Abstract
Cystathionine γ-lyase (CSE) is the last key enzyme in the trans-sulphuration pathway for biosynthesis of cysteine from methionine. Cysteine could be provided through diet; however, CSE has been shown to be important for the adequate supply of cysteine to synthesize glutathione, a major intracellular antioxidant. With a view to determining physiological roles of CSE in mice, we report the sequence of a complete mouse CSE cDNA along with its associated genomic structure, generation of specific polyclonal antibodies, and the tissue distribution and developmental expression patterns of CSE in mice. A 1.8 kb full-length cDNA containing an open reading frame of 1197 bp, which encodes a 43.6 kDa protein, was isolated from adult mouse kidney. A 35 kb mouse genomic fragment was obtained by λ genomic library screening. It contained promoter regions, 12 exons, ranging in size from 53 to 579 bp, spanning over 30 kb, and exon/intron boundaries that were conserved with rat and human CSE. The GC-rich core promoter contained canonical TATA and CAAT motifs, and several transcription factor-binding consensus sequences. The CSE transcript, protein and enzymic activity were detected in liver, kidney, and, at much lower levels, in small intestine and stomach of both rats and mice. In developing mouse liver and kidney, the expression levels of CSE protein and activity gradually increased with age until reaching their peak value at 3 weeks of age, following which the expression levels in liver remained constant, whereas those in kidney decreased significantly. Immunohistochemical analyses revealed predominant CSE expression in hepatocytes and kidney cortical tubuli. These results suggest important physiological roles for CSE in mice.
Keywords: cystathionase, cystathionine γ-lyase, cystathioninaemia, cysteine, methionine, trans-sulphuration
Abbreviations: CBS, cystathionine β-synthase; CSE, cystathionine γ-lyase; E15.5, embryonic day 15.5; HEK-293 cells, human embryonic kidney 293 cells; MZF1, myeloid zinc finger protein 1; ORF, open reading frame; PFA, paraformaldehyde; PLP, pyridoxal 5′-phosphate; poly(A)+, polyadenylated; RACE, rapid amplification of cDNA ends; Sp1, specificity protein 1; USF-1, upstream stimulatory factor-1; UTR, untranslated region
INTRODUCTION
In mammals, cysteine is provided through the diet or the trans-sulphuration pathway in which L-cysteine is synthesized by sulphur transfer from L-methionine to L-serine. Several catalytic enzymes are involved in the transsulphuration, and cystathionine γ-lyase (CSE, γ-cystathionase; EC 4.4.1.1), a PLP (pyridoxal 5′-phosphate)-dependent enzyme, catalyses its final essential step, the conversion of L-cystathionine into L-cysteine, α-ketobutyrate and ammonia. Cysteine is further irreversibly metabolized in liver to yield glutathione, taurine or inorganic sulphate by other enzymes, although CSE itself is capable of metabolizing cyst(e)ine and produces hydrogen sulphide [1–3], a gaseous neuromodulator [4] or smooth-muscle relaxant [5–7].
Deficiency of the CSE activity in humans is presumed to cause cystathioninaemia (cystathionineuria; MIM 219500), an autosomal recessive inborn error probably with no consistent clinical consequences [8]. Multiple mutations in the human CSE gene were recently found in patients with cystathioninaemia [9]. Since the CSE activity in rat liver is five times as high as that in human liver [10,11], it is possible that CSE may play more important roles in rodents. In rats, the CSE expression is restricted to specific tissues; CSE is highly expressed in liver and kidney with very low expression in brain [12–14]. Using DL-propargylglycine, a specific irreversible CSE inhibitor [15], CSE has been shown to be essential in rat liver, kidney and cultured hepatocytes for an adequate supply of cysteine to synthesize glutathione [11,16,17], a major intracellular antioxidant that protects cells from oxidative stress. Cysteine is also utilized for biosynthesis of taurine, the most abundant intracellular free amino acid, which has numerous biological functions and also could act as an antioxidant. Regardless of its role as the precursor of such bioactive molecules, cysteine itself could up-regulate the expression of cysteine dioxygenase that mediates taurine production and down-regulate the expression of γ-glutamylcysteine synthetase, which mediates glutathione production in cultured rat hepatocytes and intact rats [18,19]. Moreover, high cysteine concentrations could be cytotoxic and neurotoxic in rats [20,21] and high plasma cysteine concentrations in humans were associated with pre-eclampsia, premature delivery, low birth weight and cardiovascular diseases [22–24]. These lines of evidence suggest important roles for CSE as the regulator of cysteine homoeostasis and the glutathione–taurine rheostat.
Unfortunately, very little is known about mouse CSE at present. This study was performed to characterize CSE gene and protein in mice. We first cloned, sequenced and characterized the full-length mouse CSE cDNA and the complete mouse CSE gene. Anti-CSE polyclonal antibodies were generated and tissue distribution of the CSE transcript, protein and enzymic activity was examined in rats and mice. Developmental expression was investigated and specific CSE localization was revealed by immunohistochemistry, in both mouse liver and kidney. These results should contribute to uncover novel physiological functions of CSE and the transsulphuration pathway in mice.
EXPERIMENTAL
Materials
Molecular biology and cell-culture reagents were purchased from Invitrogen. All other reagents were obtained from Sigma, unless otherwise mentioned. Mice (C57BL/6J Jcl) and rats [Jcl:SD (Sprague–Dawley)] were purchased from Clea Japan (Tokyo, Japan). The use of animals was in compliance with the guidelines established by the Animal Care Committee of our Institute.
Mouse CSE cDNA cloning
Kidney was quickly removed from an 8-week-old male mouse and homogenized in TRIzol® with the Polytron homogenizer (Kinematica AG, Lucerne, Switzerland). Total RNA was isolated according to the manufacturer's instructions, and the first-strand cDNA was synthesized from 10 μg of total RNA using the avian myeloblastosis virus Reverse Transcriptase First-strand cDNA Synthesis kit (Invitrogen) and the NotI-adaptor primer (5′-AACTGGAAGAATTCGCGGCCGCAGGAATTTTTTTTTTTTTTTTTT-3′). This cDNA was used as a template for the first PCR with the adaptor primer (5′-TGGAAGAATTCGCGGCCGCAG-3′) and the CSE-e0 primer (5′-GCAAGACGTCGCACTCCTGCC-3′), which contains the 5′-UTR (untranslated region) sequences conserved within several mouse expressed sequence tags (GenBank® accession numbers AI891806, AI099398, AI226268 and AI316238); the PCR condition was 25 cycles of 94 °C for 15 s, 55 °C for 30 s and 68 °C for 4 min. The PCR product was further used as a template for the second PCR to obtain an entire ORF (open reading frame) with the primers CSE-e1-XhoI (5′-ATGCCTCGAGATGCAGAAGGACGCCTCTTTGAG-3′) and CSE-NotI-Cter (5′-ATGCATGCGCGGCCGCTTAAGGGTGCGCTGCCTTCAA-3′). The second PCR product was digested with XhoI and NotI, and subcloned into XhoI–NotI sites of the pME18S mammalian expression vector [25], producing the pME18S-mCSE vector. The full-length rat CSE ORF (a gift from Dr Nishi, Kagawa Medical School, Kagawa, Japan) [12] was similarly subcloned into the pME18S, producing the pME18S-rCSE vector. The 5′- and 3′-UTRs were isolated by 5′- and 3′-RACE (rapid amplification of cDNA ends) respectively using the Gene Racer Superscript II Reverse Transcriptase kit (Invitrogen). The following two primer sets were used: Gene Racer 5′-primer (Invitrogen) and CSE-e2-2 (5′-GTGCTTTGCCCCATCCAACGCAG-3′) for 5′-RACE or CSE-e9-1 (5′-TCTCACCCTCAGCATGAGCT-3′) and Gene Racer 3′-primer (Invitrogen) for 3′-RACE. More than ten independent cDNA clones from each PCR were confirmed by sequencing and the overlapping sequences were identical within all those clones.
Isolation of mouse CSE gene
Two probes, probes A and B, were used to screen total 5×105 independent plaques from the 129/SvJ mouse λ genomic library (Stratagene) with a conventional plaque hybridization method. Both probes were prepared by PCR using R1 embryonic stem cell genomic DNA [26,27] as a template. The 1.7 kb probe A, which spans exons 3–4, was amplified by PCR with the primers CSE-e3-1 (5′-GGCCTTTGCATCGGGTCTTGCTGC-3′) and CSE-e4-2 (5′-GTAATCGCTGCCTCTAGCAATTTG-3′). For the preparation of probe B, the 1.3 kb fragment that resides in exons 11–12, was amplified by PCR with the primers CSE-e11-3 (5′-TGTCACTTGCTTGTCAACACTG-3′) and CSE-rev-2 (5′-CAGAACAACCTGTTAGTTAGAAGA-3′) and subcloned into the pCR-TOPO vector (Invitrogen). The 406 bp EcoRI–XhoI fragment neighbouring exon 12 was excised as the probe B. Two overlapping clones (λCSE-1 and λCSE-2) and a single clone (λCSE-3) were isolated after the tertiary screening with probes A and B respectively. The λ phage DNA was prepared with QIAGEN λ system (Qiagen) and sequenced.
CSE promoter analyses
A 3.5 kb genomic DNA fragment upstream of the transcriptional start site (−3498 to+18) was isolated by PCR using λCSE-1 as a template with the two primers CSE-pro-1 (5′-ATGGGTACCACTTAGCATAATACTTAGAC-3′) and CSE-pro-rev (5′-ATGCCTCGAGGTGTTGCTTTGGCTAA-3′). The PCR product was digested with KpnI and XhoI, subcloned into the promoterless pGL3-Enhancer vector (Promega) that contains a firefly luciferase gene driven by the promoter activity of inserted sequences, and sequenced for confirmation, producing the pGL3-CSE-pro-1 vector. This vector was used as the PCR template to generate pGL3-CSE-pro-2–22 vectors that contain different lengths within the −3498 bp sequences. Reporter gene assay was conducted in transiently transfected Cos-7 and HEK-293 cells (human embryonic kidney 293 cells) that were maintained in phenol red-free Dulbecco's modified Eagle's medium supplemented with 10% (v/v) heat-inactivated foetal bovine serum (Hyclone Laboratories, Logan, UT, U.S.A.) and antibiotics. At 24 h before the transfection, 2×104 (for Cos-7) or 4×104 (for HEK-293) cells were seeded into each well of the ViewPlate-96 White (Packard, Meriden, CT, U.S.A.). On the day of transfection, 0.175 μg (12.6 nmol) of the pRL-TK vector (Promega) that contains a Renilla luciferase gene driven by the herpes simplex virus thymidine kinase promoter and 0.852 nmol (equivalent to 0.025 μg of the pGL3-CSE-pro-1 vector) of the pGL3-Enhancer (or pGL3-CSE-pro-1–22 vectors) were combined, and then the DNA mixture was incubated with LIPOFECTAMINE™ 2000 (Invitrogen) at the ratios of 2 μl of LIPOFECTAMINE™ 2000/1 μg of DNA. Transfection was performed according to the manufacturer's instructions and the transfected cells were assayed for both firefly and Renilla luciferase activities after 48 h of incubation. The luminescence was measured using the FireLite Dual Luminescence Reporter Gene Assay System (PerkinElmer) with the Fusion Universal Multiplate Analyzer (PerkinElmer). Both the (firefly and Renilla) luciferase activities were measured and the promoter activity was expressed as multiples of induction relative to the activity (the ratio between firefly and Renilla luciferase activities) when promoterless pGL3-Enhancer was transfected.
The TFSearch program (established by Y. Akiyama, Real World Computing Partnership, Japan) was used to search the Transfac 4.0 database [28] for transcriptional factor-binding sites within the sequences. The transactivating factors with threshold scores over 88.0 were considered to be important for the transcriptional regulation.
Northern-blot analyses
Total RNA was isolated from various tissues of an 8-week-old rat or mouse with TRIzol® and 10 μg of each was separated on 6% (w/v) formaldehyde/1% agarose gels. After transferring on to the Hybond-XL nylon membrane (Amersham Biosciences), hybridization was performed as described previously [29]. The entire ORFs of rat [12] and mouse CSE cDNAs were used as the specific probes against rat and mouse CSE respectively. The probes were radiolabelled with [α-32P]dCTP (Amersham Biosciences) and the hybridized blots were scanned with the Bio-Imaging Analyzer Bas2500 (Fuji Photo Film, Tokyo, Japan). As loading controls, 18 S rRNA was stained with ethidium bromide.
Anti-CSE polyclonal antibody production
Two different anti-rat CSE rabbit polyclonal antibodies were constructed against the N- (amino acid numbers 1–193) or C-terminal (amino acid numbers 194–398) portions of the recombinant rat CSE protein consisting of 398 amino acids. The N-terminal coding sequence was obtained by PCR using the entire rat CSE cDNA as a template with the primers rCSE-BamHI-Nter1 (5′-ATGCGGATCCGCAGGAAGGACGCCTCCTCCAGC-3′) and rCSE-XhoI-Cter1 (5′-ATGCCTCGAGATATGCAGACATGAAAGTGTT-3′), digested with BamHI and XhoI, and subcloned into the pET-21b(+) vector (Novagen), producing the pET-21b-rCSE-Nter vector. The C-terminal coding sequence was also isolated using the same template by PCR with the primers rCSE-BamHI-Nter2 (5′-ATGCGGATCCGTTCCAGAGACCTTTGGCTCTG-3′) and rCSE-XhoI-Cter2 (5′-ATGCCTCGAGAGGGTGAGATGCCTTTAAAGC-3′), digested with BamHI and XhoI, and subcloned into the same vector, producing the pET-21b-rCSE-Cter vector. Each expression vector was transformed into the BL21(DE3) Escherichia coli strain (Novagen) and the recombinant protein was overexpressed in isopropyl β-D-galactopyranoside-treated bacterial culture. Both recombinant proteins contain the His Tag (Novagen) at their N-termini, and were affinity-purified with the TALON Metal Affinity Resins (ClonTech) under guanidine-denaturing conditions according to the manufacturer's instructions. Rabbits were immunized with the purified proteins three times and the anti-serum was prepared by conventional procedures. Anti-CSE N-terminal serum was used for Western blotting. Anti-CSE C-terminal serum was further purified with HiTrap Protein G HP columns (Amersham Biosciences), and the IgG fraction was used for immunohistochemistry.
Western-blot analyses
Tissues were quickly removed from an 8-week-old SD rat or C57BL/6J mice of different ages, homogenized with a Teflon tissue grinder in ice-cold buffer (100 mM sodium phosphate, pH 7.8/1 mM PMSF), and sonicated with the Sonifier 450 (Branson Ultrasonics, Danbury, CT, U.S.A.). The homogenates were centrifuged at 10900 g for 5 min at 4 °C and the supernatants were further centrifuged at 17400 g for 20 min at 4 °C. The resulting supernatants were quickly frozen in liquid nitrogen and stored at −80 °C until use. Tissue samples (5 μg) or transfected Cos-7 samples (2 μg) were solubilized in the SDS-sample buffer, boiled for 5 min, separated on a 10% SDS/polyacrylamide gel, and transferred on to the Immobilon PVDF transfer membrane (0.45 μm, Millipore). The CSE protein was detected with anti-CSE N-terminal serum (1:3000 dilution), horseradish-peroxidase-conjugated anti-rabbit IgG antibody and the ECL® Western-blotting system (Amersham Biosciences).
CSE activity measurement
CSE activity was determined by a sensitive method recently reported by Ogasawara et al. [30] with minor modifications; DL-propargylglycine (final 1 mM), instead of 4,4′-dithiodipyridine (final 3 mM), was used to inactivate CSE. This method utilizes colorimetry for the determination of pyruvate produced from β-chloro-L-alanine by a CSE-catalysed β-elimination reaction, coupling a colour enzymic reaction with pyruvate oxidase and peroxidase [30]. Briefly, CSE catalyses the pyruvate formation from β-chloro-L-alanine. This reaction is terminated by the addition of DL-propargylglycine. The produced pyruvate is oxidized by pyruvate oxidase in the presence of thiamine pyrophosphate and bivalent magnesium to liberate H2O2. A leuco dye, N-(carboxymethylamino)-4,4′-bis(dimethylamino)-diphenylamine, is oxidized by H2O2 with peroxidase to produce Bindschedler's Green. The reaction was performed in 96-well dishes, and the absorbance of green dye (727 nm) was measured by a Bio-Rad Benchmark Plus microplate reader. The sample blank was similarly prepared, except that the sample was added to the substrate mixture after the addition of propargylglycine. The CSE-specific activity was expressed as the balance (between sample and sample blank) of absorbance at 727 nm/μg of protein.
Immunohistochemistry
Embryonic day 15.5 (E15.5) embryos and 8-week-old mice were analysed for CSE expression by immunohistochemistry using anti-CSE C-terminal antibody. The pregnant mice were euthanized by ether and embryos were quickly removed and fixed with 4% (w/v) PFA (paraformaldehyde) in 0.1 M PBS (pH 7.3). The adult (non-pregnant) female mice were anaesthetized with pentobarbital (50 μg/g of body weight), and perfused through the heart with PBS followed by 4% PFA in PBS. Each tissue (liver, kidney and spleen) was dissected out and postfixed overnight in 4% PFA in PBS. Embryos or adult tissues were then embedded in 30% sucrose in PBS. After sinking, they were embedded in Optimal Cutting Temperature compound (TISSUE-TEK; Sakura Finetechnical, Tokyo, Japan), frozen and sectioned in a cryostat at 10 μm. After washing with PBS, the sections were blocked with Blocking Reagent (Roche Diagnostics) in PBS, and then incubated with anti-CSE C-terminal antibody (IgG fraction; diluted 1:100) or pre-immune serum in the blocking solution. After incubation with the primary antibody, the sections were washed with PBS and reacted with the VECTASTAIN ABC peroxidase kit for rabbit antibodies (Vector Laboratories, Burlingame, CA, U.S.A.) followed by the chromogen diaminobenzidine.
RESULTS
Cloning and sequencing of the full-length mouse CSE cDNA
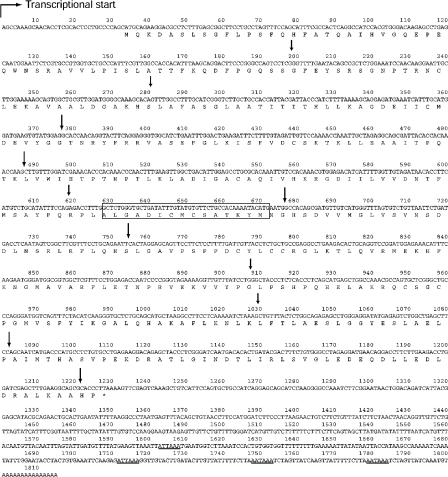
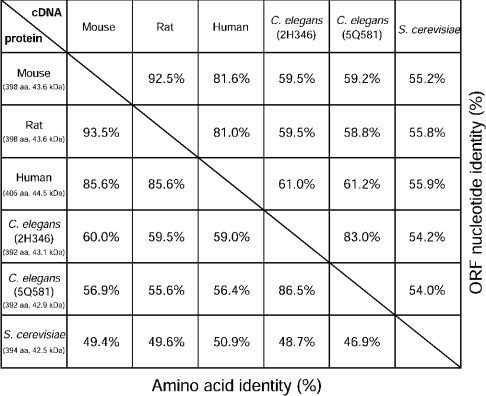
It was previously shown that the CSE gene is highly expressed in adult mouse kidney and liver [31]. Therefore we have cloned the full-length CSE cDNA from adult mouse kidney using PCR and RACE. The 1815 bp of the entire mouse CSE transcript consists of 33 bp of 5′-UTR, 1197 bp of ORF and 585 bp of 3′-UTR, of which 15 nt are poly(A)+ (polyadenylated) (Figure 1). This was the first report for mouse CSE cDNA cloning and the sequence was deposited with GenBank®, accession number AY083352. The nucleotide sequence of the mouse CSE ORF is 92.5% identical with the rat sequence and was 81.6% identical with the human sequence (Figure 2). CSE is one of the fundamental enzymes for methionine catabolism and is evolutionarily conserved from yeast to mammals (Figure 2). Possible gene duplication was reported in Caenorhabditis elegans but not in yeast or mammals (Figure 2). The mouse ORF encodes a 43.6 kDa protein, which shares high amino acid identity with those of the rat and human CSE (93.5 and 85.6% respectively) [7,32]. A putative PLP-binding site with an active lysine (Lys211) [33] is located in the middle of the ORF (Figure 1), which is fully conserved with rat CSE. Four polyadenylation consensus signal sequences (ATTAAA or AATAAA) are found in the 3′-UTR.
Figure 1. Full-length cDNA and predicted amino acid sequences of the mouse kidney CSE.
The transcriptional start site shown by a broken arrow is set at 1. The 1815 bp sequence contains a 1197 bp ORF that extends over bases 34–1230 (*, a stop codon). All the 11 exon/intron boundaries are indicated by arrows. A putative PLP-binding site and poly(A)+ consensus signal sequences (ATTAAA or AATAAA) are boxed and underlined respectively.
Figure 2. Sequence identity of CSE.
Table of percentage ORF nucleotide identity (in the upper right half) or percentage amino acid identity (in the lower left half) of CSE sequences from mouse, rat, human, C. elegans (2H346 and 5Q581) and Saccharomyces cerevisiae (GenBank® accession numbers AY083352, AB052882, NM_001902, NM_063048, NM_074652 and D14135 respectively). The number of amino acids (aa) in ORFs and estimated molecular masses (kDa) are shown.
Cloning and sequencing of the mouse CSE gene
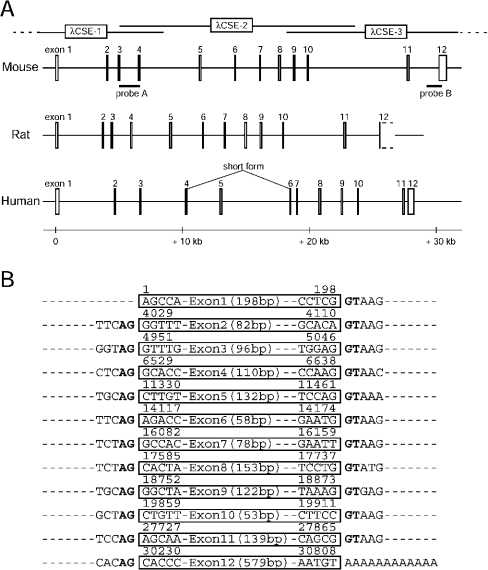
BLAST searches of the NCBI (National Center for Biotechnology Information) GenBank® for human CSE gene with the human full-length cDNA (GenBank® accession number NM_153742) revealed a 12-exon structure (Figure 3A). The searches for the rat CSE gene with rat cDNA that lacks a part of 3′-UTR (GenBank® accession number AB052882), also suggested a structure with 12 exons (Figure 3A). A similar 12-exon structure with conserved exon/intron boundaries was speculated for the mouse CSE organization. Thus two DNA fragments were amplified by PCR using the R1 cell [established from a (129/Sv×129/J)F1 origin] genomic DNA as a template and the possible exon-spanning primer sets, and two screening probes, A and B, were prepared. Southern-blot analyses using these two probes and R1 genomic DNA digested with several restriction enzymes suggested the existence of a multiexon, single copy gene for the mouse CSE (results not shown). By screening 5×105 independent plaques from a 129/SvJ mouse λ genomic library, we obtained three overlapping clones (λCSE-1–3) that cover the complete CSE gene (Figure 3A).
Figure 3. Structure of the mouse CSE gene.
(A) Schematic representation of mammalian (mouse, rat and human) CSE genes. Organization of mouse CSE gene was revealed by sequencing three overlapping λ phage clones (λCSE-1–3) that were isolated by screening the mouse genomic library with probes A and B. Sequences of rat and human CSE genes were obtained from the NCBI database. All three mammalian CSE genes consist of 12 exons with conserved exon/intron boundaries. A splice variant that skips exon 5 was reported in human liver. (B) Exon/intron boundaries in the mouse CSE gene. Exons (53–579 bp) are boxed with nucleotide numbers above when the transcriptional start site is set at 1. Invariant AG/GT sequences (splicing consensuses) are shown in bold.
Sequencing of the three clones localized 12 exons ranging in size from 53 to 579 bp that span over 30 kb in which CSE ORF is encoded within all 12 exons (Figures 1 and 3). A 35246 bp 129/SvJ mouse CSE genomic sequence, including a 4 kb sequence upstream of the transcriptional start site, 12 exons and 11 introns, was deposited with GenBank®, accession number AY262829. It contains exon/intron boundaries conserved with rat and human CSE genes that follow the GT/AG rule for intron donor/acceptor sites (Figure 3B). Genomic organization of mouse CSE gene (such as distances between exons) resembles that of the rat CSE rather than that of the human CSE (Figure 3A), reflecting the evolutionary distances. The 35246 bp sequence of the 129/SvJ origin is 99.8% identical with that of the C57BL/6J mouse gene that was recently filed in the GenBank® database. The full-length CSE cDNA sequence isolated from C57BL/6J kidney (Figure 1) is fully compatible with this 129/SvJ mouse genomic sequence.
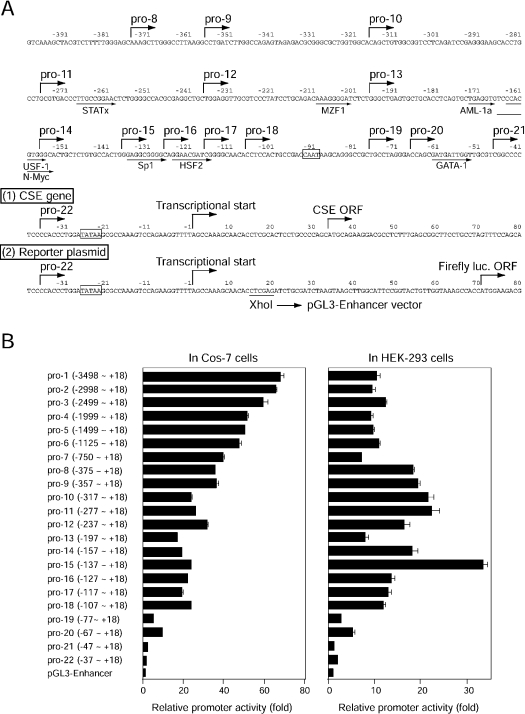
Core regions of the mouse CSE promoter
To identify important regulatory regions for CSE gene expression, deletion mutants of the 5′-flanking regions, fused to firefly luciferase reporter gene, were generated (Figure 4). The 3.5 kb genomic DNA fragment upstream from the transcriptional start site (−3498 to +18), containing canonical TATA and CAAT boxes (Figure 4A), was isolated by PCR from λCSE-1 and used as a template for further mutant constructions. A much larger amount (14.8-fold in mol) of the plasmid containing the thymidine kinase promoter-driven Renilla luciferase gene was co-transfected with the deletion constructs for the normalization of transfection efficiency into two kidney cell lines, Cos-7 and HEK-293 cells. In transfected Cos-7 cells, increasing the length of the 5′-flanking sequence up to −3498 bp enhanced the basal promoter activity with a maximum of 67-fold enhancement (Figure 4B, left panel). The −357 bp GC-rich (G, 33.7%; C, 30.1%) construct (referred to as pro-9) displayed 52.4% of the activity obtained with the −3498 bp construct (pro-1). In contrast, the −137 bp sequence conferred the highest promoter activity (33-fold enhancement) in HEK-293 cells (Figure 4B, right panel). The core regulatory regions also existed in the proximal regions (−357 to +18) in transfected HEK-293 cells (Figure 4B, right panel). The differences between the two cell lines regarding the availability of transcriptional factors may reflect the altered patterns in the deletion analyses.
Figure 4. Promoter activity of the mouse CSE gene.
(A) Nucleotide sequence of the 5′-flanking region of the mouse CSE gene. Numbering is relative to the transcriptional start site. Transcriptional start sites, translational start sites in the CSE gene (1) and the reporter plasmid (2), and the 5′-ends of the deletion constructs, are all shown by broken arrows. Putative transcriptional-factor-binding sites are indicated by arrows below the sequences. The consensus TATAA and CAAT motifs are boxed. (B) Defining core regions of the mouse CSE promoter by deletion analyses. Serial deletion constructs were inserted upstream of the firefly luciferase gene in the pGL3-Enhancer reporter plasmid. Each deletion construct was transiently transfected into Cos-7 cells (left panel) or HEK-293 cells (right panel), together with larger amounts of pRL-TK plasmid containing the Renilla luciferase gene. Both (firefly and Renilla) luciferase activities were measured 2 days later, and the promoter activity was expressed as multiples of induction relative to the activity (the ratio between firefly and Renilla luciferase activities) when promoterless pGL3-Enhancer plasmid was transfected. Results shown are means±S.E.M. for 16 samples from four experiments.
The database search for the transcriptional factor-binding consensus identified STATx (signal transducers and activators of transcription x), MZF1 (myeloid zinc finger protein 1), AML-1a (acute myeloid leukaemia-1a), USF-1 (upstream stimulatory factor-1), N-Myc, Sp1 (specificity protein 1), HSF2 (heat shock factor 2) and GATA-1 (GATA-binding factor 1) as important transactivating factors in the −357 bp sequence (Figure 4A). Deletion of MZF-1 (in both cells) or Sp1 consensus (in HEK-293 cells) significantly decreased the promoter activity (Figure 4B), suggesting the involvement of these factors in the basal transcriptional activity. In contrast, removal of AML-1a, USF-1, or N-Myc consensus sequences increased the transcriptional activity in HEK-293 cells, suggesting a role for these factors as repressive elements. In both cells, deletion of the CAAX motif or GATA-1 consensus significantly reduced the basal promoter activity. Various cell activators, including PMA (100 nM), dibutyryl cAMP (1 mM), dexamethasone (100 nM) or glucagon (100 ng/ml) [34], were tested to see whether they could regulate the CSE transcription in pGL3-CSE-pro-1 vector-transfected Cos-7 cells, but none of the tested reagents affected the transcriptional activity (results not shown).
Distribution of the CSE transcript, protein and enzymic activity in rat and mouse tissues
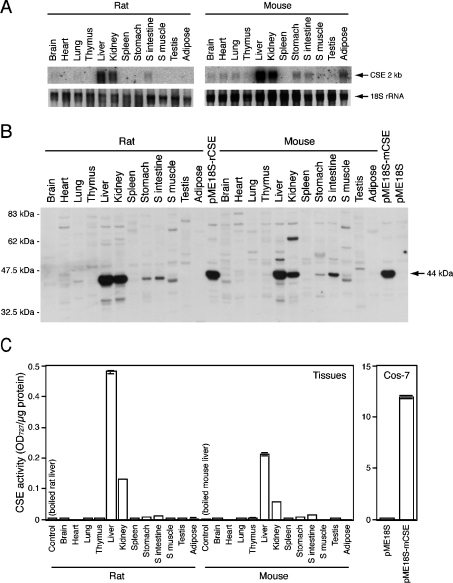
To determine CSE gene expression in rat and mouse tissues, total RNA isolated from 12 different adult tissues was analysed by Northern blot, using rat and mouse CSE cDNA fragments as probes (Figure 5A). The 2 kb single transcripts were observed in rat liver and kidney, and at a much lower level, in small intestine. In mouse, the 2 kb transcripts were found mainly in liver and kidney, and in lower abundance in adipose tissue, stomach and small intestine. A faint expression of the 2 kb single transcripts was observed in mouse brain, heart and lung. This transcript size is in good agreement with that predicted from the full-length cDNA (Figure 1) by adding approx. 200 bp poly(A)+, suggesting the existence of the CSE single transcript with no splice variant in those mouse tissues. Next, expression of the CSE protein was investigated using anti-CSE N-terminal antibody developed in this study. This antibody detected the approx. 44 kDa protein in Cos-7 cells transfected with rat and mouse CSE expression plasmids (pME18S-rCSE and pME18S-mCSE respectively), but not in control vector (pME18S)-transfected cells (Figure 5B). Thus it recognized both recombinant rat and mouse CSE. In both rodents, major 44 kDa proteins were detected mainly in liver and kidney, with much reduced levels in small intestine and stomach (Figure 5B). Finally, tissue distribution of the CSE activity was examined with a sensitive method utilizing colorimetric reaction. No significant activity was detected in control vector-transfected Cos-7 cells but transient expression of mouse CSE cDNA conferred the CSE activity (Figure 5C, right panel). In both rats and mice, the highest levels of CSE activity were detected in liver followed by kidney although very low but significant levels of activity were detected in small intestine and stomach (Figure 5C, left panel). The activities in rat liver and kidney are approx. 2.5-fold higher than those in mouse liver and kidney respectively. Heat-denatured (boiled at 100 °C for 10 min) liver samples did not show any CSE activity (Figure 5C, left panel). The 2 kb transcript was not detected in rat stomach (Figure 5A); however, more sensitive reverse transcriptase–PCR revealed low expression of the CSE transcripts in this tissue (results not shown), which might explain the presence of CSE protein and activity in this tissue (Figures 5B and 5C).
Figure 5. Tissue distribution of the CSE transcript, protein and enzymic activity in rat and mouse.
(A) Distribution of the CSE transcript. Total RNA isolated from each tissue of an adult male rat or mouse (both 8 weeks old) was analysed by Northern blot (10 μg/lane), and approx. 2 kb single bands were detected. As a loading control, 18 S rRNA was stained with ethidium bromide. (B) Distribution of the CSE protein. Tissue extracts (5 μg/lane) or Cos-7 cell extracts (2 μg/lane) were fractioned on SDS/PAGE (10% gel). The 44 kDa proteins specific to CSE were detected by Western-blot analyses. (C) Distribution of CSE enzyme activity in tissue extracts. Boiled liver samples were loaded as negative controls or reaction backgrounds. Results shown are means±S.E.M. for triplicate samples from a representative experiment. Cell extracts were prepared from Cos-7 cells transfected with CSE expression vectors and used as positive controls in both Western-blot analyses (B) and enzyme assay (C). OD727=A727.
Developmental expression of CSE in mouse
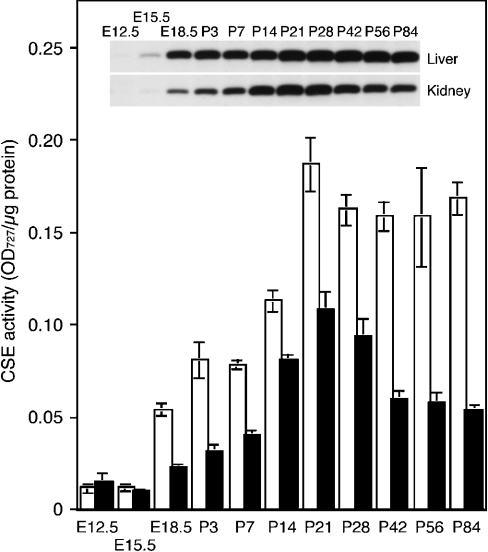
Expression of CSE protein and activity were examined in mouse liver and kidney at various developmental stages: E12.5, E15.5, E18.5, postnatal day 3 (P3), P7, P14, P21, P28, P42, P56 and P84 (Figure 6). Embryonic liver and kidney could be identified and removed as early as E12.5. Very low levels of the CSE activity were detected at E12.5 in both liver and kidney; however, the 44 kDa proteins specific to CSE were detected by Western-blot analyses only at very low levels (Figure 6). The expression of CSE protein became apparent at E15.5, although there was no simultaneous increase in CSE activity. After E18.5, the expression of CSE protein and activity significantly increased in both tissues, and thereafter, the CSE activity showed a good correlation with the protein expression level. The CSE expression in liver gradually increased with age until P21, the weaning age, and thereafter remained constant at least by P84. In contrast, the CSE expression in kidney gradually increased with age until reaching its peak at P21 but then decreased to approx. 50% of the peak level at P21.
Figure 6. Developmental expression of the CSE protein and enzyme activity in mouse liver (□) and kidney (▪).
Cell extracts prepared from both tissues at various developmental stages (E12.5–P84) were analysed for CSE protein expression by Western blotting (inset) and CSE enzyme activity (histogram). Bars shown represent means±S.E.M. for samples from three to four mice. For E12.5 liver and kidney, samples from three to four embryos were pooled because of their small yields. OD727=A727.
CSE immunostaining of mouse liver and kidney
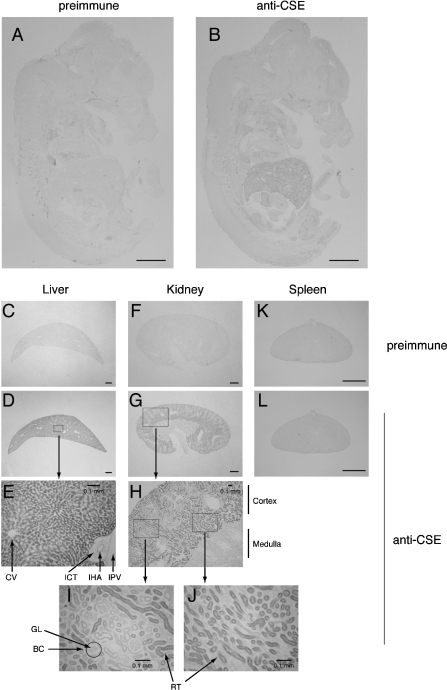
Cryosections from E15.5 embryos and adult tissues (liver, kidney and spleen) were stained with anti-CSE C-terminal antibody using pre-immune serum as negative control (Figures 7A, 7C, 7F and 7K). Immunostaining of sagittal sections from an E15.5 embryo detected weak but significant levels of CSE expression in liver (Figure 7B), which contrasts with the negative control (Figure 7A). The CSE expression was much higher in adult liver and kidney (Figures 7D, 7E and 7G–7J). The liver mainly consists of hepatocytes that were highly stained, whereas interlobular connective tissues surrounding interlobular hepatic artery were not stained (Figure 7E). In adult kidney, the CSE protein was expressed in cortex rather than in medulla (Figures 7G and 7H), especially in cortical renal tubules of the inner cortex (Figures 7I and 7J). CSE was absent in renal corpuscles that consist of glomeruli and Bowman's capsules (Figure 7I) or in adult spleen (Figure 7L).
Figure 7. Expression of the CSE protein in E15.5 embryos and adult mouse tissues: liver, kidney and spleen.
Midsagittal sections of E15.5 embryos (A, B), 8-week-old male liver (C–E), kidney (F–J) and spleen (K, L). Sections were stained with anti-CSE N-terminal antibody (B, D, E, G–J, L) or with pre-immune serum as negative controls (A, C, F, K). BC, Bowman's capsule; CV, central vein; GL, glomerulus; IHA, interlobular hepatic artery; ICT, interlobular connective tissue; IPV, interlobular portal vein; RT, renal tubules. Scale bars, 1 mm (and 0.1 mm in E, H, I and J).
DISCUSSION
Mammalian cells are capable of synthesizing cysteine from methionine by the trans-sulphuration pathway, giving the basis for the nutritional concept that cysteine is one of the non-essential amino acids among sulphur-containing amino acids. However, several investigators have suggested that infants, especially premature infants, may require the dietary supplement of cysteine because of lower CSE activity in infants when compared with that in adults [10,11,35,36]; cysteine deficiency might contribute to the low survival rates for extremely premature infants of very low birth weight. This had led to concerns about the use of cow's milk protein in infant formulas, which has a lower ratio of cysteine to methionine when compared with human milk [11]. In addition, CSE activity could be impaired in certain clinical conditions such as hepatic cirrhosis [37,38] and sepsis [39] or under surgical stress [40]. Therefore cysteine could be considered as one of the conditionally essential amino acids [41]. In the course of generating genetically engineered CSE-deficient mice to reveal in vivo roles of CSE, this is the first study in mouse of CSE cDNA and gene cloning, identification of the core promoter regions, distribution of the transcript, protein and enzymic activity within the 12 major organs, developmental expression in liver and kidney and the immunohistochemistry on entire embryos, liver and kidney sections.
The 1.8 kb full-length mouse kidney CSE cDNA contains a 1197 bp ORF encoding a 43.6 kDa protein and includes PLP-binding consensus sequences (Figure 1). The CSE ORF is encoded by all the 12 exons of the single CSE gene expanding over 30 kb (Figures 1 and 3A). It shares high homology (81–92% identity) with rat and human CSE ORF nucleotide sequences (Figure 2) and the exon/intron boundaries are all conserved within those three mammalian species (Figure 3B), suggesting an identical evolutionary origin. The mammalian CSE genes and proteins share substantial identity (47–61%) with those of C. elegans and yeast (Figure 2), indicating the evolutionary conservation of this enzyme.
The core promoter existed in the 5′-flanking region proximal to the transcriptional start site (−357 and −137 bp in Cos-7 and HEK-293 cells respectively) that contains several putative transcriptional factor-binding sites (Figure 4), although the regulation of basal activity was cell type specific. MZF-1 and Sp1 seem to play major roles in the basal transcriptional activity. Regulatory mechanisms for CSE transcription remain to be determined. Interestingly, promoter analyses of the human CBS (cystathionine β-synthase) gene revealed that the binding of MZF-1, Sp1 and USF-1 to their consensus sequences within the CBS core promoter was important for the transcriptional activity [42–44]. CBS is another PLP-dependent key enzyme of the transsulphuration pathway, which is located just upstream of CSE and catalyses the condensation of homocysteine and serine to form cystathionine. Since CBS is also highly and mainly expressed in mouse liver and kidney (results not shown), the transcription of these two transsulphuration genes might be co-regulated by the similar sets of transcriptional factors in vivo if the CBS promoter sequences in human and mouse are similar.
In adult rat and mouse tissues, the CSE transcript was detected mainly in liver followed by kidney as the 2 kb single band (Figure 5A). Occurrence of the splice variant with altered functions (the short form that skips exon 5; Figure 3A) has been reported in human liver [32,45], but such shorter transcripts were not observed in mouse tissues, as described previously [46]. Recent crystallographic analyses of the yeast CSE [47] suggest that the putative PLP-binding sites (boxed in Figure 1) are essential for the formation of active CSE tetramer, and the deletion of exon 5 coding sequences (next to the PLP-binding consensus) in human CSE may produce the loss-of-function variant [45]. We also did not detect the 4.3 kb band that Nishi et al. [12] observed in rat liver samples.
Two sets of anti-rat CSE polyclonal antibodies were generated and utilized for the Western-blot analyses (Figures 5B and 6) and immunohistochemistry (Figure 7). The anti-rat CSE N-terminal antibody specifically recognized the recombinant 44 kDa rat and mouse CSE in CSE expression vector-transfected Cos-7 cells but not in control cells, and detected the major 44 kDa proteins in several rat and mouse tissue extracts (Figure 5B). The estimated molecular masses of rat and mouse CSE are both 43.6 kDa (Figure 2), and match with the observed band mass in this study (Figure 5B) and another [48] but not with the one (approx. 40 kDa) detected by other groups [12,49]. The reason for this is unknown but anti-rat CSE C-terminal antibody also detected the 44 kDa proteins as major bands in liver and kidney of both rats and mice (results not shown). A faint level of expression of the CSE transcript, protein and enzymic activity was observed in the small intestine and stomach of both rodents. Previous studies have shown that CSE is expressed in rat vascular smooth-muscle cells [7] and, thus, the CSE gene may be expressed in tissues of smooth-muscle cell lineages. Hydrogen sulphide produced by CSE from cyst(e)ine [1–3] may regulate the relaxation of the smooth-muscle cells [5–7]. The 2 kb transcripts were also detected in rat brain, mouse brain, heart, lung and adipose tissue, all of which were not accompanied by expression of CSE protein and activity (Figure 5). Whether or not they are the CSE transcripts is unknown, although some previous reports suggested low levels of CSE expression in rat brain [13,14].
Developmental expression of CSE in liver was previously evaluated in human [10,45] and rat [14] but not in mouse. In rat liver, the CSE activity was detected at very low levels around E16, which was followed by gradual increases until it peaked just after birth [14]. Thereafter, it remained constant in adulthood [14]. In contrast, the CSE activity in mouse liver increased significantly after birth to its peak at P21, and thereafter remained constant in adulthood (Figure 6); the hepatic CSE activity at P21 was 3.4-fold higher than that at E18.5 in mice (Figure 6), but only 1.4-fold in rats [14]. In mouse kidney, the CSE activity increased until the peak at P21 and then decreased to half of the peak level in adulthood (Figure 6). Considering that the hepatic and kidney CSE activities in adult rats were both 2.5-fold higher than those in adult mice (Figure 5C), CSE may play more important roles in mice than in rats during the early postnatal period. A low level of the CSE activity was detected at E12.5 in liver and kidney, which was not accompanied by CSE protein expression (Figure 6), but the reason for this is unclear. Except for CSE, propargylglycine has been shown to inhibit some enzymes such as aspartate and alanine aminotransferases [3]. Whether or not these enzymes could metabolize β-chloro-L-alanine to pyruvate in our assay system is unknown. Interestingly, the hepatic rat CSE activity increased during lactation with the peak at 2 weeks after delivery [49] and the inhibition of CSE by propargylglycine administration was followed by the significant decrease in lactation associated with apoptosis of lactating mammary gland [50,51]. Although it is still unknown whether hepatic CSE activity is also high in lactating mice, the adequate supply of cysteine through lactation might be more important for the newborn of mice than for those of rats.
Finally, immunohistochemistry revealed enriched CSE expression in liver as early as E15.5 (Figure 7B), which is consistent with our Western-blot results (Figure 6). The CSE expression was much more evident in adult liver (Figures 7D and 7E). The CSE in adult kidney was localized to the cortex rather than medulla, especially to the renal tubule in the inner cortex. This is generally consistent with the previous results obtained for rats by House et al. [52]; they fractionated rat kidney and detected an enriched distribution of the CSE activity in the inner cortex or cortical tubule fractions. Kidney is a major locus for the removal and subsequent metabolism of plasma homocysteine that is an intermediate amino acid in the transsulphuration as well as an endogenous substrate of CBS. An increased level of plasma homocysteine is a potential risk factor for cardiovascular diseases such as atherosclerosis and thrombosis. The CSE, in combination with the CBS (that is localized to the outer cortex [52]), may influence the renal clearance of homocysteine.
In conclusion, we have cloned and characterized mouse CSE cDNA and gene, generated specific anti-CSE antibodies and examined its tissue distribution and developmental expression in mice. This study has demonstrated that (i) CSE is a fundamental enzyme conserved through evolution; (ii) CSE core promoter is located within the 5′-flanking region proximal (−357 bp) to the transcriptional start site; (iii) CSE is expressed mainly in liver and kidney, and, at much lower levels, in small intestine and stomach; (iv) CSE expression in liver and kidney is developmentally regulated; and (v) CSE is highly expressed in adult hepatocytes and kidney cortical tubuli. Such information is essential in understanding physiological roles of CSE/transsulphuration, and also provides useful information to generate and analyse CSE-deficient mice as an animal model of cystathioninaemia.
Acknowledgments
We thank K. Saito, E. Yoshida, M. Kawabata, and other animal care staff at the Division of Laboratory Animal Resources, NCNP. This work was supported in part by a grant-in-aid for Young Scientists (No. 15790066) from the Ministry of Education, Culture, Sports, Science and Technology (Japan), research grants from Uehara Memorial Foundation, Yamanouchi Foundation for Research on Metabolic Disorders, ONO Medical Research Foundation, Naito Foundation, Tokyo Biochemical Research Foundation (to I.I.) and National Institute of Neuroscience (NCNP, Japan to H.K.).
References
- 1.Stipanuk M. H., Beck P. W. Characterization of the enzymic capacity for cysteine desulphydration in liver and kidney of the rat. Biochem. J. 1982;206:267–277. doi: 10.1042/bj2060267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamanishi T., Tuboi S. The mechanism of the L-cystine cleavage reaction catalyzed by rat liver γ-cystathionase. J. Biochem. (Tokyo) 1981;89:1913–1921. doi: 10.1093/oxfordjournals.jbchem.a133393. [DOI] [PubMed] [Google Scholar]
- 3.Drake M. R., De La Rosa J., Stipanuk M. H. Metabolism of cysteine in rat hepatocytes. Evidence for cysteinesulphinate-independent pathways. Biochem. J. 1987;244:279–286. doi: 10.1042/bj2440279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abe K., Kimura H. The possible role of hydrogen sulfide as an endogenous neuromodulator. J. Neurosci. 1996;16:1066–1071. doi: 10.1523/JNEUROSCI.16-03-01066.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hosoki R., Matsuki N., Kimura H. The possible role of hydrogen sulfide as an endogenous smooth muscle relaxant in synergy with nitric oxide. Biochem. Biophys. Res. Commun. 1997;237:527–531. doi: 10.1006/bbrc.1997.6878. [DOI] [PubMed] [Google Scholar]
- 6.Teague B., Asiedu S., Moore P. K. The smooth muscle relaxant effect of hydrogen sulphide in vitro: evidence for a physiological role to control intestinal contractility. Br. J. Pharmacol. 2002;137:139–145. doi: 10.1038/sj.bjp.0704858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao W., Zhang J., Lu Y., Wang R. The vasorelaxant effect of H(2)S as a novel endogenous gaseous K(ATP) channel opener. EMBO J. 2001;20:6008–6016. doi: 10.1093/emboj/20.21.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vargas J. E., Mudd S. H., Waisbren S. E., Levy H. L. Maternal γ-cystathionase deficiency: absence of both teratogenic effects and pregnancy complications. Am. J. Obstet. Gynecol. 1999;181:753–755. doi: 10.1016/s0002-9378(99)70525-9. [DOI] [PubMed] [Google Scholar]
- 9.Wang J., Hegele R. A. Genomic basis of cystathioninuria (MIM 219500) revealed by multiple mutations in cystathionine γ-lyase (CTH) Hum. Genet. 2003;112:404–408. doi: 10.1007/s00439-003-0906-8. [DOI] [PubMed] [Google Scholar]
- 10.Zlotkin S. H., Anderson G. H. The development of cystathionase activity during the first year of life. Pediatr. Res. 1982;16:65–68. doi: 10.1203/00006450-198201001-00013. [DOI] [PubMed] [Google Scholar]
- 11.Rao A. M., Drake M. R., Stipanuk M. H. Role of the transsulfuration pathway and of γ-cystathionase activity in the formation of cysteine and sulfate from methionine in rat hepatocytes. J. Nutr. 1990;120:837–845. doi: 10.1093/jn/120.8.837. [DOI] [PubMed] [Google Scholar]
- 12.Nishi N., Tanabe H., Oya H., Urushihara M., Miyanaka H., Wada F. Identification of probasin-related antigen as cystathionine γ-lyase by molecular cloning. J. Biol. Chem. 1994;269:1015–1019. [PubMed] [Google Scholar]
- 13.Awata S., Nakayama K., Suzuki I., Sugahara K., Kodama H. Changes in cystathionine γ-lyase in various regions of rat brain during development. Biochem. Mol. Biol. Int. 1995;35:1331–1338. [PubMed] [Google Scholar]
- 14.Heinonen K. Studies on cystathionase activity in rat liver and brain during development. Effects of hormones and amino acids in vivo. Biochem. J. 1973;136:1011–1015. doi: 10.1042/bj1361011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Washtien W., Abeles R. H. Mechanism of inactivation of γ-cystathionase by the acetylenic substrate analogue propargylglycine. Biochemistry. 1977;16:2485–2491. doi: 10.1021/bi00630a026. [DOI] [PubMed] [Google Scholar]
- 16.Kim S. K., Choi K. H., Kim Y. C. Effect of acute betaine administration on hepatic metabolism of S-amino acids in rats and mice. Biochem. Pharmacol. 2003;65:1565–1574. doi: 10.1016/s0006-2952(03)00115-1. [DOI] [PubMed] [Google Scholar]
- 17.Triguero A., Barber T., Garcia C., Puertes I. R., Sastre J., Vina J. R. Liver intracellular L-cysteine concentration is maintained after inhibition of the trans-sulfuration pathway by propargylglycine in rats. Br. J. Nutr. 1997;78:823–831. doi: 10.1079/bjn19970198. [DOI] [PubMed] [Google Scholar]
- 18.Kwon Y. H., Stipanuk M. H. Cysteine regulates expression of cysteine dioxygenase and γ-glutamylcysteine synthetase in cultured rat hepatocytes. Am. J. Physiol. Endocrinol. Metab. 2001;280:E804–E815. doi: 10.1152/ajpendo.2001.280.5.E804. [DOI] [PubMed] [Google Scholar]
- 19.Cresenzi C. L., Lee J. I., Stipanuk M. H. Cysteine is the metabolic signal responsible for dietary regulation of hepatic cysteine dioxygenase and glutamate cysteine ligase in intact rats. J. Nutr. 2003;133:2697–2702. doi: 10.1093/jn/133.9.2697. [DOI] [PubMed] [Google Scholar]
- 20.Mathisen G. A., Fonnum F., Paulsen R. E. Contributing mechanisms for cysteine excitotoxicity in cultured cerebellar granule cells. Neurochem. Res. 1996;21:293–298. doi: 10.1007/BF02531643. [DOI] [PubMed] [Google Scholar]
- 21.Puka-Sundvall M., Eriksson P., Nilsson M., Sandberg M., Lehmann A. Neurotoxicity of cysteine: interaction with glutamate. Brain Res. 1995;705:65–70. doi: 10.1016/0006-8993(95)01139-0. [DOI] [PubMed] [Google Scholar]
- 22.El-Khairy L., Vollset S. E., Refsum H., Ueland P. M. Plasma total cysteine, mortality, and cardiovascular disease hospitalizations: the Hordaland homocysteine study. Clin. Chem. 2003;49:895–900. doi: 10.1373/49.6.895. [DOI] [PubMed] [Google Scholar]
- 23.El-Khairy L., Vollset S. E., Refsum H., Ueland P. M. Plasma total cysteine, pregnancy complications, and adverse pregnancy outcomes: the Hordaland homocysteine study. Am. J. Clin. Nutr. 2003;77:467–472. doi: 10.1093/ajcn/77.2.467. [DOI] [PubMed] [Google Scholar]
- 24.El-Khairy L., Ueland P. M., Refsum H., Graham I. M., Vollset S. E. Plasma total cysteine as a risk factor for vascular disease: the European Concerted Action Project. Circulation. 2001;103:2544–2549. doi: 10.1161/01.cir.103.21.2544. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y. C., Kawagishi M., Mikayama T., Inagaki Y., Takeuchi T., Ohashi H. Processing of a fusion protein by endoprotease in COS-1 cells for secretion of mature peptide by using a chimeric expression vector. Proc. Natl. Acad. Sci. U.S.A. 1993;90:8957–8961. doi: 10.1073/pnas.90.19.8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishii I., Friedman B., Ye X., Kawamura S., McGiffert C., Contos J. J., Kingsbury M. A., Zhang G., Brown J. H., Chun J. Selective loss of sphingosine 1-phosphate signaling with no obvious phenotypic abnormality in mice lacking its G protein-coupled receptor, LP(B3)/EDG-3. J. Biol. Chem. 2001;276:33697–33704. doi: 10.1074/jbc.M104441200. [DOI] [PubMed] [Google Scholar]
- 27.Ishii I., Ye X., Friedman B., Kawamura S., Contos J. J., Kingsbury M. A., Yang A. H., Zhang G., Brown J. H., Chun J. Marked perinatal lethality and cellular signaling deficits in mice null for the two sphingosine 1-phosphate (S1P) receptors, S1P(2)/LP(B2)/EDG-5 and S1P(3)/LP(B3)/EDG-3. J. Biol. Chem. 2002;277:25152–25159. doi: 10.1074/jbc.M200137200. [DOI] [PubMed] [Google Scholar]
- 28.Heinemeyer T., Wingender E., Reuter I., Hermjakob H., Kel A. E., Kel O. V., Ignatieva E. V., Ananko E. A., Podkolodnaya O. A., Kolpakov F. A., et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishii I., Contos J. J., Fukushima N., Chun J. Functional comparisons of the lysophosphatidic acid receptors, LP(A1)/VZG-1/EDG-2, LP(A2)/EDG-4, and LP(A3)/EDG-7 in neuronal cell lines using a retrovirus expression system. Mol. Pharmacol. 2000;58:895–902. doi: 10.1124/mol.58.5.895. [DOI] [PubMed] [Google Scholar]
- 30.Ogasawara Y., Ishii K., Tanabe S. Enzymatic assay of γ-cystathionase activity using pyruvate oxidase–peroxidase sequential reaction. J. Biochem. Biophys. Methods. 2002;51:139–150. doi: 10.1016/s0165-022x(02)00010-6. [DOI] [PubMed] [Google Scholar]
- 31.Habib G. M., Shi Z. Z., Ou C. N., Kala G., Kala S. V., Lieberman M. W. Altered gene expression in the liver of γ-glutamyl transpeptidase-deficient mice. Hepatology. 2000;32:556–562. doi: 10.1053/jhep.2000.9715. [DOI] [PubMed] [Google Scholar]
- 32.Lu Y., O'Dowd B. F., Orrego H., Israel Y. Cloning and nucleotide sequence of human liver cDNA encoding for cystathionine γ-lyase. Biochem. Biophys. Res. Commun. 1992;189:749–758. doi: 10.1016/0006-291x(92)92265-y. [DOI] [PubMed] [Google Scholar]
- 33.Erickson P. F., Maxwell I. H., Su L. J., Baumann M., Glode L. M. Sequence of cDNA for rat cystathionine γ-lyase and comparison of deduced amino acid sequence with related Escherichia coli enzymes. Biochem. J. 1990;269:335–340. doi: 10.1042/bj2690335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinonen K., Raiha N. C. Induction of cystathionase in human foetal liver. Biochem. J. 1974;144:607–609. doi: 10.1042/bj1440607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sturman J. A., Gaull G., Raiha N. C. Absence of cystathionase in human fetal liver: is cystine essential? Science. 1970;169:74–76. doi: 10.1126/science.169.3940.74. [DOI] [PubMed] [Google Scholar]
- 36.Vina J., Vento M., Garcia-Sala F., Puertes I. R., Gasco E., Sastre J., Asensi M., Pallardo F. V. L-cysteine and glutathione metabolism are impaired in premature infants due to cystathionase deficiency. Am. J. Clin. Nutr. 1995;61:1067–1069. doi: 10.1093/ajcn/61.4.1067. [DOI] [PubMed] [Google Scholar]
- 37.Chipponi J. X., Bleier J. C., Santi M. T., Rudman D. Deficiencies of essential and conditionally essential nutrients. Am. J. Clin. Nutr. 1982;35:1112–1116. doi: 10.1093/ajcn/35.5.1112. [DOI] [PubMed] [Google Scholar]
- 38.Look M. P., Riezler R., Reichel C., Brensing K. A., Rockstroh J. K., Stabler S. P., Spengler U., Berthold H. K., Sauerbruch T. Is the increase in serum cystathionine levels in patients with liver cirrhosis a consequence of impaired homocysteine transsulfuration at the level of γ-cystathionase? Scand. J. Gastroenterol. 2000;35:866–872. doi: 10.1080/003655200750023255. [DOI] [PubMed] [Google Scholar]
- 39.Malmezat T., Breuille D., Pouyet C., Buffiere C., Denis P., Mirand P. P., Obled C. Methionine transsulfuration is increased during sepsis in rats. Am. J. Physiol. Endocrinol. Metab. 2000;279:E1391–E1397. doi: 10.1152/ajpendo.2000.279.6.E1391. [DOI] [PubMed] [Google Scholar]
- 40.Vina J., Gimenez A., Puertes I. R., Gasco E., Vina J. R. Impairment of cysteine synthesis from methionine in rats exposed to surgical stress. Br. J. Nutr. 1992;68:421–429. doi: 10.1079/bjn19920099. [DOI] [PubMed] [Google Scholar]
- 41.Laidlaw S. A., Kopple J. D. Newer concepts of the indispensable amino acids. Am. J. Clin. Nutr. 1987;46:593–605. doi: 10.1093/ajcn/46.4.593. [DOI] [PubMed] [Google Scholar]
- 42.Ge Y., Jensen T. L., Matherly L. H., Taub J. W. Synergistic regulation of human cystathionine-β-synthase-1b promoter by transcription factors NF-YA isoforms and Sp1. Biochim. Biophys. Acta. 2002;1579:73–80. doi: 10.1016/s0167-4781(02)00509-2. [DOI] [PubMed] [Google Scholar]
- 43.Ge Y., Matherly L. H., Taub J. W. Transcriptional regulation of cell-specific expression of the human cystathionine β-synthase gene by differential binding of Sp1/Sp3 to the -1b promoter. J. Biol. Chem. 2001;276:43570–43579. doi: 10.1074/jbc.M104930200. [DOI] [PubMed] [Google Scholar]
- 44.Ge Y., Konrad M. A., Matherly L. H., Taub J. W. Transcriptional regulation of the human cystathionine β-synthase-1b basal promoter: synergistic transactivation by transcription factors NF-Y and Sp1/Sp3. Biochem. J. 2001;357:97–105. doi: 10.1042/0264-6021:3570097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levonen A. L., Lapatto R., Saksela M., Raivio K. O. Human cystathionine γ-lyase: developmental and in vitro expression of two isoforms. Biochem. J. 2000;347:291–295. [PMC free article] [PubMed] [Google Scholar]
- 46.Steegborn C., Clausen T., Sondermann P., Jacob U., Worbs M., Marinkovic S., Huber R., Wahl M. C. Kinetics and inhibition of recombinant human cystathionine γ-lyase. Toward the rational control of transsulfuration. J. Biol. Chem. 1999;274:12675–12684. doi: 10.1074/jbc.274.18.12675. [DOI] [PubMed] [Google Scholar]
- 47.Messerschmidt A., Worbs M., Steegborn C., Wahl M. C., Huber R., Laber B., Clausen T. Determinants of enzymatic specificity in the Cys-Met-metabolism PLP-dependent enzymes family: crystal structure of cystathionine γ-lyase from yeast and intrafamiliar structure comparison. Biol. Chem. 2003;384:373–386. doi: 10.1515/BC.2003.043. [DOI] [PubMed] [Google Scholar]
- 48.Sato A., Nishioka M., Awata S., Nakayama K., Okada M., Horiuchi S., Okabe N., Sassa T., Oka T., Natori Y. Vitamin B6 deficiency accelerates metabolic turnover of cystathionase in rat liver. Arch. Biochem. Biophys. 1996;330:409–413. doi: 10.1006/abbi.1996.0269. [DOI] [PubMed] [Google Scholar]
- 49.Awata S., Nakayama K., Sato A., Kawamura M., Suzuki I., Kodama H. Changes in cystathionine γ-lyase levels in rat liver during lactation. Biochem. Mol. Biol. Int. 1993;31:185–191. [PubMed] [Google Scholar]
- 50.Barber T., Triguero A., Martinez-Lopez I., Torres L., Garcia C., Miralles V. J., Vina J. R. Elevated expression of liver γ-cystathionase is required for the maintenance of lactation in rats. J. Nutr. 1999;129:928–933. doi: 10.1093/jn/129.5.928. [DOI] [PubMed] [Google Scholar]
- 51.Zaragoza R., Garcia C., Rus A. D., Pallardo F. V., Barber T., Torres L., Miralles V. J., Vina J. R. Inhibition of liver trans-sulphuration pathway by propargylglycine mimics gene expression changes found in the mammary gland of weaned lactating rats: role of glutathione. Biochem. J. 2003;373:825–834. doi: 10.1042/BJ20030387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.House J. D., Brosnan M. E., Brosnan J. T. Characterization of homocysteine metabolism in the rat kidney. Biochem. J. 1997;328:287–292. doi: 10.1042/bj3280287. [DOI] [PMC free article] [PubMed] [Google Scholar]