Abstract
Before delivery to endosomes, portions of proCD (procathepsin D) and proSAP (prosaposin) are assembled into complexes. We demonstrate that such complexes are also present in secretions of cultured cells. To study the formation and properties of the complexes, we purified proCD and proSAP from culture media of Spodoptera frugiperda cells that were infected with baculoviruses bearing the respective cDNAs. The biological activity of proCD was demonstrated by its pH-dependent autoactivation to pseudocathepsin D and that of proSAP was demonstrated by feeding to saposin-deficient cultured cells that corrected the storage of radioactive glycolipids. In gel filtration, proSAP behaved as an oligomer and proCD as a monomer. ProSAP altered the elution of proCD such that the latter was shifted into proSAP-containing fractions. ProSAP did not change the elution of mature cathepsin D. Using surface plasmon resonance and an immobilized biotinylated proCD, binding of proSAP was demonstrated under neutral and weakly acidic conditions. At pH 6.8, specific binding appeared to involve more than one binding site on a proSAP oligomer. The dissociation of the first site was characterized by a KD1 of 5.8±2.9×10−8 M−1 (calculated for the monomer). ProSAP stimulated the autoactivation of proCD and also the activity of pseudocathepsin D. Concomitant with the activation, proSAP behaved as a substrate yielding tri- and disaposins and smaller fragments. Our results demonstrate that proSAP forms oligomers that are capable of binding proCD spontaneously and independent of the mammalian type N-glycosylation but not capable of binding mature cathepsin D. In addition to binding proSAP, proCD behaves as an autoactivable and processing enzyme and its binding partner as an activator and substrate.
Keywords: complex, procathepsin D, prosaposin, pseudocathepsin D
Abbreviations: AMCA, 7-amino-4-methylcoumarin; proCD, procathepsin D; proSAP, prosaposin; pseudoCD, pseudocathepsin D
INTRODUCTION
proSAP (prosaposin) is a 60–73 kDa precursor of four proteinaceous cofactors, saposins A–D, which are indispensable for the hydrolysis of membrane-integrated sphingolipids by lysosomal enzymes [1,2]. The term saposins refers to sphingolipid activator proteins (SAPs). These activators support the degradation of ceramide (saposin D), glucocerebroside (saposin C), sulphatide (saposin B) and galactosylceramide (saposin A) and perhaps also other complex lipids [2]. Saposins participate in lipid presentation; e.g., saposin C is required for lipid loading on to CD1b molecules in T cells [3]. In addition to the saposins that consist of approx. 80 amino acids each, proSAP is composed of N- and C-terminal peptides and three intersaposin peptides. The four saposin domains of proSAP seem to have evolved from a common ancestral molecule through internal duplications and successive diversification [4]. The three-dimensional structure of saposin B has been elucidated recently [5], and the result confirmed earlier assumptions on similarity of its structure to that of NK-lysin as well as to that of a ‘swaposin’ domain [6] that is an integral part of plant vacuolar aspartic proteinase precursors [7]. We have reported that, in cultured human cells, cross-linking of protein with dithio-bis-succinimidyl propionate entraps a 60 kDa protein in complexes with the precursor form of cathepsin D, proCD (procathepsin D) [8]. Later, in human hepatoma cells [9], mouse fibroblasts [10] and human breast cancer cells [11], the protein that was cross-linked to proCD using the same reagent was identified as proSAP. The development of a post-translational mechanism for joining proSAP with an aspartic proteinase (proCD) in mammalian cells and the integration of a homologous swaposin moiety in the gene of a plant aspartic proteinase, phytepsin [7], suggest that these syndicates may play as yet unknown but important roles. Before the identification of proSAP as the complexing partner of proCD, it was observed that, in hepatoma HepG2 cells, portions of both proteins are associated with membranes in transit compartments such as Golgi and that, to a large extent, this binding is independent of mannose 6-phosphate receptors [12]. It is conceivable that the formation of the complex plays a role in mannose 6-phosphate-independent targeting of the two proteins to lysosomes. The partially mannose 6-phosphate-independent binding of proSAP to membranes [12–14] may explain the finding of significant amounts of saposin C in lysosomes in I-cells, in which phosphorylation of lysosomal enzymes is lacking [15]. The transient binding of proCD to intracellular membranes may be partly explained by an association of proCD with a membrane-bound proSAP. To understand better the formation and properties of the complex of the two lysosomal and facultatively secretory protein precursors, we prepared recombinant forms of these proteins and examined their association and reactions at different pH values in vitro.
MATERIALS AND METHODS
Materials
Human GM2 activator protein was described previously [16]. Methyl α-D-mannopyranoside and pepstatin A–agarose were obtained from Sigma–Aldrich (Taufkirchen, Germany). Peptide N-glycosidase F was obtained from Roche Diagnostics (Mannheim, Germany). Cathepsin D was purified from a homogenate of human trophoblastic tissue using a procedure similar to that described for the purification of proCD below. Normal-term placentas were obtained from local hospitals. Antisera against human saposins are described in [17,18]. Rabbit antisera against synthetic peptides were obtained from Genosys Biotechnologies (Cambridge, U.K.). For Western blots, secondary horseradish peroxidase-coupled antibodies (rabbit anti-goat immunoglobulins from Dako Diagnostika, Hamburg, Germany, and goat anti-rabbit heavy+light chains from Bio-Rad, München, Germany) were used. All standard chemicals were of analytical grade purity.
Metabolic labelling, immunoprecipitation and detection of cross-linkable complexes in conditioned media
Human U937 promonocytes were cultured in RPMI 1640 medium containing 10% (v/v) heat-inactivated fetal calf serum (both from Gibco BRL, Eggenstein, Germany), 100 units/ml penicillin and 100 μg/ml streptomycin under air/CO2 (19:1). The cells were cultured for 3 days in the presence of 2×10−7 M calcitriol (Sigma–Aldrich) to enhance the rate of synthesis of proCD [19]. Metabolic labelling was performed with Tran35S-label (11–18 MBq/ml) in methionine/cysteine-deficient RPMI 1640 medium. The labelling media contained 4% heat-inactivated fetal calf serum that was dialysed against 0.9% NaCl. The labelling was performed in the presence of 10 mM NH4Cl and 0.1 μM PMA to enhance the secretion of proCD [19]. After labelling for 15 h, the medium was harvested and incubated with 1 mM dithio-bis-succinimidyl propionate for 10 min at 37 °C. The precipitation of proCD was performed as described previously [20]. ProSAP was precipitated analogously using rabbit anti-human saposin C antiserum [15]. The labelled, precipitated proteins were solubilized in the presence of 20 mM dithiothreitol (cleaving the cross-linking reagent), separated by SDS/PAGE as above and visualized by fluorography [21].
Biosynthesis of recombinant proSAP and proCD
Insect cells, Spodoptera frugiperda Sf21, were cultured in roller bottles at 28 °C in Grace's medium (Invitrogen, Karlsruhe, Germany; composition: as defined in the supplier's catalogue) that was supplemented with 10% (v/v) heat-inactivated fetal bovine serum. The serum supplement has been shown to prevent proteinolysis of proCD in the medium [22]. The cells were infected with baculovirus harbouring human proSAP cDNA [23] or human proCD cDNA [22]. The proSAP cDNA encoded the isoform without exon 8 [18] with a His6 tag upstream of the stop codon and an affinity insert including the enterokinase recognition sequence (DPSHGDYKDDDK) downstream of the signal sequence. Four days post-infection, the medium was separated from detached cells by a low-speed centrifugation and stored frozen at −20 °C.
Purification of proSAP and protein characterization by gel electrophoresis
The medium containing proSAP (500 ml) was thawed, clarified by centrifugation at 12000 g for 30 min, dialysed against 5 mM Tris/HCl buffer (pH 7.0) at 4 °C and the centrifugation was repeated. The supernatant was applied to a 20 cm×2 cm Q-Sepharose (Sigma, Deisenhofen, Germany) column, which was operated manually. It was washed with 1 litre of 0.2 M NaCl and 10 mM Tris/HCl (pH 7.0) and proSAP was eluted with a linear gradient up to 1 M NaCl in the same buffer. At this step and at subsequent steps, proSAP was determined semi-quantitatively using a dot-blot on a nitrocellulose membrane (0.45 μm pore size; from Sartorius AG, Göttingen, Germany) with a goat anti-human saposin C antiserum and a horseradish peroxidase-conjugated donkey anti-goat IgG (H+L) antibody (Dianova, Hamburg, Germany), both diluted 1:4000. The peroxidase activity was detected with an ECL® Western-blotting detection reagent kit (Amersham Biosciences, Freiburg i. Br., Germany) as a substrate according to the manufacturer's instructions.
The pooled peak fractions were supplemented with 2% (v/v) 0.5 M sodium phosphate (pH 7.0) and 0.5% (v/v) 0.2 M PMSF (Sigma) in DMSO and applied to a 3 cm×1.5 cm concanavalin A–Sepharose column (Amersham Biosciences). The column was washed with 8 vol. of 0.15 M NaCl, 1 mM PMSF, 0.05% NaN3 and 10 mM sodium phosphate (pH 7.0). The elution of proSAP was achieved at 37 °C with 50 ml of 0.75 M methyl-α-D-mannopyranoside in 0.5 M NaCl, 1 mM PMSF and 10 mM sodium phosphate (pH 7.0). The eluate was diluted with water (1:1) and applied on to a 5 cm×1 cm FPLC hydroxyapatite column (Merck, Darmstadt, Germany). The elution was performed with a linear 10–300 mM gradient of sodium phosphate (pH 7.0). The peak fractions were pooled, concentrated in a UH 25/100 ultra-thimble (Schleicher and Schuell, Dassel, Germany) and taken up in 150 mM NaCl and 10 mM sodium phosphate (pH 6.8). The final step was a gel filtration using a 30 cm×1 cm Superdex 200 FPLC column (Amersham Biosciences). In general, FPLC was performed using the ÄKTAexplorer apparatus and Unicorn 3.1 software, both from Amersham Biosciences. In aliquots of fractions, the protein was characterized by electrophoresis (SDS/PAGE as described by Laemmli [24]) and silver nitrate staining [25].
Sphingolipid loading of cells and TLC analysis of lipid metabolites
Control and proSAP-deficient human fibroblasts were obtained from Dr K. Harzer (Neurometabolisches Labor, Universitats-kinderklinik Tübinger, Germany). The cells were grown in minimal essential medium with 10% fetal bovine serum. Where indicated, the cells were preincubated with proSAP (25 μg/ml) for 24 h, washed with minimal essential medium (Gibco BRL), incubated for 60 min at 37 °C in the fresh medium and, thereafter, loaded with L-[3-14C]serine (1 μCi/ml; 18.7 nmol/ml) in minimal essential medium containing 0.3% fetal bovine serum for 24 h, as described previously [18]. The medium was replaced by a chase medium, containing unlabelled L-serine (185 μM), 0.6% fetal bovine serum and proSAP (25 μg/ml). After 120 h, the cells were washed three times with 0.15 M NaCl, buffered with 10 mM sodium phosphate (pH 7.4), detached with 0.25% (w/v) trypsin in this buffer at 37 °C for 15 min, centrifuged, washed once with 1 ml of buffer and collected by centrifugation. The cells were homogenized in 0.4 ml of water, sonicated three times for 15 s and then mixed with 4 ml of methanol and sonicated again for 15 min.
Total lipids were extracted with 3 ml of chloroform/methanol/water/pyridine (60:30:6:1, by vol.) for 24 h at 50 °C. After drying, phospholipids were degraded by mild alkaline hydrolysis with 100 mM methanolic NaOH for 2 h at 37 °C. The solutions were neutralized with acetic acid, and the lipid extracts were desalted by reversed-phase chromatography on LiChroprep RP18 and separated into acidic and neutral sphingolipid fractions by using DEAE-Sepharose CL6B (Amersham Biosciences) columns [26]; the sphingolipids were applied in chloroform/methanol/water (3:7:1, by vol.) and the absorbed acidic fraction was eluted with the solvent mixture in which water was replaced by 0.8 M ammonium acetate, and desalted as above. The unbound and bound fractions were applied to TLC silica gel 60 plates (Merck), developed with chloroform/methanol/0.22% (w/v) aqueous CaCl2 (60:35:8, by vol.). The lipids were visualized by autoradiography (Bio-Imaging analyzer Fuji Bas 1000; Fuji, Tokyo, Japan) and identified by their RF values. Quantification was performed with the picture analysis system TINA 2.07 (Raytest, Straubenhardt, Germany).
Purification and autoactivation of proCD, determination of proteinolytic activity and determination of the fragmentation of proSAP
Aliquots of the medium containing proCD were stored frozen. After thawing, the medium was clarified by centrifugation at 12000 g for 10 min and subjected to affinity chromatography using a 1.5 cm×2.5 cm (diameter) pepstatin A–4% beaded agarose (Sigma) column at 4 °C. On the top of the column, the medium was continuously mixed (1:1) with a binding buffer containing 0.7 M NaCl, 0.2% Triton X-100 and 0.2 M sodium formiate (pH 3.5). The column was washed with 1 vol. of the binding buffer and 2 vol. each of (i) 0.4 M NaCl, 0.1% Triton X-100 and 10 mM sodium formiate (pH 3.5), (ii) the same buffer without the detergent and (iii) 0.4 M NaCl and 10 mM sodium acetate (pH 5.0). The elution was performed with 0.4 M NaCl and 20 mM Tris/HCl (pH 8.3). The eluted proCD was concentrated and the buffer was exchanged for 2 mM Tris/HCl (pH 8.3; buffer A) by ultrafiltration in a UH 25/100 ultra-thimble. After application to a 7 mm×35 mm Bio-Rad UNO-Q1 anion-exchange FPLC column, proCD was eluted with a linear gradient (40 ml) using buffer A and 150 mM NaCl/25 mM Tris/HCl (pH 8.0) as the initial and final buffers respectively at a flow rate of 0.5 ml/min. The fractions containing proCD were pooled, the protein was concentrated as above and the buffer was exchanged for 2 mM Na-Mops (pH 6.6; buffer A1) by ultrafiltration as described above. Next, FPLC was performed using a 7 mm×35 mm Bio-Rad UNO-S1R cation-exchange column. A linear gradient of buffers A1 and 150 mM NaCl and 10 mM Na-Mops (pH 6.6) with a total volume of 27 ml and a flow rate of 0.5 ml/min was used. The proenzyme elution reached a peak at approx. 50 mM NaCl. Subsequent experiments were performed with the peak fraction.
Autoactivation of proCD was examined by incubating aliquots of the proenzyme in 5 μl of 50 mM sodium formiate and 50 mM sodium acetate buffers (pH 3.65) at 37 °C for up to 1 h. The same conditions were used to prepare pseudoCD (pseudocathepsin D). The enzymic activity was detected with AMCA (7-amino-4-methylcoumarin)-haemoglobin as a substrate [27] using a SPECTRAfluor reader (Tecan, Crailsheim, Germany) at 365 and 460 nm excitation and emission wavelengths respectively.
Fragmentation of proSAP was performed by incubating proCD with proSAP in 140 mM NaCl and 10 mM sodium phosphate buffer (pH 6.8) for 1 h at room temperature; subsequent to acidification with a pH 3.65 buffer, the incubation was continued at 37 °C as specified in the legends to the Figures. The fragments were precipitated by the addition of 4 vol. of cold acetone, pelleted by centrifugation, dissolved and deglycosylated in a detergent mixture with peptide N-glycosidase F [8] and separated by SDS/PAGE. Western blotting was performed using PVDF membranes (Hybond-P; Amersham Biosciences) and the ECL® detection kit as mentioned above.
Gel filtration of proSAP–proCD complexes
The precursors and their mixtures were incubated in PBS (pH 6.8) at 4 °C for 16 h and applied to a 30 cm×1 cm Superdex 200 (Amersham Biosciences) FPLC column. The proteins were eluted with the same buffer at 0.5 ml/min and fractions of 0.25 ml were collected. Aliquots of 10 μl were analysed by SDS/PAGE and silver nitrate staining (see above).
Biotinylation of proCD and surface plasmon resonance measurements
ProCD (70 μg) was incubated in 220 μl of 110 mM NaCl and 74 mM sodium phosphate (pH 8.0) in the presence of 50 μM succinimidyl 6-(biotinamido)hexanoate (Perbio Science Deutschland, Bonn, Germany) on ice for 2 h. The reagent was added from a 2 mM stock solution in DMSO. The reaction was terminated with an excess of ethanolamine and the unbound biotin species were removed by dialysis. The autoactivability of proCD and the activity of the modified pseudoCD were not impaired by the derivatization.
Surface plasmon resonance measurements were performed in the BIAcore 3000 system (Biacore, Freiburg i. Br., Germany). Biotinylated proCD (0.5 μg/ml) was immobilized on a streptavidin-coated sensor chip SA at a flow rate of 5 μl/min, using 0.15 M NaCl, 0.005% (v/v) surfactant P-20 and 10 mM Hepes (pH 7.4) as running buffer, to provide an appropriate starting signal of 1500–2000 response units. The buffer was changed to the Theorell and Stenhagen's citrate-phosphate-borate buffers containing 150 mM NaCl, and the binding and desorption of proSAP, each for 6 min at a flow rate of 10 μl/min, were examined. Signals generated in a control cell without biotinylated proCD were subtracted from the experimental data and the initial values were set to zero. Quantitative evaluation of the binding and dissociation reactions was performed using the software BIAevaluation version 3.1, which was provided with the apparatus.
RESULTS
Presence of proSAP–proCD complexes in secretions
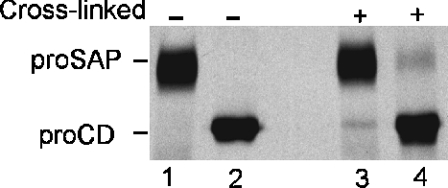
Using dithio-bis-succinimidyl propionate as a cross-linking reagent, the existence of proSAP–proCD complexes has been demonstrated in cells of various types [8–11]. Since the two precursors are known to be secreted by different cell types of epithelial [28,29] and mesenchymal origin [15,30], we decided to examine the presence of complexes in the medium of cultured cells. To enhance the concentration of precursors, we stimulated the secretion of lysosomal proteins with 10 mM NH4Cl and 0.2 μM PMA [19]. After metabolic labelling, an aliquot of the conditioned medium was subjected to cross-linking, and proCD and proSAP were isolated by immunoprecipitation. As shown in Figure 1, in the cross-linked sample, a portion of proCD was precipitated along with proSAP in the presence of antibodies against the latter protein. Similarly, in the presence of anti-cathepsin D antibodies, a portion of proSAP was precipitated along with proCD. In both cases, after the cross-linking, approx. 10% of the non-antigen precursors were co-precipitated. In additional experiments (not shown), we observed the cross-linking of a portion of the precursors in conditioned media of metabolically labelled CaCo-2, THP-1 and human fibroblasts. The amount of cross-linked complexes was increased when the secretion of the precursors was enhanced in the presence of 10 mM NH4Cl.
Figure 1. Presence of cross-linkable proSAP–proCD complexes in secretions of PMA-treated U937 cells.
U937 cells were metabolically labelled in the presence of Tran35S-label, 10 mM NH4Cl and 0.2 μM PMA for 15 h. Aliquots of the medium from 6×107 cells were either left untreated (–) or incubated with 1 mM dithio-bis-succinimidyl propionate cross-linking reagent for 5 min at room temperature (+) and processed for immunoprecipitation with anti-saposin C (lanes 1 and 3) and anti-proCD antibodies (lanes 2 and 4). The labelled precursors were separated by SDS/PAGE and visualized by fluorography.
Isolation and biological activity of recombinant proCD and proSAP
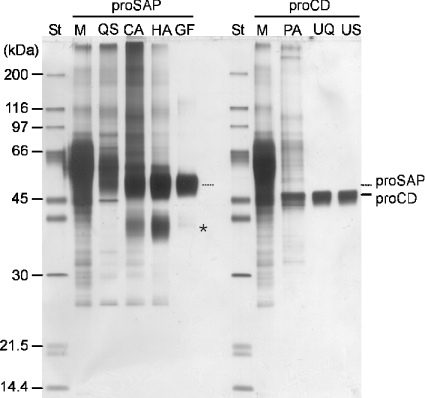
Two lysosomal precursor proteins were purified from the culture media of baculovirus-infected S. frugiperda Sf21 cells. Figure 2 presents a qualitative characterization of the proteins through the purification stages and the final preparations by SDS/PAGE and silver staining. ProSAP appeared as a broad band with an apparent molecular mass of 60 kDa and proCD with an apparent molecular mass of 46 kDa. During the production of proSAP, a fraction of the precursor was fragmented to a trisaposin, which was not completely removed from proSAP (Figure 2). The apparent heterogeneity of the precursor bands was partially due to a variable glycosylation since the heterogeneity was reduced after a digestion with peptide N-glycosidase F (see further below). Using MALDI-MS (matrix-assisted laser-desorption mass spectrometry), the molecular mass of the major species of proSAP was determined to be 60.329 kDa.
Figure 2. Qualitative characterization of protein enrichment through purification steps of proSAP and proCD.
Standard proteins and aliquots of the starting media and pools obtained through successive purification steps as indicated in the Figure were heated in the presence of SDS and dithiothreitol and subjected to SDS/PAGE. Staining of the protein with a silver nitrate method indicated that albumin was the major constituent in the starting material, whereas the purification resulted in a gradual enrichment of the recombinant proteins proSAP and proCD. ‘St’ refers to a protein standard mixture. Lane M denotes the medium and the other symbols refer to the purification steps using Q-Sepharose (QS), concanavalin A–Sepharose (CA), hydroxyapatite (HA), gel filtration (GF), pepstatin A-affinity chromatography (PA), UNO-Q ion-exchange chromatography (UQ) and UNO-S-1R ion-exchange chromatography (US) as described in the Materials and methods section. The asterisk indicates the position of trisaposin-like fragments that accompany proSAP. The standards (St), in the order of decreasing apparent masses, were myosin, β-galactosidase, phosphorylase b, BSA, ovalbumin, carbonic anhydrase, soya-bean trypsin inhibitor and lysozyme.
Biological activity of the recombinant proSAP was demonstrated by its ability to stimulate the degradation of glycosphingolipids in cultured human fibroblasts defective in the synthesis of the endogenous proSAP. After incubating with or without the recombinant proSAP, the cells were subjected to metabolic labelling with radioactive serine to examine the accumulation of several lipids. Ceramide, glucosylceramide, lactosylceramide, GM3 and GM2 gangliosides and long-chain fatty acids accumulated significantly in the saposin-precursor-deficient fibroblasts when compared with the wild-type. This storage was corrected after feeding the cells with proSAP (Figure 3; see also [31]).
Figure 3. Stimulation of the degradation of glycosphingolipids in saposin-deficient fibroblasts with recombinant proSAP added to the medium.
Control (Co) and proSAP-deficient fibroblasts (proSAP–/–) were incubated with [14C]serine (1 μCi/ml) for 24 h (pulse) followed by a chase phase of 120 h. During the pulse and chase periods, the culture medium in one proSAP(–/–) culture dish was supplemented with 25 μg/ml recombinant proSAP. Cells were harvested, and lipids were extracted and separated into acidic and neutral fractions. After chromatography, the lipids were visualized as described in the Materials and methods section. The positions of ceramide (Cer), glucosylceramide (GlcCer), lactosylceramide (LacCer), globotriaosylceramide (Globo3), globotetraosylceramide (Globo4), sphingomyelin (SM), monosialogangliosides 1–3 (GM1–GM3), disialoganglioside (GD3) and fatty acids (FA) are indicated.
In several experiments, we treated the precursors with peptide N-glycosidase F and observed a decrease in the apparent mass heterogeneity, which indicated a variable glycosylation of the precursors in Sf21 cells. After deglycosylation, proCD appeared as a sharp band at 44 kDa as shown in the first lane in Figure 4. A minor component that had a smaller mass was not further characterized. A trimming of carbohydrate side chains also occurred in proSAP. However, the broad appearance of the protein band in SDS/PAGE was less affected in this case than in proCD.
Figure 4. Autocatalytic processing of recombinant proCD at acidic pH.
ProCD (100 ng) was either left untreated (lane 0) or incubated for 10–320 min as indicated in the presence of 50 mM each of acetate and formiate/NaOH buffers (pH 3.65) in 5 μl at 37 °C. The reaction was terminated by adding 1 μl of 0.5 M Tris/HCl buffer (pH 8.0) containing 10 μM pepstatin A. The samples were denatured, deglycosylated with peptide N-glycosidase F and analysed by SDS/PAGE and silver nitrate staining.
In Figure 4, the biological activity of proCD was demonstrated by an autocatalytic conversion of the precursor into a processing intermediate during an incubation at pH 3.65. The conversion has been shown to occur under acidic conditions [30] and its product is known as pseudoCD [32]. In the present study, the removal of an N-terminal peptide was confirmed by a Western-blot analysis using rabbit antisera that were raised to synthetic peptides upstream (His-7 to Gly-21) and downstream (Ser-33 to Pro-46) of the autocatalytic cleavage site that localizes between Leu-26 and Ile-27 [33]. The latter antiserum reacted with proCD as well as with the product of its autocatalytic processing, whereas the former recognized solely the apparently larger proCD polypeptide (results not shown). The numbering of the amino acid residues in proCD used in the present study corresponds to the amino acid sequence as deduced from the nucleotide sequence by Faust et al. [34].
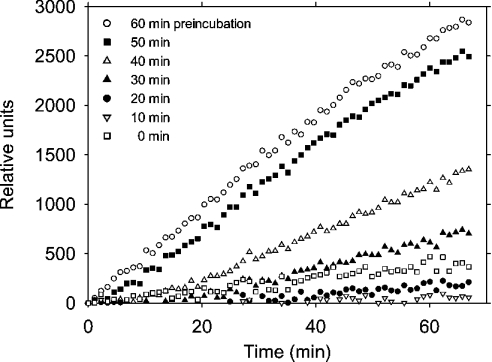
ProCD possessed low proteinolytic activity towards AMCA-haemoglobin at pH 3.65. However, this activity increased during the incubation with the substrate, indicating an autoactivation. The substrate AMCA-haemoglobin was likely to impede the autocatalytic cleavage of proCD during the assay and, therefore, preincubation was performed in the acidic buffer before the addition of the substrate. As shown in Figure 5, the activity increased significantly if the preincubation was performed for 30 min or longer. After this and shorter preincubations, the rate of AMCA-haemoglobin hydrolysis tended to increase during the assay, indicating that the activation of proCD may continue at a slow rate after diluting the proenzyme with the substrate solution. Usually the activity reached a plateau within a few hours of preincubation and was enhanced severalfold compared with the control with no preincubation. The apparent activity of proCD might be explained by an intrinsic activity of the precursor and/or the presence of a portion of processed forms in the examined precursor preparation. We observed a gradual increase in the enzymic activity after repeated freezing and thawing of the proCD sample. This limited the availability of purified proCD and precluded a quantitative examination of the intrinsic activity in the present study. The progression of the autocatalytic trimming (representatively shown in Figure 4) moderately varied between the individual experiments. A similar observation was made on the autoactivation that was monitored by the haemoglobin hydrolysis assay (Figure 5). From the results shown in Figures 4 and 5, we concluded that the proteinolytic cleavage of the precursor was a prerequisite for the gain in proteinolytic activity towards AMCA-haemoglobin.
Figure 5. Autocatalytic activation of recombinant proCD at acidic pH.
Aliquots of proCD were incubated at pH 3.65 and 37 °C as described in the legend to Figure 4. After incubation for the periods indicated, the samples were assayed for cathepsin D activity and fluorescence was recorded under standard conditions. Untreated control and the samples that were incubated in the acidic buffer for 10, 20, 30, 40, 50 and 60 min are shown, as indicated in the plot.
In vitro formation of proSAP–proCD complexes
Recombinant proSAP formed oligomers at pH 6.8, since, in the gel filtration, it eluted in a broad volume corresponding to a molecular mass equivalent to oligomers consisting of between two and six monomers (Figure 6). In contrast, proCD eluted as expected for a monomer with an apparent molecular mass of 46 kDa. After preincubation of proCD with proSAP at pH 6.8, the elution volume of the former was decreased substantially, indicating an association of the two precusors. This resembled their characterization in cultured cells [8]. From a qualitative point of view, we concluded that the association occurred spontaneously in the absence of other macromolecules. Unlike proCD, single- and double-chain forms of mature cathepsin D did not co-chromatograph with proSAP at pH 6.8 (Figure 6E). In the presence of proSAP, the bulk of single- and double-chain forms of mature cathepsin D eluted in fractions 44–50. A minor portion of mature cathepsin D eluted in and near fraction 34. In contrast with proCD, which bound heterophilically to proSAP, mature cathepsin D appeared not to bind to proSAP. In a control run (not shown) with mature cathepsin D alone, a small fraction of the protein fractionated ahead of the main peak similar to the result presented in Figure 6(E), indicating a weak homophilic interaction.
Figure 6. Demonstration of complex formation between proSAP and proCD by gel-filtration analysis.
ProSAP, proCD and their mixtures were incubated in 0.1 ml of PBS (pH 6.8) with 0.1 mM leupeptin for 1 h at room temperature and for 16 h at 4 °C. The samples were fractionated on a Superdex 200 FPLC gel-filtration column and fractions (0.25 ml each) were collected. The proteins were precipitated by mixing with 1 ml of cold acetone. The precipitates were collected by centrifugation and analysed by SDS/PAGE and silver nitrate staining of protein. In (A–D), four different runs with proCD or proSAP alone and their combinations are shown. These were performed with 0/1, 0.7/1, 1.4/1 and 1.4/0 nmol of proSAP/nmol of proCD respectively. In (E), separation of a mixture of 0.7 nmol each of proSAP and placental cathepsin D (preincubated as above at pH 6.8 in the presence of 0.1 mM leupeptin and 2 μM pepstatin A) is shown. The positions of single-chain cathepsin D (scCD) and of large subunit of mature double-chain (lmCD) cathepsin D are indicated. In the left margin lane, ovalbumin (Mr 45000) and carbonic anhydrase (Mr 30000) standards are shown. The elution of Dextran Blue (Mr 2000000), β-amylase (Mr 200000), BSA (Mr 66000), carbonic anhydrase (Mr 30000) and cytochrome c (Mr 12400), which was examined separately, is indicated by arrows that refer to Mr in thousands.
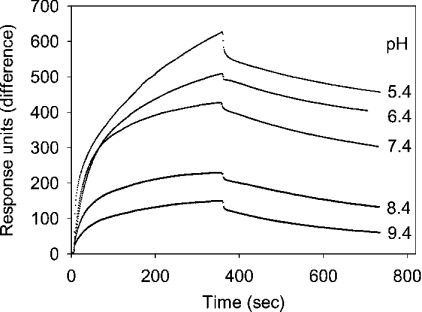
To characterize the conditions and kinetics of the complex formation, biotinylated proCD was immobilized on a streptavidin-coated surface plasmon resonance chip. A stable basal resonance signal was expected with this ligand rather than with proSAP, since, in the gel filtration, proCD behaved as a monomer. With the immobilized proCD, the binding of proSAP was examined at different pH values and concentrations of this analyte. As shown in Figure 7, a rapid binding of proSAP occurred near neutral pH. Less binding was observed at alkaline pH values. At pH values less than 5, an increased injection noise that varied between the buffer control and the sample precluded a detailed examination of the reaction. No significant binding was observed with two unrelated proteins (GM2 activator protein and BSA) that were used as controls at 0.8 μM concentration each.
Figure 7. Binding of proSAP to immobilized proCD at various pH values.
Biotinylated proCD was adsorbed on a streptavidin–dextran-coated chip and surface plasmon resonance was recorded for 6 min with solutions of proSAP (730 nM monomer) at pH values from 5.4 to 9.4 as indicated in the Figure, and with the corresponding buffers without proSAP for another 6 min.
At pH 6.8, dependence of the binding on the concentration was examined over the range of 0.24–1.47 μM proSAP monomer. Representative results of these measurements are shown in Figure 8. Among the binding models available in the standard software, the best-fit was observed with the ‘bivalent analyte’. This suggested that proSAP oligomers possess more than one binding site for proCD. Owing to the immobilized nature of the ligand (proCD), its simultaneous binding to two or more binding sites on proSAP oligomer was probably less than the optimum. Therefore only the first binding (ka1) and dissociation (kd1) constants that describe a part of the following modelled reaction were considered:
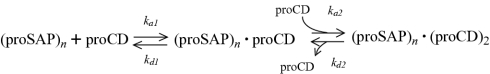 From the average values ka1=1.54±1.0×104 M−1·s−1 (4) and kd1=9.0±0.3×10−4 s−1 (4), we calculated the average values KA1=ka1kd1−1=1.7×107 M−1 and KD1=kd1ka1−1=5.8×10−8 M. The velocity constants are presented along with the S.D. values that were calculated from the results of four independent measurements.
From the average values ka1=1.54±1.0×104 M−1·s−1 (4) and kd1=9.0±0.3×10−4 s−1 (4), we calculated the average values KA1=ka1kd1−1=1.7×107 M−1 and KD1=kd1ka1−1=5.8×10−8 M. The velocity constants are presented along with the S.D. values that were calculated from the results of four independent measurements.
Figure 8. Binding and desorption of proSAP at different concentrations to biotinylated proCD on the surface of a streptavidin–dextran-coated chip.
The reactions were examined with 1.47, 0.73, 0.37 and 0.24 μM proSAP (concentrations refer to those of the monomer) as indicated in the plot. Surface plasmon resonance in the presence and absence of proSAP was recorded as described in the legend to Figure 6, but all measurements were performed at pH 6.8. The best-fit (indicated by the dotted lines) was obtained by using the bivalent analyte model for proSAP.
Proteinolysis of proSAP and formation of pseudoCD after acidification of the proSAP–proCD complexes
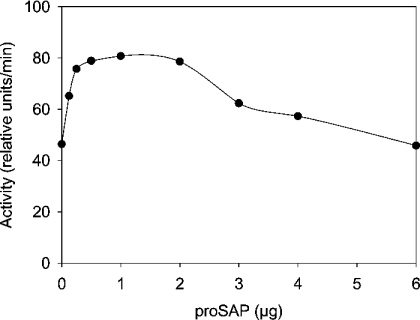
We prepared complexes of proSAP with proCD by incubating their mixtures at room temperature at pH 6.8, acidified the medium to pH 3.65 and incubated the samples for specified periods. The in vitro processing of the two precursors was examined by SDS/PAGE and silver staining (Figure 9). A major portion of proSAP was digested within 3 h of incubation. At the expense of the precursor, smaller polypeptides were formed that probably represent ‘trisaposins’ and ‘disaposins’, referring to fragments of proSAP that contain three and two neighbouring saposin domains respectively. After 1 h of incubation of proCD alone, a major portion of the precursor was converted into a smaller molecule resembling pseudoCD. The relative intensity of the two stained bands in this experiment and other similar experiments revealed that the conversion was faster in the presence of proSAP. Thus, after a 3 h digestion in the absence of proSAP, a significant portion of proCD remained undigested, whereas in the presence of proSAP, the conversion of proCD appeared to have been completed. In this situation, proCD rapidly executed both the autoactivation and fragmentation of proSAP. A possible explanation for the results is that proCD, when complexed with proSAP, is rendered a better substrate for the autoactivation. Alternatively, proSAP could enhance the activity of pseudoCD that is produced during the incubation. To test the latter possibility, the effect of proSAP on pseudoCD was examined using labelled haemoglobin as a substrate. A representative result of this test is shown in Figure 10. At an approximately stoichiometric ratio, the stimulation with proSAP was nearly 2-fold. A decrease in the activity was apparent at higher concentrations of proSAP. It is probable that proSAP plays two roles in this system. First, it complexes and activates pseudoCD and, secondly, it is a substrate of pseudoCD and competes with haemoglobin as a substrate.
Figure 9. Processing of proCD in the presence of proSAP after the acidification of the incubation medium.
Four groups of samples containing 0.2 μg of proCD and 0.1, 0.2 and 0.4 μg of proSAP or either precursor alone (0.2 and 0.4 μg respectively) in 4 μl of PBS (pH 6.8) were incubated for 1 h at room temperature. After incubation for 0, 1, 2 or 3 h with 1 μl of 0.25 M acetate and 0.25 M sodium formate buffer (pH 3.65), the reactions were terminated by adding 1 vol. of 0.1 M Tris/HCl buffer (pH 8.0) containing 10 μM pepstatin A. The polypeptides were precipitated with 80% (v/v) acetone and solubilized, deglycosylated using peptide N-glycosidase F (PNGase F) and analysed by SDS/PAGE and silver nitrate staining. Notice the acceleration by proSAP in the processing of proCD that is apparent from the relative intensities of the proCD and pseudoCD bands. This effect was reproduced in all experiments performed (n=3).
Figure 10. Activation of pseudoCD in the presence of proSAP.
PseudoCD was prepared by incubating proCD at pH 3.65 with a conversion of 95% of the precursor. Aliquots (100 ng each) were incubated with the indicated amounts of proSAP in 10 mM sodium phosphate buffer (pH 6.8) at room temperature for 30 min. The solutions were then mixed with the fluorescent haemoglobin substrate under standard cathepsin D assay conditions to determine the enzyme activity.
Incubation of proSAP with a higher amount of proCD resulted in the formation of smaller fragments. In SDS/PAGE, apparent masses of the digestion products varied between that of proSAP and those of peptides <9 kDa that may have represented incomplete saposins (Figure 11A). Apparently, starting in a complex with proSAP, proCD can be activated to pseudoCD and the latter can hydrolyse proSAP preferentially although not exclusively within the connecting (intersaposin) and terminal peptides. After the acidification, tri- and disaposins as well as monosaposin-like fragments were formed that reacted characteristically with specific antisera. In a Western blot of the digestion products, we observed that, in these artificial mixtures, significant amounts of products resembling saposins C and D were formed (Figure 11B).
Figure 11. Processing of proSAP in the presence of proCD.
(A) Samples containing 0.2, 0.4 or 0.8 μg of proSAP were incubated without or with up to 80 ng of proCD in 2 μl of 10 mM sodium phosphate buffer (pH 6.8) for 1 h at room temperature. After adding 2 μl of 0.25 M acetate and 0.25 M sodium formiate buffer (pH 3.65), the incubation was continued for 14 h at 37 °C. The reactions were terminated and the polypeptide fragments were analysed as described in the legend to Figure 9. (B) Mixtures of the two precursors were incubated at pH 3.65 for 18 h and analysed by Western blotting using antisera against saposins A–D as indicated in the Figure. Incubations were performed with 200 ng of proSAP and 20 ng of proCD (lanes 1), 400 ng of proSAP and 40 ng of proCD (lanes 2) and 400 ng of proSAP and 160 ng of proCD (lanes 3). The protein standards shown include aprotinin (6.5 kDa).
In subsequent experiments that included Western blotting, fragments with masses intermediary between those of disaposins and monosaposins, as shown in Figure 11(A), reacted with anti-saposin A antibodies. This indicated a rapid cleavage within the intersaposin AB in the vicinity of the saposin B domain or within the latter. The results suggested that (i) proCD contained within the complexes can be autoactivated to pseudoCD if the pH is decreased, (ii) proSAP in the complexes as well as its free excess is a substrate of pseudoCD, (iii) proSAP is cleaved by pseudoCD within the connecting and the terminal peptides at several positions, yielding transiently tri- and disaposins and (iv) saposin D and, less so, saposin C temporarily resist further degradation, whereas saposin A and, even faster, saposin B are degraded with little or no accumulation of related disaposin and monosaposin.
DISCUSSION
proSAP and proCD are glycoproteins that originate in the endoplasmic reticulum and reach the lysosomes by intracellular trafficking. A series of cellular studies indicated that the two precursors travel together and form protein–protein complexes with each other [8–11]. In endosomal and/or lysosomal compartments, they are proteinolytically processed to mature active lysosomal proteins, saposins A–D, and the so-called double-chain cathepsin D. As will be discussed further below, cathepsin D participates in the processing of proSAP. Hence, it is remarkable that, soon after the synthesis [8–11], the precursor (proCD) of this processing enzyme is captured by its potential substrate (proSAP) or vice versa, to be joined for targeting to the same destinations. In the present study, we demonstrated the occurrence of proSAP–proCD complexes in secretions of cultured cells. We assumed that the complexes participate in novel functions at or near the cell surface and, therefore, we described the preparation of biologically active recombinant proSAP and proCD and characterized their interactions in vitro.
Activation of proCD is modulated by proSAP
Initially, an autocatalytic processing of proCD has been observed with immunoprecipitates of this precursor as prepared from secretions of human fibroblasts and incubated at acidic pH [30]. Later, using an in vitro folded recombinant human proCD, it has been shown that the autocatalytic cleavage results in the removal of 26 amino acids from the N-terminus, thus forming pseudoCD, a homologue of pseudopepsin [32,33]. Wittlin et al. [35] demonstrated autoactivation with an immunoprecipitate of proCD from a baculovirus expression system similar to the proCD secreted by fibroblasts [30]. The proCD described in the present study was isolated after an intracellular folding and is free of antibodies, and can be quantitatively converted into pseudoCD (Figure 4). It is reported for the first time that proSAP accelerates the activation of proCD (Figure 9). Our findings suggest that proSAP interacts with both proCD and its trimmed form, pseudoCD, and stimulates their catalytic activity. Thus the interaction does not require the presence of the N-terminal hexadocosapeptide in the cathepsin moiety.
ProSAP is an oligomer that associates with proCD
Hiraiwa et al. [36] have reported that human proSAP purified from either milk or the medium of infected Spodoptera cells forms oligomers of varied masses. With proSAP from the baculovirus expression system, we observed, using gel filtration, that oligomers were the predominant form of the purified precursor at neutral pH and that these oligomers spontaneously bound proCD. Practically all proCD associated with proSAP if an excess of the latter was provided (Figure 6). The elution in the high-molecular-mass region contrasted with the behaviour of proCD alone, which fractionated mainly as a monomer.
ProSAP–proCD complexes formed in the absence of other human proteins and the binding of proSAP to immobilized proCD occurred at submicromolar concentrations of the former. In experiments with cultured cells, we observed that, after metabolic labelling, a portion of the secreted proCD was co-precipitated with proSAP if the medium was treated with the cross-linker dithio-bis-succinimidyl propionate before the addition of the antibody. Similarly, a portion of the secreted proSAP was co-precipitated with proCD. Apparently, after the secretion, in spite of the vast dilution, at least a portion of the complexes remained associated.
We suggest that, in cells, the complexes are formed spontaneously after folding of the two precursors in the endoplasmic reticulum and that portions of proSAP and proCD are transported in a tandem not only to lysosomes but also into the extracellular space. Since the proSAP used in the present study was represented by the isoform lacking exon 8 [18] and contained extensions at both the N- and C-termini, the exon 8-encoded peptide and the native termini were unlikely to play an important role in the oligomerization and complex formation. From our observations on the binding of proSAP to immobilized biotinylated pseudoCD and on the lack of a stable association of mature single- and double-chain cathepsin D with proSAP in solution, it may be speculated that, in proCD and pseudoCD, amino acid residues comprising the N-terminal region of pseudoCD are involved in association with proSAP.
Fragmentation of proSAP
Previously, a fragmentation of proSAP has been achieved in the presence of human testicular glycoprotein fraction and shown to be sensitive to pepstatin A [36] as well as to antibodies raised against cathepsin D [37], thus implicating this proteinase in the processing. In baculovirus-infected insect cells [38] and their medium [36], as well as in incubations of purified proSAP with cathepsin D [37], formation of various tri- and disaposins as well as monosaposins with N- and C-terminal extensions has been reported. Metabolic labelling and immunoprecipitation of the fragments indicated that, in a major path, intersaposin AB is severed first and that disaposins BC and CD are more abundant than disaposin AB [38,39]. We examined the processing of proSAP–proCD complexes and observed that pseudoCD was formed after acidification and it hydrolysed proSAP at multiple sites similar to what had been observed with mature cathepsin D [37].
The yield of the saposin D-like fragment appeared to be highest among the saposins, so that the distal saposin domain was least sensitive to cleavage by pseudoCD. We assume that the stability of saposins depends on their interactions with lipids and/or proteins that were omitted in the present study. Recently, it has been reported that acid β-glucosidase is rapidly degraded in fibroblasts lacking the cognate saposin C, suggesting that the latter is protecting the enzyme [40]. The saposins, in turn, may be protected by lysosomal enzymes and lipids.
Does the complex formation matter in the lysosomal targeting of the precursors?
We have not addressed this question experimentally. However, owing to the apparently strong interaction of the precursors with each other, this point is noteworthy. Major fractions of proSAP and proCD are targeted to lysosomes. The remainder is secreted and the extent of the secretion varies with cells and conditions. Both proCD and proSAP contain phosphorylated oligosaccharide side chains that may participate in the lysosomal targeting. However, when compared with normal cells, in I-cells, the absence of oligosaccharide phosphorylation results in a rather moderate decrease in the lysosomal concentration of saposins, which is consistent with an alternative, mannose 6-phosphate-independent targeting [15]. Interestingly, in I-cell fibroblasts, a significant residual lysosomal targeting of proCD takes places as well [41]. Proximal to the lysosomal segregation compartments, still in the Golgi cisternae, a significant portion of proSAP appears to be insoluble and to bind to membranes independent of mannose 6-phosphate residues [12]. Similarly, a mannose 6-phosphate-independent association of proCD with intracellular membranes has been described in [13,42,43]. This interaction depends on acidic pH and is saturable, indicating the presence of an as yet unidentified receptor [42]. In rabbit macrophages, membrane association has been detected in endosomes but not in lysosomes [43]. It is conceivable that the described interactions with membranes apply not only to the single precursors but also to their complexes.
In addition to a certain degree of carbohydrate-independent lysosomal targeting, proCD and proSAP share the ability to bind lipids. ProSAP is known to bind gangliosides [37] and, through its saposin domains, it may interact with other membrane constituents, such as phosphatidyl serine [1]. In the lysosomal targeting of proSAP, sphingomyelin was reported to be indispensable [44], with saposin D and the adjacent C-terminal moieties in proSAP playing a crucial role. Another lipid interaction was described for proCD: it bound to and was activated by ceramides [45]. In cells, not only the proteinolytic maturation but also the lysosomal targeting of proCD were accelerated [46].
Binding to lipids or membranes alone does not explain targeting to an organelle. ProSAP was shown to bind to low-density lipoprotein receptor-related protein [23] at the cell surface and to sortilin intracellularly [47]. Sortilin is a transmembrane protein with multiple ligand-binding domains and a cytosolic tail that associates with adapter proteins and, hence, it can participate in the lysosomal targeting. It should be of interest, therefore, to examine the binding of the proSAP–proCD complexes to receptors known for either moiety and also the influence of various lipids on these binding reactions.
Acknowledgments
We are indebted to G. Jarosch and H. Kaiser (Marburg) for assistance with HPLC and the baculovirus expression system respectively, Dr J. Hoernschemeyer and H. Hupfer (Bonn) for a MALDI-MS analysis of proSAP, M. Wendeler (Bonn) for providing recombinant GM2 activator protein and E. Heinemann (Marburg) for typing the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (grant Ha 915/7-1).
References
- 1.Qi X., Grabowski G. A. Differential membrane interactions of saposins A and C: implications for the functional specificity. J. Biol. Chem. 2001;276:27010–27017. doi: 10.1074/jbc.M101075200. [DOI] [PubMed] [Google Scholar]
- 2.Sandhoff K., Kolter T. Biosynthesis and degradation of mammalian glycosphingolipids. Philos. Trans. R. Soc. Lond. B. 2003;358:847–861. doi: 10.1098/rstb.2003.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winau F., Schwierzeck V., Hurwitz R., Remmel N., Sieling P. A., Modlin R. L., Porcelli S. A., Brinkmann V., Sugita M., Sandhoff K., et al. Saposin C is required for lipid presentation by human CD1b. Nature Immunol. 2004;5:169–174. doi: 10.1038/ni1035. [DOI] [PubMed] [Google Scholar]
- 4.Hazkani-Covo E., Altman N., Horowitz M., Graur D. The evolutionary history of prosaposin: two successive tandem-duplication events gave rise to the four saposin domains in vertebrates. J. Mol. Evol. 2002;54:30–34. doi: 10.1007/s00239-001-0014-0. [DOI] [PubMed] [Google Scholar]
- 5.Ahn V. E., Faull K. F., Whitelegge J. P., Fluharty A. L., Privé G. G. Crystal structure of saposin B reveals a dimeric shell for lipid binding. Proc. Natl. Acad. Sci. U.S.A. 2003;100:38–43. doi: 10.1073/pnas.0136947100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ponting C. P., Russel R. B. Swaposins: circular permutations within genes encoding saposin homologues. Trends Biochem. Sci. 1995;20:179–180. doi: 10.1016/s0968-0004(00)89003-9. [DOI] [PubMed] [Google Scholar]
- 7.Kervinen J., Tobin G. J., Costa J., Waugh D. S., Wlodawer A., Zdanov A. Crystal structure of plant aspartic proteinase prophytepsin: inactivation and vacuolar targeting. EMBO J. 1999;18:3947–3955. doi: 10.1093/emboj/18.14.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grässel S., Hasilik A. Human cathepsin D precursor is associated with a 60 kDa glycosylated polypeptide. Biochem. Biophys. Res. Commun. 1992;182:276–282. doi: 10.1016/s0006-291x(05)80141-x. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Y., Conner G. E. Intermolecular association of lysosomal protein precursors during biosynthesis. J. Biol. Chem. 1994;269:3846–3851. [PubMed] [Google Scholar]
- 10.Godbold G. D., Kyujeong A., Yeyeodu S., Lee L. F., Ting J. P.-Y., Erickson A. H. Biosynthesis and intracellular targeting of the lysosomal aspartic proteinase cathepsin D. Adv. Exp. Med. Biol. 1998;436:153–162. doi: 10.1007/978-1-4615-5373-1_21. [DOI] [PubMed] [Google Scholar]
- 11.Laurent-Matha A. L., Hüttler S., Sandhoff K., Garcia M., Rochefort H. Procathepsin D interacts with prosaposin in cancer cells but its internalization is not mediated by LDL receptor-related protein. Exp. Cell Res. 2002;277:210–219. doi: 10.1006/excr.2002.5556. [DOI] [PubMed] [Google Scholar]
- 12.Rijnboutt S., Aerts H. M. F. G., Geuze H. J., Tager J. M., Strous G. J. Mannose 6-phosphate-independent membrane association of cathepsin D, glucocerebrosidase, and sphingolipid-activating protein in HepG2 cells. J. Biol. Chem. 1991;266:4862–4868. [PubMed] [Google Scholar]
- 13.Rijnboutt S., Kal A. J., Geuze H. J., Aerts H., Strous G. J. Mannose 6-phosphate-independent targeting of cathepsin D to lysosomes in HepG2 cells. J. Biol. Chem. 1991;266:23586–23592. [PubMed] [Google Scholar]
- 14.Igdoura S. A., Rasky A., Morales C. R. Trafficking of sulfated glycoprotein-1 (prosaposin) to lysosomes or to the extracellular space in rat Sertoli cells. Cell Tissue Res. 1996;283:385–394. doi: 10.1007/s004410050549. [DOI] [PubMed] [Google Scholar]
- 15.Vielhaber G., Hurwitz R., Sandhoff K. Biosynthesis, processing, and targeting of sphingolipid activator protein (SAP) precursor in cultured human fibroblasts. J. Biol. Chem. 1996;271:32438–32446. doi: 10.1074/jbc.271.50.32438. [DOI] [PubMed] [Google Scholar]
- 16.Fürst W., Schubert J., Machleidt W., Meyer H. E., Sandhoff K. The complete amino-acid sequences of human ganglioside GM2 activator protein and cerebroside sulfate activator protein. Eur. J. Biochem. 1990;192:709–714. doi: 10.1111/j.1432-1033.1990.tb19280.x. [DOI] [PubMed] [Google Scholar]
- 17.Klein A., Henseler M., Klein C., Suzuki K., Harzer K., Sandhoff K. Sphingolipid activator protein D (sap-D) stimulates the lysosomal degradation of ceramide in vivo. Biochem. Biophys. Res. Commun. 1994;200:1440–1448. doi: 10.1006/bbrc.1994.1612. [DOI] [PubMed] [Google Scholar]
- 18.Henseler M., Klein A., Glombitza G. J., Suzuki K., Sandhoff K. Expression of the three alternative forms of the sphingolipid activator protein precursor in baby hamster kidney cells and functional assays in a cell culture system. J. Biol. Chem. 1996;271:8416–8423. doi: 10.1074/jbc.271.14.8416. [DOI] [PubMed] [Google Scholar]
- 19.Radons J., Biewusch U., Grässel S., Geuze H. J., Hasilik A. Distinctive inhibition of the lysosomal targeting of lysozyme and cathepsin D by drugs affecting pH gradients and protein kinase C. Biochem. J. 1994;302:581–586. doi: 10.1042/bj3020581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemansky P., Hasilik A. Chondroitin sulfate is involved in lysosomal transport of lysozyme in U937 cells. J. Cell Sci. 2000;114:345–352. doi: 10.1242/jcs.114.2.345. [DOI] [PubMed] [Google Scholar]
- 21.Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur. J. Biochem. 1975;56:335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- 22.Grosch H. W., Hasilik A. Protection of proteolysis-prone recombinant proteins in baculovirus expression systems. BioTechniques. 1998;24:934–936. doi: 10.2144/98246bm05. [DOI] [PubMed] [Google Scholar]
- 23.Hiesberger T., Hüttler S., Rohlmann A., Schneider W., Sandhoff K., Herz J. Cellular uptake of saposin (SAP) precursor and lysosomal delivery by the low density lipoprotein receptor-related protein (LRP) EMBO J. 1998;17:4617–4625. doi: 10.1093/emboj/17.16.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Heukeshoven J., Dernick R. Improved silver staining procedure for fast staining in PhastSystem Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis. 1988;9:28–32. doi: 10.1002/elps.1150090106. [DOI] [PubMed] [Google Scholar]
- 26.Momoi T., Ando S., Magai Y. High resolution preparative column chromatographic system for gangliosides using DEAE-sephadex and a new porus silica, latrobeads. Biochim. Biophys. Acta. 1976;441:488–497. [PubMed] [Google Scholar]
- 27.Partanen S., Storch S., Loeffler H. G., Hasilik A., Tyynelä J., Braulke T. A replacement of the active site aspartic acid residue-293 in mouse cathepsin D affects its intracellular stability, processing, and transport in HEK 293 cells. Biochem. J. 2003;369:55–62. doi: 10.1042/BJ20021226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hineno T., Sano A., Kondoh K., Ueno S., Kakimoto Y., Yoshida K. Secretion of sphingolipid hydrolase activator precursor, prosaposin. Biochem. Biophys. Res. Commun. 1991;176:668–674. doi: 10.1016/s0006-291x(05)80236-0. [DOI] [PubMed] [Google Scholar]
- 29.Vetvicka V., Vagner J., Baudys M., Tang J., Foundling S. I., Fusek M. Human breast milk contains procathepsin D – detection by specific antibodies. Biochem. Mol. Biol. Int. 1993;30:921–928. [PubMed] [Google Scholar]
- 30.Hasilik A., von Figura K., Conzelmann E., Nehrkorn H., Sandhoff K. Lysosomal enzyme precursors in human fibroblasts. Eur. J. Biochem. 1982;125:317–321. doi: 10.1111/j.1432-1033.1982.tb06685.x. [DOI] [PubMed] [Google Scholar]
- 31.Burkhardt J. K., Hüttler S., Klein A., Möbius W., Habermann A., Griffiths G., Sandhoff S. Accumulation of sphingolipids in SAP-precursor (prosaposin)-deficient fibroblasts occurs as intralysosomal membrane structures and can be completely reversed by treatment with human SAP-precursor. Eur. J. Cell Biol. 1997;73:10–18. [PubMed] [Google Scholar]
- 32.Richo G., Conner G. E. Proteolytic activation of human procathepsin D. Adv. Exp. Med. Biol. 1991;306:289–296. doi: 10.1007/978-1-4684-6012-4_35. [DOI] [PubMed] [Google Scholar]
- 33.Conner G. E., Richo G. Isolation and characterization of a stable activation intermediate of the lysosomal aspartyl protease cathepsin D. Biochemistry. 1992;31:1142–1147. doi: 10.1021/bi00119a024. [DOI] [PubMed] [Google Scholar]
- 34.Faust P. L., Kornfeld S., Chirgwin J. M. Cloning and sequence analysis of cDNA for human cathepsin D. Proc. Natl. Acad. Sci. U.S.A. 1985;82:4910–4914. doi: 10.1073/pnas.82.15.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wittlin S., Rösel J., Hofmann F., Stover D. R. Mechanisms and kinetics of procathepsin D activation. Eur. J. Biochem. 1999;265:384–393. doi: 10.1046/j.1432-1327.1999.00747.x. [DOI] [PubMed] [Google Scholar]
- 36.Hiraiwa M., O'Brien J. S., Kishimoto Y., Galdzicka M., Fluharty A. L., Ginns E. I., Martin B. M. Isolation, characterization, and proteolysis of human prosaposin, the precursor of saposins (sphingolipid activator proteins) Arch. Biochem. Biophys. 1993;304:110–116. doi: 10.1006/abbi.1993.1328. [DOI] [PubMed] [Google Scholar]
- 37.Hiraiwa M., Martin B. M., Kishimoto Y., Conner G. E., Tsuji S., O'Brien J. S. Lysosomal proteolysis of prosaposin, the precursor of saposins (sphingolipid activator proteins): its mechanism and inhibition by ganglioside. Arch. Biochem. Biophys. 1997;341:17–24. doi: 10.1006/abbi.1997.9958. [DOI] [PubMed] [Google Scholar]
- 38.Leonowa T., Qi X., Bencosme A., Ponce E., Sun Y., Grabowski G. A. Proteolytic processing patterns of prosaposin in insect and mammalian cells. J. Biol. Chem. 1996;271:17312–17320. doi: 10.1074/jbc.271.29.17312. [DOI] [PubMed] [Google Scholar]
- 39.Qi X., Grabowski G. A. Molecular and cell biology of acid β-glucosidase and prosaposin. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:203–239. doi: 10.1016/s0079-6603(00)66030-0. [DOI] [PubMed] [Google Scholar]
- 40.Sun Y., Qi X., Grabowski G. A. Saposin C is required for normal resistance of acid beta-glucosidase to proteolytic degradation. J. Biol. Chem. 2003;278:31918–31923. doi: 10.1074/jbc.M302752200. [DOI] [PubMed] [Google Scholar]
- 41.Hasilik A., Waheed A., Cantz M., von Figura K. Impaired phosphorylation of lysosomal enzymes in fibroblasts of patients with mucolipidosis III. Eur. J. Biochem. 1982;122:119–123. doi: 10.1111/j.1432-1033.1982.tb05856.x. [DOI] [PubMed] [Google Scholar]
- 42.McIntyre G. F., Erickson A. H. Procathepsins L and D are membrane-bound in acidic microsomal vesicles. J. Biol. Chem. 1991;266:15438–15445. [PubMed] [Google Scholar]
- 43.Diment S., Leech M. S., Stahl P. D. Cathepsin D is membrane-associated in macrophage endosomes. J. Biol. Chem. 1998;263:6901–6907. [PubMed] [Google Scholar]
- 44.Lefrancois S., May T., Knight C., Bourbeau D., Morales C. R. The lysosomal transport of prosaposin requires the conditional interaction of its highly conserved d domain with sphingomyelin. J. Biol. Chem. 2002;277:17188–17199. doi: 10.1074/jbc.M200343200. [DOI] [PubMed] [Google Scholar]
- 45.Heinrich M., Wickel M., Schneider-Brachert W., Sandberg C., Gahr J., Schwandner R., Weber T., Saftig P., Peters C., Brunner J., et al. Cathepsin D targeted by acid sphingomyelinase-derived ceramide. EMBO J. 1999;18:5252–5263. doi: 10.1093/emboj/18.19.5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Stefanis D., Reffo P., Bonelli G., Baccino F. M., Sala G., Ghidoni R., Codogno P., Isidoro C. Increase in ceramide level alters the lysosomal targeting of cathepsin D prior to onset of apoptosis in HT-29 colon. Biol. Chem. 2002;383:989–999. doi: 10.1515/BC.2002.106. [DOI] [PubMed] [Google Scholar]
- 47.Lefrancois S., Zeng J., Hassan A. J., Canuel M., Morales C. R. The lysosomal trafficking of sphingolipid activator proteins (SAPs) is mediated by sortilin. EMBO J. 2003;22:6430–6437. doi: 10.1093/emboj/cdg629. [DOI] [PMC free article] [PubMed] [Google Scholar]