Abstract
MZT2A is a novel core component in the γ-tubulin ring complex and aberrantly expressed in some types of tumors. However, MZT2A expression pattern across different cancers and its role in kidney renal clear cell carcinoma have not been sufficiently investigated. A thorough analysis of MZT2A expression landscape across 33 cancer types was conducted, utilizing 712 normal samples and 9807 tumor samples from TCGA (version 37.0), as well as 5112 normal samples from the GTEx databases. MZT2A's impact on KIRC cell viability and proliferation were evaluated through BrdU assays and monitored by cell imaging readers in MZT2A-expressing plasmid or siRNA-transfected cells. Additionally, the effects of MZT2A on cell apoptosis and cell cycle were detected using flow cytometry and Western blot. WGCNA analysis was employed to construct a co-expression gene network associated with MZT2A expression in KIRC, and Pearson correlation coefficient was utilized to examine the relationships between pairs of genes. MZT2A is overexpressed in 25 out of 33 types of cancer, including KIRC. In KIRC, high level of MZT2A was associated with higher clinical stage progression and poorer patients' survival. Downregulation of MZT2A decreased KIRC cell proliferation, retarded cell cycle and promoted apoptosis, while upregulation of MZT2A produced the opposite effects. WGCNA analysis and in vitro experiments revealed that MZT2A activated PI3K/AKT signaling pathway in KIRC. In all, MZT2A was overexpressed in most types of tumors. MZT2A served as an oncogene in KIRC and might be a potential target for guiding future treatments.
Keywords: MZT2A, Prognostic biomarker, Renal cell carcinoma, Proliferation, Apoptosis
1. Introduction
Cancer is a major contributor of morbidity and mortality globally, posing a substantial health and economic challenges to society [1]. In recent decades, medical techniques for treating tumors have been continuously developed and have achieved specific clinical successes [2]. However, most patients suffered with malignant tumors were diagnosed at the advanced stages, for whom the application and the effectiveness of radical resection and other routine regimens such as chemotherapy and radiotherapy were limited. Besides, for cancer patients underwent chemotherapy, radiotherapy, targeted therapy or immunotherapy, the occurrence of resistance or insensitivity and the side effects heavily restricted the therapeutic benefits. Researchers increasingly focused on developing novel immunotherapy or targeted therapy strategies based on the identification and verification of key targets. It is therefore urgent to mine novel, sensitive and reliable biomarkers with a view to therapeutic purposes.
The mitotic spindle organizing protein 2A(MZT2A) gene, alternatively referred to FAM128A, GCP8, or MOZART2A, is located at 2q21.1. The MZT2A protein consists of 158 amino acids and was first identified by Hutchins et al., in 2010 [3]. MZT2A is a cell separation-associated protein, acting as a new core subunit in the gamma-tubulin ring complex, facilitating microtubule polymerization [[4], [5], [6]]. Neus's study suggested that MZT2A depletion could disrupt γTuRC recruitment and microtubule nucleation, but didn't affect the centrosomes' overall structure [7]. The role of MZT2A has not been extensively studied in the context of cancer, although it has been mentioned in several articles [[8], [9], [10]]. Only a few studies reported a functional correlation between MZT2A and tumorigenesis in lung cancer [11,12] and gastric cancer [13]. The published studies showed that MZT2A was abnormally expressed and exerted oncogene function in lung cancer and gastric cancer, promising a potential biomarker and therapeutic target in cancers. Nevertheless, its expression patterns in many other types of cancers remain to be studied. Pan-cancer analysis is a cost-effective way to analyze the clinical significance of MZT2A in cancers. In this work, we aim to investigate MZT2A expression across different cancers, and explore the bio-function and mechanism of MZT2A in KIRC to identify the potentiality of MZT2A as a new bio-marker for KIRC.
Here, MZT2A expression pattern in malignancies was examined by analyzing data from the TCGA and GTEx databases, the role and the associated molecular pathway of MZT2A were investigated using bio-informatic tools and in vitro experiments, and MZT2A was found as a potential oncogene functioning via PI3K-Akt pathway in KIRC, and might be a new therapeutic target for KIRC treatment.
2. Materials and methods
2.1. Analysis of gene expression
To evaluate MZT2A mRNA levels, we initially mapped the gene expression across distinct cancer cell lines using the Cancer Cell Line Encyclopedia (https://sites.broadinstitute.org/ccle/) [14]. The combined data of TCGA pan-cancer samples (version 37.0) and the GTEx resource were obtained via UCSC Xena (https://xena.ucsc.edu) to explore the differential expression of MZT2A in cancers [15]. Using R software (version 3.6.4), the original data was pre-processed into log2 (TPM + 1), and the ggpubr package was used for statistical analysis. The boxplots were drawn by ggplot2 package. The expression levels of MZT2A between tumor and normal tissues were compared using the Student's t-test. The MZT2A expressions in human tissues were conducted using the Human Protein Altas (https://www.proteinatlas.org/) [16].
2.2. Analysis of survival prognosis
The relationship between MZT2A expression and patient prognosis in various types of cancers or specific tumor subtypes was investigated using the R software package (version 3.6.4). A univariate COX regression model and Kaplan-Meier Plotter were employed to investigate the correlation between MZT2A expression and prognosis [17]. For univariate COX regression model, the R package survival (version 3.6.4) was employed to examine the association between MZT2A expression and patients’ prognosis. The Log-rank test was employed as the statistical procedure to determine prognostic significance. The best cut-off points for Kaplan-Meier Plotter were confirmed by using the max-stat function in the R package to divide patients into two groups (high-MZT2A and low-MZT2A). Subsequently, the survfit function was utilized to examine the prognostic difference between two groups, with the significance of this difference evaluated using the Log-rank test. Overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI) were evaluated, and a P-value less than 0.05 was considered statistically significant [18].
2.3. Analysis of clinical phenotypes
To delve deeper into the correlation between MZT2A expression and pathological stage, data from tumor cases and healthy controls were obtained from the UCSC Xena. The MZT2A expression levels across different stages were visualized, and the association between differential MZT2A expression and clinicopathological progression was evaluated using Student's t-test or Kruskal Wallis test [19].
2.4. Cell culture
The study utilized human kidney renal clear cell carcinoma (KIRC) cell lines from the Cell Repository of the Shanghai Institute of Life Sciences, Chinese Academy of Sciences. The cell lines HK-2, 786-O, and CAKI-1 were cultured in DMEM medium, RPMI 1640 medium or McCoy's 5A (Gibco BRL, NY, USA) supplemented with 10 % fetal bovine serum, respectively. They were maintained at 37 °C in a humidified environment with 5 % CO2. The cells in good condition were employed for further experiments.
2.5. Immunocytochemistry
786-O and CAKI-1 cells were planted in 24-well plates and then cultivated for two days after transfection. Following this, cells were washed three times using pre-cooled PBS buffer, fixed with 500 μL of 4 % paraformaldehyde at room temperature for 0.5 h. Then, 500 μL PBS-T was added to permeabilize for 10 min and washed with PBS solution. Following a 1.5 h blocking period with 500 μL PBS-B, 200 μL primary antibody was added and incubated at 4 °C overnight. The next day, cells were treated for 2 h in the dark with 200 μL fluorescent-dye conjugated secondary antibody. Finally, it was stained with 100 μL DAPI solution for 10 min, mounted with fluorescent mounting medium, and imaged using a confocal microscope (Leica, Germany).
2.6. RNA extraction and quantitative real-time reverse transcription PCR (RT-qPCR)
After being extracted from cell lines using Trizol RNA isolation reagent (Invitrogen, Eugene, OR, USA), Total RNA was reversely transcribed into cDNA by a revert aid first strand cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA). Gene expression was assessed by RT-qPCR using SYBR Premix Ex Taq (Takara, Shiga, Japan) on a CFX96 real-time PCR system. The primer sequences were as follows: GAPDH-F: GGTGAAGGTCGGTGTGAACG, GAPDH-R: CTCGCTCCTGGAAGATGGTG; MZT2A-F: GACCCCGACGTGTTCAAGAT, MZT2A-R: ATGTAGCCTTTGCTGGGGAC.
2.7. Cell lysis, protein extraction, and Western blot analysis
After being washed three times with PBS buffer, the cells were lysed with RIPA buffer and transferred to 1.5 mL centrifuge tubes. Following a 30 min lysis period, the samples were centrifugated for 20 min at 12,000 rpm at 4 °C. Next, BCA protein detection kit (ThermoFisher Scientific, Waltham, USA) was used to quantify the supernatants. Furthermore, proteins were separated on suitably concentrated SDS-PAGE gels, which were then transferred to the PVDF membrane at constant pressure, and blocked with skim milk (5 %) for 2 h. After cutting the membrane based on the protein molecular weight, the blots were incubated with MZT2A (1:1000, Santa Cruz Biotechnology, sc-514516), Bax (1:5000, Proteintech Group, 50599-2-Ig), Bcl-xL (1:1000, Cell Signaling Technology, 2764S), CDK4(1:1000, Proteintech Group, 11026-1-AP), CDK6(1:1000, Proteintech Group, 14052-1-AP), Cyclin D1 (1:1000, Proteintech Group, 60186-1-Ig), AKT(1:1000, Proteintech Group, 60203-2-Ig), p-AKT(Ser473) (1:1000, Cell Signaling Technology, 4060S), PI3 Kinase p85 alpha Antibody (1:1000, Abmart, T40115), p-PI3-kinase p85-alpha/gamma(Tyr467/199) (1:1000, Abmart, T40116), and β-actin(1:5000, Proteintech Group, 66009-1-Ig) antibodies at 4 °C overnight. Subsequently, after washing 30 min with TBST, the membranes were incubated for 1 h with appropriate secondary antibodies (Dako, Glostrup, Denmark) at room temperature. Finally, membranes were washed with TBST and visualized using the Syngene GBox (Syngene Europe).
2.8. Transfection
The siRNA and overexpression plasmid (overMZT2A) were transfected into cells using Liposome 2000 transfection reagent (Invitrogen, Eugene, OR, USA) according to the manufacturer's instructions. The siRNA sequences were as follows: siRNA-1: TCCAGATGCTCAAGTCCAT; siRNA-2: GATGCTCAAGTCCATGTGT; siCtrl: ACGUGACACGUUCGGAGAATT.
2.9. Cell proliferation assay
786-O and CAKI-1 cells were implanted into 96-well plates at a density of 5 × 103 cells per well and then cultured for 1 day. Afterwards, cells were treated with siRNA or plasmid and incubated for 4 h before medium replacement. Cell proliferation was monitored at 6 h, 30 h, 54 h and 78 h using a Cytation™5 Cell Imaging Multi-Mode Reader (Biotech, Winooski, VT, USA). The Gen5 Image program (Version 3.08) was used to count suspension cells independently.
2.10. BrdU assays
5 × 104 cells were planted into each well of 48-well plates, and the cells were cultivated for 2 days after transfection. First, 3 μg/mL BrdU was combined into medium and incubated for 2 h at 37 °C. Then, following being fixed for 30 min with 4 % paraformaldehyde, the cells were treated at 37 °C for 30 min with 2 M HCl, and neutralized with 0.1 M sodium tetraborate. Next, cells were exposed to 40 μL anti-BrdU antibody (1:500, Santa Cruz Biotechnology, USA) at 4 °C for a whole night. The next day, after three washings with PBS, the cells were incubated with Alexa Fluor@ 594 AffiniPure Donkey Anti-Mouse secondary antibodies (1:500, Jackson ImmunoResearch, USA) at room temperature for 1 h. Ultimately, DAPI dihydrochloride (Solarbio, China) was used to stain the nucleus, and cells were observed using a fluorescence microscope (Leica, Germany).
2.11. Cell apoptosis assay
The cells were collected after plasmids or siRNAs transfection for 2 days. Trypsin was used to digest the cells after washing with PBS buffer and placed into 15 mL tubes. The cells were harvested by centrifugation at 1,000 rpm for 5 min, and then washed twice with PBS. Following staining with Annexin-V-FITC apoptosis detection reagent as per the manufacturer's recommendations, apoptosis levels were measured and analyzed by flow cytometry (NovoCyte, ACEA Biosciences, USA).
2.12. Cell cycle assay
786-O and CAKI-1 cells were treated with plasmids or siRNAs, and cell pellet was collected by centrifugation as described above. Then, an equal volume of RNase and PI dyes were added to stain the cells according to the instructions, followed by incubation in dark for 15 min. After that, flow cytometry (NovoCyte, ACEA Biosciences, USA) was used to examine the cell cycle profile.
2.13. MZT2A-related gene enrichment analysis
Weighted gene co-expression network analysis (WGCNA) is a commonly used bioinformatics analytic method to find modules and genes with comparable expression patterns, thereby revealing potential regulatory relationships [20]. By clustering comparable genes and eliminating important regulatory genes, the R package was utilized to identify MZT2A-related genes that were differently expressed in KIRC. Firstly, the samples were clustered to remove apparent outliers, and the correlation coefficients between MZT2A and the KIRC FPKM expression matrix were analyzed using Spearman correlation analysis, and a similarity matrix was constructed based on these correlations. Furthermore, the similarity matrix was converted to the adjacency matrix based on an optimal soft threshold power (β). Additionally, using TOM as the input of gene hierarchical clustering analysis, hierarchical clustering and clustering tree were generated to find the feature genes of MZT2A module. Furthermore, the SangerBox tool (http://www.sangerbox.com/tool) was used to visualize the unique biological characteristics of genes in modules of interest and identify their functional attributes [21].
2.14. Statistical analysis
The bioinformatics data were statistically analyzed using R software with a P value less than 0.05 being statistically significant. Prism GraphPad 7.0 software (GraphPad Software Inc., CA, USA) was used to analyze experimental data, and Student's t-test or one-way ANOVA were used to evaluate group differences. All experiments were presented as mean ± SD from at least 3 separate experiments.
3. Results
3.1. MZT2A is aberrantly expressed in pan-cancer
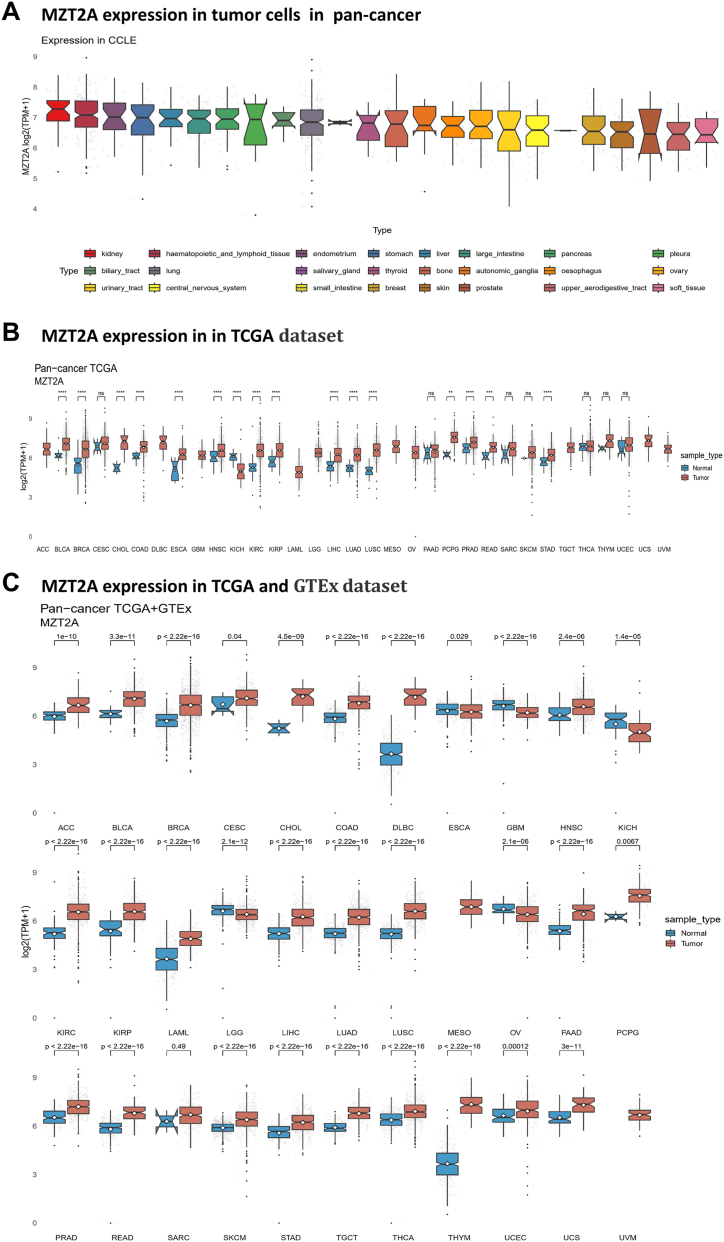
We first examined the mRNA expression level of MZT2A in various cancer cell lines using CCLE database. MZT2A expression levels in 24 tumors (1739 samples) were displayed, indicating that MZT2A expression was generally elevated in cancer cell lines. MZT2A was significantly expressed in kidney tumor cell lines, followed by hematopoietic and lymphoid tissue, endometrium, and stomach cell lines (Fig. 1A). MZT2A expression in each specific tumor cell line was listed in Table S1. Then, the pan-cancer expression pattern of MZT2A was depicted to evaluate differential expression of MZT2A across TCGA cancer types, and the samples size of each cancer type was as follows: ACC (normal 128, tumor 77), BLCA (normal 28, tumor 407), BRCA(normal 292, tumor 1099), CESC(normal 13, tumor 306), CHOL(normal 9, tumor 36), COAD(normal 349, tumor 290), DLBC(normal 337, tumor 47), ESCA(normal 286, tumor 182), GBM(normal 212, tumor 166), HNSC(normal 44, tumor 520), KICH(normal 53, tumor 66), KIRP(normal 100, tumor 531), LAML(normal 337, tumor 173), LGG(normal 207, tumor 523), LIHC(normal 160, tumor 371), LUAD(normal 347, tumor 515), LUSC(normal 338, tumor 498), MESO(tumor 87), OV(normal 88, tumor 427), PAAD(normal 171, tumor 179), PCPG(normal 3, tumor 182), PRAD(normal 152, tumor 496), READ(normal 318, tumor 93), SARC(normal 2, tumor 262), SKCM(normal 558, tumor 469), STAD(normal 211, tumor 414), TGCT(normal 165, tumor 154), THCA(normal 338, tumor 512), THYM(normal 339, tumor 119),UCEC(normal 101, tumor 181),UCS(normal 78, tumor 57), and UVM(tumor 79). Most types of cancers showed upregulated expression levels of MZT2A compared to their corresponding adjacent tissues. Cancers with the most significant increased expression of MZT2A were observed in BRCA (P = 4.28e-32), followed by KIRC (P = 1.02e-29) and LUSC (P = 1.37e-28). In 15 out of 33 tumor types, MZT2A was found to be overexpressed in tumor tissues compared to controls, but downregulated in KICH in comparison to the matched healthy tissues (Fig. 1B). Furthermore, we used the combination from the GTEx and TCGA database to analyze MZT2A expression between tumor tissues and normal controls, and the results showed MZT2A was consistently overexpressed in 25 out of 33 tumor types (Fig. 1C). In 5 types of cancer, MZT2A was significantly downregulated in tumor tissues. Interestingly, the expression of MZT2A was consistently increased in both kidney renal papillary cell carcinoma and kidney renal clear cell carcinoma, and the P value was much less than 0.001, showing a similar expression pattern for kidney cancer (Fig. 1C). In all, we discovered that the MZT2A gene is extensively dysregulated in tumors in contrast to normal tissues.
Fig. 1.
Expression profile of MZT2A in pan-cancer. (A) MZT2A mRNA expression in tumor cells in pan-cancer. (B) MZT2A's pan-cancer expression profile based on data from the TCGA database. (C) Analysis of MZT2A expression in 33 cancers using combined information from the TCGA and GTEx datasets. (*P < 0.05, **P < 0.01, ***P < 0.001).
3.2. Prognostic analysis of MZT2A expression in pan-cancer
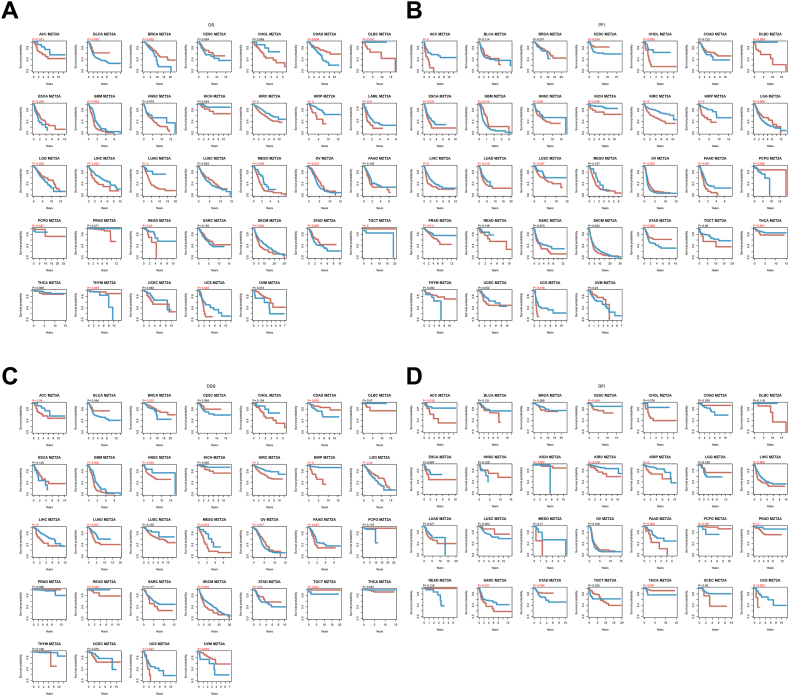
Cox univariate analysis and Kaplan-Meier plotting were employed to explore the potential prognostic value of MZT2A across different cancer types. In the OS analysis, patients with KIRP, LUAD, SKCM, KIRC, KICH, and LAML presented high expression of MZT2A based on Cox regression model analysis (Fig. S1A). Further study found significant associations between MZT2A expression and PFI in several cancer, such as KIRC, KIRP, ACC, and LIHC (Fig. S1B). Moreover, increased MZT2A expression was associated with poorer DFI in patients with LIHC, KIRP, PRAD, and ACC (Fig. S1C). Besides, greater MZT2A expression was correlated with lower DSS in KIRP, KIRC, LUAD, KICH, and SKCM (Fig. S1D). Furthermore, the predictive significance of MZT2A was explored by Kaplan-Meier plotter used for survival analysis of MZT2A in pan-cancer. All P-values and optimal cut-off points data were shown in Table S2. The findings demonstrated that the expression of MZT2A was positively correlated with 8 cancers types and negatively correlated with 14 cancers types with P value less than 0.05(Fig. 2A). Further study indicated a strong correlation between MZT2A expression and PFI in KIRP, KIRC, LIHC, ACC, PAAD, and others (Fig. 2B). The investigation of the correlation between MZT2A expression and DSS using Kaplan-Meier survival curves revealed that the poorer outcomes in 12 tumor types were related to the higher MZT2A expression, including KIRP (P = 3.78e-11), KIRC (P = 3e-06), LIHC (P = 2.81e-04), and LUAD (P = 5.26e-04) (Fig. 2C). In addition, increased MZT2A expression resulted in lower DFI in PRAD, KIRP, LIHC, PAAD, ACC, KIRC, UCS, SARC and CESC (Fig. 2D). All the presented data indicated that high MZT2A expression represented a poorer prognosis in different cancers.
Fig. 2.
Survival analysis of MZT2A in pan-cancer. Kaplan-Meier survival analysis depicting the association of MZT2A expression with OS (A), PFI (B), DSS (C), DFI (D).
3.3. Pan-cancer analysis of the correlation between MZT2A expression and Clinicopathology
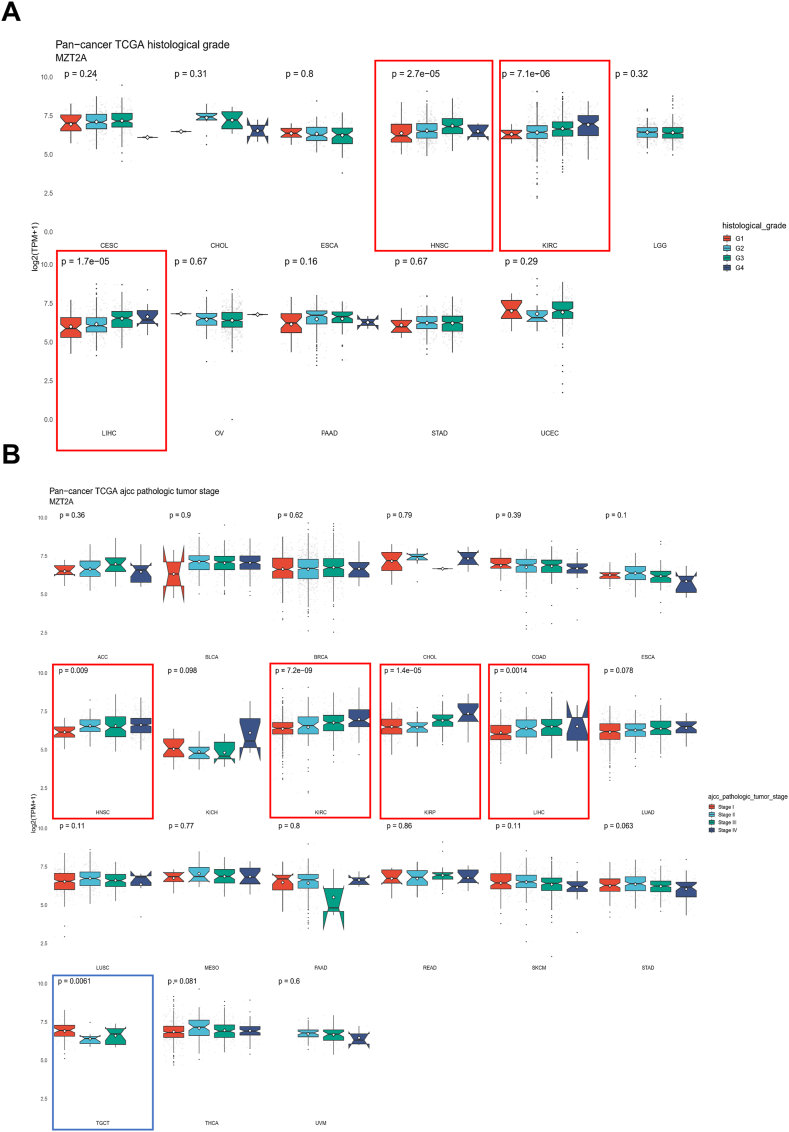
MZT2A expressions in patients with different grades and stages of cancers were compared to evaluate the relationship between MZT2A expression and clinicopathological features in multiple malignancies. The results revealed that MZT2A expression gradually increased in KIRC (P = 7.1e−06), LIHC (P = 1.7e−05) and HNSC (P = 2.7e−05) with the deterioration of tumor cell differentiation (Fig. 3A). Furthermore, the expression of MZT2A at distinct stages was significantly upregulated in KIRC, KIRP, LIHC and HNSC, while downregulated in TGCT (Fig. 3B). In addition, the correlation analysis of MZT2A with age or gender showed that MZT2A had a positive correlation with age in KIPAN, HNSC, THCA, SKCM and KICH, while had a negative association with age in SARC and LUSC. And the correlation analysis showed MZT2A was associated with gender in BRCA, SARC, THCA, and HNSC (Figs. S2A–2B). These results indicated that MZT2A expression was related to the progression of some malignancies, such as KIRC and LIHC.
Fig. 3.
Expression levels of MZT2A in different pathological grades or tumor stages. (A) Relationship between the histological grade and MZT2A expression of 11 tumors. (B) Correlation between tumor stage and MZT2A expression in 21 malignancies. Log2 (TPM+1) was used for logarithmic scaling. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
3.4. Expression analysis of MZT2A in KIRC
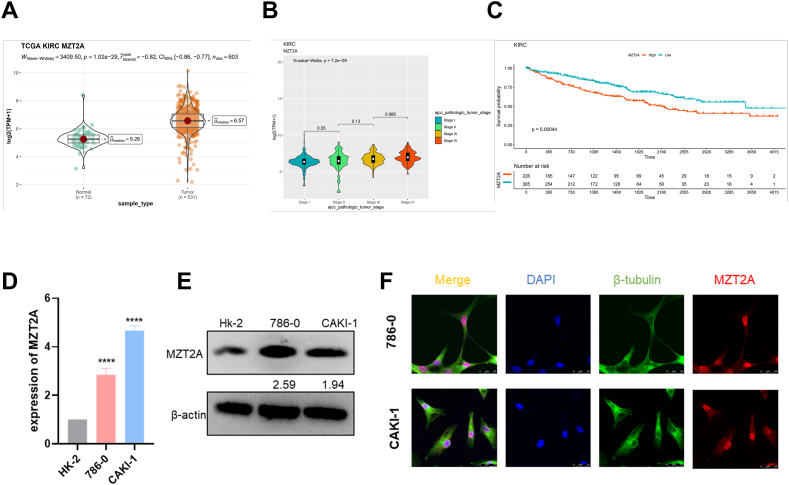
Since the expression of MZT2A was correlated with the clinical stage, histological grade, and survival prognosis of multiple tumors, and its role in KIRC has not been revealed, MZT2A was then studied in KIRC. The expression levels of MZT2A in KIRC were significantly elevated in comparison to normal tissues (P = 1.02e-29) (Fig. 4A). MZT2A was elevated with the progress of the clinical stage of KIRC, and the elevation of MZT2A was related to a worse OS (Fig. 4B–C). To further define the relationship between MZT2A and kidney cancer, the function of MZT2A in KIRC cell lines was examined. RT-qPCR and Western blot showed the expression of MZT2A in 786-O and CAKI-1 cells was higher than normal renal cell line HK-2 (Fig. 4D–E). An immunofluorescent staining and confocal microscopy observation were used to clarify the location of MZT2A in the cells, and the findings showed that MZT2A was mostly located in the nucleus, with a minor quantity also in the cytoplasm (Fig. 4F). In summary, based on bioinformatics analysis and relevant experiments, we found that MZT2A was substantially expressed in KIRC and was linked to a poor prognosis.
Fig. 4.
The expression of MZT2A in KIRC. (A) MZT2A expression levels in KIRC tissues (n = 531) vs. normal tissues (n = 72) according to the TCGA database. (B, C) Clinical stage and survival prognosis analysis of MZT2A in KIRC. (D, E) MZT2A mRNA and protein expression levels of 786-O and CAKI-1 cells in comparison to HK-2 cells. All uncropped images were presented in Fig. S5. (F) Immunofluorescence staining exhibited nuclear localization of MZT2A in cells.
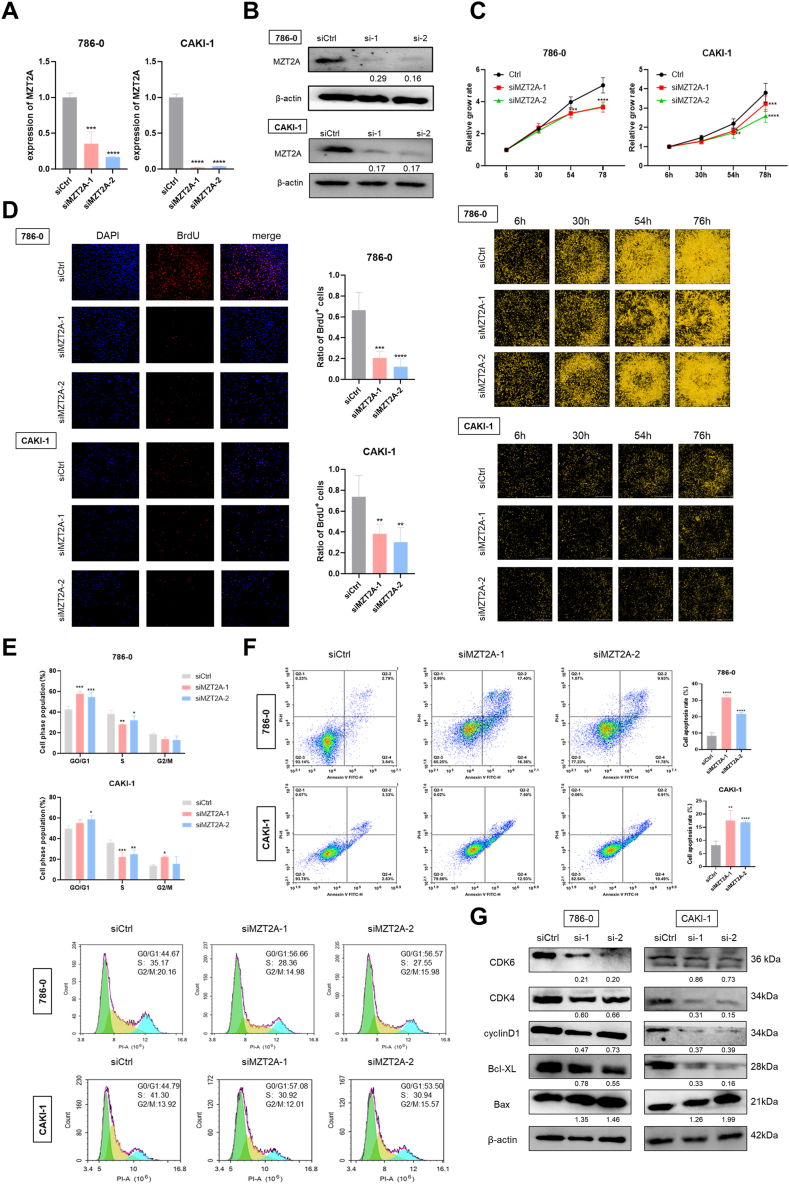
3.5. Knockdown of MZT2A promotes apoptosis and influences the cell cycle of kidney cancer cell lines
Two specific siRNAs targeting MZT2A were designed and transfected into cancer cells, resulting in downregulation of MZT2A at both RNA and protein levels (Fig. 5A–B). Cell proliferation monitoring assay and BrdU assay revealed that knocking down of MZT2A markedly decreased the rate of cell growth and BrdU incorporation (Fig. 5C–D). Additionally, cell cycle measured by flow cytometry showed that downregulation of MZT2A resulted in a rise of cell proportion in the G0/G1 phase and a decrease in the S phase (Fig. 5E). To further define that MZT2A could regulate cell apoptosis in renal carcinoma cells, cells were stained with Annexin V and FITC and analyzed by flow cytometry. The results demonstrated that downregulation of MZT2A could significantly promote 786-O and CAKI-1 cell apoptosis (Fig. 5F). The expression levels of proteins associated with cell cycle and apoptosis were also changed in kidney cancer cells, as illustrated by Western blot results that CDK4, CDK6, cyclin D1 and Bcl-XL were reduced, while Bax was increased (Fig. 5G). In summary, MZT2A promoted the malignant phenotypes of KIRC cells.
Fig. 5.
The effect of MZT2A downregulation onbiological functions ofkidney cancer cells. (A, B) The mRNA and protein levels of MZT2A in kidney cancer cells transfected with si-MZT2A. (C) Cell numbers counted by cell imaging readers. (D) Cell proliferation evaluated by BrdU incorporation assay. Detection of cell cycle (E) and cell apoptosis (F) using flow cytometry. (G) The expression of cell apoptosis and cell cycle markers detected by Western blot. Uncropped images were provided in Fig. S5. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
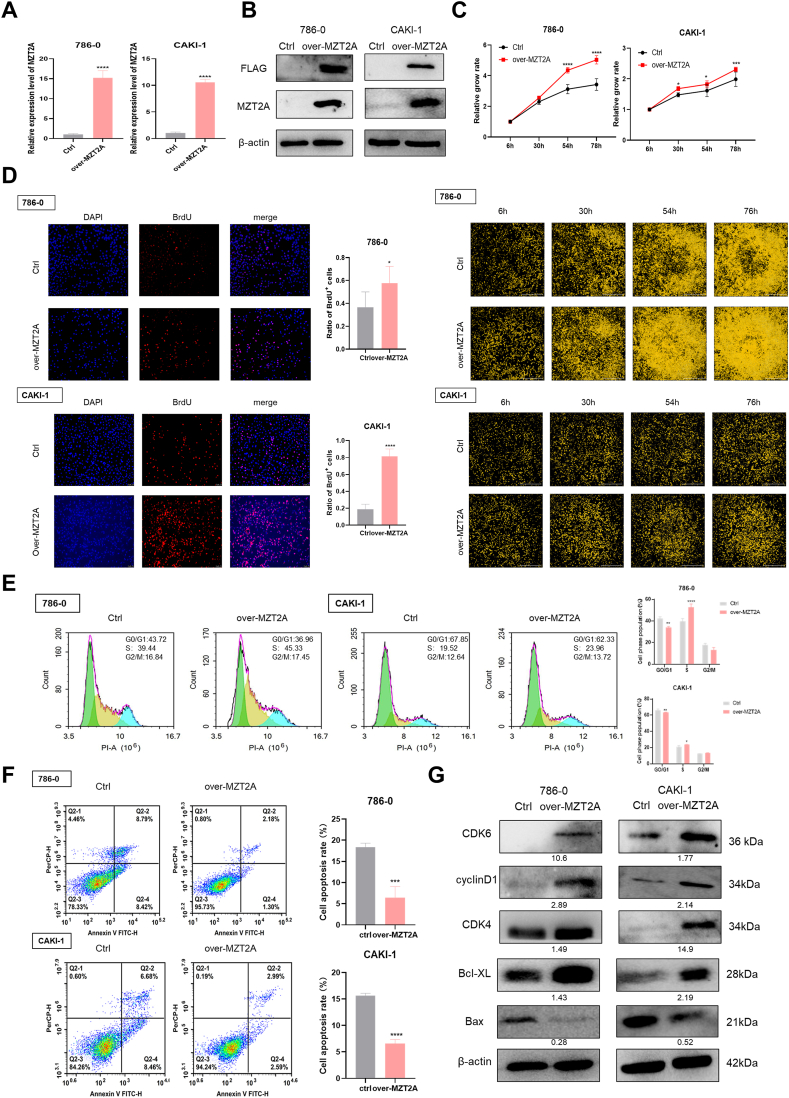
3.6. Overexpression of MZT2A enhances proliferation and reduces apoptosis of kidney cancer cells
The overexpression plasmid (overMZT2A) was transfected into 786-O and CAKI-1 in order to investigate the function of MZT2A in KIRC. Both RNA and protein levels of MZT2A were increased in MZT2A-expressing plasmid-transfected cells (Fig. 6A–B). Cell proliferation monitoring and BrdU incorporation assays confirmed MZT2A overexpression could promote proliferation compared to control groups (Fig. 6C–D). Flow cytometry assay displayed that overexpression of MZT2A accelerated the G1 to S phase transition, resulting a greater percentage of cells aggregating in the S phase (Fig. 6E). Furthermore, the results of cell apoptosis assay showed less apoptosis occurred in MZT2A-overexpressed cells compared with control groups, suggesting that MZT2A inhibited cell apoptosis (Fig. 6F). Western blot analysis revealed that increasing MZT2A enhanced the expression levels of CDK6, CDK4, cyclin D1, and Bcl-XL while decreased Bax (Fig. 6G). Moreover, neither overexpression nor inhibition of MZT2A affected cell proliferation, viability, cell cycle or apoptosis in normal kidney cells (Fig. S3). Collectively, these findings suggested that MZT2A played a pro-cancer role in KIRC.
Fig. 6.
Functional effects of MZT2A overexpression in Kidney Cancer Cell cells. (A, B) Relative mRNA and protein expression of overMZT2A-transfected kidney cancer cells. (C) Cell proliferation monitoring images. (D) BrdU incorporation assay showed increase of cell proliferation in MZT2A-upregulated KIRC cells. Flow cytometry analysis of Cell cycle (E) and cell apoptosis (F). (G) Western blot analysis of CDK6, CDK4, cyclinD1, Bax, and Bcl-XL expression in MZT2A-overexpressing cells. The original images were shown in Fig. S6. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
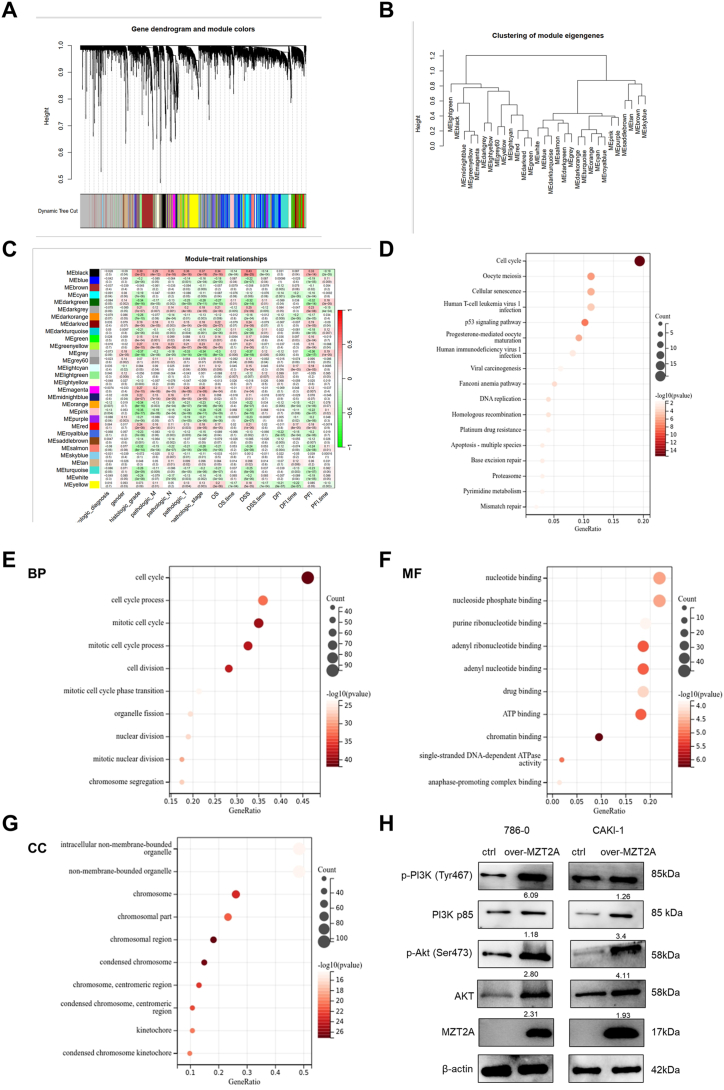
3.7. Prediction of MZT2A downstream genes and their functions roles
As MZT2A overexpression significantly changed cell apoptosis and cell cycle in kidney cancer cells, MZT2A-associated molecules needed to be further analyzed. All genes related to the MZT2A KIRC FPKM expression matrix were obtained and analyzed by WGCNA to investigate the downstream genes of MZT2A and predict the possible mechanism. In the WGCNA network, after removing the obvious outliers, the correlation cluster tree was drawn according to the selected samples (Figs. S4A–4B, Table S3). After determining the soft threshold power to be 11, the fitting index of the scale-free topology model reached 0.9, and the average connectivity approached 0, suggesting a high level of average connectivity (Fig. S4C). Subsequently, adjacency and topological overlap matrices were constructed based on this optimal soft threshold (Fig. S4D), and the genes were clustered and divided into 30 different modules with assigning different colors (Fig. 7A–B). Furthermore, a phenotypic and gene correlation heat map was obtained. Meblack and MEgrey appeared to be associated with tumor pathology (Fig. 7C). As the most positively significant area, 227 genes in the MEblack module were further analyzed by Sangerbox Tools. KEGG analysis of the MEblack module's gene revealed MZT2A was enriched with cell cycle, cellular senescence, DNA replication and mismatch repair with P value < 0.05(Fig. 7D). As for biological process (BP), the genes predominantly participated in regulating cell cycle, cell division, and organelle fission (Fig. 7E). In view of molecular function (MF), the genes were prominently associated with chromatin binding, adenyl ribonucleotide binding, and ATP binding (Fig. 7F). Regarding to cellular component (CC), chromosome, centromeric region and kinetochore were mainly enriched (Fig. 7G). After screening several signal transduction molecules, AKT signaling pathway appeared to be involved in the influences of MZT2A on cell cycle, apoptosis, and proliferation in KIRC. The results showed that significant up-regulation of PI3K p85 and AKT expression in MZT2A-overexpressed cells, and the level of phosphorylated‐AKT and phosphorylated‐PI3K p85 protein had the same effect, suggesting that MZT2A might regulate cell malignant behaviors through PI3K/AKT signaling pathway(Fig. 7H). To sum up, these data suggested that MZT2A promoted the malignant behaviors of KIRC via the PI3K/AKT signaling pathway.
Fig. 7.
WGANA analysis and Gene prediction related to MZT2A. (A) Gene dendrogram and module colors. (B) Clustering of module eigengenes showing the relationship between gene modules. (C) Relationships between module eigengenes and clinical traits in KIRC. (D) KEGG enrichment analysis of MEblack module's genes. Gene Ontology analysis was conducted for MEblack module's genes, focusing on biological processes (E), molecular functions (F), and cellular components (G). (H) Western blot analysis showing protein levels of PI3K p85, AKT, and phosphorylated proteins. Uncropped images were provided in Fig. S7. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
MZT2A is a cell separation-associated protein that may be crucial in cancer progression [22]. MZT2A is required for interphase microtubule organization. Loss of MZT2A impaired microtubule nucleation and retarded cell cycle progression. Since microtubule-targeting therapeutics have been tested effective in cancers, the overexpressed MZT2A in tumors might be a promising target for cancer treatment [11,13]. But its precise molecular role in tumorigenesis, metastasis, and recurrence of various cancers is still unknown. This study examined the expression of MZT2A in pan-cancers and investigated the correlation between MZT2A expression and the survival prognosis or clinicopathological progression. This was the first comprehensive analysis of MZT2A's differential expression at the pan-cancer level. The findings illustrated that MZT2A was abnormally overexpressed in most cancers, especially in KIRC, and the higher level of MZT2A related to the poorer prognosis across multiple tumors. MZT2A therefore emerged as a potential target gene across diverse cancers.
We initially analyzed the expression landscape of MZT2A across different cancer tissues or cell lines in TCGA database. Results confirmed that MZT2A expressed in multiple types of tumors, and its expression varied depending on the tumor types. Based on the analysis of bioinformatics data, our study discovered that MZT2A was upregulated in LUSC, KIRC, and 23 other common cancers, while downregulated in 5 cancers, including ESCA, OV, and others. The previous study of MZT2A in NSCLC showed that the mRNA and protein levels of MZT2A were upregulated, which was consistent with our analysis [11]. Then, the correlation analysis of MZT2A with survival prognosis, pathological stage, and histological grade were used to evaluate the predictive value of MZT2A. According to overall survival analysis, the expression of MZT2A was positively connected with the prognosis of 8 tumors, but adversely correlated with the prognosis of 14 cancers, including KIRP, LUAD, KIRC, GBM, and others. Similarly, expression of MZT2A was demonstrated associated with DSS, PFI, and DFI that MZT2A expression was generally adversely correlated with KIRC, KIRP, PRAD, LIHC, PAAD, ACC, UCS, and CESC, while positively correlated with PCPG and STAD. According to the histological grade analysis of KIRC, LIHC, and HNSC, MZT2A expression gradually increased with deterioration of cell differentiation. This was consistent with the trend of pathological staging analysis. Our results suggested that the MZT2A expression was related to survival prognosis, clinical stage, and histological grade. The experimental results of MZT2A in lung cancer were consistent with our analysis, but we were still uncertain about the biological role of MZT2A in other cancers [11]. As far as we are aware, this paper is the first pan-cancer study of MZT2A. Considering different expressions could bring different prognostic outcomes, the particular role of MZT2A in each tumor needs to be further studied.
To learn more about MZT2A's role, in vitro experiments were performed in KIRC cell lines. According to the functional results, MZT2A had a major impact on the viability and proliferation of KIRC cells. Downregulation of MZT2A inhibited cell proliferation, promoted apoptosis, and regulated the cell cycle, while overexpression of MZT2A brought about the opposite effects in 786-O and CAKI-1 cells. In line with these prior investigations, our study further supported the notion that MZT2A exerted pro-cancer function in a variety of tumors. Different types of cancer exhibited high expression of MZT2A, indicating MZT2A as a potential tumor marker. We found that inhibition of MZT2A impaired the malignancy of KIRC cells but rendered insignificant changes in bio-function of the normal renal cell line HK-2. Therefore, targeting MZT2A might bring about tumor suppressive effect without affecting physical function of normal cells.
Although the significant involvement of MZT2A in KIRC has been shown, more researches are still needed to understand the molecular mechanism by which it promotes tumor progression. Based on this, we initially presented biological functions and possible regulatory pathways in KIRC via WGCNA analysis and the Sangerbox tools. The results discovered that the MZT2A was related to cell process, localization, and biological regulation and played a role in regulating cell malignant behaviors, which was consistent with the biological function we found. Likewise, our study found that MZT2A promoted malignant phenotypes of KIRC, and MZT2A activated PI3K/AKT signaling pathway. Of note, our results showed that MZT2A-related co-expression gene network may be involved in chromatin assembly and organelle fission, which might be further mined and validated in future research works. MZT2A has garnered increased attention, since bioinformatic analyses offer meaningful insights into its involvement in cancers and the function experiments imply that MZT2A plays an oncogene role in KIRC.
However, although we investigated the MZT2A expression in multiple cancers via integrating information from different databases, the sizes of normal samples for some cancers were relatively small, and tumor samples of Asian patients in the TCGA database were relatively less. The examination of the expression pattern of MZT2A in clinical KIRC samples will verify the bioinformatic analysis results. In addition, MZT2A function needs to be examined in vivo. Rescue experiments would strengthen its function via PI3K/AKT pathway, and its molecular mechanisms in KIRC are worthy of being clarified.
5. Conclusion
In conclusion, MZT2A was abnormally overexpressed in most cancers and correlated with the prognosis in a variety of tumor types. MZT2A is anticipated to be a valuable target gene in pan-cancers. Moreover, MZT2A induced cancerous behavior in KIRC via PI3K/AKT signaling pathway, indicating that MZT2A was a critical oncogene in KIRC and that it could be a potential target for cancer detection and therapy.
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors of the participants are informed and have provided consent for publication.
Funding
Not applicable.
Additional information
None.
Data availability statement
The datasets generated and analyzed during the current study are publicly available. All data generated or analyzed in this study are available from the corresponding author on reasonable request.
CRediT authorship contribution statement
Li Cao: Writing – original draft, Validation, Conceptualization. Xintao Jing: Conceptualization. Lijuan Liu: Conceptualization. Hui Wang: Conceptualization. Jiaqi Zhu: Validation. Jing Zhou: Conceptualization. Hang Peng: Conceptualization. Jinyuan Zhang: Conceptualization. Fang Li: Project administration, Methodology. Xiaofei Wang: Project administration, Methodology, Conceptualization. Le Zhao: Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors extend their sincere thanks to all the participants. Furthermore, we express our gratitude to TCGA, GTEx, CCLE, and Metascape for their provision of open access resources.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e35695.
Contributor Information
Xiaofei Wang, Email: wxiaofei@xjtu.edu.cn.
Le Zhao, Email: zhaole2@mail.xjtu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Mao J.J., Pillai G.G., Andrade C.J., et al. Integrative oncology: addressing the global challenges of cancer prevention and treatment. CA Cancer J Clin. 2022;72(2):144–164. doi: 10.3322/caac.21706. [DOI] [PubMed] [Google Scholar]
- 3.Hutchins J.R., Toyoda Y., Hegemann B., et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328(5978):593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang P., Zheng S., Lu L. Mitotic-spindle organizing protein MztA mediates septation signaling by suppressing the regulatory subunit of protein phosphatase 2A-ParA in Aspergillus nidulans. Front. Microbiol. 2018;9:988. doi: 10.3389/fmicb.2018.00988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fava F., Raynaud-Messina B., Leung-Tack J., et al. Human 76p:: a new member of the γ-tubulin-associated protein family. J. Cell Biol. 1999;147(4):857–868. doi: 10.1083/jcb.147.4.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oegema K., Wiese C., Martin O.C., et al. Characterization of two related γ-tubulin complexes that differ in their ability to nucleate microtubdes. J. Cell Biol. 1999;144(4):721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teixido-Travesa N., Villen J., Lacasa C., et al. The gammaTuRC revisited: a comparative analysis of interphase and mitotic human gammaTuRC redefines the set of core components and identifies the novel subunit GCP8. Mol. Biol. Cell. 2010;21(22):3963–3972. doi: 10.1091/mbc.E10-05-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Correia J.J., Wilson L. Microtubules and microtubule-associated proteins. Preface. Methods Cell Biol. 2013;115:xix–xx. doi: 10.1016/B978-0-12-407757-7.10000-3. [DOI] [PubMed] [Google Scholar]
- 9.Song L., Qian G., Huang J., et al. AZD9291-resistant non-small cell lung cancer cell-derived exosomal lnc-MZT2A-5:1 induces the activation of fibroblasts. Ann. Transl. Med. 2021;9(20):1593. doi: 10.21037/atm-21-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DU M.J., Liang Y.C., Liu Z.X., et al. Identification of key genes related to cd8+t-cell infiltration as prognostic biomarkers for lung adenocarcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.693353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H., Jiang X., Cheng Y., et al. MZT2A promotes NSCLC viability and invasion by increasing Akt phosphorylation via the MOZART2 domain. Cancer Sci. 2021;112(6):2210–2222. doi: 10.1111/cas.14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DU M., Liang Y., Liu Z., et al. Identification of key genes related to CD8+ T-cell infiltration as prognostic biomarkers for lung adenocarcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.693353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heitor D.A., Silva Maues J., Ferreira Ribeiro H., DE Maria Maues Sacramento R., et al. Downregulated genes by silencing MYC pathway identified with RNA-SEQ analysis as potential prognostic biomarkers in gastric adenocarcinoma. Aging (Albany NY) 2020;12(24):24651–24670. doi: 10.18632/aging.202260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J.J., Aksoy B.A., Dogrusoz U., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6(269) doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vivian J., Rao A.A., Nothaft F.A., et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 2017;35(4):314–316. doi: 10.1038/nbt.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhlen M., Fagerberg L., HallströM B.M., et al. Tissue-based map of the human proteome. Science. 2015;347(6220) doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 17.Wang L., Wang X., Sun H., et al. A pan-cancer analysis of the role of HOXD1, HOXD3, and HOXD4 and validation in renal cell carcinoma. Aging (Albany NY) 2023;15(19):10746–10766. doi: 10.18632/aging.205116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang N., Dou X.L., You X., et al. Pan-cancer analysis of the oncogenic role of discs large homolog associated protein 5 (DLGAP5) in human tumors. Cancer Cell Int. 2021;21(1) doi: 10.1186/s12935-021-02155-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgel P.R., Paillasseur J.L., Caillaud D., et al. Clinical COPD phenotypes: a novel approach using principal component and cluster analyses. Eur. Respir. J. 2010;36(3):531–539. doi: 10.1183/09031936.00175109. [DOI] [PubMed] [Google Scholar]
- 20.Tian Z.L., He W.X., Tang J.N., et al. Identification of important modules and biomarkers in breast cancer based on WGCNA. OncoTargets Ther. 2020;13:6805–6817. doi: 10.2147/OTT.S258439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen W., Song Z., Zhong X., et al. Sangerbox: a comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta. 2022;1(3):e36. doi: 10.1002/imt2.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teixidó-Travesa N., VilléN J., Lacasa C., et al. The γTuRC revisited: a comparative analysis of interphase and mitotic human γTuRC redefines the set of core components and identifies the novel subunit GCP8. Mol. Biol. Cell. 2010;21(22):3963–3972. doi: 10.1091/mbc.E10-05-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are publicly available. All data generated or analyzed in this study are available from the corresponding author on reasonable request.