Abstract
Mitochondria-encoded ND (NADH dehydrogenase) subunits, as components of the hydrophobic part of complex I, are essential for NADH:ubiquinone oxidoreductase activity. Mutations or lack of expression of these subunits have significant pathogenic consequences in humans. However, the way these events affect complex I assembly is poorly documented. To understand the effects of particular mutations in ND subunits on complex I assembly, we studied four human cell lines: ND4 non-expressing cells, ND5 non-expressing cells, and rho° cells that do not express any ND subunits, in comparison with normal complex I control cells. In control cells, all the seven analysed nuclear-encoded complex I subunits were found to be attached to the mitochondrial inner membrane, except for the 24 kDa subunit, which was nearly equally partitioned between the membranes and the matrix. Absence of a single ND subunit, or even all the seven ND subunits, caused no major changes in the nuclear-encoded complex I subunit content of mitochondria. However, in cells lacking ND4 or ND5, very low amounts of 24 kDa subunit were found associated with the membranes, whereas most of the other nuclear-encoded subunits remained attached. In contrast, membrane association of most of the nuclear subunits was significantly reduced in the absence of all seven ND proteins. Immunopurification detected several subcomplexes. One of these, containing the 23, 30 and 49 kDa subunits, also contained prohibitin. This is the first description of prohibitin interaction with complex I subunits and suggests that this protein might play a role in the assembly or degradation of mitochondrial complex I.
Keywords: chaperone, complex I assembly, MS, NADH dehydrogenase, prohibitin
Abbreviations: BN-PAGE, Blue-Native PAGE; cyt c, cytochrome c; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; FP, flavoprotein; HP, hydrophobic; IP, iron protein; LC-MS/MS, liquid chromatography-tandem MS; MnSOD, manganese superoxide dismutase; mtDNA, mitochondrial DNA; NBT, Nitro Blue Tetrazolium; ND, NADH dehydrogenase; OXPHOS, oxidative phosphorylation
INTRODUCTION
NADH:ubiquinone oxidoreductase (complex I, EC 1.6.5.3) catalyses the oxidation of NADH, with the concomitant reduction of ubiquinone and ejection of protons out of the mitochondria. It is the largest, most complicated and least understood of the mitochondrial respiratory-chain enzymes [1] and consists of a multimeric assembly of seven mitochondria-encoded subunits named ND (NADH dehydrogenase) subunits and approx. 38 nuclear-encoded subunits [2]. By electron microscopy, the enzyme from Neurospora crassa, Escherichia coli and bovine has been shown to have an overall L-shaped structure with two arms perpendicular to each other [3]. It has also been proposed recently that the complex I from E. coli can exhibit, under specific conditions, a horseshoe-shaped structure [4]. In the L-shaped structure, one arm is embedded in the inner-mitochondrial membrane, whereas the other protrudes into the matrix. Structural studies aimed at defining the overall architecture of complex I have been performed using chaotropic agents or detergents to split the complex into specific subcomplexes which are easier to analyse [5,6]. Using chaotropic agents the complex can be divided into three parts: FP (flavoprotein), IP (iron protein) and HP (hydrophobic) fractions [5]. The FP fraction contains three nuclear-encoded subunits: 10, 24 and 51 kDa. The IP fraction is characterized by a large number of Fe–S clusters with seven or nine nuclear-encoded subunits: 13, 15, 18, 30, 49 and 75 kDa and B13. The 20 and 23 kDa subunits can be found either in IP or HP fractions depending on the purification procedure used. The HP fraction contains few nuclear-encoded subunits but all of the seven mitochondria-encoded ND subunits. Therefore the isolated membrane arm contains all the mtDNA (mitochondrial DNA)-encoded subunits, whereas the matrix arm contains most of the nuclear-encoded subunits.
Complex I architecture and assembly have been the subject of numerous studies performed on lower eukaryotic cellular models, and several studies have reported disruptions of some specific nuclear genes coding for complex I subunits in N. crassa that resulted in partial assembly of complex I or severe fragmentation into smaller inactive matricial or membranous subcomplexes [7]. Antonicka et al. [8] have recently proposed a rough assembly pathway for the human complex I on the basis of their detection of seven subcomplexes in mitochondria from four different patients with complex I deficiency. Nevertheless, the detailed assembly pathway is yet to be defined. Furthermore, despite the fact that the primary structure of the mammalian mtDNA-encoded subunits has been determined [9], and that transmembrane topography of the ND2, ND4, ND5 and ND6 subunits has been predicted in Rhodobacter capsulatus [10] and Paracoccus denitrificans [11], little is known about their function and their assembly in the course of complex I biogenesis. ND subunits may quite probably play important roles in the proton translocation machinery of complex I [12], and ND1 in particular may be involved, because it is targeted by N,N′-dicyclohexylcarbodi-imide, a chemical cross-linker known to inhibit proton pumping in complex I [13]. Other studies with inhibitors also suggested that ND1 and ND4 may be capable of interacting with ubiquinone, the natural electron acceptor [14].
Whereas the existence of some chaperone proteins necessary for the assembly of other human mitochondrial respiratory complexes has been shown (e.g. Surf1 for complex IV [15]), no chaperone has yet been identified for human complex I assembly. In the specific case of N. crassa, two complex I assembly chaperones, CIA30 and CIA84, have been discovered [16]. The human homologue of CIA30 has been recently characterized, but its function in complex I assembly remains to be clarified [17]. Alterations of the assembly or structure of complex I are particularly interesting events to analyse as they are expected to elicit pathological status in humans [18].
In fact, numerous clinical syndromes have been associated with complex I deficiency [19], and several mutations in the genes encoding for mtDNA-encoded subunits have been correlated with human diseases. In particular, it has been shown that Leber's hereditary optic neuropathy can be associated with mutations in the ND1, ND4, ND5 or ND6 genes [20]. Another mutation in the ND6 gene has been identified as the cause of Leber's hereditary optic neuropathy associated with dystonia [21]. In addition, mutations in the ND1 or ND5 genes have been described in MELAS (Mitochondrial Encephalopathy Lactic Acidosis Stroke-like episodes) [22,23], and Leigh syndrome [24].
A deeper understanding of the functions of the mtDNA-encoded complex I subunits is necessary to get an appreciation, at the molecular level, of the pathogenesis. In the present study, we examined complex I assembly and the role of the ND4 and ND5 subunits. Four human cell lines were investigated: 143B TK− osteosarcoma cell line (parental cells), the daughter 143B206 rho° cells, completely depleted of mtDNA by long-term exposure to ethidium bromide [25], and C4T and C9T complex I-deficient cells [26,27]. Owing to a frameshift mutation in the ND4 or ND5 genes, C4T and C9T cells lack the ND4 or ND5 gene products respectively.
We performed immunodetection experiments to determine the complex I subunit composition of matrix or membrane mitochondrial subfractions from these model cell lines. Finally, MS analysis of immunopurified subcomplexes revealed the presence of the associated prohibitin. It is proposed that prohibitin may act as a complex I assembly chaperone.
EXPERIMENTAL
Cell lines and culture conditions
All cells were cultured in a 5% CO2 atmosphere at 37 °C. The 143B TK− osteosarcoma cell line (CRL 8303) was obtained from the A.T.C.C. (Rockville, MD, U.S.A.). These cells were grown in high-glucose DMEM (Dulbecco's modified Eagle's medium), supplemented with 10% (v/v) FBS (fetal bovine serum), 100 μg/ml pyruvate and 100 μg/ml 5-bromo-2′-deoxyuridine. Both 143B206 rho° and C4T cell lines, derived from 143B TK− cells, were cultured in DMEM supplemented with 10% FBS, 100 μg/ml pyruvate, 100 μg/ml 5-bromo-2′-deoxyuridine and 50 μg/ml uridine. C9T cell line, another derivative of 143B TK− cells, was grown in DMEM supplemented with 10% FBS and 100 μg/ml pyruvate. The cell lines derived from 143B TK− cells were provided by G. Attardi and A. Chomyn (California Institute of Technology, Pasadena, CA, U.S.A.).
DNA and RNA analyses
Total DNA was extracted from cells and subjected to PCR amplification using appropriate primers and standard methods. For DNA sequencing, three different regions of the mtDNA were amplified by PCR. These three fragments individually encompass the D loop (nt 16024–576), the ND4 gene (nt 10760–12136), or the ND5 gene (nt 12337–14145), according to mtDNA base numbering [9]. Total RNA was extracted from cultured cells and harvested at a non-confluent state, using the RNeasy kit (Qiagen, Valencia, CA, U.S.A.). Reverse transcriptase–PCR was performed as described previously [28].
For all cell types, fragments of the ND4 and ND5 genes were amplified and PCR products were extensively sequenced. The mutations originally discovered by Hofhaus and Attardi have been confirmed: in C4T cells, an insertion of one cytosine residue in a stretch of six cytosines at positions 10947–10952 in ND4 and, in C9T cells, the presence of an insertion of an adenine residue in a stretch of eight adenines at positions 12417–12425 in ND5 [26,27]. Moreover, we have found two silent mutations (T10873C and A11002G) in C4T and C9T ND4 gene fragments, and one silent mutation (C12705T) in the 143B, C4T and C9T ND5 gene fragment.
Fluorescence microscopy
Cells were fixed with 3.7% formaldehyde on coverglass (Lab-Tek™; Nalge Nunc International, Naperville, IL, U.S.A.) for 15 min at 37 °C. Mitochondria were then dyed by incubating with 100 nM MitoTracker Green FM™ (Molecular Probes, Eugene, OR, U.S.A.) for 25 min at room temperature (21 °C), and nuclei dyed with 0.25% Hoëchst 33258 (Molecular Probes) for 4 min at room temperature. Cells were then examined by fluorescence microscopy.
Electron microscopy
A flask of confluent cells was fixed with 2% glutaraldehyde in cell culture medium for 15 min at room temperature, then post-fixed with 2% glutaraldehyde/0.1 M sodium cacodylate/HCl (pH 7.4) for 30 min at room temperature and, finally, with 1% OsO4/0.15 M sodium cacodylate/HCl (pH 7.4) for 30 min at 4 °C. Cells were then dehydrated by successive baths in 30–100% (v/v) ethanol at room temperature, and embedded in Epon resin which was polymerized at 60 °C for 72 h. Ultrathin sections (60–80 nm) were obtained with an RMC MTX ultramicrotome, stained by uranyl acetate and lead citrate and examined at 80 kV with a JEOL 1200CX electron microscope equipped with a digital camera MegaviewII and AnalySIS software.
Preparation and fractionation of mitochondria
Mitochondria-enriched preparations were obtained as described previously [18]. The method for preparation of submitochondrial fractions (membranes and matrix) was adapted from that described by Fessenden and Racker [29]. Mitochondria samples, kept on ice, were exposed to sonic oscillation (200 W) for eight 10-s periods. Matrix and membrane fractions were then separated by four centrifugation steps. After sonication, the solution was centrifuged for 20 min at 11000 g to remove intact mitochondria. The supernatant was centrifuged for 30 min at 100000 g. The 100000 g supernatant was collected and centrifuged again for 30 min at 100000 g. The resulting 100000 g supernatant contains the mitochondrial matrix proteins. The 100000 g pellet was resuspended in 0.01 M Tris/HCl (pH 7.4) and centrifuged again at 100000 g for 30 min. The resulting pellet containing membrane proteins was resuspended in a medium containing 0.25 M sucrose, 1 mM EDTA, 10 mM Tris/HCl (pH 7.4) and protease inhibitors (2 μg/ml aprotinin, 1 μg/ml antipain, chymostatin and leupeptin).
Enzymic measurements
Spectrophotometric analysis of the respiratory-chain complexes was performed on mitochondrial preparations from cultured cells. At a protein concentration of 1–1.5 mg/ml, mitochondria were disrupted by three freeze–thaw cycles in liquid nitrogen and then briefly sonicated. The following enzymic activities were assayed at 30 °C: citrate synthase, NADH–ubiquinone reductase (complex I), succinate–cyt c (where cyt c stands for cytochrome c) reductase (complex II+III), succinate dehydrogenase, succinate–ubiquinone reductase (complex II), ubiquinol–cyt c reductase (complex III) and cyt c oxidase (complex IV). The methodology followed was as reported previously [18], with the following modifications: complex IV activity was measured in 10 mM phosphate buffer (pH 7.0) containing 2.5 mg/ml BSA using 80 μM cyt c reduced by dithionite as described by Trounce et al. [30]; complex II+III was assayed in 50 mM phosphate buffer (pH 7.5) containing 2.5 mg/ml defatted BSA, 1 mM KCN using 100 μM cyt c and 10 mM succinate; complex III was measured in 50 mM phosphate buffer (pH 7.5) containing 2.5 mg/ml defatted BSA, 1 mM KCN, 5 μM rotenone using 100 μM cyt c and 60 μM reduced decylubiquinone; complex II was measured in the complex III phosphate buffer supplemented with 2 μg/ml antimycin using 20 mM succinate, 75 μM 2,6-dichlorophenol-indophenol and 75 μM decylubiquinone as substrates. Activity of complex I was assayed in 10 mM phosphate buffer (pH 7.5) containing 3.75 mg/ml defatted BSA, 1.8 mM KCN, 2 μg/ml antimycin and 0.1 mM decylubiquinone. The reaction was initiated with 0.2 mM NADH, and NADH oxidation was monitored at 340 nm. Rotenone-sensitive complex I activity was calculated from the difference between two parallel rates measured with and without 5 μM rotenone.
BN-PAGE (Blue-Native PAGE) analysis
Complex I was analysed by BN-PAGE, using a method described by Duby et al. [31]. Briefly, ACA buffer containing 750 mM aminocaproic acid, 0.5 mM EDTA and 50 mM Bis/Tris (pH 7.0) was added to each sample of mitochondria (1:1, v/v), and the membrane proteins were solubilized by the addition of 0.45% n-dodecyl-β-D-maltoside. The solution was centrifuged for 20 min at 15000 g, and 0.3% Coomassie Blue was added to the supernatant. Mitochondrial proteins (200 μg) were separated on a gradient of polyacrylamide (4–12%) with Coomassie Blue. Blue-Native gels were stained for ND activity using NBT (Nitro Blue Tetrazolium) as an electron acceptor. This activity was revealed with 1 mg/ml NBT, 0.2 mM NADH and 100 mM Mops (pH 8). Blue-Native gels were electroblotted on to PVDF membranes for immunodetection (see below).
Immunodetection
Charge solution composed of 0.25 M Tris/HCl (pH 2.6), 1.5 mg of Bromophenol Blue, 40% (v/v) glycerol and 8% SDS was added to each sample (cells, mitochondria-enriched suspension, matrix or membrane proteins; 1:5, v/v), and then 14 M 2-mercaptoethanol was added (1:20, v/v). The samples were heated for 5 min at 90 °C. Mitochondrial proteins were separated on 12% SDS/polyacrylamide gel, electroblotted on to PVDF membranes and incubated with antibodies as described in [18]. Polyclonal antisera were raised in rabbit against peptides corresponding to the major complex I subunits [32,33] and the cyt c oxidase subunit II (COII). The polyclonal antiserum against the 24 kDa subunit of complex I was raised in rabbit against the whole subunit. Four commercial monoclonal antibodies were used: they were raised in mouse against the 15, 20 and 39 kDa subunits of complex I, and the 30 kDa subunit of complex II (Molecular Probes) [34].
Immunopurification experiments
The method used for immunopurification was adapted from Duborjal et al. [35] and Murray et al. [36]. Swollen Protein A–Sepharose beads (150 μl) (ImmunoPure® Protein A IgG Orientation Kit; Pierce, Rockford, IL, U.S.A.) were washed with 350 μl of 50 mM sodium borate (pH 8.2). Antiserum (10 μl) was added with 70 μl of 50 mM sodium borate (pH 8.2) and bound to the beads. After 1 h at room temperature, the beads were washed twice with 350 μl of 50 mM sodium borate (pH 8.2). The antibody was cross-linked to the beads for 1 h at room temperature with 1 mg of dimethylpimelimidate in 150 μl of 0.2 M triethanolamine (pH 8.2). After a wash with 350 μl of 0.2 M triethanolamine (pH 8.2), cross-linking was blocked with 150 μl of 0.1 M ethanolamine solution (pH 8.2), for 10 min at room temperature. Beads were washed with 350 μl of 10 mM PBS solution supplemented with 0.2% n-dodecyl-β-D-maltoside, and then three times with 450 μl of 0.1 M glycine (pH 2.8), supplemented with 0.2% n-dodecyl-β-D-maltoside. The gel was then equilibrated with 350 μl of 10 mM PBS solution supplemented with 0.2% n-dodecyl-β-D-maltoside. Intact cells (approx. 1 mg of proteins) were disrupted for 1 h at room temperature with 480 μl of 10 mM PBS solution supplemented with 0.2% n-dodecyl-β-D-maltoside and 10 μl of protease inhibitors (Immunocatcher™ kit; Cytosignal Research Products, Irvine, CA, U.S.A.). Cells were centrifuged for 17 min at 13000 g, and the supernatant was incubated with the antibodycross-linked beads for 1 h at room temperature. Beads were washed three times with 350 μl of 10 mM PBS solution supplemented with 0.2% n-dodecyl-β-D-maltoside. Immunocaptured subunits were eluted twice with 50 μl of 0.1 M glycine (pH 2.8) supplemented with 0.2% n-dodecyl-β-D-maltoside, by gentle centrifugation at 12000 g in a microfuge, followed by the addition of 2.5 μl of 1 M Tris/HCl (pH 9.5) to each supernatant for neutralization.
MS and protein identification by LC-MS/MS (liquid chromatography-tandem MS)
After immunopurification, the samples were loaded on to an SDS/polyacrylamide gel; electrophoretic migration of the samples was stopped when proteins reached the separating gel and concentrated as a fine band which was then excised from the Coomassie Blue-stained gel [37]. The excised band was washed several times with destaining solutions [first with 25 mM NH4HCO3 for 15 min, and then with 50% (v/v) CH3CN containing 25 mM NH4HCO3 for 15 min]. Proteins in the band were reduced by incubation in a reducing solution containing 10 mM dithiothreitol for 45 min at 57 °C and, subsequently, alkylated by a solution containing 55 mM iodoacetamide for 45 min in the dark. In-gel proteolysis of the sample was performed for 5 h at 37 °C using sequencing-grade-modified trypsin resuspended in 25 mM NH4HCO3. A trypsin/protein ratio of 1:20 (w/w) was used.
Peptides were then extracted from the gel by passive diffusion with a 5% formic acid solution and CH3CN. After drying, tryptic peptides were resuspended in 0.5% aqueous trifluoroacetic acid and injected into a CapLC (Waters, Milford, MA, U.S.A.) nano-LC system directly coupled with QTOF Ultima mass spectrometer (Waters). MS/MS data were collected and processed automatically using Masslynx 3.5 software. Protein searches were performed in SwissProt and TrEMBL databases (http://www.expasy.org/sprot/) using the Mascot 1.7 program (Matrix Science: http://www.matrixscience.com/); the sequences of peptides identified by Mascot were all confirmed manually. The parameters for Mascot searches were as follows: two missed cleavages; 0.002 and 0.0004 kDa mass accuracy allowed for the parent and the fragment ions respectively, carbamidomethylcysteine as fixed modification, whereas oxidized methionine, acetylation of the peptide or of the N-terminus of the protein were set as variable modifications; 100 hits were allowed.
RESULTS AND DISCUSSION
To investigate the human complex I assembly pathway and the role played by the ND subunits in the overall complex I architecture, four human cell lines have been studied: 143B osteosarcoma cell line (parental cells), ND4 non-expressing C4T cells, ND5 non-expressing C9T cells, and rho° cells that do not express any of the ND subunits.
Genotypic background of the cell lines and overall shapes of their mitochondria
Hofhaus and Attardi [26,27] have shown that the mutation in the mitochondrial ND4 gene of the C4T cells and the ND5 gene of the C9T cells (described in the Experimental section) resulted, in both cases, in a translation frameshift and generation of a premature stop codon leading to a severe truncation of 346 and 546 amino acid residues in the ND4 and ND5 proteins, respectively. Normal lengths for ND4 and ND5 proteins are 459 and 603 amino acid residues respectively. Radiolabelling of mtDNA-encoded subunits with [35S]methionine in the presence of emetine revealed that ND4 and ND5 subunits are not detectable in the C4T and C9T mitochondria respectively. In contrast, the six other mitochondria-encoded subunits are all expressed in normal amounts in these cell lines [26,27].
In the present study, the mtDNA D loop as well as the ND4 and ND5 genes were amplified and sequenced for all the cell lines to ascertain their genetic features. In particular, absence of mtDNA in rho° cells was confirmed (results not shown). Surprisingly, PCR amplification of the ND4 and ND5 regions using rho° DNA as the substrate resulted in the generation of fragments. Sizes of these PCR fragments were similar to those amplified from the 143B mtDNA (not shown). As rho° cells are devoid of mtDNA, nuclear ND4 and ND5 pseudogenes may be responsible for the observed amplification. Amplification of nucleus-embedded mtDNA sequences, reported previously by Parfait et al. [38] and Tourmen et al. [39], have confirmed the existence of several mtDNA pseudogenes by in silico analysis of the human genome sequence. Sequencing of the two unexpected fragments revealed a great number of mutations in comparison with the normal mitochondrial ND4 and ND5 genes. The mutations lead to premature stop codons and subsequent significant shortening of the ND4 and ND5 proteins respectively (not shown). As a consequence, these two nuclear pseudogenes will not be translated into functional ND4 or ND5 subunits, which could complement the defective C4T or C9T mitochondria respectively.
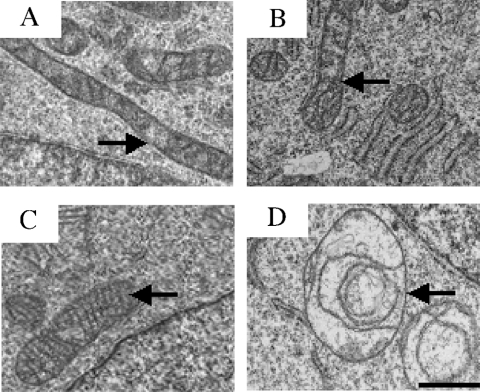
Morphological analysis of the mitochondria in the different cell lines was performed by two different methods: after fluorescent-dye labelling of the organelles and by electron microscopic observation of cell sections. Fluorescence microscopy failed to detect any significant difference in the cellular number of mitochondria and their cytoplasmic localization between the cell lines (results not shown). Electron microscopy revealed two different mitochondrial morphotypes. Mitochondria in 143B, C4T and C9T cells are rod-shaped with a dense matrix and well-structured cristae (Figures 1A, 1B and 1C respectively). In contrast, rho° cells have enlarged spherical mitochondria with rare abnormal cristae and a less dense matrix (Figure 1D) as described previously by Marusich et al. [40] and Procaccio et al. [18]. From these observations, it can be concluded that the mitochondrial nucleic acids and, probably, the mtDNA-encoded proteins are indispensable for a normal mitochondrial structure. However, the absence of either the ND4 or the ND5 subunit and the subsequent loss of complex I activity (see below) has no effect on mitochondrial morphology.
Figure 1. Electron micrographs of the cultured cells.
(A) 143B. (B) C4T. (C) C9T and (D) rho° cells. Mitochondria are indicated by arrows. Scale bar, 0.5 μm. Ultrathin sections of cells were generated and stained as described in the Experimental section.
Detection of enzymically active OXPHOS (oxidative phosphorylation) complexes in cell lines
Complex I activity and other respiratory-chain complex activities were assayed in mitochondria-enriched preparations from all the cell lines by spectrophotometric analysis (Table 1). Activity of the citrate synthase was also measured as a control of mitochondrial integrity.
Table 1. Respiratory-chain complex activities in 143B, C4T, C9T and rho° cells.
Enzymic activities were measured in isolated mitochondria prepared from each cell line as described in the Experimental section. Values are expressed in nmol of substrate (donor or acceptor) consumed·min−1·(mg of protein)−1. Assays were performed on two or three different mitochondrial preparations. Mean values are shown with the range in parentheses, and mean ratios comparing complex activities are also shown. Activities are not normalized with respect to citrate synthase activity.
| Enzyme activities | 143B cells | C4T cells | C9T cells | rho° cells |
|---|---|---|---|---|
| Complex I | 26 | 0.3 | 0.9 | ∼0 |
| (24–28), n=2 | (0–0.5), n=2 | (0–1.4), n=3 | n=2 | |
| Succinate dehydrogenase | 48 | 21 | 35 | 22 |
| (40–63), n=3 | (19–23), n=2 | (21–43), n=3 | (17–27), n=2 | |
| Complex II | 105 | 40 | 99 | 40 |
| (77–128), n=3 | (37–42), n=3 | (85–107), n=3 | (35–43), n=3 | |
| Complex II+III | 71 | 32 | 75 | ∼0 |
| (67–78), n=3 | (27–41), n=3 | (57–93), n=3 | (0–1.6), n=3 | |
| Complex III | 354 | 399 | 426 | ∼0 |
| (251–469), n=3 | (286–513), n=3 | (321–485), n=3 | (0–3.7), n=3 | |
| Complex IV | 251 | 180 | 311 | ∼0 |
| (219–287), n=3 | (157–208), n=3 | (280–336), n=3 | (0–2.6), n=3 | |
| Citrate synthase | 302 | 343 | 376 | 292 |
| (241–354), n=3 | (292–425), n=3 | (346–392), n=3 | (205–365), n=3 | |
| Ratios | ||||
| I/II+III | 0.37 | 0.01 | 0.01 | |
| I/III | 0.07 | 0.001 | 0.002 | |
| I/IV | 0.10 | 0.002 | 0.003 | |
| II/III | 0.30 | 0.10 | 0.23 | |
| II/IV | 0.42 | 0.22 | 0.32 | |
| III/IV | 1.41 | 2.22 | 1.37 | |
| IV/II+III | 3.54 | 5.63 | 4.15 |
The NADH:ubiquinone oxidoreductase activity of complex I was nearly absent from C4T, C9T and rho°. The ratios between the activities of the different complexes are consistent with an almost completely isolated complex I defect in C4T and C9T cells. As expected, in the rho° cells, activity of the two other complexes containing mtDNA-encoded subunits (complexes III and IV) was also undetectable. Therefore the absence of either ND4 or ND5 proteins leads to a severe loss of NADH:ubiquinone oxidoreductase activity, but has no major effect on the other respiratory-chain complex activities. However, in C4T cells, measurement of succinate dehydrogenase, complex II and complex II+III activities revealed that the nuclear-encoded complex II activity was reduced by approx. 50% in comparison with 143B and C9T activities. Abnormal activity ratios (II/III and II/IV) provided further support for this partial defect of complex II activity in C4T cells. In addition, complex IV activity was also partially reduced in C4T cells (64% of the average activities obtained for 143B and C9T cells). This partial reduction of complex II and IV activities in C4T cells compared with C9T and 143B cells was also detected by oxygraphy on permeabilized cells by Hofhaus and Attardi [27]. Owing to the lack of ND4, which prevents complex I activity, other respiratory-chain activities may be affected in C4T cells.
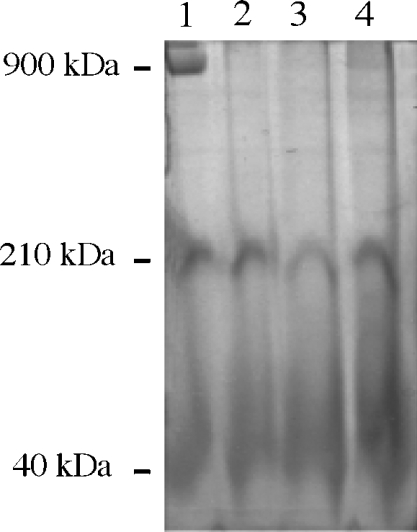
In further experiments, we checked whether any subcomplex I structures could be detected in mitochondrial samples through their ND activity, which may be catalysed by the complex I hydrophilic part. Mitochondrial samples were run in Blue-Native gel, and ND activity was detected as described in the Experimental section. The 143B mitochondria showed a signal close to 900 kDa with an intense NADH–NBT oxidoreductase activity (Figure 2, lane 1). In contrast, this signal was absent from the C4T, C9T and rho° mitochondria samples (Figure 2, lanes 2–4).
Figure 2. Detection of mitochondrial ND activity in mitochondria from 143B, rho°, C4T and C9T cells by BN-PAGE.
The gel was stained for NADH–NBT oxidoreductase activity as described in the Experimental section. Identical amounts of mitochondrial proteins (200 μg) were loaded. Lane 1, 143B cells; lane 2, rho° cells; lane 3, C4T cells; lane 4, C9T cells. The molecular masses of the proteins, indicated on the left, were calculated by comparison with known standards (urease and apoferritin from Sigma).
Therefore the absence of either the ND4 or ND5 subunits leads to an almost complete loss of both NADH:ubiquinone oxidoreductase activity and ND activity. A similar result is also observed in the absence of all the ND subunits. Hence, ND4 and ND5 are essential for complex I activity, but the effects of the absence of these subunits on the complex I assembly remain to be elucidated.
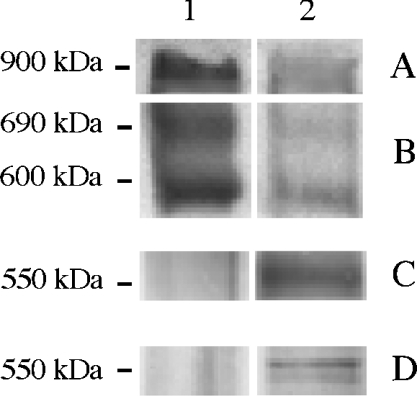
The identity of the 900 kDa ND complex as a fully assembled complex I was confirmed by immunodetection with antibodies against the 24, 39 and 49 kDa complex I subunits. These three subunits were present in the 900 kDa band of 143B mitochondria (Figure 3, lane 1). Similar signals were not immunodetected in C4T, C9T and rho° cells (Figure 3, lane 2 for rho°; results for C4T and C9T are not shown). This led to the conclusion that, in the absence of ND4, ND5 or all ND subunits, assembly of whole active complex I is impaired. As shown in Figure 2, no subcomplex with molecular mass lower than 900 kDa and exhibiting NADH–NBT activity can be detected exclusively in C4T and C9T mitochondrial samples. However, two diffuse signals at approx. 210 and 40 kDa, with similar intensity, were found in all the samples. The identity of these two enzymically active components remains unknown. Immunodetection with the 24, 39 and 49 kDa subunit antibodies failed to detect any subunit in these bands (not shown). Therefore major complex I subunits seem to be absent from these two active components. This observation is reminiscent of the results obtained with Chlamydomonas reinhardtii mitochondria, where two low-molecular-mass complexes of 160–210 and 40 kDa have been detected by their ND activity in the BN-PAGE of C. reinhardtii mitochondrial samples [41].
Figure 3. Immunological detection of complex I subunits after BN-PAGE and blotting on to PVDF membrane.
BN-PAGE experiments were performed as described in Figure 2. Identical amounts of protein from mitochondrial preparations (200 μg) were loaded. Lane 1, 143B; lane 2, rho°. After electrophoresis, proteins were transferred on to PVDF membranes and immunodetected (A, B) with an antiserum against 24 kDa subunit, (C) with an antiserum against 39 kDa subunit and (D) an antibody against the 49 kDa subunit. The molecular mass of the proteins, indicated on the left, was calculated by comparison with known standards (urease and apoferritin were from Sigma). In the 900 kDa region, the immunodetection gave essentially identical signals with those observed in (A) whether the antibodies used were directed against the 39 kDa subunit or the 49 kDa subunit.
Several inactive complexes containing complex I subunits were immunodetected in the Blue-Native gel. For example, the antiserum against the 24 kDa subunit revealed the presence of two subcomplexes of 600 and 690 kDa in all 143B and rho° cell lines, whereas the antibodies against the 39 and 49 kDa subunits detected a subcomplex of 550 kDa in rho° mitochondria (Figure 3). The two subcomplexes of 600 and 690 kDa can be linked with the human subcomplex of 650 kDa detected by Antonicka et al. [8] that contains the 24 kDa subunit, and the subcomplex of 550 kDa can be linked with the human subcomplex of 480 kDa, that contains the 39 and 49 kDa subunits also described by Antonicka et al. [8]. The formation of such subcomplexes was first described in Zea mays, where the loss of ND4 induces the formation of an altered complex missing several subunits [42], and in C. reinhardtii, where the loss of ND4 or ND4/ND5 leads to the formation of a subcomplex of 650 kDa [41].
Mitochondrial content of complex I subunits in the cell lines
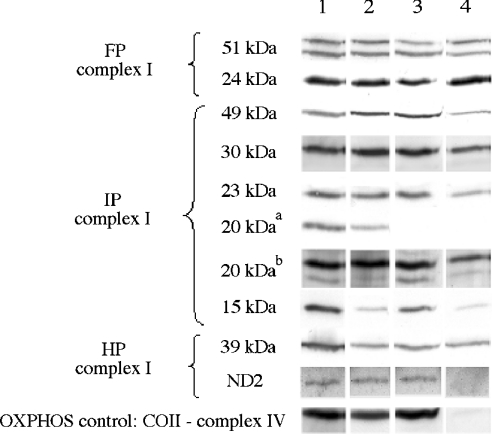
We have shown that without expression of ND4, ND5 or all the ND subunits, complex I activity is lost and assembly of complex I is impaired in the C4T, C9T and rho° cells. Consequently, first it is of interest to establish whether the complex I subunits are still present in these cell lines. We began the present study by conducting an immunological analysis of complex I subunits on both total cellular extracts and purified mitochondria preparations for all model cells (Figure 4). Identical results were obtained for the two sample types. Antibodies against subunits forming the three different complex I fractions have been tested (15, 20, 23, 24, 30, 39, 49, 51 kDa and ND2 subunits), and an antibody against a complex IV subunit (COII) was also used as a control. Mitochondrial samples showed that mitochondria-encoded ND2 of the HP part of the enzyme is detected in 143B, C4T and C9T mitochondria, but is, as expected, absent from rho° mitochondria. Therefore, even in the absence of ND4 or ND5, the ND2 subunit is still present at nearly normal amounts. No significant quantitative variation was observed in HP, IP or FP fractions between the cell lines for the nine complex I nuclear-encoded subunits tested (Figure 4), except for the 20 kDa subunit (as detected with the anti-20 kDa commercial antibody). Therefore in the absence of ND4 or ND5, the cellular qualitative and quantitative content of complex I nuclear subunits was not significantly modified. However, it was also observed that the 20 kDa subunit was detected in all cell lines by the polyclonal antiserum, but was not detected by the commercial antibody in C9T and rho° cells, i.e. in the absence of the ND5 subunit. Consequently, ND5 and, presumably, the membrane arm might play a role in the 20 kDa subunit binding to complex I. However, the differing reactivities of our polyclonal anti-20 kDa serum and the commercial anti-20 kDa antibody remain puzzling. Either these two antibodies bind to two different subunits of similar molecular masses, or some specific post-translational modification of the 20 kDa subunit in the C9T and rho° cells prevents the binding of the commercial antibody to the epitope.
Figure 4. Immunological detection of complex I subunits in mitochondria.
Identical amounts of protein from mitochondrial preparations (8 μg) were loaded. Representative Western-blot signals show the level of each indicated subunit for complexes I and IV in different cell lines. Lane 1, 143B; lane 2, C4T; lane 3, C9T; lane 4, rho°. FP, flavoprotein domain; IP, iron protein domain; HP, hydrophobic part. Assignment of the immunodetected subunits to the three complex I domains was consistent with the results from Hatefi [5]. aDetection with the commercial anti-20 kDa antibody; bDetection with a serum containing antibodies raised against the C-terminal peptide of the 20 kDa subunit. Note that, under our electrophoretic conditions, the 20 and 51 kDa subunits could be detected as doublets by their respective polyclonal antisera. Immunodetection of the same subunits on whole cell samples gave similar results (not shown).
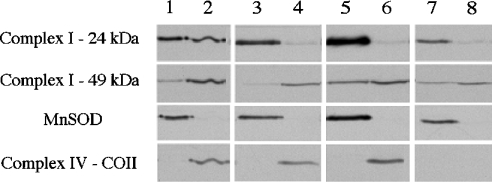
As a second step, immunological analysis was performed with submitochondrial fractions to study nuclear complex I subunit partitioning between the mitochondrial matrix and the membrane. Matrix and membrane fractions of all the cell lines were analysed using antibodies against the 15, 23, 24, 30, 39, 49 and 51 kDa subunits of complex I (Figure 5). Two proteins were also immunodetected as standards to confirm the quality of the purified fractions: MnSOD (manganese superoxide dismutase), as a matrix soluble protein, and the COII subunit of complex IV, as a highly HP protein exclusively embedded in the mitochondrial inner membrane. As shown in Figure 5, MnSOD was detected only in matrix fractions (lanes 1, 3, 5 and 7), whereas the COII subunit was found in the membrane fractions (lanes 2, 4 and 6).
Figure 5. Immunological detection of complex I subunits in mitochondrial subfractions.
Membrane or matrix fractions were obtained after sonication and differential centrifugations of mitochondrial preparations as described in the Experimental section. Identical amounts of protein (10 μg) were loaded. Representative Western-blot signals show the levels of each indicated subunit for complexes I and IV, and of MnSOD in different cell lines. Lanes 1, 3, 5 and 7, matrix fractions; lanes 2, 4, 6 and 8, membrane fractions; lanes 1 and 2, 143B fractions; lanes 3 and 4, C4T fractions; lanes 5 and 6, C9T fractions; lanes 7 and 8, rho° fractions. For simplicity, immunodetection using antibodies against the 15, 23, 30, 39 and 51 kDa subunits of complex I is not shown, but results almost identical with those obtained using the anti-49 kDa antiserum were observed.
The signal intensities were quantified and normalized to the respective total volumes of each fraction. The ratio between the normalized signal values for the membrane and those for the matrix was calculated for each subunit in all cell lines. For 143B, C4T and C9T, this ratio is >1 for all complex I subunits detected except the 24 kDa subunit. A ratio >1 clearly indicates that the subunits are mostly associated with membranes. Therefore the nuclear subunits, and putatively the peripheral arm, are all associated with a membranous domain or membranous subdomains of the enzyme even in the absence of ND4 or ND5 subunits. For rho° cells, this ratio is <1 for most of the subunits tested, revealing that they are preferentially distributed in the matrix. This suggests that the anchorage of the nuclear subunits to the membrane is significantly unfavoured in the total absence of the major HP components of the membrane arm of the enzyme. Interestingly, the 24 kDa subunit was found to exhibit a specific behaviour. For the 143B cell line, the ratio for the 24 kDa subunit is close to 0.5, indicating that this subunit is almost equally abundant in the matrix and in the membranes. Moreover, when only one ND subunit is absent (ND4 or ND5), or when all the seven ND subunits are absent (in rho° cells), the membrane/matrix ratio is close to 0. This suggests that almost all the 24 kDa subunit molecules are located in the matrix and no longer associated with the membranes in the C4T and C9T cell lines.
Detection and analysis of partially assembled complex I forms
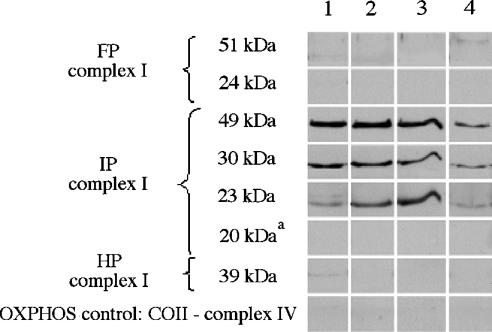
In the absence of ND4, ND5 or all the ND subunits, we found that the loss of complex I activity is not correlated with a total absence of complex I subunits in the mitochondria. In C4T and C9T cells, previous immunoprecipitation experiments on mitochondrial lysates from [35S]methionine-pulse–chased cells, using an antiserum against the 49 kDa subunit, revealed that absence of ND4 prevents the normal assembly of the complex I membrane arm [26], whereas absence of ND5 leads to a lower efficiency of assembly or to instability of the membrane arm [27,43]. In these cell lines, however, putative misassembly of the nuclear subunits in the complex I peripheral arm was not studied. We analysed the role that mitochondrial ND subunits may play in the aggregation of the nuclear-encoded complex I subunits into the inner-mitochondrial membrane. As shown above, BN-PAGE experiments revealed the existence of several subcomplexes, i.e. partial assemblies of incomplete complex I. To ascertain their composition, immunopurification experiments were performed with antisera against the 30 kDa complex I subunit using n-dodecyl-β-D-maltoside-solubilized fractions of the 143B, C4T, C9T and rho° cells (Figure 6). For all cell lines, three complex I subunits, the 23, 30 and 49 kDa subunits, were shown by Western blotting to be in the immunopurified samples. All other immunodetectable complex I subunits tested were absent from the various samples, indicating that only subcomplex I was immunopurified. The detection in all the cell lines of a subcomplex containing at least these three subunits suggests that their association is not impaired by the absence of ND4, ND5 or all ND subunits. This result indicates that, in the absence of the membrane arm, these three nuclear subunits are still capable of interacting and forming subcomplexes that remain soluble in the matrix (at least for the rho° cells). The three subunits 23, 30 and 49 kDa are supposed to constitute the connecting of complex I module between the membrane arm and the peripheral arm, which has been proposed in the bacterium R. capsulatus [14]. As a control, the absence of signal with the antibody against the COII subunit of complex IV confirmed that the immunopurified subcomplex I was free of contaminating complex IV and mitochondrial membrane patches (Figure 6). Trace amounts of other complex I subunits detected by immunoblotting are probably due to co-purification of either whole complex I in 143B cell extracts or other subcomplexes containing the 30 kDa subunit.
Figure 6. Complex I subunit composition of immunopurified subcomplex I.
Immunopurification was performed with antiserum against the 30 kDa subunit of complex I. Representative Western-blot signals show the levels of each indicated subunit for complexes I and IV in different cell lines. Lane 1, 143B; lane 2, C4T; lane 3, C9T; lane 4, rho°. FP, flavoprotein domain; IP, iron protein domain; HP, hydrophobic part. aDetection with the commercial anti-20 kDa antibody.
Subunit composition of subcomplex I assessed by MS and identification of the associated prohibitin
Owing to the limited number of available antisera directed against the individual complex I subunits, the information generated in the above immunodetection approach is restricted exclusively to the small number of immunodetectable complex I subunits. Hence, we decided to also investigate subcomplex composition by an MS-based approach. Two different types of immunopurification were performed, using antibodies directed against either the 30 or the 49 kDa subunit in 143B cells, C4T or C9T cells. Since very low amounts of total protein were collected, this made the exhaustive detection of all the polypeptides comprising the complex I subfractions unfavourable. Therefore samples were concentrated on SDS/polyacrylamide gel (as described in the Experimental section), trypsin-digested, and the resulting products were analysed by nano-LC-MS/MS (see the Experimental section). This approach allowed us to collect numerous data, both from the analysis of the elution profile (MS Survey) and from the peptide fragmentation mass spectra (see Figure 7A for an example). The main conclusions of this approach are described below.
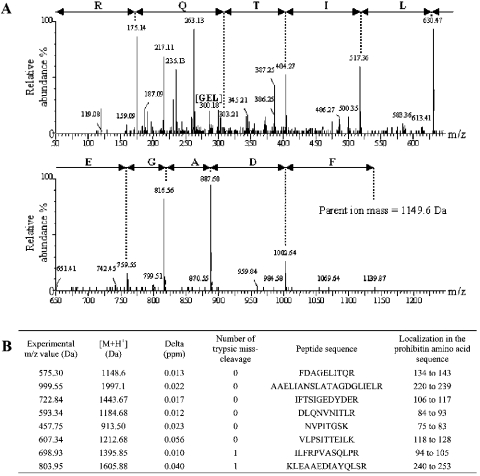
Figure 7. Prohibitin identification by nano-LC-MS/MS in the immunopurified subcomplex I with antibodies directed against either the 30 kDa subunit or the 49 kDa subunit of complex I in 143B, C4T or C9T cells.
(A) MS/MS spectrum of the prohibitin tryptic decapeptide FDAGELITQR. The peak of an internal tripeptide fragment of the identified peptide is indicated by [GEL]. (B) Description of the tryptic peptides of prohibitin protein identified by MS. Prohibitin protein in SwissProt+TrEMBL databases has the accession number P35232. Experimental m/z value (Da)=mass over the charge state of the observed precursor peptide submitted to fragmentation. [M+H+]=peptide mass (Da). δ (p.p.m.) is the error window on experimental peptide mass values, expressed in p.p.m.
Several peptides were found unambiguously attributable to the 49-, 30- and 23-kDa subunit sequences (four, three and one peptides respectively). Therefore MS analysis of the subcomplex fractions, independently immunopurified with the 30- or 49-kDa subunit antibody, confirmed the presence of the 49, 30 and 23 kDa subunits in a complex I subfraction. Interestingly, in both types of immunopurified sample, we were also able to detect one unexpected protein, prohibitin, which is not a complex I subunit. The presence of prohibitin was clearly evident from mass spectra signatures corresponding to tryptic peptides of this protein and up to eight peptides were conclusively identified (Figure 7B). Unexpectedly, it can be concluded that prohibitin interacts with complex I subunits or subcomplex I domains.
Prohibitin (272 amino acids with molecular mass of 29.8 kDa) is a mitochondrial internal membrane protein and is known to be a potential regulator of growth arrest and a tumour suppressor in human fibroblasts [44]. It is also implicated in increased replicative lifespan in Saccharomyces cerevisiae [45]. In yeast, it was shown that prohibitin is capable of interacting with two complex IV subunits (Cox2p and Cox3p) [46] to prevent their proteolysis by m-AAA proteases [47]. It is supposed that prohibitin may stabilize newly synthesized mitochondrial translation products and, hence, could act as a chaperone in the subunit assembly of mitochondrial respiratory-chain complexes [48,49]. Moreover, Taylor et al. [50] have shown that prohibitin is present in human mitochondria.
Our study demonstrates for the first time that prohibitin interacts with complex I subunits. Prohibitin might protect complex I subunits before assembly and so may be a complex I assembly chaperone. Consequently, any prohibitin alteration could result in complex I misassembly with possible pathological consequences.
Finally, we wondered whether this protein could play a role in some complex I pathologies. We amplified and sequenced the prohibitin cDNA from the fibroblasts of two patients with complex I defects of unknown genetic origin, which are related to two specific misassembly phenotypes: reduced amounts of 24 and 51 kDa subunits or reduced amounts of all investigated complex I subunits [18]. No mutation was found in the prohibitin cDNA. Therefore this protein is not responsible for the complex I defects observed in these two patients.
The present study clearly shows that our experimental approach coupling immunopurification of mitochondrial fractions and analysis by nano-LC-MS/MS could be very valuable in the study of the human mitochondrial complex I. Additional investigations are now required to identify all the ancillary catalytic components involved in complex I assembly. More specifically, it also remains to elucidate the role each of these proteins plays in the following stages: subunit folding, cofactor binding, multimeric interactions, proteolytic protection of the subunits during assembly or, in contrast, proteolytic enhancement to participate in complex I biosynthesis turnover.
Acknowledgments
We are extremely grateful to G. Attardi and A. Chomyn for providing the cell lines derived from 143B TK− cells. We thank Y. Saoudi (CS-DRDC, CEA Grenoble, France) for fluorescence microscopy, S. Peyrol and S. Balvay (CeCIL, Claude Bernard University, Lyon, France) for electron microscopy experiments and S. Padet (Laboratoire de Biochimie, Hôpital Debrousse) for expert technical assistance. Financial support was provided by the Association Française contre la Myopathie and a grant from the Ministère de la Recherche (I.B.). We thank S. Horan for a careful reading of the paper.
References
- 1.Yagi T., Matsuno-Yagi A. The proton-translocating NADH-quinone oxidoreductase in the respiratory chain: the secret unlocked. Biochemistry. 2003;42:2266–2274. doi: 10.1021/bi027158b. [DOI] [PubMed] [Google Scholar]
- 2.Hirst J., Carroll J., Fearnley I. M., Shannon R. J., Walker J. E. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim. Biophys. Acta. 2003;1604:135–150. doi: 10.1016/s0005-2728(03)00059-8. [DOI] [PubMed] [Google Scholar]
- 3.Grigorieff N. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 A in ice. J. Mol. Biol. 1998;277:1033–1046. doi: 10.1006/jmbi.1998.1668. [DOI] [PubMed] [Google Scholar]
- 4.Bottcher B., Scheide D., Hesterberg M., Nagel-Steger L., Friedrich T. A novel, enzymatically active conformation of the Escherichia coli NADH:ubiquinone oxidoreductase (complex I) J. Biol. Chem. 2002;277:17970–17977. doi: 10.1074/jbc.M112357200. [DOI] [PubMed] [Google Scholar]
- 5.Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu. Rev. Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- 6.Finel M., Majander A. S., Tyynela J., De Jong A. M., Albracht S. P., Wikstrom M. Isolation and characterisation of subcomplexes of the mitochondrial NADH:ubiquinone oxidoreductase (complex I) Eur. J. Biochem. 1994;226:237–242. doi: 10.1111/j.1432-1033.1994.tb20046.x. [DOI] [PubMed] [Google Scholar]
- 7.Videira A., Duarte M. On complex I and other NADH:ubiquinone reductases of Neurospora crassa mitochondria. J. Bioenerg. Biomemb. 2001;33:197–203. doi: 10.1023/a:1010778802236. [DOI] [PubMed] [Google Scholar]
- 8.Antonicka H., Ogilvie I., Taivassalo T., Anitori R. P., Haller R. G., Vissing J., Kennaway N. G., Shoubridge E. A. Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. J. Biol. Chem. 2003;278:43081–43088. doi: 10.1074/jbc.M304998200. [DOI] [PubMed] [Google Scholar]
- 9.Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F., et al. Sequence and organization of the human mitochondrial genome. Nature (London) 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 10.Mathiesen C., Hagerhall C. Transmembrane topology of the NuoL, M and N subunits of NADH:quinone oxidoreductase and their homologues among membrane-bound hydrogenases and bona fide antiporters. Biochim. Biophys. Acta. 2002;1556:121–132. doi: 10.1016/s0005-2728(02)00343-2. [DOI] [PubMed] [Google Scholar]
- 11.Kao M. C., Di Bernardo S., Matsuno-Yagi A., Yagi T. Characterization and topology of the membrane domain Nqo10 subunit of the proton-translocating NADH-quinone oxidoreductase of Paracoccus denitrificans. Biochemistry. 2003;42:4534–4543. doi: 10.1021/bi034166z. [DOI] [PubMed] [Google Scholar]
- 12.Friedrich T. Complex I: a chimaera of a redox and conformation-driven proton pump? J. Bioenerg. Biomembr. 2001;33:169–177. doi: 10.1023/a:1010722717257. [DOI] [PubMed] [Google Scholar]
- 13.Yagi T., Hatefi Y. Identification of the dicyclohexylcarbodiimide-binding subunit of NADH-ubiquinone oxidoreductase (complex I) J. Biol. Chem. 1988;263:16150–16155. [PubMed] [Google Scholar]
- 14.Dupuis A., Prieur I., Lunardi J. Toward a characterization of the connecting module of complex I. J. Bioenerg. Biomembr. 2001;33:159–168. doi: 10.1023/a:1010770600418. [DOI] [PubMed] [Google Scholar]
- 15.Zhu Z., Yao J., Johns T., Fu K., De Bie I., Macmillan C., Cuthbert A. P., Newbold R. F., Wang J., Chevrette M., et al. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 1998;20:337–343. doi: 10.1038/3804. [DOI] [PubMed] [Google Scholar]
- 16.Kuffner R., Rohr A., Schmiede A., Krull C., Schulte U. Involvement of two novel chaperones in the assembly of mitochondrial NADH:ubiquinone oxidoreductase (complex I) J. Mol. Biol. 1998;283:409–417. doi: 10.1006/jmbi.1998.2114. [DOI] [PubMed] [Google Scholar]
- 17.Janssen R., Smeitink J., Smeets R., van den Heuvel L. CIA30 complex I assembly factor: a candidate for human complex I deficiency? Hum. Genet. 2002;110:264–270. doi: 10.1007/s00439-001-0673-3. [DOI] [PubMed] [Google Scholar]
- 18.Procaccio V., Mousson B., Beugnot R., Duborjal H., Feillet F., Putet G., Pignot-Paintrand I., Lombes A., De Coo R., Smeets H., et al. Nuclear DNA origin of mitochondrial complex I deficiency in fatal infantile lactic acidosis evidenced by transnuclear complementation of cultured fibroblasts. J. Clin. Invest. 1999;104:83–92. doi: 10.1172/JCI6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Triepels R. H., van den Heuvel L. P., Trijbels J. M., Smeitink J. A. Respiratory chain complex I deficiency. Am. J. Med. Genet. 2001;106:37–45. doi: 10.1002/ajmg.1397. [DOI] [PubMed] [Google Scholar]
- 20.Man P. Y. W., Turnbull D. M., Chinnery P. F. Leber hereditary optic neuropathy. J. Med. Genet. 2002;39:162–169. doi: 10.1136/jmg.39.3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jun A. S., Brown M. D., Wallace D. C. A mitochondrial DNA mutation at nucleotide pair 14459 of the NADH dehydrogenase subunit 6 gene associated with maternally inherited Leber hereditary optic neuropathy and dystonia. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6206–6210. doi: 10.1073/pnas.91.13.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campos Y., Martin M. A., Rubio J. C., Gutierrez del Olmo M. C., Cabello A., Arenas J. Bilateral striatal necrosis and MELAS associated with a new T3308C mutation in the mitochondrial ND1 gene. Biochem. Biophys. Res. Commun. 1997;238:323–325. doi: 10.1006/bbrc.1997.7166. [DOI] [PubMed] [Google Scholar]
- 23.Pulkes T., Eunson L., Patterson V., Siddiqui A., Wood N. W., Nelson I. P., Morgan-Hughes J. A., Hanna M. G. The mitochondrial DNA G13513A transition in ND5 is associated with a LHON/MELAS overlap syndrome and may be a frequent cause of MELAS. Ann. Neurol. 1999;46:916–919. doi: 10.1002/1531-8249(199912)46:6<916::aid-ana16>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 24.Taylor R. W., Morris A. A., Hutchinson M., Turnbull D. M. Leigh disease associated with a novel mitochondrial DNA ND5 mutation. Eur. J. Hum. Genet. 2002;10:141–144. doi: 10.1038/sj.ejhg.5200773. [DOI] [PubMed] [Google Scholar]
- 25.King M. P., Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 26.Hofhaus G., Attardi G. Lack of assembly of mitochondrial DNA-encoded subunits of respiratory NADH dehydrogenase and loss of enzyme activity in a human cell mutant lacking the mitochondrial ND4 gene product. EMBO J. 1993;12:3043–3048. doi: 10.1002/j.1460-2075.1993.tb05973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofhaus G., Attardi G. Efficient selection and characterization of mutants of a human cell line which are defective in mitochondrial DNA-encoded subunits of respiratory NADH dehydrogenase. Mol. Cell. Biol. 1995;15:964–974. doi: 10.1128/mcb.15.2.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duborjal H., Beugnot R., de Camaret B. M., Issartel J. P. Large functional range of steady-state levels of nuclear and mitochondrial transcripts coding for the subunits of the human mitochondrial OXPHOS system. Genome Res. 2002;12:1901–1909. doi: 10.1101/gr.194102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fessenden J. M., Racker E. Partial resolution of the enzymes catalyzing oxidative phosphorylation. XI. Stimulation of oxidative phosphorylation by coupling factors and oligomycin; inhibition by an antibody against coupling factor 1. J. Biol. Chem. 1966;241:2483–2489. [PubMed] [Google Scholar]
- 30.Trounce I. A., Kim Y. L., Jun A. S., Wallace D. C. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- 31.Duby F., Cardol P., Matagne R. F., Remacle C. Structure of the telomeric ends of mt DNA, transcriptional analysis and complex I assembly in the dum24 mitochondrial mutant of Chlamydomonas reinhardtii. Mol. Genet. Genomics. 2001;266:109–114. doi: 10.1007/s004380100529. [DOI] [PubMed] [Google Scholar]
- 32.de Sury R., Martinez P., Procaccio V., Lunardi J., Issartel J. P. Genomic structure of the human NDUFS8 gene coding for the iron-sulfur TYKY subunit of the mitochondrial NADH:ubiquinone oxidoreductase. Gene. 1998;215:1–10. doi: 10.1016/s0378-1119(98)00275-3. [DOI] [PubMed] [Google Scholar]
- 33.Procaccio V., de Sury R., Martinez P., Depetris D., Rabilloud T., Soularue P., Lunardi J., Issartel J. P. Mapping to 1q23 of the human gene (NDUFS2) encoding the 49-kDa subunit of the mitochondrial respiratory Complex I and immunodetection of the mature protein in mitochondria. Mamm. Genome. 1998;9:482–484. doi: 10.1007/s003359900803. [DOI] [PubMed] [Google Scholar]
- 34.Triepels R. H., Hanson B. J., van den Heuvel L. P., Sundell L., Marusich M. F., Smeitink J. A., Capaldi R. A. Human complex I defects can be resolved by monoclonal antibody analysis into distinct subunit assembly patterns. J. Biol. Chem. 2001;276:8892–8897. doi: 10.1074/jbc.M009903200. [DOI] [PubMed] [Google Scholar]
- 35.Duborjal H., Dupuis A., Chapel A., Kieffer S., Lunardi J., Issartel J. P. Immuno-purification of a dimeric subcomplex of the respiratory NADH-CoQ reductase of Rhodobacter capsulatus equivalent to the FP fraction of the mitochondrial complex I. FEBS Lett. 1997;405:345–350. doi: 10.1016/s0014-5793(97)00212-3. [DOI] [PubMed] [Google Scholar]
- 36.Murray J., Zhang B., Taylor S. W., Oglesbee D., Fahy E., Marusich M. F., Ghosh S. S., Capaldi R. A. Oxidative damage to mitochondrial complex I due to peroxynitrite: identification of reactive tyrosines by mass spectrometry. J. Biol. Chem. 2003;278:13619–13622. doi: 10.1074/jbc.M305694200. [DOI] [PubMed] [Google Scholar]
- 37.Ferro M., Salvi D., Brugiere S., Miras S., Kowalski S., Louwagie M., Garin J., Joyard J., Rolland N. Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteomics. 2003;2:325–345. doi: 10.1074/mcp.M300030-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Parfait B., Rustin P., Munnich A., Rötig A. Co-amplification of nuclear pseudogenes and assessment of heteroplasmy of mitochondrial DNA mutations. Biochem. Biophys. Res. Commun. 1998;247:57–59. doi: 10.1006/bbrc.1998.8666. [DOI] [PubMed] [Google Scholar]
- 39.Tourmen Y., Baris O., Dessen P., Jacques C., Malthiery Y., Reynier P. Structure and chromosomal distribution of human mitochondrial pseudogenes. Genomics. 2002;80:71–77. doi: 10.1006/geno.2002.6798. [DOI] [PubMed] [Google Scholar]
- 40.Marusich M. F., Robinson B. H., Taanman J. W., Kim S. J., Schillace R., Smith J. L., Capaldi R. A. Expression of mtDNA and nDNA encoded respiratory chain proteins in chemically and genetically-derived Rho0 human fibroblasts: a comparison of subunit proteins in normal fibroblasts treated with ethidium bromide and fibroblasts from a patient with mtDNA depletion syndrome. Biochim. Biophys. Acta. 1997;1362:145–159. doi: 10.1016/s0925-4439(97)00061-6. [DOI] [PubMed] [Google Scholar]
- 41.Cardol P., Matagne R. F., Remacle C. Impact of mutations affecting ND mitochondria-encoded subunits on the activity and assembly of complex I in Chlamydomonas. Implication for the structural organization of the enzyme. J. Mol. Biol. 2002;319:1211–1221. doi: 10.1016/S0022-2836(02)00407-2. [DOI] [PubMed] [Google Scholar]
- 42.Karpova O. V., Newton K. J. A partially assembled complex I in NAD4-deficient mitochondria of maize. Plant J. 1999;17:511–521. [Google Scholar]
- 43.Chomyn A. Mitochondrial genetic control of assembly and function of complex I in mammalian cells. J. Bioenerg. Biomembr. 2001;33:251–257. doi: 10.1023/a:1010791204961. [DOI] [PubMed] [Google Scholar]
- 44.Nuell M. J., Stewart D. A., Walker L., Friedman V., Wood C. M., Owens G. A., Smith J. R., Schneider E. L., Dell' Orco R., Lumpkin C. K., et al. Prohibitin, an evolutionarily conserved intracellular protein that blocks DNA synthesis in normal fibroblasts and HeLa cells. Mol. Cell. Biol. 1991;11:1372–1381. doi: 10.1128/mcb.11.3.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coates P. J., Jamieson D. J., Smart K., Prescott A. R., Hall P. A. The prohibitin family of mitochondrial proteins regulate replicative lifespan. Curr. Biol. 1997;7:607–610. doi: 10.1016/s0960-9822(06)00261-2. [DOI] [PubMed] [Google Scholar]
- 46.Nijtmans L. G., de Jong L., Artal Sanz M., Coates P. J., Berden J. A., Back J. W., Muijsers A. O., van der Spek H., Grivell L. A. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 2000;19:2444–2451. doi: 10.1093/emboj/19.11.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steglich G., Neupert W., Langer T. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol. Cell. Biol. 1999;19:3435–3442. doi: 10.1128/mcb.19.5.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langer T., Kaser M., Klanner C., Leonhard K. AAA proteases of mitochondria: quality control of membrane proteins and regulatory functions during mitochondrial biogenesis. Biochem. Soc. Trans. 2001;29:431–436. doi: 10.1042/bst0290431. [DOI] [PubMed] [Google Scholar]
- 49.Nijtmans L. G., Artal S. M., Grivell L. A., Coates P. J. The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell. Mol. Life. Sci. 2002;59:143–155. doi: 10.1007/s00018-002-8411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor S. W., Warnock D. E., Glenn G. M., Zhang B., Fahy E., Gaucher S. P., Capaldi R. A., Gibson B. W., Ghosh S. S. An alternative strategy to determine the mitochondrial proteome using sucrose gradient fractionation and 1D PAGE on highly purified human heart mitochondria. J. Proteome Res. 2002;1:451–458. doi: 10.1021/pr025533g. [DOI] [PubMed] [Google Scholar]