Abstract
Although significant progress has been made in the development of novel targeted drugs for the treatment of acute myeloid leukemia (AML) in recent years, chemotherapy still remains the mainstay of treatment and the overall survival is poor in most patients. Here, we demonstrated the antileukemia activity of a novel small molecular compound NL101, which is formed through the modification on bendamustine with a suberanilohydroxamic acid (SAHA) radical. NL101 suppresses the proliferation of myeloid malignancy cells and primary AML cells. It induces DNA damage and caspase 3-mediated apoptosis. A genome-wide clustered regularly interspaced short palindromic repeats (CRISPR) library screen revealed that phosphatase and tensin homologous (PTEN) gene is critical for the regulation of cell survival upon NL101 treatment. The knockout or inhibition of PTEN significantly reduced NL101-induced apoptosis in AML and myelodysplastic syndrome (MDS) cells, accompanied by the activation of protein kinase B (AKT) signaling pathway. The inhibition of mammalian target of rapamycin (mTOR) by rapamycin enhanced the sensitivity of AML cells to NL101-induced cell death. These findings uncover PTEN protein expression as a major determinant of chemosensitivity to NL101 and provide a novel strategy to treat AML with the combination of NL101 and rapamycin.
Keywords: Genome-wide clustered regularly interspaced short palindromic repeats (CRISPR) library, Phosphatase and tensin homologous (PTEN), Rapamycin, Acute myeloid leukemia (AML), Chemotherapy
Abstract
近年来,在急性髓系白血病(AML)治疗的新型靶向药物开发方面虽取得显著进展,但目前化疗仍然是主要的治疗手段,大多数患者的总体生存率仍较低。本研究展示了一种新型小分子化合物NL101的抗白血病活性,该化合物是通过在苯达莫司汀的侧链加上辛二酰苯胺异羟肟酸(SAHA)基团修饰形成。NL101能抑制髓系恶性肿瘤细胞和原发性AML细胞的增殖,并可诱导DNA损伤和半胱天冬酶3介导的细胞凋亡。全基因组CRISPR文库筛选发现,PTEN基因在NL101处理后调节细胞存活中起到关键作用。PTEN敲除或抑制伴随着AKT信号通路的激活,可显著降低AML和骨髓增生异常综合征(MDS)细胞对NL101诱导的凋亡作用。雷帕霉素联用可通过抑制mTOR增强AML细胞对NL101诱导细胞死亡的敏感性。上述发现揭示了PTEN蛋白的表达是NL101化疗敏感性的主要决定因素,这提供了一种新的治疗策略,为NL101与雷帕霉素联合治疗AML提供了理论依据。
Keywords: CRISPR/Case9全基因组敲除文库, PTEN, 雷帕霉素, 急性髓系白血病, 化疗
1. Introduction
Acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) are myeloid malignancies arising from hematopoietic stem cells, with generally poor clinical outcomes, especially in patients who are unfit for intensive therapy because they are elderly or present major comorbidities. Under traditional intensive chemotherapy, only 40% of AML patients younger than 60 years can survive for more than five years, and even those with favorable core-binding factor leukemia have a mortality rate of 56% at ten years (Saygin and Carraway, 2017). With a deeper understanding of the genomic and epigenomic landscapes of AML, rapid progress has been made in targeted therapy for AML, with the involvement of FMS-like tyrosine kinase 3 (FLT3) inhibitors, isocitrate dehydrogenase (IDH) inhibitors, and B-cell lymphoma-2 (Bcl-2) inhibitors (Kantarjian et al., 2021).
Epigenetic dysregulation has emerged as a hallmark of AML (Ley et al., 2013), including genes involved in histone acetylation, an important and well-characterized epigenetic modification. However, histone deacetylase (HDAC) inhibitors exhibit limited efficacy in myeloid malignancies (Contieri et al., 2020). NL101 is a novel DNA damage/HDAC inhibition-dual targeting small molecule (Jiang et al., 2011). Previously, we demonstrated that NL101 can effectively inhibit the growth of B-cell lymphoma by blocking the cellular-myelocytomatosis viral oncogene (c-Myc)/microRNA-21 (miR-21)/MAX dimerization protein 1 (Mxd1) loop (Li et al., 2021).
Human genome-scale clustered regularly interspaced short palindromic repeats (CRISPR) knockout (GeCKO) lentiviral pooled libraries comprise a method of gene identification based on functional screening and high-throughput sequencing analysis (Shalem et al., 2014). Whole genome CRISPR libraries are powerful tools for genome-scale loss-of-function screening. Ruiz et al. (2016) identified cell division cycle 25 homolog A (CDC25A) as a determinant of sensitivity to ataxia-telangiectasia and Rad3-related (ATR) inhibitors, while Kurata et al. (2016) proposed new sources of Ara-C drug resistance in AML. In addition, Hou et al. (2017) revealed genes critical for resistance to the FLT3 inhibitor quizartinib (AC220) in AML.
In this study, we aimed to identify the genes playing a crucial role in AML sensitivity to NL101 by utilizing genome-wide CRISPR library screening in AML cells. The results offer valuable insights into the development of effective chemotherapeutic treatments for AML, specifically in terms of overcoming NL101 resistance.
2. Materials and methods
2.1. Cell lines and primary samples
KG-1a and MOLM-13 human AML cells and MUTZ-1 and SKM-1 MDS cells were analyzed by different assays. MOLM-13 and MUTZ-1 cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM), and KG-1a and SKM-1 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium at 37 ℃ with 5% CO2. Both media were supplemented with 10% (volume fraction) fetal bovine serum (FBS) and 1% (volume fraction) penicillin/streptomycin solution. Bone marrow or peripheral blood was obtained from AML patients prior to treatment. Mononuclear cells were isolated by Ficoll density gradient centrifugation and cultured in IMDM with 20% (volume fraction) FBS.
As described previously (Zheng et al., 2013), the cells infected with lentiviruses expressing single-guide RNA (sgRNA) were selected by treatment with puromycin (2 μg/mL) or blasticidin (10 μg/mL) for 7‒10 d. After treatment, antibiotic resistant cells were selected under selective conditions, collected, and then expanded for further analysis.
2.2. Reagents
NL101 was provided by Hangzhou Minsheng Institute of Pharmaceutical Research (Hangzhou, China). Rapamycin and VO-OHpic trihydrate were purchased from Selleck Chemicals (Shanghai, China). The compounds were dissolved in dimethyl sulfoxide (DMSO) and diluted with cell culture medium. For administration to non-obese diabetic/severe combined immunodeficiency (NOD/SCID)-gamma (NSG) mice, NL101 was dissolved in phosphate-buffered saline (PBS).
2.3. CRISPR/Cas9 screening
The human GeCKOv2 pooled library (two-vector system) was purchased from Addgene (Watertown, USA). CRISPR screening was conducted as described previously (Sanjana et al., 2014). Briefly, the GeCKOv2.0 DNA plasmid lentiCas9-Blast and lentiGuide-Puro were amplified and prepared on a larger scale. As described previously (Shao et al., 2022), the library plasmid lentiCas9-Blast or lentiGuide-Puro was cotransfected with two viral packaging plasmids, psPAX2 and pMD2.G, into HEK 293T cells to produce lentivirus. For the generation of CRISPR-associated protein 9 (Cas9)-expressing cells, MUTZ-1 cells were infected with lentiCas9-Blast virus for 3 d and then selected with blasticidin for 7 d. The established cell line was designated MUTZ-1-Cas9. To perform a CRISPR screen for genes critical for NL101 sensitivity, we infected MUTZ-1-Cas9 cells with lentiGuide-Puro virus at a multiplicity of infection (MOI) of 0.3 and treated the cells with puromycin. MUTZ-1-Cas9 cells were then cultured in medium containing 3 μmol/L NL101. At Days 0, 7, and 14 post-NL101 treatment, the surviving cells were split into two replicates: one was passaged until Day 14, and the other was harvested for genomic DNA extraction. Approximately 3×107 harvested cells were subjected to genomic DNA extraction and polymerase chain reaction (PCR) amplification while using Hi-Seq and double-terminal PE125 sequencing. The original image data files obtained from Genome Analyzer II (Illumina, San Diego, USA) were analyzed and converted into sequenced reads by Consensus Assessment of Sequence and Variation (CASAVA) software and the results were stored in FASTQ (fq) file format. The FASTQ file contains the name of each sequencing sequence (Read), the base sequence, and its corresponding sequencing quality information. In a FASTQ format file, each base corresponds to a base mass character, and the ASCII code value for each base mass character minus 33 yields the sequencing quality score for that base.
2.4. CRISPR knockout
The lentiCRISPRv2 plasmid (Addgene) was digested, dephosphorylated, and ligated with the sgRNA-targeting PTEN gene (5'-CTGTCACTCTCTTAGAACGT-3'; 5'-GCAGCCGCAGAAATGGATAC-3'). For the production of lentivirus, a total of 4×106 HEK 293T cells were seeded in a 100 mm cell culture dish, and transfection was performed using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, USA). The packaging plasmids, including pMD2G (3 μg) and psPA2X (6 μg), were diluted in OptiMEM and mixed with 60 μL of Lipofectamine 2000. The mixture was incubated for 20 min before being added to HEK 293T cells. After 36 and 60 h, the medium was centrifuged to pellet cell debris, and the supernatant was concentrated with an ultrafiltration centrifugation tube. Cells were infected with the lentivirus, added with polybrene, and subjected to horizontal centrifugation for 1 h.
2.5. Cell viability assay
As described previously (Lee et al., 2017), cells were seeded in 96-well plates at 1×104 (AML or MDS) or 5×104 (primary AML) cells per well. At the end of drug treatment, a final concentration of 1 mmol/L 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution was added to each well to further incubate the cells at 37 ℃ for 4 h. After centrifugation for 15 min at 1000 r/min, the supernatant was carefully taken out, and 150 μL of dimethyl sulfoxide (DMSO) was added to solubilize the formazan crystals. The absorbance was analyzed at 562 nm using a microplate reader.
2.6. Immunoblotting analysis
Cells were lysed and proteins were extracted using a modified buffer according to a previously described protocol (Zheng et al., 2011). The immunoblot analyses were performed using antibodies. Briefly, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer with 1% (volume fraction) protease inhibitor and phosphatase inhibitors on ice for 30 min. The bicinchoninic acid (BCA) reagent kit (Thermo Fisher Scientific) was employed to determine the protein concentration. The protein samples were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) before being transferred to polyvinylidene fluoride (PVDF) membranes (Merck KGaA, Darmstadt, Germany). Membranes were blocked using milk at room temperature and incubated with primary antibodies overnight at 4 ℃ and secondary antibodies for 1 h. Antibodies against caspase-3, poly-(ADP-ribose) polymerase-1 (PARP-1), phosphorylated ribosomal S6 protein (p-S6) (S240/244), phosphorylated mammalian target of rapamycin (p-mTOR) (S2448), protein kinase B (AKT) (pan), p-AKT (S473), and phosphatase and tensin homologous (PTEN) were purchased from CST (Cell Signaling Technology, Danvers, USA). The γ-H2A.X (S139) antibody was purchased from HUABIO (Hangzhou, China)
2.7. Animal models
The NSG mice at the age of four weeks were purchased from Shanghai Laboratory Animal Center (Shanghai, China). To establish an AML xenograft model, 5×106 MOLM-13 cells were injected subcutaneously. Five days after cell injection, the mice were intraperitoneally injected with PBS or NL101 (15 or 25 mg/kg) every other day and euthanized 5 d after treatment. The tumor volume was calculated using the formula (L×W 2)/2, where L is length and W is width.
2.8. Statistical analysis
Statistical analyses were based on a two-tailed unpaired t-test or multi-group analysis of variance (ANOVA). The half maximal inhibitory concentration (IC50) was determined using GraphPad Prism 6.
3. Results
3.1. Effects of NL101 on the proliferation of myeloid cancer cells in vitro and in vivo
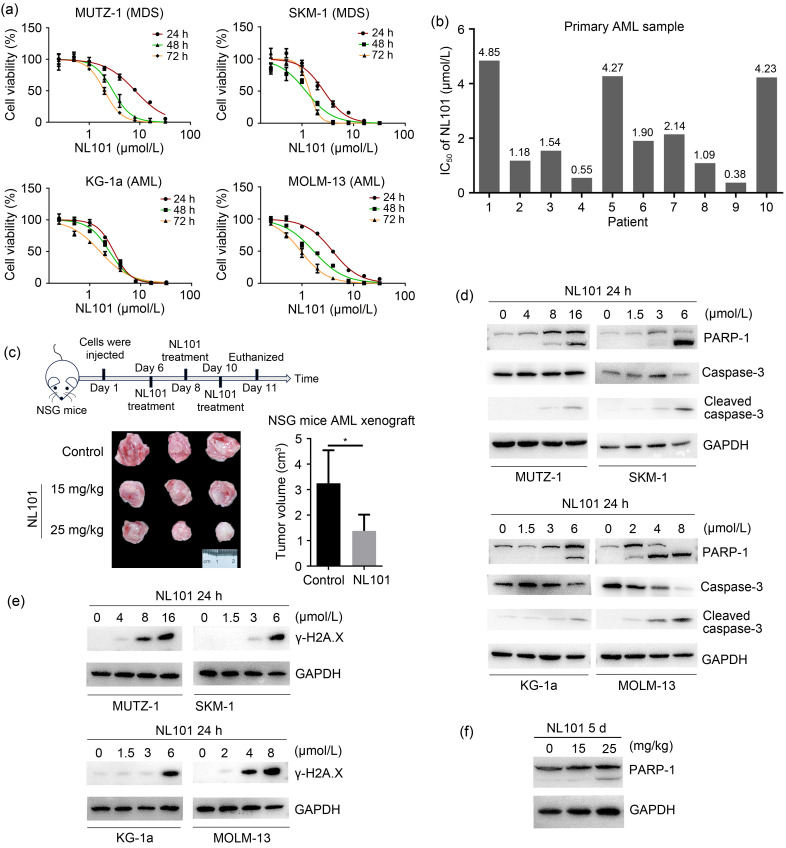
In order to establish the inhibitory effect of NL101 on the proliferation of AML or MDS cells, we performed MTT assays to calculate the IC50 of NL101 for AML or MDS cells. NL101 inhibited cell proliferation in a time- or dose-dependent manner (Fig. 1a), with 48 h IC50 values of 2.94 μmol/L for MUTZ-1 cells, 1.19 μmol/L for SKM-1 MDS cells, 2.45 μmol/L for KG-1a cells, and 1.69 μmol/L for MOLM-13 AML cells (Table 1). Importantly, we also demonstrated the cytotoxicity of NL101 against primary leukemia cells (Table S1), with IC50 values ranging from 0.38 to 4.85 μmol/L (Fig. 1b).
Fig. 1. Effects of NL101 on the proliferation of myeloid cancer cells in vitro and in vivo. (a) The time- and dose-dependent effects of NL101 on cell proliferation in myelodysplastic syndrome (MDS) cell lines (MUTZ-1 and SKM-1) and acute myeloid leukemia (AML) cell lines (MOLM-13 and KG-1a) were measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays. (b) The half maximal inhibitory concentration (IC50) of NL101 in primary leukemia cells. (c) The timeline indicating the tumor injection and treatment schedule (top) and antitumor potency of NL101 in AML xenografts (bottom). (d, e) Western blot analysis of poly-(ADP-ribose) polymerase-1 (PARP-1), cleaved caspase-3, and phosphorylated γ-H2A.X in cell lines exposed to NL101 for 24 h. (f) Western blot analysis of cleaved PARP-1 in AML xenografts. Data (a, c) are expressed as mean±standard deviation (SD) (n=3). NSG: NOD-SCID-gamma; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
Table 1.
IC50 values of the indicated cell lines
| Cell line | IC50 (95% CI) (μmol/L) | ||
|---|---|---|---|
| 24 h | 48 h | 72 h | |
| MUTZ-1 | 8.11 (7.39‒8.91) | 2.94 (2.70‒3.21) | 2.00 (1.86‒2.15) |
| KG-1a | 2.98 (2.85‒3.11) | 2.45 (2.30‒2.61) | 1.60 (1.46‒1.76) |
| MOLM-13 | 3.93 (3.73‒4.15) | 1.69 (1.52‒1.89) | 0.95 (0.90‒1.00) |
| SKM-1 | 2.66 (2.32‒3.05) | 1.19 (1.04‒1.37) | 1.40 (1.32‒1.49) |
IC50: half maximal inhibitory concentration; CI: confidence interval.
In order to explore the effectiveness of NL101 against AML in vivo, we established AML xenograft models by the subcutaneous inoculation of MOLM-13 cells into NSG mice. The mice were treated with either vehicle or NL101 at a dose of 15 or 25 mg/kg. NL101 significantly inhibited AML growth, as the excised tumors after NL101 treatment were much smaller than those of the control mice (Fig. 1c). Thus, NL101 inhibited the proliferation of myeloid cancer cells in vitro and in vivo.
3.2. Effects of NL101 on DNA damage and apoptosis
In order to determine whether NL101 can induce cell apoptosis, we treated myeloid cell lines with NL101 at variable concentrations of 2‒12 µmol/L. The PARP and caspase-3 are cleaved during apoptosis. We found that NL101 induced PARP and caspase-3 cleavage in a dose-dependent manner (Fig. 1d). Moreover, phosphorylated γ-H2A.X (S139) was increased following NL101 treatment (Fig. 1e). We also detected increased cleaved PARP in AML xenografts after NL101 treatment (Fig. 1f). These results suggest that NL101 induces DNA damage and myeloid cell apoptosis.
3.3. CRISPR library screening for identifying genes critical for drug resistance to NL101
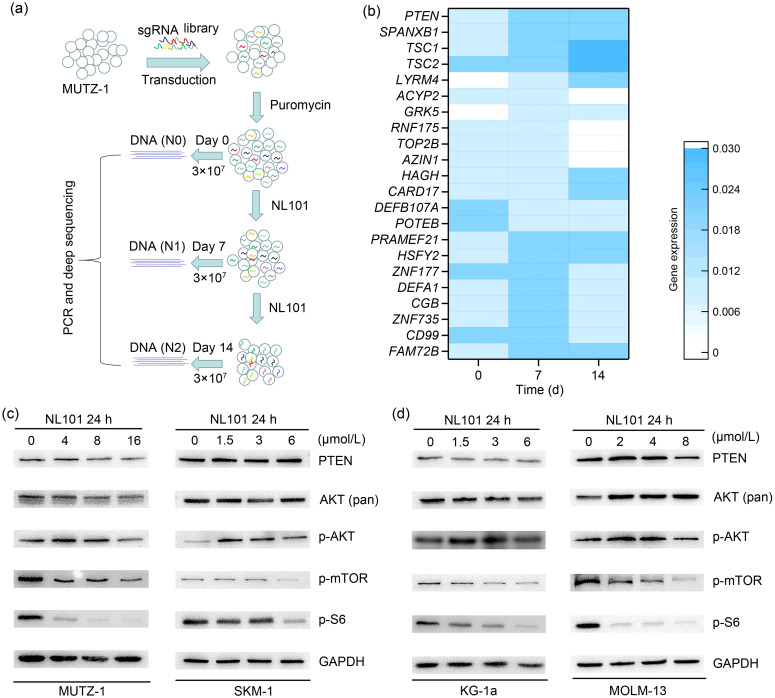
In order to gain insights into the antileukemia activity of NL101, we conducted CRISPR screening to identify genes whose deficiency confers NL101 resistance in MUTZ-1 cells, which were established from a patient with MDS who progressed to AML (Steube et al., 1997). We infected MUTZ-1 cells with lentivirus containing a pooled genome-scale CRISPR-Cas9 knockout library targeting 21 913 human genes with 122 411 unique guide sequences (six sgRNAs per gene). After puromycin selection for 5 d, we treated MUTZ-1 cells with NL101 for 14 d and collected half of the surviving cells at Days 0, 7, and 14 for genomic DNA extraction (Fig. 2a). The sgRNA regions were amplified from genomic DNA, and then analyzed by next-generation sequencing followed by statistical analyses (Fig. S1a). We found that multiple sgRNAs were enriched in NL101-resistant cells, including PTEN, LYR motif-containing protein 4 (LYRM4), tuberous sclerosis complex-1 (TSC1), TSC2, acylphosphatase-2 (ACYP2), and G-protein-coupled receptor kinase 5 (GRK5) (Fig. 2b). To examine the functions of genes related to NL101 resistance, we performed reactome pathway analysis (https://reactome.org) and found strong enrichment for fundamental biological processes, such as the tumor protein p53 (TP53)-regulated metabolic gene pathway and AKT/mTOR signaling. Not-ably, 3 of the top 5 hits were involved in the mTOR pathway, suggesting that the PTEN/AKT/mTOR pathway is associated with NL101 drug resistance in AML and MDS.
Fig. 2. Genes critical for drug resistance to NL101 identified by clustered regularly interspaced short palindromic repeats (CRISPR) library screen. (a) Landscape of a CRISPR library screen. (b) Heatmap of genes identified by genome-wide CRISPR library screen. (c, d) Western blot analyses of mammalian target of rapamycin (mTOR) signaling pathway in cell lines exposed to NL101 for 24 h. sgRNA: single-guide RNA; PCR: polymerase chain reaction; PTEN: phosphatase and tensin homologous; p: phosphorylated; AKT: protein kinase B; S6: ribosomal S6 protein; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
3.4. Effect of NL101 on mTOR
The phosphoinositide 3-kinase (PI3K)/AKT/mTOR signaling plays a central role in cancer development. To determine whether NL101 affects AKT/mTOR signaling, we analyzed phosphorylated proteins by western blot. The phosphorylated forms of mTOR (S2448) and S6 (S240/244) were decreased after NL101 treatment, while p-AKT (S473) increased, possibly due to the negative feedback loop of inhibited mTOR (Nogueira et al., 2008). In addition, total AKT (pan) and PTEN expression showed little changes upon NL101 treatment (Figs. 2c and 2d). These data suggest that NL101 inhibits mTOR phosphorylation and its downstream signaling pathway.
3.5. Effect of PTEN knockout on the resistance to NL101
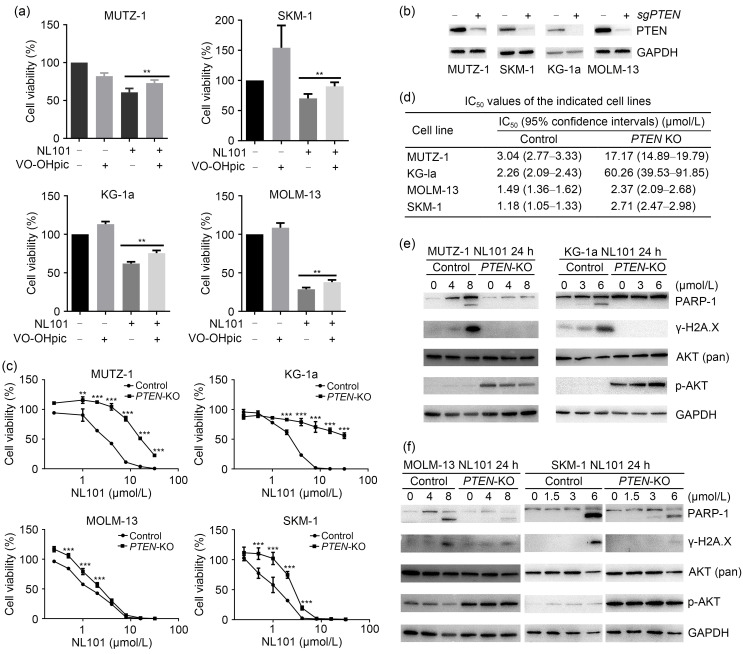
CRISPR screening identified PTEN as a potential determinant of NL101 sensitivity in myeloid leukemia cells. Survival analysis using OncoLnc (http://www.oncolnc.org) showed that higher expression of PTEN is associated with better prognosis in patients with leukemia (Fig. S1b). VO-OHpic trihydrate is a small molecule inhibitor of PTEN phosphatase activity (Mak et al., 2010). After treatment with VO-OHpic alone, cell viabilities were enhanced in SKM-1, KG-1a, and MOLM-13 cells (Fig. 3a); NL101 alone significantly inhibited cell viabilities, while the addition of VO-OHpic led to mitigated NL101-induced cell death, suggesting that PTEN inhibition alleviates NL101 cytotoxicity. To further confirm the role of PTEN in NL101 resistance, PTEN was genetically ablated using CRISPR/Cas9 in myeloid cancer cells, which exhibited significantly reduced PTEN expression (Fig. 3b). PTEN-knockout (PTEN-KO) alone did not considerably affect cell proliferation (Fig. S2). MTT assays showed that PTEN-KO cells were generally more resistant to NL101, especially MUTZ-1, KG-1a, and SKM-1 cells (Fig. 3c), and the NL101 IC50 values of the PTEN-KO cells were higher than those of the parental cells (Fig. 3d). Accordingly, compared with PTEN-wild type (PTEN-WT), PTEN-KO resulted in reduced PARP-1 cleavage and γ-H2A.X phosphorylation with increased p-AKT (S473) without the alteration of total AKT expression (Figs. 3e and 3f). These results suggest that PTEN deficiency increases myeloid cancer cell resistance to NL101 treatment.
Fig. 3. Effect of phosphatase and tensin homologous (PTEN) knockout (PTEN-KO) on the resistance to NL101. (a) The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay in cell lines exposed to NL101 and/or VO-OHpic trihydrate (PTEN inhibitor) for 48 h. (b) The clustered regularly interspaced short palindromic repeats (CRISPR)-mediated PTEN depletion was confirmed by western blot. (c, d) MTT assays of PTEN-wild type (WT) (control) and knockout (KO) cell lines exposed to NL101 for 48 h at the indicated doses, and the corresponding the half maximal inhibitory concentration (IC50) values. (e) The levels of poly-(ADP-ribose) polymerase-1 (PARP-1), phosphorylated γ-H2A.X, and phosphorylated protein kinase B (p-AKT) (S473) in PTEN-WT and PTEN-KO cell lines treated with NL101 for 24 h were determined by western blot. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and total AKT were used as controls. Data (a, c) are expressed as mean±standard deviation (SD) (n=4). ** P<0.01, *** P<0.001.
3.6. Sensitization of myeloid cancer cells to NL101-induced cytotoxicity by rapamycin
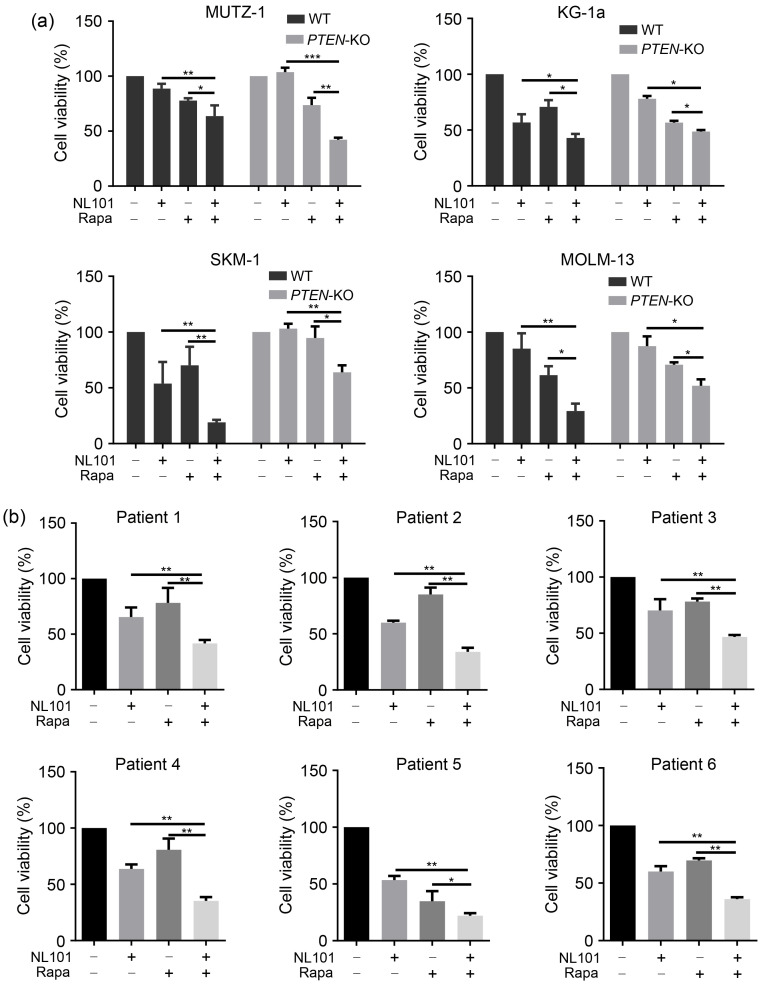
After revealing that PTEN inactivation contributes to NL101 resistance in myeloid cancer cells, we aimed to determine whether the inhibition of mTOR signaling sensitizes myeloid cancer cells to NL101-induced cell death. Rapamycin, an mTOR inhibitor widely used in the clinic, was combined with NL101 to treat cancer cells. Rapamycin treatment significantly increased the number of NL101-induced cells in both PTEN-WT and PTEN-KO cells (Fig. 4a). In addition, we collected primary AML cells from six patients and found that the combination of NL101 and rapamycin consistently led to stronger cytotoxicity than NL101 alone (Fig. 4b). These data suggest that mTOR inhibitors can sensitize myeloid cancer cells to NL101-induced cytotoxicity.
Fig. 4. Sensitization of myeloid cancer cells to NL101-induced cytotoxicity by rapamycin. (a) The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays of phosphatase and tensin homologous (PTEN) knockout (PTEN-KO) cell lines exposed to NL101 and/or rapamycin for 48 h. (b) MTT assays of primary acute myeloid leukemia (AML) sample cells exposed to NL101 and/or rapamycin for 24 h. Data are expressed as mean±standard deviation (SD) (n=3). * P<0.05, ** P<0.01, *** P<0.001; WT: wide type; Rapa: rapamycin.
4. Discussion
AML is a heterogeneous and aggressive myeloid cancer type with high rates of relapse and treatment resistance as well as poor prognosis. Targeted therapies have been increasingly applied in patients with AML (Kayser and Levis, 2018). NL101 is derived from bendamustine, a compound that induces both DNA damage and HDAC inhibition. Bendamustine is licensed for the treatment of lymphoma both alone and in combination with other drugs. Researchers of this field are committed to the development and re-establishment of bendamustine derivatives to optimize their genotoxicity in AML and MDS. Liu et al. (2015) reported a novel DNA/HDAC dual-targeting drug, CY190602, which exhibited significantly enhanced anticancer potency. López-Iglesias et al. (2017) found that EDO-S101, which is the fusion of an HDAC inhibitor radical to bendamustine, displayed a potent HDAC-inhibiting effect and a DNA-damaging effect in multiple myeloma (MM). Congruously, our results also demonstrate that NL101 induces apoptosis and DNA damage in AML and MDS cells, inhibits the proliferation of these cells in vitro, and suppresses AML xenograft growth in vivo.
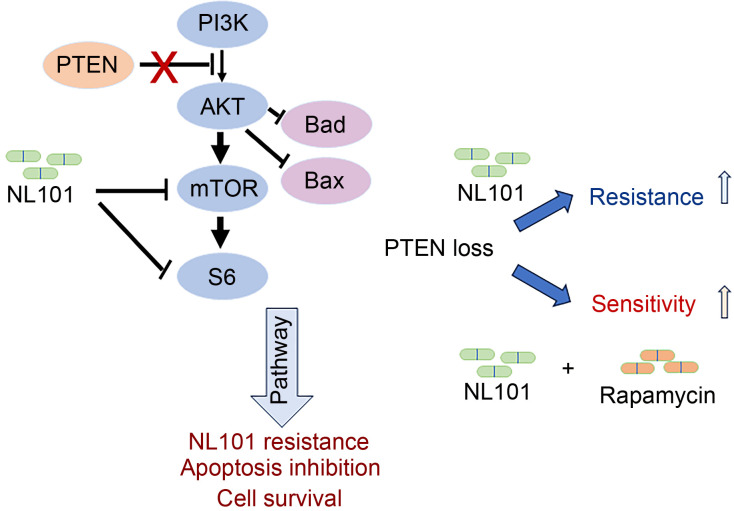
PTEN is a tumor suppressor that dephosphorylates the lipid second messenger phosphatidylinositol 3,4,5-trisphosphate (PIP3) to phosphatidylinositol 4,5-biphosphate (PIP2). PTEN antagonizes PI3K/AKT signaling, whereas activated AKT phosphorylates and inhibits pro-apoptotic Bcl-2 family members Bcl-2-associated agonist of cell death (Bad) and Bcl-2-associated X (Bax) (del Peso et al., 1997; Yamaguchi and Wang, 2001). The PI3K/AKT/mTOR pathway plays critical roles in many cellular functions, such as protein synthesis, DNA repair, cell survival, apoptosis, angiogenesis, and drug resistance (Zheng et al., 2022). We demonstrated that NL101 inhibits the activation of mTOR, which is a downstream effector of AKT activation. Thus, the expression of PTEN likely promotes cell apoptosis by the alleviation of AKT-mediated inhibition of Bad and Bax. This effect potentially enhances the sensitivity of NL101-induced cell death via the inhibition of mTOR signaling. In contrast, PTEN deficiency in tumor cells, which promotes cell survival via inhibition of Bad and Bax (del Peso et al., 1997), confers resistance to NL101-induced apoptosis. In addition, PTEN deficiency substantially increases AKT activity and subsequently activates more mTOR proteins, which may require a higher dose of NL101 for inhibition (Fig. 5).
Fig. 5. Schematic of the roles of NL101 in phosphatase and tensin homologous (PTEN) and mammalian target of rapamycin (mTOR) pathways. PI3K: phosphoinositide 3-kinase; AKT: protein kinase B; mTOR: mammalian target of rapamycin; S6: ribosomal S6 protein; Bad: B-cell lymphoma-2 (Bcl-2)-associated agonist of cell death; Bax: Bcl-2-associated X.
PTEN mutation and deficiency are frequently detected in a variety of cancers (Morotti et al., 2015). Mutated or deleted PTEN was reported to be highly linked to drug resistance in leukemia, breast cancer, colorectal cancer, and many other types of cancers (Liu et al., 2018). Conversely, extensive genetic sequences of AML did not reveal recurrent PTEN mutations and deletions (Morotti et al., 2015). Interestingly, our genome-wide CRISPR library screening results showed that PTEN expression was associated with NL101 sensitivity in AML and MDS cells, uncovering the possibility to intervene in PTEN-regulated PI3K/AKT/mTOR signaling for AML treatment.
There is an increasing number of clinical trials in hematological malignancies using mTOR inhibitors as single agents or as components of combination regimens (Teachey et al., 2009; Calimeri and Ferreri, 2017). Importantly, increasing evidence has indicated that mTOR inhibitors and HDAC inhibitors have synergistic antitumor effects in Hodgkin lymphoma, renal cell carcinoma, and RAS-driven tumors (Mahalingam et al., 2010). In this study, we showed that NL101 and rapamycin have additive or synergistic anti-AML activity (Fig. 5). However, commercial mTOR inhibitors, including everolimus and temisrolimus, are not approved by the U.S. Food and Drug Administration (FDA) for the treatment of AML; moreover, a randomized trial found no evidence that adding mTOR inhibitors to consolidation chemotherapy would decrease the risk of AML relapse (Burnett et al., 2018). Thus, many challenges, such as inadequate clinical efficacy, increased toxicities, unexpected immunosuppression, and adverse drug interactions, need to be addressed. Improved animal models that can recapitulate human AML, effective management of adverse events, as well as biomarker-based patient selection may help translate preclinical efficacies into clinical benefit. In summary, our study provides new insights into the potential of NL101-rapamycin combined treatment in hematological malignancies.
5. Conclusions
Our findings demonstrate the potential of NL101 as a selective agent for AML treatment. The identification of PTEN as a major determinant of chemo-sensitivity to NL101 provides the rationale of the combination of NL101 with mTOR inhibitor for AML treatment.
Supplementary information
Acknowledgments
This study was supported by the Zhejiang Provincial Natural Science Foundation of China (No. LY21H080005) and the National Natural Science Foundation of China (Nos. 81572920 and 82100171).
Author contributions
Yang XU, Zhimin LU, and Liming LIN conceived and designed the experiments. Liming LIN, Jingjing TAO, Yichao GAN, Xin HE, Shu LI, Jiawei ZHANG, Feiqiong GAO, and Dijia XIN performed the experiments. Liming LIN, Jingjing TAO, Ying MENG, and Jiawei ZHANG analyzed the data. Luyao WANG, Boxiao CHEN, and Yili FAN contributed reagents/materials/analysis tools. Liming LIN and Zhimin LU wrote the paper. All authors have read and approved the final manuscript, and therefore, have full access to all the data in the study and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines
Liming LIN, Jingjing TAO, Ying MENG, Yichao GAN, Xin HE, Shu LI, Jiawei ZHANG, Feiqiong GAO, Dijia XIN, Luyao WANG, Yili FAN, Boxiao CHEN, Zhimin LU, and Yang XU declare that they have no competing interests in the research.
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (the Institutional Review Board of the Second Affiliated Hospital, Zhejiang University School of Medicine, China) (No. I20180134). Informed consent was obtained from all patients for being included in the study. Additional informed consent was obtained from all patients for which identifying information is included in this article. All institutional and national guidelines for the care and use of laboratory animals were followed. The animal experiments were carried out according to the guidelines of the Institutional Review Board of Zhejiang Chinese Medical University (protocol code IACUC-20210927-23).
Data availability statement
All statistical data supporting the findings of this study are available within the paper, and the detailed questionnaire is available from the corresponding author upon reasonable request.
References
- Burnett AK, das Gupta E, Knapper S, et al. , 2018. Addition of the mammalian target of rapamycin inhibitor, everolimus, to consolidation therapy in acute myeloid leukemia: experience from the UK NCRI AML17 trial. Haematologica, 103(10): 1654-1661. 10.3324/haematol.2018.189514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calimeri T, Ferreri AJM, 2017. m-TOR inhibitors and their potential role in haematological malignancies. Br J Haematol, 177(5): 684-702. 10.1111/bjh.14529 [DOI] [PubMed] [Google Scholar]
- Contieri B, Duarte BKL, Lazarini M, 2020. Updates on DNA methylation modifiers in acute myeloid leukemia. Ann Hematol, 99(4): 693-701. 10.1007/s00277-020-03938-2 [DOI] [PubMed] [Google Scholar]
- del Peso L, González-García M, Page C, et al. , 1997. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science, 278(5338): 687-689. 10.1126/science.278.5338.687 [DOI] [PubMed] [Google Scholar]
- Hou PP, Wu C, Wang YC, et al. , 2017. A genome-wide CRISPR screen identifies genes critical for resistance to FLT3 inhibitor AC220. Cancer Res, 77(16): 4402-4413. 10.1158/0008-5472.CAN-16-1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Pritchard JR, Williams RT, et al. , 2011. A mammalian functional-genetic approach to characterizing cancer therapeutics. Nat Chem Biol, 7(2): 92-100. 10.1038/nchembio.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian HM, Kadia TM, DiNardo CD, et al. , 2021. Acute myeloid leukemia: treatment and research outlook for 2021 and the MD Anderson approach. Cancer, 127(8): 1186-1207. 10.1002/cncr.33477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser S, Levis MJ, 2018. Advances in targeted therapy for acute myeloid leukaemia. Br J Haematol, 180(4): 484-500. 10.1111/bjh.15032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata M, Rathe SK, Bailey NJ, et al. , 2016. Using genome-wide CRISPR library screening with library resistant DCK to find new sources of Ara-C drug resistance in AML. Sci Rep, 6: 36199. 10.1038/srep36199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Liu R, Li J, et al. , 2017. Stabilization of phosphofructokinase 1 platelet isoform by AKT promotes tumorigenesis. Nat Commun, 8: 949. 10.1038/s41467-017-00906-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Miller C, Ding L, et al. , 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med, 368(22): 2059-2074. 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, He X, Gan YC, et al. , 2021. Targeting miR-21 with NL101 blocks c-Myc/Mxd1 loop and inhibits the growth of B cell lymphoma. Theranostics, 11(7): 3439-3451. 10.7150/thno.53561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Ding HY, Li XX, et al. , 2015. A DNA/HDAC dual-targeting drug CY190602 with significantly enhanced anticancer potency. EMBO Mol Med, 7(4): 438-449. 10.15252/emmm.201404580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YF, Yang EJ, Zhang BY, et al. , 2018. PTEN deficiency confers colorectal cancer cell resistance to dual inhibitors of FLT3 and aurora kinase A. Cancer Lett, 436: 28-37. 10.1016/j.canlet.2018.08.011 [DOI] [PubMed] [Google Scholar]
- López-Iglesias AA, Herrero AB, Chesi M, et al. , 2017. Preclinical anti-myeloma activity of EDO-S101, a new bendamustine-derived molecule with added HDACi activity, through potent DNA damage induction and impairment of DNA repair. J Hematol Oncol, 10: 127. 10.1186/s13045-017-0495-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam D, Medina EC, Esquivel JA, et al. , 2010. Vorinostat enhances the activity of temsirolimus in renal cell carcinoma through suppression of survivin levels. Clin Cancer Res, 16(1): 141-153. 10.1158/1078-0432.Ccr-09-1385 [DOI] [PubMed] [Google Scholar]
- Mak LH, Vilar R, Woscholski R, 2010. Characterisation of the PTEN inhibitor VO-OHpic. J Chem Biol, 3(4): 157-163. 10.1007/s12154-010-0041-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morotti A, Panuzzo C, Crivellaro S, et al. , 2015. The role of PTEN in myeloid malignancies. Hematol Rep, 7(4): 6027. 10.4081/hr.2015.6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira V, Park Y, Chen CC, et al. , 2008. Akt determines replicative senescence and oxidative or oncogenic premature senescence and sensitizes cells to oxidative apoptosis. Cancer Cell, 14(6): 458-470. 10.1016/j.ccr.2008.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S, Mayor-Ruiz C, Lafarga V, et al. , 2016. A genome-wide CRISPR screen identifies CDC25A as a determinant of sensitivity to ATR inhibitors. Molecular Cell, 62(2): 307-313. 10.1016/j.molcel.2016.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F, 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods, 11(8): 783-784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saygin C, Carraway HE, 2017. Emerging therapies for acute myeloid leukemia. J Hematol Oncol, 10: 93. 10.1186/s13045-017-0463-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, et al. , 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science, 343(6166): 84-87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F, Gao YB, Wang W, et al. , 2022. Silencing EGFR-upregulated expression of CD55 and CD59 activates the complement system and sensitizes lung cancer to checkpoint blockade. Nat Cancer, 3(10): 1192-1210. 10.1038/s43018-022-00444-4 [DOI] [PubMed] [Google Scholar]
- Steube KG, Gignac SM, Hu ZB, et al. , 1997. In vitro culture studies of childhood myelodysplastic syndrome: establishment of the cell line MUTZ-1. Leuk Lymphoma, 25(3-4): 345-363. 10.3109/10428199709114174 [DOI] [PubMed] [Google Scholar]
- Teachey DT, Grupp SA, Brown VI, 2009. Mammalian target of rapamycin inhibitors and their potential role in therapy in leukaemia and other haematological malignancies. Br J Haematol, 145(5): 569-580. 10.1111/j.1365-2141.2009.07657.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Wang HG, 2001. The protein kinase PKB/Akt regulates cell survival and apoptosis by inhibiting Bax conformational change. Oncogene, 20(53): 7779-7786. 10.1038/sj.onc.1204984 [DOI] [PubMed] [Google Scholar]
- Zheng L, Liang H, Zhang QL, et al. , 2022. circPTEN1, a circular RNA generated from PTEN, suppresses cancer progression through inhibition of TGF-β/Smad signaling. Mol Cancer, 21: 41. 10.1186/s12943-022-01495-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YH, Yang WW, Xia Y, et al. , 2011. Ras-induced and extracellular signal-regulated kinase 1 and 2 phosphorylation-dependent isomerization of protein tyrosine phosphatase (PTP)-PEST by PIN1 promotes FAK dephosphorylation by PTP-PEST. Mol Cell Biol, 31(21): 4258-4269. 10.1128/Mcb.05547-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YH, Yang WW, Aldape K, et al. , 2013. Epidermal growth factor (EGF)-enhanced vascular cell adhesion molecule-1 (VCAM-1) expression promotes macrophage and glioblastoma cell interaction and tumor cell invasion. J Biol Chem, 288(44): 31488-31495. 10.1074/jbc.M113.499020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All statistical data supporting the findings of this study are available within the paper, and the detailed questionnaire is available from the corresponding author upon reasonable request.