Abstract
We studied whether ethanol is sulphonated in humans with the perspective of using the urinary excretion of ethyl sulphate after ethanol consumption as a biomarker for SULT (sulphotransferase) activity. We developed a sensitive and selective HPLC–MS/MS method for determining ethyl sulphate in urine. Ten volunteers received a low dose of ethanol (0.1 g/kg of body mass). In general, excretion of ethyl sulphate was maximal in the first or second hour after dosage. Within 8 h, 2.5–6.8 μmol of ethyl sulphate was excreted. A 5-fold increase in the dose of ethanol led to an increase in the amount of ethyl sulphate excreted within 8 h (28–95 μmol) and the presence of this metabolite in urine for at least 24 h. Since ethyl sulphate was still being excreted for a substantial period after the elimination of ethanol, it might be used as a medium-time biomarker for preceding ethanol consumption. We have expressed previously all human SULT forms identified in Salmonella typhimurium. Ethanol sulphonation was studied in cytosolic preparations of these strains. The highest activities were observed with SULT1A2, 1B1 and 1C2, followed by 1A3. Activities were markedly lower with SULT1E1, 1A1 and 2A1, and were negligible with SULT1C1, 2B1a, 2B1b and 4A1. If the expression levels in tissues are additionally taken into account, SULT1A3 might be the predominant form for the sulphonation of ethanol in vivo, although a robust estimate requires further studies. With this limitation, urinary ethyl sulphate excretion appears very promising as a biomarker for SULT activity in vivo.
Keywords: alcohol, biomarker, ethyl sulphate, phenotyping, sulpho-conjugation, sulphotransferase (SULT)
Abbreviations: MRM, multiple reaction monitoring; PAPS, 3′-phosphoadenosine-5′-phosphosulphate; SULT, sulphotransferase; UGT, UDP–glucuronosyltransferase
INTRODUCTION
Sulpho-conjugation is an important biotransformation reaction for drugs and other xenobiotics [1–3]. It involves the enzyme-mediated transfer of the sulpho group from the co-substrate PAPS (3′-phosphoadenosine-5′-phosphosulphate) to a nucleophilic site (most often an hydroxy group) of the substrate. The conjugation often terminates the pharmacological activity of the drug and usually facilitates the excretion of the acceptor molecule. However, a few pro-drugs and a substantial number of pro-carcinogens/promutagens are bioactivated via sulpho-conjugation. The sulpho-conjugation of small molecules (as opposed to proteins and polysaccharides) is mediated by soluble sulphotransferases, which are members of a common gene/enzyme superfamily, termed SULT. A total of 11 SULT forms have been identified in humans. Some forms have demonstrated functional genetic polymorphisms [4,5]. Furthermore, it was shown in animal experiments that a number of physiological and dietary factors (such as hormonal state, menstrual cycle, blood pressure, obesity, food deprivation and selenium deficiency) affect the expression of SULTs [2]. Furthermore, food contains numerous substrates and inhibitors of SULTs, such as polyphenolic plant metabolites and environmental contaminants [2,6,7]. For these various reasons, biomarkers reflecting the in vivo SULT activity would be interesting. In principle, a potential biomarker for sulpho-conjugation is a metabolite that is formed in humans after uptake of the respective substrate. In practice, such a compound must be safe and well accepted by the study subjects.
The analgesic drug, paracetamol, was frequently used for studying sulpho-conjugation in vivo (e.g. [8–10]). This drug is chiefly excreted in urine as sulpho- and glucuronic acid-conjugates. In general, the ratio of these paracetamol metabolites was used for phenotyping. However, the rate of paracetamol glucuronidation in liver microsomes is subjected to large inter-individual variation [11]. Some UGTs (UDP–glucuronosyltransferases) involved in this reaction in vitro are genetically polymorphic and/or inducible by various xenobiotics. Likewise, the use of certain drugs (such as anticonvulsives) and smoking were shown to enhance paracetamol glucuronidation, but not its sulphonation, in vivo [8,10]. This varying influence of UGTs affects and complicates the use of the ratio of sulpho- and glucuronic acid-conjugates of paracetamol as a measure of sulpho-conjugation. Other substrates (salicylamide, methyldopa, diflunisal and ritodrine) have been used less often than paracetamol for studying sulpho-conjugation in vivo, but share the same problem of the occurrence of variable competing pathways, i.e. glucuronidation or methylation [12–15].
In the present study, we explored whether ethanol could be used as a SULT substrate in humans in vivo, since the elimination kinetics of this compound are well known and show low inter-individual variation. Moreover, the actual substrate concentration can be followed readily. Ethanol is chiefly metabolized by oxidation via ethanal (acetaldehyde) and ethanoic acid (acetic acid). A minor pathway (<0.1% of the dose) has been detected, leading to the urinary excretion of ethyl glucuronide in humans [16–19]. This pathway was studied extensively as a mid-term biomarker for monitoring preceding ethanol consumption. Urinary ethyl glucuronide was detected up to 85 h after consumption of ethanol. Ethanol contains an hydroxy group and therefore is also a potential substrate of SULTs, although this pathway has not yet been described in man. However, a small number of studies reported the sulpho-conjugation of ethanol in subcelluar preparations from the rat, dog and rabbit or in the rat in vivo [20–23].
Our study goal required the development of a sensitive method for detecting ethyl sulphate in a urinary matrix, the actual analysis of human urine before and after consumption of ethanol and, in the case of a positive result, the identification of the human SULT forms involved in this conjugation reaction.
EXPERIMENTAL
Chemicals
Potassium ethyl sulphate (<98%) was obtained from ABCR (Karlsruhe, Germany). 2-Propyl sulphate was synthesized as described previously [24]. All solvents were of HPLC gradient grade and were filtered through 0.2-μm-pore-size membrane filters. All other chemicals were at least of p.a. grade.
PAPS was prepared using human PAPS synthetase 2 expressed in Escherichia coli, and was purified using anion-exchange chromatography (purity >99.5%) [25].
Chromatographic system
A Waters 2960 HPLC system (Eschborn, Germany) was connected to a triple-quadrupole mass spectrometer fitted with an electrospray source (Quattro II with Z-spray source; Micromass, Manchester, U.K.). Samples were separated on a Hypersil Duet SAX column (150 mm×4.6 mm, 5 μm; Thermoquest, Egelsbach, Germany). A Hypersil BDS C18 (10 mm×3 mm) was used as a guard column. Separation was performed isocratically with water/methanol (7:3, v/v) containing 0.05% (w/v) triethylamine and adjusted to pH 4.0. The elution rate was 0.6 ml/min and the column temperature was 25 °C.
The MS/MS methods were worked out with ethyl and 2-propyl sulphate dissolved in water or the mobile phase. After optimization, detection was carried out using negative electrospray ionization in the MRM (multiple reaction monitoring) mode. The cone voltage was 15 V and the collision energy was 15 eV. The source block and the desolvation temperature were maintained at 80 and 350 °C respectively.
Validation of the determination of ethyl sulphate in urinary samples
Urine of subjects abstaining from ethanol for 2 days was spiked with 5, 20 and 50 μM ethyl sulphate and 20 μM 2-propyl sulphate (used as an internal standard). Samples and blank urine were treated as described below. Recoveries were calculated using the standards directly dissolved in the mobile phase at the same final concentration. Standard stock solutions were stable at 4 °C for at least 1 week.
Preparation of urinary samples
Stratagene NH2-cartridges (100 mg; Phenomenex, Darmstadt, Germany) were conditioned with 1 ml of methanol and 2 ml of ammonium acetate buffer (100 μM, pH 5). A 0.75 ml aliquot of urine, supplemented with 20 μM internal standard 2-propyl sulphate, was added to the column followed by 0.75 ml of buffer. Unlike common solid-phase extractions, the eluate and the elution buffer were collected directly without any washing step. Neither ion-exchange nor RP-18 phases were able to retain ethyl sulphate.
Studies in humans
Urinary excretion of ethyl sulphate was tested in ten healthy 25- to 52-year-old volunteers. Seven subjects were female (subjects 1–7), the others were male. All but one of the subjects (no. 7) were non-smokers. The study subjects did not consume any alcoholic beverages for at least 48 h and refrained from excessive consumption for more than 1 week before the trial. Abstinence was verified with blank urine sampled before ethanol consumption. The subjects consumed sparkling wine corresponding to 0.1 g (2.2 mmol) of ethanol per kg of body mass within 5 min. Two subjects received 0.5 g (10.9 mmol) of ethanol per kg of body mass on further occasions. Total urine was collected at time intervals specified in Figures 3 and 4 over 8–12 h. After 24 h, urine was sampled randomly. Samples were stored at −80 °C until analysis.
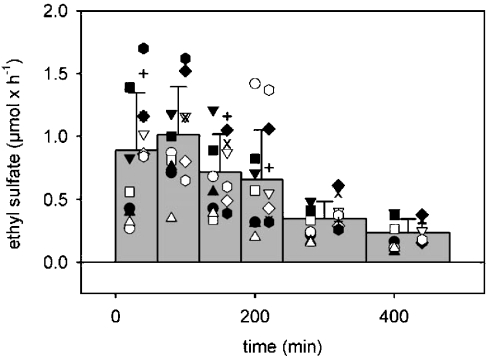
Figure 3. Time course of the urinary excretion of ethyl sulphate after consumption of 0.1 g of ethanol per kg of body mass.
Ethanol was consumed as sparkling wine within 5 min. Total urine was collected in time intervals indicated by the width of the bars. The bars indicate the means of all subjects and repeat experiments presented in Table 2. The symbols represent the individual subjects and experiments.
Figure 4. Time course of the urinary excretion of ethyl sulphate after consumption of 0.5 g of ethanol per kg of body mass.
Ethanol was consumed as sparkling wine within 5 min. Total urine was collected in time intervals indicated by the width of the bars. The bars indicate the means of both subjects and repeat experiments (see Table 2). Open and solid circles: subject 6, first and second experiment respectively; open and solid squares: subject 10, first and second experiment respectively.
Determination of the level of ethanol in urine
Urinary ethanol was determined enzymically as described elsewhere [26] with minor modification. Briefly, up to 50 μl of urine was incubated in a total volume of 1000 μl of phosphate buffer (75 mM, pH 8.7) containing 0.75 mM NAD+, 75 mM semicarbazide as a trapping agent, 20 μM glycine and 88 units of alcohol dehydrogenase from yeast. Ethanol concentrations were estimated from the formation of NADH measured photometrically at 340 nm after 20 min.
cDNA-expressed human SULTs
The expression of human SULTs in Salmonella typhimurium strain TA1538 has been described elsewhere [27]. The amino acid sequences of all enzymes used in the present study represent the wild-type (reference) forms. In the case of SULT1A2, we used a strain (TA1538-SULT1A2*1Z) containing silent mutations in the 5′-terminal region to increase the expression level [27]. The same approach was used to enhance the expression of SULT2B1b in strain TA1538-SULT2B1b*1X (M. Osterloh, W. Meinl and H. R. Glatt, unpublished work). Using purified enzyme protein as a standard, the expression levels could be estimated in some strains by immunoblotting. SULT1A1 [27] and SULT1B1 (W. Teubner, W. Meinl and H. R. Glatt, unpublished work) constitute approx. 0.7–1.4% of the cytosolic protein of the corresponding strains. No purified standards were available for the other SULTs. However, after electrophoresis of cytosolic preparations on polyacrylamide gels under denaturing conditions and Coomassie Blue staining, an additional protein band (which also reacted with the appropriate anti-SULT antibody) was detected in each recombinant strain. We estimate that the intensity of the additional band varied from 3-fold weaker to 3-fold stronger compared with SULT1A1 and SULT1B1 in strains TA1538-SULT1A1 and TA1538-SULT1B1.
Determination of the sulphonation of ethanol in the presence of cDNA-expressed SULTs
Cytosolic fractions were prepared from recombinant S. typhimurium strains as described previously [28]. Incubations contained cytosolic fraction (0.1 mg of protein), the cofactor PAPS (100 μM), the substrate ethanol (0–1000 mM), potassium phosphate buffer (50 mM, pH 7.4) and MgCl2 (5 mM) in a total volume of 100 μl. After an incubation at 37 °C for 60 min, the reaction was stopped by adding 200 μl of propan-2-ol. After centrifugation at 2000 g for 5 min, aliquots (20 μl) were analysed directly by HPLC–MS/MS. External standards of ethyl sulphate were prepared in the same incubation mixture, but without ethanol and with the use of cytosolic preparation from the parental S. typhimurium strain rather than SULT-expressing strains.
RESULTS
Development of the method for detecting ethyl sulphate
Simple negative ionization of ethyl sulphate resulted in a characteristic mass spectrum, showing two major signals, representing the molecular ion (M-H, m/z=125) and HSO4− (m/z=97) (Figure 1, upper trace). Likewise, 2-propyl sulphate produced analogous signals, reflecting the molecular ion (M-H, m/z=139) and HSO4− (m/z=97) (Figure 1, lower trace). These fragments were used for detecting ethyl and 2-propyl sulphate in the MRM mode. They were specific for these compounds in the matrices investigated, as no other signals, even with other retention times, were observed in the HPLC runs. Representative chromatograms with spiked and non-spiked control urines are shown in Figures 2(A) and 2(B). However, a strong signal representing ethyl sulphate was detected in urine collected in the 2–3 h interval after the consumption of 0.1 g of ethanol per kg of body mass (Figure 2C, upper trace). The signal was nearly 50 times above the limit of detection. 2-Propyl sulphate, which had a similar retention time to that of ethyl sulphate, did not interfere with the detection of the latter compound. It was found in non-spiked urine neither before nor after consumption of alcoholic beverages (results not shown).
Figure 1. Negative electrospray ionization mass spectra of ethyl sulphate (upper trace) and 2-propyl sulphate (lower trace).
Two main ions, the molecular ion (m/z=125 and 139 respectively) and HSO4− (m/z=97) were formed under the conditions selected (cone voltage, 25 V; source temperature, 80 °C; desolvation temperature, 350 °C). They were then used for detecting ethyl sulphate and 2-propyl sulphate in the MRM mode.
Figure 2. HPLC–MS/MS chromatograms for detecting ethyl sulphate and 2-propyl sulphate.
(A) Blank urine was spiked with 20 μM ethyl sulphate and 10 μM 2-propyl sulphate respectively; (B) a blank urine taken before consumption of ethanol spiked with the internal standard 2-propyl sulphate; (C) a urinary sample of the same subject collected between 2 and 3 h after ethanol intake (0.1 g/kg of body mass). Peaks are labelled with the retention time and the absolute area under the peak. In all cases, 20 μl of sample (equivalent to 10 μl of urine) was injected. Upper traces in each panel show ethyl sulphate (m/z for molecular and daughter ions 125 and 97 respectively); lower traces in each panel show 2-propyl sulphate (m/z for molecule and daughter ions 139 and 97 respectively).
It was not possible to retain ethyl sulphate in any of the solidphase extraction columns tested. Although the solid-phase extraction step was not critical for our method, we used it to remove urinary components that might interfere with the ionization and fragmentation of ethyl and 2-propyl sulphate in the HPLC–MS/MS system and/or lead to an enhanced aging of the HPLC columns.
The intra- and inter-day accuracy and precision of this method were very good (Table 1).
Table 1. Intra-day and inter-day accuracy and precision of the determination of ethyl sulphate in spiked urine.
After solid-phase extraction, ethyl sulphate was analysed by HPLC–MS/MS. The resulting signal was compared with the signal observed after direct injection of a solution of ethyl sulphate into the mobile phase. For the intra-day comparison, six urine samples were used with each spike-concentration level. For the inter-day comparison, three urine samples were used on each of 3 days for each condition.
| Intra-day comparison | Inter-day comparison | |||
|---|---|---|---|---|
| Concentration (μM) | Accuracy* | Precision† | Accuracy* | Precision† |
| 50 | 102.3 | 6.0 | 100.5 | 7.1 |
| 20 | 100.7 | 1.6 | 95.6 | 5.6 |
| 5 | 90.4 | 4.0 | 88.1 | 5.5 |
* Recovery (%).
† S.D. as a percentage of the mean.
Urinary excretion of ethyl sulphate after consumption of ethanol
Ethyl sulphate was not detected in any urinary samples taken from the test subjects immediately before alcohol consumption. However, it appeared shortly afterwards it both dose groups, 0.1 g of ethanol per kg of body mass (Figure 3) and 0.5 g of ethanol per kg of body mass (Figure 4). Maximal excretion rates were usually reached in the urine sample collected in the 0 –1 h or 1–2 h periods after alcohol consumption, and sporadically in the 2–3 h or 3 – 4 h samples. In all subjects, total urine was systematically collected for at least 8 h. The mean excretion rate in the last collection period (6 –8 h after alcohol intake) still was 22 and 32% of the maximal rate in the 0.1 and 0.5 g/kg dose groups respectively.
The total amount of ethyl sulphate excreted in the initial 8 h varied from 2.5 to 6.8 μmol in the ten subjects of the 0.1 g/kg dose group (Table 2). This is only 14–48 p.p.m. of the total dose. The corresponding values for the higher dose level (0.5 g/kg, two subjects, each studied on two separate occasions) amounted to 28–95 μmol for the absolute amount of ethyl sulphate excreted and 43–111 p.p.m. in relation to the dose of ethanol used. Thus the fraction of ethanol metabolized to its sulpho-conjugate may be slightly higher at the higher dose level.
Table 2. Urinary excretion of ethyl sulphate in various volunteers after consuming 0.1 or 0.5 g of ethanol per kg of body mass.
Study subjects were 24–52-year-old female (nos. 1–7) and male (nos. 8–10) Caucasians. The experiments with subjects 6 and 10 were conducted on several different occasions and using two different dose levels. All urine was collected within 8 h of consumption of ethanol, using the sampling periods indicated in Figures 3 and 4. In addition, spot urine collected after 24 h was analysed.
| Urinary ethyl sulphate | |||||
|---|---|---|---|---|---|
| Amount excreted within 8 h | Concentration (μM) | ||||
| Ethanol dose (g/kg of body mass) | Subject no./experiment no. | (μmol) | (p.p.m. of dose of ethanol) | Maximal | 24 h spot urine |
| 0.1* | 1 | 2.7 | 20 | 5.4 | 0.2 |
| 2 | 4.0 | 33 | 5.0 | −‡ | |
| 3 | 5.0 | 27 | 3.5 | 0.1 | |
| 4 | 3.5 | 25 | 7.8 | − | |
| 5 | 2.7 | 26 | 9.5 | − | |
| 6/1 | 1.8 | 14 | 4.2 | − | |
| 6/2 | 4.9 | 37 | 8.7 | − | |
| 6/3 | 6.3 | 48 | 5.5 | − | |
| 7 | 3.5 | 20 | 5.4 | − | |
| 8 | 4.0 | 25 | 6.3 | − | |
| 9 | 4.9 | 35 | 11.0 | 0.1 | |
| 10/1 | 6.8 | 40 | 8.6 | − | |
| 10/2 | 4.6 | 27 | 7.9 | − | |
| 10/3 | 5.4 | 32 | 2.2 | 0.1 | |
| 0.5† | 6/1 | 28 | 43 | 21 | 0.8 |
| 6/2 | 67 | 103 | 68 | 0.4 | |
| 10/1 | 38 | 44 | 32 | 4.5 | |
| 10/2 | 95 | 111 | 44 | 1.4 | |
* Equivalent to an absolute dose of 105–185 mmol, depending on the body mass of the subject.
† Equivalent to a dose of 665 and 850 mmol with subjects 6 and 10 respectively.
‡ Below limit of detection (0.1 μM).
The experiments with a female test subject (no. 6) and a male subject (no. 10) were conducted on several separate occasions. The results revealed 3-fold intra-individual day-by-day variation (Table 2). This variation was in the same order of magnitude as the inter-individual variation in the relatively small group studied.
In addition to the systematic collection of all urine for at least 8 h after the alcohol intake, we analysed 24 h spot urine for the presence of ethyl sulphate. Substantial levels of the metabolite were still detected at this time in the 0.5 g/kg dose group, whereas the corresponding values in the 0.1 g/kg dose group were below or slightly above the limit of detection (Table 2).
Ethanol itself was detected in all 0–1 h urinary samples and in most 1–2 h samples, but not in any later samples of the 0.1 g/kg dose group (limit of detection, 0.05 mM). In the high-dose group (0.5 g/kg), it was detected in all samples collected within the first 4 h, with a nearly linear decrease (from 7–11 mM in the 0–1 h urine to 0.2–1.6 mM in the 3–4 h urine), but was absent in any later sample.
Conjugation of ethanol by individual human SULT forms
Cytosolic preparations of the parental S. typhimurium strain and derivative strains engineered for the expression of individual human SULT forms were incubated with PAPS and varying concentrations of ethanol. No formation of ethyl sulphate was found under any conditions with the parental strain or with the strains expressing SULT2B1b or 4A1 (Table 3). With all other forms, some sulpho-conjugation of ethanol was observed. The activities of SULT1C1 and 2B1a were very low, just above the limit of detection. Slightly higher activities were detected with SULT2A1, 1A1 and 1E1. Substantially higher activities were observed with SULT1A3, 1C2, 1B1 and 1A2. With all forms, the rate of formation of ethyl sulphate was highest at the highest substrate concentration used (1000 mM), indicating a very low affinity of the substrate for the enzymes.
Table 3. Sulpho-conjugation of ethanol by individual cDNA-expressed human forms.
Cytosolic preparations (0.1 mg of protein) of a S. typhimurium strain expressing the indicated human SULT form, the cofactor PAPS (100 μM) and the substrate ethanol (0–1000 mM) in a total volume of 100 μl were incubated at 37 °C for 1 h. The reaction was stopped by the addition of a two-fold volume of propan-2-ol. After centrifugation, samples were directly analysed by HPLC–MS/MS. Values are means of at least four incubations. In general, S.E.M. was less than 10% of the mean.
| SULT activity (pmol of ethyl sulphate/min per mg of protein) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ethanol (mM) | 1A1 | 1A2 | 1A3 | 1B1 | 1C1 | 1C2 | 1E1 | 2A1 | 2B1a | 2B1b | 4A1 |
| 0 | −* | − | − | − | − | − | − | − | − | − | − |
| 1 | − | − | − | − | − | − | − | − | − | − | − |
| 10 | − | 7 | 7 | 9 | − | 9 | − | − | − | − | − |
| 100 | 6 | 64 | 28 | 26 | − | 51 | 7 | − | − | − | − |
| 1000 | 23 | 355 | 155 | 322 | 5 | 317 | 25 | 15 | 5 | − | − |
* Below limit of detection (3 pmol/mg per min).
DISCUSSION
To the best of our knowledge, this is the first report demonstrating the formation of ethyl sulphate from ethanol in humans in vivo and by human enzymes in vitro. Since only a very small fraction of ethanol is sulphonated in vivo, this pathway should not measurably affect the elimination kinetics of ethanol. However, the pathway may be useful for studying SULT activity in vivo and as a mid-term biomarker for previous ethanol consumption.
The development of a sensitive analysis of ethyl sulphate was the prerequisite for detecting this novel biotransformation pathway of ethanol in man. The HPLC–MS/MS method used is simple (apart from the requirement of expensive equipment), sensitive and highly selective. Ethyl sulphate was readily detected in micromolar concentrations, as found in urine within the initial 8 h of ingestion of a low amount of alcohol and much longer (>24 h) after the consumption of a higher-still common dose (0.5 g of ethanol per kg of body mass).
Urinary ethyl sulphate was found in all individuals after consumption of ethanol, but not when they abstained from alcoholic beverages for 2 days. Thus there is no detectable endogenous formation of ethyl sulphate and its excretion is specific for ethanol consumption. Ethanol, at the doses used (0.1 and 0.5 g of ethanol per kg of body mass) was completely eliminated within 2 and 4 h, but ethyl sulphate was readily detected even 8 and 24 h, respectively, after intake of these doses. Thus the excretion of ethyl sulphate is delayed, as has been previously reported for another minor metabolite of ethanol, ethyl glucuronide [16]. Ethyl glucuronide is utilized as a marker for demonstrating recent alcohol consumption, in the time period when ethanol itself has disappeared from the body [17,29]. It is probable that ethyl sulphate can be used for the same purpose.
However, our main reason for conducting this study was to explore whether ethanol could be used as a substrate to determine SULT activity in vivo. The results are very promising. Ethanol has the advantage that its elimination kinetics only exhibit low inter-individual variation. Therefore variation in the activity of competing pathways should affect the rate of sulpho-conjugation much less than with alternative substrates, such as paracetamol. The fact that the rate of sulphonation of ethanol is very low (but nevertheless well detectable) may be an advantage rather than a disadvantage, since it renders it unlikely that the supply of the cofactor PAPS, rather the enzyme activity, limits the rate of sulpho-conjugation, a phenomenon observed with various good SULT substrates used at high dose levels in animal models [30]. Of course, it would be important to identify the SULT forms whose activity is reflected by the sulpho-conjugation of ethanol in vivo. In order to identify candidate forms, we studied the sulpho-conjugation of ethanol in the presence of the individual cDNA-expressed human forms in vitro. Although the members of the SULT2 family are often named ‘alcohol sulphotransferases’, they showed much lower activity towards ethanol than several SULT1 forms, which are sometimes termed ‘phenol sulphotransferases’. Likewise, SULT1A1, the SULT form with the broadest substrate tolerance [1–3], only exhibited modest activity. The highest activities were found with SULT1B1, 1A2, 1A3 and 1C2. In a recent review, we have compiled information on the tissue distribution of the various human SULTs [31]. To date, SULT1C2 protein has only been detected in foetal tissues, but not in adult tissues, and even its RNA was detected only in very few adult tissues [3,31]. Likewise, the expression of SULT1A2 protein in human tissues has not yet been demonstrated in any published study. Using modified Western blotting techniques our laboratory has demonstrated the presence of SULT1A2 protein in the Caco-2 cell line and in some (but not all) colon mucosa samples; however, the level was always much lower than that of other SULT forms (W. Meinl, W. Teubner, M. Kretzschmar, A. Lampen and H. R. Glatt, unpublished work). SULT1B1 is highly expressed throughout the different sections of the gastrointestinal tract, is present at lower levels in the liver, but is low or absent in many other tissues. SULT1A3 is expressed in most tissues, with the noticeable exception of the adult liver. Particularly high levels of SULT1A3 are found in the gut. Thus we would predict that SULT1A3 is the major form involved in ethanol sulphonation in vivo. SULT1B1 may also significantly contribute to the reaction. Although SULT1A1 showed only modest activity in vitro, it might be of some importance in vivo due to its abundance, including a very high hepatic expression. Of course, these predictions are hypothetical and require corroboration.
Sulphonation is a metabolic step converting hydrophobic substrates into water-soluble conjugates. In vitro assays often involve the addition of substrates dissolved in ethanol. Furthermore, ethanol is sometimes used for stabilizing solutions of the co-substrate PAPS [32]. Our observation that ethanol is a substrate of various SULT forms implies that it could competitively inhibit the conjugation of the substrate studied. However, our results also indicate that the affinity of ethanol for SULTs is very low, as the rate of conjugation was still increased several fold when the substrate concentration was raised from 100 to 1000 mM [equivalent to 0.55 and 5.5% (v/v) respectively in the incubation medium]. Therefore a concentration of 1% ethanol should be clearly below its Km value and should not substantially affect the conjugation of the substrate actually studied, if this is used at a high concentration (in the saturation range). More caution is required when kinetics are investigated in detail. Artifacts may also be produced if the [35S]PAPS/barium salt precipitation assay is used for determining conjugation rates without any further analysis of the product. This popular assay exploits the fact that numerous sulpho-conjugates, unlike PAPS, are soluble in the presence of barium and zinc, added after the incubation period [32]. Thus, if low rates of conjugation are observed with this assay, we recommend the conductance of negative controls with and without addition of the solvent. Apart from acting as a competing substrate, ethanol may affect the activity of an enzyme via various other mechanisms. Ma et al. [33] investigated the influence of several solvents, including ethanol, on the activity of cDNA-expressed SULT1A1, 1A3, 1E1 and 2A1. In general, ethanol and methanol had less effect than the other solvents studied. At a concentration of 0.5% (87 mM), ethanol inhibited SULT activities up to 20%. A concentration of 2% was sufficient to inhibit SULT1A1 and 1E1 by 40 and 60% respectively, whereas the other forms were less susceptible.
In the present study, we observed considerable intra-individual variations in ethyl sulphate excretion. This variation was in the same range as the inter-individual variation. Indeed, no genetic polymorphisms have been described for SULT1A3 and 1B1, the prime candidates for a major role of sulpho-conjugation of ethanol in vivo, in Caucasian populations [5]. These findings indicate that variation in enzyme quality (at least as determined by the primary sequence) cannot be the critical factor, if these forms are indeed important for ethanol sulphonation in vivo. This would direct to a possible role of temporal changes in enzyme levels and/or the presence of competing substrates and inhibitors. Little is known about the regulation of the expression of SULT1A3 and 1B1. The fact that these forms show their highest expression in the gut suggests that they have a role in the elimination of food constituents, such as food-borne catecholamines (which are characteristic substrates of SULT1A3) and hydroxylated secondary plant metabolites (being substrates and/or inhibitors for many SULT forms). Supposing that the elimination of food constituents is a major function of these enzymes in the gut, it would also make sense if dietary factors (in addition to possible other factors) were involved in the regulation of their expression. We will test the hypothesis that dietary factors are causal factors of the observed variation of ethanol sulphonation in vivo and try to elucidate the mechanisms involved.
Shortly before the present paper was completed, Helander and Beck [34] also identified ethyl sulphate as an ethanol metabolite in human urine. They focused their study on the use of ethyl sulphate as a specific biomarker of recent alcohol intake with clinical and forensic applications. They used simple HPLC–MS monitoring the pseudomolecular ion at m/z=125, whereas we worked in the MRM mode (with a daughter ion at m/z=97). The latter method is potentially more specific, but requires more complicated equipment than simple HPLC–MS. In the present case, the enhanced specificity was not required, as no interfering urinary components were observed. Helander and Beck [34] only used a single subject receiving a single dose of ethanol (0.5 g/kg of body mass) to study the excretion of ethanol and its metabolites. The results were similar to those obtained in the present study with the same dose level. Helander and Beck [34] additionally studied the excretion of ethyl glucuronide. Both conjugates were still detected in the sample collected at 29 h, but not at 32 h. Furthermore, they analysed 54 clinical urine samples sent to the laboratory for testing of recent alcohol consumption. All 31 samples with detectable ethyl glucuronide were also positive for ethyl sulphate. Two other samples were only positive for ethyl sulphate. These findings support the idea that urinary ethyl sulphate could be a promising candidate marker to disclose recent alcohol consumption. To come back to our study concept, concurrent determination of ethyl glucuronide and sulphate after administration of ethanol may give information about the in vivo activities of two important classes of conjugating enzymes, UGTs and SULTs.
Acknowledgments
We thank Martina Scholtyssek for excellent technical assistance.
References
- 1.Falany C. N. Sulfation and sulfotransferases: 3. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997;11:206–216. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- 2.Glatt H. R. Sulphotransferases. In: Ioannides C., editor. Handbook of Enzyme Systems that Metabolise Drugs and Other Xenobiotics. Chichester: John Wiley & Sons; 2002. pp. 353–439. [Google Scholar]
- 3.Coughtrie M. W. H. Sulfation through the looking glass – recent advances in sulfotransferase research for the curious. Pharmacogenomics J. 2002;2:297–308. doi: 10.1038/sj.tpj.6500117. [DOI] [PubMed] [Google Scholar]
- 4.Raftogianis R. B., Wood T. C., Weinshilboum R. M. Human phenol sulfotransferases SULT1A2 and SULT1A1: genetic polymorphisms, allozyme properties, and human liver genotype-phenotype correlations. Biochem. Pharmacol. 1999;58:605–616. doi: 10.1016/s0006-2952(99)00145-8. [DOI] [PubMed] [Google Scholar]
- 5.Glatt H. R., Meinl W. Pharmacogenetics of soluble sulfotransferases (SULTs) Naunyn-Schmiedebergs Arch. Pharmacol. 2004;369:55–68. doi: 10.1007/s00210-003-0826-0. [DOI] [PubMed] [Google Scholar]
- 6.Kester M. H., Bulduk S., van Toor H., Tibboel D., Meinl W., Glatt H. R., Falany C. N., Coughtrie M. W. H., Schuur A. G., Brouwer A., Visser T. J. Potent inhibition of estrogen sulfotransferase by hydroxylated metabolites of polyhalogenated aromatic hydrocarbons reveals alternative mechanism for estrogenic activity of endocrine disrupters. J. Clin. Endocrinol. Metab. 2002;87:1142–1150. doi: 10.1210/jcem.87.3.8311. [DOI] [PubMed] [Google Scholar]
- 7.Ghazali R. A., Waring R. H. The effects of flavonoids on human phenolsulphotransferases: potential in drug metabolism and chemoprevention. Life Sci. 1999;65:1625–1632. doi: 10.1016/s0024-3205(99)00423-3. [DOI] [PubMed] [Google Scholar]
- 8.Miners J. O., Attwood J., Birkett D. J. Determinants of acetaminophen metabolism: effect of inducers and inhibitors of drug metabolism on acetaminophen's metabolic pathways. Clin. Pharmacol. Ther. 1984;35:480–486. doi: 10.1038/clpt.1984.64. [DOI] [PubMed] [Google Scholar]
- 9.Nash R. M., Stein L., Penno M. B., Passananti G. T., Vesell E. S. Sources of interindividual variations in acetaminophen and antipyrine metabolism. Clin. Pharmacol. Ther. 1984;36:417–430. doi: 10.1038/clpt.1984.199. [DOI] [PubMed] [Google Scholar]
- 10.Bock K. W., Wiltfang J., Blume R., Ullrich D., Bircher J. Paracetamol as a test drug to determine glucuronide formation in man: effects of inducers and of smoking. Eur. J. Clin. Pharmacol. 1987;31:677–683. doi: 10.1007/BF00541295. [DOI] [PubMed] [Google Scholar]
- 11.Court M. H., Duan S. X., von Moltke L. L., Greenblatt D. J., Patten C. J., Miners J. O., Mackenzie P. I. Interindividual variability in acetaminophen glucuronidation by human liver microsomes: identification of relevant acetaminophen UDP–glucuronosyltransferase isoforms. J. Pharmacol. Exp. Ther. 2001;299:998–1006. [PubMed] [Google Scholar]
- 12.Herman R. J., Loewen G. R., Antosh D. M., Taillon M. R., Hussein S., Verbeeck R. K. Analysis of polymorphic variation in drug metabolism: III. Glucuronidation and sulfation of diflunisal in man. Clin. Invest. Med. 1994;17:297–307. [PubMed] [Google Scholar]
- 13.Brashear W. T., Kuhnert B. R., Wei R. Maternal and neonatal urinary excretion of sulfate and glucuronide ritodrine conjugates. Clin. Pharmacol. Ther. 1988;44:634–641. doi: 10.1038/clpt.1988.205. [DOI] [PubMed] [Google Scholar]
- 14.Levy G., Regardh C. G. Drug biotransformation interactions in man: 5. Acetaminophen and salicylic acid. J. Pharm. Sci. 1971;60:608–611. doi: 10.1002/jps.2600600423. [DOI] [PubMed] [Google Scholar]
- 15.Campbell N. R., Sundaram R. S., Werness P. G., van Loon J., Weinshilboum R. M. Sulfate and methyldopa metabolism: metabolite patterns and platelet phenol sulfotransferase activity. Clin. Pharmacol. Ther. 1985;37:308–315. doi: 10.1038/clpt.1985.45. [DOI] [PubMed] [Google Scholar]
- 16.Schmitt G., Aderjan R., Keller T., Wu M. Ethyl glucuronide: an unusual ethanol metabolite in humans. Synthesis, analytical data, and determination in serum and urine. J. Anal. Toxicol. 1995;19:91–94. doi: 10.1093/jat/19.2.91. [DOI] [PubMed] [Google Scholar]
- 17.Wurst F. M., Kempter C., Seidl S., Alt A. Ethyl glucuronide: a marker of alcohol consumption and a relapse marker with clinical and forensic implications. Alcohol Alcoholism. 1999;34:71–77. doi: 10.1093/alcalc/34.1.71. [DOI] [PubMed] [Google Scholar]
- 18.Nishikawa M., Tsuchihashi H., Miki A., Katagi M., Schmitt G., Zimmer H., Keller T., Aderjan R. Determination of ethyl glucuronide, a minor metabolite of ethanol, in human serum by liquid chromatography–electrospray ionization mass spectrometry. J. Chromatogr. B Biomed. Sci. Appl. 1999;726:105–110. doi: 10.1016/s0378-4347(99)00008-0. [DOI] [PubMed] [Google Scholar]
- 19.Stephanson N., Dahl H., Helander A., Beck O. Direct quantification of ethyl glucuronide in clinical urine samples by liquid chromatography–mass spectrometry. Ther. Drug Monit. 2002;24:645–651. doi: 10.1097/00007691-200210000-00011. [DOI] [PubMed] [Google Scholar]
- 20.Boström H., Vestermark A. Some aspects on sulphate conjugation. Biochem. Pharmacol. 1961;6:72–81. [Google Scholar]
- 21.Bernstein J., Meneses P., Basilio C., Martinez B. Further characterization of the pulmonary ethanol metabolizing system (PET) Res. Commun. Chem. Pathol. Pharmacol. 1984;46:121–136. [PubMed] [Google Scholar]
- 22.Bernstein J., Basilio C., Martinez B. Ethanol sulfation by the pulmonary ethanol metabolizing system (PET) Res. Commun. Chem. Pathol. Pharmacol. 1990;68:219–234. [PubMed] [Google Scholar]
- 23.Manautou J. E., Carlson G. P. Comparison of pulmonary and hepatic glucuronidation and sulphation of ethanol in rat and rabbit in vitro. Xenobiotica. 1992;22:1309–1319. doi: 10.3109/00498259209053159. [DOI] [PubMed] [Google Scholar]
- 24.Calhoun G. M., Burwell R. L., Jr The hydrolysis of sodium alkyl sulfates in basic aqueous solution. J. Am. Chem. Soc. 1955;77:6441–6447. [Google Scholar]
- 25.Muckel E., Landsiedel R., Glatt H. R. Preparation of highly pure 3′-phosphoadenosine-5′-phosphosulfate (PAPS) using recombinant human PAPS synthetase and purification by ion exchange high performance liquid chromatography. Naunyn-Schmiedebergs Arch. Pharmacol. 2001;363:R137. [Google Scholar]
- 26.Beutler H. O. Ethanol. In: Bergmeyer J., Grassl M., editors. Methods of Enzymatic Analysis, vol. 4. Weinheim: Verlag Chemie GmbH; 1984. pp. 598–606. [Google Scholar]
- 27.Meinl W., Meerman J. H., Glatt H. R. Differential activation of promutagens by alloenzymes of human sulfotransferase 1A2 expressed in Salmonella typhimurium. Pharmacogenetics. 2002;12:677–689. doi: 10.1097/00008571-200212000-00002. [DOI] [PubMed] [Google Scholar]
- 28.Landsiedel R., Pabel U., Engst W., Ploschke J., Seidel A., Glatt H. R. Chiral inversion of 1-hydroxyethylpyrene enantiomers mediated by enantioselective sulfotransferases. Biochem. Biophys. Res. Commun. 1998;247:181–185. doi: 10.1006/bbrc.1998.8756. [DOI] [PubMed] [Google Scholar]
- 29.Sarkola T., Dahl H., Eriksson C. J., Helander A. Urinary ethyl glucuronide and 5-hydroxytryptophol levels during repeated ethanol ingestion in healthy human subjects. Alcohol Alcoholism. 2003;38:347–351. doi: 10.1093/alcalc/agg083. [DOI] [PubMed] [Google Scholar]
- 30.Klaassen C. D., Boles J. W. Sulfation and sulfotransferases: 5. The importance of 3′-phosphoadenosine 5′-phosphosulfate (PAPS) in the regulation of sulfation. FASEB J. 1997;11:404–418. doi: 10.1096/fasebj.11.6.9194521. [DOI] [PubMed] [Google Scholar]
- 31.Glatt H. R., Boeing H., Engelke C. E., Ma L., Kuhlow A., Pabel U., Pomplun D., Teubner W., Meinl W. Human cytosolic sulphotransferases: genetics, characteristics, toxicological aspects. Mutat. Res. 2001;482:27–40. doi: 10.1016/s0027-5107(01)00207-x. [DOI] [PubMed] [Google Scholar]
- 32.Foldes A., Meek J. L. Rat brain phenolsulfotransferase: partial purification and some properties. Biochim. Biophys. Acta. 1973;327:365–374. doi: 10.1016/0005-2744(73)90419-1. [DOI] [PubMed] [Google Scholar]
- 33.Ma B., Shou M., Schrag M. L. Solvent effect on cDNA-expressed human sulfotransferase (SULT) activities in vitro. Drug Metab. Dispos. 2003;31:1300–1305. doi: 10.1124/dmd.31.11.1300. [DOI] [PubMed] [Google Scholar]
- 34.Helander A., Beck O. Mass spectrometric identification of ethyl sulfate as an ethanol metabolite in humans. Clin. Chem. 2004;50:936–937. doi: 10.1373/clinchem.2004.031252. [DOI] [PubMed] [Google Scholar]