Abstract
Dysfunction of mitochondrial ATPase (F1Fo-ATP synthase) due to missense mutations in ATP6 [mtDNA (mitochondrial DNA)-encoded subunit a] is a frequent cause of severe mitochondrial encephalomyopathies. We have investigated a rare mtDNA mutation, i.e. a 2 bp deletion of TA at positions 9205 and 9206 (9205ΔTA), which affects the STOP codon of the ATP6 gene and the cleavage site between the RNAs for ATP6 and COX3 (cytochrome c oxidase 3). The mutation was present at increasing load in a three-generation family (in blood: 16%/82%/>98%). In the affected boy with severe encephalopathy, a homoplasmic mutation was present in blood, fibroblasts and muscle. The fibroblasts from the patient showed normal aurovertin-sensitive ATPase hydrolytic activity, a 70% decrease in ATP synthesis and an 85% decrease in COX activity. ADP-stimulated respiration and the ADP-induced decrease in the mitochondrial membrane potential at state 4 were decreased by 50%. The content of subunit a was decreased 10-fold compared with other ATPase subunits, and [35S]-methionine labelling showed a 9-fold decrease in subunit a biosynthesis. The content of COX subunits 1, 4 and 6c was decreased by 30–60%. Northern Blot and quantitative real-time reverse transcription–PCR analysis further demonstrated that the primary ATP6 – COX3 transcript is cleaved to the ATP6 and COX3 mRNAs 2–3-fold less efficiently. Structural studies by Blue-Native and two-dimensional electrophoresis revealed an altered pattern of COX assembly and instability of the ATPase complex, which dissociated into subcomplexes. The results indicate that the 9205ΔTA mutation prevents the synthesis of ATPase subunit a, and causes the formation of incomplete ATPase complexes that are capable of ATP hydrolysis but not ATP synthesis. The mutation also affects the biogenesis of COX, which is present in a decreased amount in cells from affected individuals.
Keywords: ATP6, ATP synthase, COX3, cytochrome c oxidase, mitochondrial disease, mitochondrial DNA (mtDNA)
Abbreviations: ATPase, F1Fo-ATP synthase; BN-PAGE, Blue-Native PAGE; COX, cytochrome c oxidase; CS, citrate synthase; 2D, two-dimensional; DDM, dodecyl maltoside; FCCP, carbonyl cyanide 4-trifluoromethoxyphenylhydrazone; LRPPRC, leucine-rich pentatricopeptide repeat cassette; mtDNA, mitochondrial DNA; OXPHOS, oxidative phosphorylation; ΔΨm, mitochondrial membrane potential; RFLP, restriction fragment length polymorphism; RT-PCR, reverse transcription–PCR; SDH, succinate dehydrogenase; TMPD, N,N,N′,N′-tetramethyl-p-phenylenediamine; TMRM, tetramethylrhodamine methyl ester; WB, Western blot
INTRODUCTION
The mammalian ATPase (F1Fo-ATP synthase) complex catalyses the synthesis of ATP from ADP and Pi, the final step of the OXPHOS (oxidative phosphorylation) pathway. The ATPase complex consists of 16 different subunits [1], and is composed of the globular F1 catalytic part connected by two stalks to the membrane-embedded Fo moiety that translocates protons across the mitochondrial inner membrane. Only two Fo subunits, subunit a (ATP6) and A6L (ATP8), are encoded by mtDNA (mitochondrial DNA) [2]. Both are essential for the biogenesis and assembly of the ATPase complex [3].
Mutations in mtDNA represent a frequent cause of mitochondrial diseases. They can involve tRNAs, rRNAs or protein-encoding genes (for review see [4]) and are associated with a wide variety of clinical pictures, ranging from isolated myopathy to multisystem disorders, affecting primarily tissues with high energy demands, such as skeletal muscle, heart and nervous system. Most pathogenic mtDNA mutations are in tRNAs. Mutations in protein-encoding genes are much less frequent, with the exception of those associated with complex I or ATPase dysfunction. All maternally inherited ATPase diseases are caused by mutations in the ATP6 gene; no mutation has been reported in the ATP8 (A6L) gene. Mutations in the ATP6 gene disturb the function of the ATPase proton channel, which consists of subunit a and multiple copies of subunit c. The most frequent are heteroplasmic T8993G [5] or less severe T8993C mutations [6], which result in replacement of Leu156 by Arg or Pro in subunit a, and often present as a NARP (neurogenic muscle weakness, ataxia, retinitis pigmentosa) [5] or MILS (maternally inherited Leigh syndrome) [7] phenotype. Several other, less frequent, mutations of ATP6 at positions 9176 or 8851 have also been described (for review see [8]), resulting in similar lesions in brain, particularly in the striatum (familiar bilateral striatal necrosis). The T8993G mutation results in a decrease in mitochondrial ATP production [9] without a significant effect on ATP hydrolysis [7], and in structural changes in the ATPase complex [10], which, however, could not be found in some cases [11]. It has been observed that the ATPase deficiency is associated with a decreased ability of cells from affected individuals to assemble correctly the ATPase complex, which shows instability in BN-PAGE (Blue-Native PAGE) experiments [10,12].
In the present paper we have studied a very rare mtDNA mutation in the ATP6 gene – a 2 bp microdeletion at positions 9205 and 9206 (9205ΔTA). This mutation cancels the STOP codon of ATP6 gene and changes the cleavage site between the ATP6 and COX3 (cytochrome c oxidase subunit 3) transcripts. It was originally discovered in a newborn with transient lactic acidosis [13]. Recently we found a second case of a 9205ΔTA mutation that was present in a child with severe encephalopathy and hyperlactacidaemia [14]. Here we present the results of molecular and biochemical studies of ATPase and COX that focus on the biosynthesis of ATPase subunit a and the structural and functional consequences of the 9205ΔTA mutation.
EXPERIMENTAL
Ethics
This study was carried out in accordance with the Declaration of Helsinki of the World Medical Association, and was approved by the Committees of Medical Ethics at all collaborating institutions. Informed consent was obtained from the parents of the child.
Case report
The boy was born at term from a second, uncomplicated pregnancy, with birth weight 3450 g and length 52 cm. Failure to thrive, spastic quadruparesis and microcephalia were observed from the 3rd month of life, followed by practical arrest of any psychomotor development. Metabolic investigations revealed intermittent hyperlactacidaemia (B-lactate, 0.95–3.4 mmol/l; controls <2.1 mmol/l), with increased levels of lactate and alanine in the cerebrospinal fluid [lactate, 4.8 mmol/l (controls <1.8 mmol/l); alanine, 36 μmol/l (controls <34 μmol/l)]. He is 5 years old at present. Both parents are healthy, but an older brother (from the first marriage of the mother) died due to a respiratory failure at the age of 3 years. He presented with fatal infantile encephalopathy, severe psychomotor delay, frontal lobe atrophy and lactic acidosis.
Cell cultures and isolation of mitochondria
Fibroblast cultures were established from skin biopsies, and cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal calf serum (Sigma) at 37 °C in 5% CO2 in air. Cells were grown to approx. 90% confluence and harvested using 0.05% (w/v) trypsin and 0.02% (w/v) EDTA. Detached cells were diluted with an ice-cold culture medium, sedimented by centrifugation and washed twice in PBS.
Mitochondria were isolated from fibroblasts by a hypo-osmotic shock method [15]. The freshly harvested cells were disrupted in 10 mM Tris buffer (pH 7.4) and quickly homogenized in a Teflon/glass homogenizer at 4 °C. Sucrose was added to a final concentration of 0.25 M immediately after homogenization. The nuclei were removed by centrifugation for 10 min at 4 °C and 600 g and the mitochondrial fraction was isolated from the postnuclear supernatant by centrifugation for 10 min at 4 °C and 10000 g. The mitochondrial pellet was washed and finally resuspended in 0.25 M sucrose, 2 mM EGTA, 40 mM KCl and 20 mM Tris, pH 7.4, and stored at −70 °C.
Mitoplasts were prepared from fibroblasts as described previously [16]. In brief, trypsinized cells suspended in an STE medium (0.25 M sucrose, 10 mM Tris, 1 mM EDTA, pH 7.4) were treated with digitonin (0.4 mg/mg of protein; Fluka) on ice for 15 min. The suspension was diluted 10-fold with STE and centrifuged for 10 min at 4 °C and 12000 g. The pellet was washed by centrifugation and resuspended in STE to a final concentration of 1–2 mg of protein/ml. Based on immunodetection and enzyme activity measurements, >95% of the mitochondrial inner membrane proteins were recovered in this fraction.
Muscle mitochondria were isolated according to [17], but without use of protease. Tissue samples were homogenized at 4 °C in a KCl medium (100 mM KCl, 50 mM Tris, 2 mM EDTA, 10 mg/ml aprotinin, pH 7.5). The homogenate was centrifuged for 10 min at 4 °C and 600 g, the supernatant was filtered through a 100 μm nylon screen, and mitochondria were sedimented by centrifugation for 10 min at 4 °C and 10000 g. The mitochondrial pellet was washed by centrifugation and resuspended to a final protein concentration of 20–25 mg/ml.
DNA analysis and sequencing
Total genomic DNA from muscle and cultured fibroblasts was isolated by phenol extraction. The entire mtDNA was amplified in six overlapping fragments by PCR (7–3148, 2073–5719, 5645–8815, 8403–11132, 11005–14684 and 13863–136). Purified fragments were sequenced on the automatic sequencer ALFExpress II (Amersham Biosciences) using cycle sequencing with 41 Cy5-labelled internal sequencing primers.
Restriction analysis
To determine the amount of mtDNA containing the microdeletion, the PCR/RFLP (restriction fragment length polymorphism) analysis method was performed according to [18] using the mismatched (bold) primers 5′-CCT CTA CCT GCA CGA CAA TGC A-3′ (forward) and 5′-CGT TAT GCA TTG GAA GTG AAA TCA C-3′ (reverse), which introduce two novel NsiI restriction sites in the case of the wild-type mtDNA and one NsiI restriction site in the case of mutant mtDNA. PCR products were radioactively labelled with [α-32P]dCTP in the final cycle of PCR and run on a non-denaturing 13% (w/v) polyacrylamide gel after complete digestion. The proportions of wild-type and mutant mtDNA were measured using a PhosphorImager and ImageQuant software (Molecular Dynamics).
Northern blot analysis
Total RNA was isolated from cultured fibroblasts by phenol/guanidium thiocyanate/chloroform extraction [19]. Approx. 20 μg of total RNA per lane was separated through a 1.2% (w/v) agarose/formaldehyde gel and transferred to a Hybond-N nylon membrane (Amersham) in 20×SSC (1×SSC is 0.15 M NaCl/0.015 M sodium citrate). The membrane was prehybridized for 2 h at 45 °C in 5×SSC, Denhardt's solution, 0.5% SDS and 100 μg/ml sonicated herring sperm. The membranes were hybridized overnight at 45 °C with [α-32P]dCTP-labelled probes corresponding to regions of the genes ATP6 (8361–9060), COX3 (9269–9912), ND1 (3313–4252) and COX1 (6120–6960). The radioactivity was detected by phosphor imaging (as above).
Quantitative RT-PCR (reverse transcription–PCR) analysis
Total RNA was isolated from cultured fibroblasts using RNA Blue reagent (Top-Bio, Prague, Czech Republic). Following DNase I treatment (Invitrogen), first-strand cDNA was synthesized from 1 μg RNA aliquots with 200 units SuperScript II reverse transcriptase using either 200 ng of random hexamer primers or 500 ng of oligo(dT)12–18 (all Invitrogen) according to the manufacturer's instructions. Real-time quantitative RT-PCR was performed on a LightCycler instrument (Roche Diagnostics) using a QuantiTect SYBR Green PCR kit (Qiagen). PCR reactions were performed on cDNAs using primer pairs specific for the ATP6 gene transcript (forward, 5′-CCT TAT GAG CGG GCA CAG T-3′; reverse, 5′-CAG GGC TAT TGG AA-3′; nt 8846–8994), for the COX3 gene transcript (forward, 5′-GCC CTC TCA GCC CTC CTA ATG-3′; reverse, 5′-GTG GCC TTG GTA TGT GCT TTC TCG-3′; nt 9267–9416), for the ATP6–COX3 gene transcript (forward, 5′-AAT CCA AGC CTA CGT TTT CAC ACT-3′; reverse, 5′-TAG GCC GGA GGT CAT TAG G-3′; nt 9150–9299), for the CYTB gene transcript (forward, 5′-GAC CTC CCC ACC CCA TCC A-3′; reverse, 5′-AAA GGC GGT TGA GGC GTC TG-3′; nt 14804–14935) and for the ND1 gene transcript (forward, 5′-CAA CCT CAA CCT AGG CCT CCT-3′; reverse, 5′-ACG GCT AGG CTA GAG GTG GC-3′; nt 3595–3644). The primer pair for ATP6–COX3 was designed to flank the splice site of the ATP6 – COX3 transcript. Amplified regions of the ATP6 or COX3 transcript were present in both processed and unprocessed RNAs. All reactions were run at an annealing temperature of 60 °C. The PCR mixture contained 5 μl of 2×SYBR Green PCR Master Mix, 2 μl of 100×diluted reverse transcription product and 200 nM of each primer in a total volume of 10 μl. All reactions were performed in triplicate. For each primer pair, non-template controls were included to check for the absence of contaminants and primer-dimers that would interfere with quantification when SYBR Green is used. The external standard curve was generated in parallel for all reactions using serial dilutions of cDNA synthesized from control RNA. For each sample, the relative amounts of ATP6, COX3, CYTB, ND1 and unprocessed ATP6 – COX3 transcripts were determined from the standard curves. Each sample was analysed in two separate experiments.
Electrophoresis and WB (Western blot) analysis
BN-PAGE [20] was used for the separation of native mitochondrial OXPHOS complexes on 6–15% (w/v) polyacrylamide gradient minigels (Mini Protean system; Bio-Rad) as described previously [16]. Mitoplasts were pelleted by centrifugation for 10 min at 4 °C and 10000 g, and solubilized using 1 g of DDM (dodecyl maltoside)/g of protein for 20 min on ice in a buffer containing 1.75 M aminocaproic acid, 2 mM EDTA and 75 mM Bis-Tris (pH 7.0). Samples were centrifuged for 20 min at 20000 g and Serva Blue G dye was added to collected supernatants at a concentration of 0.1 g/g of detergent. Electrophoresis was performed at 50 V for 30 min and then at 90 V.
SDS/PAGE [21] was performed on 10% (w/v) polyacrylamide slab minigels, and analysis of [35S]methionine-labelled proteins was performed on a 16 cm long 15–20% (w/v) gradient polyacrylamide slab gels (Protean system; Bio-Rad). The samples were boiled for 3 min in sample lysis buffer [2% (v/v) mercaptoethanol, 4% (w/v) SDS, 10 mM Tris/HCl, 10% (v/v) glycerol]. For 2D (two-dimensional) analysis, strips of the first-dimension BN-PAGE gel were incubated for 1 h in 1% (w/v) SDS and 1% (v/v) mercaptoethanol and then subjected to SDS/PAGE (10% polyacrylamide) for separation in the second dimension [21].
Gels were blotted on to Hybond C-extra nitrocellulose membranes (Amersham) by semi-dry electrotransfer for 1 h at 0.8 mA/cm2 and the membrane was blocked in PBS containing 0.2% (v/v) Tween 20 (PBST). The membranes were used whole or were cut according to molecular mass markers into portions containing individual OXPHOS complexes or their subunits. Membranes were incubated for 3 h with primary antibodies diluted in PBS containing 2% (w/v) BSA (PBSTA). Previously characterized polyclonal antibodies were used at the indicated titres: those against the Foc subunit of ATPase (1:900) [22] and those against the Foa (ATP6) subunit of ATPase (1:500) [23]. In addition, we used monoclonal antibodies against subunits COX1 (1:330; Molecular Probes A-6403), COX4 (1:670; Molecular Probes A-6409), COX6c (1:200; Molecular Probes A-6401), NADH39 (1:250; Molecular Probes A-11140), SDH70 (succinate dehydrogenase 70 kDa subunit; 1:2000; Molecular Probes A-11142) and subunit α of F1-ATPase (1:200000; lot 20D6 [24]). Incubation with peroxidase-labelled secondary antibodies in PBSTA was performed for 1 h using either goat anti-mouse IgG (1:1000; Sigma A8924) or goat anti-rabbit IgG (1:1000; Sigma F0382). The chemiluminescence reaction with an ECL® kit (Amersham) was detected on an LAS 1000 instrument (Fuji) and the signal was quantified using Aida 2.11 Image Analyser software.
Spectrophotometric assays
The activities of the mitochondrial enzymes NADH:CoQ reductase (complex I), succinate:CoQ reductase (complex II), CoQ:cytochrome c reductase (complex III), COX (complex IV), NADH:cytochrome c reductase (complex I+III), succinate:cytochrome c reductase (complex II+III) and CS (citrate synthase) were measured spectrophotometrically by standard methods at 37 °C in muscle homogenate and isolated muscle mitochondria [17,25–28] or in cultured fibroblasts [29].
ATPase hydrolytic activity was measured in a ATP-regenerating system as described in [30]. Mitochondria (8–22 μg of protein/ml) were incubated in a medium containing 40 mM Tris (pH 7.4), 5 mM MgCl2, 10 mM KCl, 2 mM phosphoenolpyruvate, 0.2 mM NADH, 1 μg/ml rotenone, 0.1% (w/v) BSA, 5 units of pyruvate kinase and 5 units of lactate dehydrogenase for 2 min. The reaction was started by the addition of 1 mM ATP and the rate of NADH oxidation, equimolar to ATP hydrolysis, was monitored as the decrease in absorbance at 340 nm. Sensitivity to aurovertin was determined by parallel measurements in the presence of 2 μM inhibitor.
High-resolution oxygraphy
Oxygen consumption by cultured fibroblasts was determined at 30 °C as described previously [29,31] using an Oxygraph-2k (Oroboros, Innsbruck, Austria). Freshly harvested fibroblasts were suspended in a KCl medium (80 mM KCl, 10 mM Tris/HCl, 3 mM MgCl2, 1 mM EDTA, 5 mM potassium phosphate, pH 7.4) and cells were permeabilized by digitonin (0.1 mg of detergent/mg of protein). Respiratory substrates and inhibitors were used at the concentrations indicated. Oxygen consumption was expressed in pmol of oxygen/s per mg of protein. COX activity was measured with 5 mM ascorbate and 1 mM TMPD (N, N, N′, N′-tetramethyl-p-phenylenediamine), and was corrected for substrate auto-oxidation (determined as oxygen uptake insensitive to 0.33 mM KCN).
Cytofluorimetric analysis of ΔΨm (mitochondrial membrane potential)
Cytofluorimetric measurements were performed on a FACSort flow cytometer (Becton Dickinson) according to [32]. Fibroblasts were suspended in a KCl medium containing 10 mM succinate to a protein concentration of 1 mg/ml and permeabilized by 0.1 mg of digitonin/mg of protein. Permeabilized fibroblasts were diluted to 0.2 mg of protein/ml and incubated with 20 nM TMRM (tetramethylrhodamine methyl ester; Molecular Probes) for 15 min. OXPHOS inhibitors and ADP at the concentrations indicated were added 1 min before cytofluorimetric analysis. Approx. 10000 cells were used for each measurement. Data were acquired on a logarithmic scale using CellQuest (Becton Dickinson) and analysed with WinMDI 2.8 software (J. Trotter, TSRI, La Jolla, CA, U.S.A.). Arithmetic mean values of fluorescence signal in arbitrary units were determined for each sample for subsequent graphic presentation.
ATP synthesis
The rate of ATP synthesis was measured at 37 °C in 150 mM KCl, 25 mM Tris/HCl, 10 mM potassium phosphate, 2 mM EDTA and 1% (w/v) BSA, pH 7.2, using 0.5 mM ADP and 10 mM succinate or 10 mM pyruvate+10 mM malate as substrate, as described previously [33]. Protein concentration was 1 mg/ml. For permeabilization of fibroblasts, 0.1 mg of digitonin/mg of protein was used. The reaction was started by addition of fibroblasts and performed for the indicated time intervals. Reaction mixture aliquots of 200 μl were added to 200 μl of DMSO, and ATP content was determined in DMSO-quenched samples by a luciferase assay according to [34]. ATP production was expressed in nmol of ATP/min per mg of protein.
Biosynthesis of mitochondrial proteins
Growth medium was removed from cultured fibroblasts, and the cells were rinsed with methionine-free medium without serum (Gibco medium 21013; 1 mM pyruvate, 2 mM glutamine and 30 mg/l cysteine) and incubated in the same medium containing 10% (v/v) dialysed fetal calf serum and 100 μg/ml emetine for 10 min. The cells were labelled for 3 h with 300 μCi/ml L-[35S]methionine, as described in [35]. The products were separated by 15–20% (w/v) polyacrylamide gradient SDS/PAGE. A small aliquot of the samples prepared for electrophoresis was used to measure the total incorporation of radioactivity in the mitochondrial fraction as trichloroacetic acid-precipitable counts. The radioactivity of proteins was quantified in dried gels using a BAS-5000 system (Fuji). Labelled proteins were identified according to their molecular mass as reported previously in ex vivo translation assays [35].
Protein determination
The protein content was measured by the Bradford or Micro BCA protein kit assays (Bio-Rad), using BSA as a standard. Samples were sonicated for 20 s prior to protein determination.
RESULTS
Activities of respiratory chain enzymes and mitochondrial ATPase
The activities of respiratory chain enzymes (Table 1A) in a muscle homogenate from a patient with encephalopathy and lactic acidosis showed a decrease in the COX/CS ratio. In isolated muscle mitochondria, both COX activity and the COX/CS ratio were at the lower end of the control range, complex I activity was below the control range and complex III activity was moderately decreased. In fibroblasts from the patient, we found a pronounced decrease in COX activity to 15% of the control, CS was just below the control range, and the activities of other respiratory chain enzymes were normal. ATPase hydrolytic activity was determined in isolated mitochondria from the patient's fibroblasts. It was measured as aurovertin-sensitive activity at a constant ATP concentration and was found to be normal.
Table 1. Respiratory chain enzyme and mitochondrial ATPase activities in isolated muscle mitochondria and cultured skin fibroblasts.
SCCR, succinate:cytochrome c reductase; NCCR, NADH:cytochrome c reductase; SQR, succinate:CoQ reductase; QCCR, CoQ:cytochrome c reductase; NQR, NADH:CoQ reductase; ND, not determined. Values for age-related controls are presented as means±S.D. and as a control range (parentheses).
| (A) | |||||||
|---|---|---|---|---|---|---|---|
| Activity (nmol/min per mg of protein) | |||||||
| Muscle homogenate | Muscle mitochondria | Fibroblasts (mitochondria*) | |||||
| Enzyme | Patient | Controls (n=20) | Patient | Controls (n=20) | Patient | Controls (n=30) | |
| COX | 122 | 125±72 (53–197) | 331 | 682±395 (287–1077) | 5.1 | 29±11 (18–40) | |
| CS | 241 | 114±29 (85–143) | 284 | 419±221 (200–640) | 40.6 | 58±12 (46–70) | |
| COX/CS | 0.50 | 1.04±0.44 (0.60–1.48) | 1.16 | 1.66±0.57 (1.09–2.23) | 0.12 | 0.51±0.18 (0.33–0.69) | |
| SCCR | ND | 98 | 127±77 (50–206) | ND | |||
| NCCR | ND | 76 | 99±57 (42–156) | 16.3 | 28±14 (14–42) | ||
| SQR | ND | 43 | 64±44 (20–108) | 10.6 | 13±8 (5–21) | ||
| QCCR | ND | 54 | 178±79 (100–257) | 14.1 | 17±10 (7–27) | ||
| NQR | ND | 192 | 274±80 (194–354) | 33.8 | 23±10 (15–50) | ||
| ATPase | ND | ND | 320±33.1* | 254±16.0* | |||
| (B) | |||||||
| Activity (nmol/min per mg of protein) | |||||||
| Patient | Mother | Grandmother | Controls (n=30) | ||||
| Enzyme | Expt 1 | Expt 2 | |||||
| COX | 7.9 | 2.4 | 16.9 | 34.4 | 29±11 | ||
| CS | 45.5 | 33.8 | 50.6 | 52.2 | 58±12 | ||
| COX/CS | 0.17 | 0.07 | 0.33 | 0.66 | 0.51±0.18 | ||
When COX activity was measured in fibroblasts from the patient's asymptomatic mother and grandmother, decreased COX activity and a COX/CS ratio near the lower limit of the reference range were found in the mother, but not in the grandmother (Table 1B).
mtDNA 9205ΔTA mutation in the affected family
Sequencing of the patient's mtDNA revealed a very rare mtDNA mutation – a heteroplasmic 2 bp deletion TA at positions 9205 and 9206 (9205ΔTA) in the ATP6–COX3 genes. Analysis of mutation heteroplasmy in tissues of the affected boy by radioactive RFLP (using NsiI restrictase; Table 2) showed practically homoplasmic 9205ΔTA mtDNA mutation in fibroblasts, muscle and blood (>98%). Analysis of blood DNA confirmed the presence of the 9205ΔTA mutation at lower loads both in the mother (82%) and in the grandmother (16%). Interestingly, a similar mutation load was found in their fibroblasts (mother 92%; grandmother 9%), in correspondence with the observed decrease in COX activity in the mother's fibroblasts (see above).
Table 2. 9205ΔTA mutation load in family members.
| Sample | Heteroplasmy (%) |
|---|---|
| mtDNA | |
| Grandmother – blood DNA | 16 |
| Grandmother – fibroblast DNA | 9 |
| Mother – blood DNA | 82 |
| Mother – fibroblast DNA | 92 |
| Patient – blood DNA | >98 |
| Patient – muscle DNA | >98 |
| Patient – fibroblasts DNA | >98 |
| cDNA | |
| Patient – fibroblasts cDNA oligo(dT) | >98 |
| Patient – fibroblasts cDNA oligo(dT)+random primers | >98 |
An equal mutation load in the patient's fibroblasts was found using RFLP of isolated DNA as well as of cDNA reverse-transcribed from isolated RNA using either oligo(dT) or oligo (dT)+random primers, indicating that the mutation is fully retained in ATP6–COX3 RNA and poly(A) RNA (Table 2).
Oxygraphic analysis of cultured fibroblasts
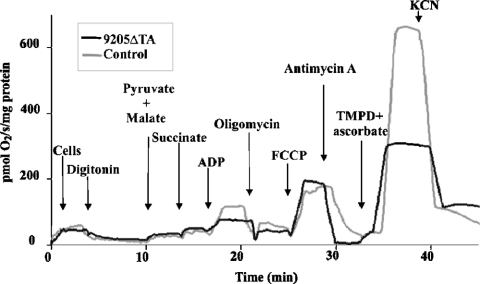
The functional consequences of the 9205ΔTA mutation were analysed by high-resolution oxygraphy of the patient's fibroblasts that had been permeabilized by a low concentration of digitonin. The 9205ΔTA cells showed a pronounced decrease in ADP-stimulated oxygen consumption using pyruvate, malate and succinate as substrate, which contrasted with a normal rate of state 3 respiration in the presence of the uncoupler FCCP (carbonyl cyanide 4-trifluoromethoxyphenylhydrazone). We also observed a decrease in COX activity determined with TMPD and ascorbate (Figure 1). In 9205ΔTA cells from the patient, ADP-stimulated respiration was 50–60%, FCCP-stimulated respiration was 100–110% and COX activity was 40–50% of values in control cells. Although well coupled, the mitochondria of 9205ΔTA cells thus showed a strongly impaired effect of ADP, indicating a decreased ability to synthesize ATP.
Figure 1. Oxygen consumption by digitonin-permeabilized skin fibroblasts.
Measurements were performed using 0.45–0.62 mg of cell protein/ml and 0.1 mg of digitonin/mg of protein. Subsequent additions of pyruvate (5 mM), malate (1.5 mM), succinate (10 mM), ADP (0.5 mM), oligomycin (1 μM), FCCP (1 μM), antimycin A (0.2 μg/ml), ascorbate (5 mM), TMPD (1 mM) and KCN (0.33 mM) are indicated. Oxygen consumption is expressed as negative values of the first time derivative of changes in oxygen tension (pmol of O2/s per mg of protein).
Low ATP production in 9205ΔTA cells
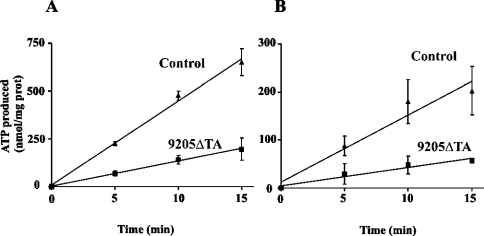
The ability of fibroblasts to synthesize ATP by the mitochondrial OXPHOS pathway was measured directly in cells suspended in mitochondrial medium, permeabilized by digitonin and supplied with respiratory substrates and ADP, as in the oxygraphic experiments. With either succinate or pyruvate+malate as substrate, linear production of ATP was observed in control cells for 15 min, corresponding to ATP production of 16.4±1.8 and 45.4±1.5 nmol/min per mg of protein respectively, in accordance with the oxygraphic measurements. In 9205ΔTA fibroblasts, a steadily diminished ATP production was observed during the 15 min interval with both types of substrate, giving values of 4.8±0.7 and 13.7±0.4 nmol/min per mg of protein respectively, which are approx. 30% of control values (Figure 2).
Figure 2. Production of ATP by control and 9205ΔTA fibroblasts.
The rate of ATP synthesis was measured in the presence of 0.5 mM ADP in 9205ΔTA (▪) and control (▴) cultured skin fibroblasts treated with 0.1 mg of digitonin/mg of protein. Substrates were 10 mM pyruvate+10 mM malate (A) or 10 mM succinate (B). ATP was determined in DMSO-quenched aliquots by a luciferase assay. Data represent means±S.D. for three experiments.
Changes in ΔΨm
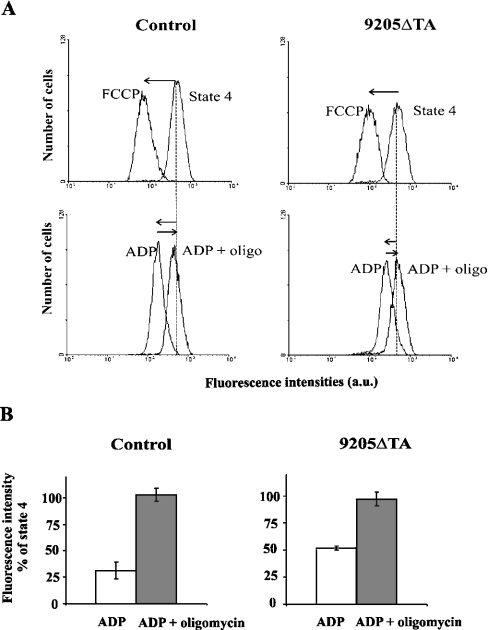
ΔΨm was analysed cytofluorimetrically in digitonin-permeabilized fibroblasts using the membrane potential-sensitive cationic probe TMRM. We found that the ΔΨm of the patient's fibroblasts was practically normal at state 4 using succinate as a substrate, but the effect of the addition of ADP on ΔΨm was significantly decreased in 9205ΔTA cells (Figure 3). Whereas in control cells the TMRM fluorescence was decreased by the addition of ADP (state 3 ADP) to approx. 28% of the state 4 value, in the 9205ΔTA cells the TMRM fluorescence decreased to only 50%. The observed 2-fold difference in TMRM fluorescence would correspond to an even more pronounced difference in ΔΨm values in mV, because the accumulation of the fluorophore obeys the Nernst equation and depends exponentially on ΔΨm [36]. In both types of cells, the effect of ADP was fully reversible by oligomycin (Figure 3), atractyloside or aurovertin (results not shown), indicating that the effect of ADP requires the transport of ADP to the mitochondrial matrix as well as the catalytic activity of ATPase. In accordance with the measurements of ATP production and respirometry, these results suggest that ATP synthesis by mitochondrial ATPase is strongly impaired in 9205ΔTA cells from the affected individual.
Figure 3. Cytofluorimetric analysis of control and 9205ΔTA fibroblasts.
Cytofluorimetric analysis was performed in digitonin-treated fibroblasts (0.1 mg of digitonin/mg of protein) and stained with 20 nM TMRM in a KCl medium containing 10 mM succinate. (A) Typical reading of TMRM fluorescence at state 4 and effect of 1 μM FCCP is shown in the upper panels. Lower panels show the effect of 0.1 mM ADP and its sensitivity to 1 μM oligomycin (oligo). (B) TMRM fluorescence of state 3-ADP and effect of oligomycin is expressed as percentage of the state 4 signal. The ΔΨm-independent signal (after addition of FCCP) was subtracted from all data. The data represent means±S.D. for four independent experiments.
Altered composition and increased lability of ATPase and COX
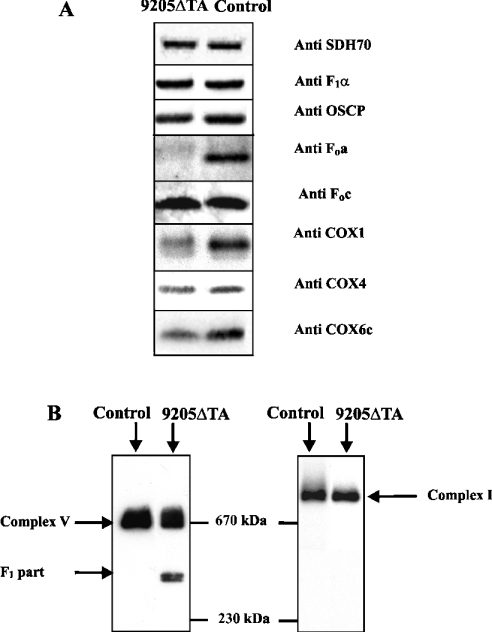
In order to assess the cellular content of ATPase and COX subunits, we analysed the mitochondrial proteins of isolated fibroblasts by SDS/PAGE and WB. It is clearly apparent from Figure 4(A) that 9205ΔTA fibroblasts had a selectively diminished content of ATPase subunit a and a significantly decreased content of COX subunits. In comparison with control cells, 9205ΔTA mitochondria contained 11±5.9% of Fo subunit a, 127±13.0% of F1 α subunit, 85% of OSCP (oligomycin-sensitivity-conferring protein), 108±18.0% of Fo subunit c, 47±2.8% of COX1 subunit, 69±8.5% of COX4 subunit and 36±21.2% of COX6c subunit (average values of four experiments; data normalized to the content of the SDH 70 kDa subunit). The same pattern was obtained when analysing complete cell protein extracts (results not shown).
Figure 4. Electrophoretic analysis of OXPHOS complexes.
(A) SDS/PAGE WB of ATPase, COX and SDH subunits was performed using aliquots of 7.5 μg of protein of mitochondria from control and 9205ΔTA cells. Detection was done with monoclonal antibodies against subunits SDH70, F1α, OSCP (oligomycin-sensitivity-conferring protein), COX1, COX4 and COX6c, and with polyclonal antibodies against ATPase subunits Foa and Foc. (B) BN-PAGE WB with monoclonal antibody against F1α and NADH39 subunit was performed using 15 μg aliquots of protein solubilized using 1 g of DDM/g of protein from mitoplasts of control and 9205ΔTA cells. The migration of molecular mass standards is indicated.
To resolve the native ATPase complex, we solubilized mitochondrial OXPHOS complexes from mitoplasts by DDM and analysed them by BN-PAGE and WB using an antibody against ATPase F1 α subunit. As shown in Figure 4(B), in 9205ΔTA cells we found a decreased content of the full-size ATPase and the accumulation of an incomplete form of the ATPase with a molecular mass of approx. 390 kDa. A parallel WB analysis of complex I using an antibody to the NADH39 subunit showed an identical pattern in control and 9205ΔTA cells – we observed only intact complex with a molecular mass above 800 kDa (Figure 4B) that was present in the same amounts in 9205ΔTA and control cells.
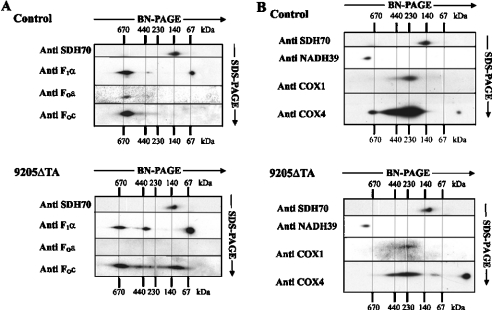
2D electrophoretic analysis of ATPase and COX
Detailed analysis of the different forms of ATPase present in DDM-solubilized enzyme complexes was performed by 2D electrophoresis, whereby proteins resolved by BN-PAGE in the first dimension were separated by SDS/PAGE in the second dimension and detected by WB. The ATPase complex with an apparent molecular mass of approx. 620 kDa was significantly decreased in 9205ΔTA cells, but several other, smaller ATPase subcomplexes were detected by F1 subunit α- and Fo subunit c-immunoreactive signals (Figure 5A). In addition to the 390 kDa F1 subcomplex containing the α subunit, which was present in the 9205ΔTA cells in increased amounts, a larger 460 kDa complex containing both subunits α and c was also found in the 9205ΔTA cells. Moreover, an additional, subunit c-containing complex with a molecular mass of approx. 120 kDa and a markedly increased amount of free subunit α (approx. 60 kDa) were seen. However, none of the above complexes showed a detectable reaction with an antibody against subunit a in the 9205ΔTA cells.
Figure 5. 2D electrophoretic analysis and immunodetection of OXPHOS complexes.
Aliquots (15 μg of protein) of DDM-solubilized mitoplasts from 9205ΔTA and control fibroblasts were separated in the first dimension by BN-PAGE and in the second dimension by SDS/PAGE. WB analysis was performed with an antibody against the SDH70 subunit and with antibodies (A) against ATPase subunits F1α, Foa and Foc, or (B) against complex I subunit NADH39 and against COX subunits COX1 and COX4. The migration of molecular mass standards is indicated.
2D analysis of COX was performed using antibodies to COX1 and COX4 subunits (Figure 5B). A pronounced decrease in the amount of COX was observed in the DDM solubilizate of 9205ΔTA cells. In control cells, most of the COX was present as a monomer, but a significant amount of COX dimers (approx. 420–440 kDa) and small amounts of COX supercomplex (approx. 670 kDa; probably COX-bc1 supercomplex) as well as some monomeric COX4 subunits were clearly seen. In the 9205ΔTA cells the pattern was different. An additional band of molecular mass approx. 100 kDa was present, and the relative content of the free COX4 subunit was much higher (approx. 15-fold) than in control cells.
WB analysis of complex I on 2D gels using a monoclonal antibody to the NADH39 subunit showed no difference between control and 9205ΔTA cells. Only the full-size, assembled complex I was found, and was present in similar amounts in both types of cells. No assembly intermediates were detected. Similarly, no difference was found in the case of complex II using the anti-SDH70 antibody. Thus, unlike ATPase and COX, the biosynthesis and/or stability of complex I were not affected by the 9205ΔTA mutation.
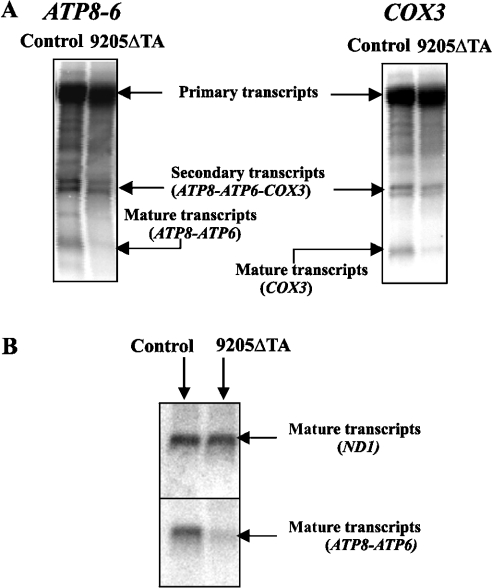
Steady-state levels and processing of ATP6 and COX3 mRNAs
The 9205ΔTA microdeletion is situated between the genes ATP6 and COX3, which are transcribed in one polycistronic transcript that is subsequently cleaved stepwise and polyadenylated into mature mRNA. Therefore we studied the steady-state levels and processing of ATP6 and COX3 mRNAs. Northern blot analysis using cDNA complementary to mtDNA sequences 8361–9060 and 9269–9912 as a probe was performed to determine whether the mutation disturbs the RNA processing of ATP6 and COX3 transcripts (Figure 6A). The primary, unprocessed, and the secondary ATP8–ATP6 – COX3 transcripts occurred in both 9205ΔTA cells and control cells in similar amounts. The steady-state levels of processed COX3 and ATP8–ATP6 transcripts were decreased in 9205ΔTA cells. The relative values of the ATP8–ATP6/ATP8–ATP6 – COX3 and COX3/ATP8–ATP6 – COX3 RNA ratios revealed approx. 40% and 50% decreases respectively in ATP8–ATP6 and COX3 RNA levels. In parallel, we also determined the steady-state levels of the transcripts of two other genes encoded by mtDNA, i.e. ND1 and COX1. We observed the same content of the mature ND1 transcript (Figure 6B) in control and 9205ΔTA cells. Similarly, there was no difference in the content of COX1 transcripts (results not shown).
Figure 6. Northern blot analysis of ATP6 and COX3 transcripts.
Northern blot analysis of COX3, ATP8–ATP6 and ND1 transcripts was performed in control and 9205ΔTA cells. The mRNA species representing (A) the primary mtDNA polycistron transcript, partially processed ATP8–ATP6–COX3 transcripts and mature COX3 and ATP8–ATP6 transcripts and (B) mature ND1 and ATP8–ATP6 transcripts are indicated.
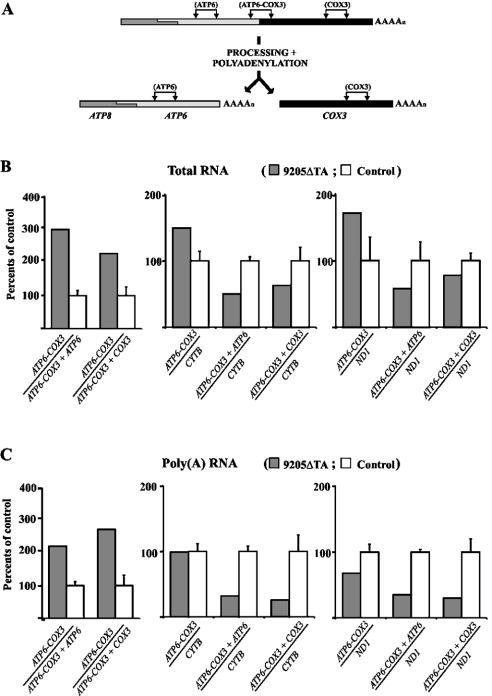
These Northern blot data were fully confirmed by quantitative RT-PCR experiments (Figure 7). Using PCR products corresponding to ATP6 (nt 8846–8994), COX3 (nt 9267–9416) and the ATP6 – COX3 cleavage site (nt 9150–9299), we found that ATP6 as well as COX3 RNA levels were decreased 2–3-fold relative to those of the uncleaved polycistronic ATP8–ATP6–COX3 transcript. The decrease was also confirmed when relating ATP6 and COX3 RNA levels to CYTB and ND1 expression (PCR product of 14804–14935 for CYTB and 3595–3644 for ND1). As also shown in Figure 7, similar results were obtained using cDNA prepared by oligo(dT) or random primers, indicating that polyadenylation is neither impaired nor affects transcript processing.
Figure 7. Quantitative real-time RT-PCR analysis of ATP6, COX3, cytochrome b and ND1 transcripts.
(A) Scheme of ATP6–COX3 RNA processing and PCR products analysed. For experiments, cDNAs obtained from (B) total RNA (reverse transcription with random primers) or (C) mRNA [reverse transcription with oligo(dT) primers] were used. The relative amount of uncleaved ATP6–COX3 transcript and the amounts of all ATP6 or COX3 transcripts present either in unprocessed form or in mature form were measured and correlated. The values were also correlated to the expression of mitochondrial CYTB and ND1 as reference genes. Graphs depict results obtained in 9205ΔTA fibroblasts expressed as a percentage of the mean±S.D. for three controls.
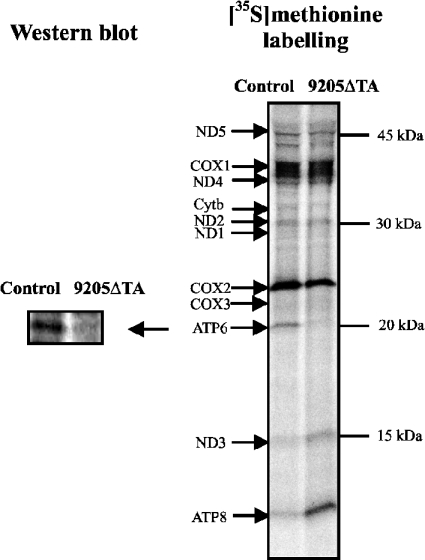
Specific decrease in the biosynthesis of ATPase subunit a
To assess how the 9205ΔTA mutation affects the biosynthesis of subunit a and other mtDNA-encoded proteins, [35S]methionine labelling of proteins in 9205ΔTA cells was performed in the presence of emetine (an inhibitor of cytosolic proteosynthesis). The extent of labelling (Figure 8) was comparable in 9205ΔTA and control cells, and the pattern of labelled proteins was also similar, with two exceptions – markedly decreased labelling of a band of approx. 22 kDa and increased labelling of a band of approx. 12 kDa. The first band was identified as ATPase subunit a, based on WB analysis performed on the same gel with an antibody against subunit a (Figure 8), in accordance with the size and mobility of subunit a in this type of SDS/PAGE [37]. On the basis of its size, the second band was identified as ATPase subunit 8 (A6L). The COX3 subunit was weakly labelled in both types of cells, and no significant difference could be seen between the 9205ΔTA and control cells. Three independent experiments gave essentially the same result, and clearly showed, in accordance with all WB data, that mutation of the ATP6 STOP codon greatly decreases the synthesis of ATPase subunit a (to 12.7±0.73% of the control). In turn, up-regulation of ATP8 synthesis is apparent.
Figure 8. Incorporation of [35S]methionine into proteins encoded by mtDNA.
mtDNA-specific translation was performed in control and 9205ΔTA fibroblasts in the presence of emetine. Radioactive proteins were separated by SDS/PAGE on a 15–20% (w/v) gradient polyacrylamide gel and detected by phosphorimaging. On the left side, the WB from the same SDS/PAGE run using an antibody against subunit Foa is shown. The migration of mtDNA-encoded polypeptides and of molecular mass standards is indicated.
DISCUSSION
We present a detailed analysis of the impact of a maternally inherited 2 bp microdeletion TA at position 9205 and 9206 in the mtDNA (9205ΔTA) on the respiratory chain complexes, with the aim of disclosing the pathogenic mechanism in a patient with severe encephalomyopathy, spastic quadruparesis, microcephalia and hyperlactacidaemia.
Biochemically, the patient presented with a pronounced selective decrease in COX activity in his fibroblasts (15% of control), while a lowered COX/CS ratio and a mild decrease in complex III activity were found in muscle. Sequencing of mtDNA revealed a 2 bp microdeletion, which disrupts the STOP codon of the ATP6 gene. This mutation has been described only once previously, in a patient with seizures and repetitive bouts of lactic acidaemia [13]. It was reported as being homoplasmic (lymphocytes and fibroblasts), but was not detected in that patient's mother or grandmother. In our patient, the mutation was also found to be homoplasmic (>98%) in all tissues tested. In addition, the heteroplasmic mutation 9205ΔTA was present in the asymptomatic mother (82%) and grandmother (16%). Importantly, a high mutation load in the mother's fibroblasts was also associated with a decrease in COX activity.
The mtDNA mutation 9205ΔTA lies on the boundary of two genes, ATP6 and COX3, and it is highly probable that it influences the transcription and post-transcriptional modification of these genes. Assuming that this is the main cause of the patient's phenotype, insufficient energy provision is to be expected in the tissues, particularly in brain (explaining the dominating encephalopathy). Our measurements in the patient's fibroblasts showed a 3-fold decrease in mitochondrial ATP production. In fact, the observed decrease was even higher than that observed in cases of missense mutations in the ATP6 gene (T9176G, T8993G and T8993C), where higher values of residual mitochondrial ATP production have been reported [10–12,38]. By analogy with these mutations, we found completely normal ATPase hydrolytic activity. Thus it appears that only a fraction of ATPase complexes can utilize ΔΨm to drive ATP synthesis in 9205ΔTA cells. This view is further supported by the normal ΔΨm observed at state 4 and its decrease by FCCP, while ADP (state 3-ADP) caused a much smaller decrease in ΔΨm in the patient. These data indicate that mitochondria in 9205ΔTA cells can be fully energized and that the mutation does not enhance the passive H+ transport at state 4 via the Fo proton channel of the ATPase complexes, which are known to contribute to the proton conductivity of the mitochondrial inner membrane [39]. When, however, ADP becomes available, the discharge by ATP synthesis of ΔΨm in 9205ΔTA cells is significantly decreased.
Our functional data support the view that the Fo part has an altered function in the majority of ATPase complexes of the patient's fibroblasts, due to the absence of subunit a (an essential component of the ATPase proton channel) or an alteration of its structure (for review, see [40]). The absence of the ATP6 STOP codon could interfere with synthesis of the subunit by decreasing the translational efficacy of the ATP6 mRNA, resulting in a low amount of subunit a being produced and in the formation of ATPase complexes lacking subunit a and incapable of synthesizing ATP. Another possibility might be production of subunit a modified in the C-terminal part, as a poly(A) tail in the absence of a regular STOP codon could cause extension of several lysine residues at the C-terminus. Indeed, several amino acid residues involved in H+ translocation are located in this part of the protein [8].
To delineate the subunit composition and native structure of ATPase, we employed electrophoretic/WB analysis using subunit-specific antibodies, including a polyclonal antibody against subunit a [23]. We have found that the content of subunit a is greatly decreased (to 11% of control) in the patient's fibroblasts, whereas other ATPase subunits are unchanged. The antibody was raised against 10 amino acids at the N-terminal part of human subunit a, and most probably would react with a putative form of subunit a elongated at the C-terminus. However, no larger forms of subunit a could be detected by WB. A low content of subunit a was confirmed by 9-fold decreased labelling of this subunit by [35S]methionine, and the labelling pattern also did not indicate the accumulation of a larger form of this subunit in 9205ΔTA cells.
Under native electrophoretic conditions, we found in the 9205ΔTA cells a decreased content of DDM-solubilized, normal-sized F1Fo-ATPase, together with an increased content of an F1 subcomplex of 390 kDa. There was no apparent difference between the mobility of the 620 kDa ATPase complex in the 9205ΔTA and control samples; however, subunit a, present in the ATPase in one copy, represents only 4% of the total ATPase mass, and the expected difference in mobility of the whole, subunit a-lacking complex is below the resolution of BN-PAGE. Furthermore, we found yet another larger, F1-containing subcomplex of 460 kDa that consists of F1-ATPase and some Fo subunits, including subunit c. This pattern closely resembles that in cells with a T8993G mutation in the ATP6 gene [10,12], or in cells with doxycycline-inhibited mitochondrial protein synthesis [3]. Interestingly, these three different mechanisms that affect the biosynthesis of subunit a gave a very similar picture at the level of ATPase structure. In the case of the 9205ΔTA mutation, the ATPase complexes exert lability upon DDM extraction that is not observed in those from control cells; however, even the subunit a-lacking ATPase complexes maintain structural interactions between the F1 and Fo parts of the enzyme, although these are weaker. It appears that incomplete Fo is also able to ‘gate’ the F1 in these complexes, as no H+ leak has been detected by measurement of membrane potential.
In addition to changes in ATPase, we also found changes in the structure and function of COX. The pronounced decrease in COX activity observed in spectrophotometric assays was confirmed by the oxidation of TMPD+ascorbate in 9205ΔTA cells. The remaining COX activity, however, did not limit the generation of ΔΨm, and it was also sufficient for substrate oxidation in the uncoupled state. The functional consequences at the level of COX resembled to some extent COX assembly defects due to SURF1 mutations [41]. The total amount of COX subunits detected by WB was also decreased, indicating a 30–50% decrease in enzyme content. Some of the COX complexes showed a normal size, but some COX subunits detected by 2D/WB analysis were found to be present in COX subcomplexes or as free subunits. The 2D pattern resembled the pattern observed in cells with a 15 bp deletion (9480Δ15) in COX3 [37] that show a failure to assemble the holoenzyme complex and instability of the COX1–COX2 interaction [42].
When comparing the changes in ATPase and COX, it appears that 9205ΔTA cells contain approx. 10% of the control content of normal ATPase complexes and 30–50% of that of normal COX complexes. Both enzymes are present in mitochondria at normal, physiological conditions in excess in most tissues, and their threshold values are known to vary in individual cell types [43]. With regard to the dominating encephalopathy in our patient, it is interesting that the reserve capacity of COX appears to be rather high in brain, while the reserve capacity of ATPase appears to be much lower.
All of the data presented herein are in accordance with the hypothesis that the mechanism by which the 9205ΔTA mutation affects mitochondrial function is associated with changes in the transcription of the ATP6 and COX3 genes and their translational competence and efficacy. Using two independent approaches, i.e. Northern blot and quantitative real-time PCR analysis of cDNA, we found that processing of the primary ATP8–ATP6 – COX3 transcript is impaired in our patient. Both methods revealed a 2–3-fold decrease in the content of mature ATP6 and COX3 transcripts, while the content of ND1 and COX1 transcripts was unaffected. The same picture emerged when analysing polyadenylated forms of ATP6 and COX3 RNAs. These differences found in the patient's cells were also confirmed when relating the steady-state levels of ATP6 and COX3 RNAs and mRNAs to the levels of CYTB or ND1 transcripts.
The decrease in the amount of the mature ATP6 transcript agreed well with the decreased synthesis and content of subunit a. Interestingly, the labelling of subunit 8 was increased, indicating up-regulated translation of the ATP8 gene, which precedes and partially overlaps the ATP6 gene. The translation of the ATP8 and ATP6 mRNAs is well described in yeast, but the structure of these genes and its regulation differs completely from that in mammalian mitochondria, where the mechanism of ATP8 and ATP6 biosynthesis is largely unknown. The question arises whether increased labelling of subunit 8 could be caused by translation of ATP8 from an unspliced form of the ATP8–ATP6 – COX3 transcript, part of which is, according to our results, polyadenylated [cDNA synthesis with random primers and oligo(dT) primer] and could be therefore subjected to translation.
Recently, Seneca and co-workers continued an analysis of the fibroblasts from the first patient identified with the 9205ΔTA deletion [18,44]. They investigated the levels and fate of ATP6 and COX3 mRNAs and did not find any difference in primary transcript processing in their patient, but some differences in the deadenylation of mRNAs were present. In the second paper they investigated the biochemical consequences of the mutation, and the only difference they found was an accumulation of the F1-ATPase intermediate and increased ATP8 labelling. Therefore two patients with the same 9205ΔTA homoplasmic mutation differ dramatically, and this difference is difficult to understand. There appears to be a good correlation between changes in mature transcripts, pronounced biochemical consequences and severe encephalopathy in our patient, compared with unchanged RNA processing, insignificant biochemical changes and much weaker clinical presentation in the Seneca study. On the other hand, both patients harbour the same homoplasmic 9205ΔTA mutation. Thus in one case the mutation is pathogenic, and in the other it has little effect. We have no explanation for this difference at the moment, but several aspects might be important. In both cases numerous other changes in mtDNA sequence were found. In our case all were identified as more or less frequent polymorphisms, one of which was present in the ATP6 gene (A8860G) and one in the COX3 gene (G9477A). In the Seneca case mtDNA sequencing revealed 42 changes from the revised Cambridge Reference Sequence, including three rare polymorphisms (C9335T, A11362G and A12822G) and two novel transitions (T15287C and T15705C) in the CYTB gene. The two types of mtDNA also belong to different haplogroups.
The question arises as to whether in the Seneca case the absence of pathogenicity might be due to some compensatory mechanism affecting the post-transcriptional processing and translation of the ATP6 and COX3 mRNAs. Two revertants have been described in human mtDNA, in cells carrying tRNA mutations. A suppressor mutation at position 12300 was found, generating tRNALeu (CUN), which compensated for the 3243 (MELAS; mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes) mutation of tRNALeu (UUR) [45]. In the second case, a G5703A mutation in tRNAAsn was functionally rescued without changes in mtDNA upon cultivation in a galactose medium [46]. A compensatory import of surrogate tRNA due to nuclear mutation was proposed, but not shown. Moreover, a similar compensatory import of RNAs or proteins is highly unlikely in 9205ΔTA cells.
A possible compensatory mechanism could be connected with factors that affect (and/or correct) the processing of mitochondrial transcripts and their translation in 9205ΔTA cells. Interestingly, several nuclear-encoded factors have been described in yeast (NCA2, NCA3, NAM1/MTF2, Aep3p) that are essential for proper processing of mitochondrial RNAs, namely the ATP8–ATP6 co-transcript (for references see [47]), but their mammalian orthologues have not been found, possibly reflecting differences in the structure of mitochondrial RNAs between yeast and mammals. Another group of factors is represented by proteins mediating the mRNA–ribosome interaction. The search for mammalian orthologues in this group was more successful. A LRPPRC (leucine-rich pentatricopeptide repeat cassette) protein was identified using a functional genomics approach [48], and it was shown that mutation in the LRPPRC gene causes the Leigh syndrome of French–Canadian type, which is a human mitochondrial COX deficiency [49]. However, the divergence of the LRPPRC protein sequence from that of the analogous yeast protein is very large and, therefore, it has not yet been possible to identify other mammalian proteins by a sequence similarity approach.
Acknowledgments
This work was supported by grants from the Grant Agency of the Ministry of Health of the Czech Republic (NR/7790-3, 8065-3), the Grant Agency of Charles University (GAUK 14/2004), the Grant Agency of the Czech Republic (303/03/0749) and by institutional projects (AVOZ5011922, VZ 111100003). The expert technical assistance of V. Fialová and V. Brožková is gratefully acknowledged.
References
- 1.Walker J. E., Collinson I. R. The role of the stalk in the coupling mechanism of F1F0-ATPases. FEBS Lett. 1994;346:39–43. doi: 10.1016/0014-5793(94)00368-8. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H. L., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F., et al. Sequence and organization of the human mitochondrial genome. Nature (London) 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 3.Nijtmans L. G., Klement P., Houstek J., van den Bogert C. Assembly of mitochondrial ATP synthase in cultured human cells: implications for mitochondrial diseases. Biochim. Biophys. Acta. 1995;1272:190–198. doi: 10.1016/0925-4439(95)00087-9. [DOI] [PubMed] [Google Scholar]
- 4.DiMauro S., Schon E. A. Mitochondrial DNA mutations in human disease. Am. J. Med. Genet. 2001;106:18–26. doi: 10.1002/ajmg.1392. [DOI] [PubMed] [Google Scholar]
- 5.Holt I. J., Harding A. E., Petty R. K. H., Morgan-Hughes J. A. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am. J. Hum. Genet. 1990;46:428–433. [PMC free article] [PubMed] [Google Scholar]
- 6.de Vries D. D., van Engelen B. G., Gabreels F. J., Ruitenbeek W., van Oost B. A. A second missense mutation in the mitochondrial ATPase 6 gene in Leigh's syndrome. Ann. Neurol. 1993;34:410–412. doi: 10.1002/ana.410340319. [DOI] [PubMed] [Google Scholar]
- 7.Tatuch Y., Christodoulou J., Feigenbaum A., Clarke J. T., Wherret J., Smith C., Rudd N., Petrova Benedict R., Robinson B. H. Heteroplasmic mtDNA mutation (T→G) at 8993 can cause Leigh disease when the percentage of abnormal mtDNA is high. Am. J. Hum. Genet. 1992;50:852–858. [PMC free article] [PubMed] [Google Scholar]
- 8.Schon E. A., Santra S., Pallotti F., Girvin M. E. Pathogenesis of primary defects in mitochondrial ATP synthesis. Semin. Cell Dev. Biol. 2001;12:441–448. doi: 10.1006/scdb.2001.0281. [DOI] [PubMed] [Google Scholar]
- 9.Tatuch Y., Robinson B. H. The mitochondrial DNA mutation at 8993 associated with NARP slows the rate of ATP synthesis in isolated lymphoblast mitochondria. Biochem. Biophys. Res. Commun. 1993;192:124–128. doi: 10.1006/bbrc.1993.1390. [DOI] [PubMed] [Google Scholar]
- 10.Houstek J., Klement P., Hermanska J., Houstkova H., Hansikova H., van den Bogert C., Zeman J. Altered properties of mitochondrial ATP-synthase in patients with a T→G mutation in the ATPase 6 (subunit a) gene at position 8993 of mtDNA. Biochim. Biophys. Acta. 1995;1271:349–357. doi: 10.1016/0925-4439(95)00063-a. [DOI] [PubMed] [Google Scholar]
- 11.Garcia J. J., Ogilvie I., Robinson B. H., Capaldi R. A. Structure, functioning, and assembly of the ATP synthase in cells from patients with the T8993G mitochondrial DNA mutation. Comparison with the enzyme in Rho0 cells completely lacking mtDNA. J. Biol. Chem. 2000;275:11075–11081. doi: 10.1074/jbc.275.15.11075. [DOI] [PubMed] [Google Scholar]
- 12.Nijtmans L. G., Henderson N. S., Attardi G., Holt I. J. Impaired ATP synthase assembly associated with a mutation in the human ATP synthase subunit 6 gene. J. Biol. Chem. 2001;276:6755–6762. doi: 10.1074/jbc.M008114200. [DOI] [PubMed] [Google Scholar]
- 13.Seneca S., Abramowicz M., Lissens W., Muller M. F., Vamos E., de Meirleir L. A mitochondrial DNA microdeletion in a newborn girl with transient lactic acidosis. J. Inherit. Metab. Dis. 1996;19:115–118. doi: 10.1007/BF01799407. [DOI] [PubMed] [Google Scholar]
- 14.Fornuskova D., Tesarova M., Hansikova H., Zeman J. New mtDNA mutation 9204delTA in a family with mitochondrial encephalopathy and ATP synthase defect. Cas. Lek. Cesk. 2003;142:313. [Google Scholar]
- 15.Bentlage H. A., Wendel U., Schagger H., ter Laak H. J., Janssen A. J., Trijbels J. M. Lethal infantile mitochondrial disease with isolated complex I deficiency in fibroblasts but with combined complex I and IV deficiencies in muscle. Neurology. 1996;47:243–248. doi: 10.1212/wnl.47.1.243. [DOI] [PubMed] [Google Scholar]
- 16.Klement P., Nijtmans L. G., Van den Bogert C., Houstek J. Analysis of oxidative phosphorylation complexes in cultured human fibroblasts and amniocytes by blue-native-electrophoresis using mitoplasts isolated with the help of digitonin. Anal. Biochem. 1995;231:218–224. doi: 10.1006/abio.1995.1523. [DOI] [PubMed] [Google Scholar]
- 17.Makinen M., Lee C. P. Biochemical studies of skeletal muscle mitochondria: I. Microanalysis of cytochrome content, oxidative and phosphorylative activities of mammalian skeletal muscle mitochondria. Arch. Biochem. Biophys. 1968;126:75–82. doi: 10.1016/0003-9861(68)90561-4. [DOI] [PubMed] [Google Scholar]
- 18.Chrzanowska-Lightowlers Z. M., Temperley R. J., Smith P. M., Seneca S. H., Lightowlers R. N. Functional polypeptides can be synthesized from human mitochondrial transcripts lacking termination codons. Biochem. J. 2004;377:725–731. doi: 10.1042/BJ20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J., Russell D. W. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 20.Schagger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 21.Schagger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 22.Houstek J., Andersson U., Tvrdik P., Nedergaard J., Cannon B. The expression of subunit c correlates with and thus may limit the biosynthesis of the mitochondrial F0F1-ATPase in brown adipose tissue. J. Biol. Chem. 1995;270:7689–7694. doi: 10.1074/jbc.270.13.7689. [DOI] [PubMed] [Google Scholar]
- 23.Dubot A., Godinot C., Dumur V., Sablonniere B., Stojkovic T., Cuisset J. M., Vojtiskova A., Pecina P., Jesina P., Houstek J. GUG is an efficient initiation codon to translate the human mitochondrial ATP6 gene. Biochem. Biophys. Res. Commun. 2004;313:687–693. doi: 10.1016/j.bbrc.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 24.Moradi-Ameli M., Godinot C. Characterization of monoclonal antibodies against mitochondrial F1-ATPase. Proc. Natl. Acad. Sci. U.S.A. 1983;80:6167–6171. doi: 10.1073/pnas.80.20.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wharton D. C., Tzagoloff A. Cytochrome oxidase from beef heart mitochondria. Methods Enzymol. 1967;10:245–253. [Google Scholar]
- 26.Rustin P., Chretien D., Bourgeron T., Gerard B., Rotig A., Saudubray J. M., Munnich A. Biochemical and molecular investigations in respiratory chain deficiencies. Clin. Chim. Acta. 1994;228:35–51. doi: 10.1016/0009-8981(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 27.Fischer J. C., Ruitenbeek W., Trijbels J. M. F., Veerkamp J. H., Stadhouders A. M., Sengers R. C. A., Janssen A. J. M. Differential investigation of the capacity of succinate oxidation in human skeletal muscle. Clin. Chim. Acta. 1985;153:37–42. doi: 10.1016/0009-8981(85)90135-4. [DOI] [PubMed] [Google Scholar]
- 28.Fischer J. C., Ruitenbeek W., Trijbels J. M. F., Veerkamp J. H., Stadhouders A. M., Sengers R. C. A., Janssen A. J. M. Estimation of NADH oxidation in human skeletal muscle mitochondria. Clin. Chim. Acta. 1986;155:263–274. doi: 10.1016/0009-8981(86)90246-9. [DOI] [PubMed] [Google Scholar]
- 29.Chowdhury S. K., Drahota Z., Floryk D., Calda P., Houstek J. Activities of mitochondrial oxidative phosphorylation enzymes in cultured amniocytes. Clin. Chim. Acta. 2000;298:157–173. doi: 10.1016/s0009-8981(00)00300-4. [DOI] [PubMed] [Google Scholar]
- 30.Baracca A., Amler E., Solaini G., Parenti-Castelli G., Lenaz G., Houstek J. Temperature-induced states of isolated F1-ATPase affect catalysis, enzyme conformation and high-affinity nucleotide binding sites. Biochim. Biophys. Acta. 1989;976:77–84. doi: 10.1016/s0005-2728(89)80191-4. [DOI] [PubMed] [Google Scholar]
- 31.Pecina P., Capkova M., Chowdhury S. K., Drahota Z., Dubot A., Vojtiskova A., Hansikova H., Houst'kova H., Zeman J., Godinot C., Houstek J. Functional alteration of cytochrome c oxidase by SURF1 mutations in Leigh syndrome. Biochim. Biophys. Acta. 2003;1639:53–63. doi: 10.1016/s0925-4439(03)00127-3. [DOI] [PubMed] [Google Scholar]
- 32.Floryk D., Houstek J. Tetramethyl rhodamine methyl ester (TMRM) is suitable for cytofluorometric measurements of mitochondrial membrane potential in cells treated with digitonin. Biosci. Rep. 1999;19:27–34. doi: 10.1023/a:1020193906974. [DOI] [PubMed] [Google Scholar]
- 33.Wanders R. J. A., Ruiter J. P. N., Wijburg F. A., Zeman J., Klement P., Houstek J. Prenatal diagnosis of systemic disorders of the respiratory chain in cultured chorionic villus fibrolasts by study of ATP synthesis in digitonin-permeabilized cells. J. Inherit. Metab. Dis. 1996;19:133–136. doi: 10.1007/BF01799412. [DOI] [PubMed] [Google Scholar]
- 34.Ouhabi R., Boue-Grabot M., Mazat J. P. Mitochondrial ATP synthesis in permeabilized cells: assessment of the ATP/O values in situ. Anal. Biochem. 1998;263:169–175. doi: 10.1006/abio.1998.2776. [DOI] [PubMed] [Google Scholar]
- 35.Chomyn A. In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol. 1996;264:197–211. doi: 10.1016/s0076-6879(96)64020-8. [DOI] [PubMed] [Google Scholar]
- 36.Plasek J., Sigler K. Slow fluorescent indicators of membrane potential: a survey of different approaches to probe response analysis. J. Photochem. Photobiol. B. 1996;33:101–124. doi: 10.1016/1011-1344(96)07283-1. [DOI] [PubMed] [Google Scholar]
- 37.Hoffbuhr K. C., Davidson E., Filiano B. A., Davidson M., Kennaway N. G., King M. P. A pathogenic 15-base pair deletion in mitochondrial DNA-encoded cytochrome c oxidase subunit III results in the absence of functional cytochrome c oxidase. J. Biol. Chem. 2000;275:13994–14003. doi: 10.1074/jbc.275.18.13994. [DOI] [PubMed] [Google Scholar]
- 38.Carrozzo R., Murray J., Santorelli F. M., Capaldi R. A. The T9176G mutation of human mtDNA gives a fully assembled but inactive ATP synthase when modeled in Escherichia coli. FEBS Lett. 2000;486:297–299. doi: 10.1016/s0014-5793(00)02244-4. [DOI] [PubMed] [Google Scholar]
- 39.Pansini A., Guerrieri F., Papa S. Control of proton conduction by the H+-ATPase in the inner mitochondrial membrane. Eur. J. Biochem. 1978;92:545–551. doi: 10.1111/j.1432-1033.1978.tb12776.x. [DOI] [PubMed] [Google Scholar]
- 40.Hutcheon M. L., Duncan T. M., Ngai H., Cross R. L. Energy-driven subunit rotation at the interface between subunit a and the c oligomer in the FO sector of Escherichia coli ATP synthase. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8519–8524. doi: 10.1073/pnas.151236798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buchet K., Godinot C. Functional F1-ATPase is essential in maintaining growth and membrane potential of human mitochondrial DNA-depleted rho degrees cells. J. Biol. Chem. 1998;273:22983–22989. doi: 10.1074/jbc.273.36.22983. [DOI] [PubMed] [Google Scholar]
- 42.Shoubridge E. A. Cytochrome c oxidase deficiency. Am. J. Med. Genet. 2001;106:46–52. doi: 10.1002/ajmg.1378. [DOI] [PubMed] [Google Scholar]
- 43.Rossignol R., Faustin B., Rocher C., Malgat M., Mazat J. P., Letellier T. Mitochondrial threshold effects. Biochem. J. 2003;370:751–762. doi: 10.1042/BJ20021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Temperley R. J., Seneca S. H., Tonska K., Bartnik E., Bindoff L. A., Lightowlers R. N., Chrzanowska-Lightowlers Z. M. Investigation of a pathogenic mtDNA microdeletion reveals a translation-dependent deadenylation decay pathway in human mitochondria. Hum. Mol. Genet. 2003;12:2341–2348. doi: 10.1093/hmg/ddg238. [DOI] [PubMed] [Google Scholar]
- 45.El Meziane A., Lehtinen S. K., Hance N., Nijtmans L. G., Dunbar D., Holt I. J., Jacobs H. T. A tRNA suppressor mutation in human mitochondria. Nat. Genet. 1998;18:350–353. doi: 10.1038/ng0498-350. [DOI] [PubMed] [Google Scholar]
- 46.Hao H., Morrison L. E., Moraes C. T. Suppression of a mitochondrial tRNA gene mutation phenotype associated with changes in the nuclear background. Hum. Mol. Genet. 1999;8:1117–1124. doi: 10.1093/hmg/8.6.1117. [DOI] [PubMed] [Google Scholar]
- 47.Ellis T. P., Helfenbein K. G., Tzagoloff A., Dieckmann C. L. Aep3p stabilizes the mitochondrial bicistronic mRNA encoding subunits 6 and 8 of the H+-translocating ATP synthase of Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:15728–15733. doi: 10.1074/jbc.M314162200. [DOI] [PubMed] [Google Scholar]
- 48.Mili S., Pinol-Roma S. LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol. Cell. Biol. 2003;23:4972–4982. doi: 10.1128/MCB.23.14.4972-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mootha V. K., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F., et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. U.S.A. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]