Abstract
CPPs (cell-penetrating peptides) facilitate the cellular uptake of covalently attached oligonucleotides, proteins and other macromolecules, but the mechanism of their uptake is disputed. Two models are proposed: direct movement through the phospholipid bilayer and endocytic uptake. Mitochondria are a good model system to distinguish between these possibilities, since they have no vesicular transport systems. Furthermore, CPP-mediated delivery of macromolecules to the mitochondrial matrix would be a significant breakthrough in the study of mitochondrial function and dysfunction, and could also lead to new therapies for diseases caused by mitochondrial damage. Therefore we investigated whether two CPPs, penetratin and Tat, could act as mitochondrial delivery vectors. We also determined whether conjugation of the lipophilic cation TPP (triphenylphosphonium) to penetratin or Tat facilitated their uptake into mitochondria, since TPP leads to uptake of attached molecules into mitochondria driven by the membrane potential. Neither penetratin nor Tat, nor their TPP conjugates, are internalized by isolated mitochondria, indicating that these CPPs cannot cross mitochondrial phospholipid bilayers. Tat and TPP–Tat are taken up by cells, but they accumulate in endosomes and do not reach mitochondria. We conclude that CPPs cannot cross mitochondrial phospholipid bilayers, and therefore cannot deliver macromolecules directly to mitochondria. Our findings shed light on the mechanism of uptake of CPPs by cells. The lack of direct movement of CPPs through mitochondrial phospholipid bilayers, along with the observed endosomal accumulation of Tat and TPP–Tat in cells, makes it unlikely that CPPs enter cells by direct membrane passage, and instead favours cellular uptake via an endocytic pathway.
Keywords: cell-penetrating peptide, mitochondrion, penetratin, phospholipid bilayer permeation, protein transduction domain, targeting
Abbreviations: CPP, cell-penetrating peptide; DIPEA, di-isopropylethylamine; DMEM, Dulbecco's modified Eagle's medium; DMF, N,N-dimethylformamide; ER, endoplasmic reticulum; ESI, electrospray ionization; FCCP, carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone; FCS, foetal calf serum; KLH, keyhole limpet haemocyanin; MALDI–TOF, matrix-assisted laser-desorption/ionization–time-of-flight; MBS, m-maleimidobenzoic acid N-hydroxysuccinimide ester; mtDNA, mitochondrial DNA; Δψm, mitochondrial membrane potential; Pen, penetratin; PyBOP, benzotriazol-1-yloxytris(pyrrolidino)phosphonium hexafluorophosphate; RP, reverse-phase; TFA, trifluoroacetic acid; TPMP, methyltriphenylphosphonium; TPP, triphenylphosphonium; Twvp, Trimeresurus wagleri venom peptide II
INTRODUCTION
Delivery of large polar molecules, such as nucleic acids or proteins, to cells enables the selective manipulation of cell function. One of the most useful strategies for macromolecule delivery is the use of CPPs (cell-penetrating peptides) [1], which are 10–30-amino-acid peptides that mediate the cellular uptake of large and polar molecules. Covalent attachment of CPPs to proteins [2], oligonucleotides [3,4] and other molecules [5,6] leads to their intracellular uptake and to the expression of their biological activity within the cell. This enables manipulation of the cellular environment for investigative purposes, as well as the development of potential therapies [7,8]. The most intensively studied CPPs are Pen (penetratin), which is the DNA-binding domain of the Drosophila melanogaster homeoprotein Antennapedia [9], and Tat, which is the DNA-binding helix of the HIV-1 transactivator protein [10–12].
CPPs have been used to deliver cargo molecules to the cytosol and nucleus; however, the mitochondrion is another important potential target for the delivery of macromolecules within the cell [13]. Mitochondria are at the heart of many crucial cellular processes, including oxidative phosphorylation, iron–sulphur centre and haem synthesis, the TCA (tricarboxylic acid) cycle, fatty acid oxidation, calcium homoeostasis and apoptosis. Consequently, damage to mitochondria contributes to the pathophysiology of a wide range of human diseases, including diabetes [14], Parkinson's disease [15] and Friedreich's ataxia [16]. Because assembly of functional mitochondria requires expression of mtDNA (mitochondrial DNA), there are also many diseases caused by mtDNA mutation [17], and our knowledge of the expression and maintenance of the mitochondrial genome is limited [18]. It would therefore be advantageous to deliver macromolecules such as proteins, peptides and nucleic acids directly to mitochondria in vivo and in vitro, in order to protect, repair or report on aspects of mitochondrial function and dysfunction. However, this has proven difficult in mammalian systems because in addition to the challenges of delivering the macromolecule to the cytoplasm, it must then be targeted further across the mitochondrial outer and inner membranes to the matrix [19]. The only method that has worked to date for proteins [20], oligonucleotides [21] and oligonucleotide analogues [22] is targeting by conjugation to a mitochondrial protein import sequence. However, this approach is limited by the difficulties of transporting macromolecules to the cytoplasm of cells in culture and in vivo. Therefore what is required is a vector that can deliver large, polar molecules to mitochondria in cells in vitro and in vivo. CPPs are promising candidates for such a vector; however, there is uncertainty about their mechanism of uptake into cells, which has an impact on their potential utility as mitochondrial vectors.
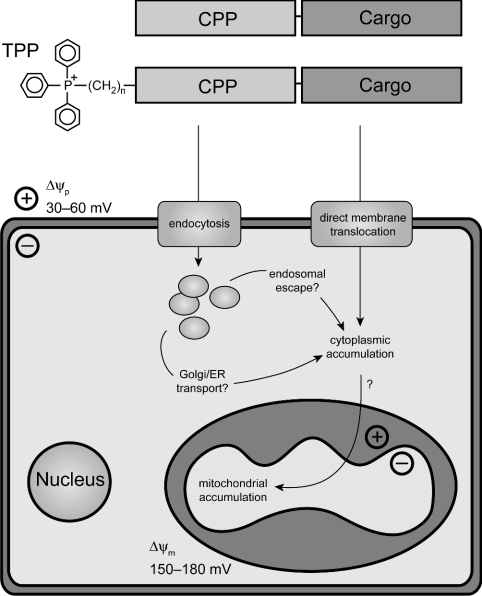
While it is clear that the uptake into cells of molecular cargo attached to CPPs occurs by a non-receptor-mediated process, the mechanism remains controversial. Two main models have been advanced (Figure 1): either CPPs destabilize the phospholipid bilayer, to form inverted micelles that enable the CPP and its cargo to enter cells without leaving an aqueous environment, or, alternatively, the mechanism of uptake involves an endocytic pathway. The inverted-micelle mechanism of CPP uptake was initially proposed because cellular uptake of Pen and Tat, measured by flow cytometry and by fluorescence microscopy, was rapid, temperature- and receptor-independent, and insensitive to disruptors of endocytosis [12,23]. However, these findings have been called into question by the artifactual cytoplasmic and nuclear localization on fixation of cells for fluorescence microscopy and by the adherence of CPPs to the cell surface [24,25]. Recent experiments, which have avoided these fixation artifacts by analysing the uptake of fluorescent peptide derivatives into live cells by confocal fluorescence microscopy, indicate an endocytic mode of CPP uptake [24,26–30]. The endocytic pathway by which CPPs are internalized is uncertain, and may differ according to the attached cargo. CPPs without cargo have been shown to co-localize with transferrin, a marker of clathrin-dependent endocytosis [24,28]. In contrast, caveolar uptake was observed for a Tat–EGFP (enhanced green fluorescent protein) fusion, while a Tat–Cre recombinase fusion protein was internalized by macropinocytosis [26,27]. However, even if uptake is via endocytosis, the CPPs must still escape from endocytic vesicles, as their cargos are known to exert biological activity in the cytosol. How this occurs is not known, but the release of CPPs from endocytic vesicles [28] or from the Golgi/ER (endoplasmic reticulum) [26,29] without disrupting the phospholipid bilayer may indicate that CPPs can cross intracellular membranes as the final step of internalization, possibly facilitated by the lower endosomal pH [28]; alternatively, high concentrations of CPPs within endosomes may destabilize the endosome, leading to leakage into the cytosol [30]. However, there is still considerable uncertainty about the mechanism of uptake into cells, and it has also been shown that CPPs can cross simple phospholipid bilayers in artificial liposomes, and that the rate of translocation is increased by a transmembrane potential [31,32].
Figure 1. A modular vector to target polar macromolecules to mitochondria.
The CPPs Pen and Tat were synthesized with and without TPP to form vectors for delivery of a macromolecular cargo. The vector–cargo complexes would first be taken up into the cytoplasm of cells, either through direct transport across the plasma membrane in inverted micelles [23,31], or via endocytosis, followed by release of the complex into the cytoplasm by endosomal escape [27,28] or by retrograde transport to the Golgi/ER and subsequent release [29]. The vector–cargo complexes should then have the ability to cross mitochondrial membranes directly, possibly facilitated by the action of the Δψm on the fixed positive charge of the TPP.
As well as being an important cellular target for delivery of macromolecules, mitochondria are a good model system to study the mechanism of cellular uptake of CPPs. If uptake is via inverted micelle formation, then the CPP vector and its cargo should cross all phospholipid bilayers and be able to enter mitochondria. In contrast, if uptake is endocytic, then CPPs will not deliver their cargo to mitochondria, which do not participate in intracellular vesicular transport. Therefore, as a topologically closed biological membrane that contains membrane proteins, but does not participate in endocytosis, isolated mitochondria are a better model than pure phospholipid liposomes for studying the mechanism of CPP internalization [31,32].
If CPPs were to cross phospholipid bilayers through inverted micelles, once in the cell the CPP–cargo would distribute randomly to other membrane-bound compartments such as the Golgi, ER and mitochondria. Ideally, a mitochondrial delivery vector would show selectivity over other organelles by bringing about its own mitochondrial accumulation. One way to achieve this may be by exploiting the large Δψm (mitochondrial membrane potential), which drives the accumulation of lipophilic cations such as TPP (triphenylphosphonium) into mitochondria in cells in vitro and in vivo [33,34]. These cations pass easily through phospholipid bilayers, and conjugation to TPP has been used to target non-polar molecules such as Vitamin E and ubiquinol derivatives [35,36], thiol probes [37,38] and peptide nucleic acids [39] to mitochondria. Therefore the addition of TPP to a CPP might yield a vector that both transported large polar molecules across membranes and also facilitated their accumulation in mitochondria; such a vector would be a useful tool for mitochondrial research, with potential therapeutic applications. For simple lipophilic cations such as TPMP (methyltriphenylphosphonium), mitochondrial uptake is Nernstian, with approx. 10-fold accumulation for every 61.5 mV of Δψm, leading to the several hundred-fold accumulation in mitochondria within cells [19,33]. Although it is unclear whether TPP conjugation to a highly basic CPP would affect CPP distribution in response to Δψm, the fixed positive charge of the TPP might facilitate mitochondrial uptake. Furthermore, a membrane potential (negative inside) has been shown to enable membrane translocation by Pen into liposomes, increasing the likelihood of successful mitochondrial uptake by CPPs [32].
In the present study, we determined whether the CPPs Tat and Pen crossed mitochondrial phospholipid bilayers, and whether conjugation of CPPs to TPP facilitated uptake by isolated mitochondria and by mitochondria in cells. We found no evidence for mitochondrial uptake of CPPs, suggesting that CPPs cannot cross mitochondrial phospholipid bilayers. Furthermore, the accumulation of Tat and TPP–Tat within endosomes suggests that CPP uptake into cells is not by direct passage through inverted micelles within the phospholipid bilayer, and that uptake is by an endocytic mechanism.
MATERIALS AND METHODS
Peptide synthesis, derivatization, purification and analysis
Peptide synthesis and purification
Syntheses of Pen (H2N-RQIKIWFQNRRMKWKK-COOH), Tat (H2N-YGRKKRRQRRRP-COOH), Tat-Cys (H2N-YGRKKRRQRRRPC-COOH), Cys-Tat (H2N-CYGRKKRRQRRRP-COOH) and Cys-Pen (H2N-CRQIKIWFQNRRMKWKK-COOH) were carried out in the solid phase using a Pioneer peptide synthesizer (Applied Biosystems) following the Fmoc (fluoren-9-ylmethoxycarbonyl) chemistry protocol, using PyBOP [benzotriazol-1-yloxytris(pyrrolidino)phosphonium hexafluorophosphate]/DIPEA (di-isopropylethylamine)/DMF (N,N-dimethylformamide) coupling reactions. Underivatized peptides were cleaved and deprotected in 10 ml of 94:3:3 TFA (trifluoroacetic acid)/TIS (triisopropylsilane)/water for 2 h. The volume was reduced to <5 ml by sparging with N2, then peptides were precipitated with excess diethyl ether (50 ml), harvested by centrifugation (3000 g for 5 min) and washed four times in diethyl ether. Peptides were then purified by RP (reverse-phase) HPLC (Gilson 321 pump, 1 ml·min−1) on a C18 column (Jupiter 300 Å; Phenomenex), using a linear gradient of 4.5–90% acetonitrile in 0.1% TFA over 30 min, and detected at 220 nm using a Gilson UV/VIS 151 spectrophotometer. The major peak was collected, lyophilized and resuspended in water. The identity and purity of the peptides was confirmed by MALDI–TOF (matrix-assisted laser-desorption/ionization–time-of-flight) or ESI (electrospray ionization) MS. MALDI–TOF spectra were obtained with a TofSpec2E mass spectrometer (MicroMass, Manchester, U.K.) in reflectron mode, using α-cyano-4-hydroxycinnamic acid as a matrix; ESI spectra were obtained by the Protein and Nucleic Acid Chemistry Facility (Department of Biochemistry, University of Cambridge).
TPP conjugation to peptides
A series of TPP–carboxylic acids with 4-, 8- and 12-carbon alkyl chains was synthesized as follows: (3-carboxypropyl)triphenylphosphonium iodide (1; [40]) was synthesized by heating 4-iodobutanoic acid (600–1000 mg, 3–5 mmol) and 1 equivalent of triphenylphosphine at 85 °C for 1 h, under N2. After cooling, the white solid was triturated with diethyl ether and subjected to three rounds of dissolution in ethanol followed by precipitation and trituration in diethyl ether. The crude product was then stirred with excess ether at room temperature (21 °C) until a white crystalline solid was formed (approx. 2 h). The solid was isolated and washed, and traces of solvent were removed under vacuum. (7-Carboxyheptyl)triphenylphosphonium bromide (2; [41]) and (11-carboxyundecyl)triphenylphosphonium bromide (3; [40]) were synthesized by heating 8-bromooctanoic acid (600–1000 mg, 3–5 mmol) or 12-bromododecanoic acid (800–1400 mg, 3–5 mmol) respectively with 1 equivalent of triphenylphosphine in a minimal volume (3–5 ml) of toluene at 110 °C for 4 h, under N2 with refluxing. Subsequent steps were as described for 1, except that dichloromethane was used in place of ethanol to dissolve the precipitated product. The identity of all three TPP–carboxylic acids was confirmed by 1H-NMR (299.9 MHz; using tetramethylsilane as a standard) and 31P-NMR (121.4 MHz; using 85% phosphoric acid as a standard) in 2H2O (1) or [2H]chloroform (2, 3), using a Varian Inova-300 spectrometer. Data (in p.p.m.) were as follows: 1: 1H-NMR: 1.82 (m, 2 H, -CH2-CH2-CH2-), 2.47 (t, J=6.75 Hz, 2 H, -CH2-CH2-COOH), 3.22 (m, 2 H, PPh3-CH2-CH2-), 7.65 (m, ∼15 H, -PPh3). 31P-NMR: 24.1. 2: 1H-NMR: 1.31 (m, 4 H, 2× -CH2-), 1.60 (m, 6 H, 3× -CH2-), 2.49 (t, J=7.20 Hz, 2 H, -CH2-CH2-COOH), 3.64 (m, 2 H, PPh3-CH2-CH2-), 7.78 (m, ∼15 H, -PPh3). 31P-NMR: 25.6. 3: 1H-NMR: 1.22 (m, 12 H, 6× -CH2-), 1.60 (m, 6 H, 3× -CH2-), 2.40 (t, J=7.20 Hz, 2 H, -CH2-CH2-COOH), 3.75 (m, 2 H, PPh3-CH2-CH2-), 7.80 (m, ∼15 H, -PPh3). 31P-NMR: 25.6. Ph indicates phenyl.
TPP–carboxylic acids were coupled manually to synthetic peptides in the solid phase. A 4-fold molar excess over peptide of 1, 2 or 3 and in situ activator PyBOP were dissolved in minimal DMF, and DIPEA (8-fold molar excess over peptide) was added. The mixture was reacted with peptidated resin at room temperature for 8 h. The resin was then washed with DMF followed by propan-2-ol, and stored in vacuo. Deprotection, cleavage, purification and analysis were as described above for underivatized peptides. RP-HPLC purification demonstrated a characteristic retardation (2–3 min) of TPP–peptides on RP-HPLC with respect to underivatized peptides, consistent with the expected increase in hydrophobicity on TPP conjugation.
Peptide quantification
An online Biopolymer Calculator (http://paris.chem.yale.edu/extinct.html) was used to calculate ε280 (M−1·cm−1) for Pen and Tat. To obtain ε268, A268 was plotted against A280 for a dilution series of each peptide, showing that ε268 was approx. 80% of ε280. These ε268 values were added to the ε268 values of the TPP–carboxylic acids (approx. 3000 M−1·cm−1) which were obtained by plotting A268 against the concentration of a dilution series of the compound. The resulting ε268 values for the modified peptides are shown in Table 1. An ε490 of 81000 M−1·cm−1 (Molecular Probes; http://www.probes.com) was used to quantify Oregon-Green-labelled peptides.
Table 1. Peptides and derivatives used in the present study.
The molar absorption coefficients are for 268 nm unless otherwise indicated. Calculated molecular masses are given; values determined experimentally by MS were within 0.05% of the calculated mass. The Trimeresurus wagleri venom peptide II (Twvp), a likely competitive antagonist of nicotinic acetylcholine receptors [51], was used as a control. OG, Oregon Green.
| Peptide | Sequence | Molecular mass (Da) | Molar absorption coefficient (M−1·cm−1) |
|---|---|---|---|
| Tat | YGRKKRRQRRRP | 1657.0 | 1304 |
| TPP4Tat | TPP-C4-YGRKKRRQRRRP | 1988.3 | 4669 |
| TPP8Tat | TPP-C8-YGRKKRRQRRRP | 2044.5 | 4669 |
| TPP12Tat | TPP-C12-YGRKKRRQRRRP | 2100.6 | 4669 |
| Tat-Cys | YGRKKRRQRRRPC | 1760.1 | 1304 |
| TPP4Tat-Cys | TPP-C4-YGRKKRRQRRRPC | 2147.6 | 4669 |
| Tat-Cys–OG | YGRKKRRQRRRPC–OG | 2223.5 | ∼81000 (491 nm) |
| TPP4Tat-Cys–OG | TPP-C4-YGRKKRRQRRRPC–OG | 2610.9 | ∼81000 (491 nm) |
| Cys-Tat | CYGRKKRRQRRRP | 1760.1 | 1304 |
| Pen | RQIKIWFQNRRMKWKK | 2246.8 | 9665 |
| TPP4Pen | TPP-C4-RQIKIWFQNRRMKWKK | 2578.2 | 13030 |
| TPP8Pen | TPP-C8-RQIKIWFQNRRMKWKK | 2634.3 | 13030 |
| TPP12Pen | TPP-C12-RQIKIWFQNRRMKWKK | 2690.4 | 13030 |
| Cys-Pen | CRQIKIWFQNRRMKWKK | 2349.9 | 9665 |
| Twvp | GGKPDLRPCYPPCHYIPRPKPR | 2546.0 | 3040 (280 nm) |
Peptide labelling with the Oregon Green fluorophore
Tat-Cys was derivatized with Oregon Green maleimide (Molecular Probes) as follows: equimolar peptide and Oregon Green maleimide (typically 100–500 μM) were reacted in PBS (pH 7.2) for 2 h in the dark at room temperature, and the reaction was then quenched with excess 2-mercaptoethanol (20–60 molar equivalent). Derivatization of TPP4Tat-Cys was carried out similarly, except that the solvent was 50% acetonitrile/PBS. Labelled peptides were separated from unreacted components by RP-HPLC as described above.
Antiserum generation
Rabbit antisera against the Cys-Pen and Cys-Tat peptides and the TPP moiety [37] were generated as follows: the thiol–amine crosslinker MBS (m-maleimidobenzoic acid N-hydroxysuccinimide ester; 15 mg·ml−1) in 50–100 μl of DMSO was diluted 1:10 in PBS, pH 6, containing KLH (keyhole limpet haemocyanin; 10 mg·ml−1) and reacted at room temperature for 30 min. MBS–KLH was purified on Sephadex G-25 and reacted at room temperature with 2.5 mg of Cys-Pen or 5 mg of Cys-Tat for 1 h, or with 20 mg of (4-thiobutyl)triphenylphosphonium overnight. Antigens were emulsified with an equal volume of Freund's Complete Adjuvant and injected subcutaneously into two New Zealand White rabbits at multiple sites. These were followed by between one and three booster injections of antigen emulsified in Freund's Incomplete Adjuvant, exsanguination and collection of serum. For Cys-Tat, emulsification, injection and exsanguination were carried out by CovalAb (Ouillins Cedex, France).
Isolation of mitochondria and mitochondrial membranes
Rat liver mitochondria were prepared by homogenization and differential centrifugation [42], in STE [250 mM sucrose, 5 mM Tris/HCl (pH 7.4) and 1 mM EGTA]. Rat heart mitochondria were prepared in STE with initial subtilisin treatment (Type VIII from Bacillus licheniformis; Sigma) followed by homogenization and differential centrifugation [43]. Protein concentration was measured by the biuret assay using BSA as a standard [44], and was typically 40–70 mg of protein·ml−1. Mitochondrial membranes were prepared by subjecting rat liver mitochondria [10 mg of protein·ml−1 in KCl buffer (120 mM KCl, 10 mM Hepes (pH 7.2) and 1 mM EGTA)] to three cycles of freeze–thawing in solid CO2/ethanol and a 30 °C water bath. Mitochondrial lysis (>90%) was confirmed by assaying for release of the matrix enzyme citrate synthase [45].
Mitochondrial incubations
Mitochondrial incubations to investigate peptide uptake were adapted from measurements of mitochondrial protein import [46]. Peptides (1–10 μM; typically 2 μM) were incubated at 25 °C in a 250 μl reaction volume with mitochondria (400 μg of protein·ml−1) in KCl buffer, supplemented with succinate (10 mM) and rotenone (4 μg·ml−1), and in the presence or absence of 0.5 μM FCCP [carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone]. After 30 min of incubation, samples were treated on ice with trypsin (100 μg·ml−1) or carrier, and 20 min later, soybean trypsin inhibitor (200 μg·ml−1) was added. Mitochondria were pelleted (4000 g for 1 min), the supernatant was carefully removed, and the mitochondria were resuspended in loading buffer [4% SDS, 12% (w/v) glycerol, 50 mM Tris/HCl (pH 6.8), 0.025% Serva Blue G, 4% (w/v) dithiothreitol]. All samples were heated to 95 °C for 5 min, and centrifuged at 10000 g for 2 min before loading.
Measurement of Δψm
For Δψm determination, mitochondrial incubations were carried out as above, with [3H]TPMP (0.5 μM/50 nCi·ml−1; American Radiolabeled Chemicals). At t=5, 30 and 50 min, mitochondria were pelleted (4000 g for 1 min in a benchtop microfuge) and 200 μl of supernatant was removed to a scintillation vial. The remaining medium was aspirated and the pellet was resuspended in 40 μl of 20% (v/v) Triton X-100 and transferred to a scintillation vial. Scintillant (3 ml of Fluoransafe; BDH) was added and the radioactivity was quantified using a Packard TriCarb 2100R liquid-scintillation analyser with appropriate quench corrections. After correcting for pellet accumulation of [3H]TPMP in the presence of FCCP, the Δψm was estimated using the Nernst equation, assuming a mitochondrial volume of 0.5 μl·mg of protein−1 and that 60% of intramitochondrial TPMP is membrane-bound [47].
Analysis of peptide uptake by isolated mitochondria
Samples from mitochondrial incubations were separated on Tris-Tricine SDS/20%-PAGE gels [48], and electroblotted on to a 0.2-μm-pore-size nitrocellulose membrane (BioRad) using a Bio-Rad Mini Trans-Blot apparatus and high-pH transfer buffer [10 mM NaHCO3, 30 mM Na2CO3 (pH 9.9) and 20% (v/v) methanol]. Transfer was carried out either at 30 V overnight at 4 °C or at 100 V for 1 h at room temperature. Blots were blocked with PBS/1% (w/v) milk powder/0.05% (v/v) Tween 20 and then incubated with antiserum diluted 1:200–1000 in PBS/1% (w/v) milk powder/0.05% (v/v) Tween 20 for 1–3 h, washed five times for 5 min in PBS, incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (Sigma; 1:5000), washed a further five times for 5 min, and visualized by ECL (enhanced chemiluminescence; Amersham Biosciences). Densitometry was performed using NIH Image 1.63.
Cell incubations and microscopy
Cells were cultivated at 37 °C in a humidified atmosphere containing 95% air/5% CO2. Human fibroblasts from the foreskin of a 4-year-old Caucasian male were cultured in DMEM (Dulbecco's modified Eagle's medium; GibcoBRL) supplemented with 10% FCS (foetal calf serum). Rat basophilic leukaemia cells (RBL-2H3; A.T.C.C.) were grown in MEM (minimal essential medium) with Earle's salts (GibcoBRL) supplemented with 15% FCS. For immunocytochemistry and live-cell visualization, cells were plated on 22 mm diameter glass coverslips and grown to approx. 50% confluence.
Cells destined for live-cell visualization were incubated with Oregon-Green-labelled peptide (2–10 μM) with or without an organelle-specific marker (20 nM MitoTracker Orange or 50 μg·ml−1 Alexa Fluor 555–transferrin; both Molecular Probes). MitoTracker Orange was added 5 min before the end of the incubation, while labelled transferrin was added with the peptide. Cells were incubated at 37 °C for 30 min to 24 h, in DMEM lacking FCS; for incubations >2 h, FCS (10%) was added after 2 h. After washing in PBS, coverslips were mounted in pre-warmed DMEM lacking Phenol Red and FCS, and visualized immediately. Images were acquired using a Bio-Rad Radiance 2000 laser scanning microscope with a Nikon Eclipse E800 microscope and a Nikon ×60 (numerical aperture 1.4) oil-immersion Plan-Apochromat objective. An argon laser (laser line, 488 nm) and a green helium–neon laser (laser line, 543 nm) with AG-2 filter block were used at identical gain and black settings. Magnification was ×1200.
For immunocytochemistry, cells were incubated with unlabelled peptide (1–10 μM) or IBTP [(4-iodobutyl)triphenylphosphonium; 2 μM] with or without MitoTracker Orange (20 nM). Incubations were at 37 °C for 0.5–3 h, in DMEM lacking FCS. After washing in PBS, cells were fixed in paraformaldehyde (4% in PBS) for 1 h at room temperature, then permeabilized, and non-specific antigenic-binding sites blocked, by incubation in PBS/10% FCS/0.1% Triton X-100. Cells were incubated with primary antibody (anti-TPP; 1:1000 in PBS/10% FCS) overnight at 4 °C, washed, incubated with secondary antibody (Oregon-Green-conjugated anti-rabbit IgG (Molecular Probes; 1:5000 in PBS/10% FCS) for 30 min at room temperature, washed and mounted in DABCO (1,4-diazabicyclo[2.2.2]octane)/PVA [poly(vinyl alcohol)] antifade agent. Image acquisition was as described for live cells.
RESULTS AND DISCUSSION
Synthesis and TPP derivatization of CPPs
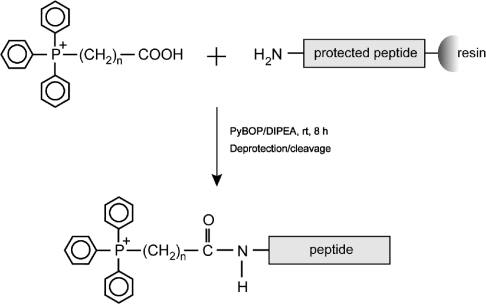
The CPPs Pen and Tat, and a Tat derivative with an extra C-terminal cysteine residue (Tat-Cys), were synthesized by standard solid-phase procedures (Table 1). TPP–peptide derivatives with a range of carbon linker chain lengths were made because hydrophobicity of the linker may affect the mitochondrial binding and uptake of the TPP–peptide conjugates. TPP–carboxylic acids with 4-, 8- and 12-carbon aliphatic alkyl chains (1, 2 and 3) were coupled to the N-terminal amino acid residues of the peptides via an amide bond during the final round of a conventional solid-phase peptide synthesis reaction (Figure 2). This procedure is a convenient method for linking the N-termini of peptides to the TPP cation, and can be adapted easily to attach other moieties. Following deprotection and release from the resin, fluorescent derivatives of the peptides were made by reaction of Oregon Green maleimide with the free thiols of Tat-Cys and TPP4Tat-Cys. For all peptides, the identity, successful conjugation to TPP and addition of Oregon Green were confirmed by MS (Table 1). As a control in Δψm measurements, we used the commercially available peptide II (Twvp) from Wagler's pit viper, Trimeresurus wagleri, because it is of similar size to Pen and Tat, contains several basic residues, and does not disrupt membranes (Table 1). Therefore we have assembled a range of modified Pen and Tat peptides to enable us to determine whether these peptides enter isolated mitochondria or mitochondria in cells, and whether TPP conjugation facilitates this process.
Figure 2. Chemistry of conjugation of TPP to the N-terminus of CPPs.
Interaction of Tat and Pen with isolated mitochondria
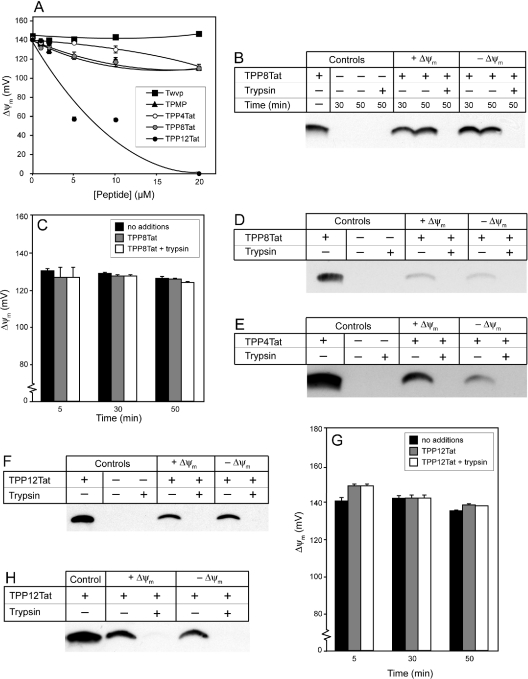
To see if CPPs could act as mitochondria-targeting vectors, we first determined whether the Tat and Pen peptides were taken up into isolated mitochondria through the inner membrane. Measurement of peptide uptake depends on mitochondrial integrity, and accumulation may also be affected by the magnitude of the Δψm [19]. However, as CPPs interact with phospholipid bilayers, they might affect the permeability of the mitochondrial inner membrane and dissipate Δψm. Therefore we measured their effect on Δψm. Pen and Tat both started to disrupt Δψm at concentrations above 10 μM, although their effect on Δψm at 20 μM was still relatively modest (Figure 3A). In contrast, the control peptide Twvp did not affect Δψm at these concentrations. Therefore, to avoid affecting uptake experiments by decreasing Δψm, 2 μM was used for mitochondrial incubations with Tat, whereas for Pen, it was necessary to use 10 μM to enable its visualization with anti-Pen antiserum. At these concentrations, the disruption of Δψm is minimal and is unlikely to affect peptide-uptake experiments.
Figure 3. Tat and Pen are not protected from trypsin digestion by isolated mitochondria.
(A) The effects of Tat and Pen on Δψm. Liver mitochondria (400 μg of protein·ml−1) were incubated at 25 °C for 30 min with 0–20 μM Tat, Pen or Twvp, in KCl buffer supplemented with 10 mM succinate, 4 μg·ml−1 rotenone and 0.5 μM [3H]TPMP with or without 0.5 μM FCCP, and Δψm values were determined. (B) Tat bound to liver mitochondria is not protected from trypsin digestion. Mitochondria were incubated in the presence of 2 μM Tat with or without FCCP as described in (A). All samples were incubated for 30 min at 25 °C, and were analysed then and at 50 min, after 20 min subsequent incubation at 4 °C with or without 0.01% trypsin. Mitochondria were then pelleted, processed and run on Tris-Tricine SDS/PAGE alongside an amount of Tat equivalent to that added to the mitochondrial incubations. Electrophoresed proteins and peptides were transferred on to nitrocellulose and probed with anti-Tat antiserum. (C) Δψm remains stable during mitochondrial incubation in the presence of Tat and trypsin. Liver mitochondria were incubated at 25 °C with or without 2 μM Tat in KCl buffer supplemented with 10 mM succinate, 4 μg·ml−1 rotenone and 0.5 μM [3H]TPMP. After a 30 min incubation, trypsin (0.01%) was added where indicated, and samples were incubated on ice for a further 20 min. Samples were taken at 5, 30 and 50 min to measure Δψm. Results are the means±range for duplicate samples in a typical experiment. (D) Tat bound to heart mitochondria is not protected from trypsin digestion. Mitochondria were incubated with or without 2 μM Tat with or without FCCP, as described for liver mitochondria in (B). Samples were taken after 20 min on ice with or without 0.01% trypsin. (E) Pen bound to liver mitochondria is not protected from trypsin digestion. Mitochondria were incubated with or without 10 μM Pen with or without FCCP, as described for Tat in (B). After processing, mitochondria were run on Tris-Tricine SDS/PAGE alongside an amount of isolated Pen peptide equivalent to 10% of that added in the assay. The Δψm remained stable over the course of the experiment; therefore the lack of uptake is not due to disruption of mitochondria by Pen or trypsin (results not shown).
Lack of uptake of Pen and Tat by isolated mitochondria
To determine whether Tat crossed the mitochondrial inner membrane and entered the matrix, we incubated it with isolated mitochondria, then harvested the mitochondria and assayed them for Tat association by immunoblotting (Figure 3B). Under these conditions, approx. 40–65% of the Tat peptide added to the incubation was mitochondria-bound. However, this mitochondrial association was not affected by the uncoupler FCCP and was therefore independent of Δψm. The mitochondria-associated Tat could either be localized in the mitochondrial matrix, or be bound to the outer surface of the mitochondria, as CPPs are known to bind strongly to biological membranes. Therefore we carried out protease-protection experiments, using trypsin [46], which is known to remove membrane-bound CPPs [24]. When mitochondria were incubated with Tat for 30 min at 25 °C, followed by 20 min on ice, there was significant association of Tat (Figure 3B). However, when trypsin was present during the incubation on ice, mitochondria-associated Tat was completely digested. That none of the mitochondria-associated peptide was protected from trypsinization implied that Tat was bound non-specifically to mitochondria, and was not internalized in the matrix. Trypsin treatment did not affect mitochondrial integrity, as was confirmed by the maintenance of Δψm (Figure 3C). Therefore, during the trypsin incubation, the mitochondria were intact, and the digestion of the added Tat by trypsin was not due to disruption of the mitochondria allowing the proteolysis of internalized peptide.
To see whether the non-specific binding of Tat to mitochondria was a general phenomenon, we analysed Tat binding to heart mitochondria. Approx. 20–35% of the Tat peptide added to the incubation bound to heart mitochondria (Figure 3D). This is less than for liver mitochondria, presumably due to the higher protein/lipid ratio of heart mitochondria, which would diminish the amount of Tat bound per mg of mitochondrial protein. The nature of the association of Tat with heart mitochondria was the same as for liver mitochondria, because it was not diminished by dissipating Δψm with FCCP, and bound Tat was digested by trypsin (Figure 3D).
To determine whether the Pen peptide interacted with mitochondria in a similar manner to Tat, we incubated it with liver mitochondria. Approx. 3–8% of the added Pen was bound to mitochondria (Figure 3E). However, as Pen incubations were carried out at a 5-fold higher concentration (10 μM) than Tat (2 μM), similar absolute amounts of Tat and Pen bound to mitochondria. In analysing the uptake of Pen by mitochondria, we found that, while purified Pen ran as a single band during Tris-Tricine SDS/PAGE, Pen incubated with mitochondria gave rise to an additional, slower-moving band (Figure 3E). This may represent an aggregated form of Pen, which runs anomalously under certain SDS/PAGE conditions [9], or a complex of Pen with a mitochondrial phospholipid; we cannot eliminate the possibility that it is a contaminant of the Pen preparation that has been enriched by stronger mitochondrial binding. While both the Pen bands were associated with mitochondria, this binding was unaffected by Δψm, and was completely removed by trypsin. Therefore both Tat and Pen bind to isolated mitochondria, but this binding is independent of Δψm and is trypsin-sensitive. This indicates that, although there is extensive non-specific binding of Pen and Tat to mitochondria, there is negligible accumulation of these CPPs in the mitochondrial matrix.
Lack of uptake of TPP-modified Pen and Tat by isolated mitochondria
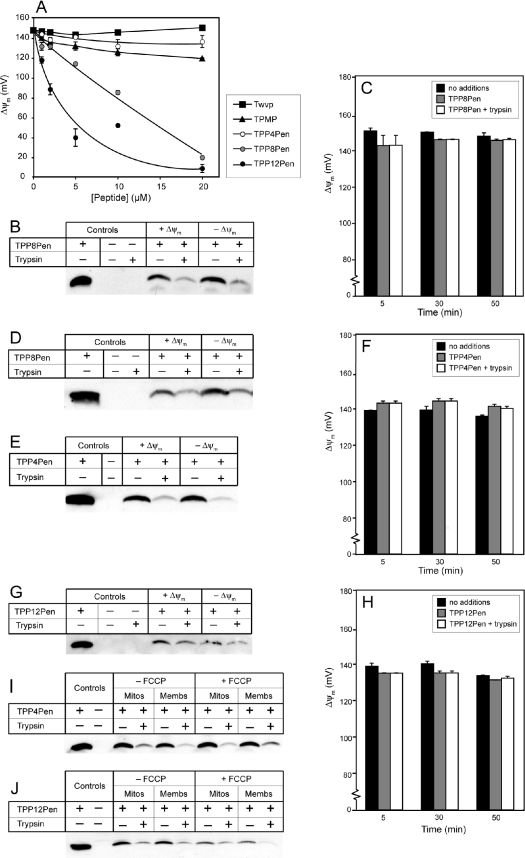
We next determined whether addition of the mitochondria-targeting TPP moiety to the N-terminus of Tat or Pen via an aliphatic carbon chain facilitated mitochondrial uptake. To see whether the TPP-modified peptides disrupted isolated mitochondria, we compared their effects on Δψm with those of the control peptide Twvp and with the simple TPP cation TPMP (Figures 4A and 5A). TPP4Tat and TPP8Tat only affected Δψm at concentrations ≥10 μM, which was similar to unmodified Tat (Figure 3A). In contrast, TPP12Tat lowered Δψm at 5 μM (Figure 4A). TPMP began to diminish the Δψm at 10–20 μM, suggesting that the greater effect of TPP12Tat on membrane integrity was due to its hydrophobicity and not to the TPP moiety alone. TPP4Pen had a similar effect on Δψm to TPMP, while TPP8Pen and TPP12Pen began to affect Δψm at approx. 5 μM and 1 μM respectively (Figure 5A). The greater effects on Δψm of increasing hydrophobicity are consistent with those of the TPP–carboxylic acids, where the C4 and C8 TPP–carboxylic acids had a similar effect to TPMP, while the C12 TPP–carboxylic acid started to diminish Δψm at 5 μM (results not shown). In summary, disruption of Δψm increased with the length of carbon linker between the TPP and peptide, with TPP(Cn)Pen peptides generally having a greater effect on Δψm than TPP(Cn)Tat peptides. To avoid potential adverse effects of the TPP–peptides on mitochondrial integrity and Δψm, concentrations of 1–2 μM were used for uptake experiments.
Figure 4. TPP-coupled Tat is not protected from trypsin digestion by isolated mitochondria.
(A) The effect of increasing TPP–Tat concentration on Δψm. Mitochondria (400 μg of protein·ml−1) were incubated at 25 °C for 30 min with 0–20 μM Twvp, TPMP, TPP4Tat, TPP8Tat or TPP12Tat in KCl buffer supplemented with succinate (10 mM), rotenone (4 μg·ml−1) and [3H]TPMP (0.5 μM), with or without FCCP (0.5 μM). Samples were taken after 30 min and analysed to generate Δψm values. Results are the means±range for duplicate samples in a typical experiment. (B) TPP8Tat bound to liver mitochondria is not protected from trypsin digestion. Mitochondria were incubated in KCl buffer with or without 2 μM TPP8Tat supplemented with succinate (10 mM), rotenone (4 μg·ml−1) with or without FCCP (0.5 μM). After 30 min incubation at 25 °C, trypsin was added to 0.01% and samples were incubated on ice for a further 20 min. Samples were taken at 30 min (before trypsin digestion), and at 50 min (after 20 min on ice with or without 0.01% trypsin). Mitochondria were pelleted, processed and run on Tris-Tricine SDS/PAGE alongside an amount of isolated TPP8Tat peptide equivalent to that in the assay. Electrophoresed proteins and peptides were transferred on to nitrocellulose and probed with anti-Tat antiserum. (C, G) Δψm remains stable over the incubations, and is not adversely affected by TPP–Tat peptide or 0.01% trypsin. Liver mitochondria were incubated with or without 2 μM TPP8Tat (C) or 1 μM TPP12Tat (G) in KCl buffer supplemented as in (B). After 30 min of incubation at 25 °C, samples were incubated on ice for a further 20 min with or without 0.01% trypsin. Samples were taken at 5, 30 and 50 min to measure Δψm. Results are the means±range for duplicate samples in a typical experiment. (D–F, H) TPP–Tat peptides bound to mitochondria are not protected from trypsin digestion. Rat heart mitochondria were incubated with 2 μM TPP8Tat (D), 2 μM TPP4Tat (E) or 1 μM TPP12Tat (F) or rat liver mitochondria incubated with 1 μM TPP12Tat (H), as described in (B). Samples were taken at 50 min with or without 0.01% trypsin. Electrophoresed proteins were transferred on to nitrocellulose and probed with anti-Tat antiserum.
Figure 5. TPP-coupled Pen is partially protected from trypsin digestion by isolated mitochondria.
(A) The effect of increasing TPP–Pen concentration on Δψm. Liver mitochondria were incubated with 0–20 μM Twvp, TPMP, TPP4Pen, TPP8Pen or TPP12Pen as described in Figure 4(A). Results are the means±range for duplicate samples from a typical experiment. (B) TPP8Pen bound to liver mitochondria is partially protected from trypsin digestion, in a Δψm-independent manner. Mitochondria were incubated at 25 °C with or without 2 μM TPP8Pen, with or without FCCP (0.5 μM). After 30 min of incubation, trypsin was added to 0.01% and samples incubated on ice for a further 20 min. Samples were taken at 50 min with or without 0.01% trypsin. Mitochondria were pelleted, processed and run on Tris-Tricine SDS/PAGE alongside an amount of TPP8Pen peptide equivalent to that in the assay. Electrophoresed proteins were transferred on to nitrocellulose and probed with anti-TPP antiserum. (C, F, H) Δψm remains stable over the mitochondrial incubation, and is not affected by the TPP–Pen peptide or 0.01% trypsin. Liver mitochondria were incubated with 2 μM TPP8Pen (C), 2 μM TPP4Pen (F) or 1 μM TPP12Pen (H) in KCl buffer supplemented with 10 mM succinate, 4 μg·ml−1 rotenone and 0.5 μM [3H]TPMP, with or without FCCP. After a 30 min incubation at 25 °C, trypsin was added at 0.01%, and samples incubated on ice for a further 20 min. Samples were taken at 5, 30 and 50 min, and analysed to generate Δψm values as described in the Materials and methods section. Results are the means±range for duplicate samples in a typical experiment. (D, E, G) TPP–Pen peptides bound to mitochondria are partially protected from trypsin digestion, in a Δψm-independent manner. Rat heart mitochondria were incubated with 2 μM TPP8Pen (D), or rat liver mitochondria with 2 μM TPP4Pen (E) or 1 μM TPP12Pen (G), as described in (B). Samples were taken at 50 min with or without 0.01% trypsin. (I, J) Mitochondrial membranes protect TPP–Pen peptides against trypsin digestion as effectively as intact mitochondria. TPP4Pen (2 μM; I) or TPP12Pen (1 μM; J) were incubated with rat liver mitochondria or an equivalent amount of rat liver mitochondrial membranes, in the presence and absence of FCCP, as described for intact mitochondria in (B).
We next tested whether conjugation to TPP facilitated uptake of Pen or Tat into isolated liver mitochondria. When TPP8Tat was incubated with isolated mitochondria (Figure 4B), approx. 70–85% of the added TPP8Tat bound to mitochondria. This association was independent of Δψm, and all the mitochondria-associated TPP8Tat was digested by trypsin. Δψm remained stable over the course of the experiment (Figure 4C), indicating that the lack of uptake was not due to mitochondrial disruption. Experiments on heart mitochondria showed that approx. 8–13% of the added TPP8Tat bound to mitochondria, but again this association was independent of Δψm and was trypsin-sensitive (Figure 4D). To see if varying the hydrophobicity of the TPP linker affected uptake, the mitochondrial accumulation of TPP4Tat and TPP12Tat was also measured. Incubating TPP4Tat with heart mitochondria (Figure 4E), and TPP12Tat with liver or heart mitochondria (Figures 4F–4H), demonstrated extensive mitochondrial binding; however, in all cases, this binding was independent of Δψm and was completely trypsin-sensitive. These results imply that the TPP(Cn)Tat peptides bind exclusively to the outside of isolated mitochondria, and do not accumulate within the matrix.
We next determined whether the conjugation of TPP to Pen mediated its mitochondrial uptake. On incubation with isolated liver mitochondria, 40–55% of the added TPP8Pen associated with the mitochondria (Figure 5B). The proportion of bound peptide was similar in the presence and absence of Δψm, but in contrast with Tat, approx. 10% of the mitochondria-associated TPP8Pen was trypsin-insensitive. The Δψm remained stable during the experiment (Figure 5C). Heart mitochondria also showed trypsin-insensitive TPP8Pen association that was independent of FCCP (Figure 5D).
To see how the trypsin-insensitive association of TPP(Cn)Pen with mitochondria was affected by hydrophobicity, we next tested the uptake of TPP4Pen and TPP12Pen. On incubation with liver mitochondria, approx. 40–45% of the added TPP4Pen was bound to mitochondria, and approx. 5–10% of the bound TPP4Pen was trypsin-insensitive in the presence and absence of Δψm (Figures 5E and 5F). For TPP12Pen, 16–20% of the added peptide bound to mitochondria, and 7–12% of the bound peptide was trypsin-insensitive. However, both TPP12Pen binding and the extent of trypsin protection were variable, probably due to its tendency to disrupt mitochondrial membranes (Figures 5G and 5H). Therefore TPP(Cn)Pen peptides show some trypsin insensitivity on incubation with mitochondria. Trypsin fully digests mitochondria-associated Pen (Figure 3E), and all TPP(Cn)Pen peptides in the absence of mitochondria (results not shown). Therefore this finding suggests that on interaction with mitochondria, the TPP(Cn)Pen peptides are protected from trypsin digestion. This could be due to uptake of the peptide into the mitochondrial matrix; however, the fact that protease protection was independent of Δψm suggests that TPP was not stimulating Δψm-dependent uptake into the matrix. Another possibility is that the TPP(Cn)Pen peptides were protected from digestion by interaction with mitochondrial membranes, and not by accumulation in the matrix. To test this hypothesis, we incubated TPP4Pen and TPP12Pen with mitochondrial membrane fragments (Figures 5I and 5J) in which both faces of the membrane are open to the solvent and therefore any protection from protease digestion would be due to interactions with the membranes themselves. Mitochondrial membrane fragments protected TPP4Pen (Figure 5I) and TPP12Pen (Figure 5J) from protease digestion to a similar extent to intact mitochondria. This suggests that the mitochondrial protection against TPP(Cn)Pen peptide digestion is likely to be a result of these peptides interacting with phospholipid bilayers, rather than peptide transport through the inner membrane leading to sequestration within the matrix. Therefore, in the case of the TPP(Cn)Pen peptides, protease protection is not an indication of mitochondrial uptake.
Neither Tat nor TPP4Tat reach mitochondria in live cells
Experiments with isolated mitochondria and membrane fragments gave no evidence of membrane transport and matrix localization of Tat, Pen or their TPP-conjugated derivatives. However, all of the tested peptides bound strongly to isolated mitochondria, and, in the case of TPP(Cn)Pen, peptides interacted with mitochondrial membranes so as to protect them from protease digestion. We were therefore interested to find out whether the peptides localized to mitochondria within intact cells. There are also a number of advantages in studying peptide uptake in whole cells over isolated mitochondria: in cells, the duration of the experiment can be longer than the 30–50 min incubation used for isolated mitochondria, which is limited by mitochondrial durability. It is also possible that cytoplasmic factors that are missing from experiments with isolated mitochondria may facilitate the mitochondrial uptake of peptides within cells. Finally, the peptides may move easily back and forth across the mitochondrial membranes, in which case external trypsin could still lead to the complete digestion of the peptide despite its uptake into the matrix. Such a scenario could lead to a false-negative protease-protection result with isolated mitochondria, but experiments in intact cells avoid this problem.
We initially studied cellular uptake of CPPs by fixing cells after CPP incubation, then using an antibody against the TPP moiety to analyse the intracellular localization of TPP(Cn)Tat and TPP(Cn)Pen by confocal immunofluorescence microscopy. In fixed cells, all the TPP(Cn)Tat peptides showed a very low level of apparent mitochondrial localization; in contrast, there was no evidence of mitochondrial localization by the TPP(Cn)Pen peptides (results not shown). We therefore chose to investigate only Tat and TPP4Tat further. However, we were concerned that the mitochondrial localization might be an artifact, as it has been shown that some CPPs distribute anomalously upon fixation [24,25]. Even relatively mild fixation reagents such as 4% paraformaldehyde can cause dramatic redistribution of CPPs into the cytoplasm and nucleus from endosomes and the cell surface [24]. To avoid this potential artifact, we studied the localization of Tat and TPP4Tat in live cells. To do this, we synthesized Tat and TPP4Tat derivatives with a C-terminal cysteine residue to enable conjugation to the fluorophore Oregon Green via a maleimide–thiol linkage. This fluorophore enables visualization of the conjugates within living cells without fixation, thereby avoiding potential artifactual mitochondrial association. Furthermore, its position at the peptide's C-terminus, distal to the TPP, eliminates the possibility of apparent mitochondrial localization arising from the accumulation of partially degraded TPP–Tat fragments in mitochondria; this is a significant concern as it has recently been reported that Tat is degraded swiftly by proteases after reaching the cytosol [29].
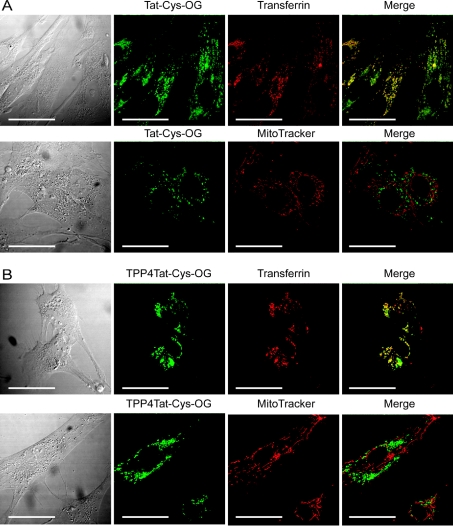
We incubated Tat and TPP4Tat with live fibroblasts, either alone or in tandem with MitoTracker Orange (to visualize mitochondria) or Alexa Fluor 555–transferrin (to visualize endosomes). Tat accumulated within endosomes, as shown by its strong co-localization with transferrin (Figure 6A). Furthermore, the lack of co-localization with MitoTracker Orange showed clearly that the peptide did not localize to mitochondria. There was also no evidence for gross changes in intracellular localization of the Tat on conjugation to TPP, with TPP4Tat, like Tat, accumulating exclusively within endosomes, and exhibiting no mitochondrial localization (Figure 6B). Shorter (30 min–12 h) and longer incubation times (48–72 h) gave qualitatively similar results; although peptide accumulation within endosomes increased over time, it was not detected in the cytoplasm at any stage (results not shown). Higher concentrations of peptide (≥10 μM) also had no effect on the intracellular localization. These findings are not specific to skin fibroblasts, as incubation of Tat and TPP4Tat with rat basophilic leukaemia cells also showed only endosomal accumulation, and exclusion from mitochondria (results not shown). Throughout these incubations there was no evidence of cell toxicity on TPP–peptide treatment, as indicated by the lack of gross changes to cellular or mitochondrial morphology, as visualized by MitoTracker Orange staining. Together these results demonstrate a lack of mitochondrial uptake of Tat within live cells, even when conjugated to the mitochondria-targeting moiety TPP. They provide strong support for a vesicular/endosomal means of uptake of CPPs, and no evidence that direct transport of CPPs across phospholipid bilayers occurs.
Figure 6. Tat and TPP4Tat accumulate within endosomes in cultured fibroblasts, and are not delivered to mitochondria.
(A) Tat-Cys–OG (Oregon Green) accumulates in endosomes. Fibroblasts were incubated for 24 h with Tat-Cys-OG (2 μM), with and without Alexa Fluor 555–transferrin (50 μg·ml−1) and MitoTracker Orange (20 nM), and visualized without fixation. Co-localization of Tat-Cys–OG and transferrin is indicated by yellow staining in the merged image. (B) TPP4Tat-Cys–OG accumulates in endosomes in a similar manner to Tat-Cys–OG. Fibroblasts were incubated with TPP4Tat-Cys–OG as described above for Tat-Cys–OG, mounted and visualized without fixation. Scale bar, 50 μm.
Concluding remarks
We have investigated the ability of the two CPPs Pen and Tat to act as potential vectors for the delivery of macromolecules to mitochondria in cells. Neither the peptides nor the peptides modified by the lipophilic cation TPP accumulated in the matrix of isolated mitochondria, even though TPP(Cn)Pen peptides were partially protected from protease digestion by association with mitochondria. This indicates that the direct passage of CPPs through phospholipid bilayers is not a general property of CPPs, and suggests that CPPs are unlikely to act as targeting vectors to mitochondria. This finding also sheds light on the potential mechanism of uptake of CPPs into cells. It has been claimed that CPPs are taken up through the plasma membrane by inverted micelles, but this evidently does not apply to the mitochondrial inner membrane. The lack of uptake could be due to a difference in phospholipid composition or higher protein content of the mitochondrial inner membrane, although the more likely conclusion is that CPPs cannot cross biological phospholipid bilayers via inverted micelles, and that the uptake into the cell is instead via a vesicular (endocytic) pathway. Although the uptake of CPPs by artificial liposomes of a range of phospholipid compositions has been reported [31,32], synthetic lipid bilayers may not be a good model for biological membranes because they lack membrane proteins. It is therefore possible that uptake into liposomes does occur by inverted micelle formation, but that in mitochondria, the greater membrane protein content makes the formation of inverted micelles impossible.
An endocytic uptake pathway of CPPs is also supported by our observation of the endosomal localization of Tat and TPP4Tat following uptake by two cell lines. Co-localization with transferrin indicated that both peptides were taken up by clathrin-dependent endocytosis, in agreement with other studies of the Tat peptide in live cells [24,28]. It is notable that the addition of TPP to Tat had no effect on intracellular localization of the peptide; therefore the increased hydrophobicity of TPP–Tat over Tat does not promote its release from endosomes.
Our data suggest that the initial hope of a vector that would pass through the plasma membrane and then the mitochondrial inner membrane, carrying DNA and proteins to mitochondria, is unlikely to be achievable. However, it may still be possible to implement CPPs as part of a mitochondria-targeting construct including a mitochondrial import sequence, as has been shown recently for a Tat-mitochondrial import sequence–GFP construct. The authors demonstrated mitochondrial uptake of the construct, and its subsequent entrapment within mitochondria upon removal of the import sequence and Tat by the matrix processing peptidase [49]. These results were interpreted to show that Tat could facilitate the uptake into the mitochondrial matrix of attached proteins. However, inclusion of the full mitochondrial targeting sequence in their construct makes it difficult to draw conclusions about its mode of uptake. An alternative hypothesis is that the Tat facilitated cellular uptake and endosomal escape, but that the mitochondrial targeting sequence directed uptake by the classical TOM (translocase of the outer membrane)/TIM (translocase of the inner membrane) protein import pathway. In further work, the authors also showed apparent partial uptake of Tat–GFP into isolated mitochondria, as measured by protease insensitivity [50]. It may be that Tat-fusion proteins can cross mitochondrial membranes, even though we have shown that the unconjugated Tat peptide cannot. However, exposure of the Tat–GFP-treated mitochondria to protease over a range of osmolarities, in conjunction with analysis of the protease sensitivity of mitochondrial intermembrane space and matrix marker proteins, is required to confirm the mitochondrial localization of Tat–GFP.
In summary, CPPs cannot enter isolated mitochondria or mitochondria within cells. Instead, we see clear evidence of endosomal uptake of CPPs in live cells. Together these results indicate that the normal mode of CPP uptake into cells is not by membrane permeation by inverted micelles, but by an endocytic pathway.
Acknowledgments
We thank David Owen, Michael Harbour and Frances Blaikie for technical assistance with peptide synthesis, MS and NMR. M.F.R. is a Top Achiever Doctoral Scholar, and A.F. is a New Zealand Science and Technology Postdoctoral Fellow, of the Foundation for Research, Science and Technology of New Zealand.
References
- 1.Langel U. Boca Raton: CRC Press; 2002. Cell-Penetrating Peptides: Processes and Applications. [Google Scholar]
- 2.Wadia J. S., Dowdy S. F. Modulation of cellular function by TAT mediated transduction of full length proteins. Curr. Protein Pept. Sci. 2003;4:97–104. doi: 10.2174/1389203033487289. [DOI] [PubMed] [Google Scholar]
- 3.Astriab-Fisher A., Sergueev D., Fisher M., Shaw B. R., Juliano R. L. Conjugates of antisense oligonucleotides with the Tat and antennapedia cell-penetrating peptides: effects on cellular uptake, binding to target sequences, and biologic actions. Pharm. Res. 2002;19:744–754. doi: 10.1023/a:1016136328329. [DOI] [PubMed] [Google Scholar]
- 4.Gait M. J. Peptide-mediated cellular delivery of antisense oligonucleotides and their analogues. Cell. Mol. Life Sci. 2003;60:844–853. doi: 10.1007/s00018-003-3044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polyakov V., Sharma V., Dahlheimer J. L., Pica C. M., Luker G. D., Piwnica-Worms D. Novel Tat-peptide chelates for direct transduction of technetium-99m and rhenium into human cells for imaging and radiotherapy. Bioconjug. Chem. 2000;11:762–771. doi: 10.1021/bc000008y. [DOI] [PubMed] [Google Scholar]
- 6.Braun K., Peschke P., Pipkorn R., Lampel S., Wachsmuth M., Waldeck W., Friedrich E., Debus J. A biological transporter for the delivery of peptide nucleic acids (PNAs) to the nuclear compartment of living cells. J. Mol. Biol. 2002;318:237–243. doi: 10.1016/S0022-2836(02)00031-1. [DOI] [PubMed] [Google Scholar]
- 7.Dietz G. P., Kilic E., Bahr M. Inhibition of neuronal apoptosis in vitro and in vivo using TAT-mediated protein transduction. Mol. Cell. Neurosci. 2002;21:29–37. doi: 10.1006/mcne.2002.1165. [DOI] [PubMed] [Google Scholar]
- 8.Vocero-Akbani A. M., Heyden N. V., Lissy N. A., Ratner L., Dowdy S. F. Killing HIV-infected cells by transduction with an HIV protease-activated caspase-3 protein. Nat. Med. 1999;5:29–33. doi: 10.1038/4710. [DOI] [PubMed] [Google Scholar]
- 9.Derossi D., Joliot A. H., Chassaing G., Prochiantz A. The third helix of the antennapedia homeodomain translocates through biological membranes. J. Biol. Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- 10.Frankel A. D., Pabo C. O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 11.Green M., Loewenstein P. M. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell. 1988;55:1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 12.Vives E., Brodin P., Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 13.Muratovska A., Lightowlers R. N., Taylor R. W., Wilce J. A., Murphy M. P. Targeting large molecules to mitochondria. Adv. Drug Delivery Rev. 2001;49:189–198. doi: 10.1016/s0169-409x(01)00134-x. [DOI] [PubMed] [Google Scholar]
- 14.Green K., Brand M. D., Murphy M. P. Prevention of mitochondrial oxidative damage as a therapeutic strategy in diabetes. Diabetes. 2004;53(Suppl. 1):S110–S118. doi: 10.2337/diabetes.53.2007.s110. [DOI] [PubMed] [Google Scholar]
- 15.Orth M., Schapira A. H. Mitochondria and degenerative disorders. Am. J. Med. Genet. 2001;106:27–35. doi: 10.1002/ajmg.1425. [DOI] [PubMed] [Google Scholar]
- 16.Puccio H., Koenig M. Recent advances in the molecular pathogenesis of Friedreich ataxia. Hum. Mol. Genet. 2000;9:887–892. doi: 10.1093/hmg/9.6.887. [DOI] [PubMed] [Google Scholar]
- 17.Leonard J. V., Schapira A. H. Mitochondrial respiratory chain disorders I: mitochondrial DNA defects. Lancet. 2000;355:299–304. doi: 10.1016/s0140-6736(99)05225-3. [DOI] [PubMed] [Google Scholar]
- 18.Scheffler I. E. New York: Wiley-Liss; 1999. Mitochondria. [Google Scholar]
- 19.Murphy M. P. Selective targeting of bioactive compounds to mitochondria. Trends Biotechnol. 1997;15:326–330. doi: 10.1016/S0167-7799(97)01068-8. [DOI] [PubMed] [Google Scholar]
- 20.Mihara K., Omura T. Protein import into mammalian mitochondria. Methods Enzymol. 1995;260:302–310. doi: 10.1016/0076-6879(95)60147-3. [DOI] [PubMed] [Google Scholar]
- 21.Flierl A., Jackson C., Cottrell B., Murdock D., Seibel P., Wallace D. C. Targeted delivery of DNA to the mitochondrial compartment via import sequence-conjugated peptide nucleic acid. Mol. Ther. 2003;7:550–557. doi: 10.1016/s1525-0016(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 22.Chinnery P. F., Taylor R. W., Diekert K., Lill R., Turnbull D. M., Lightowlers R. N. Peptide nucleic acid delivery to human mitochondria. Gene Ther. 1999;6:1919–1928. doi: 10.1038/sj.gt.3301061. [DOI] [PubMed] [Google Scholar]
- 23.Derossi D., Calvet S., Trembleau A., Brunissen A., Chassaing G., Prochiantz A. Cell internalization of the third helix of the antennapedia homeodomain is receptor-independent. J. Biol. Chem. 1996;271:18188–18193. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 24.Richard J. P., Melikov K., Vives E., Ramos C., Verbeure B., Gait M. J., Chernomordik L. V., Lebleu B. Cell-penetrating peptides: a reevaluation of the mechanism of cellular uptake. J. Biol. Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 25.Lundberg M., Johansson M. Positively charged DNA-binding proteins cause apparent cell membrane translocation. Biochem. Biophys. Res. Commun. 2002;291:367–371. doi: 10.1006/bbrc.2002.6450. [DOI] [PubMed] [Google Scholar]
- 26.Fittipaldi A., Ferrari A., Zoppe M., Arcangeli C., Pellegrini V., Beltram F., Giacca M. Cell membrane lipid rafts mediate caveolar endocytosis of HIV-1 Tat fusion proteins. J. Biol. Chem. 2003;278:34141–34149. doi: 10.1074/jbc.M303045200. [DOI] [PubMed] [Google Scholar]
- 27.Wadia J. S., Stan R. V., Dowdy S. F. Transducible TAT–HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat. Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 28.Potocky T. B., Menon A. K., Gellman S. H. Cytoplasmic and nuclear delivery of a TAT-derived peptide and a β-peptide after endocytic uptake into HeLa cells. J. Biol. Chem. 2003;278:50188–50194. doi: 10.1074/jbc.M308719200. [DOI] [PubMed] [Google Scholar]
- 29.Fischer R., Kohler K., Fotin-Mleczek M., Brock R. A stepwise dissection of the intracellular fate of cationic cell-penetrating peptides. J. Biol. Chem. 2004;279:12625–12635. doi: 10.1074/jbc.M311461200. [DOI] [PubMed] [Google Scholar]
- 30.Fuchs S. M., Raines R. T. Pathway for polyarginine entry into mammalian cells. Biochemistry. 2004;43:2438–2444. doi: 10.1021/bi035933x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thoren P. E., Persson D., Karlsson M., Norden B. The antennapedia peptide penetratin translocates across lipid bilayers – the first direct observation. FEBS Lett. 2000;482:265–268. doi: 10.1016/s0014-5793(00)02072-x. [DOI] [PubMed] [Google Scholar]
- 32.Terrone D., Sang S. L., Roudaia L., Silvius J. R. Penetratin and related cell-penetrating cationic peptides can translocate across lipid bilayers in the presence of a transbilayer potential. Biochemistry. 2003;42:13787–13799. doi: 10.1021/bi035293y. [DOI] [PubMed] [Google Scholar]
- 33.Liberman E. A., Topaly V. P., Tsofina L. M., Jasaitis A. A., Skulachev V. P. Mechanism of coupling of oxidative phosphorylation and the membrane potential of mitochondria. Nature (London) 1969;222:1076–1078. doi: 10.1038/2221076a0. [DOI] [PubMed] [Google Scholar]
- 34.Smith R. A., Kelso G. F., Blaikie F. H., Porteous C. M., Ledgerwood E. C., Hughes G., James A. M., Ross M. F., Asin-Cayuela J., Cocheme H. M., et al. Using mitochondria-targeted molecules to study mitochondrial radical production and its consequences. Biochem. Soc. Trans. 2003;31:1295–1299. doi: 10.1042/bst0311295. [DOI] [PubMed] [Google Scholar]
- 35.Kelso G. F., Porteous C. M., Coulter C. V., Hughes G., Porteous W. K., Ledgerwood E. C., Smith R. A., Murphy M. P. Selective targeting of a redox-active ubiquinone to mitochondria within cells: antioxidant and antiapoptotic properties. J. Biol. Chem. 2001;276:4588–4596. doi: 10.1074/jbc.M009093200. [DOI] [PubMed] [Google Scholar]
- 36.Smith R. A., Porteous C. M., Coulter C. V., Murphy M. P. Selective targeting of an antioxidant to mitochondria. Eur. J. Biochem. 1999;263:709–716. doi: 10.1046/j.1432-1327.1999.00543.x. [DOI] [PubMed] [Google Scholar]
- 37.Lin T. K., Hughes G., Muratovska A., Blaikie F. H., Brookes P. S., Darley-Usmar V., Smith R. A., Murphy M. P. Specific modification of mitochondrial protein thiols in response to oxidative stress: a proteomics approach. J. Biol. Chem. 2002;277:17048–17056. doi: 10.1074/jbc.M110797200. [DOI] [PubMed] [Google Scholar]
- 38.Burns R. J., Murphy M. P. Labeling of mitochondrial proteins in living cells by the thiol probe thiobutyltriphenylphosphonium bromide. Arch. Biochem. Biophys. 1997;339:33–39. doi: 10.1006/abbi.1996.9861. [DOI] [PubMed] [Google Scholar]
- 39.Muratovska A., Lightowlers R. N., Taylor R. W., Turnbull D. M., Smith R. A., Wilce J. A., Martin S. W., Murphy M. P. Targeting peptide nucleic acid (PNA) oligomers to mitochondria within cells by conjugation to lipophilic cations: implications for mitochondrial DNA replication, expression and disease. Nucleic Acids Res. 2001;29:1852–1863. doi: 10.1093/nar/29.9.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narayanan K. S., Berlin K. D. Novel synthesis of ω-(diphenylphosphinyl)alkylcarboxylic acids from triphenyl-ω-carboxyalkylphosphonium salts. J. Org. Chem. 1980;45:2240–2243. [Google Scholar]
- 41.Dawson M. I., Vasser M. Synthesis of prostaglandin synthetase substrate analogs. 1. (Z)-14-Hydroxy-12,13-methano-8-nonadecenoic acid. J. Org. Chem. 1977;42:2783–2785. doi: 10.1021/jo00436a030. [DOI] [PubMed] [Google Scholar]
- 42.Chappell J., Hansford R. Preparation of mitochondria from animal tissues and yeasts. In: Birnie G., editor. Subcellular Components: Preparation and Fractionation. London: Butterworths; 1972. pp. 77–91. [Google Scholar]
- 43.Tyler D. D., Gonze J. The preparation of heart mitochondria from laboratory animals. Methods Enzymol. 1967;10:81–86. [Google Scholar]
- 44.Gornall A., Bardawill C., David M. Determination of serum protein by means of the biuret reaction. J. Biol. Chem. 1949;177:751–766. [PubMed] [Google Scholar]
- 45.Robinson J. B., Jr, Brent L. G., Sumegi B., Srere P. A. An enzymatic approach to the study of the Krebs tricarboxylic acid cycle. In: Darley-Usmar V. M., Rickwood D., Wilson M. T., editors. Mitochondria: a Practical Approach. Oxford: IRL Press Ltd; 1987. pp. 153–170. [Google Scholar]
- 46.Rospert S., Schatz G. Protein translocation into mitochondria. In: Celis J. E., editor. Cell Biology: a Laboratory Handbook. 2nd edn. San Diego: Academic Press Inc.; 1998. pp. 277–285. [Google Scholar]
- 47.Brown G. C., Brand M. D. Thermodynamic control of electron flux through mitochondrial cytochrome bc1 complex. Biochem. J. 1985;225:399–405. doi: 10.1042/bj2250399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 49.Del Gaizo V., Payne R. M. A novel TAT-mitochondrial signal sequence fusion protein is processed, stays in mitochondria, and crosses the placenta. Mol. Ther. 2003;7:720–730. doi: 10.1016/s1525-0016(03)00130-8. [DOI] [PubMed] [Google Scholar]
- 50.Del Gaizo V., MacKenzie J. A., Payne R. M. Targeting proteins to mitochondria using TAT. Mol. Genet. Metab. 2003;80:170–180. doi: 10.1016/j.ymgme.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 51.Molles B. E., Rezai P., Kline E. F., McArdle J. J., Sine S. M., Taylor P. Identification of residues at the α and ε subunit interfaces mediating species selectivity of Waglerin-1 for nicotinic acetylcholine receptors. J. Biol. Chem. 2002;277:5433–5440. doi: 10.1074/jbc.M109232200. [DOI] [PubMed] [Google Scholar]