Abstract
NusB is a prokaryotic transcription factor involved in antitermination processes, during which it interacts with the boxA portion of the mRNA nut site. Previous studies have shown that NusB exhibits an all-helical fold, and that the protein from Escherichia coli forms monomers, while Mycobacterium tuberculosis NusB is a dimer. The functional significance of NusB dimerization is unknown. We have determined five crystal structures of NusB from Thermotoga maritima. In three crystal forms the protein appeared monomeric, whereas the two other crystal forms contained assemblies, which resembled the M. tuberculosis dimers. In solution, T. maritima NusB could be cross-linked as dimers, but it migrated as a monomer in gel-filtration analyses, suggesting a monomer/dimer equilibrium with a preference for the monomer. Binding to boxA-like RNA sequences could be detected by gel-shift analyses and UV-induced cross-linking. An N-terminal arginine-rich sequence is a probable RNA binding site of the protein, exhibiting aromatic residues as potential stacking partners for the RNA bases. Anions located in various structures support the assignment of this RNA binding site. The proposed RNA binding region is hidden in the subunit interface of dimeric NusB proteins, such as NusB from M. tuberculosis, suggesting that such dimers have to undergo a considerable conformational change or dissociate for engagement with RNA. Therefore, in certain organisms, dimerization may be employed to package NusB in an inactive form until recruitment into antitermination complexes.
Keywords: NusB, N-utilization substance, RNA–protein interaction, transcriptional antitermination, transcription regulation, X-ray crystallography
Abbreviations: Nus, N-utilization substance; rmsd, root-mean-square deviation; rrn, rRNA operon; the prefixes eco, mtu and tma denote proteins from Escherichia coli, Mycobacterium tuberculosis and Thermotoga maritima respectively
INTRODUCTION
The decision by RNA polymerases to terminate transcription at specific sites or to read through terminators (antitermination) is of particular importance for the regulation of gene expression (reviewed in [1,2]). The mechanisms underlying termination/antitermination competition have been intensively studied in phage λ and related systems (reviewed in [3–6]). In the protein N-dependent antitermination of phage λ (reviewed in [2,5,7,8]), λN enlists four host-encoded Nus (N-utilization substance) factors (NusA, NusB, NusE and NusG; NusE corresponds to ribosomal protein S10) upon encountering nut (N-utilization) sites on the mRNA transcript. The λ nut sites consist of a short conserved ∼12-nucleotide stretch (boxA) followed by a stem–loop structure (boxB) [9]. The Nus factors augment the basic λN antitermination activity [10] and confer longevity and long-range efficiency to the antitermination complexes [10–12]. A closely related antitermination system underlies the transcriptional regulation of rRNA (rrn) operons [13–18].
The λN antitermination complexes rely on intricate protein–protein and protein–RNA networks dominated by multiple weak interactions [9,12,19]. Among these, a complex of NusB and NusE [20] recognizes the boxA element [21]. In Escherichia coli, NusB alone can bind to boxA [19,22,23], but NusE significantly enhances the association [21] by an as yet unknown mechanism. The interaction of NusB and boxA may be of particular importance, as it has been suggested that it precludes the action of an antitermination inhibitor [24]. Surprisingly, despite the prominence of the NusB–NusE interaction in λN antitermination, only NusB has so far been found to be essential in rrn antitermination [16,18].
In order to improve our understanding of antitermination mechanisms, detailed structure–function analyses are necessary. The structures of ecoNusB (NusB from E. coli) [25] and mtuNusB (NusB from Mycobacterium tuberculosis) [26] have been determined previously. While ecoNusB was found to be monomeric, mtuNusB is a dimer. The significance of the mtuNusB dimers and the existence of NusB dimers in other organisms are unknown. We have therefore initiated a crystallographic study of tmaNusB (NusB from Thermotoga maritima) and obtained five different crystal forms of the protein. Three crystal forms harboured monomeric NusB, whereas two forms showed dimeric tmaNusB arrangements similar to the previously described mtuNusB dimers. Solution studies in turn suggested a preference of tmaNusB for the monomeric form. Sequence and structural characteristics pointed to an important RNA binding site in the N-terminal region of NusB. Because this RNA binding site is occluded in dimeric forms of the protein, some organisms may use NusB dimerization to package the protein into an inactive form.
EXPERIMENTAL
Materials
DNA oligonucleotides were synthesized by Thermo-Hybaid (Ulm, Germany). Restriction enzymes were obtained from Gibco BRL (Karlsruhe, Germany) and New England Biolabs (Schwalbach, Germany). T4 DNA ligase was from Roche Diagnostics (Penzberg, Germany). DNA fragments and plasmids were purified with the Cycle Pure Kit and plasmid isolation kits from Peqlab (Erlangen, Germany) respectively. All chemicals were from Sigma-Aldrich (Taufkirchen, Germany) or Eurogentec (Seraing, Belgium).
RNA oligomers were synthesized on a modified ABI 394 automated nucleic acid synthesizer (ABI, Foster City, CA, U.S.A.) employing 5′-silyl,2′-O-bis(2-acetoxyethoxy)methyl orthoester phosphoramidites (Dharmacon, Lafayette, CO, U.S.A.) [27]. The oligos were liberated from the support and deprotected according to the manufacturer's instructions. To check the efficiency of each synthetic run, aliquots of the product were analysed on an 18% (w/v) polyacrylamide/7 M urea gel and detected by UV shadowing. In each case, the full-length sequence was the only product detectable. After deprotection, the oligomers were precipitated by addition of 0.1 vol. of 3.5 M ammonium acetate, pH 5.2, and 3 vol. of ethanol, dried, dissolved in water and stored at −70 °C until use.
Cloning and expression
The tmanusB gene was amplified by PCR on chromosomal DNA from T. maritima strain MSB8 (forward primer, 5′-GAG GAG AAA TTA ACC ATG AAA ACA CCG AGG CGA AGA ATG-3′; reverse primer, 5′-ATA GGA TCC TCA AAG TTC GAA TTT TTC TTT TGG AGC-3′; start codon in boldface; BamHI restriction site underlined; sequences corresponding to the genomic copy of tmanusB in italics). The product served as a template for a second PCR step, employing the previous reverse primer and a forward primer which introduced an MfeI restriction site (5′-ACA CAC AAT TGA TTA AAG AGG AGA AAT TAA CCA TG-3′). The second product was inserted into vector pNCO113. To confirm the desired construct, the promoter and insert region of the resulting plasmid, pNCO113-tmanusB, were sequenced (MWG Biotech, Ebersberg, Germany).
Plasmid pNCO113-tmanusB was transformed into E. coli strain M15, and the cells were grown to an attenuance of 0.7 at 600 nm in LB (Luria–Bertani) medium, induced with 1 mM isopropyl β-D-thiogalactopyranoside and shaken at 37 °C for 3 h. After harvesting at 4 °C, the cell pellet was washed with 0.9% NaCl and stored at −20 °C.
Purification of tmaNusB
An aliquot of 5 g of frozen cells was thawed in 40 ml of buffer A (75 mM potassium phosphate, pH 7.0, 0.05% NaN3), sonicated and centrifuged at 30000 g for 45 min. The supernatant was diluted to 100 ml with buffer A, heated to 80 °C for 30 min and cooled on ice for 10 min. Denatured E. coli proteins were removed by centrifugation at 10000 g for 30 min. The heat-resistant fraction was applied to a Superdex G75 size exclusion column (2.6 cm×26 cm; Amersham-Pharmacia, Uppsala, Sweden) and chromatographed with buffer B (70 mM potassium phosphate, pH 7.0, 0.05% NaN3; flow rate 3 ml/min). The eluate was analysed by SDS/PAGE, and peak fractions were combined and concentrated by ultrafiltration (YM5 membrane; Millipore, Eschborn, Germany). Aliquots of 3 ml were placed on a 5 ml High Trap desalting column (Amersham-Pharmacia), eluted with crystallization buffer (10 mM Tris/HCl, pH 7.0), concentrated by ultrafiltration and stored at −70 °C. The protein concentration was determined photometrically using the molar absorption coefficient ε280=12090 M−1·cm−1 of the tmaNusB protein, estimated on the basis of amino acid composition. The N-terminal sequence of the purified protein was determined by the automated Edman method using a 471A protein sequencer (PerkinElmer, Foster City, CA, U.S.A.).
Crystallization and data collection
All crystallization experiments were performed in the sitting-drop vapour diffusion format with drop volumes of between 2 and 4 μl and 500–1000 μl reservoirs. Crystallization conditions for tmaNusB were initially probed with a number of commercial and home-made incomplete factorial solutions at 20 °C. After refinement of pH and precipitant concentrations, optimized recipes for five crystal forms were obtained (Table 1). All crystal forms could be frozen in a liquid nitrogen stream (100 K; Oxford Cryosystems, Oxford, U.K.) after transfer into perfluoropolyether. Data were collected at beamline BW6 of DESY (Hamburg, Germany; http://www-hasylab.desy.de/facility/experimental_stations/stations/BW6.htm) (Table 1). Indexing, integration and reduction of the diffraction data were carried out with DENZO/SCALEPACK [28] and programs from the CCP4 suite [29].
Table 1. Data collection and refinement statistics.
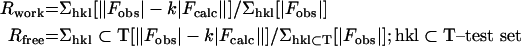 |
| Crystal form… | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Crystallization | |||||
| Crystallization buffer | pH 6.3 | pH 7.4 | pH 7.0 | pH 6.6 | pH 7.9 |
| 0.2 M AS | 10% propan-2-ol | 0.2 M CaCl2 | 0.2 M AA | 1.2M NaCit | |
| 30% PEG8000 | 20% PEG4000 | 25% PEG4000 | 2.0M K/NaPi | ||
| Data collection | |||||
| Space group | P21 | P212121 | P3121 | P3121 | P3221 |
| Unit cell (Å, °) | a=59.7, b=34.4, c=74.3; β=94.2 | a=33.8, b=59.6, c=64.3 | a=b=46.0, c=122.8 | a=b=60.2, c=87.7 | a=b=60.0, c=172.4 |
| Resolution (Å) | 30.0–1.55 | 30.0–1.85 | 30.0–1.60 | 30.0–1.35 | 30.0–1.72 |
| Reflections | |||||
| Unique | 43301 | 11016 | 20371 | 41043 | 39263 |
| Redundancy | 3.8 | 2.5 | 3.5 | 5.4 | 4.7 |
| Completeness (%) | 97.9 (95.2) | 94.6 (94.8) | 98.4 (95.6) | 99.9 (99.9) | 99.6 (99.5) |
| I/σ(I) | 28.7 (1.6) | 20.6 (3.1) | 21.4 (3.5) | 38.1 (3.4) | 37.6 (2.9) |
| Rsym | 0.072 (0.616) | 0.068 (0.253) | 0.071 (0.323) | 0.072 (0.358) | 0.069 (0.373) |
| Refinement | |||||
| Resolution (Å) | 20.0–1.55 | 20.0–1.85 | 20.0–1.60 | 20.0–1.35 | 20.0–1.72 |
| Reflections (number/%) | 40560/96.7 | 10471/94.9 | 20345/98.5 | 40933/99.7 | 39095/99.6 |
| Test set (%) | 5 | 5 | 5 | 5 | 5 |
| Rworkd | 0.235 | 0.224 | 0.187 | 0.199 | 0.217 |
| Rfreed | 0.251 | 0.253 | 0.223 | 0.214 | 0.249 |
| Content of a.u. | |||||
| Protein molecules | 2 | 1 | 1 | 1 | 2 |
| Protein atoms | 2362 | 1175 | 1196 | 1188 | 2376 |
| Water atoms | 392 | 137 | 264 | 288 | 238 |
| Ligands | 2 sulphate ions | 1 sulphate ion | 1 calcium ion | – | 2 citrate ions |
| Mean B-factors (Å2) | |||||
| Wilson | 26.2 | 27.6 | 19.4 | 19.4 | 33.3 |
| Protein | 24.7 | 27.9 | 19.3 | 22.7 | 26.5 |
| Water | 45.8 | 49.8 | 35.2 | 39.3 | 56.8 |
| Ligands | 80.2 | 79.3 | 14.7 | – | 91.4 |
| Ramachandran plot | |||||
| Preferred | 0.992 | 0.984 | 0.992 | 0.992 | 1.000 |
| Additions allowed | 0.8 | 0.8 | 0.8 | 0.0 | 0.0 |
| Disallowed | 0.0 | 0.8 | 0.0 | 0.8 | 0.0 |
| Rmsd geometry | |||||
| Bond length (Å) | 0.009 | 0.010 | 0.007 | 0.008 | 0.008 |
| Bond angle (°) | 1.18 | 1.02 | 1.40 | 1.27 | 1.08 |
| Rmsd B-factors (Å2) | |||||
| Main-chain bonds | 1.4 | 1.27 | 2.7 | 2.4 | 1.3 |
| Main-chain angles | 2.2 | 2.42 | 3.3 | 3.3 | 2.2 |
| Side-chain bonds | 1.6 | 3.40 | 5.3 | 4.6 | 1.5 |
| Side-chain angles | 2.3 | 5.83 | 6.6 | 5.9 | 2.5 |
| PDB entry | 1TZT | 1TZU | 1TZW | 1TZV | 1TZX |
Structure solution and refinement
The structure of tmaNusB in the most highly resolved P3121 form (form 4, Table 1) could be solved by a rotation-translation search (MOLREP, CCP4) with a monomeric polyalanine model of mtuNusB (Protein Databank entry 1EYV). The refined tmaNusB structure, including side chains but excluding water oxygens, was subsequently employed to solve the other four structures by molecular replacement. In all cases, rigid body refinement in CNS [30] produced initial 2Fo–Fc electron density maps, which allowed the stepwise building or repositioning of side chains in MAIN ([31]; http://www-bmb.ijs.si/doc/index.html). Refinement continued with standard protocols (positional-, simulated annealing- and B-factor refinement) in CNS. For space groups, which contained two molecules per asymmetric unit, the final round included a TLS-refinement step with REFMAC5 (CCP4) with different anisotropic temperature factor corrections for the two molecules. The structure co-ordinates have been deposited with the Protein Databank (http://www.rcsb.org/pdb; for accession numbers see Table 1) and will be released upon publication.
Glutaraldehyde cross-linking
tmaNusB at a concentration of 20 μM in buffer C (20 mM Tris/HCl, pH 7.0, 150 mM NaCl) or PBS, pH 7.4, was mixed with glutaraldehyde (final concentration 1 mM). At defined times, aliquots of the reactions were mixed with an equal volume of SDS loading buffer, heated briefly to 95 °C and frozen in liquid nitrogen. Samples were analysed on a SDS/15% (w/v)-polyacrylamide gel.
Analytical gel-filtration chromatography
tmaNusB was analysed by analytical gel-filtration chromatography on a Superdex 75 PC 3.2/3.0 column (Amersham-Pharmacia; 2.4 ml gel bed; column dimensions 3.2 mm×300 mm). Aliquots of 50 μl (between 0.1 and 0.4 mM) in buffer C, PBS, 20 mM Tris/HCl (pH 7.0)/500 mM NaCl or 20 mM Tris/HCl (pH 7.0)/10% (v/v) ethanol were incubated at 50 °C for 15 min and chromatographed at a flow rate of 40 μl/min in the respective buffers. The column was calibrated with bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), cyclophilin H (20 kDa), aprotinin (6.5 kDa) and pyridoxal phosphate (1.35 kDa). The void volume (Vo) was determined with blue dextran (2MD), and the total volume of the liquid phase (Vt) was determined with water. Apparent molecular mass values were extracted from plots of log\(molecular mass) against Kd [(Ve−Vo)/(Vt−Vo), in which Ve is the analyte elution volume].
Gel mobility shift assays
RNA oligomers were 5′-end-labelled in 10 μl reaction mixtures, employing T4 polynucleotide kinase, [γ-32P]ATP (5 μl, 5000 Ci/mmol) and standard procedures. The labelled fragments were purified and buffer was exchanged with MicroSpin G25 columns (Amersham-Pharmacia). Samples for the gel-shift experiments were prepared by incubating 50000 c.p.m. of RNA oligomer (4 pmol), 5–25 pmol of tmaNusB, 10 units of RNAsin (Promega, Mannheim, Germany), various amounts of NaCl and various amounts of heparin in a volume of 10 μl for 1 h on ice. The basic incubation buffer was PBS. The samples were loaded on 8% (v/v) native polyacrylamide gels supplemented with 8% (v/v) glycerol, which were run in 0.5×TBE buffer (1×TBE=45 mM Tris/borate/1 mM EDTA) at 10 W and 4 °C. Gels were developed on a Typhoon 8600 phosphorimager (Molecular Dynamics/Amersham-Pharmacia).
UV-induced cross-linking
Analyses were carried out in 10 μl volumes in buffer C or in PBS, employing standard 1.5 ml reaction vials. Each reaction contained 60–300 pmol of tmaNusB, 4 pmol of radiolabelled RNA and various amounts (0, 20, 250 and 4000 pmol) of unlabelled competitor RNA (same sequence). The mixtures were exposed for 1 min to UV radiation (254 nm), boiled in SDS loading buffer and analysed on SDS/15%-polyacrylamide gels. Gels were developed on a Typhoon 8600 phosphorimager.
RESULTS AND DISCUSSION
Structure solution and quality of the models
Purified tmaNusB could be crystallized under various conditions in a monoclinic (P21), an orthorhombic (P212121) and three trigonal (two different P3121 forms and one P3221 form; Table 1) space groups. The crystal structures were solved by molecular replacement, employing a monomeric polyalanine model of mtuNusB [26]. During all refinement steps, all experimental data better than 20.0 Å (1 Å≡0.1 nm) resolution without intensity cut-offs were included, and the Rfree-factors were monitored continuously with 5% of the reflections (Table 1). In all structures, the main chains and the vast majority of side chains, except for some surface-exposed flexible residues, were completely enveloped in the final 2Fo–Fc electron density maps at the 1σ level. The final models contained all residues of the protein, some lacking the N-terminal methionine or the C-terminal leucine, and displayed good stereochemistry (Table 1) with low residual co-ordinate errors (approx. 0.1 Å [32]). The models portray the structure of tmaNusB at resolutions between 1.85 and 1.35 Å in different crystalline environments.
Overall fold
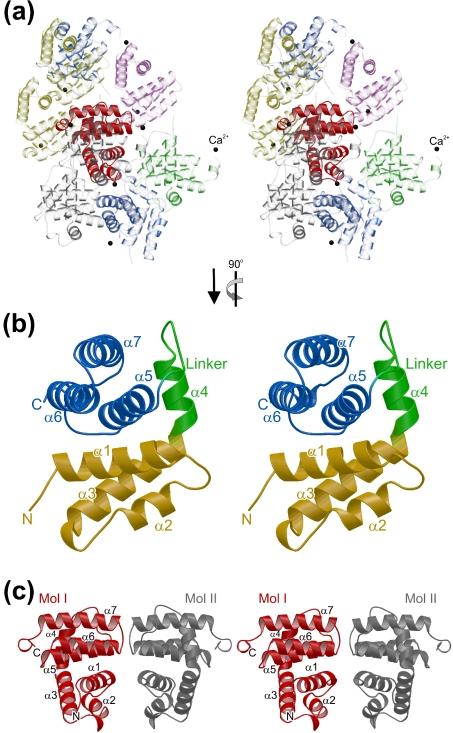
In all crystal forms, tmaNusB adopts a globular, all-helical fold (seven α-helices, two 310-helices) with approximate overall dimensions of 21 Å×22 Å×24 Å (Figure 1). All eight crystallographically independent molecules observed (Table 1) can be superimposed with pairwise rmsds (root-mean-square deviations) of approx. 0.7 Å for all 142 Cα-atoms and thus exhibit no major conformational differences. The structure of tmaNusB is therefore rather independent of the crystalline environment and will presumably be similar in solution.
Figure 1. Structures of tmaNusB.
(a) Stereo ribbon plot of a tmaNusB monomer (crystal form 3). Molecules surrounding a reference (solid red) are transparent and in various colours. Black spheres denote calcium ions, which mediate packing interactions in this crystal form but are not seen in the other structures. Unless mentioned otherwise, the orientation of the reference molecule in this figure is the default setting for all subsequent structural figures. By default, structural figures were prepared with BOBSCRIPT (http://www.strubi.ox.ac.uk/bobscript) and rendered with Raster3D [40]. (b) Close-up stereo view of the monomer rotated 90° counterclockwise around the vertical axis. The two subdomains are shown in yellow (N-terminal three-helix bundle) and blue (C-terminal three-helix bundle), with the intervening linker in green. Secondary structure elements are labelled. (c) Stereo ribbon plot of a tmaNusB dimeric arrangement as seen in crystal form 4. One monomer is shown in red and the other in grey. Secondary structure elements and the protein termini are labelled. The orientation of the left molecule is the same as that of the reference molecules in (a).
The tmaNusB structure can be divided into two subdomains, both of which comprise three-helix bundles, which can be superimposed on one another with an rmsd of 1.5 Å (37 matching Cα positions). Bundle I is built from helices α1 (Arg5–His19), α2 (Leu27–Ile34) and α3 (Lys41–Lys56), and bundle II is constructed from helices α5 (Val79–Phe94), α6 (Ile100–Tyr114) and α7 (Glu117–His134). Helix α4 (Leu58–Ser66), the following loop (Arg67–Ser73), helix 3101 (Leu74–Arg76) and another short loop (Leu77–Ser78) connect the two subdomains. The two helical bundles are stacked on each other at approximately right angles, with helices α1 and α3 of bundle I lying on top of helices α5 and α6 of bundle II (Figure 1b). Upon association, they cover ∼1560 Å2 of combined surface area. They interact through a hydrophobic patch in the centre and through multiple salt bridges and hydrogen bonds at the periphery of their contact surfaces.
Phylogenetic comparisons
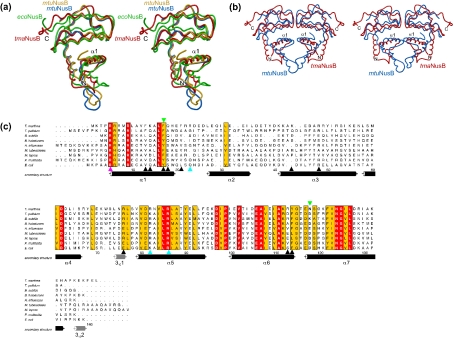
ecoNusB and mtuNusB, whose structures have been determined previously [25,26], contain respectively 40.9% and 41.7% identical residues with tmaNusB and consistently maintain globally quite similar folds. tmaNusB and mtuNusB can be superimposed with an rmsd of 1.44 Å for 127 matching Cα atoms (Figure 2a). ecoNusB fits on tmaNusB with an rmsd of 1.80 Å for 88 matching residues (Figure 2a). Local differences among these proteins are pronounced in the region around helices α2 and α3, which show relatively high sequence variation among NusB proteins (Figure 2c). Helix α2 in tmaNusB is shorter by one turn than the corresponding element in mtuNusB, and the directions of the α2 helices relative to the bulk of the proteins deviate by ∼15° from each other (Figure 2a). Whereas in tmaNusB and ecoNusB helix α2 is in close contact with helix α1, the element is more closely associated with helix α3 in mtuNusB. Overall, helix α2 and the following loop are further removed from the remainder of the molecule in mtuNusB, giving this protein a more elongated shape than tmaNusB.
Figure 2. Phylogenetic comparison of NusB proteins.
(a) Superimposition of known NusB structures. For clarity, only one tmaNusB model is shown (red). Both crystallographically independent molecules of the mtuNusB crystal structure (blue and gold; [26]) and the NMR structure of ecoNusB (green; [25]) are displayed. (b) Superimposition of the dimeric mtuNusB structure (blue) on a tmaNusB dimer as seen in crystal form 4 (red). More extensive dimer contacts in the mtuNusB molecules are apparent at the bottom, at the tip of helix α2. For orientation, N- and C-termini as well as helices α1 are labelled. (c) Alignment of nine bacterial NusB sequences. Numbering corresponds to tmaNusB. The secondary structure elements indicated below the sequences denote the situation seen in the present tmaNusB crystal structures. α-Helices are shown in black, and 310 helices in grey. The background of highly conserved amino acids is red, and that of intermediately conserved amino acids is yellow. Black arrows denote residues involved in binding anions in the various tmaNusB crystal forms. The magenta arrowhead indicates arginine, which in mtuNusB is involved in binding a phosphate ion. Cyan arrowheads indicate residues forming a positive surface patch, which may constitute an RNA binding site [25]. Green arrowheads indicate mutations analysed in the E. coli protein. Abbreviations: T. maritima, Thermotoga maritima; T. pallidum, Treponema pallidum; B. subtilis, Bacillus subtilis; B. halodurans, Bacillus halodurans; H. influenzae, Haemophilus influenzae; M. tuberculosis, Mycobacterium tuberculosis; M. leprae, Mycobacterium leprae; P. multocida, Pasteurella multocida; E. coli, Escherichia coli. The figure was prepared with ALSCRIPT [41].
Quarternary structures
In light of the previously observed mtuNusB dimers [26], we analysed the tmaNusB crystal structures for possible dimeric contacts. Important features that characterize dimerization motifs in proteins are a significant size of the dimer interface, the involvement of specific atomic interactions, the participation of conserved residues in the interchain contacts and the saturation of the interaction potential of the monomers, i.e. the formation of isologous contacts [33]. In crystal forms 1–3 (Table 1), we did not discern associations between neighbouring molecules which qualified as bona fide dimerization motifs (Figure 1a). In contrast, two molecules in the form 4 crystals were related through a crystallographic two-fold axis in an isologous fashion, resembling the subunit arrangement of the mtuNusB dimer [26] (Figures 1c and 2b). Moreover, crystal form 5 maintained an identical and crystallographically independent apposition of two molecules.
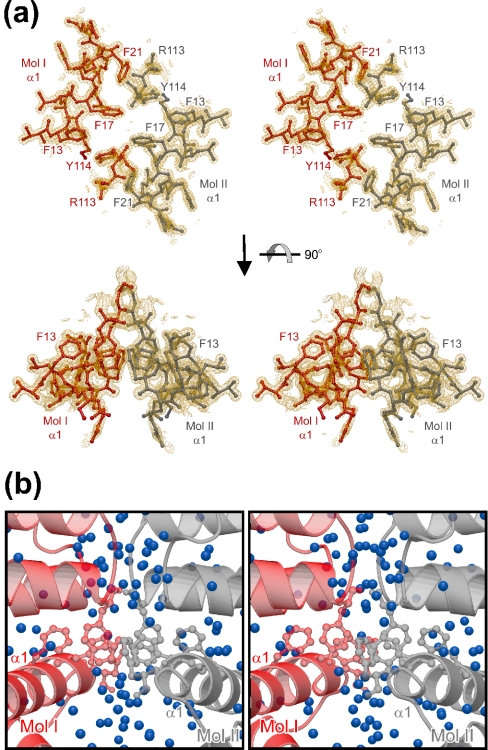
Two contact regions sustain the crystallographic tmaNusB dimers. First, the loop between helices α6 and α7 of one molecule lies alongside the loop preceding helix α5 of the other, and vice versa (Figure 1c). The shape complementarity of these regions leads to van der Waals contacts, especially around the side chains of Trp72 and Thr116. In addition, weak electrostatic interactions between Arg76 and Glu117 and between Lys71 and Asn118 can be discerned. Arg113 and Tyr114 from the C-terminus of helix α6 reach into the second contact site. Here, the N-terminal α1 helices of the two subunits run in antiparallel fashion through the body of the assembly, engaging in a number of side-chain to side-chain interactions (Figures 1c and 3a). Phe13, Phe17 and Phe21 project from one side of helix α1 into the direction of the other monomer and partially stack on each other. The Phe21 ring also faces Tyr114 from the opposite monomer. In total, eight aromatic residues are arranged in a circular interaction network at the core of the dimer (Figure 3a). The Arg113 side chain hydrogen bonds to the Tyr114 hydroxy group of its own monomer, partially intervening into the intermolecular Phe21–Tyr114 association. It thus reinforces the Phe21–Tyr114 interaction through cation–π contacts to the two aromatic ring systems. In addition, the electrostatic surface potential of tmaNusB reveals a charge complementarity at the dimer interface, which should lead to an attraction between the monomers (Figure 4a).
Figure 3. tmaNusB dimerization in the crystal.
(a) Stereo plot of the aromatic interactions in the dimer interface of crystal form 4. The orientation of the upper panel is that of Figure 1(a) rotated 90° into the plane of the paper around the vertical axis. Residues originating from one monomer are in red, and those from the other monomer are in grey. The aromatic residues participating in the interaction network are labelled. Arg113 may support the arrangement by additional cation–π interactions. A 2Fo–Fc electron density map for the final model contoured at the 2σ level is superimposed. (b) Close-up view of the dimer interface in default orientation. Molecules are coloured as in (a). Blue spheres denote water molecules in the structure, mediating a large number of indirect interactions between the monomers. The side chains of the critical aromatic amino acids are drawn in ball-and-stick notation.
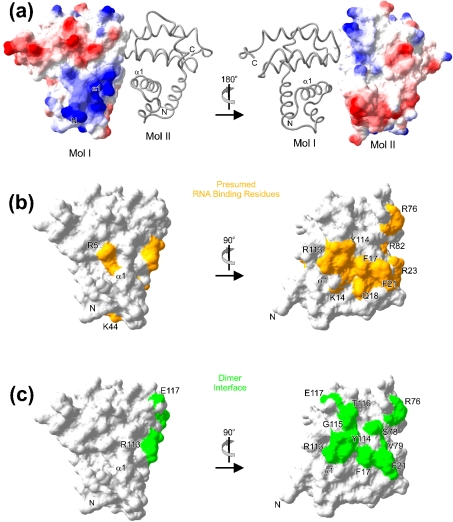
Figure 4. Surface topology of tmaNusB.
(a) Electrostatic surface potential calculated for one monomer, with the other displayed as a grey ribbon. Comparison of the 180° views illustrates a charge complementarity at the dimer interface. In panels depicting the electrostatic surface potential, negative potential is in red and positive potential is in blue. Surface images were prepared with the Swiss PDB Viewer (http://www.expasy.org/spdbv/mainpage.html). (b) Mapping of residues on the surface of tmaNusB which in the present or former structures have been implicated in RNA binding. Positions of individual residues are indicated. (c) Two orthogonal views of the dimer interface residues (green), as seen in the tmaNusB crystalline dimers, mapped on the surface of tmaNusB. A similar but more extended dimer interface is seen in mtuNusB (not shown). Residues are labelled and the positions of helix α1 and the N-terminus are indicated. Comparison with (b) indicates that the dimerization region and the RNA interaction region are largely coincident.
Notably, there is a gap of more than 30000 Å3 between the two subunits, mainly due to a deep cleft below the aromatic interaction motif (Figure 1c). Voids between the subunits are filled with well-ordered water molecules (Figure 3b), which mediate a considerable number of indirect interchain links. In the 1.35 Å structure of crystal form 4, there are ten single-water bridges and numerous additional connections through longer water chains. In addition, the tmaNusB dimer arrangement buries only ∼1050 Å2 of accessible surface area. This number falls at the lower margin for typical dimeric proteins [33]. The comparably small contact area, the sparse number of direct interactions, the large number of water-mediated interactions and the charged residues surrounding the hydrophobic patches suggest that tmaNusB dimers are facultative or unstable in solution [33]. This conclusion is consistent with our observation of both monomers and dimers in different crystal forms.
In mtuNusB, the monomers interact more intimately, burying approximately double the accessible surface area [26]. The different positioning of helix α2 in mtuNusB leads to additional contacts at the bottom of the dimers through the C-termini of these helices (Figure 2b). However, there is a large channel (∼5000 Å3) between the monomers. Similar to tmaNusB, there are 16 direct water bridges between the monomers, as well as one salt bridge and seven hydrogen bonds. These contacts again surround a central stacking interaction, which in mtuNusB involves only the two Phe22 residues of the α1 helices (Phe17 in tmaNusB).
tmaNusB is monomeric in solution
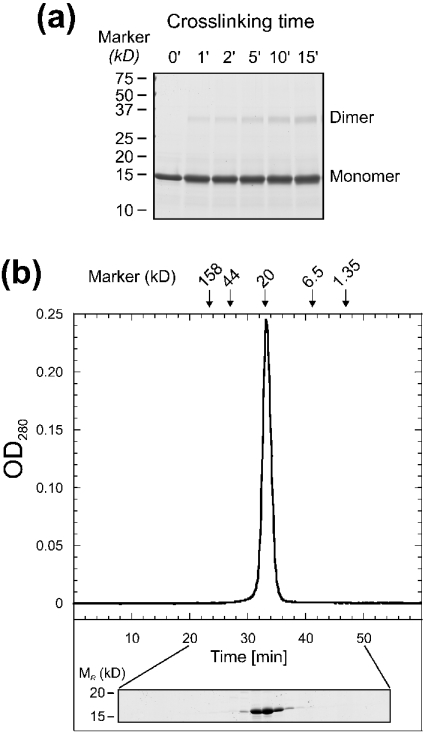
The similarity to the mtuNusB dimers suggested that the dimeric associations in the form 4 and 5 crystals of tmaNusB could be physiologically relevant. We therefore tested the potential of tmaNusB to dimerize in solution by glutaraldehyde cross-linking under conditions where monomeric proteins did not produce covalently bonded aggregates in our hands [34]. A continuous accumulation of covalent tmaNusB dimers was observed throughout the reaction (Figure 5a). However, in analytical gel filtration analyses with various buffer systems, tmaNusB clearly migrated as a monomer (Figure 5b). We therefore conclude that, in solution, tmaNusB does not assemble into stable dimers, but molecules may associate transiently in the fashion seen in crystal forms 4 and 5.
Figure 5. Oligomeric state of tmaNusB in solution.
(a) SDS/PAGE showing the time course of a glutaraldehyde cross-linking experiment with tmaNusB. Samples were taken between 1 and 15 min after the onset of the reaction, as indicated. (b) Representative gel-filtration analysis of tmaNusB. The elution time of the single peak corresponds to a monomer molecular mass (∼18 kDa). Arrows indicate elution peaks of reference molecules. OD280=A280. The protein profile of the run as seen on an SDS/polyacrylamide gel is shown below the graph.
Binding of tmaNusB to boxA-like RNA
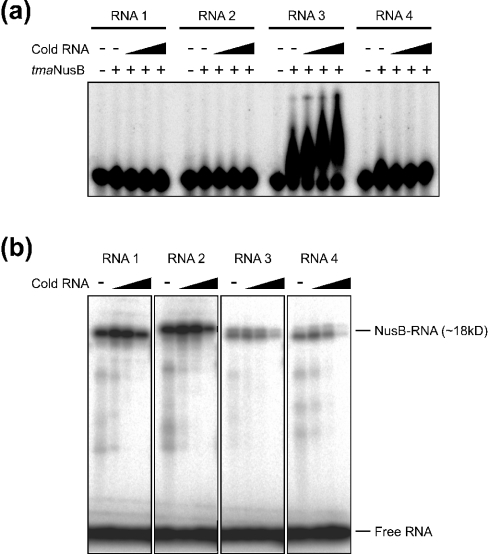
Whereas ecoNusB has been shown to bind boxA RNA [19,22,23], Gopal et al. [35] failed to detect binding of mtuNusB to boxA RNA in vitro. In order to test RNA binding by tmaNusB, two variants of the E. coli rrn-boxA sequence (5′-UGCUCUUUA-3′ and 5′-UGCUCUUUAACA-3′, termed RNA1 and RNA2 respectively) and two sequences from the T. maritima rrn leader and spacer regions (5′GGGUCUUCA-3′ and 5′-GGCUCAUUG-3′; RNA3 and RNA4 respectively), which had been suggested previously to constitute boxA elements in Thermotoga [15], were chemically synthesized and probed in electrophoretic gel mobility shift assays. RNA3 was seen to bind to tmaNusB better than the other RNAs (Figure 6a), indicating some specificity of the interaction. However, binding was weak or transient, because the complex seemed to dissociate during electrophoresis. Binding was enhanced when unlabelled RNA of the same type was added to the reactions (Figure 6a). Such co-operative effects could be due to a conformational change in the protein upon binding or to the uncovering of another binding site after association with the first RNA molecule. In any case, the RNA binding capacity of NusB molecules seems to parallel their oligomeric state in solution: ecoNusB or tmaNusB, which exist preferentially as monomers, exhibit binding capacity, while dimeric mtuNusB does not.
Figure 6. RNA binding of tmaNusB.
(a) Electrophoretic gel mobility shift analysis of tmaNusB–boxA complexes. Only RNA3 was shifted by the protein under the experimental conditions (see the Experimental section). The shapes of the retarded bands suggest that the tmaNusB–RNA3 complex is not stable, but partially dissociates during the run. Complex stability is increased by the addition of unlabelled RNA3. (b) SDS/PAGE analysis of tmaNusB UV-cross-linked to various boxA-like RNA oligomers. Each reaction contained 300 pmol of NusB and 4 pmol of radiolabelled RNA. The first to fourth lanes of each block contained increasing amounts (0, 20, 250 and 4000 pmol) of unlabelled (‘cold’) competitor RNA (same sequence) as indicated above the gel. Because the short RNAs do not markedly modulate the migration behaviour of the proteins on a denaturing SDS/polacrylamide gel, the cross-link at ∼18 kDa presumably corresponds to monomeric NusB–boxA complexes.
In contrast with the results of the electrophoretic gel-mobility shift assays, all RNA sequences tested could be covalently attached to tmaNusB by UV-induced cross-linking (Figure 6b). These results confirm that tmaNusB associates transiently with various RNA sequences. The cross-link to labelled RNA was enhanced by the addition of small amounts (in the order of the protein concentration) of unlabelled oligomer; larger amounts of unlabelled RNA led to a decrease in the cross-linking to labelled RNA (Figure 6b). These results confirm that boxA-like sequences bind co-operatively to NusB, e.g. by eliciting a conformational change in the protein.
A putative RNA binding region at the NusB N-terminus
The above RNA interaction studies validate an interpretation of the tmaNusB structures in terms of an RNA binding fold. Approximately one-third of the first 30 residues of tmaNusB are Arg and Lys residues. This arginine-rich motif is conserved in other NusB molecules (Figure 2c). It gives rise to a broad stretch of positive surface potential, which extends along the centre of the molecules (Figure 4a) and appears well suited for interaction with the negatively charged sugar phosphate backbone of a nucleic acid.
We located several anions bound to this region in various tmaNusB crystal structures, which mark possible attachment sites for the RNA sugar phosphate backbone. In the form 5 crystals, two citrate ions are bound to the surfaces of the two α1 helices of the dimer. Each citrate ion lies along the aliphatic portion of the Lys14 side chain. One of the terminal carboxy groups forms salt bridges with Arg113 and a hydrogen bond to the Tyr114 hydroxy group. The 2-carboxy group is hydrogen-bonded to the amide group of Gln18 of the other monomer. In the P21 and P212121 crystal forms, several sulphate ions were bound to Lys44, Arg48 and Arg76, which flank the binding site for the citrate ions.
In other RNA binding proteins, aromatic residues are often essential for stacking to nucleic acid bases [36]. As pointed out above, residues Phe13, Phe17, Phe21 and the side chain of Tyr114 lie along the surface of helix α1 and could thus play a similar role in tmaNusB. Among these aromatics, Phe17 and Tyr114 show the highest degree of conservation and are particularly interesting candidates for RNA contact sites. Consistent with this interpretation, the ecoNusB5 mutant (Y18D in ecoNusB; Phe17 in tmaNusB, Figure 2c) abrogates λN-mediated antitermination activity [22]. NusB has been found to act as a termination enhancer at a suboptimal Rho site. To perform this function, it has to interact with a variant boxA sequence upstream of the terminator [37]. The data presented by Carlomagno and Nappo [37] strongly support the suggestion that Phe17 (Tyr18 in ecoNusB) is directly involved in boxA RNA binding.
The above localization of a NusB–RNA interaction site is also consistent with previous structural studies. Chemical shift changes upon titration of ecoNusB with a boxA RNA dodecamer [38] suggested contacts of the RNA with the N-terminal region of the protein. Furthermore, Altieri et al. [25] pointed out that the region around Arg82 and Arg86 exhibits features typical of an RNA binding site. These two highly conserved arginines form a positively charged surface patch, which in tmaNusB is enlarged by Arg23. This patch lies above the C-terminal end of helix α1. Finally, the mtuNusB crystal structure includes a phosphate ion bound in the N-terminal region to Arg10 (Arg5 in tmaNusB) [26], located at the entrance to helix α1. Taken together, the sequence of and anion binding by tmaNusB, as well as previous structural analyses of NusB proteins, suggest that NusB harbours a continuous RNA interaction region along the centre of the protein, largely following the trace of helix α1 (Figure 4b).
Hypothesis – dimerization as a silencing mechanism for some NusB proteins
In stable NusB dimers, such as mtuNusB, the region implicated above in RNA binding overlaps with the dimerization interface (Figures 4b and 4c). Thus concomitant dimerization and interaction with the RNA are seemingly incompatible. The dilemma could be resolved if NusB proteins that form dimers underwent a large conformational change to expose helix α1 or were converted into monomers for RNA interaction. Such structural changes of NusB may depend on other factors within antitermination complexes. In this scenario, dimerization could be exploited in some bacteria to inactivate NusB until it is needed for transcriptional regulation. The crystalline dimers of tmaNusB feature a large gap below the putative RNA binding site and thus still allow access for small anions, but may also obstruct binding of larger RNA molecules. Conspicuously, mechanisms of tuneable RNA recognition have been identified previously in NusA [39] and have been suggested for NusG [34]. They therefore may emerge as a general theme in nucleic acid binding by the Nus factors, exploited for regulating termination and antitermination processes.
Acknowledgments
We thank Gleb P. Bourenkov (DESY, Hamburg, Germany) for help during synchrotron data collection, and Heiko Manninga (Max-Planck Insitut für Biophysikalische Chemie) for his assistance with RNA synthesis. This work was supported by the Max-Planck-Society and the Stiftung Volkswagenwerk.
References
- 1.Roberts J. W. Trancription termination and its control. In: Lin E., Lynch A., editors. Regulation of Gene Expression in E. coli. Austin, TX: R.G. Landes Co.; 1996. pp. 27–44. [Google Scholar]
- 2.Weisberg R. A., Gottesman M. E. Processive antitermination. J. Bacteriol. 1999;181:359–367. doi: 10.1128/jb.181.2.359-367.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman D. I. Regulation of phage gene expression by termination and antitermination of transcription. In: Calender R., editor. The Bacteriophages. New York: Plenum Press; 1988. pp. 263–319. [Google Scholar]
- 4.Greenblatt J., Nodwell J. R., Mason S. W. Transcriptional antitermination. Nature (London) 1993;364:401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- 5.Friedman D. I., Court D. L. Transcription antitermination: the lambda paradigm updated. Mol. Microbiol. 1995;18:191–200. doi: 10.1111/j.1365-2958.1995.mmi_18020191.x. [DOI] [PubMed] [Google Scholar]
- 6.Richardson J. P., Greenblatt J. Control of RNA chain elongation and termination. In: Neidhardt F. C., Curtiss R. III, Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., Reznikoff W. S., Riley M., Schaechter M., Umbarger H. E., editors. Escherichia coli and Salmonella typhimirium: Cellular and Molecular Biology. Washington, DC: American Society for Microbiology; 1996. pp. 822–848. [Google Scholar]
- 7.von Hippel P. H., Rees W. A., Rippe K., Wilson K. S. Specificity mechanisms in the control of transcription. Biophys. Chem. 1996;59:231–246. doi: 10.1016/0301-4622(96)00006-3. [DOI] [PubMed] [Google Scholar]
- 8.Greenblatt J., Mah T. F., Legault P., Mogridge J., Li J., Kay L. E. Structure and mechanism in transcriptional antitermination by the bacteriophage lambda N protein. Cold Spring Harbor Symp. Quant. Biol. 1998;63:327–336. doi: 10.1101/sqb.1998.63.327. [DOI] [PubMed] [Google Scholar]
- 9.Nodwell J. R., Greenblatt J. The nut site of bacteriophage lambda is made of RNA and is bound by transcription antitermination factors on the surface of RNA polymerase. Genes Dev. 1991;5:2141–2151. doi: 10.1101/gad.5.11.2141. [DOI] [PubMed] [Google Scholar]
- 10.DeVito J., Das A. Control of transcription processivity in phage lambda: Nus factors strengthen the termination-resistant state of RNA polymerase induced by N antiterminator. Proc. Natl. Acad. Sci. U.S.A. 1994;91:8660–8664. doi: 10.1073/pnas.91.18.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mason S. W., Li J., Greenblatt J. Host factor requirements for processive antitermination of transcription and suppression of pausing by the N protein of bacteriophage lambda. J. Biol. Chem. 1992;267:19418–19426. [PubMed] [Google Scholar]
- 12.Mogridge J., Mah T. F., Greenblatt J. Involvement of boxA nucleotides in the formation of a stable ribonucleoprotein complex containing the bacteriophage lambda N protein. J. Biol. Chem. 1998;273:4143–4148. doi: 10.1074/jbc.273.7.4143. [DOI] [PubMed] [Google Scholar]
- 13.Aksoy S., Squires C. L., Squires C. Evidence for antitermination in Escherichia coli rRNA transcription. J. Bacteriol. 1984;159:260–264. doi: 10.1128/jb.159.1.260-264.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S. C., Squires C. L., Squires C. Antitermination of E. coli rRNA transcription is caused by a control region segment containing lambda nut-like sequences. Cell. 1984;38:851–860. doi: 10.1016/0092-8674(84)90280-0. [DOI] [PubMed] [Google Scholar]
- 15.Berg K. L., Squires C., Squires C. L. Ribosomal RNA operon anti-termination. Function of leader and spacer region box B-box A sequences and their conservation in diverse micro-organisms. J. Mol. Biol. 1989;209:345–358. doi: 10.1016/0022-2836(89)90002-8. [DOI] [PubMed] [Google Scholar]
- 16.Squires C. L., Greenblatt J., Li J., Condon C. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc. Natl. Acad. Sci. U.S.A. 1993;90:970–974. doi: 10.1073/pnas.90.3.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Condon C., Squires C., Squires C. L. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zellars M., Squires C. L. Antiterminator-dependent modulation of transcription elongation rates by NusB and NusG. Mol. Microbiol. 1999;32:1296–1304. doi: 10.1046/j.1365-2958.1999.01442.x. [DOI] [PubMed] [Google Scholar]
- 19.Mogridge J., Mah T. F., Greenblatt J. A protein-RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the lambda N protein. Genes Dev. 1995;9:2831–2845. doi: 10.1101/gad.9.22.2831. [DOI] [PubMed] [Google Scholar]
- 20.Mason S. W., Li J., Greenblatt J. Direct interaction between two Escherichia coli transcription antitermination factors, NusB and ribosomal protein S10. J. Mol. Biol. 1992;223:55–66. doi: 10.1016/0022-2836(92)90715-v. [DOI] [PubMed] [Google Scholar]
- 21.Nodwell J. R., Greenblatt J. Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell. 1993;72:261–268. doi: 10.1016/0092-8674(93)90665-d. [DOI] [PubMed] [Google Scholar]
- 22.Court D. L., Patterson T. A., Baker T., Costantino N., Mao X., Friedman D. I. Structural and functional analyses of the transcription-translation proteins NusB and NusE. J. Bacteriol. 1995;177:2589–2591. doi: 10.1128/jb.177.9.2589-2591.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luttgen H., Robelek R., Muhlberger R., Diercks T., Schuster S. C., Kohler P., Kessler H., Bacher A., Richter G. Transcriptional regulation by antitermination. Interaction of RNA with NusB protein and NusB/NusE protein complex of Escherichia coli. J. Mol. Biol. 2002;316:875–885. doi: 10.1006/jmbi.2001.5388. [DOI] [PubMed] [Google Scholar]
- 24.Patterson T. A., Zhang Z., Baker T., Johnson L. L., Friedman D. I., Court D. L. Bacteriophage lambda N-dependent transcription antitermination. Competition for an RNA site may regulate antitermination. J. Mol. Biol. 1994;236:217–228. doi: 10.1006/jmbi.1994.1131. [DOI] [PubMed] [Google Scholar]
- 25.Altieri A. S., Mazzulla M. J., Horita D. A., Heath Coats R., Wingfield P. T., Das A., Court D. L., Byrd A. R. The structure of the transcriptional antiterminator NusB from Escherichia coli. Nat. Struct. Biol. 2000;7:470–474. doi: 10.1038/75869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gopal B., Haire L. F., Cox R. A., Jo Colston M., Major S., Brannigan J. A., Smerdon S. J., Dodson G. The crystal structure of NusB from Mycobacterium tuberculosis. Nat. Struct. Biol. 2000;7:475–478. doi: 10.1038/75876. [DOI] [PubMed] [Google Scholar]
- 27.Scaringe S. A. RNA oligonucleotide synthesis via 5′-silyl-2′-orthoester chemistry. Methods. 2001;23:206–217. doi: 10.1006/meth.2000.1132. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z., Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1996;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project No. 4. The CCP4 Suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Turk D. MAIN 96: An interactive software for density modifications, model building, structure refinement and analysis. In: Bourne P. E., Watenpaugh K., editors. Meeting of the International Union of Crystallography Macromolecular Macromolecular Computing School. WA: International Union of Crystallography, Western Washington University; 1996. [Google Scholar]
- 32.Luzzati V. Traitement statistique des erreurs dans la détermination des structures cristallines. Acta Crystallogr. A. 1952;5:802–810. [Google Scholar]
- 33.Jones S., Thornton J. M. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steiner T., Kaiser J. T., Marinkovic S., Huber R., Wahl M. C. Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. EMBO J. 2002;21:4641–4653. doi: 10.1093/emboj/cdf455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gopal B., Papavinasasundaram K. G., Dodson G., Colston M. J., Major S. A., Lane A. N. Spectroscopic and thermodynamic characterization of the transcription antitermination factor NusE and its interaction with NusB from Mycobacterium tuberculosis. Biochemistry. 2001;40:920–928. doi: 10.1021/bi0018279. [DOI] [PubMed] [Google Scholar]
- 36.Oubridge C., Ito N., Evans P. R., Teo C. H., Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature (London) 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 37.Carlomagno M. S., Nappo A. The antiterminator NusB enhances termination at a sub-optimal Rho site. J. Mol. Biol. 2001;309:19–28. doi: 10.1006/jmbi.2001.4678. [DOI] [PubMed] [Google Scholar]
- 38.Huenges M., Rolz C., Gschwind R., Peteranderl R., Berglechner F., Richter G., Bacher A., Kessler H., Gemmecker G. Solution structure of the antitermination protein NusB of Escherichia coli: a novel all-helical fold for an RNA-binding protein. EMBO J. 1998;17:4092–4100. doi: 10.1093/emboj/17.14.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mah T. F., Kuznedelov K., Mushegian A., Severinov K., Greenblatt J. The alpha subunit of E. coli RNA polymerase activates RNA binding by NusA. Genes Dev. 2000;14:2664–2675. doi: 10.1101/gad.822900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merritt E. A., Bacon D. J. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 41.Barton G. J. ALSCRIPT – A tool for multiple sequence alignments. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]