Abstract
One of the foremost challenges in the post-genomic era will be to chart the gene regulatory networks of cells, including aspects such as genome annotation, identification of cis-regulatory elements and transcription factors, information on protein–DNA and protein–protein interactions, and data mining and integration. Some of these broad sets of data have already been assembled for building networks of gene regulation. Even though these datasets are still far from comprehensive, and the approach faces many important and difficult challenges, some strategies have begun to make connections between disparate regulatory events and to foster new hypotheses. In this article we review several different genomics and proteomics technologies, and present bioinformatics methods for exploring these data in order to make novel discoveries.
Keywords: bioinformatics, cis-regulatory code, data processing, functional genomics and proteomics, gene regulatory network, protein–DNA and protein–protein interaction
Abbreviations: ChIP-chip, chromatin immunoprecipitation–DNA microarray; FFL, feed-forward loop; GRN, gene regulatory network; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; ORF, open reading frame; NF-κB, nuclear factor-κB; SBML, System Biology Markup Language; SBW, System Biology Workbench; TAP, tandem affinity purification; TF, transcription factor; XML, eXtensible Makeup Language; Y2H, yeast two-hybrid
INTRODUCTION
Charting GRNs (gene regulatory networks) – that is, how all the functional molecules of these networks exist, interact and react spatio-temporally – is a major focus of interest in modern biology. In the networks, TFs (transcription factors) receive input information from upstream signal transduction cascades and bind, directly or indirectly via other TFs, to target sequences on so-called cis-regulatory regions of genes. Bound TFs stimulate or repress the assembly of pre-initiation complexes on the gene promoter, thereby promoting or inhibiting RNA polymerase assembly. Thus the information is transferred downstream to other regulatory genes and to the structural genes whose products account for the catalytic and structural versatility of the cell. This is an intricate and precise regulatory process that provides living cells with their remarkable properties. Biological results clearly show that the mechanism of regulation is a multi-level system of high complexity formed by genes and TFs, which determines how organisms develop and respond to environmental stimuli. This involves physical, informational, proximal, distal, upstream and downstream cis-regulatory elements on the DNA, as well as protein–protein, protein–DNA and other ‘component–component’ interaction events.
GRNs must be based in the genomic DNA sequence and, in experimental terms, the relevant sequence is that containing the genes in networks and their cis-regulatory control elements [1,2]. With the high-throughput sequencing of the complete genomes of a large variety of species, new experimental strategies combined with information technology and computational modelling have been developed for exploring these rapidly accumulating new data, allowing biologists to accelerate the pace of understanding of the logic of GRNs in a systematic manner [3,4]. Using DNA microarray technology, for example, patterns of similar expression profiles under various conditions have been linked to shared regulatory mechanisms [5,6]. The computational approaches used to analyse and elucidate these control mechanisms are various [7]. Popular approaches include searching for novel cis-elements using the program PROJECTION [8], or the more recent Gibbs Recursive Sampler [9] and YMF [10].
Studies of GRNs in yeast [11], and of those for the development of the endoderm in a sea urchin embryo [3], illustrate the power of combining genomic techniques with computational analysis, and also indicate additional challenges. Recently, a genome-wide analysis of the binding sites of TFs in the yeast Saccharomyces cerevisiae was achieved [12]. That study not only documents potential pathways used by yeast cells to regulate gene expression, but also identifies network motifs, the simplest units of network architecture. Integration of such datasets with other information, such as protein–protein interactions, can provide detailed insight into specific cellular processes, such as GRNs [12]. However, to create meaningful output, the information collected from each approach or their combination should be of high quality [13]. Substantial effort must be devoted to organizing the information into databases in structured formats that can be interrogated computationally in order to manage, integrate, analyse and visualize all of the data.
In the present review, we discuss several novel functional genomic and proteomic strategies in conjunction with some bioinformatic approaches for elucidation of the components of GRNs and links between these components. We will review work completed and in progress to chart GRNs, focusing on hypothesis- and discovery-driven data mining and integration, and construction of regulatory network motifs in cells.
GENOME ANNOTATION
The mapping, sequencing and dissecting of genomes provides an invaluable resource for the study of regulatory networks. At present, the annotation of whole genome sequences for functional elements is clearly one of the most formidable challenges facing the bioscience community. Despite extensive research in the area of gene prediction, current predictors do not provide a complete solution to the problem of gene identification [14]. For example, micro-exons and small genes remain difficult to locate, because discriminatory statistical characteristics are less likely to appear in short strands. Furthermore, some genes do not possess the characteristic features that identify most genes, and hence it is impossible to track them by using gene predictors that rely on these features. Consequently they often can be missed and designated as hypothetical ORFs (open reading frames) [14].
An additional challenge to genome annotation efforts lies in the prediction of genes for non-coding RNA, including genes of rRNAs, tRNAs and small RNAs. The small RNA subfamily contains siRNAs (small interfering RNAs) and microRNAs that have been revealed recently, as well as snRNAs (small nuclear RNAs) and snoRNAs (small nucleolar RNAs), each with their own properties and functions, from structural through regulatory to catalytic [15]. These types of genes have been hard to detect both experimentally and computationally because of their small size, lack of an ORF and diverse nature. Even in the Escherichia coli genome, only a proportion of the population of genes encoding small RNAs was predicted [16].
In addition to finding new genes, the refinement and verification of the results of gene prediction are also extremely important. Both comparative genomics [17] and genome-wide functional analyses [18] show that the S. cerevisiae genome, despite its low content of introns, requires annotation improvements. A number of approaches, including large-scale sequencing of random cDNAs, or ESTs (expressed sequence tags) [19,20], recent analyses of the genomic sequences of a number of related yeast species [21,22], Gateway-based ORFeome cloning [23,24] and proteomics-based protein expression [25], have been used to distinguish between real and misannotated ORFs. Through continual refinement, the false genes can be removed and novel ORFs added. Correct information from the corresponding protein-coding gene annotation is critical for constructing tools such as DNA chips, protein arrays [26] and reverse transfection strategies [27], allowing researchers to study the activity of thousands of genes at a time.
DECIPHERING CIS-REGULATORY CODES
The heart of GRNs consists of genes encoding TFs and the cis-regulatory elements that control the expression of those genes [1]. Each of these cis-regulatory elements receives multiple inputs from other genes in the network; these inputs are the TFs for which the element contains the specific target site sequences. The functional linkages of which the network is composed are those between the outputs of regulatory genes and the sets of genomic target sites to which their products bind [3,28]. These cis-regulatory elements act like a kind of ‘code’, transforming a set of input information into a second set as output [1].
Genome annotation requires not only the identification of coding segments of genes and the mapping of novel transcripts, but also information on how individual genes are separately regulated. Coding regions in the genomes of higher eukaryotes occupy only a small fraction of the total genome. In the case of the human genome, protein coding sequences account for less than 2% of the total. These genomes contain vast amounts of cis-regulatory sequences responsible for directing spatial and temporal patterns of gene expression in response to metabolic requirements, developmental programmes and a plethora of external stimuli [29]. Therefore, identifying and characterizing these cis-regulatory sequences represents the first step towards building complex models of regulatory networks [30]. Accordingly, many high-throughput experimental and computational strategies are emerging.
ChIP-chip (chromatin immunoprecipitation–DNA microarray) is one such approach, which can efficiently map global binding sites for TFs and chromatin proteins on a genome-wide scale in vivo. The approach combines a modified ChIP procedure, which had been used previously to study protein–DNA complexes. The purified protein-bound DNA is amplified, labelled and hybridized to intergenic DNA microarrays. This approach was first used successfully in yeast [12,31–34], was also developed in Drosophila [35,36], and has been used more recently in a limited fashion to identify TF binding sites in mammalian cells [37–40]. For example, the ChIP-chip assay has been used with human DNA microarrays to identify binding sites for GATA-1 in the 75 kb sequence of the β-globin locus [39], binding sites for E2F in promoters of genes expressed during cell cycle entry [37], and binding sites for NF-κB (nuclear factor-κB) across the whole of chromosome 22 [40].
ChIP-chip assays provide a treasure trove of experimental data. However, it is difficult to confidently assign TFs to genes solely on the basis of these binding data because, although the data are obviously very good, there clearly exists a significant degree of error of uncertain magnitude. Taking two yeast cell cycleassociated studies, for example, the agreement between the binding data of Simon et al. [32] for Mbp1, Swi4 and Swi6 (the components of the transcription factors MBF and SBF) and those of Iyer et al. [33] for the same proteins is only moderate [7]. In addition, the technique can only map the probable protein–DNA interaction loci within ∼1–2 kb resolution. Moreover, binding does not prove that there is regulation and, importantly, does not distinguish between positive and negative regulation. The use of both binding information and large-scale expression data from DNA microarrays should prove to be an important and powerful combination of analyses that can be expected to result in the reliable assignment of a TF to a gene [32,41].
Like DNA microarrays in the late 1990s, it is almost certain that the new ChIP-chip technology will quickly catch on with researchers worldwide, and before long large numbers of high-throughput DNA-binding datasets will be available. Powerful and sophisticated computer algorithms, such as GRAM (Genetic Regulatory Module) [42], REDUCE [43,44] and modified MOTIF REGRESSOR [45], will be needed to analyse these data.
The development of the ChIP-chip technique has experienced two important stages: from yeast to multicellular organisms such as Drosophila, and then to mammalian cells. With the development of ChIP-chip assays for use in cells from higher eukaryotes, some inherent challenges still exist, including the large size of the genome and intergenic sequences, the complexity of gene regulation and chromatin structure, and the high proportion of repetitive elements. Can the ChIP-chip assay be applied to an entire mammalian genome? This will be a new developing trend.
Compared with the above experimental strategies, computational methods for deciphering cis-regulatory regions have a longer history and greater power [30,46,47]. Many algorithms for the large-scale discovery of candidate regulatory regions have been developed. Detailed consideration of these algorithms is beyond the scope of this review, and more specific descriptions are found elsewhere [30,46–48]. Although these methods are unable to assign specific TFs to their cognate binding sites, they can still be of tremendous use in identifying relevant binding site motifs. Other types of programs can currently be obtained from internet resources. Here, with the availability of some whole genomes, we will mainly discuss the strategy of comparative genomics [49]. This method is a powerful approach for dissecting the complexities of cis-regulatory codes [50]. The rationale behind this approach is that evolutionary conservation of a feature implies that it has been retained by selection, which means that it is likely to have a function (also referred to as ‘phylogenetic footprinting’) [51]. Indeed, several studies [50,52] have shown that putative TF binding sites are enriched in conserved non-coding genomic sequences (footprints). This approach has proved valuable not only on a gene-by-gene scale, but also on a genomic basis [48,52], and several algorithms have been developed for cross-species sequence comparisons, complete with gene, in some cases, TF binding site and annotation [53,54]. Two recent studies are very attractive. Cliften et al. [55] sequenced the genomes of five different Saccharomyces species and aligned them with the S. cerevisiae genome sequence, thereby identifying hundreds of sequences. They estimated that there are around 5500 different conserved upstream motifs, and that 73% of these are made up of combinations of the known binding sites of 37 TFs [55]. Nobrega et al. [56] compared human DACH (Dachsund) flanking sequences with mouse genomic DNA and, by combining additional genome comparison information from distantly related vertebrates such as frog, zebrafish and pufferfish, reported that they contain several important enhancers.
Despite the potential power of comparative sequence-based approaches, they still have some limitations when conducting genome-wide searches for regulatory sites. For example, it is not possible to identify, merely from alignment data, the functional role of an identified conserved sequence motif. Moreover, some conserved elements may be found in intergenic regions far from any coding sequence, so it is not always clear what gene is subject to regulation by the elements in question.
Using a purely computational approach, uncertainty remains as to whether a predicted cis-regulatory element actually possesses the expected function [57]. With a purely experimental method, on the other hand, difficulty remains in predicting cis-regulatory elements on a large scale. Thus the union of experimental and computational strategies represents a new approach to deciphering cis-regulatory codes [58]. The most successful of these approaches to date appear to be those that rely on gene expression profiles from DNA microarrays. Some of these are currently being employed to study genome-wide transcriptional regulation [59–62]. Typical analyses include clustering of binding sites and finding DNA sequence regions where their local density is high [59,60], or predicting the targets of a TF using support vector machines [61]. Other methods have combined transcriptional profiling data with additional information such as shared DNA binding motifs, and been applied to identify novel TF combinations in the promoters of yeast genes [62]. Among the more commonly used of these programs, REDUCE is an excellent algorithm and is designed for analysis of a single transcriptome [5,64]. Furthermore, the algorithm has been successfully applied to analyse genome-wide protein–DNA interaction data in Drosophila [43,44]. In addition, other functional genomics and proteomics data, rather than genome sequence and expression profiling information, have been used to facilitate the identification of TF binding sites using appropriate computational tools. Recently, Ettwiller et al. [65] combined functional information such as protein–protein interactions and metabolic networks with genome information in S. cerevisiae and developed a new scoring method to predict cis-regulatory motifs in the upstream regions of genes. Roulet et al. [66] coupled appropriate bioinformatics tools with a high-throughput SELEX/SAGE (systematic evolution of ligands by exponential enrichment/serial analysis of gene expression) method for quantitative modelling of mammalian TF binding sites.
As mentioned above, although many high-throughput and powerful methods have emerged, deciphering cis-regulatory codes still faces some formidable challenges. First, several regulatory DNAs, enhancers, silencers and insulators are scattered within tens of kilobases of transcription start sites, upstream or downstream of the gene, or within its introns in higher eukaryotes [67], thus introducing significant complexity into such approaches, and false positive rates can be high. Secondly, a typical promoter or enhancer usually contains multiple TF binding sites and receives input from a number of different signalling cascades [28]. Thirdly, but not least, most TF binding sites are short sequence elements (6–20 bp) and extremely difficult to use for sequence comparisons. A large number of such motifs may occur randomly in the genome, and the vast majority of these have no role to play in gene regulation. An additional aspect is that the genomes of eukaryotic cells usually contain a wealth of information not encoded directly in their DNA sequence (termed ‘epigenetic regulatory information’), such as DNA methylation and the histone code (i.e. various post-translational modifications of histones). How to mine and integrate this information into GRNs will be an enormous challenge.
IDENTIFICATION OF TFs
TFs lie at the centre of gene regulation [29]. The regulation of specific genes by TFs can be defined as a direct transcriptional regulatory interaction. When all such interactions in a living cell are considered, what we see is a complex set of interactions, which is known as the GRN. These regulatory networks form the framework for gene expression, determining which genes should be expressed in a cell, and when. Precise control of gene expression is achieved by combinatorial and concerted interactions of various TFs with their cognate binding sites, with each other and with the transcription initiation complex [68,69]. Therefore their identification is crucial to the understanding of gene regulatory mechanisms. Emerging evidence suggests that organism complexity correlates with increases in both the proportions and absolute numbers of TFs per genome [67].
Compared with the identification of cis-regulatory elements, the development of strategies for isolating and identifying TFs lags behind. Current TF coding information is predicted mainly from genome analysis based on computational methods. For example, through sequence similarity or structural comparison, Riechmann et al. [70] characterized the entire complement of TFs encoded by the genomes of Arabidopsis, Drosophila, Caenorhabditis elegans and S. cerevisiae. Recently, several in silico analyses identified a total of 326 putative C2H2 zinc-finger proteins in the genome of Drosophila [71], and 147 bHLH (basic helix–loop–helix) [72] and 107 MADS-box protein-coding genes [73] in the Arabidopsis genome.
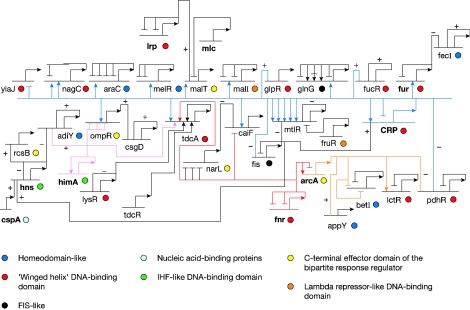
Madan Babu and Teichmann [74] have reported a novel method for identifying TFs. In this approach, structural domains are first assigned to sequences, and then sequences that have known DNA binding domains are identified as potential TFs. Importantly, the authors ensured that DNA binding domains seen in DNA repair proteins and some enzymes were not included in the analysis. This is a better approach, because the HMM (Hidden Markov Model)-based structural domain assignment method picks up more distant homologues in a reliable manner than sequence comparison methods [74]. Using this method, Madan Babu and Teichmann identified a total of 271 E. coli TFs from the SUPERFAMILY database. For 121 of these 271 TFs, experimental information about the genes that they regulate had been provided in previous studies. Individually, these 121 TFs each regulate from 1 to 197 genes; altogether, there are 1302 genes and 303 operons in the regulatory network [75]. To investigate the regulation of TFs, the authors integrated the information available from other studies, to produce a diagram of the TF regulatory network in E. coli [74]. Figure 1 shows the network of 35 TFs currently known to regulate each other in E. coli. For example, FNR and ArcA regulate four TF genes, and the tdcA gene is regulated by a combination of several TFs. Using the current information on the E. coli GRN shown in Figure 1, it appears that TFs vary with regard to the number of genes they regulate. The majority of TFs are ‘fine tuners’ that control a limited, specific set of genes, while a small number of TFs are ‘dominant TFs’, that regulate a large number of genes, and also interact with a large number of TFs to amplify their influence [74,76,77]. Figure 1 shows one central part of the GRN currently known in E. coli [74]. From these events we can see that, even in simpler organisms such as the prokaryote E. coli, there are also cascades of TFs that regulate each other in order to amplify or diversify the effect of a signal on gene regulation. It is estimated that roughly 5–10% of the total coding capacity of the metazoan genome is dedicated to the coding of TFs [67]. Therefore the TF regulatory networks in such organisms would be more intricate than in E. coli. We can envision that computational methods similar to that described above could be developed to identify TFs and to dissect the complicated GRNs of TFs in metazoans.
Figure 1. Regulatory network of TFs in E. coli.
This figure illustrates the core of the GRN in E. coli, where TFs regulate other TFs [74]. Short horizontal lines from which bent arrows extend represent cis-regulatory elements responsible for the expression of the genes named below the line. When more than one TF regulates a gene, the order of their binding sites is as given in the figure. An arrowhead indicates activation and a horizontal bar indicates repression when the position of the binding site is known. If only the nature of TF regulation is known, without binding site information, ‘+’ and ‘−’ symbols indicate activation and repression respectively. These examples may be indirect rather than direct regulation. The circles with the different colours as given in the key represent the different families of DNA binding domains. The names of dominant regulators are in bold. FIS, factor for inversion stimulation; IHF, integration host factor. Modified with permission, from Madan Babu, M. and Teichmann, S. A., (2003), Nucleic Acids Res. 31, 1234–1244. © Oxford University Press.
The traditional experimental approaches for the identification of TFs, such as DNA affinity chromatography [78] and DNA binding assays [79], are usually time-consuming, labour-intensive and low-throughput. One of the main reasons is that TFs are normally present at very low concentrations in cells. To understand gene regulation, the development of high-throughput methods is needed. The first method for the identification of DNA binding proteins on a genome-wide basis was provided by Hazbun and Fields [80]. They combined a gel shift assay with pools of 6144 glutathione S-transferase fusion proteins in yeast and identified several TFs that bound to a specific cis-regulatory sequence. This method demonstrates the feasibility of identifying DNA binding activities by rapidly assaying a large fraction of the predicted ORFs of an organism for binding to a regulatory DNA motif.
Recent advances in proteomics and MS have created unprecedented power for the identification of DNA binding proteins [81]. Nordhoff et al. [82] reported a method that relies on MALDI-TOF (matrix-assisted laser desorption/ionization time-of-flight) MS to identify DNA binding proteins that bound to a DNA probe harbouring specific cis-regulatory motifs immobilized on to small paramagnetic particles. Bound proteins were analysed directly by MALDI-TOF MS and then identified by retrieving databases [82]. In another report, Woo et al. [83] developed a powerful method for the identification of DNA binding proteins seen in electrophoretic mobility shift assays by utilizing high-resolution two-dimensional electrophoresis coupled with MS [83]. These two methods, combined with DNA chip technology, could be applied to identify TFs systematically.
MAPPING PROTEIN–DNA INTERACTIONS
Protein–DNA interactions – that is, physical binding of trans-acting regulatory gene products to specific cis-acting elements of the regulated genes – are at the heart of the regulatory mechanisms that control gene expression in the GRN. A more complete understanding of these protein–DNA interactions will permit more comprehensive and quantitative mapping of the regulatory pathways within cells, as well as a deeper understanding of the potential functions of individual genes regulated by newly identified TF binding sites [29]. Numerous techniques have been employed for studying protein–DNA binding interactions, including several methods in vitro, sensitive fluorescence-based approaches and high-throughput array-based assays in vivo.
The first class of methods, such as gel mobility shift assays, is the most commonly used [79]. For example, using electrophoretic mobility shift assays, the protein–DNA complex results in a decrease in the electrophoretic mobility of the DNA fragment in non-denaturing polyacrylamide or agrose gels. The assay usually involves the addition of a binding protein to the DNA sample and separation of the free and complexed DNA by gel electrophoresis with autoradiography or fluorescence detection. The whole process requires a large amount of sample and is too laborious and time-consuming to be used for the analysis of a large number of protein–DNA interactions.
Fluorescence-based approaches for measuring specific protein–DNA interactions have been developed to circumvent the deficiencies of such methods [84]. For example, fluorescence detection eliminates environmental concerns related to the disposal of radioactive waste and provides outstanding sensitivity of detection, even to the level of single molecules [85]. However, the methodologies are not practical for gathering data on vast numbers of protein–DNA interaction pairings.
Methodologies to map protein–DNA interactions using array-based assays have been developed in yeast [12,31,33], Drosophila [35,36] and mammalian cells [37,39,86]. To date, the interactions of over 100 yeast TFs with cognate DNA regulatory sequences have been mapped on a genome-wide scale. For example, Lee et al. [12] constructed a series of yeast strains in which each of the 141 known yeast regulators was epitope-tagged at its C-terminus and expressed under the control of its normal promoter at its appropriate chromosomal locus. After the growth of each strain, ChIP analysis was carried out, in which each tagged protein was purified along with its population of bound DNA, and the identity and amount of DNA was determined using intergenic microarrays. Of the 141 TFs, 106 were identified, and the study allowed not only a genomic view of protein–DNA interactions, but also the description of a number of different networks of transcription regulation in the cell, and a functional assessment of the role of each TF in yeast [12]. Recently, Sun et al. [36] demonstrated the use of genomic DNA tiling path microarrays to map protein–DNA interactions at high resolution along large segments of genomic DNA from Drosophila.
DISSECTING PROTEIN INTERACTION NETWORKS
Networks of protein interactions mediate many cellular responses to environmental stimuli and direct the execution of developmental programmes. Each protein typically interacts and reacts with other interaction partners to execute their functions. The selectivity of these interactions determines the developmental potential of the cell and its response to extracellular stimuli.
No individual factor is capable of playing a dominant role in generating the immense specificity required to regulate transcription, especially in eukaryotes. The GRN is composed of and mediated by cascades of interacting components or complexes, which bind to promoters and enhancers, or communicate between activators and repressors and sites of transcription initiation. Each of these complexes might be a key player in regulating a given gene. Complexes of TFs, co-repressors and chromatin binding proteins maintain normal cells in a quiescent state, and disruption of these protein interactions may be significant in permitting the unregulated growth of cancer cells [87]. A major challenge is to determine how all of these complexes work together to ensure proper regulation.
For example, in a gene regulatory pathway, the transcriptional regulatory proteins receive input information from upstream signal transduction cascades that are regulated by specific protein interactions. Then the proteins will bind to short cis-DNA sequence motifs found in the promoter and enhancer regions of downstream genes and, through interactions with other components of the transcription machinery, promote access to DNA and facilitate the recruitment of RNA polymerase enzymes to the transcriptional start site [88]. Therefore protein interactions provide the mechanistic basis for much of gene regulation in all organisms. Comprehensive analysis of protein interaction events, integrated with cis-regulatory and TF binding information, will provide a powerful first step towards charting GRNs.
An enormous amount of protein–protein interaction information has been obtained for some organisms using high-throughput Y2H (yeast two-hybrid) systems [89], MS-based proteomics [90–92], protein arrays [93] and fluorescence-based interaction assays [94,95]. These large-scale datasets have provided a wealth of new leads in many areas of biology, such as global protein function prediction [96] and functional module discoveries [97]. Some work is obviously not directly related to the subject of this review, but the results are testimonies that these methodologies are scalable and beneficial to the understanding and charting of GRNs. Of particular relevance to gene regulation, for example, Yatherajam et al. [89] performed a systematic Y2H analysis of TAF–TAF (TATA-binding protein associated factor) interactions and their topological arrangements within TFIID. Many studies have shown that TFIID plays important roles in many aspects of the regulation of gene expression [98]. Newman and Keating [93] used protein arrays to test 492 pairings of a nearly complete set of coiled-coil strands from human bZIP (basic-region leucine zipper) TFs.
The scaling-up of protein interaction screens using the Y2H system has made it possible to analyse complete proteomes and identify thousands of interactions. Surprisingly, several proteome-wide screens in the yeast S. cerevisiae have yielded very little overlap in the interactions detected. Several analyses of genomic Y2H results suggest that about 50% represent valid interactions [99–102]. These results are largely unexpected, and lead to speculation on high error rates in large-scale interaction screens and the need for an upward revision and reliability assessment of the number of protein interactions in yeast and other organisms. For example, integrating Y2H data on protein interactions with data on protein complex composition from affinity chromatography plus MS and co-expression data from transcriptome analyses allows the production of a list of validated protein interactions [99,100,102].
Since many protein complexes participate in gene regulation, affinity tagging coupled with MS-based proteomics may have a significant impact on the dissection of gene regulatory mechanisms [90]. Considerable efforts have been devoted to developing tagging systems optimized for the analysis of protein complexes [103]. Here we will highlight the current state of one popular tagging system – the TAP (tandem affinity purification) method – and its role in the identification of transcription complexes. The TAP method is a protein tag-based affinity purification technique originally developed and successfully employed in yeast [104,105]. Technically, two affinity tags, Protein A and calmodulin-binding peptide, separated by a TEV (tobacco etch virus) protease cleavage site, are fused the protein of interest [105]. The TAP-tagged protein is expressed in yeast cells to physiological concentrations to form a complex with endogenous components. Extracts prepared from cells expressing the TAP-tagged protein are subjected to two successive high-stringency purification steps. Once the purified complex is available in soluble form, it is resolved by SDS/PAGE, and the protein bands are digested in-gel and identified by MS [104,105]. One application of this methodology for analysing transcription complexes was described by Mueller and Jaehning [106]. To further characterize the composition and function of the complex between Paf1 and RNA polymerase II, and to compare it with the Srb–mediator complex in yeast, they TAP-tagged the products of four chromosomal genes: CDC73, SRB5, HPR1 and CTR9. MS analyses of the associated complexes revealed that the two complexes were biochemically different [106]. Another two fascinating studies demonstrating the power of the method will also be mentioned here [107,108]. In the first study, Chung et al. [107] combined the TAP-tagging method with cryo-electron microscopy and determined the structure of a complex between RNA polymerase II and TFIIF [107]. In another work, Rodriguez-Navarro et al. [108] TAP-tagged a novel nuclear protein, Sus1, and found that it is physically associated with SAGA, a histone acetylase complex, and the Sac3–Thp complex, which is involved in mRNA export. The results partially elucidated the physical nature of transcription-coupled mRNA export [108]. Although most applications of the approach have thus far been described for yeast complexes, the TAP method can be modified and used for the retrieval of protein complexes from higher eukaryotes, such as human [109–111] and Drosophila [112]. For instance, more recently, Bouwmeester et al. [109] analysed the human tumour necrosis factor-α/NF-κB signal transduction pathway, and identified receptor, kinase and TF-associated complexes.
Compared with Y2H and array-based approaches, this strategy has the advantages that the fully processed and modified protein can serve as the bait, that the interactions take place in the native environment and cellular location, and that multi-component complexes can be isolated and analysed in a single operation [111,113]. In addition, the sensitivity of the method is very high, and it is able to identify proteins characterized by low levels of expression, such as TFs and TF-associated complexes [111,114]. Therefore the amount of sample purified by this method is usually limited, and the electrophoretic step is not desirable. Accordingly, the TAP-MudPIT (multidimensional chromatography–MS) approach and other variant methodologies are emerging [115,116]. However, the TAP MS method also has some drawbacks. For example, the strategy does not provide information on the orientation of complex components; thus complex characterization and Y2H analyses are ideally complementary.
As demonstrated above, each method for identifying protein interactions has its drawbacks, and none gives complete or unambiguous data. Side-by-side comparisons of data obtained by different methods show limited reproducibility and a prevalence of false positives and false negatives [99]. Many biologically relevant protein interactions are of low affinity, transient and generally dependent on the specific cellular environment in which they occur. Thus a straightforward affinity experiment will detect only a subset of the protein interactions that actually occur. The development of quantitative methods based on stable-isotope labelling [117] is likely to revolutionize the study of stable or transient interactions and interactions dependent on post-translational modifications. In such experiments, accurate quantification by means of stable-isotope labelling is not used for protein quantification per se; instead, the stable isotope ratios distinguish between the protein compositions of two or more protein complexes. In the case of a sample containing a complex and a control sample containing only contaminating proteins, the stable-isotope method can distinguish between true complex components and non-specifically associated proteins. In situations where complexes are isolated from cells in different states, the method can identify dynamic changes in the composition of a protein complex [91,92]. For example, Ranish et al. [91] employed the method to guide the identification of the genuine components of a large RNA polymerase II pre-initiation complex (approx. 68 subunits) within a high background of co-purifying proteins following a simple one-step DNA affinity procedure. The method increases the tolerance of high background levels and allows for fewer purification steps and less stringent washing conditions, thus increasing the chance of finding transient and weak interactions.
NETWORK MOTIFS OF GRNs
Recent advances in data connection and analysis are generating unprecedented amounts of information about GRNs. However, it is still extremely difficult to construct GRNs based on this information, due to network complexity. Some studies have proposed that such networks can be dissected into small functional modules [118]. Therefore the notion of motifs widely used for sequence analysis is generalized to the level of networks. Of particular relevance to GRNs, specific building blocks of complex networks, or network motifs [12,76], have been identified in GRNs of E. coli and yeast. Network motifs are regulatory circuit patterns that occur in the network far more often than in randomized networks with the same degree sequence [76,119]. Each network motif can perform a specific information-processing task, such as filtering out spurious input function, generating temporal programmes of expression or accelerating the throughput of the network [12,76,119].
Lee et al. [12] have developed a high-throughput method to identify six frequently appearing network motifs, ranging from multi-input motifs (in which a group of regulators binds to the same set of promoters) to regulatory chains (alternating regulator–promoter sequences generating a clear temporal succession of information transfer). They assembled these motifs into larger network structures, and constructed the regulatory logic of the cell cycle in yeast from the location and expression data [12]. A similar set of highly significant regulatory motifs was uncovered previously in the bacterium E. coli by Alon and co-workers [76]. The significance of these structures raises the question of whether they have specific information-processing roles in the network. If they do, they might be useful for understanding network dynamics in terms of elementary computational building blocks. One of the most significant motifs in both E. coli and yeast is the FFL (feed-forward loop) [12,76]. The FFL, a three-gene pattern, is composed of two input TFs, one of which regulates the other, and both jointly regulating a target gene. Lee et al. [12] found that 39 TFs are involved in 49 FFLs potentially controlling 240 genes in the yeast network. Recently, Mangan and Alon [120] analysed the structure and functions of the FFL on the basis of mathematical modelling and simulations. The results showed that the FFL has eight possible structural types; half are termed incoherent FFLs and the other half coherent FFLs. These authors found that the incoherent FFLs speed up the response time of target gene expression following stimulus steps in transcription networks. On the other hand, the coherent FFLs serve as a sign-sensitive delay element: a circuit that responds rapidly to step-like stimuli in one direction, and as a delay to steps in the opposite direction [121].
Network motifs are emerging as our knowledge of GRNs become complete [119]. It would be fascinating to study the function of additional regulatory network motifs to determine whether GRNs can be understood in terms of recurring circuit elements, each with a defined information-processing role. Once a dictionary of network motifs and their function is established, one can envision researchers detecting new network motifs. These motifs can be used as building blocks to construct large network structures through a computational approach that combines genome-wide binding information with large-scale transcriptome data in the absence of original knowledge of regulator functions [122]. From the study of Lee et al. [12] we can deduce that the network of transcriptional regulators that control genes encoding other transcriptional regulators is highly connected. Such a deduction implies that the network substructures for cellular functions such as the cell cycle and development are themselves co-ordinated at a transcriptional regulatory level. We can envision mapping the regulatory networks that control gene expression programmes in further depth in yeast and in other higher eukaryotes. Knowledge of these networks will be of significance for understanding human health and designing new strategies to resist diseases [123].
DISCOVERY BY DATA MINING AND INTEGRATION
As described above, vast amounts of valuable data have been generated by large-scale functional genomic and proteomic experiments. These include profiling of mRNA and protein expression at the whole-genome level, locating the binding sites of given TFs along the genome, and proteome-wide identification of interacting proteins. Each dataset by itself calls for the application of appropriate computational tools for data processing, let alone the integration of different types of information [123]. These integrative analyses provide new molecular insights that could not be revealed using each type of information alone. Of particular relevance to gene regulation, several such studies have been reported, most of which involve the integration of mRNA profiling data in the yeast S. cerevisiae with other types of data [62,124]. Although data relating to the unravelling of transcriptional profiling are not discussed as one of the main points of this article, several examples in this section will also include the mining and integration of such information and other kinds of data resources.
First, the combination of genome-wide data for TFs and their target genes and data for protein–protein interactions based on classical graph algorithms has identified a large number of gene regulatory circuits [125]. These datasets consists of 5976 protein pairs connected as protein–DNA interactions [12,31–33] and 8184 protein pairs connected by protein–protein interactions. A total of 746 statistically significant circuits are obtained by the cellular process assessment, and by either the cellular localization assessment or the knockout results assessment (or by both). Such circuits can be used for complex regulatory tasks. For example, some circuits regulate genes participating in metabolism; some may function as an efficient positive or negative feedback loop, a key component of various control systems.
In a second example of functional genomic insight, it has been shown that the effect of the transcriptional regulatory network in S. cerevisiae on the expression of targeted genes can be determined by integrating gene expression and TF binding data [126]. The gene expression dataset originates from a genome-wide transcriptional profile of the mitotic cell cycle in yeast [127] and analysis of the expressed genes using a local clustering method [128]. TF binding data are explored by merging the results of genetic, biochemical and ChIP-chip assays [12,129–131]. The analysis contains 7419 interactions connecting 180 TFs with their 3474 target genes. Previous studies of the data discovered six basic motifs. The study of Yu et al. [126] found significant connections between the two kinds of datasets. Genes targeted by the same TF tend to be co-expressed, and the correlation is stronger for genes targeted by multiple, common TFs. In addition, target genes of the same TFs are more likely to share similar functions than expected randomly. Relationships between TFs and target genes are more complex than just co-expression. The degree of complexity is different in different motifs.
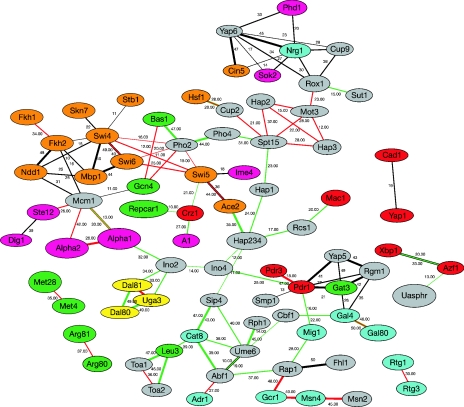
As a final example of the value of large-scale datasets, Manke et al. [132] integrated high-throughput protein–DNA interaction data into the overall network of experimental protein interactions, resulting in the production of a graphic representation of synergistic TF interactions (Figure 2). Two main types of datasets for yeast were used: (1) large collections of protein–DNA binding information obtained from genome-wide location analyses [12], the TRANSFAC database [131] and putative binding sites of TFs based on computational prediction algorithms [132]; and (2) collections of proteome-wide Y2H protein–protein interactions [133,134] and whole protein complex analysis [111,113]. The overlap of the corresponding datasets is very small. For example, of the 106 TFs investigated by Lee at al. [12], only 12 were found in the purified complexes identified by Gavin et al. [111], 35 in the sets reported by Ho et al. [113] and 50 in TRANSFAC. Through high-stringent theoretical analysis and computational extraction, a large number of prevalent co-occurring TF pairs were identified from large-scale protein–DNA binding data [12,131,132]. To increase the reliability of TF pair predictions, several sources of physical protein–protein interaction information were mined [111,113,133,134]. The Y2H screens provide valuable information on possible pairwise interaction; the protein complexes can be interpreted as complete sub-graphs in which each protein is linked to every other. Finally, according to synergy and their co-occurrence frequency, the 50 highest-ranking synergistic TF pairs were obtained, and are represented in Figure 2 [132]. The figure illustrates the complementary character of the available DNA binding data and highlights the significance of a given TF pair, as measured by large-scale protein interaction networks. This is very similar in spirit to previous efforts correlating transcriptome and interactome mapping data [135]. Although these original large-scale data contain inherent imperfections, they provide a valuable opportunity to systematically search additional information. Using the frequently occurring transcriptional module Mcm1–Fkh2–Ndd1, for example, several new target genes involved in cell-cycle control and filament formation were identified [132]. Such results particularly encourage researchers to computationally integrate these diverse datasets and observe significant commonalities in them. Such integration may allow the investigators to identify many well known regulatory modules and extract biologically relevant sub-networks [132,135]. Through finding commonalities in the datasets, the reliability of network predictions can be increased [132].
Figure 2. Synergy graph of TFs in S. cerevisiae.
The figure shows the results of joint analysis of large-scale protein–DNA binding information and protein–protein interaction data [132]. It can be visualized as a network in which the TFs are nodes and TF pairs are weighted links (edges) between the nodes. Black edges indicate results based on in vivo genome-wide location analysis data [12], red edges correspond to TRANSFAC database information [131], and green links are results of in silico predictions. There are always two lines with different colours aligning with each other between two TFs. The number on each edge serves as a rank measure, as described in [132]. The colour scheme for the nodes illustrates the predominant functional category (where known) of regulated genes: orange, mitotic cell cycle; pink, budding and filament formation; green, amino acid metabolism; yellow, nitrogen and sulphur utilization; blue, C-compound and carbohydrate utilization; red, TFs; grey, unspecific or several functional categories.
Taken together, the observations described above suggest that the large-scale datasets discussed in this review can be correlated and integrated for the unicellular yeast S. cerevisiae, and new discoveries made. For multicellular organisms, this approach remains difficult. However, with the development of assays such as the ChIP-chip method applied to mammalian cells and the recent appearance of the first multicellular protein interaction networks for Drosophila [136] and C. elegans [137], we believe that the notion of such data integration can be extended to high eukaryotes, and even to whole animals.
DATABASES AND SOFTWARE TOOLS
Charting a complicated network of gene regulation is a major challenge [3], which will require the integration of many layers of systematic cell and molecular biology and many direct lines of research. The conventional methods for creating a network model include performing a series of experiments to identify specific interactions and conducting extensive literature surveys. Recent developments of high-throughput strategies have resulted in the accumulation of large amounts of data, including protein–protein, protein–DNA and genetic interactions [138], as well as information on cis-acting and trans-acting factors. These data will require powerful information storage, and query and analysis engines to handle data manipulation computationally. Current representational models of GRNs will need to evolve substantially in order to manage these data in a meaningful way.
Several large-scale, comprehensive databases on GRNs have been created. Databases such as AraC-XylS [139], Regulon DB [75], PlantCARE [140], AGRIS [141], EPD [142], TRRD [143] and TRANSFAC [131] are intended to serve as repositories for information on the regulation of gene transcription. A database of transcriptional start sites for human genes has been created and can thus provide a rich source of raw data for searches for promoter-proximal regulatory sequences [144]. Other databases such as TRANSCompel® [145], MIPS [134], BIND [146], DIP [133], MINT [147], GRID [148] and GeneNet [149] serve as repositories for protein and genetic interactions and associated regulatory events. These databases, when cross-referred to gene expression databases, which already have stored huge amounts of DNA microarray information from many organisms [150,151], will generate comprehensive and large-scale raw data for charting GRNs of different organisms. Constructing and maintaining a high-quality database requires a substantial amount of effort. Thus, creating a database large enough to capture gene regulatory information will require massive community investment and commitment, ranging from the individual researcher to funding agencies and journals, as well as innovation from database developers. Important goals of these databases include minimal redundancy, maximal annotation and integration with other databases [152].
Although such databases are useful sources of knowledge, there are numerous examples in information management and processing where the existence of multiple and/or specialized file formats has hindered accessibility, information exchange and integration [153]. Therefore it will also be important to standardize these data. It is crucial that software development is linked at an early stage through agreed documentation, XML (eXtensible Makeup Language)-based definitions and controlled vocabularies that allow different tools to exchange primary datasets. Considerable effort has already gone into interaction databases [133,134,146–149] and system biology software infrastructure [154], which should be built upon by current and future functional genomics and proteomics initiatives/researchers. Lessons learned from the analysis of DNA microarray data, including clustering, compendium and pattern-matching approaches, should be transportable to analysis of GRNs [155,156]. A proteomics standards initiative is currently developing formats for MS and protein–protein interaction data and annotation [157]. SBML (System Biology Markup Language; see http://sbw-sbml.org), along with CellML, represent attempts to define a standard for an XML-based computer-readable format that enables models to be shared and used even in a different software environment. SBW (System Biology Workbench) is built on SBML and provides a modular, broker-based and message-passing framework for system biology research. Both SBML and SBW represent the collective efforts of a number of research groups sharing the same vision [154].
Data standardization will make it easy to retrieve information from different databases [153]. A variety of software tools will be necessary in order to process and analyse the resulting large-scale data, as well as to understand data relationships quickly and to make biologically relevant predictions [158]. For molecular interactions, general-purpose graph viewers such as Pajek [159] are available to organize and display the data as a two-dimensional network; specialized tools such as Cytoscape [160] and Osprey [161] provide these capabilities and also link the network to molecular interaction and functional databases such as BIND [146], DIP [133] and TRANSFAC [131]. Indeed, these visualization software tools perhaps could be developed as the interactive entry point to the integrated network of gene regulation, where a gene of interest connects directly to the latest information about that gene and its relationships. Using Cytoscape, for example, the software is able to integrate both molecular interactions (such as protein–protein, protein–DNA and genetic interaction data) and state measurements together in a common framework, and to then bridge these data with a wide assortment of whole parameters and other biological attributes [160]. Cytoscape focuses on the high-level representation of components and interactions. Discoveries and hypothesis generation prompted by large-scale datasets generated across all manner of model systems will depend on data assembly tools such as Cytoscape or Osprey.
CONCLUSIONS AND FUTURE PERSPECTIVES
The past few years have witnessed a number of functional genomics, proteomics and bioinformatics approaches to dissecting the structure and function of GRNs at a genomic level. These have helped to identify and characterize components of GRNs and links between them, as well as to elucidate the important gene regulatory events in E. coli, yeast and sea urchin. Yet there is still a need both for additional high-throughput technologies and for computational methods with which to analyse large datasets and to integrate complex and disparate kinds of protein–protein, protein–DNA and genetic interactions, as well as expression profiling information. As is often the case, advances in technology have driven scientific breakthroughs. More recently, the development of a computational method to analyse a gene co-expression network should provide a novel point of view for understanding GRNs from evolutionary and conserved points of view [162].
Another perspective will be for the field of gene regulatory research to work hand in hand with those focused on pivotal biological processes, such as the cell cycle and development, in order to best convert the broad but shallow gene regulatory information available into a deeper understanding. Looking ahead, genomics, quantitative proteomics and computing sciences will be integrated into a comprehensive strategy for designing, modelling and analysing experiments to investigate complex biological networks: a new endeavour in the multidisciplinary field of bioinformatics. Therefore, in the near future, we might have a reasonably complete picture of the GRN of a simple model organism, such as E. coli or yeast. This picture, in turn, will provide a blueprint for understanding the GRNs of other, more complex, model organisms and of humans.
Acknowledgments
We thank Dr X. Lv, Dr L. Xin, Dr X. S. Wu and other colleagues for helpful discussion and critical reading of the manuscript. We are especially grateful to Dr M. Madan Babu, Dr T. Manke and Professor M. Vingron for providing the figures and related analyses. We apologize for any errors or omissions in this review. This work was supported by a grant from the National Natural Science Foundation of China (no. 30393110).
References
- 1.Hood L., Galas D. The digital code of DNA. Nature (London) 2003;421:444–448. doi: 10.1038/nature01410. [DOI] [PubMed] [Google Scholar]
- 2.Bolouri H., Davidson E. H. Modeling DNA sequence-based cis-regulatory gene networks. Dev. Biol. 2002;246:2–13. doi: 10.1006/dbio.2002.0617. [DOI] [PubMed] [Google Scholar]
- 3.Davidson E. H., Rast J. P., Oliveri P., Ransick A., Calestani C., Yuh C. H., Minokawa T., Amore G., Hinman V., Arenas-Mena C., et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 4.Brown C. T., Rust A. G., Clarke P. J., Pan Z., Schilstra M. J., De Buysscher T., Griffin G., Wold B. J., Cameron R. A., Davidson E. H., Bolouri H. New computational approaches for analysis of cis-regulatory networks. Dev. Biol. 2002;246:86–102. doi: 10.1006/dbio.2002.0619. [DOI] [PubMed] [Google Scholar]
- 5.Bussemaker H. J., Li H., Siggia E. D. Regulatory element detection using correlation with expression. Nat. Genet. 2001;27:167–171. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- 6.Ueda H. R., Chen W., Adachi A., Wakamatsu H., Hayashi S., Takasugi T., Nagano M., Nakahama K., Suzuki Y., Sugano S., et al. A transcription factor response element for gene expression during circadian night. Nature (London) 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 7.Futcher B. Transcriptional regulatory networks and the yeast cell cycle. Curr. Opin. Cell Biol. 2002;14:676–683. doi: 10.1016/s0955-0674(02)00391-5. [DOI] [PubMed] [Google Scholar]
- 8.Buhler J., Tompa M. Finding motifs using random projections. J. Comput. Biol. 2002;9:225–242. doi: 10.1089/10665270252935430. [DOI] [PubMed] [Google Scholar]
- 9.Thompson W., Rouchka E. C., Lawrence C. E. Gibbs Recursive Sampler: finding transcription factor binding sites. Nucleic Acids Res. 2003;31:3580–3585. doi: 10.1093/nar/gkg608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinha S., Tompa M. YMF: A program for discovery of novel transcription factor binding sites by statistical overrepresentation. Nucleic Acids Res. 2003;31:3586–3588. doi: 10.1093/nar/gkg618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyrick J. J., Young R. A. Deciphering gene expression regulatory networks. Curr. Opin. Genet. Dev. 2002;12:130–136. doi: 10.1016/s0959-437x(02)00277-0. [DOI] [PubMed] [Google Scholar]
- 12.Lee T. I., Rinaldi N. J., Robert F., Odom D. T., Bar-Joseph Z., Gerber G. K., Hannett N. M., Harbison C. T., Thompson C. M., Simon I., et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 13.Grunenfelder B., Winzeler E. A. Treasures and traps in genome-wide data sets: case examples from yeast. Nat. Rev. Genet. 2002;3:653–661. doi: 10.1038/nrg886. [DOI] [PubMed] [Google Scholar]
- 14.Mathe C., Sagot M. F., Schiex T., Rouze P. Current methods of gene prediction, their strengths and weaknesses. Nucleic Acids Res. 2002;30:4103–4117. doi: 10.1093/nar/gkf543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finnegan E. J., Matzke M. A. The small RNA world. J. Cell Sci. 2003;116:4689–4693. doi: 10.1242/jcs.00838. [DOI] [PubMed] [Google Scholar]
- 16.Hershberg R., Altuvia S., Margalit H. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 2003;31:1813–1820. doi: 10.1093/nar/gkg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blandin G., Durrens P., Tekaia F., Aigle M., Bolotin-Fukuhara M., Bon E., Casaregola S., de Montigny J., Gaillardin C., Lepingle A., et al. Genomic exploration of the hemiascomycetous yeasts: 4. The genome of Saccharomyces cerevisiae revisited. FEBS Lett. 2000;487:31–36. doi: 10.1016/s0014-5793(00)02275-4. [DOI] [PubMed] [Google Scholar]
- 18.Oshiro G., Wodicka L. M., Washburn M. P., Yates J. R., III, Lockhart D. J., Winzeler E. A. Parallel identification of new genes in Saccharomyces cerevisiae. Genome Res. 2002;12:1210–1220. doi: 10.1101/gr.226802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kikuchi S., Satoh K., Nagata T., Kawagashira N., Doi K., Kishimoto N., Yazaki J., Ishikawa M., Yamada H., Ooka H., et al. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- 20.Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 21.Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature (London) 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 22.Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B. A., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 23.Reboul J., Vaglio P., Tzellas N., Thierry-Mieg N., Moore T., Jackson C., Shin-I T., Kohara Y., Thierry-Mieg D., Thierry-Mieg J., et al. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat. Genet. 2001;27:332–336. doi: 10.1038/85913. [DOI] [PubMed] [Google Scholar]
- 24.Reboul J., Vaglio P., Rual J. F., Lamesch P., Martinez M., Armstrong C. M., Li S., Jacotot L., Bertin N., Janky R., et al. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- 25.Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. Global analysis of protein expression in yeast. Nature (London) 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 26.Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 27.Ziauddin J., Sabatini D. M. Microarrays of cells expressing defined cDNAs. Nature (London) 2001;411:107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 28.Yuh C. H., Bolouri H., Davidson E. H. Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- 29.Lemon B., Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- 30.Ohler U., Niemann H. Identification and analysis of eukaryotic promoters: recent computational approaches. Trends Genet. 2001;17:56–60. doi: 10.1016/s0168-9525(00)02174-0. [DOI] [PubMed] [Google Scholar]
- 31.Ren B., Robert F., Wyrick J. J., Aparicio O., Jennings E. G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 32.Simon I., Barnett J., Hannett N., Harbison C. T., Rinaldi N. J., Volkert T. L., Wyrick J. J., Zeitlinger J., Gifford D. K., Jaakkola T. S., Young R. A. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- 33.Iyer V. R., Horak C. E., Scafe C. S., Botstein D., Snyder M., Brown P. O. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature (London) 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 34.Harismendy O., Gendrel C. G., Soularue P., Gidrol X., Sentenac A., Werner M., Lefebvre O. Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J. 2003;22:4738–4747. doi: 10.1093/emboj/cdg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Steensel B., Delrow J., Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- 36.Sun L. V., Chen L., Greil F., Negre N., Li T. R., Cavalli G., Zhao H., Van Steensel B., White K. P. Protein-DNA interaction mapping using genomic tiling path microarrays in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9428–9433. doi: 10.1073/pnas.1533393100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinmann A. S., Yan P. S., Oberley M. J., Huang T. H., Farnham P. J. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wells J., Graveel C. R., Bartley S. M., Madore S. J., Farnham P. J. The identification of E2F1-specific target genes. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3890–3895. doi: 10.1073/pnas.062047499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horak C. E., Mahajan M. C., Luscombe N. M., Gerstein M., Weissman S. M., Snyder M. GATA-1 binding sites mapped in the beta-globin locus by using mammalian chIp-chip analysis. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2924–2929. doi: 10.1073/pnas.052706999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martone R., Euskirchen G., Bertone P., Hartman S., Royce T. E., Luscombe N. M., Rinn J. L., Nelson F. K., Miller P., Gerstein M., et al. Distribution of NF-kappaB-binding sites across human chromosome 22. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu G., Spellman P. T., Volpe T., Brown P. O., Botstein D., Davis T. N., Futcher B. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature (London) 2000;406:90–94. doi: 10.1038/35017581. [DOI] [PubMed] [Google Scholar]
- 42.Bar-Joseph Z., Gerber G. K., Lee T. I., Rinaldi N. J., Yoo J. Y., Robert F., Gordon D. B., Fraenkel E., Jaakkola T. S., Young R. A., Gifford D. K. Computational discovery of gene modules and regulatory networks. Nat. Biotechnol. 2003;21:1337–1342. doi: 10.1038/nbt890. [DOI] [PubMed] [Google Scholar]
- 43.Orian A., van Steensel B., Delrow J., Bussemaker H. J., Li L., Sawado T., Williams E., Loo L. W., Cowley S. M., Yost C., et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Steensel B., Delrow J., Bussemaker H. J. Genomewide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2580–2585. doi: 10.1073/pnas.0438000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conlon E. M., Liu X. S., Lieb J. D., Liu J. S. Integrating regulatory motif discovery and genome-wide expression analysis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3339–3344. doi: 10.1073/pnas.0630591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu P. Recent advances in computational promoter analysis in understanding the transcriptional regulatory network. Biochem. Biophys. Res. Commun. 2003;309:495–501. doi: 10.1016/j.bbrc.2003.08.052. [DOI] [PubMed] [Google Scholar]
- 47.Rombauts S., Florquin K., Lescot M., Marchal K., Rouze P., van de Peer Y. Computational approaches to identify promoters and cis-regulatory elements in plant genomes. Plant Physiol. 2003;132:1162–1176. doi: 10.1104/pp.102.017715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pennacchio L. A., Rubin E. M. Genomic strategies to identify mammalian regulatory sequences. Nat. Rev. Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- 49.Ureta-Vidal A., Ettwiller L., Birney E. Comparative genomics: genome-wide analysis in metazoan eukaryotes. Nat. Rev. Genet. 2003;4:251–262. doi: 10.1038/nrg1043. [DOI] [PubMed] [Google Scholar]
- 50.Wasserman W. W., Palumbo M., Thompson W., Fickett J. W., Lawrence C. E. Human-mouse genome comparisons to locate regulatory sites. Nat. Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- 51.Xuan Z., Wang J., Zhang M. Q. Computational comparison of two mouse draft genomes and the human golden path. Genome Biol. 2003;4:R1. doi: 10.1186/gb-2002-4-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lenhard B., Sandelin A., Mendoza L., Engstrom P., Jareborg N., Wasserman W. W. Identification of conserved regulatory elements by comparative genome analysis. J. Biol. 2003;2:13. doi: 10.1186/1475-4924-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loots G. G., Ovcharenko I., Pachter L., Dubchak I., Rubin E. M. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 2002;12:832–839. doi: 10.1101/gr.225502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz S., Elnitski L., Li M., Weirauch M., Riemer C., Smit A., Green E. D., Hardison R. C., Miller W. (NISC Comparative Sequencing Program) MultiPipMaker and supporting tools: Alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 2003;31:3518–3524. doi: 10.1093/nar/gkg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B. A., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 56.Nobrega M. A., Ovcharenko I., Afzal V., Rubin E. M. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 57.Michelson A. M. Deciphering genetic regulatory codes: A challenge for functional genomics. Proc. Natl. Acad. Sci. U.S.A. 2002;99:546–548. doi: 10.1073/pnas.032685999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hasty J., McMillen D., Isaacs F., Collins J. J. Computational studies of gene regulatory networks:in numero molecular biology. Nat. Rev. Genet. 2001;2:268–279. doi: 10.1038/35066056. [DOI] [PubMed] [Google Scholar]
- 59.Berman B. P., Nibu Y., Pfeiffer B. D., Tomancak P., Celniker S. E., Levine M., Rubin G. M., Eisen M. B. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. U.S.A. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Markstein M., Markstein P., Markstein V., Levine M. S. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. U.S.A. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qian J., Lin J., Luscombe N. M., Yu H., Gerstein M. Prediction of regulatory networks: genome-wide identification of transcription factor targets from gene expression data. Bioinformatics. 2003;19:1917–1926. doi: 10.1093/bioinformatics/btg347. [DOI] [PubMed] [Google Scholar]
- 62.Pilpel Y., Sudarsanam P., Church G. M. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat. Genet. 2001;29:153–159. doi: 10.1038/ng724. [DOI] [PubMed] [Google Scholar]
- 63.Reference deleted.
- 64.Roven C., Bussemaker H. J. REDUCE: An online tool for inferring cis-regulatory elements and transcriptional module activities from microarray data. Nucleic Acids Res. 2003;31:3487–3490. doi: 10.1093/nar/gkg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ettwiller L. M., Rung J., Birney E. Discovering novel cis-regulatory motifs using functional networks. Genome Res. 2003;13:883–895. doi: 10.1101/gr.866403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roulet E., Busso S., Camargo A. A., Simpson A. J., Mermod N., Bucher P. High-throughput SELEX SAGE method for quantitative modeling of transcription-factor binding sites. Nat. Biotechnol. 2002;20:831–835. doi: 10.1038/nbt718. [DOI] [PubMed] [Google Scholar]
- 67.Levine M., Tjian R. Transcription regulation and animal diversity. Nature (London) 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 68.Hannenhalli S., Levy S. Predicting transcription factor synergism. Nucleic Acids Res. 2002;30:4278–4284. doi: 10.1093/nar/gkf535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kadonaga J. T. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 70.Riechmann J. L., Heard J., Martin G., Reuber L., Jiang C., Keddie J., Adam L., Pineda O., Ratcliffe O. J., Samaha R. R., et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 71.Chung H. R., Schafer U., Jackle H., Bohm S. Genomic expansion and clustering of ZAD-containing C2H2 zinc-finger genes in Drosophila. EMBO Rep. 2002;3:1158–1162. doi: 10.1093/embo-reports/kvf243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toledo-Ortiz G., Huq E., Quail P. H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parenicova L., de Folter S., Kieffer M., Horner D. S., Favalli C., Busscher J., Cook H. E., Ingram R. M., Kater M. M., Davies B., et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell. 2003;15:1538–1551. doi: 10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Madan Babu M., Teichmann S. A. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 2003;31:1234–1244. doi: 10.1093/nar/gkg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salgado H., Gama-Castro S., Martinez-Antonio A., Diaz-Peredo E., Sanchez-Solano F., Peralta-Gil M., Garcia-Alonso D., Jimenez-Jacinto V., Santos-Zavaleta A., Bonavides-Martinez C., et al. RegulonDB (version 4.0): transcriptional regulation, operon organization and growth conditions in Escherichia coli K-12. Nucleic Acids Res. 2004;32:D303–D306. doi: 10.1093/nar/gkh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen-Orr S. S., Milo R., Mangan S., Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 77.Martinez-Antonio A., Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 2003;6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 78.Gadgil H., Jurado L. A., Jarrett H. W. DNA affinity chromatography of transcription factors. Anal. Biochem. 2001;290:147–178. doi: 10.1006/abio.2000.4912. [DOI] [PubMed] [Google Scholar]
- 79.Yang V. W. Eukaryotic transcription factors:identification, characterization and functions. J. Nutr. 1998;128:2045–2051. doi: 10.1093/jn/128.11.2045. [DOI] [PubMed] [Google Scholar]
- 80.Hazbun T. R., Fields S. A genome-wide screen for site-specific DNA-binding proteins. Mol. Cell. Proteomics. 2002;1:538–543. doi: 10.1074/mcp.t200002-mcp200. [DOI] [PubMed] [Google Scholar]
- 81.Forde C. E., McCutchen-Maloney S. L. Characterization of transcription factors by mass spectrometry and the role of SELDI-MS. Mass Spectrom. Rev. 2002;21:419–439. doi: 10.1002/mas.10040. [DOI] [PubMed] [Google Scholar]
- 82.Nordhoff E., Krogsdam A. M., Jorgensen H. F., Kallipolitis B. H., Clark B. F., Roepstorff P., Kristiansen K. Rapid identification of DNA-binding proteins by mass spectrometry. Nat. Biotechnol. 1999;17:884–888. doi: 10.1038/12873. [DOI] [PubMed] [Google Scholar]
- 83.Woo A. J., Dods J. S., Susanto E., Ulgiati D., Abraham L. J. A proteomics approach for the identification of DNA binding activities observed in the electrophoretic mobility shift assay. Mol. Cell. Proteomics. 2002;1:472–478. doi: 10.1074/mcp.t200003-mcp200. [DOI] [PubMed] [Google Scholar]
- 84.Weiss S. Fluorescence spectroscopy of single biomolecules. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 85.Heyduk T., Heyduk E. Molecular beacons for detecting DNA binding proteins. Nat. Biotechnol. 2002;20:171–176. doi: 10.1038/nbt0202-171. [DOI] [PubMed] [Google Scholar]
- 86.Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., Young R. A., Dynlacht B. D. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ogawa H., Ishiguro K., Gaubatz S., Livingston D. M., Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 88.McKenna N. J., O'Malley B. W. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 89.Yatherajam G., Zhang L., Kraemer S. M., Stargell L. A. Protein-protein interaction map for yeast TFIID. Nucleic Acids Res. 2003;31:1252–1260. doi: 10.1093/nar/gkg204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aebersold R., Mann M. Mass spectrometry-based proteomics. Nature (London) 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 91.Ranish J. A., Yi E. C., Leslie D. M., Purvine S. O., Goodlett D. R., Eng J., Aebersold R. The study of macromolecular complexes by quantitative proteomics. Nat. Genet. 2003;33:349–355. doi: 10.1038/ng1101. [DOI] [PubMed] [Google Scholar]
- 92.Blagoev B., Kratchmarova I., Ong S. E., Nielsen M., Foster L. J., Mann M. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol. 2003;21:315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 93.Newman J. R., Keating A. E. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 94.Hu C. D., Chinenov Y., Kerppola T. K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 95.Hu C. D., Kerppola T. K. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Samanta M. P., Liang S. Predicting protein functions from redundancies in large-scale protein interaction networks. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12579–12583. doi: 10.1073/pnas.2132527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Spirin V., Mirny L. A. Protein complexes and functional modules in molecular networks. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Davidson I. The genetics of TBP and TBP-related factors. Trends Biochem. Sci. 2003;28:391–398. doi: 10.1016/S0968-0004(03)00117-8. [DOI] [PubMed] [Google Scholar]
- 99.von Mering C., Krause R., Snel B., Cornell M., Oliver S. G., Fields S., Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature (London) 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 100.Kemmeren P., van Berkum N. L., Vilo J., Bijma T., Donders R., Brazma A., Holstege F. C. Protein interaction verification and functional annotation by integrated analysis of genome-scale data. Mol. Cell. 2002;9:1133–1143. doi: 10.1016/s1097-2765(02)00531-2. [DOI] [PubMed] [Google Scholar]
- 101.Deane C. M., Salwinski L., Xenarios I., Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol. Cell. Proteomics. 2002;1:349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 102.Jansen R., Greenbaum D., Gerstein M. Relating whole-genome expression data with protein-protein interactions. Genome Res. 2002;12:37–46. doi: 10.1101/gr.205602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Phizicky E., Bastiaens P. I. H., Zhu H., Snyder M., Fields S. Protein analysis on a proteomic scale. Nature (London) 2003;422:208–215. doi: 10.1038/nature01512. [DOI] [PubMed] [Google Scholar]
- 104.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 105.Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 106.Mueller C. L., Jaehning J. A. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chung W. H., Craighead J. L., Chang W. H., Ezeokonkwo C., Bareket-Samish A., Kornberg R. D., Asturias F. J. RNA polymerase II/TFIIF structure and conserved organization of the initiation complex. Mol. Cell. 2003;12:1003–1013. doi: 10.1016/s1097-2765(03)00387-3. [DOI] [PubMed] [Google Scholar]
- 108.Rodriguez-Navarro S., Fischer T., Luo M. J., Antunez O., Brettschneider S., Lechner J., Perez-Ortin J. E., Reed R., Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 109.Bouwmeester T., Bauch A., Ruffner H., Angrand P. O., Bergamini G., Croughton K., Cruciat C., Eberhard D., Gagneur J., Ghidelli S., et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat. Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 110.Knuesel M., Wan Y., Xiao Z., Holinger E., Lowe N., Wang W., Liu X. Identification of novel protein-protein interactions using a versatile mammalian tandem affinity purification expression system. Mol. Cell. Proteomics. 2003;2:1225–1233. doi: 10.1074/mcp.T300007-MCP200. [DOI] [PubMed] [Google Scholar]
- 111.Gavin A. C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature (London) 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 112.Forler D., Kocher T., Rode M., Gentzel M., Izaurralde E., Wilm M. An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat. Biotechnol. 2003;21:89–92. doi: 10.1038/nbt773. [DOI] [PubMed] [Google Scholar]
- 113.Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature (London) 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 114.Shevchenko A., Schaft D., Roguev A., Pijnappel W. W., Stewart A. F., Shevchenko A. Deciphering protein complexes and protein interaction networks by tandem affinity purification and mass spectrometry: analytical perspective. Mol. Cell. Proteomics. 2002;1:204–212. doi: 10.1074/mcp.m200005-mcp200. [DOI] [PubMed] [Google Scholar]
- 115.Ohi M. D., Link A. J., Ren L., Jennings J. L., McDonald W. H., Gould K. L. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 2002;22:2011–2024. doi: 10.1128/MCB.22.7.2011-2024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Graumann J., Dunipace L. A., Seol J. H., McDonald W. H., Yates J. R., Wold B. J., Deshaies R. J. Applicability of tandem affinity purification MudPIT to pathway proteomics in yeast. Mol. Cell. Proteomics. 2004;3:226–237. doi: 10.1074/mcp.M300099-MCP200. [DOI] [PubMed] [Google Scholar]
- 117.Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 118.Hartwell L. H., Hopfield J. J., Leibler S., Murray A. W. From molecular to modular cell biology. Nature (London) 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 119.Milo R., Shen-Orr S., Itzkovitz S., Kashtan N., Chklovskii D., Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 120.Mangan S., Alon U. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mangan S., Zaslaver A., Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J. Mol. Biol. 2003;334:197–204. doi: 10.1016/j.jmb.2003.09.049. [DOI] [PubMed] [Google Scholar]
- 122.Wall M. E., Hlavacek W. S., Savageau M. A. Design of gene circuits: lessons from bacteria. Nat. Rev. Genet. 2004;5:34–42. doi: 10.1038/nrg1244. [DOI] [PubMed] [Google Scholar]
- 123.Barabasi A. L., Oltvai Z. N. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 124.Segal E., Shapira M., Regev A., Pe'er D., Botstein D., Koller D., Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 125.Yeger-Lotem E., Margalit H. Detection of regulatory circuits by integrating the cellular networks of protein-protein interactions and transcription regulation. Nucleic Acids Res. 2003;31:6053–6061. doi: 10.1093/nar/gkg787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yu H., Luscombe N. M., Qian J., Gerstein M. Genomic analysis of gene expression relationships in transcriptional regulatory networks. Trends Genet. 2003;19:422–427. doi: 10.1016/S0168-9525(03)00175-6. [DOI] [PubMed] [Google Scholar]
- 127.Cho R. J., Campbell M. J., Winzeler E. A., Steinmetz L., Conway A., Wodicka L., Wolfsberg T. G., Gabrielian A. E., Landsman D., Lockhart D. J., Davis R. W. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 128.Qian J., Dolled-Filhart M., Lin J., Yu H., Gerstein M. Beyond synexpression relationships: local clustering of time-shifted and inverted gene expression profiles identifies new, biologically relevant interactions. J. Mol. Biol. 2001;314:1053–1066. doi: 10.1006/jmbi.2000.5219. [DOI] [PubMed] [Google Scholar]
- 129.Guelzim N., Bottani S., Bourgine P., Kepes F. Topological and causal structure of the yeast transcriptional regulatory network. Nat. Genet. 2002;31:60–63. doi: 10.1038/ng873. [DOI] [PubMed] [Google Scholar]
- 130.Horak C. E., Luscombe N. M., Qian J., Bertone P., Piccirrillo S., Gerstein M., Snyder M. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 2002;16:3017–3033. doi: 10.1101/gad.1039602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Matys V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A. E., Kel-Margoulis O. V., et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Manke T., Bringas R., Vingron M. Correlating protein-DNA and protein-protein interaction networks. J. Mol. Biol. 2003;333:75–85. doi: 10.1016/j.jmb.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 133.Salwinski L., Miller C. S., Smith A. J., Pettit F. K., Bowie J. U., Eisenberg D. The Database of interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mewes H. W., Amid C., Arnold R., Frishman D., Guldener U., Mannhaupt G., Munsterkotter M., Pagel P., Strack N., Stumpflen V., et al. MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res. 2004;32:D41–D44. doi: 10.1093/nar/gkh092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ge H., Liu Z., Church G. M., Vidal M. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat. Genet. 2001;29:482–486. doi: 10.1038/ng776. [DOI] [PubMed] [Google Scholar]
- 136.Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E., et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 137.Li S., Armstrong C. M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P. O., Han J. D., Chesneau A., Hao T., et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tong A. H., Lesage G., Bader G. D., Ding H., Xu H., Xin X., Young J., Berriz G. F., Brost R. L., Chang M., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 139.Tobes R., Ramos J. L. AraC-XylS database: a family of positive transcriptional regulators in bacteria. Nucleic Acids Res. 2002;30:318–321. doi: 10.1093/nar/30.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lescot M., Dehais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouze P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Davuluri R. V., Sun H., Palaniswamy S. K., Matthews N., Molina C., Kurtz M., Grotewold E. AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics. 2003;4:25. doi: 10.1186/1471-2105-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schmid C. D., Praz V., Delorenzi M., Perier R., Bucher P. The Eukaryotic Promoter Database EPD: the impact of in silico primer extension. Nucleic Acids Res. 2004;32:D82–D85. doi: 10.1093/nar/gkh122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kolchanov N. A., Ignatieva E. V., Ananko E. A., Podkolodnaya O. A., Stepanenko I. L., Merkulova T. I., Pozdnyakov M. A., Podkolodny N. L., Naumochkin A. N., Romashchenko A. G. Transcription Regulatory Regions Database (TRRD): its status in 2002. Nucleic Acids Res. 2002;30:312–317. doi: 10.1093/nar/30.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Suzuki Y., Yamashita R., Sugano S., Nakai K. DBTSS, DataBase of Transcriptional Start Sites: progress report 2004. Nucleic Acids Res. 2004;32:D78–D81. doi: 10.1093/nar/gkh076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kel-Margoulis O. V., Kel A. E., Reuter I., Deineko I. V., Wingender E. TRANSCompel: a database on composite regulatory elements in eukaryotic genes. Nucleic Acids Res. 2002;30:332–334. doi: 10.1093/nar/30.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bader G. D., Betel D., Hogue C. W. BIND: the Biomolecular interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., Cesareni G. MINT: a Molecular iNTeraction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 148.Breitkreutz B. J., Stark C., Tyers M. The GRID: the General Repository for interaction Datasets. Genome Biol. 2003;4:R23. doi: 10.1186/gb-2003-4-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ananko E. A., Podkolodny N. L., Stepanenko I. L., Ignatieva E. V., Podkolodnaya O. A., Kolchanov N. A. GeneNet: a database on structure and functional organisation of gene networks. Nucleic Acids Res. 2002;30:398–401. doi: 10.1093/nar/30.1.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Edgar R., Domrachev M., Lash A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gollub J., Ball C. A., Binkley G., Demeter J., Finkelstein D. B., Hebert J. M., Hernandez-Boussard T., Jin H., Kaloper M., Matese J. C., et al. The Stanford Microarray Database: data access and quality assessment tools. Nucleic Acids Res. 2003;31:94–96. doi: 10.1093/nar/gkg078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bader G. D., Heilbut A., Andrews B., Tyers M., Hughes T., Boone C. Functional genomics and proteomics: charting a multidimensional map of the yeast cell. Trends Cell Biol. 2003;13:344–356. doi: 10.1016/s0962-8924(03)00127-2. [DOI] [PubMed] [Google Scholar]
- 153.Stein L. Creating a bioinformatics nation. Nature (London) 2002;417:119–120. doi: 10.1038/417119a. [DOI] [PubMed] [Google Scholar]
- 154.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 155.Brazma A., Hingamp P., Quackenbush J., Sherlock G., Spellman P., Stoeckert C., Aach J., Ansorge W., Ball C. A., Causton H. C., et al. Minimum information about a microarray experiment (MIAME) – toward standards for microarray data. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 156.Ball C. A., Sherlock G., Parkinson H., Rocca-Sera P., Brooksbank C., Causton H. C., Cavalieri D., Gaasterland T., Hingamp P., Holstege F., et al. Standards for microarray data. Science. 2002;298:539. doi: 10.1126/science.298.5593.539b. [DOI] [PubMed] [Google Scholar]
- 157.Taylor C. F., Paton N. W., Garwood K. L., Kirby P. D., Stead D. A., Yin Z., Deutsch E. W., Selway L., Walker J., Riba-Garcia I, et al. A systematic approach to modeling, capturing, and disseminating proteomics experimental data. Nat. Biotechnol. 2003;21:247–254. doi: 10.1038/nbt0303-247. [DOI] [PubMed] [Google Scholar]
- 158.Buckingham S. Bioinformatics: Programmed for success. Nature (London) 2003;425:209–215. doi: 10.1038/425209a. [DOI] [PubMed] [Google Scholar]
- 159.Batageli V., Mrvar A. Pajek – program for large network analysis. Connections. 1998;21:47–57. [Google Scholar]
- 160.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Breitkreutz B. J., Stark C., Tyers M. Osprey: a network visualization system. Genome Biol. 2003;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Stuart J. M., Segal E., Koller D., Kim S. K. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]