Macromol. 2013;57:151–155. doi: 10.1016/j.ijbiomac.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 50.Kucuk H.B., Yusufoglu A., Matarci E., Dosler S. Synthesis and biological activity of new 1,3-dioxolanes as potential antibacterial and antifungal compounds. Molecules. 2011;16:6806–6815. doi: 10.3390/molecules16086806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bojarska J., et al. A supramolecular approach to structure-based design with a focus on synthons hierarchy in ornithine-derived ligands: review, synthesis, experimental and in silico studies. Molecules. 2020;25:1135. doi: 10.3390/molecules25051135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abubakar A.R., Haque M. Preparation of medicinal plants: basic extraction and fractionation procedures for experimental purposes. J. Pharm. BioAllied Sci. 2020;12:1–10. doi: 10.4103/jpbs.JPBS_175_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wikler M.A. vol. 26. CLSI; Wayne, Pennsylvania: 2006. pp. M7–A7. (Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically: Approved Standard). [Google Scholar]
- 54.Clinical Laboratory Standard Institute (CLSI) 33rd ed. 2023. Performance Standards for Antimicrobial Susceptibility Testing. Wayne, Pennsylvania. [Google Scholar]
- 55.Clinical Laboratory Standard Institute (CLSI) CLSI supplement M100. Wayne, Pennsylvania. 2023 [Google Scholar]
- 56.Heuser E., Becker K., Idelevich E.A. Bactericidal activity of sodium bituminosulfonate against Staphylococcus aureus. Antibiotics. 2022;11:896. doi: 10.3390/antibiotics11070896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bellio P., Fagnani L., Nazzicone L., Celenza G. New and simplified method for drug combination studies by checkerboard assay. MethodsX. 2021;8 doi: 10.1016/j.mex.2021.101543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haney E.F., Trimble J., Hancock R.E.W. Microtiter plate assays to assess antibiofilm activity against bacteria. Nat. Protoc. 2021;16:2615–2632. doi: 10.1038/s41596-021-00515-3. [DOI] [PubMed] [Google Scholar]
- 59.Isbilen O., Volkan E. Allium willeanum Holmboe exerts anticancer activities on metastatic breast cancer cells MCF-7 and MDA-MB-231. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stegger M., et al. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA (LGA251) Clin. Microbiol. Infect. 2012;18:395–400. doi: 10.1111/j.1469-0691.2011.03715.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this study are available from the corresponding author upon reasonable request.