Abstract
Background
Spontaneous intracerebral hemorrhage (sICH) is a form of stroke with high mortality rates and significant neurological implications for patients. Abnormalities in lipid metabolism have been implicated in various cardiovascular diseases, yet their relationship with sICH remains insufficiently explored, particularly concerning their association with inflammatory factors.
Methods
Employing a two-sample, two-step Mendelian Randomization approach, combined with data from GWAS datasets, to investigate the causal relationship between plasma lipid levels and sICH. Additionally, the role of inflammatory factors in this relationship was examined, and sensitivity analyses were conducted to ensure the robustness of the results.
Results
The results indicate a significant causal relationship between 19 plasma lipid metabolites and sICH. Furthermore, mediation analysis revealed that three distinct lipids, namely Sterol ester (27:1/20:2), Phosphatidylcholine (16:0_20:4), and Sphingomyelin (d34:1), exert their influence on sICH through inflammatory factors. TRAIL (OR: 1.078, 95% CI: 1.016–1.144, p = 0.013) and HGF (OR: 1.131, 95% CI: 1.001–1.279, p = 0.049) were identified as significant mediators.
Conclusion
This study provides new evidence linking abnormalities in lipid metabolism with sICH and elucidates the role of inflammatory factors as mediators. These findings contribute to a better understanding of the pathogenesis of sICH and offer novel insights and therapeutic strategies for its prevention and treatment.
Keywords: lipids, inflammatory factors, sICH, Mendelian randomization, causal effect
Introduction
Spontaneous intracerebral hemorrhage (sICH), resulting from the rupture of blood vessels in the brain, accounts for 10–15% of total stroke occurrences and represents a significant contributor to neurological morbidity and mortality, with a 30-day mortality rate ranging from 35 to 52%. Half of these fatalities occur within the initial 48 h, imposing a significant burden on families and society (1, 2). The incidence of sICH increases with age, particularly among individuals aged 65 and above. However, it can also occur in younger populations, especially in the presence of risk factors. Hypertension is one of the primary risk factors for sICH. Other risk factors include smoking, alcohol consumption, vascular diseases, diabetes, and hyperlipidemia (3–5).
Lipids constitute a fundamental category of organic compounds present in living organisms, encompassing fats, fatty acids, triglycerides, phospholipids, cholesterol, and other constituents. Lipids are indispensable for various cellular functions, including cell membrane integrity, energy storage and release, signaling cascades, and hormone synthesis (6, 7). Dyslipidemia, characterized by aberrant lipid metabolism, is intricately linked to the development and progression of numerous ailments, such as cardiovascular diseases, obesity, fatty liver disease, and metabolic syndrome (8, 9). In terms of the impact of dyslipidemia on cerebrovascular ailments, prior investigations have established that conditions like hypercholesterolemia and atherosclerosis can result in cerebral blood vessel constriction or obstruction, with hyperlipidemia emerging as a recognized risk factor for ischemic stroke (10). While previous studies have primarily focused on the relationship between lipids and ischemic stroke, emerging evidence suggests that dyslipidemia may also be associated with the risk of sICH (11–13). When lipid metabolism becomes abnormal, a series of complex biological changes may occur in the body, directly or indirectly increasing the risk of hemorrhagic stroke. Firstly, abnormal lipid metabolism may recruit immune cells and lead to the release of pro-inflammatory factors. These inflammatory responses can result in endothelial cell damage, leading to structural changes in the blood vessel walls (14, 15). Endothelial damage may constitute a fundamental basis for increased vessel susceptibility to rupture (15). Secondly, abnormal lipid metabolism may accelerate the formation and development of atherosclerotic plaques. The instability of these plaques increases the risk of rupture, particularly in complex plaques where macrophage-driven inflammatory responses and macrophage apoptosis may lead to the release of lipid contents, thereby further promoting plaque rupture (14, 16). Therefore, abnormal lipid metabolism may lead to structural changes in the blood vessel walls, reduced elasticity, and increased brittleness, thereby making the vessels more prone to rupture and bleeding. Previous studies have found that low total cholesterol levels may promote cell necrosis in the arterial intima, rendering it susceptible to microaneurysms and associated with the onset of hemorrhagic stroke (17).
Moreover, certain lipid substances, such as low-density lipoprotein cholesterol, may promote platelet activation and tissue factor expression, leading to impaired coagulation function, which may play a role in the pathogenesis of cerebral hemorrhage (18). These mechanisms may interact with each other, collectively increasing the risk of hemorrhagic stroke occurrence. In conclusion, the etiology of sICH is multifactorial, with inflammation potentially augmenting vascular permeability and exacerbating vascular pathologies such as arteriosclerosis and aneurysm formation, thereby increasing the risk of vascular rupture and subsequent intracranial bleeding (14). This finding has spurred further investigation into the role of lipid substances in the pathophysiology of stroke, offering new perspectives for the development of novel therapeutic strategies. Therefore, exploring the relationship between lipid substances and sICH holds significant importance.
Mendelian randomization (MR) is an analytical approach that leverages genetic variation to mimic randomized controlled trials (RCTs), facilitating causal inference regarding the relationship between risk factors and diseases (19). It serves to mitigate the influence of confounding variables and address issues of reverse causation. It has been extensively employed to investigate causal associations between exposures and diseases (19, 20). Based on the aforementioned conditions and background, a two-sample MR analysis will be conducted to elucidate the relationship between lipid levels and sICH. Subsequently, a mediation MR analysis will be performed to assess the potential role of inflammatory factors within this association, thereby exploring the underlying mechanistic pathways involved.
Materials and methods
Data resources for plasma lipidome, inflammatory biomarkers, and sICH
Data of plasma lipidome were obtained from a univariate and multivariate GWAS involving 179 lipid species across 13 lipid classes from 7,174 Finnish individuals in the GeneRISK cohort (21). This study performed a phenome-wide association study on identified lipid-related genetic loci among 377,277 participants from the FinnGen biobank, followed by colocalization analysis of these endpoints (accession numbers GCST90277238 to GCST90277287).
The inflammatory biomarkers data were obtained from Zhao et al.’s study (22). In this research, a thorough evaluation of genetic influences on inflammation-related proteins was conducted using a genome-wide analysis of protein quantitative trait loci. The study encompassed 14,824 participants of European descent and involved the measurement of 91 plasma proteins via the Olink panel. Accession numbers GCST90274758 to GCST90274848.
The data of sICH were acquired from the FinnGen dataset consisting of 7,040 cases and 374,631 controls of European ancestry.1 The FinnGen project is an open large-scale genetic research initiative originating from Finland, utilizing samples from extensive populations across various regions within the country. Its objective is to employ genetic research methodologies, particularly genome-wide association studies (GWAS), to elucidate the associations between genetic variations and diseases as well as health characteristics. The FinnGen dataset comprises substantial genomic, clinical, and biosample information, which can be utilized for investigating a multitude of diseases, including but not limited to cardiovascular diseases, tumors, metabolic disorders, and neurological conditions (23).
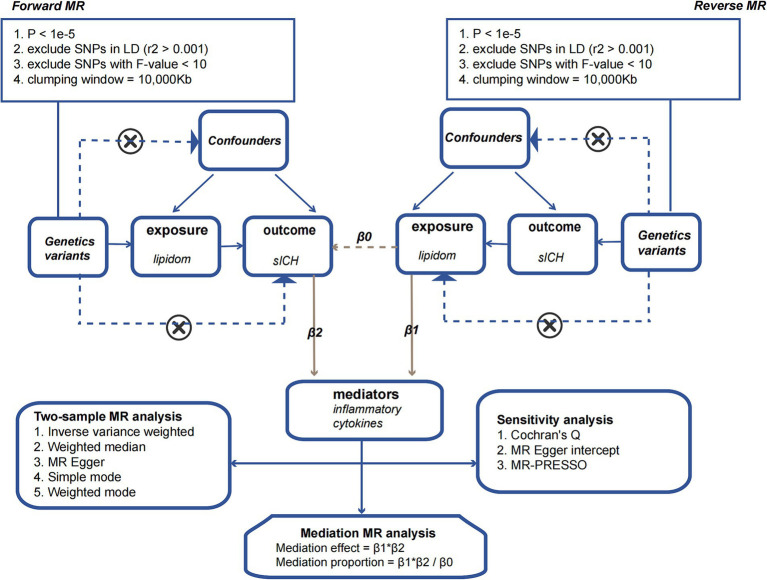
Selection of genetic instrumental variables
According to previous researches (24, 25), we employed a threshold of p < 1 × 10^-5 to select SNPs associated with the plasma lipidome. In the reverse MR analysis, SNPs associated with sICH were selected using the same criteria. Subsequently, we performed a clumping procedure to ensure independence among the chosen SNPs (r^2 < 0.001, clumping window = 10,000 kb) (26). To assess the reliability of each SNP, we computed the F-statistic and retained only those with values exceeding 10 (27). This stringent criterion ensured the robustness of our instrumental variables. The F-statistic for each SNP was calculated using the formula F = R^2 / (1 − R^2) × (N-2), where R^2 represents the variance of exposure explained by the instrumental variables (IVs), and N denotes the sample size. Furthermore, we calculated the variance of exposure explained by the instrument variable using the formula R^2 = β^2 / (β^2 + se^2 × N), where β denotes the effect size for the genetic variant of interest, she represents the standard error for β, and N indicates the sample size.
Statistical analysis
We utilized the Inverse Variance Weighting (IVW) method as our primary approach, which effectively leverages multiple single nucleotide polymorphisms (SNPs) as instrumental variables to estimate the causal impact of the exposure on the outcome. This method aggregates effect estimates of individual SNPs through inverse variance weighting, aiming to maximize the precision of the combined effect and thereby yield robust (28). Additionally, we employed weighted mode, MR-Egger, Simple mode, and weighted median tests to assess the effects, ensuring accuracy and robustness by detecting causal relationships from various aspects.
We employed a two-sample MR approach to dissect the direct and indirect effects of the plasma lipidome on sICH. In addition to the fundamental effect estimate (β1) of the plasma lipidome on inflammatory factors obtained from univariable MR analysis, two additional estimates were calculated: the causal effect of the mediator (inflammatory biomarkers) on sICH (β2), and the total effect of the plasma lipidome on sICH (β0). The mediation effect refers to the causal impact of the plasma lipidome on sICH through the mediator (inflammatory biomarkers), which can be estimated using the coefficient product method (β1 × β2). Hence, the proportion of the indirect effect can be calculated as “mediation effect/total effect” ([β1 × β2]/β0) (Figure 1).
Figure 1.
The flowchart illustrating the Mendelian randomization process.
Sensitivity analysis
Sensitivity analysis involves assessing the robustness of causal estimates to potential biases and confounding factors. We utilized Cochran’s Q test to evaluate the heterogeneity in the impact of genetic variants on the exposure factor. Additionally, MR-Egger intercept and MR-PRESSO analyses were conducted to assess the potential influence of pleiotropy on the outcomes of the MR analysis. Simultaneous utilization of scatter plots and funnel plots for data visualization and quality assurance aided in a comprehensive examination of the accuracy and reliability of the analytical results. Finally, a leave-one-out sensitivity analysis was conducted, removing one SNP at a time, to investigate if any single SNP was responsible for the causal association.
The effect estimates were reported as odds ratios (ORs) with their 95% confidence intervals (CIs). Statistical significance was defined as a two-sided p-value <0.05. All analyses were conducted using the “TwoSampleMR” package (version 0.5.8) in the R software (version 4.3.1).
Results
Effect of plasma lipidome on spontaneous intracerebral hemorrhage
Assessed primarily through the IVW method, the findings indicate significant causal associations between genetically predicted levels of 19 distinct plasma lipidome components and the risk of sICH. These components include Sterol ester (27:1/16:0) (OR: 0.938, 95% CI: 0.883–0.996, p = 0.036), Sterol ester (27:1/20:2) (OR: 1.088, 95% CI: 1.016–1.166, p = 0.016), Sterol ester (27:1/20:4) (OR: 0.925, 95% CI: 0.886–0.967, p = 0.000), Phosphatidylethanolamine (16:0_18:2) (OR: 1.066, 95% CI: 1.011–1.124, p = 0.019), Phosphatidylinositol (16:0_18:1) (OR: 1.158, 95% CI: 1.052–1.276, p = 0.003), and Sphingomyelin (d34:1) (OR: 0.929, 95% CI: 0.871–0.992, p = 0.027) (Table 1, causal effects assessed using different methods are presented in Supplementary Table S1). In subsequent reverse MR analyses, employing the IVW method, we confirmed a causal relationship between the plasma levels of Sterol ester (27:1/20:4) (OR: 0.936, 95% CI: 0.883–0.991, p = 0.024) and Phosphatidylcholine (18:0_20:4) (OR: 0.931, 95% CI: 0.879–0.986, p = 0.015) with the occurrence of sICH. The remaining 17 lipid species showed no reverse causal relationship with sICH (Table 2, detailed data in Supplementary Table S2).
Table 1.
The causal effects of plasma lipidom on sICH by IVW method.
| Plasma lipidom | nsnp | Beta | OR | or_lci95 | or_uci95 | p-value |
|---|---|---|---|---|---|---|
| Sterol ester (27:1/16:0) | 34 | −0.064 | 0.938 | 0.883 | 0.996 | 0.036 |
| Sterol ester (27:1/20:2) | 27 | 0.085 | 1.088 | 1.016 | 1.166 | 0.016 |
| Sterol ester (27:1/20:4) | 31 | −0.078 | 0.925 | 0.886 | 0.967 | 0.000 |
| Sterol ester (27:1/22:6) | 23 | −0.132 | 0.876 | 0.800 | 0.960 | 0.004 |
| Phosphatidylethanolamine (18:2_0:0) | 27 | 0.084 | 1.088 | 1.024 | 1.156 | 0.007 |
| Phosphatidylcholine (14:0_18:2) | 26 | 0.098 | 1.103 | 1.001 | 1.215 | 0.047 |
| Phosphatidylcholine (15:0_18:2) | 39 | 0.060 | 1.062 | 1.003 | 1.123 | 0.037 |
| Phosphatidylcholine (16:0_20:4) | 24 | −0.062 | 0.940 | 0.897 | 0.986 | 0.011 |
| Phosphatidylcholine (16:0_20:5) | 24 | −0.074 | 0.929 | 0.863 | 1.000 | 0.049 |
| Phosphatidylcholine (16:0_22:5) | 29 | −0.059 | 0.943 | 0.891 | 0.998 | 0.043 |
| Phosphatidylcholine (17:0_20:4) | 30 | −0.078 | 0.925 | 0.877 | 0.975 | 0.003 |
| Phosphatidylcholine (18:0_20:4) | 27 | −0.077 | 0.926 | 0.881 | 0.973 | 0.002 |
| Phosphatidylcholine (18:1_18:3) | 18 | 0.120 | 1.128 | 1.007 | 1.263 | 0.037 |
| Phosphatidylcholine (O-17:0_17:1) | 33 | 0.096 | 1.101 | 1.019 | 1.189 | 0.015 |
| Phosphatidylethanolamine (16:0_18:2) | 21 | 0.064 | 1.066 | 1.011 | 1.124 | 0.019 |
| Phosphatidylinositol (16:0_18:1) | 18 | 0.147 | 1.158 | 1.052 | 1.276 | 0.003 |
| Sphingomyelin (d34:1) | 32 | −0.074 | 0.929 | 0.871 | 0.992 | 0.027 |
| Sphingomyelin (d36:2) | 24 | −0.080 | 0.923 | 0.854 | 0.998 | 0.043 |
| Triacylglycerol (52:3) | 30 | −0.075 | 0.928 | 0.862 | 0.999 | 0.047 |
IVW, Inverse variance weighting; sICH, Spontaneous intracerebral hemorrhage.
Table 2.
The causal effects of sICH on plasma lipidom by IVW method.
| Plasma lipidom | nsnp | Beta | OR | or_lci95 | or_uci95 | p-value |
|---|---|---|---|---|---|---|
| Sterol ester (27:1/16:0) | 45 | −0.037 | 0.964 | 0.909 | 1.021 | 0.213 |
| Sterol ester (27:1/20:2) | 45 | −0.038 | 0.962 | 0.898 | 1.032 | 0.282 |
| Sterol ester (27:1/20:4) | 45 | −0.067 | 0.936 | 0.883 | 0.991 | 0.024 |
| Sterol ester (27:1/22:6) | 45 | −0.019 | 0.981 | 0.926 | 1.040 | 0.519 |
| Phosphatidylethanolamine (18:2_0:0) | 45 | 0.052 | 1.053 | 0.992 | 1.117 | 0.089 |
| Phosphatidylcholine (14:0_18:2) | 45 | 0.003 | 1.003 | 0.945 | 1.065 | 0.927 |
| Phosphatidylcholine (15:0_18:2) | 45 | −0.014 | 0.986 | 0.917 | 1.060 | 0.700 |
| Phosphatidylcholine (16:0_20:4) | 45 | −0.050 | 0.951 | 0.898 | 1.008 | 0.090 |
| Phosphatidylcholine (16:0_20:5) | 45 | −0.007 | 0.993 | 0.934 | 1.056 | 0.831 |
| Phosphatidylcholine (16:0_22:5) | 45 | −0.006 | 0.994 | 0.938 | 1.053 | 0.840 |
| Phosphatidylcholine (17:0_20:4) | 45 | −0.027 | 0.974 | 0.918 | 1.032 | 0.368 |
| Phosphatidylcholine (18:0_20:4) | 45 | −0.071 | 0.931 | 0.879 | 0.986 | 0.015 |
| Phosphatidylcholine (18:1_18:3) | 45 | 0.018 | 1.018 | 0.956 | 1.085 | 0.573 |
| Phosphatidylcholine (O-17:0_17:1) | 45 | 0.036 | 1.036 | 0.972 | 1.105 | 0.275 |
| Phosphatidylethanolamine (16:0_18:2) | 45 | 0.009 | 1.009 | 0.944 | 1.078 | 0.796 |
| Phosphatidylinositol (16:0_18:1) | 45 | 0.033 | 1.034 | 0.971 | 1.101 | 0.299 |
| Sphingomyelin (d34:1) | 45 | −0.026 | 0.974 | 0.919 | 1.032 | 0.374 |
| Sphingomyelin (d36:2) | 45 | −0.049 | 0.952 | 0.899 | 1.009 | 0.101 |
| Triacylglycerol (52:3) | 45 | −0.015 | 0.985 | 0.918 | 1.056 | 0.670 |
IVW, Inverse variance weighting; sICH, Spontaneous intracerebral hemorrhage.
Effect of plasma lipidome on inflammatory biomarkers
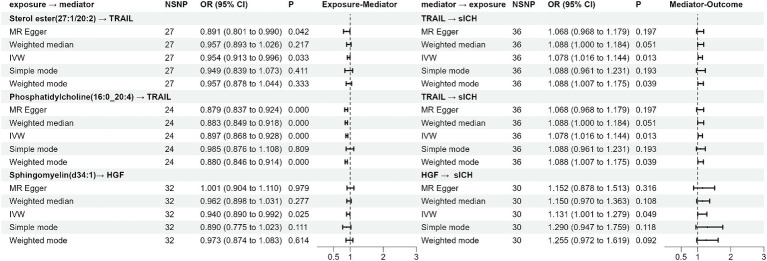
The impact of 17 different lipid substances on 91 inflammatory biomarkers was assessed. Causal relationships were found between 3 lipid species and 2 inflammatory biomarkers (Table 3; Supplementary Table S3). Sterol ester (27:1/20:2) (OR: 0.913, 95% CI: 0.913–0.996, p = 0.033) and Phosphatidylcholine (16:0_20:4) (OR: 0.897, 95% CI: 0.868–0.928, p = 0.000) were both negatively correlated with TNF-related apoptosis-inducing ligand levels (TRAIL). Additionally, plasma levels of Sphingomyelin (d34:1) were negatively correlated with Hepatocyte growth factor levels (HGF) (OR: 0.940, 95% CI: 0.890–0.992, p = 0.025).
Table 3.
The causal effects of plasma lipidom on inflammatory biomarkers by IVW method.
| Plasma lipidom | Inflammatory cytokines | nsnp | Beta | OR | 95%CI | p-value |
|---|---|---|---|---|---|---|
| Sterol ester(27:1/20:2) | TRAIL | 27 | −0.047 | 0.954 | 0.913–0.996 | 0.033 |
| Phosphatidylcholine(16:0_20:4) | TRAIL | 24 | −0.108 | 0.897 | 0.868–0.928 | 0.000 |
| Sphingomyelin(d34:1) | HGF | 32 | −0.062 | 0.940 | 0.890–0.992 | 0.025 |
IVW, Inverse variance weighting; TRAIL, TNF-related apoptosis-inducing ligand levels; HGF, Hepatocyte growth factor levels.
Effect of inflammatory biomarkers on spontaneous intracerebral hemorrhage
Increased levels of TRAIL (OR: 1.078, 95% CI: 1.016–1.144, p = 0.013) and HGF (OR: 1.131, 95% CI: 1.001–1.279, p = 0.049) were both associated with a higher risk of sICH (Table 4; Supplementary Table S4).
Table 4.
The causal effects of inflammatory biomarkers on sICH by IVW method.
| Inflammatory cytokines | nsnp | Beta | OR | or_lci95 | or_uci95 | p-value |
|---|---|---|---|---|---|---|
| TNF-related apoptosis-inducing ligand | 36 | 0.075 | 1.078 | 1.016 | 1.144 | 0.013 |
| Hepatocyte growth factor | 30 | 0.123 | 1.131 | 1.001 | 1.279 | 0.049 |
IVW, Inverse variance weighting; sICH, Spontaneous intracerebral hemorrhage.
Mediation effect of inflammatory biomarkers
TRAIL and HGF were significantly associated with both specific plasma lipidome and sICH. A mediation effect of plasma lipidome on sICH via TRAIL and HGF was observed. We found that the highest proportion was for the effect of plasma Phosphatidylcholine (16:0_20:4) concentration mediated by TRAIL on sICH, with a mediation effect of 13.2%, while the lowest was for the effect of plasma Sterol ester (27:1/20:2) concentration mediated by TRAIL on sICH, which was only 4.2%. The mediation effects of different mediators are demonstrated in Table 5. All the results of two-step MR analysis were demonstrated in Figure 2.
Table 5.
The mediation effect and proportion of inflammatory biomarkers by IVW method.
| Plasma lipidom | Inflammatory cytokines | β0 | β1 | β2 | β1*β2 | Mediation proportion (%) |
|---|---|---|---|---|---|---|
| Sterol ester(27:1/20:2) | TRAIL | 0.085 | −0.047 | 0.075 | −0.004 | 4.2 |
| Phosphatidylcholine(16:0_20:4) | TRAIL | −0.062 | −0.108 | 0.075 | −0.008 | 13.2 |
| Sphingomyelin(d34:1) | HGF | −0.074 | −0.062 | 0.123 | −0.008 | 10.4 |
IVW, Inverse variance weighting; TRAIL, TNF-related apoptosis-inducing ligand levels; HGF, Hepatocyte growth factor levels.
Figure 2.
The forest plot of the Mendelian randomization analysis results.
Sensitivity analysis
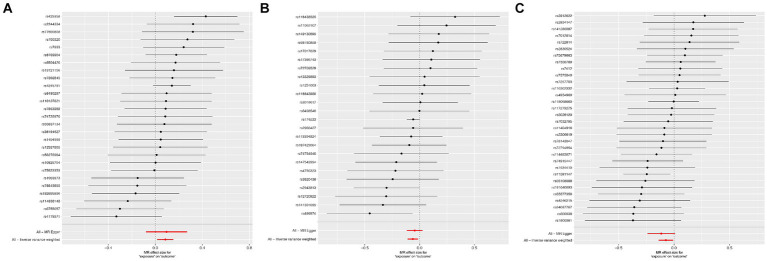
After rigorous screening, the number of eligible SNPs serving as IVs in the plasma lipidome for sICH at exposure are as follows: 27, 24, and 32, corresponding to Sterol ester (27:1/20:2), Phosphatidylcholine (16:0_20:4), and Sphingomyelin (d34:1). In the reverse MR analysis, based on our screening criteria, a total of 45 usable SNPs related to sICH were identified. When selecting SNPs to study the causal relationship between plasma lipidome as exposure and inflammatory factors as outcome, the determined range of available instrumental variables is from 24 to 32. Subsequently, In the process of screening for TRAIL and HGF instrumental variables, we found 36 and 30 variables, respectively. All SNPs had F-statistics ranging from 19.552 to 475.334. An F-statistic >10 is considered indicative of adequate instrument strength.
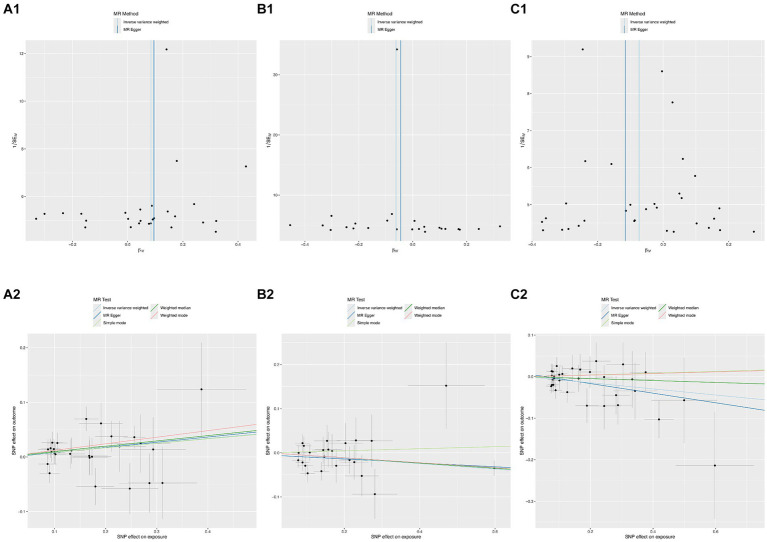
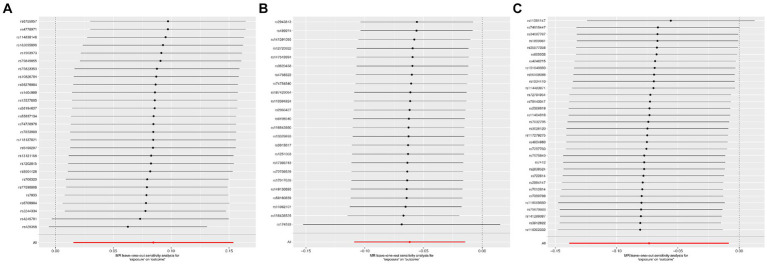
According to Cochran’s Q test, there was no evidence of heterogeneity in the instrumental variables from the plasma lipidome to sICH. To assess the potential horizontal pleiotropy of SNPs, we employed MR-Egger regression, providing a valuable assessment of its presence. Sensitivity analysis results did not reveal significant evidence of directional pleiotropy (p > 0.05). During sensitivity analysis of the association between plasma lipidome and inflammatory factors, we observed no heterogeneity or horizontal pleiotropy in plasma lipidome traits. Furthermore, leave-one-out analysis demonstrated that no SNP significantly influenced the results (Figure 3), and the Scatter Plot and Funnel Plot revealed no apparent outliers or biases (Figure 4). The forest plot demonstrates that the effect estimates all point in the same direction, with no significant heterogeneity observed (Figure 5). By the research procedure, the relevant data are presented sequentially from Supplementary Tables S5–S7.
Figure 3.
The forest plot from Sterol ester (27:1/20:2) (A), Phosphatidylcholine (16:0_20:4) (B), and Sphingomyelin (d34:1) (C) to spontaneous intracerebral hemorrhage.
Figure 4.
The funnel plots and scatter plots from Sterol ester (27:1/20:2) (A1,A2), Phosphatidylcholine (16:0_20:4) (B1,B2), and Sphingomyelin (d34:1) (C1,C2) to spontaneous intracerebral hemorrhage.
Figure 5.
The leave-one-out sensitivity analysis from Sterol ester (27:1/20:2) (A), Phosphatidylcholine (16:0_20:4) (B), and Sphingomyelin (d34:1) (C) to spontaneous intracerebral hemorrhage.
Discussion
According to the current literature, this is the first study to investigate how inflammatory factors mediate the causal pathway between plasma lipidome levels and sICH. This research discovered a possible correlation between genetically determined plasma lipidome levels and the risk of sICH. Furthermore, additional mediation analysis supported that the causal effects of plasma lipidome levels on sICH were partially mediated by inflammatory factors.
Lipids are essential for cellular function, serving as fundamental constituents of cell membranes, contributing to energy storage, maintaining equilibrium, and regulating cellular signaling pathways (7). Lipids are integral constituents of the brain, and their imbalance is correlated with disorders of the nervous system. Perturbations in lipid metabolism are connected with vascular inflammation and oxidative stress, both critical factors contributing to the development of atherosclerosis (29). We identified Sterol ester (27:1/20:2), Phosphatidylcholine (16:0_20:4), and Sphingomyelin (d34:1) as belonging to the categories of lipid esters, phospholipids, and sphingolipids, respectively. Through inflammatory mediators, they are causally associated with the onset of sICH. Sterol esters, resulting from the esterification of sterols with fatty acids, are pivotal in preserving the structural and operational integrity of cellular membranes. They modulate membrane fluidity and stability, thereby impacting cellular responsiveness to external stimuli and signal transduction. Notably, within the nervous system, sterol esters assume a critical role, particularly in the formation and sustenance of myelin sheaths. These sheaths serve as protective coatings for nerve fibers, and the presence of sterol esters aids in safeguarding nerve fibers while facilitating efficient nerve signal transmission (30). Additionally, in the context of inflammation, sterol esters may serve as signaling molecules or regulatory factors. By modulating cellular signal transduction pathways, they can affect the expression of genes related to inflammation and the release of inflammatory mediators. This modulation helps regulate the polarization state of inflammatory cells and the balance between pro-inflammatory and anti-inflammatory factors, thereby influencing the onset and progression of inflammation (31). Phosphatidylcholine constitutes a fundamental constituent within cell membranes, comprising a glycerol scaffold, dual fatty acid chains, and a phosphoric acid moiety connecting a choline unit. It serves as a critical player in upholding membrane architecture, cellular signaling cascades, choline provisioning, and transportation, alongside the pathophysiological pathways involved in disease onset (32). Among these, choline is an important nutrient involved in the synthesis of the neurotransmitter acetylcholine, maintenance of cellular membrane integrity, and metabolism of methyl (33). The metabolic pathways of choline have been associated with ischemic stroke and cognitive dysfunction after acute ischemic stroke (34, 35). Sphingomyelin is a phospholipid compound found within cell membranes, exerting a crucial role in upholding membrane structure, cellular signaling, neurological system function, and metabolic modulation (36). Within the nervous system, it serves as a primary constituent of both neuronal cell membranes and myelin sheaths, essential for maintaining the structural integrity and functionality of neurons (37). Specifically, sphingomyelin contributes to the protection of neuronal cell membranes from external environmental damage while promoting the formation and maintenance of myelin sheaths, thereby ensuring efficient neural signal transmission (37). Moreover, studies suggest that Sphingomyelin acts as a signaling molecule in modulating the initiation and progression of inflammatory responses. Upon cellular inflammation stimulation, phospholipase catalyzes the hydrolysis of Sphingomyelin into phosphoric acid and choline, with phosphoric acid playing a pivotal role as a constituent of inflammatory signals. Phosphatidic acid can activate inflammatory signaling pathways, such as NF-κB and MAPK pathways, thereby promoting the production and release of inflammatory cytokines, leading to the initiation of inflammatory responses (38). In conclusion, three of them play crucial roles in cell membrane structure and function, serve as key players in the nervous system, and are involved in modulating inflammatory responses.
TRAIL, categorized as a surface protein, is a member of the tumor necrosis factor (TNF) family and is predominantly expressed by activated immune cells, including T and B lymphocytes, neutrophils, dendritic cells, monocytes, macrophages, natural killer cells, and Natural Killer T cells (NKT) (39). Its main role involves maintaining the internal balance of the immune system, responding to infections, autoimmune diseases, and apoptosis. TRAIL exhibits pro-angiogenic activity and stimulates the proliferation of vascular smooth muscle cells (40–42). Conversely, TRAIL has also been found to inhibit vascular endothelial growth factor (VEGF)-mediated angiogenesis through both caspase-8-dependent and caspase-8-independent mechanisms (43), thus demonstrating a dual functionality. TRAIL might impact vascular development by controlling the survival and apoptosis of endothelial cells and vascular smooth muscle cells. Recent studies have indicated that CD4+ T cells derived from atherosclerotic plaques induce apoptosis of Vascular Smooth Muscle Cells (VSMCs) through a TRAIL-dependent mechanism, potentially leading to plaque instability and rupture (44). Hence, apoptosis of vascular cells triggered by TRAIL may regulate cellular turnover within the vascular wall. TRAIL has the potential to trigger the activation of the NF-κB signaling pathway, resulting in heightened activation and transcriptional potency of NF-κB (45, 46). NF-κB can modulate the expression of inflammation-related genes in both endothelial cells and vascular smooth muscle cells, promoting the onset of inflammatory responses. Activation of inflammation within endothelial cells can lead to increased vascular permeability and leukocyte adhesion, thereby contributing to the development of inflammatory vascular diseases such as atherosclerosis. Additionally, during the process of angiogenesis, NF-κB influences the formation of new blood vessels by regulating the expression of angiogenesis-related factors such as VEGF and matrix metalloproteinases (MMPs) (47, 48). Although research on the relationship between TRAIL and sICH is currently lacking, making it challenging to ascertain the precise mechanisms involved, it is hypothesized that TRAIL might affect sICH by influencing the function and integrity of the vascular wall. Hepatocyte Growth Factor, also known as Hepatotropin, is a multifunctional protein. It is a cytokine produced by fibroblasts and other cells. HGF primarily exerts its biological functions through binding to its receptor, the c-Met receptor. This binding activates the c-Met receptor, triggering a cascade of signaling pathways such as Ras-MAPK, PI3K-Akt, and STAT pathways. Consequently, these signaling pathways regulate diverse biological processes such as cell proliferation, anti-fibrotic and anti-inflammatory responses, as well as tissue repair and regeneration (49–51). In the central nervous system, HGF is recognized as a neuroprotective factor, exerting a beneficial influence on neuronal survival and repair. In the context of ischemic stroke, HGF mitigates the decline in tight junction protein levels and diminishes Blood–Brain Barrier disruption, thereby ameliorating cerebral edema following ischemia. Moreover, it reduces neuroinflammatory responses and attenuates neurotoxic damage, thus aiding in the regulation of cerebral inflammatory reactions (52–55). Furthermore, HGF is implicated in modulating angiogenesis and neuroregeneration processes. It facilitates endothelial cell migration and proliferation, regulates vascular smooth muscle cell function, and amplifies the expression and functionality of additional angiogenic factors like VEGF, thereby expediting angiogenesis, which is vital for vascular formation and restructuring. Additionally, it fosters brain tissue restoration and reshaping by augmenting neuroregeneration mechanisms (56, 57). However, current research on the role of HGF in sICH is insufficient, making it difficult to determine whether it retains its protective effects in this subtype of stroke. It is even possible that HGF may increase the risk of sICH. Further animal and clinical studies are needed to verify these potential effects. Although our study identified that plasma levels of Sterol ester (27:1/20:2), Phosphatidylcholine (16:0_20:4) are associated with a reduced risk of sICH through TRAIL, and plasma Sphingomyelin (d34:1) levels are associated with a reduced risk of sICH through HGF, their effects are limited, requiring further investigation to explore the potential molecular mechanisms involved. Additionally, it is noteworthy that the correlation analysis for Sterol ester (27:1/20:2) presents a paradox. While its overall effect increases the risk of sICH, it can also reduce the likelihood of sICH by lowering TRAIL levels. The mechanisms involved may include dysregulated lipid metabolism not only increasing the expression of pro-inflammatory mediators but also altering factors such as immune cells and oxidative stress. These combined factors ultimately lead to an overall effect of increased sICH risk.
Strengths and limitations
Our study utilized MR to infer causal relationships between exposure factors and diseases by leveraging genetic variation. Compared to observational studies, MR effectively reduces confounding factors, reverse causation, and information bias. Additionally, MR offers high statistical efficiency and minimizes reverse causation effects and information bias, making it a powerful tool for elucidating disease mechanisms. Despite these advantages, several limitations need to be addressed. MR results may be influenced by potential heterogeneity and genetic pleiotropy. Our study primarily focused on individuals of European descent, which may limit the generalizability of the findings, necessitating further investigation in diverse populations. Furthermore, although the initial threshold for exposure-related SNPs was set at p < 5×10−8, it was adjusted to p < 1×10−5 due to the limited availability of effective SNPs, potentially introducing some instability in the results. Future research should encompass populations of different ethnicities and geographical regions to validate the universality and reliability of our findings. While we identified inflammatory factors as potential mediators, we did not explore other biological pathways related to sICH mediated by lipid factors. Future studies should include additional mediators, such as immune cells and oxidative stress, and validate our findings across various populations. Further research should also investigate how inflammatory factors and lipid metabolism contribute to sICH. Integrating large-scale cohort data with multi-omics approaches could provide a more comprehensive understanding of the underlying mechanisms. Exploring potential intervention methods, including pharmacological treatments, lifestyle changes, and dietary adjustments, could help reduce sICH risk and improve outcomes.
Conclusion
Through a two-step, two-sample MR analysis, this study provides robust evidence of a causal relationship between blood lipid levels and the risk of sICH. Additionally, the findings indicate that blood lipid levels may influence the risk of sICH via inflammatory pathways, thereby contributing novel insights for future strategies in prevention and monitoring.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent from the patients/participants or patients/participants' legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author contributions
MH: Conceptualization, Data curation, Formal analysis, Writing – original draft. YL: Conceptualization, Methodology, Software, Visualization, Writing – review & editing. YC: Methodology, Writing – review & editing. WD: Project administration, Supervision, Methodology, Writing – review & editing.
Acknowledgments
The authors express their sincere gratitude to the original GWASs and the associated consortia for their efforts in gathering and overseeing the vast data resources.
Funding Statement
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Abbreviations
sICH, Spontaneous intracerebral hemorrhage; MR, Mendelian randomization; RCTs, Randomized controlled trials; GWAS, Genome-wide association studies; IVs, Instrumental variables; IVW, Inverse variance weighting; SNP, Single nucleotide polymorphisms; ORs, Odds ratios; CIs, Confidence intervals; TRAIL, TNF-related apoptosis-inducing ligand; HGF, Hepatocyte growth factor; TNF, Tumor necrosis factor; VEGF, Vascular endothelial growth factor; MMPs, Matrix metalloproteinases; VSMCs, Vascular smooth muscle cells; NKT, Natural Killer T.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fneur.2024.1411555/full#supplementary-material
References
- 1.Broderick J, Connolly S, Feldmann E, Hanley D, Kase C, Krieger D, et al. Guidelines for the management of spontaneous intracerebral hemorrhage in adults: 2007 update: a guideline from the American Heart Association/American Stroke Association stroke council, high blood pressure research council, and the quality of care and outcomes in research interdisciplinary working group. Circulation. (2007) 116:e391–413. doi: 10.1161/CIRCULATIONAHA.107.183689, PMID: [DOI] [PubMed] [Google Scholar]
- 2.Qureshi AI, Mendelow AD, Hanley DF. Intracerebral haemorrhage. Lancet. (2009) 373:1632–44. doi: 10.1016/S0140-6736(09)60371-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Asch CJ, Luitse MJ, Rinkel GJ, van der Tweel I, Algra A, Klijn CJ. Incidence, case fatality, and functional outcome of intracerebral haemorrhage over time, according to age, sex, and ethnic origin: a systematic review and meta-analysis. Lancet Neurol. (2010) 9:167–76. doi: 10.1016/S1474-4422(09)70340-0, PMID: [DOI] [PubMed] [Google Scholar]
- 4.Sarfo FS, Asowata OJ, Akpa OM, Akinyemi J, Wahab K, Singh A, et al. Stroke occurrence by hypertension treatment status in Ghana and Nigeria: a case-control study. J Neurol Sci. (2024) 459:122968. doi: 10.1016/j.jns.2024.122968, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou JF, Wang JY, Luo YE, Chen HH. Influence of hypertension, lipometabolism disorders, obesity and other lifestyles on spontaneous intracerebral hemorrhage. Biomed Environ Sci. (2003) 16:295–303. PMID: [PubMed] [Google Scholar]
- 6.Harayama T, Riezman H. Understanding the diversity of membrane lipid composition. Nat Rev Mol Cell Biol. (2018) 19:281–96. doi: 10.1038/nrm.2017.138 [DOI] [PubMed] [Google Scholar]
- 7.Han X. Lipidomics for studying metabolism. Nat Rev Endocrinol. (2016) 12:668–79. doi: 10.1038/nrendo.2016.98 [DOI] [PubMed] [Google Scholar]
- 8.Ference BA, Ginsberg HN, Graham I, Ray KK, Packard CJ, Bruckert E, et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease. 1. Evidence from genetic, epidemiologic, and clinical studies. A consensus statement from the European atherosclerosis society consensus panel. Eur Heart J. (2017) 38:2459–72. doi: 10.1093/eurheartj/ehx144, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bray GA, Kim KK, Wilding JPH. Obesity: a chronic relapsing progressive disease process. A position statement of the world obesity federation. Obes Rev. (2017) 18:715–23. doi: 10.1111/obr.12551 [DOI] [PubMed] [Google Scholar]
- 10.Tsai CF, Anderson N, Thomas B, Sudlow CL. Risk factors for ischemic stroke and its subtypes in Chinese vs. Caucasians: systematic review and meta-analysis. Int J Stroke. (2015) 10:485–93. doi: 10.1111/ijs.12508, PMID: [DOI] [PubMed] [Google Scholar]
- 11.Bonaventure A, Kurth T, Pico F, Barberger-Gateau P, Ritchie K, Stapf C, et al. Triglycerides and risk of hemorrhagic stroke vs. ischemic vascular events: the Three-City study. Atherosclerosis. (2010) 210:243–8. doi: 10.1016/j.atherosclerosis.2009.10.043, PMID: [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Tuomilehto J, Jousilahti P, Wang Y, Antikainen R, Hu G. Total and high-density lipoprotein cholesterol and stroke risk. Stroke. (2012) 43:1768–74. doi: 10.1161/STROKEAHA.111.646778 [DOI] [PubMed] [Google Scholar]
- 13.O'Donnell MJ, Xavier D, Liu L, Zhang H, Chin SL, Rao-Melacini P, et al. Risk factors for ischaemic and intracerebral haemorrhagic stroke in 22 countries (the INTERSTROKE study): a case-control study. Lancet. (2010) 376:112–23. doi: 10.1016/S0140-6736(10)60834-3, PMID: [DOI] [PubMed] [Google Scholar]
- 14.Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. (2013) 13:709–21. doi: 10.1038/nri3520, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordonnier C, Demchuk A, Ziai W, Anderson CS. Intracerebral haemorrhage: current approaches to acute management. Lancet. (2018) 392:1257–68. doi: 10.1016/S0140-6736(18)31878-6 [DOI] [PubMed] [Google Scholar]
- 16.Meier N, Nedeltchev K, Brekenfeld C, Galimanis A, Fischer U, Findling O, et al. Prior statin use, intracranial hemorrhage, and outcome after intra-arterial thrombolysis for acute ischemic stroke. Stroke. (2009) 40:1729–37. doi: 10.1161/STROKEAHA.108.532473, PMID: [DOI] [PubMed] [Google Scholar]
- 17.Tirschwell DL, Smith NL, Heckbert SR, Lemaitre RN, Longstreth WT, Jr, Psaty BM. Association of cholesterol with stroke risk varies in stroke subtypes and patient subgroups. Neurology. (2004) 63:1868–75. doi: 10.1212/01.WNL.0000144282.42222.DA, PMID: [DOI] [PubMed] [Google Scholar]
- 18.Pechlaner R, Tsimikas S, Yin X, Willeit P, Baig F, Santer P, et al. Very-low-density lipoprotein-associated apolipoproteins predict cardiovascular events and are lowered by inhibition of APOC-III. J Am Coll Cardiol. (2017) 69:789–800. doi: 10.1016/j.jacc.2016.11.065, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davey Smith G, Holmes MV, Davies NM, Ebrahim S. Mendel's laws, Mendelian randomization and causal inference in observational data: substantive and nomenclatural issues. Eur J Epidemiol. (2020) 35:99–111. doi: 10.1007/s10654-020-00622-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Kong J, Diao X, Cai J, Zheng J, Xie W, et al. Depression and prostate cancer risk: a Mendelian randomization study. Cancer Med. (2020) 9:9160–7. doi: 10.1002/cam4.3493, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ottensmann L, Tabassum R, Ruotsalainen SE, Gerl MJ, Klose C, Widén E, et al. Genome-wide association analysis of plasma lipidome identifies 495 genetic associations. Nat Commun. (2023) 14:6934. doi: 10.1038/s41467-023-42532-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao JH, Stacey D, Eriksson N, Macdonald-Dunlop E, Hedman ÅK, Kalnapenkis A, et al. Genetics of circulating inflammatory proteins identifies drivers of immune-mediated disease risk and therapeutic targets. Nat Immunol. (2023) 24:1540–51. doi: 10.1038/s41590-023-01588-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. (2023) 613:508–18. doi: 10.1038/s41586-022-05473-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orrù V, Steri M, Sidore C, Marongiu M, Serra V, Olla S, et al. Complex genetic signatures in immune cells underlie autoimmunity and inform therapy. Nat Genet. (2020) 52:1036–45. doi: 10.1038/s41588-020-0684-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Yu X, Wu X, Chai K, Wang S. Causal relationships between gut microbiota, immune cell, and non-small cell lung cancer: a two-step, two-sample Mendelian randomization study. J Cancer. (2024) 15:1890–7. doi: 10.7150/jca.92699, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies NM, Holmes MV, Davey SG. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. (2018) 362:k601. doi: 10.1136/bmj.k601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burgess S, Thompson SG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. (2011) 40:755–64. doi: 10.1093/ije/dyr036 [DOI] [PubMed] [Google Scholar]
- 28.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey SG. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. (2008) 27:1133–63. doi: 10.1002/sim.3034, PMID: [DOI] [PubMed] [Google Scholar]
- 29.Liu J, Tang L, Lu Q, Yu Y, Xu QG, Zhang S, et al. Plasma quantitative lipid profiles: identification of CarnitineC18:1-OH, CarnitineC18:2-OH and FFA (20:1) as novel biomarkers for pre-warning and prognosis in acute myocardial infarction. Front Cardiovasc Med. (2022) 9:848840. doi: 10.3389/fcvm.2022.848840, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saher G, Quintes S, Nave KA. Cholesterol: a novel regulatory role in myelin formation. Neuroscientist. (2011) 17:79–93. doi: 10.1177/1073858410373835, PMID: [DOI] [PubMed] [Google Scholar]
- 31.Oishi Y, Spann NJ, Link VM, Muse ED, Strid T, Edillor C, et al. SREBP1 contributes to resolution of pro-inflammatory TLR4 signaling by reprogramming fatty acid metabolism. Cell Metab. (2017) 25:412–27. doi: 10.1016/j.cmet.2016.11.009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vance JE, Tasseva G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim Biophys Acta. (2013) 1831:543–54. doi: 10.1016/j.bbalip.2012.08.016 [DOI] [PubMed] [Google Scholar]
- 33.Blusztajn JK, Slack BE, Mellott TJ. Neuroprotective actions of dietary choline. Nutrients. (2017) 9:815. doi: 10.3390/nu9080815, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu P, Li R, Antonov AA, Wang L, Li W, Hua Y, et al. Discovery of metabolite biomarkers for acute ischemic stroke progression. J Proteome Res. (2017) 16:773–9. doi: 10.1021/acs.jproteome.6b00779 [DOI] [PubMed] [Google Scholar]
- 35.Zhong C, Lu Z, Che B, Qian S, Zheng X, Wang A, et al. Choline pathway nutrients and metabolites and cognitive impairment after acute ischemic stroke. Stroke. (2021) 52:887–95. doi: 10.1161/STROKEAHA.120.031903, PMID: [DOI] [PubMed] [Google Scholar]
- 36.Merrill AH, Jr. Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem Rev. (2011) 111:6387–422. doi: 10.1021/cr2002917, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. (2008) 9:139–50. doi: 10.1038/nrm2329, PMID: [DOI] [PubMed] [Google Scholar]
- 38.Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature. (2014) 510:58–67. doi: 10.1038/nature13475, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ehrlich S, Infante-Duarte C, Seeger B, Zipp F. Regulation of soluble and surface-bound TRAIL in human T cells, B cells, and monocytes. Cytokine. (2003) 24:244–53. doi: 10.1016/S1043-4666(03)00094-2, PMID: [DOI] [PubMed] [Google Scholar]
- 40.Secchiero P, Gonelli A, Carnevale E, Corallini F, Rizzardi C, Zacchigna S, et al. Evidence for a proangiogenic activity of TNF-related apoptosis-inducing ligand. Neoplasia. (2004) 6:364–73. doi: 10.1593/neo.03421, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cartland SP, Genner SW, Zahoor A, Kavurma MM. Comparative evaluation of TRAIL, FGF-2 and VEGF-A-induced angiogenesis in vitro and in vivo. Int J Mol Sci. (2016) 17:2025. doi: 10.3390/ijms17122025, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kavurma MM, Schoppet M, Bobryshev YV, Khachigian LM, Bennett MR. TRAIL stimulates proliferation of vascular smooth muscle cells via activation of NF-kappaB and induction of insulin-like growth factor-1 receptor. J Biol Chem. (2008) 283:7754–62. doi: 10.1074/jbc.M706927200, PMID: [DOI] [PubMed] [Google Scholar]
- 43.Na HJ, Hwang JY, Lee KS, Choi YK, Choe J, Kim JY, et al. TRAIL negatively regulates VEGF-induced angiogenesis via caspase-8-mediated enzymatic and non-enzymatic functions. Angiogenesis. (2014) 17:179–94. doi: 10.1007/s10456-013-9387-0, PMID: [DOI] [PubMed] [Google Scholar]
- 44.Sato K, Niessner A, Kopecky SL, Frye RL, Goronzy JJ, Weyand CM. TRAIL-expressing T cells induce apoptosis of vascular smooth muscle cells in the atherosclerotic plaque. J Exp Med. (2006) 203:239–50. doi: 10.1084/jem.20051062, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trauzold A, Wermann H, Arlt A, Schütze S, Schäfer H, Oestern S, et al. CD95 and TRAIL receptor-mediated activation of protein kinase C and NF-kappaB contributes to apoptosis resistance in ductal pancreatic adenocarcinoma cells. Oncogene. (2001) 20:4258–69. doi: 10.1038/sj.onc.1204559, PMID: [DOI] [PubMed] [Google Scholar]
- 46.Wajant H. TRAIL and NFkappaB signaling--a complex relationship. Vitam Horm. (2004) 67:101–32. doi: 10.1016/S0083-6729(04)67007-5, PMID: [DOI] [PubMed] [Google Scholar]
- 47.Tong Z, Zhang Y, Guo P, Wang W, Chen Q, Jin J, et al. Steroid receptor coactivator 1 promotes human hepatocellular carcinoma invasiveness through enhancing MMP-9. J Cell Mol Med. (2024) 28:e18171. doi: 10.1111/jcmm.18171, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bakshi HA, Quinn GA, Nasef MM, Mishra V, Aljabali AAA, El-Tanani M, et al. Crocin inhibits angiogenesis and metastasis in Colon Cancer via TNF-α/NF-kB/VEGF pathways. Cells. (2022) 11:11. doi: 10.3390/cells11091502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weidner KM, Behrens J, Vandekerckhove J, Birchmeier W. Scatter factor: molecular characteristics and effect on the invasiveness of epithelial cells. J Cell Biol. (1990) 111:2097–108. doi: 10.1083/jcb.111.5.2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, Vande Woude GF, et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science. (1991) 251:802–4. doi: 10.1126/science.1846706 [DOI] [PubMed] [Google Scholar]
- 51.Naldini L, Vigna E, Narsimhan RP, Gaudino G, Zarnegar R, Michalopoulos GK, et al. Hepatocyte growth factor (HGF) stimulates the tyrosine kinase activity of the receptor encoded by the proto-oncogene c-MET. Oncogene. (1991) 6:501–4. PMID: [PubMed] [Google Scholar]
- 52.Kitamura K, Iwanami A, Nakamura M, Yamane J, Watanabe K, Suzuki Y, et al. Hepatocyte growth factor promotes endogenous repair and functional recovery after spinal cord injury. J Neurosci Res. (2007) 85:2332–42. doi: 10.1002/jnr.21372, PMID: [DOI] [PubMed] [Google Scholar]
- 53.Shimamura M, Sato N, Oshima K, Aoki M, Kurinami H, Waguri S, et al. Novel therapeutic strategy to treat brain ischemia: overexpression of hepatocyte growth factor gene reduced ischemic injury without cerebral edema in rat model. Circulation. (2004) 109:424–31. doi: 10.1161/01.CIR.0000109496.82683.49 [DOI] [PubMed] [Google Scholar]
- 54.Date I, Takagi N, Takagi K, Tanonaka K, Funakoshi H, Matsumoto K, et al. Hepatocyte growth factor attenuates cerebral ischemia-induced increase in permeability of the blood-brain barrier and decreases in expression of tight junctional proteins in cerebral vessels. Neurosci Lett. (2006) 407:141–5. doi: 10.1016/j.neulet.2006.08.050, PMID: [DOI] [PubMed] [Google Scholar]
- 55.Yamada N, Nakagawa S, Horai S, Tanaka K, Deli MA, Yatsuhashi H, et al. Hepatocyte growth factor enhances the barrier function in primary cultures of rat brain microvascular endothelial cells. Microvasc Res. (2014) 92:41–9. doi: 10.1016/j.mvr.2013.12.004, PMID: [DOI] [PubMed] [Google Scholar]
- 56.Xin X, Yang S, Ingle G, Zlot C, Rangell L, Kowalski J, et al. Hepatocyte growth factor enhances vascular endothelial growth factor-induced angiogenesis in vitro and in vivo. Am J Pathol. (2001) 158:1111–20. doi: 10.1016/S0002-9440(10)64058-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carmeliet P. Mechanisms of angiogenesis and arteriogenesis. Nat Med. (2000) 6:389–95. doi: 10.1038/74651 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.