Abstract
The c-type cytochromes are characterized by the covalent attachment of haem to the polypeptide via thioether bonds formed from haem vinyl groups and, normally, the thiols of two cysteines in a CXXCH motif. Intriguingly, the mitochondrial cytochromes c and c1 from two euglenids and the Trypanosomatidae contain only a single cysteine within the haem-binding motif (XXXCH). There are three known distinct pathways by which c-type cytochromes are matured post-translationally in different organisms. The absence of genes encoding any of these c-type cytochrome biogenesis machineries is established here by analysis of six trypanosomatid genomes, and correlates with the presence of single-cysteine cytochromes c and c1. In contrast, we have identified a comprehensive catalogue of proteins required for a typical mitochondrial oxidative phosphorylation apparatus. Neither spontaneous nor catalysed maturation of the single-cysteine Trypanosoma brucei cytochrome c occurred in Escherichia coli. However, a CXXCH variant was matured by the E. coli cytochrome c maturation machinery, confirming the proposed requirement of the latter for two cysteines in the haem-binding motif and indicating that T. brucei cytochrome c can accommodate a second cysteine in a CXXCH motif. The single-cysteine haem attachment conserved in cytochromes c and c1 of the trypanosomatids is suggested to be related to their cytochrome c maturation machinery, and the environment in the mitochondrial intermembrane space. Our genomic and biochemical studies provide very persuasive evidence that the trypanosomatid mitochondrial cytochromes c are matured by a novel biogenesis system.
Keywords: c-type cytochrome, cytochrome c biogenesis, Euglena, mitochondrial respiratory chain, oxidative phosphorylation, trypanosome
Abbreviations: Ccm system, cytochrome c maturation system; ETF, electron-transferring flavoprotein; SDH, succinate dehydrogenase
INTRODUCTION
The c-type cytochromes are widespread proteins essential for virtually all life. They are characterized by the covalent attachment of haem to the polypeptide. Thioether bonds are normally formed between the vinyl groups of the haem and the thiols of two cysteine residues in a conserved CXXCH motif, where the histidine functions as an axial ligand to the haem iron [1–3]. Mitochondrial cytochrome c is by far the best known of this type of protein. It is located in the intermembrane space and transfers electrons from the cytochrome bc1 complex to the cytochrome aa3 oxidase; cytochrome c1 of the bc1 complex is also a c-type cytochrome. There are many other distinct c-type cytochromes in bacteria; the evolution of the CXXCH attachment motif is unclear [4].
Remarkably, there are in different organisms at least three distinct pathways by which c-type cytochromes undergo the chemically difficult process of post-translational haem attachment [4]. One is the so-called Ccm (cytochrome c maturation) system (or System I), consisting of up to nine proteins, which is found in the α-, γ- and some β-proteobacteria, Deinococcus, and also in plant and red algal mitochondria. Cytochrome c biogenesis System II, involving at least four proteins, occurs in β-, δ- and ε-proteobacteria, Gram-positive bacteria, cytophagales, aquaficales, plant and algal chloroplasts and cyanobacteria. System III is the enzyme called haem lyase; it is found in the mitochondria of fungal, vertebrate and invertebrate cells and is also present in the genomes of Chlamydomonas reinhardtii and Plasmodium falciparum [5]. In many of the organisms, other than mammals, in which System III is used, it appears that specific haem lyase isoforms are preferentially required for the maturation of either cytochrome c or cytochrome c1 [5]. We have also shown in recent work that at least one c-type cytochrome can form correctly in vitro in an uncatalysed reaction, although the rate was low and for all other cytochromes c investigated to date malformed side-products were also observed [6–8].
Intriguingly, there is a small group of organisms within the phylum Euglenozoa that does not conform to the usual pattern of c-type cytochromes with a CXXCH haem-binding motif. The cytochromes c and c1 from the mitochondria of the euglenids Euglena gracilis and Euglena viridis [9–11] and the Trypanosomatidae (including Crithidia fasciculata and Trypanosoma brucei) [9,12,13] contain only a single cysteine residue within the haem-binding motif [A(A/G)QCH and FAPCH motifs in the cytochromes c and c1, respectively], and are thus matured by the formation of only one thioether bond between the haem and the apocytochrome. In Euglena the single cysteine attachment of haem to c-type cytochromes is specific to the mitochondria; the plastid c-type cytochromes f (despite its misleading name, cytochrome f is a c-type cytochrome) and c552 both contain a CXXCH motif [14,15], and, by analogy with cytochrome c biogenesis in higher plant chloroplasts and cyanobacteria, are presumably matured by a System II apparatus [16,17].
We have explored the maturation of the single-cysteine mitochondrial c-type cytochromes of the Euglenozoa using the newly available complete genome data for the medically relevant trypanosomatids T. brucei, Trypanosoma cruzi and Leishmania major, and the extensive genome coverage for Trypanosoma vivax, Trypanosoma congolense and Leishmania infantum. In the present paper we report the outcome of these investigations and complementary studies of the expression of T. brucei cytochrome c in Escherichia coli.
EXPERIMENTAL
Expression of recombinant T. brucei cytochrome c
The T. brucei cytochrome c gene was amplified by PCR using the primers 5′-TTGAATTCGCATGCCACCAAAGGAGCGTGC-3′ and 5′-GCAAGCTTTTAGTCCTTTAATGTCTCGAGG-3′ with Herculase DNA polymerase (Stratagene). The PCR product was digested with EcoRI and HindIII (sites underlined and in bold respectively within the primer sequences) and ligated into the pKK223-3 E. coli expression vector (Amersham Pharmacia) that had also been digested with EcoRI and HindIII to yield pKK223-Tbcytc. For targeting the recombinant cytochrome to the periplasm, complementary oligonucleotides encoding a periplasmic targeting signal (from Pseudomonas aeruginosa cytochrome c551) were annealed to provide a double-stranded DNA molecule with EcoRI-compatible sticky ends; this was ligated into EcoRI-digested pKK223-Tbcytc, yielding pKK223-Tbcytcperi1. Additional residues between the end of the periplasmic targeting sequence and T. brucei cytochrome c were removed by the Quik-change method (Stratagene), producing plasmid pKK223-Tbcytcperi2. Finally, both pKK223-Tbcytcperi2 and pKK223-Tbcytc were mutated using the Quikchange method to produce plasmids to express periplasmic (pKK223-TbcytcperiCXXCH) and cytoplasmic (pKK223-TbcytcCXXCH) variants respectively of T. brucei cytochrome c with a CXXCH haem-binding motif. All constructs were DNA sequenced.
Biochemical procedures
E. coli strain JCB387 [18] was used for cytochrome c expression experiments. Cells were transformed with plasmids coding for cytoplasmic wild-type, cytoplasmic CXXCH, periplasmic wild-type or periplasmic CXXCH forms of T. brucei cytochrome c. Plasmid pEC86 [19] was used to express the E. coli Ccm system. Cultures of 500 ml were grown, harvested and fractionated exactly as described previously [8,20]. Cytochrome content was determined from absorption spectra of the crude periplasmic or cytoplasmic fractions to which a few grains of disodium dithionite had been added.
C. fasciculata was cultured as described previously [12]. Cells were harvested by centrifugation, washed twice with PBS, the pellet slurried in 25 ml of PBS containing 600 mM NaCl and the cells lysed by six cycles of freeze–thawing in liquid nitrogen followed by sonication. The cell extract was centrifuged at 75000 g for 1 h; the supernatant was acidified to pH 5.0 with 2 M acetic acid and centrifuged again at 10000 g for 20 min. The supernatant after this step was retained for purification of the cytochrome c.
C. fasciculata cytochrome c and recombinant T. brucei cytochrome c (CXXCH variant) were purified using an SP-Sepharose chromatography column (Amersham Pharmacia XK26/20; bed volume 70 ml). Chromatography was done at room temperature in 50 mM Tris/HCl, pH 8.0. The column was eluted with a 0–0.5 M NaCl gradient at a flow rate of 9 ml·min−1; 7 ml fractions were collected. Electrospray ionization MS was performed as described [8]. Absorption spectra of reduced haems in the presence of hydroxide and pyridine are characteristic of the type of Fe–porphyrin present, as well as of any modifications to it (e.g. covalent attachment to the polypeptide in a c-type cytochrome); such pyridine haemochrome spectra were obtained according to the method of Bartsch [21].
Sequence similarity searches
Sequence similarity searching was performed using the publicly available databases at NCBI, and various other dedicated organism-specific genome browsers. Sequence data for the various trypanosomatid genomes were obtained from The Sanger Institute (http://www.genedb.org/genedb/tryp/index.jsp). Essential components from each of the known cytochrome c biogenesis systems, or proteins necessary for the assembly or activity of the mitochondrial oxidative phosphorylation apparatus, were used as query sequences in BLAST analyses of the trypanosomatid databases. In addition to using complete open reading frames as queries, sequence alignments were used to refine the query for each biogenesis factor down to stretches of similarity that identified without ambiguity orthologues from the widest possible variety of taxa. These regions were also used to search each of the trypanosomatid genomes. Some E. coli and Pseudomonas contaminants were identified among unassembled reads on the basis of perfect nucleotide identity with bacterial sequences already deposited in the non-redundant database.
For System III, the essential proteins are cytochrome c haem lyase and, in many eukaryotes, cytochrome c1 haem lyase [5]. For System I, the conserved and essential proteins used for our searches were CcmC, CcmE and CcmF [1,4]. We obtained eukaryotic sequences from Arabidopsis thaliana, Oryza sativa, the red alga Cyanidioschyzon merolae and the jakaboid protozoan Reclinomonas americana. From the prokaryotes, we used Rickettsia prowazekii and Magnetospirillum magnetotacticum (α-proteobacteria), Nitrosomonas europaea and Bordetella bronchiseptica (β-proteobacteria), plus E. coli and Azotobacter vinelandii from the γ-proteobacteria. For System II, attention was focused on the proteins ResB and ResC from Bacillus subtilis and their orthologues the nuclear-encoded Ccs1 and the plastid-encoded CcsA from Chlamydomonas reinhardtii [22–24].
RESULTS
XXXCH c-type cytochromes and many respiratory chain components, but absence of a cytochrome c biogenesis system, in trypanosomatids
An analysis of publicly available genome data indicated that the natural occurrence of cytochromes c and c1 with a single-cysteine (XXXCH) rather than a double-cysteine (CXXCH) haem-binding motif remains unique to the mitochondria of Euglena species and trypanosomatids. From the trypanosomatid databases (T. brucei, T. vivax, T. congolense, T. cruzi, L. major and L. infantum) we identified cytochrome c and c1 sequences (five of each) that were not previously available. Each of these cytochromes c and c1 has a conserved single-cysteine haem-binding motif (AAQCH and FAPCH respectively) (Table 1).
Table 1. Cytochromes c and c1 from trypanosomatids contain a conserved single-cysteine haem-binding motif.
A high degree of overall sequence identity is also retained between the different trypanosomatids (>80% and >90% for cytochromes c and c1 respectively).
| Haem-binding motif | ||
|---|---|---|
| Organism | Cytochrome c | Cytochrome c1 |
| Trypanosoma brucei | AAQCH | FAPCH |
| Trypanosoma congolense | Not identified | FAPCH |
| Trypanosoma vivax | AAQCH | FAPCH |
| Trypanosoma cruzi | AAQCH | FAPCH |
| Crithidia oncopelti | AAQCH | FAPCH |
| Crithidia fasciculata | AAQCH | FAPCH |
| Leishmania major | AAQCH | FAPCH |
| Leishmania infantum | AAQCH | FAPCH |
| Euglena gracilis | AAQCH | FAPCH |
| Euglena viridis | AGQCH | Not identified |
| Saccharomyces cerevisiae | CLQCH (iso-1) | CAACH |
| CQQCH (iso-2) | ||
| Schizosaccharomyces pombe | CAQCH | CSACH |
| Homo sapiens | CSQCH | CASCH |
| Mus musculus | CAQCH | CSSCH |
When we asked how these unusual single-cysteine cytochromes are matured, we were astounded to find that there was no evidence for any of the known cytochrome c biogenesis systems, i.e. the Ccm system (System I), System II or haem lyase (System III), in any of the trypanosomatid nuclear or mitochondrial genomes (bioinformatic probes as described above). The T. brucei, T. cruzi and L. major genomes are now frozen as complete, and a vast amount of sequence from six genome projects, EST databases plus trypanosomatid mitochondrial genome sequence [25] is available. Coupled with the high degree of amino acid identity conservation between homologous coding units in the different trypanosomatids, it is extremely unlikely that a conserved biogenesis component(s) remains encoded within DNA that has not been sequenced. Our ability to catalogue candidate genes that encode many of the proteins necessary for or characteristic of assembly or activity of a typical eukaryotic oxidative phosphorylation apparatus (see Supplementary Table 1 at http://www.BiochemJ.org/bj/383/bj3830537add.htm) not only confirms the exhaustive sequence coverage available for each trypanosomatid genome project, but, moreover, provides the necessary and important control analysis to substantiate our claim of the absence of any known cytochrome c biogenesis system. Haem lyase, in conjunction with cleavage of an N-terminal targeting sequence, is involved in the import and assembly of cytochrome c1 in fungal mitochondria. The absence of haem lyase from trypanosomatids may correlate with the observed absence of a cleavable N-terminal sequence on any of the trypanosomatid cytochromes c1 identified; it is notable that cytochrome c1 has recently been shown to be imported into T. brucei mitochondria along an unusual pathway [26].
For our control analysis of the trypanosome and Leishmania respiratory chains, we first identified, by reference to the published composition of Complex I isolated from bovine heart mitochondria and Neurospora crassa [27,28], nuclear-encoded catalytic subunits that are conserved between eukaryotic and prokaryotic NADH:ubiquinone oxidoreductase, and also the acyl-carrier protein subunit identified in the mammalian and fungal complexes (Supplementary Table 1; http://www.BiochemJ.org/bj/383/bj3830537add.htm). Given the paucity of data regarding Complex I function in protozoa, we did not search the trypanosomatid genomes for the presence or absence of the accessory subunits that characterize the mammalian and fungal enzymes. The roles of such subunits are ill-defined and they may not be widely conserved.
Peptide sequences from purified components of C. fasciculata cytochrome bc1 and Leishmania tarentolae cytochrome oxidase [12,29] were used to identify nuclear genes encoding Complex III and Complex IV components in the different trypanosomatid genomes (Supplementary Table 1). Subunits of C. fasciculata cytochrome bc1 [12] were mapped to trypanosome and Leishmania homologues of three peptidases required to cleave import sequences present on other subunits, the Rieske Fe–S protein, cytochrome c1, a homologue of the 14 kDa subunit conserved in other organisms [30], and a protein that has no discernable homologue outside of the Trypanosomatidae. Additionally, we identified a gene encoding the 7.8 kDa homologue of Complex III hinge protein [31]. The cytochrome b subunit is mitochondrially encoded [25]. For trypanosomatid Complex IV, as for that of other eukaryotes, three large hydrophobic subunits encoded by the mitochondrial genome constitute the catalytic core. At least 11 accessory factors are required to complete L. tarentolae cytochrome oxidase [29]. Using the 11 peptide sequences published previously [29], we identified 10 corresponding nuclear genes in the trypanosomatid genomes (peptides from bands 4 and 5 in the published study both map to the same open reading frame), including eight with no apparent homologues outside of the Trypanosomatidae.
For SDH (succinate dehydrogenase; Complex II), the large FAD-containing SDH1 subunit and the smaller Fe–S cluster-binding SDH2 subunit were readily detected in the genomes, albeit with the Fe–S-binding subunit divided such that amino acids with similarity to the N-terminus of Saccharomyces cerevisiae SDH2 were contained on one 27.2 kDa polypeptide and the amino acids that shared identity with the C-terminus of yeast SDH2 were present on a second 21 kDa polypeptide. We also identified (Supplementary Table 1) predicted trypanosomatid orthologues of the heterodimeric ETF (electron-transferring flavoprotein), ETF:ubiquinone oxidoreductase, several conserved components necessary for iron–sulphur cluster biosynthesis and a number of other respiratory chain accessory factors. Based in part on the composition of C. fasciculata F1Fo ATPase, as isolated by Benne and co-workers [32], we also identified nuclear genes for catalytic and accessory subunits of F1Fo ATP synthase (Supplementary Table 1). There was no evidence in the genomes for the presence of copper-containing soluble electron-transfer proteins homologous to azurin or plastocyanin that might provide alternatives to cytochrome c, as happens for example in thylakoids and many bacteria [33].
Expression of T. brucei cytochrome c and maturation of a CXXCH variant in E. coli
The absence of a known biogenesis system raises questions about how haem is attached to the unusual single-cysteine-attached mitochondrial c-type cytochromes of trypanosomes. For example, can the cytochromes c be matured in an uncatalysed process? Also, are there intrinsic features of the trypanosomatid cytochrome c that prevent it from being matured in a double-cysteine form (i.e. with a CXXCH haem-binding motif)? The answers to these questions will provide insight into the endogenous cytochrome c biogenesis pathway in the organisms where the single-cysteine c-type cytochromes are found.
Four expression vectors were constructed. One contained the gene for wild-type T. brucei cytochrome c and another contained the double-cysteine (CXXCH) variant. The third and fourth carried the gene for wild-type or CXXCH T. brucei cytochrome c fused with a periplasmic targeting sequence. E. coli strain JCB387 was separately transformed with each of these plasmids, and cell extracts were assayed spectrophotometrically for holocytochrome c formation. For the periplasmic cytochrome c expression vectors, E. coli were also co-transformed with a plasmid from which the periplasmically functioning Ccm system is expressed [19]; the aerobic growth conditions used largely repress expression of the endogenous Ccm apparatus. Each expression experiment was repeated at least five times.
Under our growth conditions, E. coli produces cytoplasmic b-type cytochromes and a low level of periplasmic c-type cytochromes; reference data for these endogenous cytochrome levels in strain JCB387 are available [3,20]. Attempted expression of T. brucei cytochrome c in the cytoplasm resulted in neither the wild-type nor the CXXCH variant being spectroscopically detectable above the expected background cytochrome levels. Furthermore, the spectra (absorption and pyridine haemochrome maxima) gave no indication of cytoplasmic formation of either a single- or a double-cysteine c-type cytochrome. Additionally, no wild-type T. brucei cytochrome c was observed when such E. coli cytoplasmic extracts were subjected to chromatography using SP-Sepharose to remove any endogenous proteins that may have masked the T. brucei cytochrome in these absorption spectra. Thus we conclude that neither wild-type T. brucei cytochrome c, nor its CXXCH variant, is matured in the cytoplasm of E. coli.
Similarly, when wild-type (AXXCH) T. brucei cytochrome c was directed to the E. coli periplasm, either with or without co-expression of the Ccm system, no increase in cytochrome c expression was observed relative to the expected endogenous (background) levels, while the absorption maxima gave no indication of maturation of a single-cysteine c-type cytochrome. Our previous work has shown that the Ccm apparatus could not mature variants of Hydrogenobacter thermophilus cytochrome c552 with either AXXCH or CXXAH haem-binding motifs, even though both variants formed stable holocytochromes in the E. coli cytoplasm in an exceptional uncatalysed reaction [20]. The present observations complement this earlier failure, but with a cytochrome that has evolved to have only one thioether bond between haem and protein. Exceptional features in the natural single-cysteine cytochromes c from organisms such as T. brucei may have allowed their maturation by the Ccm apparatus, but that possibility can now be excluded. In contrast, a CXXCH variant of T. brucei cytochrome c was matured in the periplasm of E. coli in the presence of the plasmid-expressed Ccm proteins. It is clear, therefore, that the Ccm pathway requires two cysteine residues in the apocytochrome haem-binding motif. Plausible explanations include the possibility that two cysteines are a required recognition feature, and/or that they are required for haem release from the chaperone CcmE, and/or that an intramolecular disulphide bond in the CXXCH motif is an obligate intermediate in haem attachment by the Ccm system.
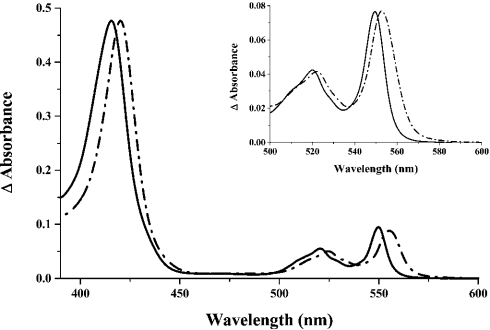
The CXXCH variant of T. brucei cytochrome c produced was purified using SP-Sepharose. Its absorption and pyridine haemochrome spectra are shown in Figure 1. The absorption maxima in the two spectra are absolutely characteristic of a c-type cytochrome with haem bound to protein through two thioether bonds. The β- and α-absorption maxima are at 520.5 and 550 nm respectively; the pyridine haemochrome has its α-band maximum at 549.5 nm, diagnostic of a double thioether linkage. For comparison, the absorption and pyridine haemochrome spectra of native C. fasciculata cytochrome c, a natural single-cysteine cytochrome c, are also shown in Figure 1. As expected for a single rather than a double thioether bond attachment, the bands are broadened and clearly red-shifted from those observed for the CXXCH variant of T. brucei cytochrome c (maxima at 524.5 and 555 nm). Single-cysteine-attached cytochromes c give the pyridine haemochrome α-band maximum at 552–3 nm [9,12], as we observed for our sample of the C. fasciculata protein, which had this peak at 553 nm (Figure 1).
Figure 1. Absorption spectra of the recombinant CXXCH variant of Trypanosoma brucei cytochrome c and of wild-type Crithidia fasciculata cytochrome c.
The CXXCH variant of T. brucei cytochrome c was purified from periplasmic extracts of E. coli cells co-transformed with the plasmid for the E. coli Ccm system. The spectrum (solid line) was recorded at 25 °C with the protein in 50 mM Tris/HCl, pH 7.5, following addition of a few grains of disodium dithionite. Wild-type C. fasciculata cytochrome c was purified from the mitochondria of a chanomastigote culture; the spectrum (broken line) was recorded under the same conditions as above, except at pH 8.0. In the absence of accurate molar absorption coefficients, these spectra have been normalized by the Soret band intensity. Inset: reduced pyridine haemochrome spectra of the CXXCH variant of T. brucei cytochrome c purified from the periplasm of E. coli (solid line) and of C. fasciculata cytochrome c (broken line). The final concentrations of NaOH and pyridine were 0.2 M and 30% (v/v) respectively; spectra were normalized by the α-band intensity.
The purified T. brucei CXXCH cytochrome c was analysed by electrospray MS. A peak was observed corresponding to a mass of 12918 Da, in agreement with the expected mass of T. brucei CXXCH cytochrome c with covalently bound haem (12920 Da). (DNA sequencing of the cytochrome c gene that we cloned from genomic DNA isolated from 427 strain trypanosomes indicated the presence of an alanine as residue 15 of the mature peptide, whereas in the genome reference strain this residue is valine, implying some natural variability at this position. Our calculated mass is for the protein with alanine-15). The N-terminal region of the protein was found to be as expected by peptide sequencing. In combination, our analytical data confirm categorically that the c-type cytochrome overexpressed in the periplasm of E. coli was complete T. brucei cytochrome c with a CXXCH motif to which haem was covalently attached through two thioether bonds. The yield of such cytochrome c produced periplasmically in E. coli with co-expression of the Ccm apparatus was approx. 1.6 mg per g of wet cells, assuming a reduced Soret band molar absorption coefficient of 130 mM−1·cm−1 (the value for horse heart mitochondrial cytochrome c). These results demonstrate that there is no intrinsic reason why T. brucei cytochrome c cannot accommodate a second cysteine in the typical CXXCH cytochrome c haem-binding motif; our CXXCH variant is clearly folded and stable. The expression of this variant also provides a positive control for our experiments in which T. brucei cytochrome c maturation was not observed in E. coli (i.e. for the periplasmic AXXCH, cytoplasmic AXXCH and cytoplasmic CXXCH forms).
DISCUSSION
To date, three cytochrome c biogenesis systems have been described, but our analysis shows that none of these operates in trypanosomatid mitochondria. This striking observation correlates with the occurrence of single-cysteine (XXXCH) mitochondrial cytochromes c and c1 exclusively within the phylum Euglenozoa, to which the trypanosomatids and euglenids, such as Euglena gracilis and Euglena viridis, belong. In contrast with the absence of a known cytochrome c biogenesis system, we readily detected the major required components and accessory proteins of the mitochondrial oxidative phosphorylation apparatus in the genomes of six different trypanosomatid parasites (Supplementary Table 1; http://www.BiochemJ.org/bj/383/bj3830537add.htm).
In the absence of a recognizable cytochrome c biogenesis apparatus in these organisms, one possibility is that the haem covalently attaches to apocytochrome c in an uncatalysed reaction. However, this seems highly unlikely even in principle, given the complexity of the maturation systems that have evolved in prokaryotes and the precise stereocontrol of attachment of the asymmetrical molecule haem that is observed in cytochromes c [2]. One might consider that stable and correct maturation in the E. coli cytoplasm of cytochrome c552 from the thermophile H. thermophilus sets a precedent for spontaneous maturation of c-type cytochromes [34,35]. However, although the reaction can also be reconstituted in vitro, the rate is very low [6], and all other attempts to produce holocytochromes c without a maturation apparatus have led to mixed products in vitro or in vivo [7,8,36–38]. The mitochondrial cytochromes c from humans and yeast, expressed as either double-cysteine (wild type) or single-cysteine (variant) forms, are stable in the E. coli cytoplasm when matured by haem lyase (e.g. [39–42]), and T. brucei CXXCH cytochrome c is stable in the periplasm (this work). Thus our failure to observe wild-type T. brucei cytochrome c formation in either the cytoplasm or periplasm of E. coli leads us to conclude that spontaneous formation is not the route taken for cytochrome c maturation by the Euglenozoa.
In combination, our biochemical and bioinformatic observations indicate that a novel fourth biogenesis system operates in trypanosomatids, and by inference in euglenids. Such a system may be further differentiated, since it needs to mature two cytochromes (c and c1). The single-cysteine haem attachment conserved in these species can be argued to be the result of a positive evolutionary process, possibly linked to the environment in the mitochondrial intermembrane space where the cytochromes are located. Perhaps there is a powerful oxidizing protein (such as the conserved Erv1) in the intermembrane space of Euglenozoid mitochondria that would irreversibly oxidize the CXXCH motif of a typical apocytochrome c to a disulphide bond, preventing haem attachment. A disulphide bond has been shown to form in several apocytochromes c [6–8], but would be avoided by the apocytochrome having only one cysteine residue. Another possibility is that acquisition of a novel cytochrome c biogenesis system by the Euglenozoa provided not just the opportunity through which an existing maturation system, such as haem lyase, could be lost, but also the evolutionary pressure to move from CXXCH to XXXCH cytochromes c. Notably, the available biophysical data suggest that single-cysteine-attached cytochromes c have reduction potentials and stabilities comparable with those of their double-cysteine-attached counterparts [9,42,43].
Crystal structures of the cytochrome b6f complexes from the Chlamydomonas reinhardtii chloroplast and the cyanobacterium Mastigocladus laminosus intriguingly show, in addition to the expected cytochrome b and cytochrome f centres, a novel covalently bound haem [44,45]. As in the mitochondrial cytochromes c of the Euglenozoa, this haem is covalently bound to the polypeptide through one thioether bond between a cysteine residue and a vinyl group. Four mutants have been identified that are unable to make this newly identified covalent link in the Chlamydomonas reinhardtii b6 f complex [46]. Originally interpreted as a requirement for b-type cytochrome biogenesis proteins, it may now transpire that within these gene loci [46] another distinct ‘c-type’ cytochrome biogenesis system is encoded [47]. Trypanosomatids share many metabolic traits with higher plants that are yet to be adequately explained [48,49]. In this context, our intriguing findings raise the possibility that the arrival in the nucleus of an euglenid/trypanosomatid progenitor of a biogenesis machinery required for the single thioether covalent haem attachment in plastid cytochromes b6f, and its subsequent co-option into the maturation pathway for mitochondrial cytochromes c, might constitute a further plant-like metabolic trait. It should be noted, however, that the cytochromes b from the trypanosomatid bc1 complexes do not contain the cysteine to which the novel haem of the b6f complex is attached, but this cysteine is universally conserved among the b6f enzymes. If the machinery for covalent attachment of haem to the b6f complex is not related to the cytochrome c biogenesis system of trypanosomatids, there are two new systems for thioether bond formation between haem and apocytochromes awaiting identification.
Online data
Acknowledgments
This work was funded by BBSRC grants C13443 and BB/C508118/1 (to S. J. F. and M. L. G.), the Royal Society (to M. L. G., who is a Royal Society University Research Fellow) and a Wellcome Trust programme grant awarded to Keith Gull. We are grateful to the E. P. Abraham Trust for support for J. W. A. A. We acknowledge The Institute for Genome Research, Seattle Biomedical Research Institute, The Karolinska Institute and The Sanger Institute for the public availability of the trypanosomatid genome sequences; support for these projects was provided by The National Institute of Allergy and Infectious Diseases (NIH) and The Wellcome Trust. We gratefully acknowledge Keith Gull for support and useful discussions, Anthony Willis for N-terminal sequencing, and Julie Stevens and Bill Wickstead for helpful discussion.
References
- 1.Thöny-Meyer L. Haem-polypeptide interactions during cytochrome c maturation. Biochim. Biophys. Acta. 2000;1459:316–324. doi: 10.1016/s0005-2728(00)00167-5. [DOI] [PubMed] [Google Scholar]
- 2.Barker P. D., Ferguson S. J. Still a puzzle: why is haem covalently attached in c-type cytochromes? Struct. Fold Des. 1999;7:R281–R290. doi: 10.1016/s0969-2126(00)88334-3. [DOI] [PubMed] [Google Scholar]
- 3.Allen J. W. A., Ferguson S. J. Variation of the axial haem ligands and haem-binding motif as a probe of the Escherichia coli c-type cytochrome maturation (Ccm) system. Biochem. J. 2003;375:721–728. doi: 10.1042/BJ20030752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen J. W. A., Daltrop O., Stevens J. M., Ferguson S. J. c-Type cytochromes: diverse structures and biogenesis systems pose evolutionary problems. Philos. Trans. R. Soc. London B Biol. Sci. 2003;358:255–266. doi: 10.1098/rstb.2002.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard D. G., Gabilly S. T., Dujardin G., Merchant S., Hamel P. P. Overlapping specificities of the mitochondrial cytochrome c and c1 heme lyases. J. Biol. Chem. 2003;278:49732–49742. doi: 10.1074/jbc.M308881200. [DOI] [PubMed] [Google Scholar]
- 6.Daltrop O., Allen J. W. A., Willis A. C., Ferguson S. J. In vitro formation of a c-type cytochrome. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7872–7876. doi: 10.1073/pnas.132259099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daltrop O., Ferguson S. J. Cytochrome c maturation. The in vitro reactions of horse heart apocytochrome c and Paracoccus dentrificans apocytochrome c550 with heme. J. Biol. Chem. 2003;278:4404–4409. doi: 10.1074/jbc.M211124200. [DOI] [PubMed] [Google Scholar]
- 8.Allen J. W. A., Barker P. D., Ferguson S. J. A cytochrome b562 variant with a c-type cytochrome CXXCH heme-binding motif as a probe of the Escherichia coli cytochrome c maturation system. J. Biol. Chem. 2003;278:52075–52083. doi: 10.1074/jbc.M307196200. [DOI] [PubMed] [Google Scholar]
- 9.Pettigrew G. W., Leaver J. L., Meyer T. E., Ryle A. P. Purification, properties and amino acid sequence of atypical cytochrome c from two protozoa, Euglena gracilis and Crithidia oncopelti. Biochem. J. 1975;147:291–302. doi: 10.1042/bj1470291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukai K., Yoshida M., Toyosaki H., Yao Y., Wakabayashi S., Matsubara H. An atypical heme-binding structure of cytochrome c1 of Euglena gracilis mitochondrial complex III. Eur. J. Biochem. 1989;178:649–656. doi: 10.1111/j.1432-1033.1989.tb14494.x. [DOI] [PubMed] [Google Scholar]
- 11.Ambler R. P., Kamen M. D., Bartsch R. G., Meyer T. E. Amino acid sequences of Euglena viridis ferredoxin and cytochromes c. Biochem. J. 1991;276:47–52. doi: 10.1042/bj2760047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Priest J. W., Hajduk S. L. Cytochrome c reductase purified from Crithidia fasciculata contains an atypical cytochrome c1. J. Biol. Chem. 1992;267:20188–20195. [PubMed] [Google Scholar]
- 13.Torri A. F., Hajduk S. L. Posttranscriptional regulation of cytochrome c expression during the developmental cycle of Trypanosoma brucei. Mol. Cell. Biol. 1988;8:4625–4633. doi: 10.1128/mcb.8.11.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santillan Torres J. L., Atteia A., Claros M. G., Gonzalez-Halphen D. Cytochrome f and subunit IV, two essential components of the photosynthetic bf complex typically encoded in the chloroplast genome, are nucleus-encoded in Euglena gracilis. Biochim. Biophys. Acta. 2003;1604:180–189. doi: 10.1016/s0005-2728(03)00058-6. [DOI] [PubMed] [Google Scholar]
- 15.Pettigrew G. W. The purification and amino acid sequence of cytochrome C-552 from Euglena gracilis. Biochem. J. 1974;139:449–459. doi: 10.1042/bj1390449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kranz R. G., Beckett C. S., Goldman B. S. Genomic analyses of bacterial respiratory and cytochrome c assembly systems: Bordetella as a model for the system II cytochrome c biogenesis pathway. Res. Microbiol. 2002;153:1–6. doi: 10.1016/s0923-2508(01)01278-5. [DOI] [PubMed] [Google Scholar]
- 17.Nakamoto S. S., Hamel P., Merchant S. Assembly of chloroplast cytochromes b and c. Biochimie. 2000;82:603–614. doi: 10.1016/s0300-9084(00)00605-2. [DOI] [PubMed] [Google Scholar]
- 18.Hussain H., Grove J., Griffiths L., Busby S., Cole J. A seven-gene operon essential for formate-dependent nitrite reduction to ammonia by enteric bacteria. Mol. Microbiol. 1994;12:153–163. doi: 10.1111/j.1365-2958.1994.tb01004.x. [DOI] [PubMed] [Google Scholar]
- 19.Arslan E., Schulz H., Zufferey R., Kunzler P., Thöny-Meyer L. Overproduction of the Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem. Biophys. Res. Commun. 1998;251:744–747. doi: 10.1006/bbrc.1998.9549. [DOI] [PubMed] [Google Scholar]
- 20.Allen J. W. A., Tomlinson E. J., Hong L., Ferguson S. J. The Escherichia coli cytochrome c maturation (Ccm) system does not detectably attach heme to single cysteine variants of an apocytochrome c. J. Biol. Chem. 2002;277:33559–33563. doi: 10.1074/jbc.M204963200. [DOI] [PubMed] [Google Scholar]
- 21.Bartsch R. G. Cytochromes: bacterial. Methods Enzymol. 1971;23:344–363. [Google Scholar]
- 22.Le Brun N. E., Bengtsson J., Hederstedt L. Genes required for cytochrome c synthesis in Bacillus subtilis. Mol. Microbiol. 2000;36:638–650. doi: 10.1046/j.1365-2958.2000.01883.x. [DOI] [PubMed] [Google Scholar]
- 23.Dreyfuss B. W., Hamel P. P., Nakamoto S. S., Merchant S. Functional analysis of a divergent system II protein, Ccs1, involved in c-type cytochrome biogenesis. J. Biol. Chem. 2003;278:2604–2613. doi: 10.1074/jbc.M208652200. [DOI] [PubMed] [Google Scholar]
- 24.Hamel P. P., Dreyfuss B. W., Xie Z., Gabilly S. T., Merchant S. Essential histidine and tryptophan residues in CcsA, a system II polytopic cytochrome c biogenesis protein. J. Biol. Chem. 2003;278:2593–2603. doi: 10.1074/jbc.M208651200. [DOI] [PubMed] [Google Scholar]
- 25.Simpson L., Wang S. H., Thiemann O. H., Alfonzo J. D., Maslov D. A., Avila H. A. U-insertion/deletion Edited Sequence Database. Nucleic Acids Res. 1998;26:170–176. doi: 10.1093/nar/26.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Priest J. W., Hajduk S. L. Trypanosoma brucei cytochrome c1 is imported into mitochondria along an unusual pathway. J. Biol. Chem. 2003;278:15084–15094. doi: 10.1074/jbc.M212956200. [DOI] [PubMed] [Google Scholar]
- 27.Hirst J., Carroll J., Fearnley I. M., Shannon R. J., Walker J. E. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim. Biophys. Acta. 2003;1604:135–150. doi: 10.1016/s0005-2728(03)00059-8. [DOI] [PubMed] [Google Scholar]
- 28.Videira A., Duarte M. From NADH to ubiquinone in Neurospora mitochondria. Biochim. Biophys. Acta. 2002;1555:187–191. doi: 10.1016/s0005-2728(02)00276-1. [DOI] [PubMed] [Google Scholar]
- 29.Horvath A., Kingan T. G., Maslov D. A. Detection of the mitochondrially encoded cytochrome c oxidase subunit I in the trypanosomatid protozoan Leishmania tarentolae. Evidence for translation of unedited mRNA in the kinetoplast. J. Biol. Chem. 2000;275:17160–17165. doi: 10.1074/jbc.M907246199. [DOI] [PubMed] [Google Scholar]
- 30.Braun H. P., Schmitz U. K. Molecular features and mitochondrial import pathway of the 14-kilodalton subunit of cytochrome c reductase from potato. Plant Physiol. 1995;107:1217–1223. doi: 10.1104/pp.107.4.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braun H. P., Jansch L., Kurft V., Schmitz U. K. The ‘Hinge’ protein of cytochrome c reductase from potato lacks the acidic domain and has no cleavable presequence. FEBS Lett. 1994;347:90–94. doi: 10.1016/0014-5793(94)00515-x. [DOI] [PubMed] [Google Scholar]
- 32.Speijer D., Breek C. K., Muijsers A. O., Hartog A. F., Berden J. A., Albracht S. P., Samyn B., Van Beeumen J., Benne R. Characterization of the respiratory chain from cultured Crithidia fasciculata. Mol. Biochem. Parasitol. 1997;85:171–186. doi: 10.1016/s0166-6851(96)02823-x. [DOI] [PubMed] [Google Scholar]
- 33.Pearson I. V., Page M. D., van Spanning R. J., Ferguson S. J. A mutant of Paracoccus denitrificans with disrupted genes coding for cytochrome c550 and pseudoazurin establishes these two proteins as the in vivo electron donors to cytochrome cd1 nitrite reductase. J. Bacteriol. 2003;185:6308–6315. doi: 10.1128/JB.185.21.6308-6315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambongi Y., Crooke H., Cole J. A., Ferguson S. J. A mutation blocking the formation of membrane or periplasmic endogenous and exogenous c-type cytochromes in Escherichia coli permits the cytoplasmic formation of Hydrogenobacter thermophilus holo cytochrome c552. FEBS Lett. 1994;344:207–210. doi: 10.1016/0014-5793(94)00399-8. [DOI] [PubMed] [Google Scholar]
- 35.Karan E. F., Russell B. S., Bren K. L. Characterization of Hydrogenobacter thermophilus cytochromes c552 expressed in the cytoplasm and periplasm of Escherichia coli. J. Biol. Inorg. Chem. 2002;7:260–272. doi: 10.1007/s007750100292. [DOI] [PubMed] [Google Scholar]
- 36.Keightley J. A., Sanders D., Todaro T. R., Pastuszyn A., Fee J. A. Cloning and expression in Escherichia coli of the cytochrome c552 gene from Thermus thermophilus HB8. Evidence for genetic linkage to an ATP-binding cassette protein and initial characterization of the cycA gene products. J. Biol. Chem. 1998;273:12006–12016. doi: 10.1074/jbc.273.20.12006. [DOI] [PubMed] [Google Scholar]
- 37.Barker P. D., Nerou E. P., Freund S. M., Fearnley I. M. Conversion of cytochrome b562 to c-type cytochromes. Biochemistry. 1995;34:15191–15203. doi: 10.1021/bi00046a027. [DOI] [PubMed] [Google Scholar]
- 38.Barker P. D., Ferrer J. C., Mylrajan M., Loehr T. M., Feng R., Konishi Y., Funk W. D., MacGillivray R. T., Mauk A. G. Transmutation of a heme protein. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6542–6546. doi: 10.1073/pnas.90.14.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pollock W. B., Rosell F. I., Twitchett M. B., Dumont M. E., Mauk A. G. Bacterial expression of a mitochondrial cytochrome c. Trimethylation of lys72 in yeast iso-1-cytochrome c and the alkaline conformational transition. Biochemistry. 1998;37:6124–6131. doi: 10.1021/bi972188d. [DOI] [PubMed] [Google Scholar]
- 40.Olteanu A., Patel C. N., Dedmon M. M., Kennedy S., Linhoff M. W., Minder C. M., Potts P. R., Deshmukh M., Pielak G. J. Stability and apoptotic activity of recombinant human cytochrome c. Biochem. Biophys. Res. Commun. 2003;312:733–740. doi: 10.1016/j.bbrc.2003.10.182. [DOI] [PubMed] [Google Scholar]
- 41.Martin A. G., Fearnhead H. O. Apocytochrome c blocks caspase-9 activation and Bax-induced apoptosis. J. Biol. Chem. 2002;277:50834–50841. doi: 10.1074/jbc.M209369200. [DOI] [PubMed] [Google Scholar]
- 42.Rosell F. I., Mauk A. G. Spectroscopic properties of a mitochondrial cytochrome c with a single thioether bond to the heme prosthetic group. Biochemistry. 2002;41:7811–7818. doi: 10.1021/bi016060e. [DOI] [PubMed] [Google Scholar]
- 43.Tomlinson E. J., Ferguson S. J. Loss of either of the two heme-binding cysteines from a class I c-type cytochrome has a surprisingly small effect on physicochemical properties. J. Biol. Chem. 2000;275:32530–32534. doi: 10.1074/jbc.M004022200. [DOI] [PubMed] [Google Scholar]
- 44.Stroebel D., Choquet Y., Popot J. L., Picot D. An atypical haem in the cytochrome b6f complex. Nature (London) 2003;426:413–418. doi: 10.1038/nature02155. [DOI] [PubMed] [Google Scholar]
- 45.Kurisu G., Zhang H., Smith J. L., Cramer W. A. Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science. 2003;302:1009–1014. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- 46.Kuras R., de Vitry C., Choquet Y., Girard-Bascou J., Culler D., Buschlen S., Merchant S., Wollman F. A. Molecular genetic identification of a pathway for heme binding to cytochrome b6. J. Biol. Chem. 1997;272:32427–32435. doi: 10.1074/jbc.272.51.32427. [DOI] [PubMed] [Google Scholar]
- 47.De Vitry C., Desbois A., Redeker V., Zito F., Wollman F. A. Biochemical and spectroscopic characterization of the covalent binding of heme to cytochrome b6. Biochemistry. 2004;43:3956–3968. doi: 10.1021/bi036093p. [DOI] [PubMed] [Google Scholar]
- 48.Waller R. F., McConville M. J., McFadden G. I. More plastids in human parasites? Trends Parasitol. 2004;20:54–57. doi: 10.1016/j.pt.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 49.Leander B. S. Did trypanosomatid parasites have photosynthetic ancestors? Trends Microbiol. 2004;12:251–258. doi: 10.1016/j.tim.2004.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.