Abstract
The nucleotide pyrophosphatases/phosphodiesterases NPP1 and NPP2/autotaxin are structurally related eukaryotic ecto-enzymes, but display a very different substrate specificity. NPP1 releases nucleoside 5′-monophosphates from various nucleotides, whereas NPP2 mainly functions as a lysophospholipase D. We have used a domain-swapping approach to map substrate-specifying determinants of NPP1 and NPP2. The catalytic domain of NPP1 fused to the N- and C-terminal domains of NPP2 was hyperactive as a nucleotide phosphodiesterase, but did not show any lysophospholipase D activity. In contrast, chimaeras of the catalytic domain of NPP2 and the N- and/or C-terminal domains of NPP1 were completely inactive. These data indicate that the catalytic domain as well as both extremities of NPP2 contain lysophospholipid-specifying sequences. Within the catalytic domain of NPP1 and NPP2, we have mapped residues close to the catalytic site that determine the activities towards nucleotides and lysophospholipids. We also show that the conserved Gly/Phe-Xaa-Gly-Xaa-Xaa-Gly (G/FXGXXG) motif near the catalytic site is required for metal binding, but is not involved in substrate-specification. Our data suggest that the distinct activities of NPP1 and NPP2 stem from multiple differences throughout the polypeptide chain.
Keywords: autotaxin, lysophospholipase D, nucleotide pyrophosphatase/phosphodiesterase (NPP), plasma-cell differentiation antigen-1 (PC-1), phosphodiesterase
Abbreviations: ECL®, enhanced chemiluminescence; EDG, endothelial cell differentiation gene; HA, haemagglutinin; HRP, horseradish peroxidase; NPP, nucleotide pyrophosphatase/phosphodiesterase; pNP-TMP, p-nitrophenyl thymidine 5′-monophosphate; TBS, Tris-buffered saline; wt, wild-type
INTRODUCTION
Nucleotide pyrophosphatases/phosphodiesterases (NPPs) represent a family of ubiquitous and conserved eukaryotic proteins that are either expressed as transmembrane ecto-enzymes or as secreted proteins [1–6]. Mammalian genomes contain at least seven NPP-encoding genes, but only three NPPs, namely NPP1, NPP2 and NPP3, have been studied in some detail. NPP1–3 show 40–50% identity at the protein level, and they all consist of an N-terminal intracellular domain (10–80 residues), a single transmembrane domain (approx. 20 residues) and a large extracellular domain (approx. 800 residues). The latter comprises consecutively two somatomedin-B-like homodimerization domains, a catalytic domain and a C-terminal ‘nuclease-like’ domain with an unknown role [7].
NPP1–3 release nucleoside 5′-monophosphates from a variety of nucleotides and nucleotide derivatives [1,4]. For example, NPP1 hydrolyses ATP into AMP and pyrophosphate, the latter being a potent inhibitor of bone mineralization and tissue calcification. This function accounts for the phenotype of NPP1-null mice, which suffer from hypermineralization abnormalities such as osteoarthritis and ossification of the posterior longitudinal ligaments of the spine [8,9]. Compared with NPP1 and NPP3, NPP2 is a rather poor NPP [10]. However, in contrast with NPP1 and NPP3, NPP2 also acts as a lipid phosphodiesterase [11–13]. Notably, NPP2 displays an intrinsic lysophospholipase D activity, and is capable of releasing lysophosphatidic acid from lysophosphatidylcholine or sphingosine 1-phosphate from lysosphingomyelin. Lysophosphatidic acid acts as both a mitogen and a motogen through G-protein-coupled EDG (endothelial cell differentiation gene) receptors. Sphingosine 1-phosphate also acts through G-protein-associated EDG receptors and modulates cell motility. The ability to release signalling molecules from lysophospholipids accounts for the well-known contribution of NPP2 to tumour metastasis and angiogenesis [14].
NPPs belong to a superfamily of phospho-/sulpho-co-ordinating metalloenzymes [2,15]. Residues that are essential for catalysis, including a threonine that forms the covalent catalytic intermediate and several residues that co-ordinate two metals in the catalytic site, are conserved among all NPPs, implicating a similar or identical catalytic mechanism. However, the molecular basis for the substrate-specificity of NPPs remains unknown. Previous efforts to address this issue have failed, in part because NPP fragments appear to be inactive [7]. For example, whereas wildtype (wt) NPP1 is active when expressed in mammalian cells, the deletion of only small fragments from either the N-terminus or the C-terminus yields a completely inactive enzyme. It is not clear from the available data whether the lack of activity of NPP1 fragments results from the deletion of substrate-recognition sequences or from protein-maturation problems. To differentiate between these possibilities and to map the substrate-specifying determinants of NPP1 and NPP2, we have used domain-swapping and site-directed mutagenesis. Our data suggest that isoform-specific sequences throughout the polypeptide chain determine the activity and substrate specificity of NPP1 and NPP2.
EXPERIMENTAL
Site-directed mutagenesis and construction of NPP1–NPP2 chimaeras
The fusions of mouse NPP1 and rat NPP2 with an N-terminal HA (haemagglutinin)-tag and a C-terminal myc-tag were described previously [10]. Site-directed mutagenesis of these constructs was performed using the QuikChange™ kit (Stratagene). All mutations were verified by restriction and sequence analysis. The mutations involved the glycine-rich motif in the catalytic domain (see Figures 6 and 7), as well as residues near the catalytic site that differed in NPP1 and NPP2 (see Figures 4 and 5). To enable the swapping of domains between NPP1 and NPP2, specific restriction sites were introduced in the regions of the HA–NPP–myc constructs that correspond to the beginning and the end of the catalytic domain (Figure 1A). Thus silent mutations were introduced in codons 186–187 of mouse NPP1 (CCAGCA→CCCGCG), creating a SacII restriction site, and in codons 568–569 (GGAAGT→GGTACC), resulting in a conservative point mutation (S569T; one-letter amino acid codes) and in the creation of a KpnI restriction site. Similarly, SacII and KpnI restriction sites were introduced in the rat NPP2 construct by the silent mutation of codons 157–158 of (CCTGCA→CCCGCG) and codons 523–524 (GGAACT→GGTACC) respectively. The digestion of the latter constructs with the appropriate restriction enzymes, and the ligation of similarly digested and purified NPP1/NPP2 fragments into these constructs enabled us to generate six NPP1–NPP2 chimaeras with various combinations of their N-terminal, catalytic and nuclease-like domains (Figure 1).
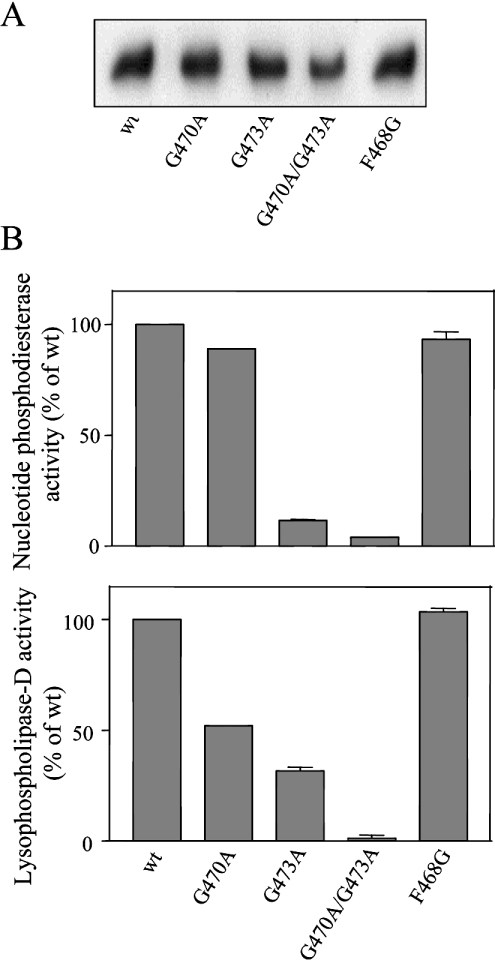
Figure 6. GXGXXG motif is essential for catalysis by NPP1.
The indicated mutants of NPP1 were expressed in COS-1 cells and immunoprecipitated from the cell lysates with anti-HA antibodies. The immunoprecipitated NPP1 variants were diluted to the same molar concentration, as detected by immunoblot analysis (A) and assayed for nucleotide phosphodiesterase activities with pNP-TMP as a substrate (B). Data are means±S.E.M. (n=3).
Figure 7. The FXGXXG motif is essential for catalysis by NPP2.
The indicated mutants of NPP2 were expressed in COS-1 cells and immunoprecipitated with anti-myc antibodies from the culture medium. The immunoprecipitated NPP2 variants were diluted to a similar molar concentration, as detected by immunoblot analysis (A) and assayed for nucleotide phosphodiesterase activities with pNP-TMP as a substrate (B, upper panel) and for lysophospholipase D activities with lysophosphatidylcholine as a substrate (B, lower panel). Data are means±S.E.M. (n=3).
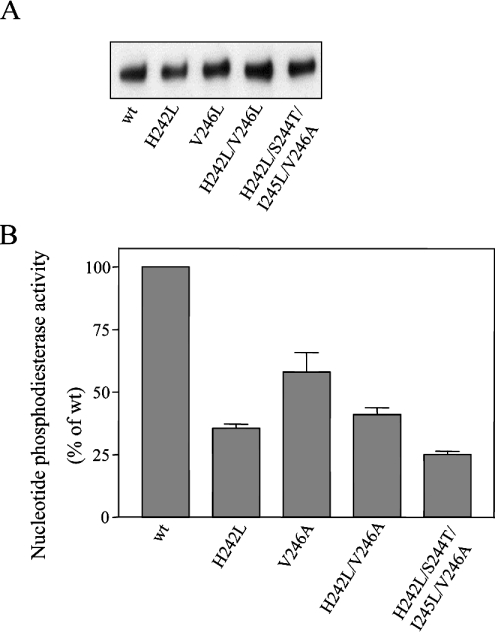
Figure 4. Mutagenesis of residues in the catalytic site of NPP1.
The indicated mutants of NPP1 were expressed in COS-1 cells and immunoprecipitated from the cell lysates with anti-HA antibodies. Immunoprecipitated NPP1 variants were assayed at the same protein concentration, as detected by immunoblot analysis (A) and were assayed for nucleotide phosphodiesterase activities with pNP-TMP as a substrate (B). Data are means±S.E.M. (n=3).
Figure 5. Mutagenesis of residues in the catalytic site of NPP2.
The indicated mutants of NPP2 were expressed in COS-1 cells and immunoprecipitated with anti-myc antibodies from the culture medium. The immunoprecipitated NPP2 variants were diluted to a similar molar concentration, as detected by immunoblot analysis (A) and assayed for nucleotide phosphodiesterase activities with pNP-TMP as a substrate (B, upper panel), and for lysophospholipase D activities with lysophosphatidylcholine as a substrate (B, lower panel). Data are means±S.E.M. (n=3).
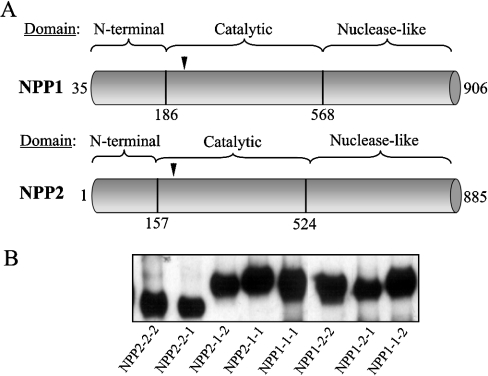
Figure 1. Expression of chimaeras of NPP1 and NPP2.
(A) Domain structure of NPP1 and NPP2. The vertical black lines indicate the position where silent sites were introduced for domain-swapping. Also indicated is the position of the catalytic-site threonine (arrowhead). (B) Western blot analysis with anti-myc antibodies of immunoprecipitates of NPP (chimaeras). HA- and myc-tagged NPP1, NPP2 and the indicated chimaeras were expressed in COS-1 cells. NPP1-1-1 (wt NPP1), NPP1-1-2, NPP1-2-2 and NPP1-2-1 were immunoprecipitated with anti-HA antibodies from the cell lysates, whereas NPP2-2-2 (wt NPP2), NPP2-1-1, NPP2-1-2 and NPP2-1-1 were immunoprecipitated from the culture medium with anti-myc antibodies. The immunoprecipitated proteins were used at the same molar concentrations for the enzymic assays shown in Figure 2.
Expression of NPP mutants
COS-1 cells were grown at 37 °C, in a humidified atmosphere containing 5% CO2, in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) heat-inactivated foetal bovine serum, penicillin (100 units/ml) and streptomycin (100 μg/ml). Cells were transiently transfected at 30–40% confluency using the FuGene™ 6 transfection system (Roche Diagnostics). The cells were harvested 48–72 h after transfection, washed twice in ice-cold PBS and lysed in 20 mM Tris/HCl at pH 7.5, 0.5 mM PMSF, 0.5 mM benzamidine, 0.3 M NaCl and 0.2% (v/v) Triton X-100. After centrifugation (5 min at 10000 g), the supernatant was used for immunoprecipitation with anti-HA-tag monoclonal antibodies (clone 12CA5) and protein A-TSK (Affiland, Liège, Belgium) for 8 h at 4 °C. The precipitates were washed once with 0.25 M LiCl and twice with ice-cold TBS (Tris-buffered saline), resuspended in TBS and used for Western blot analysis or enzymic assays. Secreted NPPs were immunoprecipitated from the medium using anti-myc-tag monoclonal antibodies (clone 9E10) and protein A-TSK for 8 h at 4 °C.
Western blot analysis
Protein samples were separated in 7.5% tricine gels (SDS/PAGE) and transferred on to a PVDF membrane (Amersham Biosciences) at 39 V in 50 mM Tris-base supplemented with 50 mM boric acid (pH 8.3). Non-specific binding sites were blocked with 5% (w/v) powdered milk and 0.2% (v/v) Triton X-100 in PBS. The NPP-fusions were visualized with anti-myc (clone 9E10) or anti-HA (clone HA-7) monoclonal antibodies, as indicated. HRP (horseradish peroxidase)–goat anti-mouse IgG (Dako) was used as a secondary antibody. The blot was treated with ECL® (enhanced chemiluminescence) reagent according to the manufacturer's protocol (Amersham Biosciences), exposed to Hyperfilm-ECL® and then developed.
Assays for the determination of NPP enzymic activities
All NPP activity assays were performed on immunoprecipitates that were quantified at least twice by Western blot analysis. A first immunoblot, often at several dilutions of the samples, was carried out to quantify differences in the amount of immunoprecipitated NPPs (results not shown). Such differences can be due to different levels of expression of the NPP isoforms/chimaeras and/or differences in the reproducibility of the immunoprecipitations. Based on the scanning results of the immunoblots, the samples were diluted to the same concentrations, which was verified again by immunoblot blot analysis (blots shown in Figures 1B, 4A, 5A, 6A and 7A) and equal volumes of the same samples were used for enzymic assays (see Figures 2, 4B, 5B, 6B and 7B). The data in Figures 1, 2, and 4-7 are representative of two to four independent immunoprecipitation experiments.
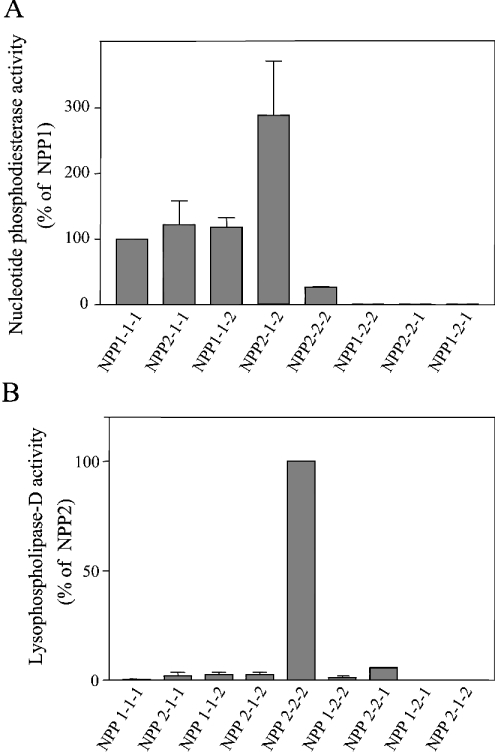
Figure 2. Effect of domain swapping on enzymic activities of NPP1 and NPP2.
Immunoprecipitated NPP1, NPP2 and the NPP chimaeras were assayed at equal molar concentrations, as detected by immunoblotting (Figure 1B). The enzymes were assayed for nucleotide phosphodiesterase activities with pNP-TMP as a substrate (A) and for lysophospholipase D activities with lysophosphatidylcholine as a substrate (B). Data are means±S.E.M. (n=3–7).
The nucleotide phosphodiesterase activity was measured with the nucleotide derivative pNP-TMP (p-nitrophenyl thymidine 5′-monophosphate; Sigma) as a substrate [16]. Briefly, the NPP constructs were incubated with 5 mM pNP-TMP at 30 °C in 100 mM Tris/HCl at pH 8.9. The enzyme reaction was stopped by a 10-fold dilution with 3% (w/v) trichloroacetic acid. Subsequently, the reaction mixture was neutralized with NaOH and p-nitrophenolate was quantified colorimetrically at 405 nm.
The nucleotide pyrophosphatase activity was measured with ATP as substrate [10]. Following incubation of the samples at 37 °C with 50 μM [γ-32P]ATP in 100 mM Tris/HCl at pH 8.9, the reaction was arrested with 100 mM EDTA. An aliquot of the reaction mixture was analysed by TLC on poly(ethyleneimine) cellulose plates (Merck). Nucleotides and degradation products were separated by ascending chromatography in 750 mM KH2PO4/HCl at pH 3.0. Radioactive spots were visualized by autoradiography.
Lysophospholipase D activity was assessed by determining the release of choline from lysophosphatidylcholine [11]. Briefly, the NPPs were incubated with 2 mM C14:0 lysophosphatidylcholine from egg (Sigma) in the presence of 100 mM Tris/HCl at pH 8.9, 5 mM MgCl2, 5 mM CaCl2, 500 mM NaCl and 0.05% (v/v) Triton X-100 at room temperature (20 °C) for 8 h. The liberated choline was detected by an enzymic colorimetric method using choline oxidase, HRP and TOOS [N-ethyl-N-(2-hydroxy-3-sulphoproryl)-3-methylaniline]. The purple-coloured reaction product was quantified at 540 nm.
RESULTS
Enzymic activities of NPP1–NPP2 chimaeras
Initial efforts to map the substrate-specifying domain(s) of NPP1 by deletion analysis yielded inconclusive data because NPP1 fragments generated by the deletion of relatively small fragments (<100 residues) from the N-terminus or the C-terminus were completely inactive when expressed in either bacteria or COS-1 cells ([7]; and results not shown). In view of the very different substrate specificities of NPP1 and NPP2, we have therefore made use of a domain-swapping approach for the initial mapping of substrate-specifying regions. We generated chimaeric NPPs with six combinations of the N-terminal, catalytic and nuclease-like domains of NPP1 and NPP2 (Figure 1). The wt and chimaeric proteins were expressed as fusions with an N-terminal HA-tag and a C-terminal myc-tag in COS-1 cells, immunoprecipitated from the culture medium or the cell lysates, and assayed at equal concentrations, as determined by immunoblotting with anti-myc antibodies (Figure 1B).
Swapping of the N- or C-terminal domains of NPP1 for those of NPP2 yielded chimaeras (NPP2-1-1 and NPP1-1-2) with a specific nucleotide phosphodiesterase activity similar to that of wt NPP1 (Figure 2A). Surprisingly, the chimaera NPP2-1-2 was nearly 3 times more active as a nucleotide phosphodiesterase than was NPP1. The latter finding was unexpected, since NPP2 is a poor nucleotide phosphodiesterase [10] and displayed in the present study a 4-fold lower specific activity towards nucleotides than did NPP1 (Figure 2A). Collectively, our data indicate that the N- and C-terminal domains of NPP1 can be functionally replaced by those of NPP2, and that either the flanking regions of the catalytic domain of NPP1 are inhibitory or that the flanking regions of the catalytic domain of NPP2 are activatory. Neither NPP1 nor the chimaeras of NPP2 with the catalytic domain of NPP1 had lysophospholipase D activity (Figure 2B), demonstrating that the catalytic domain of NPP2 is essential for the recognition of lysophospholipids as substrates. Unexpectedly, in contrast with wt NPP2, none of the chimaeras with the catalytic domain of NPP2, i.e. NPP2-1-1, NPP1-2-2 and NPP1-2-1, were capable of hydrolysing either nucleotides or lysophospholipids (Figures 2A and 2B). Our results indicate that both the N- and C-terminal domains of NPP2 contain sequences that are essential for the expression of catalytic activity and that are absent in the extremities of NPP1.
Substrate-specifying residues near the catalytic-site threonine
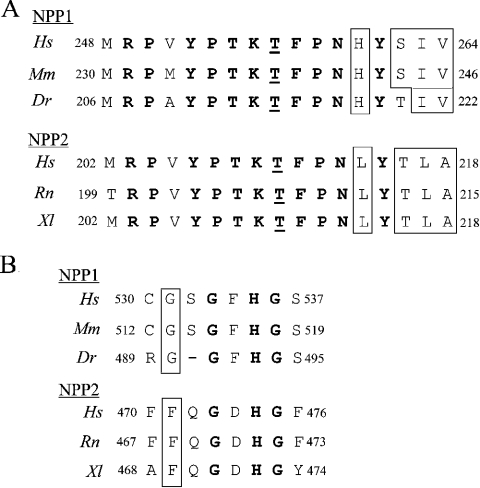
We subsequently used site-directed mutagenesis to identify substrate-specifying residues near the catalytic site. Although the sequence surrounding the catalytic-site threonine is nearly identical in NPP1 and NPP2, there are a few striking isoform-specific differences that are conserved from fish or amphibia to humans (Figure 3A). Thus the histidine residue in NPP1 (His242 of mouse NPP1) that is four residues C-terminal to the catalytic-site threonine, is replaced by a leucine residue in NPP2 (Leu211 of rat NPP2). The replacement of His242 in NPP1 by a leucine residue decreased the nucleotide phosphodiesterase activity by 60% (Figure 4B), whereas substitution of a histidine residue for the corresponding leucine residue in NPP2 (Leu211) increased the specific nucleotide phosphodiesterase activity by more than 2-fold (Figure 5B). Thus a histidine residue in this position was associated with a higher nucleotide phosphodiesterase activity of both NPP1 and NPP2. Interestingly, the L211H mutation in NPP2 decreased its lysophospholipase D activity by 70% (Figure 5B), indicating that Leu211 contributes to the high specific lysophospholipase D activity of NPP2. However, the H242L mutant of NPP1 did not show any lysophospholipase D activity (results not shown), demonstrating that additional essential determinants exist for the expression of lysophosholipase D activity.
Figure 3. Conservation of residues near the catalytic site of NPP1 and NPP2.
(A) Alignments of the catalytic site of NPP1 and NPP2 for the indicated species. The residues that are conserved in both NPP1 and NPP2 are in bold. The catalytic-site threonine is underlined and the conserved isoform-specific residues are boxed. (B) Alignments of the G/FXGXXG motif, which was predicted to be close to the catalytic site [2]. The residues that are conserved in both NPP1 and NPP2 are in bold and the first residue of G/FXGXXG motif is boxed. Dr, Danio rerio; Hs, Homo sapiens; Mm, Mus musculus; Rn, Rattus norvegicus; Xl, Xenopus laevis. GenBank® accession numbers are P22413 (HsNPP1), P06802 (MmNPP1), BX293547.6 (DrNPP1), Q13822 (HsNPP2), Q64610 (RnNPP2) and AAH44675 (XlNPP2).
Another isoform-specific difference C-terminal to the catalytic-site threonine concerns the SIV (Ser-Ile-Val) triplet in NPP1 (residues 244–246 of mouse NPP1), which is replaced by the TLA (Thr-Leu-Ala) triplet in NPP2 (residues 213–215 of rat NPP2) (Figure 3A). Replacement of Val246 in NPP1 by an alanine residue, as occurs in NPP2, decreased the nucleotide phosphodiesterase activity of NPP1 by some 40%, but did not affect further the activity of the H242L mutant of NPP1 (Figure 4B). The H242L mutation in combination with the replacement of the SIV triplet by a TLA triplet resulted in a NPP1 mutant that was slightly less active than NPP1-H242L as a nucleotide phosphodiesterase (Figure 4B), and which did not display any lysophospholipase D activity (results not shown). Conversely, mutation of the TLA triplet in NPP2, as in NPP2-A215V, had no major effect on either its nucleotide phosphodiesterase or lysophospholipase D activities (Figure 5B). Surprisingly, the mutation of the TLA triplet reversed the stimulatory effect of the L211H mutation on the nucleotide phosphodiesterase activity of NPP2, but had no effect on the lysophospholipase D activity (Figure 5B).
Role of the Gly/Phe-Xaa-Gly-Xaa-Xaa-Gly (G/FXGXXG) motif
The catalytic domain of NPP1 contains a conserved GXGXXG motif (Figure 3B) that was predicted to be positioned in the vicinity of the catalytic site [1,2], and that resembles a consensus dinucleotide-binding motif [17]. Intriguingly, none of the eukaryotic NPP2 enzymes has a GXGXXG motif, but they contain a conserved FXGXXG sequence instead, which, because of its more hydrophobic nature, could represent a lysophospholipid-binding motif. We found that the replacement of the first glycine residue of the GXGXXG motif with alanine did not affect the nucleotide phosphodiesterase activity of NPP1, whereas the replacement of this glycine residue with phenylalanine, as is present in NPP2, decreased the NPP1 activity by approx. 50% (Figure 6B). Importantly, the latter mutation did not endow NPP1 with lysophospholipase D activity (results not shown). Mutation of either the second or third glycine residue of the GXGXXG motif decreased the activity of NPP1 by 40–60% and the combined mutation of the last two or all three glycine residues abolished the activity of NPP1 completely (Figure 6B). Likewise, the mutation to alanine of both glycine residues in the FXGXXG motif of NPP2 abolished the enzymic activity towards both nucleotides and lysophospholipids (Figure 7B) completely. However, the replacement of the phenylalanine residue in this motif by glycine, as is present in NPP1, did not have any effect on these activities (Figure 7B).
The above data show clearly that the G/FXGXXG motif is essential for the expression of catalytic activity by both NPP1 and NPP2, but that it does not represent a substrate-determining motif. Since the penultimate residue of the G/FXGXXG motif is a conserved histidine residue (Figure 3B) that has been implicated in the binding of an essential metal at the catalytic site [2], we have examined whether or not mutants of the G/FXGXXG motif are inactive because of deficient metal binding. In agreement with this interpretation, we found that the activity of mouse NPP1-G513A/G515A/G518A as a nucleotide pyrophosphatase (Figure 8A) or nucleotide phosphodiesterase (Figure 8B) was partially recovered by the addition of ZnCl2. The maximal recovery by the addition of ZnCl2 amounted to 27% of wt activity (Figure 8B). Importantly, ZnCl2 did not affect the activity of wt NPP1.
Figure 8. Partial recovery of the activity of a glycine motif mutant of NPP1 by ZnCl2.
(A) The nucleotide pyrophosphatase activities of immunoprecipitated NPP1-wt and NPP1-G513A/G515A/G518A were measured with [γ-32P]ATP as substrate, in the absence or presence of ZnCl2, as indicated. The control refers to an incubation without added enzyme. The Figure shows an autoradiogram after separation of ATP and PPi by TLC. (B) The nucleotide phosphodiesterase activity of immunoprecipitated NPP1-wt (○) and NPP1-G513A/G515A/G518A (•) was measured with pNP-TMP as substrate in the presence of the indicated concentrations of ZnCl2. The activities were expressed as a percentage of the activities at 1 mM ZnCl2 (logarithmic scale). The activity of NPP1-G513A/G515A/G518A at 1 mM ZnCl2 amounted to 27% of the activity of NPP1-wt under the same conditions.
DISCUSSION
Contribution of the extremities of NPP1 and NPP2 to enzymic activities
N- or C-terminally truncated forms of NPP1 are inactive when expressed in mammalian cells, possibly because these deletion mutants are not properly targeted and, as a consequence, fail to undergo key maturation steps that are essential for the expression of catalytic activity. For example, NPP1 with a truncated C-terminus remained associated with the endoplasmic reticulum and was not transported to the plasma membrane [7]. Similarly, the N-terminal domain harbours the transmembrane domain and is therefore also essential for the intracellular trafficking of NPP1. Thus the analysis of truncation mutants does not appear to be a valid approach to examine the contribution of specific domains to the enzymic properties of NPP1 and NPP2. Since the latter enzymes have a clearly distinct substrate specificity, we have initially used a domain-swapping approach to delineate their activity- and substrate-specifying determinants.
Although our results revealed that the catalytic domains of NPP1 and NPP2 play a key role in substrate-specification, the flanking domains also emerged as important determinants of their respective enzymic activities. For example, the NPP2-1-2 chimaera was about 3-fold more active than wt NPP1, whereas all three chimaeras with the catalytic domain of NPP2 were completely inactive. One interpretation for these findings is that the N-terminal and C-terminal domains of NPP1, but not those of NPP2, are inhibitory towards the catalytic domain. An alternative explanation is that the extremities of NPP2, but not those of NPP1, represent activatory domains. In either case, the effect of the flanking regions appears to be most prominent in NPP2, since all chimaeric constructs with the catalytic domain of NPP2 were inactive.
When expressed in COS-1 cells, NPP1 is enriched in the cell lysate, whereas NPP2 is much more abundant in the culture medium [10]. The secretion of NPP2 has been explained by the proteolytic removal of an N-terminal fragment that includes the transmembrane domain [1,4]. Although we cannot currently rule out the possibility that the specific activity of cell-associated and secreted NPPs is different, our domain-swapping studies indicate that the localization of NPP1 and NPP2 is not a major determinant of their relative activity towards different substrates. Indeed, NPP2-1-1 and NPP2-1-2, like NPP2, were secreted and yet these chimaeras did not display any lysophospholipase D activity (Figures 1B and 2). Similarly, unlike NPP1, cell-associated NPP1-2-2 and NPP1-2-1 did not display any nucleotide phosphodiesterase activity.
Substrate-specifying determinants in the catalytic domain
Our domain-swapping experiments revealed that the catalytic domains of NPP1 and NPP2 harbour major activity- and substrate-specifying determinants. We reported earlier that the catalytic-site threonine is positioned at the beginning of an α-helical structure [2]. In the present study, we have identified conserved residues near the catalytic site (His242 and Val246 of NPP1) that are important for the expression of a high nucleotide phosphodiesterase activity (Figures 4B and 6B). Indeed, the mutation of these residues to the corresponding residues of NPP2, as in NPP1-H242L and NPP1-V246A, decreased the nucleotide phosphodiesterase activity of NPP1 2–3-fold, but did not endow this enzyme with a lysophospholipase D activity. Conversely, the replacement of Leu211 in NPP2 with a histidine residue, corresponding to His242 in NPP1, increased the specific nucleotide phosphodiesterase activity of NPP2 2-fold (Figure 5B, upper panel), and decreased its lysophospholipase D activity to a similar extent (Figure 5B, lower panel). Interestingly, His242 and Val246 in NPP1, and Leu213 and Ala217 in NPP2, are four and eight positions C-terminal to the catalytic-site threonine respectively (Figure 3A). Taking into account that an α-helix consists of nearly four residues/turn, this suggests that these conserved residues in NPP1 and NPP2 are positioned at the same side of the α-helix as is the catalytic-site threonine. This could explain why the mutation of these residues had a major effect on catalysis whereas mutation of flanking residues did not (Figures 4 and 5).
The GXGXXG motif of NPP1 resembles a consensus binding sequence for dinucleotides [17], raising the possibility that this glycine-rich motif was implicated in the binding of nucleotide substrates of NPP1. We have used site-directed mutagenesis to examine the role of the G/FXGXXG motif in the enzymic activities of NPP1 and NPP2. Our results show unequivocally that this motif is essential for catalysis, not because of a role in substrate-binding, but because it harbours one of the six metal-co-ordinating residues, i.e. His517 in mouse NPP1 and His471 in rat NPP2 (Figure 8).
What makes NPP2 a lipid phosphodiesterase?
Neither the point mutants of NPP1 nor any of the NPP chimaeras that we have generated displayed an important lysophospholipase D activity. This was a surprise, since we [10] and others [18] have demonstrated previously that the hydrolysis of lysophospholipids by NPP2 depends on the same catalytic-site residues that are implicated in the hydrolysis of nucleotides and which are conserved among NPPs. Our data therefore suggest that the expression of lysophospholipase D activity depends on isoform-specific determinants in the N-terminal and the catalytic, as well as the nuclease-like, domains of NPP2. Perhaps sequences in all three domains together form a lysophospholipid-binding site. In addition, we cannot currently rule out the possibility that post-translational modifications contribute to the expression of a lysophospholipase D activity.
Acknowledgments
This work was supported by the Fund for Scientific Research – Flanders Grant G.0082.02. Filip Diepvens provided excellent technical assistance. We thank Dr A. Boudrez, Dr H. Ceulemans and Dr A. Van Eynde for helpful advice.
References
- 1.Bollen M., Gijsbers R., Ceulemans H., Stalmans W., Stefan C. Nucleotide pyrophosphatases/phosphodiesterases on the move. Crit. Rev. Biochem. Mol. Biol. 2000;35:393–432. doi: 10.1080/10409230091169249. [DOI] [PubMed] [Google Scholar]
- 2.Gijsbers R., Ceulemans H., Stalmans W., Bollen M. Structural and catalytic similarities between nucleotide pyrophosphatases/phosphodiesterases and alkaline phosphatases. J. Biol. Chem. 2001;276:1361–1368. doi: 10.1074/jbc.M007552200. [DOI] [PubMed] [Google Scholar]
- 3.Duan R.-D., Bergman T., Xu N., Wu J., Cheng Y., Duan J., Nelander S., Palmberg C., Nilsson A. Identification of human intestinal alkaline sphingomyelinase as a novel ecto-enzyme related to the nucleotide phosphodiesterase family. J. Biol. Chem. 2003;278:38528–38536. doi: 10.1074/jbc.M305437200. [DOI] [PubMed] [Google Scholar]
- 4.Goding J. W., Grobben B., Slegers H. Physiological and pathophysiological functions of the ectonucleotide pyrophosphatase/phosphodiesterase family. Biochim. Biophys. Acta. 2003;1638:1–19. doi: 10.1016/s0925-4439(03)00058-9. [DOI] [PubMed] [Google Scholar]
- 5.Moolenaar W. Lysophospholipids in the limelight: autotaxin takes center stage. J. Cell Biol. 2003;158:197–199. doi: 10.1083/jcb.200206094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohe Y., Ohnishi H., Okazawa H., Tomizawa K., Kobayashi H., Okawa K., Matozaki T. Characterization of nucleotide pyrophosphatase-5 as an oligomannosidic glycoprotein in rat brain. Biochem. Biophys. Res. Commun. 2003;308:719–725. doi: 10.1016/s0006-291x(03)01454-2. [DOI] [PubMed] [Google Scholar]
- 7.Gijsbers R., Ceulemans H., Bollen M. Functional characterization of the noncatalytic ectodomains of the nucleotide pyrophosphatase/phosphodiesterase NPP1. Biochem. J. 2003;371:321–330. doi: 10.1042/BJ20021943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okawa A., Nakamura I., Goto S., Moriya H., Nakamura Y., Ikegawa S. Mutation in Npps in a mouse model of ossification of the posterior longitudinal ligament of the spine. Nat. Genet. 1998;19:271–273. doi: 10.1038/956. [DOI] [PubMed] [Google Scholar]
- 9.Sali A., Favaloro J. M., Terkeltaub R., Goding J. W. Germline deletion of the nucleoside triphosphate pyrophosphohydrolase (NTPPPH) plasma cell membrane glycoprotein (PC-1) produces abnormal calcification of periarticular tissues. In: Vanduffel L., Lemmens R., editors. Ecto-ATPases and Related Ectonucleotidases. Maastricht: Shaker Publishing B.V.; 2000. pp. 267–280. [Google Scholar]
- 10.Gijsbers R., Aoki J., Arai H., Bollen M. Hydrolysis of lysophospholipids and nucleotides by autotaxin (NPP2) involves a single catalytic site. FEBS Lett. 2003;538:60–64. doi: 10.1016/s0014-5793(03)00133-9. [DOI] [PubMed] [Google Scholar]
- 11.Umezu-Goto M., Kishi Y., Taira A., Hama K., Dohmae N., Takio K., Yamori T. Mills G. B., Inoue K., Aoki J., Arai H. Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J. Cell. Biol. 2002;158:227–233. doi: 10.1083/jcb.200204026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tokumura A., Majima E., Kariya Y., Tominaga K., Kogure K., Yasuda K., Fukuzawa K. Identification of human plasma lysophospholipase D, a lysophosphatidic acid-producing enzyme, as autotaxin, a multifunctional phosphodiesterase. J. Biol. Chem. 2002;277:39436–39442. doi: 10.1074/jbc.M205623200. [DOI] [PubMed] [Google Scholar]
- 13.Ferry G., Tellier E., Try A., Gres S., Naime I., Simon M. F., Rodriguez M., Boucher J., Tack I., Gesta S., et al. Autotaxin is released from adipocytes, catalyzes lysophosphatidic acid synthesis, and activates preadipocyte proliferation: up-regulated expression with adipocyte differentiation and obesity. J. Biol. Chem. 2003;278:18162–18169. doi: 10.1074/jbc.M301158200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nam S. W., Clair T., Kim Y., McMarlin A., Schiffman E., Liotta L. A., Stracke M. L. Autotaxin (NPP-2), a metastasis-enhancing motogen, is an angiogenic factor. Cancer Res. 2001;61:6938–6944. [PubMed] [Google Scholar]
- 15.Galperin M. Y., Bairoch A., Koonin E. V. A superfamily of metalloenzymes unifies phosphopentomutase and cofactor-independent phosphoglycerate mutase with alkaline phosphatases and sulfatases. Protein Sci. 1998;7:1829–1835. doi: 10.1002/pro.5560070819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stefan C., Stalmans W., Bollen M. Threonine autophosphorylation and nucleotidylation of the hepatic membrane protein PC-1. Eur. J. Biochem. 1996;241:338–342. doi: 10.1111/j.1432-1033.1996.00338.x. [DOI] [PubMed] [Google Scholar]
- 17.Bossemeyer D. The glycine-rich sequence of protein kinases: a multifunctional element. Trends Biochem. Sci. 1994;19:201–205. doi: 10.1016/0968-0004(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 18.Koh E., Clair T., Woodhouse E. C., Schiffmann E., Liotta L., Stracke M. Site-directed mutations in the tumor-associated cytokine, autotaxin, eliminate nucleotide phosphodiesterase I, lysophospholipase D, and motogenic activities. Cancer Res. 2003;63:2042–2045. [PubMed] [Google Scholar]