Abstract
A highly pure, chemically defined representative of a new class of antimicrobial peptide from the Atlantic white shrimp (Litopenaeus setiferus), penaeidin class 4 [Pen4-1 (penaeidin class 4 isoform 1)], was produced synthetically. Chemical synthesis was achieved by native ligation from two separate domains yielding a bioactive peptide that reflected the characteristics of native penaeidin. Synthetic Pen4-1 proved to be an effective antimicrobial peptide, particularly against the broad-spectrum pathogen Fusarium oxysporum, exhibiting a complex effect on reproductive growth at inhibitory concentrations resulting in the suppression of spore formation. Pen4-1 exhibits unique features [not previously observed for penaeidins from the Pacific white shrimp (L. vannamei)], including target-species specificity against Gram-positive bacteria, indicating a potential partitioning of antimicrobial function among this family of peptides. The proline-rich domain of penaeidin class 4 alone was an active antimicrobial peptide, having the same target range as the full-length Pen4-1. These findings indicate that the proline-rich domain of penaeidin is sufficient to confer target specificity and that divergence in this domain between classes can result in a gain in antimicrobial function as observed for the proline-rich domain of Pen4-1.
Keywords: antimicrobial peptide, Atlantic white shrimp (Litopenaeus setiferus), cysteine- and proline-rich domains, native chemical ligation, penaeidin class 4, peptide synthesis
Abbreviations: anti-MP, antimicrobial peptide; Boc, t-butyloxycarbonyl; DIPEA, N,N-di-isopropylethylamine; Fmoc, fluoren-9-ylmethoxycarbonyl; HOBt, N-hydroxybenzotriazole; MALDI-TOF, matrix-assisted laser-deionization–time-of-flight; MBHA, 4-methylbenzhydrylamine; MIC, minimum inhibitory concentration; Pen4-1, penaeidin class 4 isoform 1; PRD, proline-rich domain; TFA, trifluoroacetic acid
INTRODUCTION
Invertebrates lack the adaptive immune system that is characteristic of most vertebrate organisms and they therefore rely exclusively on the innate immune system for protection from potential pathogens [1,2]. Anti-MPs (antimicrobial peptides) constitute vital components of the innate immune system in most, if not all, multicellular organisms, acting as immune effectors by killing or inhibiting the growth of their microbial targets [3–6]. Penaeidins, anti-MPs originally isolated from the haemocytes of the Pacific white shrimp (Litopenaeus vannamei), are a diverse peptide family with a unique two-domain structure that are characterized by their antimicrobial activity [7], primarily directed against Gram-positive bacteria and fungi [8]. They are synthesized in granular haemocytes, released into the plasma after immune challenge [9,10] and localize to tissues, bound to cuticle surfaces [11]. Tissue localization of penaeidins to the cuticle is probably facilitated by their general affinity for chitin [10]. Analysis of the penaeidin class 3 three-dimensional structure [12] revealed that this peptide has a combination of characteristics that permits the classification of penaeidins into most, if not all, categories of anti-MPs. A broad range of penaeidin microbial targets, including both bacterial and fungal species, is hypothesized to be reflective of complexity inherent in the multidomain structure of the peptide [12].
Penaeidin messages represent most of the expressed genes of immunological interest in haemocytes [13,14] making them the most readily detectable gene in EST (expressed-sequence-tag) libraries from two separate species [14]. Among the expressed genes detected from shrimp haemocytes, a new class of penaeidin was discovered, designated ‘penaeidin class 4’, in separate shrimp species and multiple individuals [13]. Class-4 sequences had features consistent with the diverse family of penaeidin anti-MPs, including a highly conserved leader sequence, the presence of a PRD (proline-rich domain) and a cysteine-rich domain with a conserved cysteine array [13,15]. An extended proline-rich region that includes a Pro-Arg-Pro motif is identical between penaeidin classes 1/2 and 3 in L. vannamei (Figure 1B below), but is divergent in the class-4 sequences [15]. However, the functional domains of penaeidin class 4 show lower levels of homology with previously identified penaeidin classes from L. vannamei, even though class 4 exhibits leader-sequence conservation in common with other penaeidins [13]. In general, penaeidins exhibit only limited homology with other types of proline-rich anti-MPs [16,17]. It is hypothesized that the divergent amino acid sequence of class 4 confers functional differences when compared with the other classes. Such differences in function probably contribute to the ability of the shrimp immune system to counter an extensive array of potential environmental pathogens.
Figure 1. Amino acid sequence alignments of penaeidins.
(A) Alignment of full-length penaeidin sequences from L. setiferus. Representative full-length penaeidin sequences from L. setiferus are aligned against Litset Pen4-1. Conserved residues with Pen4-1 are shown in black and variant residues are in grey. Predicted C-terminal amidation is symbolized by asterisks (*). (B) PRD divergence in penaeidin class 4. Aligned are the PRD of PRD Pen4-1 (from L. setiferus) and PRD Pen3-1 (from L. vannamei). PRD Pen4-1 shows interesting differences in the primary sequence, such as variation in the Pro-Arg-Pro motif (underlined) that is present in other proline-rich anti-MPs [17] and conserved across many of the other penaeidin sequences, including Litvan Pen3-1, resulting in a Arg-Lys-Pro sequence. At the same time PRD Pen4-1 and PRD Pen3-1 share conserved residues that link these divergent segments, indicating that the general structure of this domain may be conserved across divergent classes.
To investigate the unique features of penaeidin class 4, an expressed isoform [Litset Pen4-1 or Pen4-1 (penaeidin class 4 isoform 1); formerly designated Ls4d], from the Atlantic white shrimp (L. setiferus), was synthesized utilizing the native chemical-ligation method [18,19]. The chemical-synthesis approach permits the production of penaeidin that retains native characteristics, including C-terminally amidation, without the introduction of unwanted modifications that can be problematic with some recombinant expression systems [8].
EXPERIMENTAL
HPLC
Preparative reversed-phase HPLC was performed using a Synchropak RP-P column (2.1 cm×25 cm) at a solvent flow rate of 5 ml/min (system 1) or on a Phenomenex JUPITER 5u C18 300A column (250 mm×10.00 mm) at a solvent flow rate of 3 ml/min (system 2). Analytical HPLC was performed on a Bakerbond Wide-Pore column (4.6 mm×250 mm) at a solvent flow rate of 1 ml/min (system 3) or Waters SymmetryShield RP18 column (4.6 mm×50 mm; pore size 10 nm; particle size 3.5 μm) at a solvent flow rate of 1 ml/min (system 4). With each system, solution A [0.1% TFA (trifluoroacetic acid) in water] was used with a linear 20–60% (v/v) gradient of solution B (0.1% TFA in 80% acetonitrile) over 30 min unless otherwise specified.
MALDI-TOF (matrix-assisted laser-deionization–time-of-flight) MS
MALDI-TOF MS was performed on either a PerSeptive Biosystems Voyager DR instrument or an Applied Biosystems (ABI) Voyager DR-STE instrument using α-cyano-4-hydroxycinnamic acid as the ionization matrix. The peptide was dissolved in 0.1% TFA in water (1 μg/μl) and diluted with 3 volumes of 50 mM α-cyano-4-hydroxycinnamic acid in 80% acetonitrile. Mass-to-charge ratios (m/z) are presented directly from the observed spectra in comparison with predicted values for each peptide examined respectively.
Amino acid analysis
Peptides were hydrolysed in the vapour phase of 6 M HCl containing 1% phenol for 1 h at 150 °C, and the amino acids derivatized with phenyl isothiocyanate followed by HPLC separation using a Waters PicoTag system.
Peptide synthesis
The proline-rich segment was constructed through t-butyloxycarbonyl (Boc) chemistry on MBHA (4-methylbenzhydrylamine) resin by solid-phase peptide synthesis using an ABI peptide synthesizer model 433A. The resin (0.5 mmol) was loaded with Boc-Gly-SCH2CH2COOH [18,19], the amino group liberated with TFA and the free amine retrieved by treatment with DIPEA (N, N-di-isopropylethylamine). For elongation, a 4-fold excess of each amino acid was activated by the dicyclohexylcarbodi-imide/N-hydroxybenzotriazole (HOBt) method and condensed to the amino group. The side chains of all trifunctional amino acids were protected by conventional HF-labile groups. After completion of the synthesis, deprotection of the side chains and simultaneous cleavage from the solid support was accomplished by HF in the presence of 5% m-cresol for 60 min at 0 °C. The crude peptide was extracted with 20% (v/v) acetic acid, freeze-dried, desalted on a Sephadex G25-sf column (2.5 cm×50 cm) in 1 M acetic acid and freeze-dried (overall yield, 190 mg; retention time in HPLC system 3, 14.7 min; m/z: 2620.46 found, 2627.98 calculated).
The cysteine-rich segment was synthesized via Fmoc (fluoren-9-ylmethoxycarbonyl) chemistry on the ABI peptide synthesizer (model 433A) using conventional TFA-labile protecting groups for the side chains of trifunctional amino acids. The 4-(2′,4′-dimethoxyphenyl-Fmoc-aminomethyl)phenoxyacetamidonorleucyl-MBHA (Rink Amide MBHA) resin (0.25 mmol) was allowed to react with a 4-fold excess of Fmoc-amino acid activated with TBTU [O-(benzotriazol-1-yl)-N, N, N′, N′-tetramethyluronium tetrafluoroborate] in the presence of HOBt and DIPEA. Liberation of the side chains and release from the solid support was carried out with 1.6 g of peptidyl-resin in a premixed solution containing 30 ml of TFA, 2.25 g of phenol, 0.75 ml of ethanedithiol, 1.5 ml of thioanisole and 1.5 ml of water for 3 h at room temperature. Thereafter the resin was filtered off, and the peptide precipitated with diethyl ether. The resultant pellet was air-dried, resuspended in 2 ml of water and freeze-dried. Purification of the cysteine-rich domain was achieved by HPLC on system 1 in portions of 50–100 mg (overall yield 130 mg). The retention time of the purified cysteine-rich domain in system 3 was 19.7 min; m/z: 2782.96 found, 2784.17 calculated.
Native chemical ligation
Native chemical ligation was carried out in batches using 22 mg (7.9 μmol) of cysteine-rich domain and 28 mg (10.7 μmol) of PRD (PRD Pen4-1) following published protocols [18,19]. After 48 h the product was purified by HPLC on system 2 (average yield per batch, 13 mg). The retention time of the final reduced product on system 3 was 21.5 min; m/z: 5303.24 found, 5304.16 calculated.
PRD
The proline-rich thioester recovered from native ligation reactions was saponified (1 mg/ml) in 0.05 M NaOH for 2 h at room temperature under an argon atmosphere, acidified with acetic acid and purified by HPLC using system 2. PRD Pen4-1 had a retention time of 14.3 min. on system 3 (m/z: 2534.32 found, 2534.98 calculated).
Oxidation of thiol groups
Reduced full-length Pen4-1 purified by HPLC was oxidized by the methods of Tam and Shen [20]. On an analytical scale the reduced peptide was dissolved in water (1 mg/ml) and diluted to 0.5 mg/ml using a 50 mM buffer at the selected pH (range 5.5–7.5) in the presence or absence of 0.15 M NaCl. DMSO (10%, v/v) was added to assist oxidation and folding proceeded at room temperature to completion (48 h). On a preparative scale, 52 mg of the reduced ligation product was oxidized at pH 7.0 in 25 mM Mops buffer in batches. The dominant isomer was isolated by preparative HPLC as described above (total yield, 31.4 mg) with a retention time of 17.8 min on system 3 (m/z: 5298.80 found, 5298.16 calculated).
CD
Molecular ellipticity was measured in the far-UV using a Jasco J-710 spectropolarimeter. The protein was dissolved in water and the concentration was determined by UV spectroscopy (the calculated absorption coefficient for Pen4-1 based on the presence of three tyrosine residues is 4020 cm−1·M−1). The peptide was diluted (1:1, v/v) to a final concentration of 5–50 μM with 50 mM phosphate buffer, pH 7.5. Measurements were performed using a cell of 0.1 cm pathlength, at a resolution of 0.2 nm and a bandwidth of 2 nm, and ten spectra were averaged.
Antimicrobial assays
The antimicrobial activity of Pen4-1 was assayed against several bacterial species, including Gram-positive Micrococcus luteus, Aerococcus viridans, Staphylococcus aureus, Bacillus megaterium and Gram-negative Escherichia coli 363, Vibrio vulnificus, Salmonella typhimurium and Klebsiella pneumoniae. In addition, marine filamentous fungi known to be pathogenic for shrimp were used, including Fusarium oxysporum, Botrytis cinerea and Penicillium crustosum. MICs (minimum inhibitory concentrations) were determined in duplicate by the liquid-growth-inhibition assay, essentially as described previously [8]. Poor broth (1% bactotryptone/0.5% NaCl, pH 7.5) nutrient medium was used for standard bacteria, and saline peptone water (1.5% peptone/1.5% NaCl, pH 7.2) was used for marine bacteria. The antifungal assay was performed in potato dextrose broth (Difco) at half strength, supplemented with tetracycline (10 μg/ml final concn.) and cefotaxim (100 μg/ml final concn.). Briefly, in a sterile microtitration plate, 10 μl of peptide, or deionized water as a control, were added to 90 μl of a midexponential-growth-phase culture of bacteria at a starting attenuance (D600) of 0.01 or with 80 μl of fungal spores (final concn. 104 spores/ml). Plates were incubated for 24 h at 30 °C with vigorous shaking or for 48 h at 25 °C in the dark without shaking and in a moist chamber, for antibacterial and antifungal assays respectively. Growth was monitored by spectrophotometry at 620 nm on a Multiscan microplate reader colorimeter (Labsystem). MICs are expressed in μM as a concentration interval [a]–[b], where [a] is the highest concentration tested at which microbial growth can be observed and [b] is the lowest concentration that causes 100% growth inhibition [21].
Chitin-binding assay
A 20 μg portion of Pen4-1 was freeze-dried, reconstituted in 800 μl of 50 mM Tris/0.1 M NaCl at pH 8.00, and incubated for 10 min at room temperature with 40 mg of chitin (Sigma–Aldrich) with gentle agitation. Samples were centrifuged (1000 g, 5 min) and the supernatant recovered. The chitin pellet was successively washed with 800 μl of a 50 mM Tris solution at pH 8.00, containing 0.1 M NaCl (wash 1), then 1 M NaCl (wash 2), and finally incubated in 10% acetic acid (elution). At every step, incubations were carried out for 10 min at room temperature under gentle agitation and supernatants were recovered after centrifugation (1000 g, 5 min). The collected fractions were acidified to pH 4.00 by addition of 500 μl of 0.5% TFA, and subjected to HPLC on a SymetryShield RP18 column (4.6 mm×50 mm, 3.5 μm; Waters). Elution was performed with a linear gradient of 0–60% (v/v) acetonitrile in water containing 0.046% TFA over 30 min at a flow rate of 1 ml/min. The control consisted in 20 μg of freeze-dried Pen4-1 reconstituted in 800 μl of 50 mM Tris/0.1 M NaCl, pH 8.00, then acidified with 500 μl of 0.5% TFA treated as for the samples.
Analysis of L. setiferus haemocytes by MALDI-TOF MS
Individual L. setiferus shrimps were bled into modified Alsever's solution containing 9 mM EDTA, 336 mM NaCl, 27 mM sodium citrate and 115 mM glucose at pH 7.0. Cells were collected by centrifugation at 2000 g for 15 min and lysed in 100 μl of acidified water (0.1% TFA) with brief vortex-mixing at room temperature. MALDI-TOF was done using 1 μl of a 1:10 dilution of lysate mixed with 3 μl of 50 mM α-cyano-4-hydroxycinnamic acid, of which 1 μl was spotted and dried for analysis of haemocyte lysate contents.
Affinity purification of Litset Pen4-1 from L. setiferus haemocytes
Serum immunoglobulins from rabbits immunized with synthetic Litset Pen4-1 (Cocalico Biologicals, Reamstown, PA, U.S.A.) were purified using Protein A–agarose (Sigma–Aldrich) and coupled to the AminoLink coupling resin (Pierce) by following the manufacturer's suggested protocols. Pooled haemocyte extracts from 37 L. setiferus individuals were concentrated using a Sep-Pak® Vac C18 cartridge (Waters) and applied to the Litset Pen4-1 affinity resin. The eluted sample was freeze-dried and resuspended in 100 μl of 0.1% TFA. MALDI-TOF MS was performed using 1 μl of affinity-purified material with 3 ml of 50 mM α-cyano-4-hydroxycinnamic acid, of which 1 μl was spotted and dried for analysis of affinity-purified material.
MALDI-TOF MS/MS analysis of native Litset Pen4-1
The immunopurified material was desalted and fractionated on an ABI 130A separation system. A single fraction contained a mass corresponding to the mature Litset Pen4-1 (m/z 5300.84) as detected by MALDI-TOF MS on an ABI4700 Proteomics Analyzer (Figure 7A below). MS/MS (tandem MS) sequencing of this mass confirmed the sequence, originally deduced from an expressed gene, which was used to design the synthetic Litset Pen4-1.
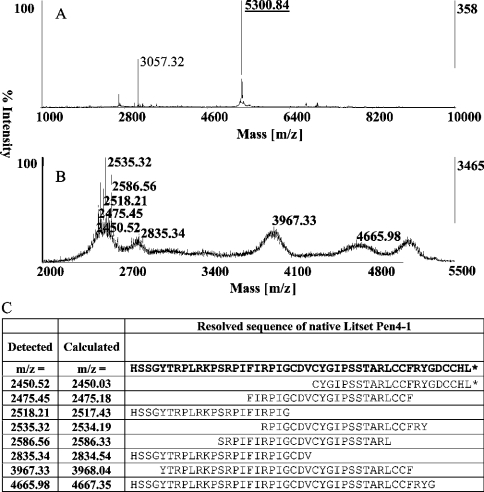
Figure 7. Peptide sequencing of native Litset Pen4-1.
(A) MALDI-TOF MS spectrum of the Litset-Pen4-1-containing fraction. A singly charged Litset Pen4-1 is detected (m/z 5300.84) that was selected for MS/MS mode fragmentation on the ABI4700 proteomics analyzer. (B) MS/MS mode fragmentation spectrum of Litset Pen4-1. Fragmentation of the parent-Litset-Pen4-1-ion-generated fragments that were most easily detectable in the range of m/z 2450.52–2586.56. Other fragmentation products were detectable at m/z 2835.34, 3967.33 and 4665.98. (C) Resolved peptide sequence of native Litset Pen4-1. The predicted mature Litset Pen4-1 peptide sequence used as the basis for chemical synthesis was used to generate a list of calculated fragmentation products. The most intense fragmentation products detected by the ABI4700 instrument matched calculated fragmentation products listed here. Representative sequence of the fragments used to confirm the native Litset Pen4-1 sequence are shown with calculated and detected m/z values.
RESULTS
Synthesis of Pen4-1
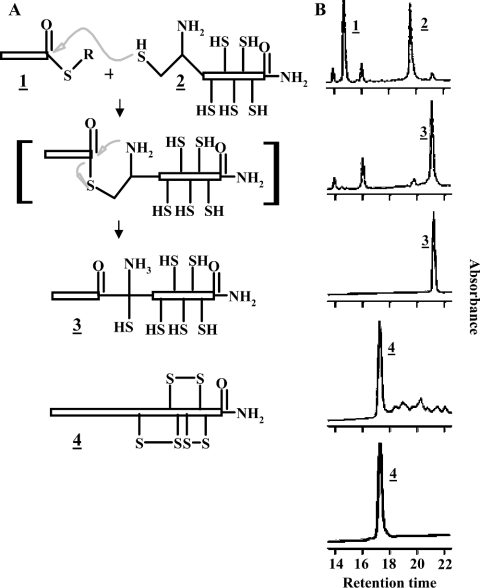
The protein sequence of penaeidin class 4 from L. setiferus (Pen4-1) deduced from the cDNA sequence was selected for synthesis. Figure 1 depicts the overall sequence alignment of Pen4-1 with representatives of the other penaeidin classes expressed in L. setiferus and a comparison of the PRDs of L. vannamei (Litvan Pen3-1) and L. setiferus (Litset Pen4-1). Segments matching the proline-rich and the cysteine-rich domains of Pen4-1 were produced and combined by native ligation (Figure 2A). After 5 h, a robust native ligation product could be observed by analytical HPLC. The reaction was allowed to proceed until completion at 48 h (Figure 2B panel 2), at which time a single uniform peak (ligation product) was collected from the reaction mixture by HPLC.
Figure 2. Synthesis approach for penaeidin class 4.
(A) Illustration of chemical synthesis of Pen4-1. Scheme of Pen4-1 synthesis, including the mechanism of native ligation joining the proline-rich (1) N-terminal segment and the cysteine-rich C-terminal segment (2) to form the full-length reduced product (3), which was subsequently oxidized in the presence of 10% DMSO (4). (B) Monitoring chemical synthesis by analytical HPLC. Analytical HPLC of class-4 synthesis steps is presented sequentially from the top (panels 1–5). Panel 1, native ligation at the initial reaction time point containing reactant PRD (1) and cysteine-rich domains (2) after initial mixing; panel 2, formation of the full-length Pen4-1 (3) after 48 h; panel 3, HPLC-purified reduced Pen4-1; panel 4, Pen4-1 after a 48 h oxidation reaction [note that the retention times (in min) shifted from 21.5 to 17.8 for disulphide isomers]; panel 5, the predominant isomer (at 17.8 min retention time) after purification (4). Additional high-resolution analytical HPLC was performed on an ABI 130A instrument using 5 μg of the final Pen4-1 oxidized product, which revealed only a single peak, which matched that shown in panel 5 (results not shown).
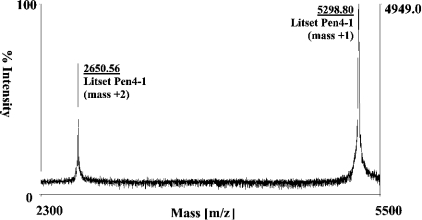
Optimization of protein folding and oxidation of the thiol groups to disulphides was assessed under a variety of conditions. In general, oxidation was enhanced by the addition of 10% (v/v) DMSO. The progress of the reaction and the final product abundance was determined by analytical HPLC (Figure 2B). At pH 7.5, the reduced form of the peptide was much less soluble and most (>80%) of the oxidized protein precipitated. In contrast, all peptides, regardless of the level of oxidation remained soluble at pH 5.5. In both cases, at pH 5.5 and pH 7.5, the yield of the desired oxidized isomer in solution was low and hence further optimization of the oxidation conditions using various pH conditions, including pH 6.0, 6.5 and 7.0, was necessary. In addition, the presence of salt at each pH condition tested resulted in a less intense product peak. Oxidation at pH 7.0 gave the most robust peak at 17.8 min, as examined by analytical HPLC (Figure 2B panel 4), and these conditions were used to scale-up the reaction for the preparation of penaeidin class 4. The HPLC profile of the final product is depicted in Figure 2(B), panel 5. The same conditions were used to refold and oxidize the cysteine-rich fragment alone, which also remained soluble; however, a mixture of disulphide isomers was generated without the appearance of a dominating product at all pH value tested. The pure oxidized full-length Pen4-1 isoform was characterized by MALDI-TOF MS, revealing an accurate mass (m/z 5298.80 found, 5298.16 calculated) and a highly pure disulphide isomer (Figure 3). Amino acid analysis and N-terminal peptide sequencing corroborated the predicted sequence of the full-length Pen4-1 oxidized peptide (Figure 1).
Figure 3. Mass analysis of Pen4-1.
MALDI-TOF MS spectrum of the purified Pen4-1 showing the determined mass (m/z 5298.80] within 0.65 mass units of the predicted mass (m/z 5298.16).
Class 4 structural stability
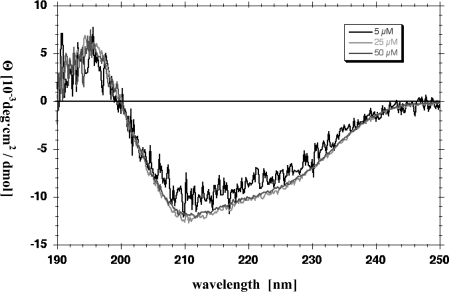
Near-UV CD spectra (Figure 4) were virtually identical across the range of concentrations used in the anti-MP activity assays. This indicates that the peptide does not significantly aggregate at higher concentrations.
Figure 4. CD analysis of Pen4-1 at different concentrations.
Concentration intervals of 5 μM, beginning at 5 μM extending up to 50 μM, were analysed for any observable shift in signal that may indicate aggregation at increased concentrations. Representative spectra for concentrations of 5, 25 and 50 μM are shown in direct comparison. The molar ellipticity observed for Pen4-1 did not appear to be affected by increased peptide concentration up to 50 μM.
Antimicrobial activity of penaeidin class 4
Liquid-culture growth-inhibition assays using a wide variety of microbes were performed to assess the range of antimicrobial activity of penaeidin class 4. Two sets of assay data were collected, including an initial direct comparison of Pen4-1 with recombinant penaeidin class 3 (Litvan Pen3-1) from L. vannamei [8], followed up by a direct comparison of Pen4-1 with PRD Pen4-1, including an expansion of the range of microbial targets (Table 1).
Table 1. Antimicrobial spectrum of activity of the chemically synthesized L. setiferus Pen4-1 and PRD alone (PRD Pen4-1) expressed as the MIC range.
Pen4-1 was compared directly in antimicrobial assays with PRD-Pen4-1, full-length penaeidin class 3 from L. vannamei (Pen3-1) and the PRD alone of Pen3-1 (PRD-Pen3-1). MIC values are expressed in this Table as the interval [a]-[b], where [a] is the highest concentration tested at which the growth of the microbe is not affected and [b] the lowest concentration that causes 100% growth inhibition
| MIC (μM) | ||||
|---|---|---|---|---|
| Micro-organism | Litset Pen4-1 | PRD-Pen4-1 | Litvan Pen3-1 | PRD-Pen3-1 |
| Filamentous fungi | ||||
| F. oxysporum | 0.84–1.26 | 2.94–4.38 | 3.12–6.25 | >50 |
| B. cinerea | 4.38–6.57 | 14.8–22.2 | Not tested | >50 |
| P. crustosum | 1.26–1.9 | 4.38–6.57 | Not tested | >50 |
| Bacteria | ||||
| Gram-positive | ||||
| A. viridans | 1.9–2.92 | 9.86–14.8 | 0.8–1.6 | >50 |
| M. luteus | 1.9–2.92 | 9.86–14.8 | 0.8–1.6 | >50 |
| B. megaterium | >50 | >50 | 3.12–6.25 | >50 |
| Staph. aureus | >50 | >50 | >50 | >50 |
| Gram-negative | ||||
| E. coli 363 | 22–33 | >50 | 6.25–12. 5 | >50 |
| V. vulnificus | >50 | >50 | >50 | >50 |
| S. thyphimurium | >50 | >50 | >50 | >50 |
| K. pneumoniae | >50 | >50 | >50 | >50 |
Antimicrobial activity was observed to varying extents for Pen4-1 against multiple fungal species, including B. cinera, P. crustosum, and F. oxysporum (Table 1). Pen4-1 exhibited a greater effectiveness than Litvan Pen3-1 against F. oxysporum, an opportunistic shrimp pathogen [22]. When exposed to Pen4-1 at growth-inhibitory concentrations (within the MIC range), F. oxysporum exhibited altered growth morphology, characterized by accentuated hyphal growth and an inhibition of sporulation, in a similar manner to what has been observed for Litvan Pen3-1 from L. vannamei [8]. At concentrations above the MIC range (>1.26 μM), no growth was observed for F. oxysporum in the presence of Pen4-1. As predicted, Pen4-1 was also active against the Gram-positive bacterial species M. luteus and A. viridans, a known crustacean pathogen. No inhibition was observed for B. megaterium with Pen4-1 from L. setiferus (up to 50 μM), even though this bacterium was inhibited by exposure to L. vannamei classes 2 and 3 [8], demonstrating target species specificity for penaeidins heretofore unobserved to an appreciable level. As is typical for penaeidins in general [7], very little activity was observed against Gram-negative bacteria, and Pen4-1 was inhibitory against E. coli only at relatively high concentrations (22–33 μM).
PRD antimicrobial activity
Unlike the PRD of Pen3-1 from L. vannamei, the PRD Pen4-1 of L. setifierus exhibited antimicrobial activity with the same target species spectrum as the full-length Pen4-1 for Gram-positive bacteria and fungi, suggesting that the PRD confers microbial target species activity. One exception was observed: PRD Pen4-1 was ineffective against E. coli at concentrations up to 50 μM, whereas the full-length Pen4-1 was inhibitory at high concentrations (Table 1). PRD Pen4-1 exhibits an effect on F. oxysporum growth morphology (Figure 5), resulting in an inhibition of sporulation and perturbation of hyphal growth, identical with that observed in the presence of the Pen4-1 (results not shown). Similar to the full-length peptide, at PRD Pen4-1 concentrations of 4.38 μM or greater, F. oxysporum growth was completely inhibited (Table 1).
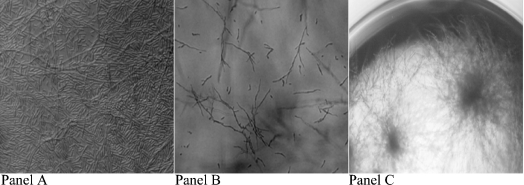
Figure 5. Effect of class 4 on Fusarium growth morphology.
F. oxysporum control growth (A) is compared here with growth in the presence of the PRD Pen4-1 (B), shown at high magnification to emphasize the suppressive effect on sporulation. Control growth (A), characterized by a field of spores that coat the bottom of the well and few hyphae, should be compared with growth in the presence of PRD-Pen4-1 (at the same magnification at the bottom of the well), where few spores are seen. Penaeidin class 4 suppresses spore formation and induces abnormal vegetative growth, highlighted at low magnification (C). In this panel, extensive hyphal growth is seen into the middle of the well, similar to the hyperbranching effect seen with certain other anti-MPs [24,25]. An indistinguishable effect was observed with the Pen4-1 at growth inhibitory (MIC) concentrations.
Pen4-1 binds chitin
Chitin was used for batch-method chitin-binding assays for Pen4-1 as previously described [9]. A high level of affinity was observed between Pen4-1 and chitin, as indicated by the resistance to elution in salt buffer (results not shown). The class 4 peptide–chitin complex was finally dissociated with 10% (v/v) acetic acid and the released peptide identified by its HPLC retention time.
Analysis of haemocyte lysates and detection of native Litset Pen4-1
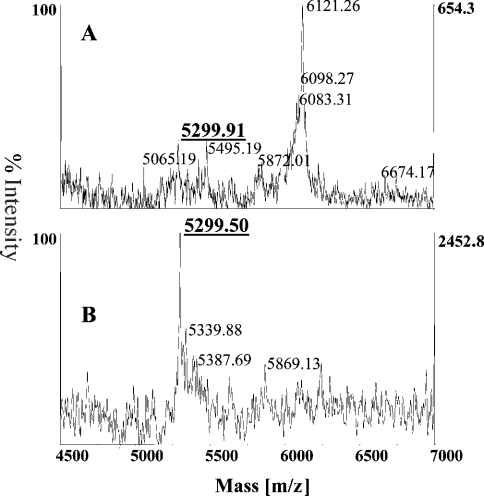
Haemocyte lysates from L. setiferus were analysed by MALDI-TOF MS after lysis, and a mass for Pen4-1 was predicted. A representative spectrum from an L. setiferus individual is presented (Figure 6A), and which exhibits a peak with a mass (m/z 5299.91) that is within 1.12 mass units of the detected mass for our synthetic Pen4-1 [Figure 3(m/z 5298.80)]. Affinity purification of a pool of L. setiferus haemocyte extracts using an rabbit anti-(Litset Pen4-1) immunoglobulin column greatly enhanced the relative abundance of the mass detectable at m/z 5299.51, indicating the specific recognition of the native Litset Pen4-1 expressed in haemocytes. The affinity-purified native penaeidin class-4 isoform from L. setiferus haemolymph was further subjected to MALDI-TOF MS/MS fragmentation on the ABI4700 Proteonomics Analyzer. Predicted fragmentation products matched the detected fragments generated. A portion of the complete spectrum is shown (Figure 7B) that contains the most easily detectable fragmentation products. The detected fragments were used to construct the native Litset Pen4-1 sequence (Figure 7C).
Figure 6. In vivo detection of Pen4-1 mass.
(A) Whole haemocyte extract from L. setiferus. Haemocytes of L. setiferus contain peptide masses that imply the presence of mature Pen4-1 protein expressed in vivo. A representative spectrum shown revealing a detected mass (m/z 5299.91) that is within 1.12 mass units of the detected mass for the synthetic Pen4-1 (m/z 5298.80). (B) Affinity purification of Litset Pen4-1. Combined haemocyte extracts were concentrated and applied to a rabbit serum anti-(Litset Pen4-1) immunoglobulin column. Examination of the eluted material revealed the enhanced percentage intensity of a peak (m/z 5299.50) that is within 1 mass unit of the known mass of Litset Pen4-1, a strong indication that mature Litset Pen4-1 is produced in L. setiferus hemocytes.
DISCUSSION
This is the first description of the use of native chemical ligation followed by chemically mediated oxidation of disulphide bonds to construct an active multidomain anti-MP such as Pen4-1. This synthetic approach was highly efficient, yielding enough material (31.5 mg) for functional assays and structural characterization. A comparable yield of Pen4-1 would not have been achievable by direct synthesis without utilization of the native ligation technique. Long contiguous synthesis approaches to making peptides of this length (47 amino acids) are inefficient, owing to the fractionation of yield at each deprotection and coupling step. An even more challenging aspect for the synthesis of Pen4-1 was the presence of six cysteine residues in the C-terminal region of the mature peptide, which lowered synthesis efficiency [23]. A general benefit of using chemical synthesis to produce penaeidins is the ability to specifically direct all aspects of peptide modification, avoiding non-native peptide modifications, such as glycosylation, a problem which was encountered in the expression of Litvan Pen3-1 in Saccharomyces cerevisiae [8]. In addition, it was also possible to design and construct Pen4-1 complete with specific characteristics found in the native peptide such as C-terminal amidation [7], a modification that has not been attainable for penaeidins through recombinant expression systems [8]. MS of haemolymph before and after affinity chromatography using a Pen4-1-specific antibody indicated that the native Pen4 was present in vivo, and MALDI-TOF MS sequencing results confirmed the expression of the native Litset Pen4-1 in L. setiferus haemocytes as predicted from previous haemocyte expressed-gene-sequence analysis.
Not only was synthetic Pen4-1 an effective antimicrobial agent, particularly against filamentous fungi, but its PRD exhibited a comparable effectiveness against F. oxysporum close to that of the full-length Litvan Pen3-1. The divergent amino acid sequence of Pen4-1, specifically in the PRD, may account for the differences observed in target species range and effectiveness with respect to class 3 from L. vannamei. While all penaeidins tested against F. oxysporum thus far demonstrate fungicidal activity, the observed differences in effectiveness between Pen4-1 and Litvan Pen3-1 were not observed when classes 2 and 3 from L. vannamei were compared directly [8]. Penaeidin 4-1 is the most effective penaeidin against the representative Fusarium species, a genus of environmental pathogens that infect plants and animals. F. oxysporum spore production and hence reproduction appears to be suppressed by Pen4-1, causing the fungus to favour hyphal growth. A similar effect known as ‘hyperbranching’ [24,25] has also been observed for some cysteine-rich anti-MPs that target fungi. However, it is noteworthy that PRD Pen4-1 has the same effect on growth morphology, clearly indicating that the PRD of Pen4-1 alone is sufficient for the observed effect on F. oxysporum proliferation. Inhibition of sporulation by penaeidins at low concentrations could, in effect, prevent the spread of a fungal infection or reinfection.
Pen4-1 was quite effective against multiple fungal species and against the Gram-positive bacteria M. luteus and A. viridans, but had no effect on B. megaterium, even at concentrations as high as 50 μM. The effective concentration of Pen4-1 exposed to the microbe (and hence its activity or lack thereof) was not affected by aggregation, since CD analysis revealed no significant changes even at the highest concentrations tested. While all penaeidins thus far characterized are cationic in character (with a pI greater than 8.0), this general property is not sufficient to promote Pen4-1 antimicrobial activity against B. megaterium, indicating that positive charge itself is insufficient to account for the antimicrobial activity of penaeidins. Though the cationic nature of penaeidins probably serves to promote interaction with the microbial membrane surface [11], this characteristic is clearly not sufficient in itself for antimicrobial activity. That Pen4-1 has a unique Gram-positive bacterial target-species range is especially interesting, since previous comparisons between penaeidin classes 2 and 3 did not reveal appreciable target-species differentiation. This is the first finding of partitioning of function within this anti-MP family.
One potential source of the observed target-species specificity is the difference in the otherwise-well-conserved primary sequence of the extended proline-rich motif between Pen4-1 and other penaeidin classes. Indeed the PRD Pen4-1 alone inhibits the same range of microbial targets as the full-length Pen4-1, unlike the PRD of class 3. Not only does the spectrum of activity for PRD Pen4-1 provide some insight into target-species specificity, but it also indicates that the PRD of Pen4-1 is sufficient for activity and that the cysteine-rich region of this peptide is not qualitatively necessary for antibiotic function.
Although the mechanism of action of penaeidins has not yet been determined, recent work with other proline-rich anti-MPs indicates that internal cytosolic proteins can be anti-MP targets. Specifically it has been observed that the proline-rich peptides from insects, such as apidaecin [17], not only have the capability to penetrate the membrane, but also target and inhibit the function of HSP 70 (heat-shock protein 70), a molecular chaperone that can actively refold proteins through an ATP-dependent mechanism [26]. A seemingly complex effect is observed for both Litvan Pen3-1 and Litset Pen4-1 against F. oxysporum that affects sporulation. At this time it is unclear whether this effect is due to targeting of an internal microbial cellular component. The effect certainly implies a more complex mechanism than the non-specific disruption of the microbial membrane [3,5,17]. The PRD of most penaeidin sequences, including Litvan Pen3-1, exhibits general homology in length and amino acid composition with these insect proline-rich peptides, but Pen4-1 lacks the canonically conserved Pro-Arg-Pro motif found in other penaeidins and is highly substituted in this domain. It is possible that substitution in this motif results in dramatic secondary-structural changes that results in the PRD Pen4-1 gain of antimicrobial function. Hence it is premature at this point to draw conclusions by direct comparison with insect proline-rich peptides, because the mechanism of activity may not depend on the level of peptide-length conservation or amino acid sequence identity. Instead, secondary- or tertiary-structural features that are not obvious from sequence analysis may account for differences in activity and specificity. In particular the placement of the proline residues may have a dramatic effect on function, owing to the structural characteristics that this particular amino acid may influence. An additional level of complexity in function is hypothesized to be characteristic of anti-MPs, which can act in a co-ordinated manner, with multiple molecules working to generate a net effect that is not achievable by a single molecule or at low peptide concentrations [3,5]. Direct comparison of the two penaeidin PRDs demonstrates the dramatic effect that changes in length and amino acid composition between classes of related molecules can have on antimicrobial function. In the future examining the potential for interaction between individual PRD molecules from the penaeidin classes may address the gain of PRD Pen4-1. The sequence of this domain clearly promotes function at relatively low concentrations, a function that may rely on greater affinity of this domain for like molecules in the presence of specific microbes that is not observed for PRD Pen3-1.
The use of direct chemical synthesis and native ligation permitted the generation of a homogeneous product with features like those of naturally occurring penaeidins. Characterization of L. setiferus Pen4-1 revealed a unique Gram-positive bacterial target specificity not previously observed for penaeidin classes 2 and 3 from L. vannamei. In addition, the activity of the PRD against the same range of microbial targets as the full-length class-4 peptide indicates that target specificity is one function of this domain, and implies that the cysteine-rich domain may have a tandem or synergistic role. Diversity in penaeidins provides the shrimp with a battery of anti-MPs that have differences in effectiveness, as observed for F. oxysporum, and microbial target activity, as seen with B. megaterium. A focused examination of the regulation of penaeidin class expression at the protein level, in vivo in response to specific pathogens, might help clarify the role of each class in contributing to the immune capabilities of the shrimp. The multidomain structure, obvious sequence diversity and increasingly apparent functional complexity for penaeidins implies the need for functional studies that focus on the mechanism of action of these anti-MPs against specific microbial target species.
Acknowledgments
We thank Dr Gregory Warr and Dr Robert Chapman for providing research direction and collaborative effort. Shrimp were caught by the Crustacean Monitoring Service of the South Carolina Department of Natural Resources (SCDNR), and we thank especially Larry Delancy. Peptide sequencing was done by Dr Christian Schwabe. This is publication number 6 of the Marine Biomedicine and Environmental Sciences program at the Medical University of South Carolina. This research was funded by NSF (National Science Foundation) grant no. MCB0110576, USDA (United States Department of Agriculture) NRICGP–CSREES (National Research Initiative Competitive Grants Program–Cooperative State Research, Education, and Extension Service) grant no. 2002-35201-11620 and the SCDNR.
References
- 1.Hoffmann J., Reichhart J. Drosophila innate immunity: an evolutionary perspective. Nat. Immunol. 2002;2:121–126. doi: 10.1038/ni0202-121. [DOI] [PubMed] [Google Scholar]
- 2.Rolff J., Siva-Jothy M. Invertebrate ecological immunology. Science. 2003;301:472–475. doi: 10.1126/science.1080623. [DOI] [PubMed] [Google Scholar]
- 3.Nissen-Meyer J., Nes I. Ribosomally synthesized antimicrobial peptides: their function, structure, biogenesis, and mechanism of action. Arch. Microbiol. 1997;167:67–77. [PubMed] [Google Scholar]
- 4.Martin E., Ganz T., Lehrer R. Defensins and other endogenous peptide antibiotics of vertebrates. J. Leukocyte Biol. 1995;58:128–136. doi: 10.1002/jlb.58.2.128. [DOI] [PubMed] [Google Scholar]
- 5.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature (London) 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 6.Tzou P., Reichhart J., Lemaitre B. Constitutive expression of a single antimicrobial peptide can restore wild-type resistance to infection in immunodeficient Drosophila mutants. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2152–2157. doi: 10.1073/pnas.042411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Destoumieux D., Bulet P., Loew D., Van Dorsselaer A., Rodriguez J., Bachère E. Penaeidins, a new family of antimicrobial peptides isolated from the shrimp Penaeus vannamei (Decapoda) J. Biol. Chem. 1997;272:28398–28406. doi: 10.1074/jbc.272.45.28398. [DOI] [PubMed] [Google Scholar]
- 8.Destoumieux D., Bulet P., Strub J., Van Dorsselaer A., Bachère E. Recombinant expression and range of activity of penaeidins, antimicrobial peptides from penaeid shrimp. Eur. J. Biochem. 1999;266:335–346. doi: 10.1046/j.1432-1327.1999.00855.x. [DOI] [PubMed] [Google Scholar]
- 9.Destoumieux D., Muñoz M., Cosseau C., Rodriguez J., Bulet P., Comps M., Bachère E. Penaeidins, antimicrobial peptides with chitin-binding activity, are produced and stored in shrimp granulocytes and released after microbial challenge. J. Cell. Sci. 2000;113:461–469. doi: 10.1242/jcs.113.3.461. [DOI] [PubMed] [Google Scholar]
- 10.Destoumieux D., Muñoz M., Bulet P., Bachère E. Penaeidins, a family of antimicrobial peptides from penaeid shrimp (Crustacea, Decapoda) Cell. Mol. Life Sci. 2000;57:1260–1271. doi: 10.1007/PL00000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muñoz M., Vandenbulcke F., Saulnier D., Bachère E. Expression and distribution of penaeidin antimicrobial peptides are regulated by haemocyte reactions in microbial challenged shrimp. Eur. J. Biochem. 2002;269:2678–2689. doi: 10.1046/j.1432-1033.2002.02934.x. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y., Poncet J., Garnier J., Zatylny C., Bachère E., Aumelas A. Solution structure of the recombinant penaeidin-3, a shrimp antimicrobial peptide. J. Biol. Chem. 2003;278:36859–36867. doi: 10.1074/jbc.M305450200. [DOI] [PubMed] [Google Scholar]
- 13.Cuthbertson B., Shepard E., Chapman R., Gross P. Diversity of the penaeidin antimicrobial peptides in two shrimp species. Immunogenetics. 2002;54:442–445. doi: 10.1007/s00251-002-0487-z. [DOI] [PubMed] [Google Scholar]
- 14.Gross P., Bartlett T., Browdy C., Chapman R., Warr G. Immune gene discovery by expressed sequence tag analysis of hemocytes and hepatopancreas in the Pacific white shrimp, Litopenaeus vannamei, and the Atlantic white shrimp, L. setiferus. Dev. Comp. Immunol. 2001;25:565–577. doi: 10.1016/s0145-305x(01)00018-0. [DOI] [PubMed] [Google Scholar]
- 15.Bachère E., Destoumieux D., Bulet P. Penaeidins, antimicrobial peptides of shrimp: a comparison with other effectors of innate immunity. Aquaculture. 2000;191:71–88. [Google Scholar]
- 16.Otvos L., Jr The short proline-rich antibacterial peptide family. Cell Mol. Life Sci. 2002;7:1138–1150. doi: 10.1007/s00018-002-8493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otvos L., Jr Antibacterial peptides isolated from insects. J. Peptide Sci. 2000;6:497–511. doi: 10.1002/1099-1387(200010)6:10<497::AID-PSC277>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 18.Tam J., Lu Y., Liu C., Shao J. Peptide synthesis using unprotected peptides through orthogonal coupling methods. Proc. Natl. Acad. Sci. U.S.A. 1995;92:12485–12489. doi: 10.1073/pnas.92.26.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson P., Muir T., Clark-Lewis I., Kent S. Synthesis of proteins by native chemical ligation. Science. 1994;266:766–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 20.Tam J. Wu C. R., Liu W., Zhang J. W. Disulfide bond formation in peptides by dimethyl sulfoxide: scope and applications. J. Am. Chem. Soc. 1991;113:6657–6662. [Google Scholar]
- 21.Casteels P., Anti-MPse C., Jacobs F., Tempst P. Functional and chemical characterization of hymenoptaecin, an antibacterial polypeptide that is infection-inducible in the honeybee (Apis mellifera) J. Biol. Chem. 1993;10:7044–7054. [PubMed] [Google Scholar]
- 22.Bian B., Egusa S. Histopathology of black gill disease caused by Fusarium solani (Martius) infection in the Kuruma prawn, Penaeus japonicus Bate. J. Fish. Dis. 1981;4:195–191. [Google Scholar]
- 23.DiMarchi R., Mayer J., Fan L., Brems D., Frank B., Green L., Hoffmann J., Howey D., Long H., Shaw W., et al. Synthesis of a fast-acting insulin based on structural homology with insulin-like growth factor I. In: Smith J. A., Rivier J. E., editors. Peptides, Chemistry and Biology: Proceedings of the 12th American Peptide Symposium. Leiden: Escom; 1992. pp. 26–28. [Google Scholar]
- 24.Nielsen K., Nielsen J., Madrid S., Mikkelsen J. Characterization of a new antifungal chitin-binding peptide from sugar beet leaves. Plant. Physiol. 1997;113:83–91. doi: 10.1104/pp.113.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van den Bergh K., Proost P., Van Damme J., Coosemans J., Van Damme E., Peumans W. Five disulfide bridges stabilize a hevein-type antimicrobial peptide from the bark of spindle tree (Euonymus europaeus L.) FEBS Lett. 2002;530:181–185. doi: 10.1016/s0014-5793(02)03474-9. [DOI] [PubMed] [Google Scholar]
- 26.Otvos L., Jr, Rogers M., Consolvo P., Condie B., Lovas S., Bulet P., Blaszczyk-Thurin M. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry. 2000;46:14150–14159. doi: 10.1021/bi0012843. [DOI] [PubMed] [Google Scholar]