Abstract
The HIV-1 accessory protein Vif was found to modulate the RNA- and DNA-dependent DNA synthesis activity of the viral RT (reverse transcriptase) in two ways: (i) it stimulated the binding of the viral RT to the primer by increasing the association rate kcat/Km and by decreasing the thermodynamic barrier ΔH[ES] for complex fomation, and (ii) it increased the polymerization rate of HIV-1 RT. A Vif mutant lacking the final 56 amino acids at the C-terminus failed to stimulate the viral RT. On the other hand, another Vif mutant lacking the first 43 amino acids at the N-terminus, which are involved in RNA binding and interaction with the viral protease, was able to stimulate RT activity. In addition, Vif was found to promote the bypass of an abasic site by HIV-1 RT.
Keywords: abasic site, AIDS, HIV, enzyme kinetics, reverse transcription, Vif
Abbreviations: AP (site), abasic site (where a pusine or pyridimine group is removed from the DNA strand); DDS, DNA-dependent DNA synthesis; FIV, feline immunodeficiency virus; HCV, hepatitis C virus; MuLV, murine leukaemia virus; PCNA, proliferating-cell nuclear antigen; RDS, RNA-dependent DNA synthesis; RP-A, replication protein-A; RT, reverse transcriptase; TLS, translesion synthesis
INTRODUCTION
The vif gene of HIV-1 was initially shown to be dispensable for virus infection of transformed T-cell lines. However, subsequent studies showed that vif-deleted virions assemble in permissive cells, and can penetrate into non-permissive cells and initiate the process of reverse transcription, but this process fails to go to completion [1]. This vif-dependent infectivity is manifested only in primary lymphocytes and macrophages and in a limited number of T-cell lines [2,3]. The vif gene is conserved in almost all lentiviruses of primate and non-primate hosts, indicating an essential role for its product in the replication of these viruses [4]. The vif gene encodes the 23 kDa basic protein Vif, a substantial fraction of which is membrane-associated in virus-producing cells and co-localizes with the protein Gag [5,6]. Analysis of the reverse transcription process in ΔVif HIV-1 mutants in non-permissive cells showed a specific block in the synthesis of minus- and plus-strand viral DNA products, resulting in an inability to produce full-length viral DNA genomes [7]. This suggests a possible role for Vif in the retrotranscription step [8–10]. Vif has been shown to bind HIV-1 RNA [11] and to form multimers in vivo through a self-association domain located at its C-terminus [12]. This domain was also found to be essential for the Vif-dependent stimulation of HIV-1 infectivity. Vif is phosphorylated by cellular kinases, and this post-translational modification has been shown to be important in regulating HIV-1 infectivity [13]. In non-permissive cells, Vif is thought to inhibit an antiviral pathway recently demonstrated to require the cellular protein CEM15, also called APOBEC3G [14,15], which is a member of the family of cytidine deaminases. The APOBEC enzyme family deaminates specific cytidine (C) residues in either DNA or mRNA, converting them into uridine (U) residues. APOBEC3G has been shown to be incorporated into virions and then to deaminate cytidine residues specifically on the minus DNA strand. As a result, when the complementary plus-strand DNA is synthesized, high levels of G→A substitutions are accumulated, leading eventually to the production of non-functional proviral DNA [16]. HIV-1 Vif has been shown to form a complex with human APOBEC3G, to impair the translation of APOBEC3G mRNA and to accelerate the post-translational degradation of the APOBEC3G protein by the 26 S proteasome [17,18]. As a result, virion encapsidation of APOBEC3G is prevented, dramatically reducing G→A substitutions in the proviral genome.
Recent results have clearly shown that Vif is also specifically packaged into virions as a component of the viral nucleoprotein and that it interacts with the genomic viral RNA, with the viral protease and with Gag. Moreover, viral genomic RNA complexed with natural initiator tRNA purified from Vif-defective virions proved to be a poor substrate for in vitro reverse transcription [19,20]. Thus we asked whether Vif, in addition to counteracting APOBEC3G activity, might play some additional role within the ribonucleoprotein core complex of HIV-1, in particular at the reverse transcription step [21]. As a first attempt to investigate the possible role(s) of Vif in the reverse transcription process, we analysed its influence on the in vitro activity of viral RT (reverse transcriptase).
MATERIALS AND METHODS
Chemicals
[3H]dTTP (40 Ci/mmol) and [γ-32P]ATP (3000 Ci/mmol) were from Amersham Biosciences, and unlabelled dNTPs were from Roche Molecular Biochemicals. Whatman was the supplier of the GF/C filters. The tetrahydrofurane (dSpacer) was from Glen Research. MuLV (murine leukaemia virus) RT was from Applied Biosystems. HCV (hepatitis C virus) NS3 recombinant protein was expressed and purified as described in [29]. All other reagents were of analytical grade and were purchased from Merck or Fluka.
Nucleic acid substrates
The homopolymeric templates poly(rA)·oligo(dT) (10:1, mol/mol) and poly(dA)·oligo(dT) (10:1) (Pharmacia) were prepared as described in [22]. The d73-mer, either undamaged or containing the synthetic (tetrahydrofurane) abasic site, and the corresponding primer were chemically synthesized and purified on a denaturing polyacrylamide gel. The sequence is 5′ GATCGGGAGGGTAGGAATATTGAG[X/G]ATGAAGGGTTGAGTTGAGTGGAGATAGTGGAGGGTAGTATGGTGGATA 3′. The sequence complementary to the 18-mer primer is underlined. The position of the lesion, or the corresponding G residue in the undamaged template, is indicated in bold (where X denotes the abasic site).
Expression and purification of recombinant HIV-1 proteins
The expression vector pD10Vif [23] was introduced into Escherichia coli strain DH5α by electroporation. Cells were grown at 37 °C in 1 litre of Luria Broth to an A600 of 0.6. Isopropyl β-D-thiogalactoside was added to a final concentration of 1 mM and growth was continued for 4 h. Cells were lysed and recombinant His-tagged Vif protein was purified by affinity chromatography on Ni2+-nitrilotriacetate–agarose and subsequently by ion-exchange chromatography using an FPLC MonoS column. For preparation of the VifΔN and VifΔC mutants, the vif gene was amplified using the linearized pD10Vif plasmid as a template. The amplified full-length gene was then subjected to another round of PCR amplification using appropriate pairs of primers to generate two fragments, one encompassing nt 1–421 (vifΔC) and the other comprising nt 138–590 (vifΔN), bearing restriction sites for the BamHI and HindIII endonucleases. The two fragments were cloned into the pD10 plasmid to generate pD10VifΔN and pD10VifΔC respectively. Expression and purification of the recombinant VifΔN and VifΔC proteins was performed as described above. Recombinant His-tagged HIV-1 RT was expressed and purified as described in [24].
Assay of HIV-1 RT RNA- and DNA-dependent DNA polymerase activity
The RNA-dependent and DNA-dependent DNA polymerase activity of RT was assayed as described in [22]. When the 5′-32P-labelled template was used, a volume of 10 μl contained 0.05 μM (3′-hydroxy ends) of the DNA template, 5 nM RT and Vif protein as indicated in the Figure legends.
Steady-state kinetic measurements
Reactions were performed under the conditions described for the HIV-1 RT activity assay. Kinetic parameters were determined by non-least-squares computer-fitting methods utilizing the equations specified in the Figure legends.
Calculation of thermodynamic parameters
Reactions were carried out at different temperatures, and three separate incubations were performed at each temperature in a thermostatted water-bath, in the absence or in the presence of 0.4, 0.8, 1.6 or 3.2 μM Vif. ΔH[ES] and ΔS[ES] values were determined according to the van't Hoff equation:
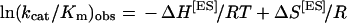 |
where R is the gas constant and T is the absolute temperature (K). ΔG[ES] was calculated using the equation:
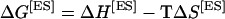 |
RESULTS
HIV-1 Vif protein increases the affinity of the viral RT for the 3′-hydroxy primer during both RDS (RNA-dependent DNA synthesis) and DDS (DNA-dependent DNA synthesis)
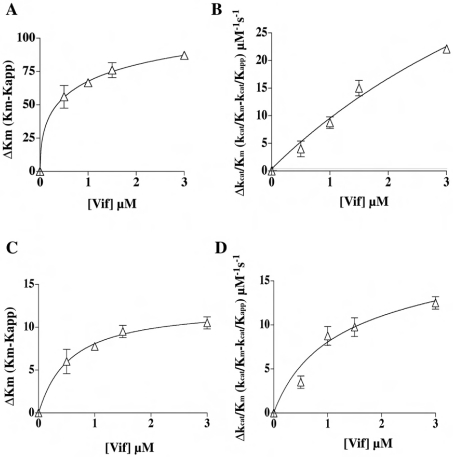
Recombinant Vif protein was titrated in assays of RDS and DDS in the presence of increasing concentrations of the 3′-hydroxy primer. Table 1 summarizes the calculated kinetic parameters in the presence or in the absence of Vif. The apparent affinity of RT for the 3′-hydroxy primer was increased by Vif by 30-fold under RDS conditions and by 26-fold under DDS conditions, as reflected by a decrease in the value of Km (Figures 1A and 1C; Table 1). A similar stimulatory effect was observed in the case of the parameter kcat/Km, which was increased 37-fold under RDS conditions and 26-fold under DDS conditions (Figures 1B and 1D; Table 1). The dependence of both Km and kcat/Km on the Vif concentration under RDS conditions allowed the estimation of values for the apparent affinity constant, Kd, of Vif for the RT·primer complex of 0.18 and 0.21 μM respectively. When the same analysis was performed under DDS conditions, the calculated Kd values for Vif were 0.27 and 0.23 μM respectively.
Table 1. Effects of Vif on the kinetic and thermodynamic parameters for RDS and DDS catalysed by HIV-1 RT.
kcat/Km values were calculated using various concentrations of the 3′-hydroxy primer. kpol values were calculated using various concentrations of RT. Kd values for Vif were calculated from variations in the Km value (*), the kcat/Km value (†) or the kpol value (‡) with Vif concentration. Values are means±S.D. Thermodynamic parameters were determined at T=310 K (1 kJ=4.184 kcal).
| (A) Kinetic | |||||
|---|---|---|---|---|---|
| RDS | DDS | ||||
| Km (nM) | kcat/Km (M−1·s−1) | kpol (s−1) | Km (nM) | kcat/Km (M−1·s−1) | |
| −Vif | 109±10 | (3.5±0.5)×106 | 0.25±0.01 | 10.6±1 | (1.9±0.1)×106 |
| +Vif | 3.6±0.5 | (1.3±0.2)×108 | 0.98±0.02 | 0.4±0.05 | (5.0±0.5)×107 |
| Kd Vif (μM) | 0.18±0.03* | 0.21±0.05† | 0.20±0.01‡ | 0.27±0.03* | 0.23±0.04† |
| (B) Thermodynamic | |||||
| ΔH[ES] (kcal·M−1) | ΔS[ES] (cal−1·M−1·K−1) | ΔG[ES] (kcal·M−1) | |||
| −Vif | 4.9±0.1 | 17.5±1 | −0.5±0.06 | ||
| +Vif | 0.3±0.05 | 9.5±1 | −2.6±0.7 | ||
Figure 1. HIV-1 Vif increases the affinity of the viral RT for a 3′-hydroxy primer during both RDS and DDS.
(A) The variation of the Km value for the 3′-hydroxy primer (ΔKm) under RDS conditions is expressed as the difference between the Km value determined in the absence of Vif and in its presence (Kapp). The points were fitted to the hyperbolic equation ΔKm=Δmax/(1+Kd,vif/[Vif]), where Δmax denotes the maximum difference in the kinetic parameters extrapolated at infinite Vif concentration. (B) The variation of the kcat/Km value (Δkcat/Km) under RDS conditions is expressed as the difference between the kcat/Km value determined in the absence of Vif and that determined in its presence (kcat/Kapp). The points were fitted to the hyperbolic equation Δkcat/Km=Δmax/(1+Kd,vif/[Vif]). The fitting parameters were r2=0.9735 and SSE (sum of squares of errors)=15.76. (C, D) As in (A) and (B) respectively, but under DDS conditions. The fitting parameters were r2=0.9559 and SSE=9.
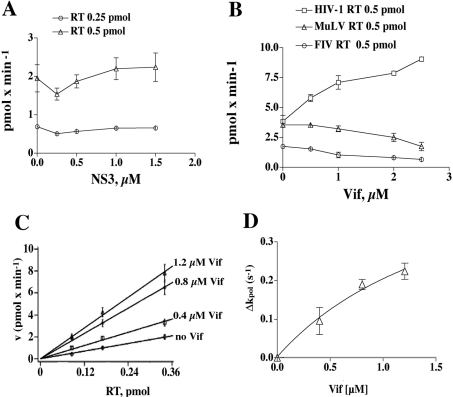
In control experiments, the RNA-binding protein NS3 of HCV was titrated in RDS assays in the presence of different amounts of HIV-1 RT. A shown in Figure 2(A), NS3 did not stimulate HIV-1 RT. Similarly, BSA, PCNA (proliferating-cell nuclear antigen), RP-A (replication protein-A) or histone H1 was titrated in the RDS and DDS assays, but no effects on RT activity were observed (results not shown). To test the specificity of Vif stimulation further, we titrated increasing concentrations of HIV-1 Vif in the presence of RT from HIV-1, FIV (feline immunodeficiency virus) or MuLV. As shown in Figure 2(B), Vif increased the activity of HIV-1 RT; however, it failed to stimulate both the FIV and MuLV RT enzymes, even inhibiting the reaction at high concentrations, indicating that its effect was specific for HIV-1.
Figure 2. HIV-1 Vif specifically stimulates the reaction rate (kpol) of HIV-1 RT.
(A) Increasing amounts of recombinant HCV NS3 were titrated in the presence of 0.25 (○) or 0.5 (▵) pmol of HIV-1 RT, under RDS conditions. The NS3 concentrations used were 0.25, 0.5, 1 and 1.5 μM. (B) Increasing amounts of recombinant Vif were titrated in the presence of HIV-1 RT (□), FIV RT (○) or MuLV RT (▵). Vif concentrations used were 0.5, 1, 2 and 2.5 μM. (C) The dependence of the reaction velocity (pmol·min−1) under RDS conditions from the RT concentration was tested in the absence or in the presence of 0.4, 0.8 or 1.2 μM Vif, as indicated. (D) The slopes of the curves shown in (A), representing the reaction rate kpol. The variation of kpol under RDS conditions (Δkpol) is expressed as the difference between the values determined in the absence and in the presence of Vif. The points were fitted to the hyperbolic equation Δkpol=Δmax/(1+Kd,vif/[Vif]), where Δmax denotes the maximum difference in the kinetic parameters extrapolated at infinite Vif concentration. Fitting parameters were r2=0.9605, SSE (sum of squares of errors)=0.0025.
HIV-1 Vif stimulates the reaction rate (kpol) of HIV-1 RT
Different amounts of recombinant RT were titrated in the RDS reaction, in the presence of increasing concentrations of Vif protein. As shown in Figure 2(C), the reaction velocity at different RT concentrations was increased by Vif. When the increase in the slopes of the curves, representing the polymerization rate kpol, was re-plotted to show dependence on the Vif concentration (Figure 2D), a maximum kpol value of 0.98 s−1, corresponding to a 4-fold stimulation of RT activity, was calculated. The apparent Kd value for Vif was 0.2 μM (Table 1).
HIV-1 Vif decreases the thermodynamic barrier for the formation of the complex of viral RT with the primer-template
Since Vif increased the rate of association (kcat/Km) of RT with the 3′-hydroxy primer, the dependence of kcat/Km on the absolute temperature of the reaction was studied in the absence or in the presence of Vif. The corresponding thermodynamic parameters ΔH[ES], ΔS[ES] and ΔG[ES] for RT·primer complex formation are reported in Table 1. HIV-1 Vif was found to decrease the thermodynamic barrier for complex formation, ΔH[ES], resulting in a 5-fold increase (i.e. shifted towards more negative values) of ΔG[ES] for the reaction.
The C-terminal domain of Vif is required for stimulation of HIV-1 RT activity
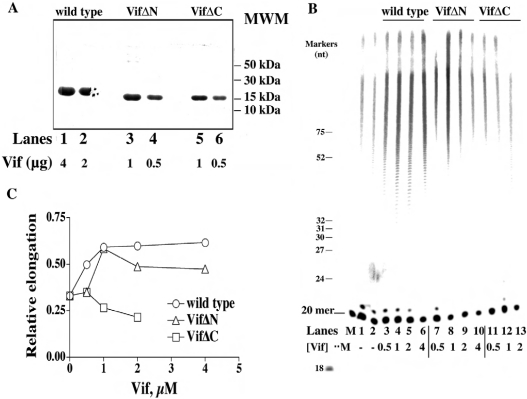
We produced two mutants of the Vif protein: one lacking the first 43 aa at the N-terminus (VifΔN) and the other lacking the final 56 aa at the C-terminus (VifΔC). An SDS/polyacrylamide gel of the purified proteins is shown in Figure 3(A). We then tested the effects of wild-type Vif and the VifΔN and VifΔC mutants on HIV-1 RT activity against the homopolymer poly(rA)·oligo(dT). By using a 5′-labelled (dT)20 oligonucleotide as primer, we could directly visualize the synthesized products on a polyacrylamide gel. As shown in Figure 3(B), addition of increasing amounts of wild-type Vif (lanes 3–6) or of VifΔN (lane 7–10) resulted in the stimulation of total DNA synthesis by HIV-1 RT, compared with control reactions without Vif (lanes 1 and 2). On the other hand, the VifΔC mutant failed to stimulate HIV-1 RT activity. Figure 3(C) shows the quantification of the products, expressed as relative amounts of elongated primer relative to the total input substrate concentration. Under the conditions used, HIV-1 RT elongated approx. 30% of the input DNA primers in the absence of Vif. The RT activity was increased by more than 2-fold by wild-type Vif and the VifΔN mutant, whereas VifΔC even slightly inhibited the reaction.
Figure 3. Purification and characterization of the N- and C-terminal domain deletion mutants of HIV-1 Vif.
(A) SDS/PAGE of samples taken after the final step of purification (Mono-S) of recombinant wild-type Vif and the mutants VifΔN and VifΔC. Increasing amounts of Vif were subjected to denaturing PAGE (10% gel) and stained with Coomassie Blue. MWM denotes molecular-mass markers (B) 7 M Urea/17% polyacrylamide sequencing gel analysis of the products synthesized by HIV-1 RT on poly(rA)·[5′-32P]oligo(dT)20 in the absence or in the presence of wild-type Vif and the VifΔN and VifΔC mutants. The concentrations of recombinant Vif proteins used are indicated. The positions corresponding to the 20-mer primer, as well as of various oligonucleotide size markers, are indicated on the left. (C) Quantification of the products synthesized in the experiment shown in (B) was performed after scanning densitometry with the program ImageQuant. The intensities of the signals corresponding to all the products above the 20-mer primer were normalized to the total intensities of the corresponding lane, and the relative incorporation (r.i..; expressed as % of total input) values were plotted against Vif concentration. Data were fitted to the hyperbolic equation r.i. (%)=%incmax/(1+Kd,vif/[Vif]), where %incmax denotes maximal percentage incorporation extrapolated at infinite Vif concentration.
The C-terminal domain of Vif is required for stimulation of binding of HIV-1 RT to the nucleic acid substrate
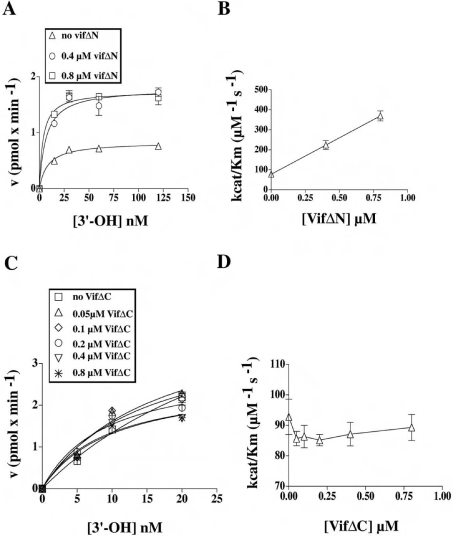
Next, the polymerization rate of HIV-1 RT was measured under RDS conditions as a function of the poly(rA)·oligo(dT) concentration, in the absence or presence of increasing amounts of the VifΔN and VifΔC mutants. As shown in Figure 4(A), VifΔN was still able to increase the efficiency of template utilization by RT in a manner similar to full-length Vif, resulting in a 4-fold stimulation of the kcat/Km of RT for the template (Figure 4B). On the other hand, when VifΔC was tested over a wide range of concentrations and in the presence of increasing amounts of template, no stimulation was observed (Figures 4C and 4D). In summary, the C-terminal domain of Vif appears to be essential to stimulate HIV-1 RT activity.
Figure 4. The C-terminal domain of Vif is required for stimulation of HIV-1 RT activity.
(A) The variation in the velocity of the reaction catalysed by HIV-1 RT under RDS conditions, in the absence or in the presence of increasing amounts of the VifΔN protein, was plotted as a function of the nucleic acid substrate concentration (expressed as available 3′-hydroxy ends). Data were fitted to the Michelis–Menten equation of the form v=Vmax/(1+Km/[3′-hydroxy]). Poly(rA)·oligo(dT) concentrations used were 15, 30, 60 and 120 nM. (B) The maximal reaction rate (kcat) was calculated from the Vmax values derived from the experiments shown in (A) according to the relationship kcat=Vmax/[E0], where [E0] is the total input enzyme concentration. These kcat values were used to calculate the kcat/Km values in the absence and in the presence of VifΔN, which were then plotted against the VifΔN concentration. (C) Experiments were performed as in (A), but in the absence or in the presence of increasing amounts of VifΔC protein. Poly(rA)·oligo(dT) concentrations used were 5, 10 and 20 nM. (D) kcat/Km values in the absence or in the presence of VifΔC were calculated as in (B) and plotted against VifΔ concentration.
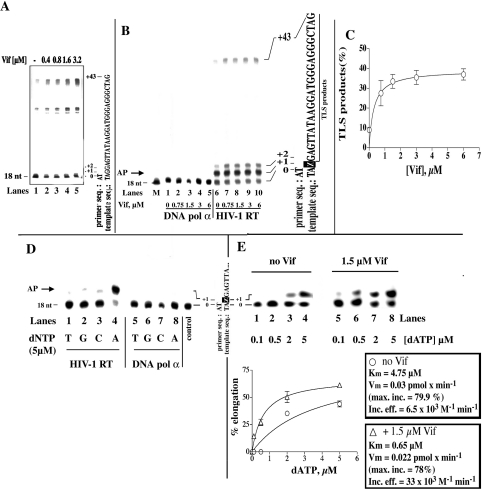
Effect of Vif on the ability of HIV-1 RT to bypass abasic sites
It has been shown that Vif counteracts the effects of a cellular cytidine deaminase (CEM15/APOBEC3G) which is responsible for massive deamination of cytosine to uracil in the first (minus)-strand DNA intermediate of HIV-1 reverse transcription. A possible consequence of the presence of uracil in DNA is triggering of the base-excision repair pathway, with excision of the base by a glycosylase and generation of an abasic (AP) site. Since abasic sites can sometimes block the progress of DNA synthesis by a DNA polymerase, we investigated the ability of HIV-1 RT to bypass this lesion in the absence or in the presence of Vif. In a first series of experiments, we tested the effects of Vif on the activity of HIV-1 RT in the presence of the DNA/DNA heteropolymeric primer-template 17/73-mer. As shown in Figure 5(A), Vif was able to stimulate DDS by HIV-1 RT on this template in a dose-dependent manner (compare lane 1 with lanes 2–5). Next, we tested the effects of Vif on the same 17/73-mer substrate, but this time with a synthetic abasic site (tetrahydrofuran moiety) at position +1 on the template strand (Figure 5B). As shown, DDS by HIV-1 RT using the AP-site-containing template was increased by Vif in a dose-dependent manner (Figure 5B, compare lane 6 with lanes 7–10). When the same Vif concentrations were tested with the damaged template in the presence of DNA polymerase α, an enzyme which is blocked by an abasic site, no stimulation was observed (Figure 5B, lanes 1–5). Quantification of the translesion synthesis (TLS) products (Figure 5C) showed a 4-fold stimulation of RT activity and a Kd value for Vif of 0.3 μM.
Figure 5. Vif stimulates TLS by HIV-1 RT in the presence of an abasic site.
(A) 7 M Urea/20% (w/v) polyacrylamide sequencing gel analysis of the products synthesized by HIV-1 RT on the undamaged 18/73-mer DNA template in the presence of Vif. (B) 7 M Urea/20% (w/v) polyacrylamide sequencing gel analysis of the products synthesized by HIV-1 RT (lanes 6–10) or human DNA polymerase α (lanes 1–5) in the absence or in the presence of Vif and in the presence of the 18/73-mer DNA template containing an abasic site at position +1. The concentrations of recombinant Vif protein used are indicated. The positions corresponding to the 18-mer primer and to the full-length products, as well as the sequence of the template strand, are indicated on the right. The arrow on the left (AP) indicates the position of the products corresponding to incorporation opposite the lesion. (C) Quantification of the TLS products synthesized in the experiment shown in (B) was performed after scanning densitometry with the program ImageQuant. The intensity of the signal corresponding to the TLS products were normalized to the total intensity of the corresponding lane, and the relative incorporation (TLS products; expressed as % of total input) was plotted against Vif concentration. Data were fitted to the hyperbolic equation TLS (%)=%TLSmax/(1+Kd,vif/[Vif]), where %TLSmax denotes maximal percentage TLS extrapolated at infinite Vif concentration. (D) 7 M Urea/20% polyacrylamide sequencing gel analysis of the products synthesized by HIV-1 RT (lanes 1–4) or human DNA polymerase α (lanes 5–8) under single nucleotide incorporation conditions. Reactions were performed in the absence of Vif and in the presence of the 18/73-mer DNA template containing an abasic site at position +1. The single nucleotide added in each reaction is indicated. The position corresponding to the 18-mer primer is indicated on the left, and the sequence of the template strand is shown on the right. The arrow on the left indicates the position of the products corresponding to incorporation opposite the lesion. (E) Upper panel: 7 M Urea/20% polyacrylamide sequencing gel analysis of the products synthesized by HIV-1 RT in the absence or in the presence of Vif and in the presence of the 18/73-mer DNA template containing an abasic site at position +1, under single nucleotide incorporation conditions. The concentrations of the single-nucleotide dATP used in each reaction are indicated below the gel. The positions corresponding to the 18-mer primer (0) and to the first incorporated nucleotide (+1), as well as the sequence of the template strand, are indicated on the left. Lower panel: quantification of products synthesized (see upper panel) was performed after scanning densitometry with the program ImageQuant. The intensity of the signal corresponding to the +1 products was normalized to the total intensity of the corresponding lane, and the relative elongation values (expressed as % of elongated primer ends compared with total input) were plotted against dATP concentration. Data were fitted to the Michelis–Menten equation in the form: v=Vmax/(1+Km/[dATP]). The calculated Km, Vmax (Vm) and Vmax/Km (incorporation efficiency) values are indicated on the right.
Next, we tested the specificity of nucleotide incorporation by HIV-1 RT in front of an abasic site. As shown in Figure 5(D), HIV-1 RT preferentially incorporated dATP at the +1 position, (compare lane 4 with lanes 1–3). In a subsequent experiment (Figure 5E, upper panel), the efficiency of nucleotide incorporation in front of the abasic site by HIV-1 RT was measured by varying the dATP concentration in the absence (lanes 1–4) or in the presence (lanes 5–8) of Vif. Quantification of the products (Figure 5E, lower panel) revealed that Vif increased by 6-fold the efficiency of dATP incorporation opposite an abasic site by HIV-1 RT.
DISCUSSION
We have analysed possible functions of Vif in the reverse transcription process. The effect of Vif, as shown by kinetic and thermodynamic analysis, was to increase the efficiency of the association of HIV-1 RT with the template-primer substrate. This effect appeared to be specific, since other unrelated RNA-binding (HCV NS3) and DNA-binding (such PCNA, RP-A and histone H1) proteins did not show any stimulation (Figure 2A and results not shown). The N-terminal domain of Vif, which is essential for RNA binding, was dispensable for stimulation, whereas the C-terminal domain was essential. Moreover, stimulation was observed both under RDS and DDS conditions. These results suggest that Vif stimulates HIV-1 RT through a direct interaction. However, we were not able to detect a physical interaction of Vif with HIV-1 RT using a variety of techniques, such as co-immunoprecipitation, gel filtration and pull-down assays (results not shown). It is possible that such an interaction might be too weak to survive direct isolation of the complex, a fact that could also explain the relatively high amount of Vif protein required to detect stimulation of RT activity in vitro (50-fold molar excess over the RT). Vif has been shown to interact with other components of the nucleoprotein complex; hence one possibility is that additional factors might be required to increase the stability of the RT–Vif complex.
Since the requirement for a functional Vif is restricted to certain cell types, it is possible that the interaction between Vif and HIV-1 RT might not be essential for the reverse transcription process per se. This is not surprising, since several other HIV-1 proteins have been implicated in contributing to the efficiency of the reverse transcription process in vivo, such as the integrase, the nucleocapsid protein and Tat [21]. The auxiliary protein Nef has also been shown to interact physically with HIV-1 RT and to stimulate its association with the viral RNA [25]. The effect of Vif, however, might become more relevant under challenging conditions, for example when a minus-strand DNA containing abasic sites has to be used as a template by HIV-1 RT. That abasic sites might be present in the proviral DNA is suggested by the fact that the cellular uracil N-glycosylase UNG2 is packaged into HIV-1 virions through physical interaction with the viral RT and integrase [26]. UNG2 removes uracil residues from double- or even single-stranded DNA, generating abasic sites. Its presence in the HIV-1 virion has been explained as part of the viral defence mechanism against the accumulation of uracil in the viral DNA. Even in the absence of APOBEC3G, that is in the presence of Vif and/or in permissive cells, there are a number of potential sources of dUTP incorporation into the minus-strand viral DNA. For example, in macrophages the pools of deoxynucleotides in general, and those of TTP and dCTP in particular, are very low (<1 pmol/100 cells), and there is no de novo deoxynucleotide synthesis [26]. Thus when a genomic RNA sequence of the type A-G-C-U is to be reverse transcribed in such an environment, some of the minus strands might look like dT-dU-dG-dA because of the lack of dCTP and the stability of the dU-G pair. Accumulation of dU in the minus-strand DNA has been shown to be dangerous for retroviruses. In particular, for HIV-1 it has been proposed that accumulation of uracil in retroviral DNA may disrupt the viral life cycle by altering the specificity of plus-strand DNA synthesis initiation during reverse transcription [27]. Owing to the presence of UNG2 physically associated with the RT, it is likely that most of the dU residues present in the minus-strand DNA will be readily converted into abasic sites. Since the viral RNA genome is degraded as soon as the first DNA strand is synthesized, there is probably a time interval between the initiation of minus-strand DNA synthesis and completion of the plus-strand DNA, when viral DNA is very sensitive to the action of uracil-processing enzymes. The presence of abasic sites in the minus-strand DNA might either trigger viral DNA cleavage by specific endonucleases or hinder the synthesis of the complementary plus-strand DNA. Indeed, HIV-1 RT has been shown to bypass abasic sites in an error-prone fashion, either by inserting an A opposite the lesion, or through primer misalignment with a nucleotide downstream of the lesion. Moreover, in both cases the incorporation efficiency opposite the abasic site was severalfold lower than with undamaged templates, resulting in marked pausing of the enzyme at the damaged site [28]. Our analysis showed that Vif can increase by 5–6-fold the efficiency with which the viral RT can perform TLS over an abasic site in vitro. However, this bypass was always error-prone, leading to incorporation of an A in front of the abasic site. Thus, independent of the mechanism by which uracil is incorporated into minus-strand DNA (either by deamination of dC or by misincorporation of dU in front of a G), the result of TLS by HIV-1 RT will be a G→A substitution in the plus-strand DNA.
We can thus envisage a scenario wherein Vif has the general function of preventing the ‘stalling’ of the reverse transcription complex at abasic sites present in the minus-strand viral DNA, but at the expense of accumulating mutations. In the infected cell, the resulting level of mutation would normally be compatible with proviral DNA function, and might even contribute to increasing the genetic variability of HIV-1. However, in the presence of APOBEC3G, massive deamination of dC residues would increase the accumulation of G→A mutations to lethal levels, hence the need for the additional role of Vif in preventing APOBEC3G incorporation into virions.
Acknowledgments
We thank Dr D. Gabuzda (Dana-Farber Cancer Institute, Boston, MA, U.S.A.) for kindly providing us with the expression plasmid pD10Vif, Professor U. Hübscher (University of Zürich, Switzerland) for sharing with us the recombinant FIV RT, and Dr G. Villani (IPBS-CNRS, Toulouse, France) for his generous donation of the abasic-site-containing oligonucleotide. This work was supported by EU Contract LSHG-CT-2003-503480-TRIoH (to G.M.) and by the Programma Nazionale di Ricerca sull'AIDS – ISS (to G.M. and S.S.). R.C. was a recipient of an ICGEB Fellowship.
References
- 1.Frankel A. D., Young J. A. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Gabuzda D. H., Li H., Lawrence K., Vasir B. S., Crawford K., Langhoff E. Essential role of vif in establishing productive HIV-1 infection in peripheral blood T lymphocytes and monocyte/macrophages. J. Acquired Immune Defic. Syndr. 1994;7:908–915. [PubMed] [Google Scholar]
- 3.Inubushi R., Adachi A. Cell-dependent function of HIV-1 Vif for virus replication. Int. J. Mol. Med. 1999;3:473–476. doi: 10.3892/ijmm.3.5.473. [DOI] [PubMed] [Google Scholar]
- 4.Simon J. H., Miller D. L., Fouchier R. A., Soares M. A., Peden K. W., Malim M. H. The regulation of primate immunodeficiency virus infectivity by Vif is cell species restricted: a role for Vif in determining virus host range and cross-species transmission. EMBO J. 1998;17:1259–1267. doi: 10.1093/emboj/17.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon J. H., Carpenter E. A., Fouchier R. A., Malim M. H. Vif and the p55(Gag) polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J. Virol. 1999;73:2667–2674. doi: 10.1128/jvi.73.4.2667-2674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouyac M., Courcoul M., Bertoia G., Baudat Y., Gabuzda D., Blanc D., Chazal N., Boulanger P., Sire J., Vigne R., Spire B. Human immunodeficiency virus type 1 Vif protein binds to the Pr55Gag precursor. J. Virol. 1997;71:9358–9365. doi: 10.1128/jvi.71.12.9358-9365.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nascimbeni M., Bouyac M., Rey F., Spire B., Clavel F. The replicative impairment of Vif-mutants of human immunodeficiency virus type 1 correlates with an overall defect in viral DNA synthesis. J. Gen. Virol. 1998;79:1945–1950. doi: 10.1099/0022-1317-79-8-1945. [DOI] [PubMed] [Google Scholar]
- 8.Goncalves J., Korin Y., Zack J., Gabuzda D. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J. Virol. 1996;70:8701–8709. doi: 10.1128/jvi.70.12.8701-8709.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dornadula G., Yang S., Pomerantz R. J., Zhang H. Partial rescue of the Vif-negative phenotype of mutant human immunodeficiency virus type 1 strains from nonpermissive cells by intravirion reverse transcription. J. Virol. 2000;74:2594–2602. doi: 10.1128/jvi.74.6.2594-2602.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohagen A., Gabuzda D. Role of Vif in stability of the human immunodeficiency virus type 1 core. J. Virol. 2000;74:11055–11066. doi: 10.1128/jvi.74.23.11055-11066.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dettenhofer M., Cen S., Carlson B. A., Kleiman L., Yu X. F. Association of human immunodeficiency virus type 1 Vif with RNA and its role in reverse transcription. J. Virol. 2000;74:8938–8945. doi: 10.1128/jvi.74.19.8938-8945.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang S., Sun Y., Zhang H. The multimerization of human immunodeficiency virus type I Vif protein: a requirement for Vif function in the viral life cycle. J. Biol. Chem. 2001;276:4889–4893. doi: 10.1074/jbc.M004895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X., Gabuzda D. Mitogen-activated protein kinase phosphorylates and regulates the HIV-1 Vif protein. J. Biol. Chem. 1998;273:29879–29887. doi: 10.1074/jbc.273.45.29879. [DOI] [PubMed] [Google Scholar]
- 14.Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature (London) 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H., Yang B., Pomerantz R. J., Zhang C., Arunachalam S. C., Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature (London) 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris R. S., Bishop K. N., Sheehy A. M., Craig H. M., Petersen-Mahrt S. K., Watt I. N., Neuberger M. S., Malim M. H. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 17.Marin M., Rose K. M., Kozak S. L., Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 18.Sheehy A. M., Gaddis N. C., Malim M. H. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 19.Khan M. A., Aberham C., Kao S., Akari H., Gorelick R., Bour S., Strebel K. Human immunodeficiency virus type 1 vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J. Virol. 2001;75:7252–7265. doi: 10.1128/JVI.75.16.7252-7265.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H., Pomerantz R. J., Dornadula G., Sun Y. Human immunodeficiency virus type 1 Vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process. J. Virol. 2000;74:8252–8261. doi: 10.1128/jvi.74.18.8252-8261.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baraz L., Kotler M. The Vif protein of human immunodeficiency virus type 1 (HIV-1): enigmas and solutions. Curr. Med. Chem. 2004;11:221–231. doi: 10.2174/0929867043456124. [DOI] [PubMed] [Google Scholar]
- 22.Maga G., Amacker M., Hubscher U., Gosselin G., Imbach J. L., Mathe C., Faraj A., Sommadossi J. P., Spadari S. Molecular basis for the enantioselectivity of HIV-1 reverse transcriptase: role of the 3′-hydroxyl group of the L-(beta)-ribose in chiral discrimination between D- and L-enantiomers of deoxy- and dideoxy-nucleoside triphosphate analogs. Nucleic Acids Res. 1999;27:972–978. doi: 10.1093/nar/27.4.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X., Goncalves J., Gabuzda D. Phosphorylation of Vif and its role in HIV-1 replication. J. Biol. Chem. 1996;271:10121–10129. doi: 10.1074/jbc.271.17.10121. [DOI] [PubMed] [Google Scholar]
- 24.Maga G., Ubiali D., Salvetti R., Pregnolato M., Spadari S. Selective interaction of the human immunodeficiency virus type 1 reverse transcriptase nonnucleoside inhibitor efavirenz and its thio-substituted analog with different enzyme-substrate complexes. Antimicrob. Agents Chemother. 2000;44:1186–1194. doi: 10.1128/aac.44.5.1186-1194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fournier C., Cortay J. C., Carbonnelle C., Ehresmann C., Marquet R., Boulanger P. The HIV-1 nef protein enhances the affinity of reverse transcriptase for RNA in vitro. Virus Genes. 2002;25:255–269. doi: 10.1023/a:1020971823562. [DOI] [PubMed] [Google Scholar]
- 26.Priet S., Navarro J. M., Gros N., Querat G., Sire J. Functional role of HIV-1 virion-associated uracil DNA glycosylase 2 in the correction of G:U mispairs to G:C pairs. J. Biol. Chem. 2003;278:4566–4571. doi: 10.1074/jbc.M209311200. [DOI] [PubMed] [Google Scholar]
- 27.Klarmann G. J., Chen X., North T. W., Preston B. D. Incorporation of uracil into minus strand DNA affects the specificity of plus strand synthesis initiation during lentiviral reverse transcription. J. Biol. Chem. 2003;278:7902–7909. doi: 10.1074/jbc.M207223200. [DOI] [PubMed] [Google Scholar]
- 28.Cai H., Bloom L. B., Eritja R., Goodman M. F. Kinetics of deoxyribonucleotide insertion and extension at abasic template lesions in different sequence contexts using HIV-1 reverse transcriptase. J. Biol. Chem. 1993;268:23567–23572. [PubMed] [Google Scholar]
- 29.Locatelli G. A., Spadari S., Maga G. Hepatitis C virus NS3 ATPase/helicase: an ATP switch regulates the cooperativity among the different substrate binding sites. Biochemistry. 2002;41:10332–10342. doi: 10.1021/bi026082g. [DOI] [PubMed] [Google Scholar]