Abstract
Proteins of the NACHT [NAIP (neuronal apoptosis inhibitory protein), CIITA (MHC class II transcription activator), HET-E (incompatibility locus protein from Podospora anserina) and TP1 (telomerase-associated protein)] family may serve as critical pathogen-sensing and signal-transducing molecules within the innate immune system. In the present paper, we show that CLAN [CARD (caspase-recruitment domain), LRR (leucine-rich repeat) and NACHT domain-containing protein], a NACHT-containing protein originally demonstrated to bind and activate pro-caspase 1, is also capable of influencing the functions of other members of the NACHT family. Through heterotypic NACHT-domain interactions, CLAN was found to associate with Nod1, Nod2 and NAC [nucleotide-binding domain and CARD-containing protein; NALP1 (NACHT, LRR and PYRIN protein 1)] when co-expressed in HEK-293T (human embryonic kidney) cells. NF-κB (nuclear factor κB) reporter assays demonstrated that co-expression of either full-length CLAN or the NACHT domain of CLAN significantly inhibited NF-κB activation induced by Nod1 or Nod2 overexpression. In addition, co-expression of CLAN or the NACHT domain of CLAN with Nod1 or Nod2 inhibited the ability of these proteins to generate active IL-1β (interleukin 1β) through their association with pro-caspase 1. The NACHT domain of CLAN was demonstrated by co-immunoprecipitation experiments to bind all NACHT domains that were tested, including the NACHT domains from CLAN itself, Nod1, Nod2, cryopyrin, NAC, PAN2 {PAAD [pyrin, AIM (absent-in-melanoma), ASC (apoptosis-associated speck-like protein containing a CARD) and death-domain-like]- and NACHT-containing protein} and NAIP (neuronal apoptosis inhibitory protein). Finally, monocyte-expressed CLAN was found to associate with Nod2 following exposure to bacterial peptidoglycan, implying a regulatory role for interaction of these NACHT proteins in the innate immune response. These studies suggest that by mediating hetero-oligomerization, NACHT domains provide a means by which various NACHT-containing proteins may interact, creating protein-interaction networks that potentially modulate immune responses to invading pathogens.
Keywords: caspase 1, CLAN, interleukin-1β, Nod2, Nod1, nuclear factor κB (NF-κB)
Abbreviations: Apaf-1, apoptotic protease-activating factor 1; CARD, caspase-recruitment domain; CARDIAK, CARD-containing interleukin-1β-converting-enzyme-associated kinase; CLAN, CARD, LRR and NACHT domain-containing protein; ECL®, enhanced chemiluminescence; HEK-293T, human embryonic kidney; IKK, inhibitory κB kinase; LPS, lipopolysaccharide; LRR, leucine-rich repeat; NAC, nucleotide-binding domain and CARD-containing protein; NACHT, NAIP (neuronal apoptosis inhibitory protein), CIITA (MHC class II transcription activator), HET-E (incompatibility locus protein from Podospora anserina) and TP1 (telomerase-associated protein); NALP, NACHT, LRR and PYRIN protein 1; NB-ARC domain, nucleotide-binding domain shared by Apaf-1, certain R gene products and CED-4; NF-κB, nuclear factor κB; ORF, open reading frame; PAAD, pyrin, AIM (absent-in-melanoma), ASC (apoptosis-associated speck-like protein containing a CARD) and death-domain-like; PAMP, pathogen-associated molecular pattern; PAN, PAAD- and NACHT-containing protein; PGN, peptidoglycan; RACE, rapid amplification of cDNA ends; RICK, RIP-like interacting CLARP kinase; RIP2, receptor-interacting protein; TLR, Toll-like receptor
INTRODUCTION
TLRs (Toll-like receptors) represent an important component of innate immunity, and are conserved among vertebrates and invertebrates [1]. These transmembrane proteins contain extracellular domains comprising LRRs (leucine-rich repeats) that recognize various PAMPs (pathogen-associated molecular patterns), transducing signals into the interior of host cells that initiate defensive responses. The innate immune system represents the initial line of defence against invading pathogens. Recently, a family of cytosolic proteins containing LRRs has been identified which also appears to participate in innate immunity, presumably sensing the presence of pathogens that have entered host cells. In addition to LRRs, many of these proteins contain a predicted nucleotide-binding fold known as the NACHT [NAIP (neuronal apoptosis inhibitory protein), CIITA (MHC class II transcription activator), HET-E (incompatibility locus protein from Podospora anserina) and TP1 (telomerase-associated protein)] domain, often in combination with additional effector domains such as CARD (caspase recruitment domain) or PYRIN/PAAD [pyrin, AIM (absent-in-melanoma), ASC (apoptosis-associated speck-like protein containing a CARD) and death-domain-like] domains.
Consistent with their postulated importance in innate immunity, hereditary mutations found in NACHT proteins have been implicated in a number of inflammatory diseases. For example, mutations within the coding sequence of the NACHT family protein Nod2 are associated with susceptibility to inflammatory bowel disorders, such as Crohn's disease and Blau syndrome [2,3], suggesting an important role for NACHT proteins in keeping pathogenic bacteria at bay in the gut. Also, the gene encoding cryopyrin (a protein containing NACHT, LRR and PYRIN/PAAD domains) has been found to be mutated in individuals suffering from FCU (familial cold urticaria) and Muckle–Wells syndrome [4].
The NACHT proteins are believed to primarily exert their effects on inflammation through two pathways, modulating the activation of NF-κB (nuclear factor κB) family transcription factors and regulating the activation of caspase 1, a protease responsible for generating the secretable, active form of the pro-inflammatory cytokine IL-1β (interleukin 1β). Both NF-κB and IL-1β are critically involved in mounting effective innate immune responses [5,6].
By analogy with the closely related NB-ARC domain (nucleotide-binding domain shared by Apaf-1, certain R gene products and CED-4) of apoptosis-regulating proteins CED-4 and Apaf-1 (apoptotic protease-activating factor 1) [7], the NACHT domain is thought to function as an oligomerization domain, creating a scaffold on which signal transduction complexes can be assembled [8,9]. For example, Nod1 and Nod2 are inducers of NF-κB by way of their interactions with the IKK (inhibitory κB kinase) γ-binding protein CARDIAK/RIP2 (receptor-interacting protein)/RICK [CARD-containing interleukin-1β-converting-enzyme (ICE)-associated kinase/RIP-like interacting CLARP kinase], mediated by CARD–CARD associations [10]. These proteins are reported to induce activation of IKK complex-associated protein kinases by an ‘induced proximity’ mechanism upon oligomerization [11]. Recently, Nod1 was also discovered to be an activator of caspase 1 as well [12]. In this regard, the adapter protein CARDIAK/RIP2/RICK binds not only the IKKγ subunit of the NF-κB-activating IKK complex, but also pro-caspase 1, via its CARD domain. Precedent for activation of caspases by an induced proximity mechanism also exists, with the Apaf-1 protein representing a quintessential example of such an activator [13].
It was originally reported that Nod1 and Nod2 served as intracellular receptors for LPS (lipopolysaccharide), a component of Gram-negative bacteria [14]. Recently, Nod2 was found to activate NF-κB following exposure to muramyl dipeptide [15,16], a PGN (peptidoglycan) motif often found as a contaminant in LPS preparations. Similarly, Nod1 was found to recognize a unique tetrapeptide motif found in Gram-negative bacterial PGN [17,18]. Indeed, Nod1 was shown to oligomerize and subsequently activate NF-κB through CARDIAK/RIP2/RICK in response to invading Shigella flexneri, independent of TLR signalling [15,16].
CLAN [CARD, LRR and NACHT domain-containing protein; also called Ipaf (interleukin-1β-converting-enzyme protease-activating factor)] is another member of the NACHT family found primarily in monocytes/macrophages [19]. CLAN was originally described as an activator of caspase 1, using its N-terminal CARD to associate with the CARD of pro-caspase 1 [20]. In the present paper, we show that CLAN, through its NACHT domain, can interact with several other NACHT-family proteins, directly affecting their functions with respect to NF-κB induction and caspase 1 activation (IL-1β secretion). In monocytes, CLAN was found to bind to at least one other NACHT family member, Nod2, in response to exposure to bacterial PGN. These heterotypic interactions represent a previously unrecognized level of regulation for the innate immune response, which must be tightly controlled to maintain a balance between effective pathogen defence and hyper-responses that are detrimental to the host organism. Given that the human genome encodes at least 20 NACHT-family proteins [21], the combinatorial opportunities for interactions may be extensive, possibly eliciting differential responses to diverse pathogens.
MATERIALS AND METHODS
cDNA cloning and plasmid construction
The full-length CLAN-A cDNA was amplified by PCR using Pfu Turbo® (Stratagene) and cloned into pcDNA3.1(−)/myc-His6(A) (Invitrogen), which places the myc tag at the C-terminus of expressed proteins. Nod2 was cloned by 5′ and 3′ RACE (rapid amplification of cDNA ends) PCR from cDNA generated from the THP-1 cell line using the SMART™ RACE cDNA amplification kit (Clontech). The following primers were used for amplification: 5′ RACE, 5′-CAAGATCAAGCAGCCTTCTTGCCCTCTGG-3′; 3′ RACE, 5′-CATCCAGGCCTCGGAGGGAAAGGACAGCAG-3′. Bands were excised from an agarose gel, purified, and subjected to nested PCR to confirm Nod2 specificity. 5′ and 3′ RACE products were then cloned into the TOPO cloning vector (Invitrogen) and multiple clones were sequenced for each. Clones of the 3′ end of Nod2 were consistent with published Nod2 sequences. Clones of the 5′ end revealed two previously unreported alternatively spliced exons (GenBank® accession number AY423901) that altered the start of the ORF (open reading frame) of the published Nod2 sequence by 78 base pairs (−26 amino acid residues). The 5′ exon of Nod2 previously cloned from mammary tissue (encoding a more proximal start site) was not found [10]. The following primers were used to amplify the full-Nod2 cDNA from normal human monocyte cDNA: forward, 5′-CGGAATTCATGTGCTCGCAGGAGGCTTTTC-3′; reverse, 5′-CAAGTTCAGCCTTAGGCAGGAC-3′. Products were again excised from an agarose gel and cloned into the TOPO cloning vector. Multiple clones were sequence-verified and the ORF encoding full-length Nod2 was cloned into the pcDNA3.1(−)/myc-His6(A) plasmid. The plasmids pcDNA3/myc-NAC, pcDNA3/myc-PAN2, and pcI-FLAG-Nod1 were previously described in [22–24].
Co-immunoprecipitation assays
HEK-293T (human embryonic kidney) cells (0.5×106) were seeded into six-well plates and were grown overnight. The following day, LIPOFECTAMINE™ Plus (Invitrogen) was used to transfect 2 μg of various expression plasmids according to the manufacturer's recommended protocol. After 24 h, cells were recovered and lysed in isotonic co-immunoprecipitation buffer [142 mM KCl, 0.2% Nonidet P40, 5 mM MgCl2, 10 mM Hepes, 0.5 mM EGTA, 12.5 mM β-glycerophosphate, 2 mM NaF, 1 mM Na3VO4, 1 mM PMSF and 1×Complete™ protease inhibitor mix (Roche)]. Lysates were clarified by centrifugation (10 min at 16000 g) and subjected to immunoprecipitation for 2–24 h at 4 °C using agarose-conjugated anti-myc antibody (Santa Cruz), anti-FLAG antibody (Sigma) or non-specific antibodies coupled to Protein G–agarose. Immune complexes were washed extensively with lysis buffer, boiled in Laemmli buffer and separated by SDS/PAGE using 8 or 10% gels. Proteins were then transferred on to nitrocellulose membranes and detected by immunoblotting using anti-c-myc or anti-FLAG antibodies conjugated to horseradish peroxidase, followed by visualization using an ECL® (enhanced chemiluminescence) method (Amersham Biosciences).
NF-κB luciferase reporter assays
Cells were seeded at 105 cells/well in 24-well plates. The following day, cells were transfected with 100 ng of pNF-κB-luc, 50 ng pTK-RL (Stratagene) and various expression plasmids (as indicated in the Figure legends) using SuperFect (Qiagen). Total DNA was kept constant using empty pcDNA3/myc plasmid. After 24 h, cells were lysed and activity from firefly and Renilla luciferases was assayed using a Dual-Luciferase Reporter System (Promega) and a luminometer.
IL-1β detection
Cells were seeded at 105 cells/per well in 24-well plates. The following day, cells were transfected using SuperFect according to the manufacturer's protocol. Amounts of each plasmid used are indicated in the Figure legends, maintaining total DNA constant using empty pcDNA3/myc plasmid. At 24 h post-transfection, supernatants were collected, clarified by centrifugation (10 min at 1000 g), and assayed for the presence of mature IL-1β using a murine IL-1β ELISA (R&D Systems).
Gel-filtration analysis
Cells were seeded at 8×106 cells/150-mm-diameter tissue culture dish and transfected 24 h later using LIPOFECTAMINE™ Plus according to the recommended protocol. After 24 h, cells were recovered and lysed in buffer containing 100 mM NaCl in combination with 0.2% Nonidet P40, 1 mM EDTA, 1 mM dithiothreitol, 0.5 mM MgCl2 and 0.5 mM EGTA. Lysates were dialysed overnight and then separated using a Superdex-200 column. Fractions were collected and subjected to anti-FLAG immunoprecipitation, then proteins were separated by SDS/PAGE (10% gels) and detected as above.
Generation of CLAN-overexpressing THP-1 cell line
The pLXRN retroviral vector (Clontech) containing a neomycin-resistance cassette and a cDNA encoding full-length CLAN (with a C-terminal c-myc tag), or empty pLXRN, was transfected along with a vector encoding the stomatitis virus glycoprotein (VSV-G) into the 293-GP packaging cell line using SuperFect. At 24 h post-transfection, virus-containing supernatants were collected, passed through 0.45 μm pore-size syringe-top filters, and applied to 5×105 THP-1 cells in the presence of 5 μg/ml polybrene in 24-well plates. Cells were sedimented by centrifugation for 45 min at 800 g. After three rounds of infection, cells were incubated at 32 °C for 24 h and were provided with fresh medium containing 800 μg/ml geneticin the following day. Overexpression of CLAN mRNA and protein was confirmed by RT-PCR and Western blotting.
CLAN/Nod2 co-immunoprecipitation
THP-1/CLAN and THP-1/Neo cells were exposed to PGN isolated from Staphylococcus aureus (Sigma) at 10 μg/ml for 30–120 min. Cells were then lysed in isotonic lysis buffer. Whole-cell extracts were sonicated, cleared by centrifugation (10 min at 16000 g and re-centrifuged until precipitates were completely removed), and subjected to immunoprecipitation with anti-myc–agarose beads overnight. Precipitates were washed extensively with cold lysis buffer and separated by SDS/PAGE (8% gels). Endogenous Nod2 protein was detected by rabbit anti-Nod2 antibodies (a gift from Dr Daniel K. Podolsky, Gastrointestinal Unit, Department of Medicine, Massachusetts General Hospital and Harvard Medical School, Boston, MA, U.S.A.) [25], using the ECL® method.
Statistical analysis
Data comparisons were made by one-way ANOVA followed by Bonferroni's comparison test.
RESULTS
Association of NACHT-containing proteins
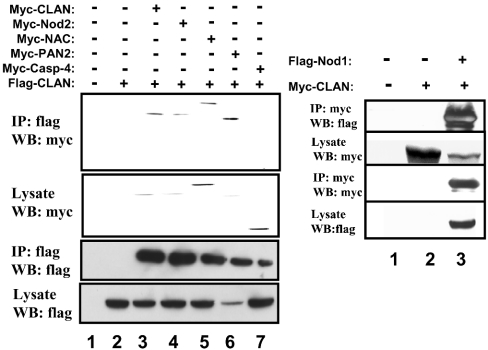
To assess associations between various NACHT-containing proteins, HEK-293T cells were transiently transfected with expression plasmids encoding full-length, epitope-tagged CLAN (also known as CLAN-A, containing CARD, NACHT, and LRR domains [19]), along with plasmids coding for other epitope-tagged NACHT-containing proteins or various control proteins. Co-immunoprecipitation studies demonstrated that CLAN associates with itself and also with Nod2 (CARD15), PAN2 (PAAD- and NACHT-containing protein) [NALP4 (NACHT, LRR and PYRIN protein 1)] and NAC (nucleotide-binding domain and CARD-containing protein) [NALP1/Defcap (death effector filament-forming CED-4-like apoptosis protein)], when co-expressed in HEK-293T cells (Figure 1). CLAN associated more weakly with Nod1 (CARD4), but did not associate with other CARD-containing proteins not carrying a NACHT domain, such as pro-caspase 4 and pro-caspase 9.
Figure 1. Interaction of NACHT-family proteins.
HEK-293T cells were transfected with epitope-tagged expression plasmids encoding the complete coding sequences for various proteins as indicated. Co-immunoprecipitations (IP) were carried out using anti-FLAG- or anti-myc-conjugated agarose beads and immune complexes were analysed by SDS/PAGE and immunoblotting (WB) using antibodies that detect myc or FLAG epitopes. As a control, some lysates were subjected to IP with normal mouse IgG (left-hand panel, lane 2). Expression of proteins was verified by analysing 10% of each lysate taken before immunoprecipitation.
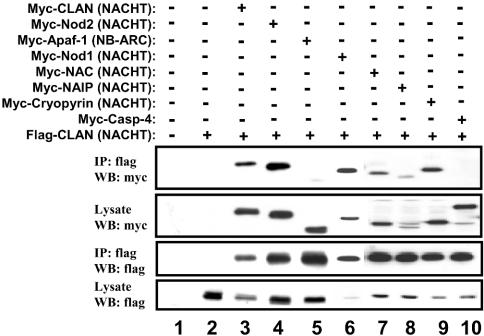
To determine if the NACHT domain by itself is capable of mediating these heterologous protein interactions, an expression plasmid encoding the epitope-tagged NACHT domain of CLAN was co-expressed with various other epitope-tagged NACHT domains in HEK-293T cells. Following immunoprecipitation and immunoblotting, the NACHT domain of CLAN was found to associate with itself and also with the NACHT domains of Nod1, Nod2, NAC, NAIP, PAN2 and cryopyrin (Figure 2). In contrast, the NACHT domain of CLAN did not bind to pro-caspase 4 and only very weakly to the NB-ARC domain of Apaf-1. The NB-ARC domain is a nucleotide binding domain containing Walker A and B boxes, but lacking several of the features of a NACHT domain [7,8].
Figure 2. Heterotypic NACHT-domain interactions.
HEK-293T cells were transfected with epitope-tagged expression plasmids encoding various NACHT domains or other proteins as indicated. Co-immunoprecipitations (IP) were performed using anti-FLAG-conjugated agarose beads and immune complexes were analysed using by SDS/PAGE and immunoblotting (WB). As a control, some samples were subjected to immunoprecipitation with normal mouse IgG (lane 2). Expression of proteins was verified by analysing 10% of each lysate taken before immunoprecipitation.
The NACHT domain of CLAN forms a large protein complex in cells
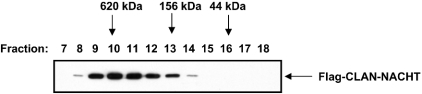
To assess whether or not the NACHT domain of CLAN exists as a monomer or as part of a larger complex within the cell, HEK-293T cells were transfected with FLAG-tagged CLAN-NACHT and lysed 24 h later. Lysates were fractionated by gel-filtration using a Superdex-200 column, fractions were collected and analysed by immunoprecipitation with agarose-conjugated anti-FLAG antibodies. Western blotting demonstrated that the NACHT domain associated with protein complexes of sizes primarily between 156 and 620 kDa (Figure 3). No evidence that CLAN-NACHT exists as a monomer was obtained in these assays, suggesting that the NACHT domain of CLAN either self-associates spontaneously or binds other proteins present endogenously in HEK-293T lysates.
Figure 3. Gel filtration analysis of the NACHT domain of CLAN.
FLAG-tagged CLAN-NACHT was transiently expressed in HEK-293T cells, followed by lysis in 100 mM NaCl, 0.1% Nonidet P40, 1 mM EDTA and 1 mM dithiothreitol. Extracts were separated on a Superdex-200 HR column, and fractions were collected and subjected to immunoprecipitation with anti-FLAG–agarose conjugate. The blot is representative of two independent experiments.
CLAN inhibits Nod-mediated NF-κB activation
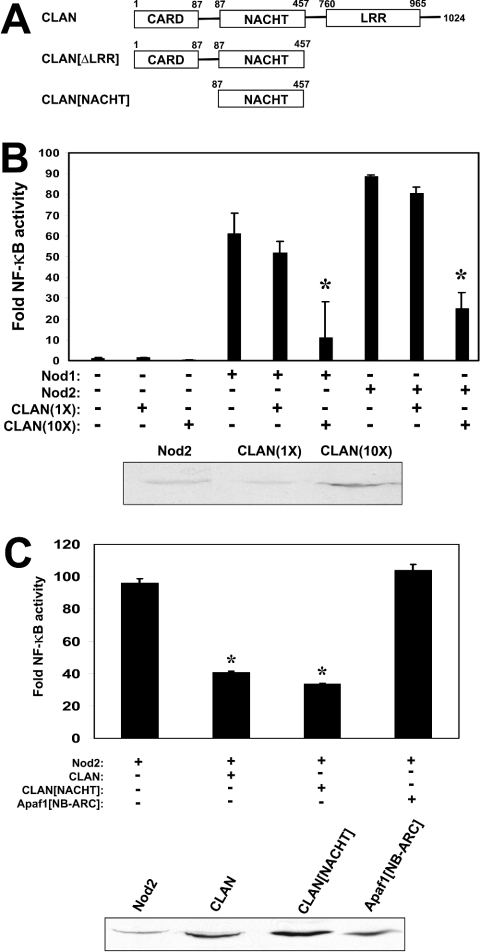
Having shown that CLAN interacts with other NACHT-family proteins, we next explored whether CLAN affects the functions of these proteins. Nod1 and Nod2 are known to bind CARDIAK/RIP2/RICK via their CARD domains, subsequently leading to IKK activation, IκBα (inhibitory κB kinase α) degradation and nuclear translocation of NF-κB transcription factors [11,24]. We used NF-κB-reporter gene assays to assess the effects of CLAN and various deletion mutants of CLAN (depicted in Figure 4A) on Nod-mediated NF-κB activation in HEK-2932T cells. Overexpression of Nod1 or Nod2 induced NF-κB activity in these transient transfections (Figure 4B). When CLAN was co-expressed with full-length Nod1 or Nod2 (at a 4:1 ratio, as determined by immunoblotting), NF-κB activity was significantly reduced.
Figure 4. CLAN inhibits NF-κB activation induced by Nod1 and Nod2.
(A) Domain architecture and amino acid composition of the CLAN deletion mutants used. (B, upper panel) HEK-293T cells were seeded into 24-well plates and transfected the following day with 100 ng of p-NF-κB-luc and 50 ng of pTK-RL reporter gene plasmids together with 100 ng plasmids encoding Nod1 or Nod2 and either 100 ng or 1 μg of plasmid encoding CLAN. Cells were lysed 24 h later and luciferase activity was determined using an automated luminometer. Results are means±S.D. of fold induction of NF-κB activity relative to control-transfected cells (n=3) after normalization for transfection efficiency based on Renilla luciferase activity. *P≤0.001, determined by one-way ANOVA and Bonferroni's comparison test, comparing Nod1 or Nod2 alone versus in combination with CLAN. (B, lower panel) Lysates from equal numbers of transfected cells were analysed by SDS/PAGE and immunoblotting, using anti-myc antibody with ECL®-based detection. Densitometry analysis indicates that the ratio of expression of CLAN(10X)/Nod2 is approx. 4:1. (C, upper panel) HEK-293T cells were transfected with Nod2 (50 ng) in conjunction with CLAN (800 ng), CLAN[NACHT] (400 ng) or Apaf-1[NB-ARC] (600 ng), as indicated. (C, lower panel) Lysates from equal numbers of transfected cells were analysed by SDS/PAGE and immunoblotting, using anti-myc antibody with ECL®-based detection, to compare the levels of expression of myc-Nod2, myc-CLAN, myc-CLAN[NACHT] or myc-Apaf-1[NB-ARC]. Densitometry analysis indicates that the ratio of expression of CLAN(10X)/Nod2 and CLAN[NACHT]/Nod2 is approx. 4:1. Results are representative of three independent experiments.
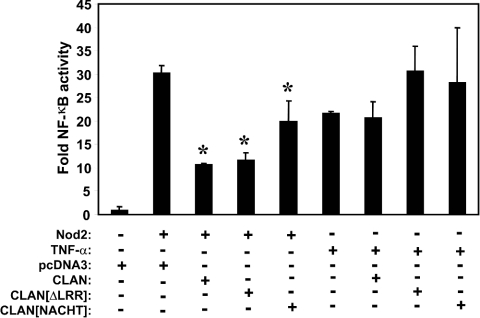
To map the domain in CLAN involved in suppression of Nod2-induced NF-κB, we contrasted full-length CLAN with a fragment of CLAN lacking the LRR, and with a fragment comprising only the NACHT domain. The amino acid residues contained within these CLAN variants are shown in Figure 4(A). In these experiments, CLAN inhibited NF-κB when expressed at a 4:1 ratio compared with Nod2 (determined by immunoblotting) (Figure 4C). These experiments also show that the NACHT domain of CLAN (when expressed in 4-fold excess compared with Nod2) is sufficient to suppress NF-κB activation by Nod2, while the NB-ARC domain of Apaf-1 had no significant inhibitory effects on Nod2-mediated NF-κB activation. Furthermore, the inhibition of Nod2 function is specific because CLAN or the NACHT domain of CLAN did not interfere with NF-κB activity induced by other stimuli such as TNFα (tumour necrosis factor α) (Figure 5).
Figure 5. CLAN-mediated suppression of Nod2-induced NF-κB is specific and requires only the NACHT domain.
The indicated expression plasmids were co-transfected into HEK-293T (Nod2, 50 ng; others, 400 ng) with a luciferase reporter plasmid and cells were lysed 24 h later. Luciferase activity was determined using an automated luminometer. Results are means±S.D. (n=3) expressed as fold activation relative to unstimulated cells transfected with pcDNA3 (=1.0). *P<0.001 relative to Nod2 alone.
Effects of CLAN on Nod-mediated IL-1β secretion
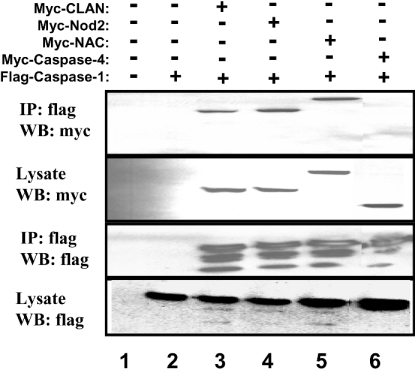
Recently, Nod1 has been shown to associate with pro-caspase 1 and induce its proteolytic activation in overexpression studies [12]. Caspase 1 cleaves pro-IL-1β, generating the mature form of this cytokine, which is subsequently secreted from cells. We have found that Nod2, CLAN and NAC also associate with pro-caspase 1 in transient transfection/co-immunoprecipitation studies of epitope-tagged proteins in HEK-293T cells (Figure 6). The multiple bands detected in FLAG–caspase 1 precipitates represent partially processed pro-caspase 1, generated through its autocatalytic activity when overexpressed.
Figure 6. CARD-carrying NACHT-family proteins bind pro-caspase 1.
HEK-293T cells were transfected with epitope-tagged expression plasmids encoding various full-length NACHT-family proteins or caspases, as indicated. Co-immunoprecipitations (IP) were performed using anti-FLAG-conjugated agarose beads and immune complexes were analysed using SDS/PAGE and immunoblotting (WB). As a control, some samples were subjected to immunoprecipitation with normal mouse IgG (lane 2). Expression of proteins was verified by analysing 10% of each lysate taken before immunoprecipitation.
Based on these experimental results, we proceeded to test the effects of CLAN on the caspase-1-dependent production of secreted IL-1β. For initial experiments, we co-transfected HEK-293T cells with plasmids encoding full-length pro-caspase 1, Nod2 or CLAN, or combinations of these proteins. The combination of pro-caspase 1 and Nod2 resulted in substantial amounts of IL-1β secretion, while CLAN co-transfection with pro-caspase 1 had a relatively smaller effect (Figure 7A). It remains to be determined whether Nod2 induces IL-1β secretion by increasing its processing through the direct activation of pro-caspase 1 or by enhancing the secretory mechanism for this cytokine, which is currently poorly understood. In contrast with the IL-1β-inducing effects of Nod2 expression, co-expressing this protein with CLAN and pro-caspase 1 resulted in less IL-1β secretion compared with pro-caspase 1 and Nod2 alone. As shown in Figure 7(B), the NACHT domain of CLAN is sufficient to inhibit Nod2-induced IL-1β secretion, while the NB-ARC domain of Apaf-1 had no inhibitory effects when co-expressed with Nod2. Co-expression of another CARD-containing protein, procaspase 4, also had no effect on Nod2-induced IL-1β secretion in these experiments.
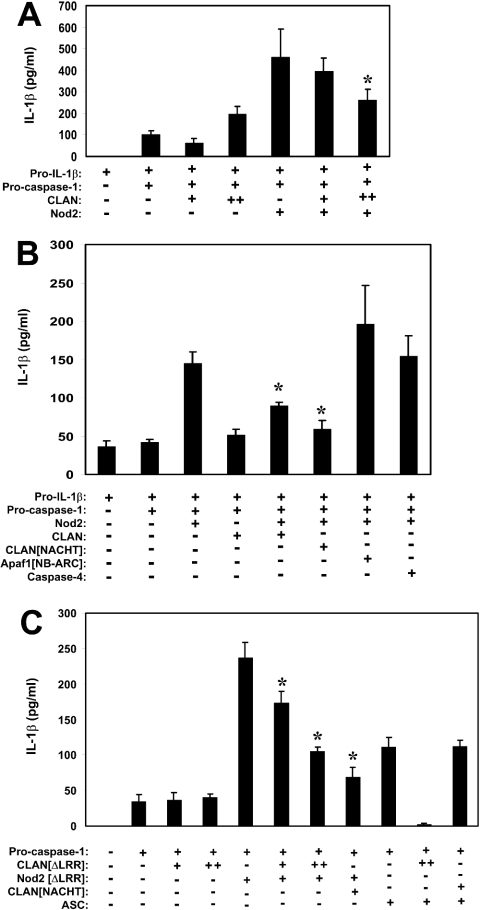
Figure 7. CLAN inhibits caspase 1 activation induced by Nod2.
(A) HEK-293T cells were transfected with plasmids encoding pro-IL-1β (400 ng) and caspase 1 (100 ng), with (+) or without (−) Nod2 (50 ng) or CLAN [+, 50 ng; ++, 500 ng]. Supernatants were analysed for IL-1β secretion by ELISA at 24 h post-transfection. *P<0.05, relative to Nod2 alone. (B) HEK-293T cells were transfected with plasmids encoding pro-IL-1β (400 ng) and caspase 1 (100 ng), with (+) or without (−) Nod2 (50 ng), CLAN (400 ng), CLAN[NACHT] (400 ng), Apaf-1[NB-ARC] (400 ng) or caspase 4 (100 ng), as indicated. Supernatants were analysed for IL-1β secretion by ELISA at 24 h post-transfection. (C) HEK-293T cells were transfected with plasmids encoding pro-IL-1β (400 ng) and caspase 1 (100 ng), with (+) or without (−) CLAN[ΔLRR] (100 ng [+] or 500 ng [++]), Nod2[ΔLRR] (100 ng), CLAN[NACHT] (500 ng) or ASC (200 ng). Supernatants were analysed for IL-1β secretion by ELISA 24 h post-transfection. *, Experimental conditions that resulted in significantly less IL-1β secretion compared with Nod2 alone (P<0.05). Results are means±S.D. representative of at least two independent experiments.
Next, we used expression plasmids encoding Nod2 lacking the LRR domains, which are known to auto-repress NACHT-family protein activity unless certain bacteria-derived ligands are present [20,26], analogous to the WD40 repeat region of Apaf-1, which represses its own activation in the absence of cytochrome c [27]. As seen in Figure 7(C), expression of Nod2[ΔLRR] enhanced caspase-1-mediated production of IL-1β in culture supernatant. Since CLAN interacts with both pro-caspase 1 and the Nod proteins, we examined its effects in this IL-1β secretion assay. CLAN[ΔLRR], a mutant of CLAN reported to be constitutively active [20], did not significantly increase IL-1β secretion at the levels expressed. Instead, CLAN inhibited the Nod2-mediated secretion of IL-1β (Figure 7C). Additionally, the NACHT domain of CLAN by itself was sufficient to inhibit Nod2-mediated IL-1β production. Similar observations were made when full-length CLAN, CLAN[ΔLRR] or the NACHT domain of CLAN was co-expressed with Nod1 (results not shown).
To explore the specificity of the effects of the NACHT domain of CLAN on Nod2-mediated IL-1β production, we contrasted its effects on Nod2 with ASC, another caspase 1-activating protein [28]. As shown in Figure 7(C), the NACHT domain of CLAN as well as a NACHT-containing fragment of CLAN lacking the LRRs (CLAN[ΔLRR]) suppressed IL-1β secretion induced by transfection of cells with Nod2 in combination with pro-caspase 1. In contrast, the NACHT domain of CLAN failed to suppress IL-1β secretion induced by transfecting cells with the combination of ASC (which lacks a NACHT domain) and pro-caspase 1 (Figure 7C). Interestingly, while the NACHT domain of CLAN did not suppress ASC-induced IL-1β secretion, the CLAN[ΔLRR] protein did, presumably because the CARD domain of CLAN is known to bind pro-caspase 1 and thus competes with ASC for access to pro-caspase 1.
CLAN transiently associates with Nod2 following peptidoglycan exposure in human monocytes
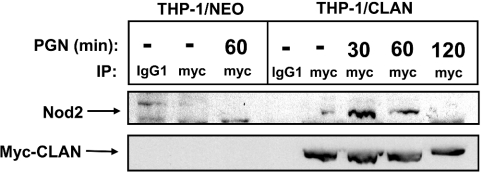
To determine whether CLAN and Nod2 associate in monocytes/macrophages, we performed co-immunoprecipitation experiments. Lacking an anti-CLAN antibody capable of immunoprecipitating the endogenous CLAN protein, we instead relied upon the use of a THP-1 monocytic cell line stably expressing myc-epitope-tagged CLAN as a result of retroviral gene transfer. This epitope-tagged version of the CLAN protein is suitable for immunoprecipitation using anti-myc epitope antibodies. To activate Nod2 and assess its association with CLAN, THP-1 cells expressing CLAN (THP-1/CLAN) or control cells selected for neomycin resistance (THP-1/Neo) were exposed to PGN (10 μg/ml), which has been previously demonstrated to activate Nod2 [15,16]. Following exposure to PGN for 30, 60 or 120 min, cells were lysed and myc-tagged CLAN was immunoprecipitated, using anti-myc–agarose. The presence of endogenous Nod2 protein in the immunoprecipitates was determined by immunoblotting using anti-Nod2 antibodies. As shown in Figure 8, little association of CLAN and Nod2 was detected in lysates from unstimulated cells (T=0). In contrast, association of CLAN and Nod2 was increased following PGN exposures of 30 or 60 min. After 120 min of PGN exposure, CLAN/Nod2 binding was no longer seen by co-immunoprecipitation, suggesting either a transient association of the proteins or translocation of the CLAN/Nod2 protein complex to a non-extractable compartment in cells. Taken together, these data provide evidence that selected NACHT family proteins can associate in response to pathogen-derived stimulatory molecules.
Figure 8. CLAN and Nod2 co-associate in monocytes following exposure to PGN.
THP-1 cells expressing epitope-tagged CLAN (THP-1/CLAN) or neomycin-resistant control cells (THP-1/Neo) were exposed to PGN (10 μg/ml) for the times indicated, then lysed, and subjected to anti-myc immunoprecipitation and immunoblotting for detection of associated Nod2 protein. To assess equivalent immunoprecipitation of CLAN, membranes were stripped and re-probed with anti-myc antibodies. As a control, some samples were subjected to immunoprecipitation with normal mouse IgG (lanes 1 and 4). The blot is representative of three independent experiments.
DISCUSSION
In the present paper, we show that the NACHT domain of CLAN has the ability to bind to NACHT domains found in other proteins of this family. Furthermore, interaction of CLAN with other NACHT-family members, such as Nod2, is induced by bacteria-derived molecules known to interact with the LRRs of these proteins. The results presented in the present study thus support the notion that CLAN, in addition to eliciting its own effects on pro-IL-1β processing, has the ability to modulate NF-κB activity and cytokine secretion mediated by other NACHT family proteins. We therefore speculate that the observation that CLAN has been demonstrated to be either an inducer [20] or an inhibitor of caspase 1 and IL-1β activation may be attributed to differences in the ratios of CLAN relative to caspase 1 or to other NACHT family proteins. Accordingly, the relative amounts of NACHT family proteins are likely to be critical to the overall effect of the CLAN protein in macrophages. Hypothetically, CLAN may act as a positive or negative regulator of the inflammatory response depending on the pathogen, the pathogen's intracellular location (cytosolic or vesicle-associated) or PAMPs presented to this and other NACHT-family members within the cell. In this regard, CLAN may have two conformational states that determine whether it functions as an inhibitor or activator, depending on whether its LRRs are engaged by microbial or cellular ligands. Further experimentation is required to determine the LRR-binding ligand for CLAN and to define the complete in vivo circumstances where CLAN functions as a modulator of other NACHT family proteins such as Nod1, Nod2 and NAC.
Although it is unknown what factors control heterologous interactions among NACHT family proteins, it can be hypothesized that they are induced by specific LRR-binding microbial ligands or by temporal alterations in protein expression levels. As evidenced by PGN-induced CLAN/Nod2 interactions, molecules produced by specific pathogens may activate collections of NACHT-family proteins via interactions with their LRRs, thus generating combinatorial NACHT–NACHT associations that specify the type of host response, the magnitude of the response or its duration. The ability of CLAN to associate with and regulate the functions of other NACHT-family proteins via NACHT–NACHT interactions represents a novel mechanism for controlling innate immune responses. With 20 NACHT-family proteins encoded in the human genome [21], the combinatorial possibilities for NACHT–NACHT interactions are large, possibly permitting specificity in cellular responses to microbial pathogens.
Acknowledgments
We thank Dr C. Stehlik and Dr L. Fiorentino for helpful discussions and the NIH (National Institutes of Health) (GM61694; AI56324) for generous support. J.D. is supported by the Department of Defense, Breast Cancer Research Program Fellowship (DAMD-17-01-1-0166).
References
- 1.Armant M. A., Fenton M. J. Toll-like receptors: a family of pattern-recognition receptors in mammals. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-8-reviews3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogura Y., Bonen D. K., Inohara N., Nicolae D. L., Chen F. F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R. H., et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature (London) 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 3.Miceli-Richard C., Lesage S., Rybojad M., Prieur A. M., Manouvrier-Hanu S., Hafner R., Chamaillard M., Zouali H., Thomas G., Hugot J. P. CARD15 mutations in Blau syndrome. Nat. Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 4.Hoffman H. M., Mueller J. L., Broide D. H., Wanderer A. A., Kolodner R. D. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat. Genet. 2001;29:301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li P., Allen H., Banerjee S., Franklin S., Herzog L., Johnston C., McDowell J., Paskind M., Rodman L., Salfeld J., et al. Mice deficient in IL-1β-converting enzyme are defective in production of mature IL-1β and resistant to endotoxic shock. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 6.Karin M., Delhase M. The IκB kinase (IKK) and NF-κB: key elements of proinflammatory signalling. Semin. Immunol. 2000;12:85–98. doi: 10.1006/smim.2000.0210. [DOI] [PubMed] [Google Scholar]
- 7.Jaroszewski L., Rychlewski L., Reed J. C., Godzik A. ATP-activated oligomerization as a mechanism for apoptosis regulation: fold and mechanism prediction for CED-4. Proteins. 2000;39:197–203. doi: 10.1002/(sici)1097-0134(20000515)39:3<197::aid-prot10>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 8.Koonin E. V., Aravind L. The NACHT family – a new group of predicted NTPases implicated in apoptosis and MHC transcription activation. Trends Biochem. Sci. 2000;25:223–224. doi: 10.1016/s0968-0004(00)01577-2. [DOI] [PubMed] [Google Scholar]
- 9.Linhoff M. W., Harton J. A., Cressman D. E., Martin B. K., Ting J. P. Two distinct domains within CIITA mediate self-association: involvement of the GTP-binding and leucine-rich repeat domains. Mol. Cell. Biol. 2001;21:3001–3011. doi: 10.1128/MCB.21.9.3001-3011.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogura Y., Inohara N., Benito A., Chen F. F., Yamaoka S., Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-κB. J. Biol. Chem. 2001;276:4812–4818. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 11.Inohara N., Koseki T., Lin J., del Peso L., Lucas P. C., Chen F. F., Ogura Y., Nunez G. An induced proximity model for NF-κB activation in the Nod1/RICK and RIP signaling pathways. J. Biol. Chem. 2000;275:27823–27831. doi: 10.1074/jbc.M003415200. [DOI] [PubMed] [Google Scholar]
- 12.Yoo N. J., Park W. S., Kim S. Y., Reed J. C., Son S. G., Lee J. Y., Lee S. H. Nod1, a CARD protein, enhances pro-interleukin-1β processing through the interaction with pro-caspase-1. Biochem. Biophys. Res. Commun. 2002;299:652–658. doi: 10.1016/s0006-291x(02)02714-6. [DOI] [PubMed] [Google Scholar]
- 13.Srinivasula S. M., Ahmad M., Fernandes-Alnemri T., Alnemri E. S. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol. Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 14.Inohara N., Ogura Y., Chen F. F., Muto A., Nunez G. Human Nod1 confers responsiveness to bacterial lipopolysaccharides. J. Biol. Chem. 2001;276:2551–2554. doi: 10.1074/jbc.M009728200. [DOI] [PubMed] [Google Scholar]
- 15.Girardin S. E., Boneca I. G., Viala J., Chamaillard M., Labigne A., Thomas G., Philpott D. J., Sansonetti P. J. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 16.Inohara N., Ogura Y., Fontalba A., Gutierrez O., Pons F., Crespo J., Fukase K., Inamura S., Kusumoto S., Hashimoto M., et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2: implications for Crohn's disease. J. Biol. Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 17.Girardin S. E., Boneca I. G., Carneiro L. A., Antignac A., Jehanno M., Viala J., Tedin K., Taha M. K., Labigne A., Zathringer U., et al. Nod1 detects a unique muropeptide from Gram-negative bacterial peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 18.Chamaillard M., Hashimoto M., Horie Y., Masumoto J., Qiu S., Saab L., Ogura Y., Kawasaki A., Fukase K., Kusumoto S., et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 19.Damiano J. S., Stehlik C., Pio F., Godzik A., Reed J. C. CLAN, a novel human CED-4-like gene. Genomics. 2001;75:77–83. doi: 10.1006/geno.2001.6579. [DOI] [PubMed] [Google Scholar]
- 20.Poyet J. L., Srinivasula S. M., Tnani M., Razmara M., Fernandes-Alnemri T., Alnemri E. S. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J. Biol. Chem. 2001;276:28309–28313. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- 21.Reed J. C., Doctor K., Rojas A., Zapata J. M., Stehlik C., Fiorentino L., Damiano J., Roth W., Matsuzawa S., Newman R., et al. Comparative analysis of apoptosis and inflammation genes of mice and humans. Genome Res. 2003;13:1376–1388. doi: 10.1101/gr.1053803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu Z. L., Pio F., Xie Z., Welsh K., Krajewska M., Krajewski S., Godzik A., Reed J. C. A novel enhancer of the Apaf1 apoptosome involved in cytochrome c-dependent caspase activation and apoptosis. J. Biol. Chem. 2001;276:9239–9245. doi: 10.1074/jbc.M006309200. [DOI] [PubMed] [Google Scholar]
- 23.Fiorentino L., Stehlik C., Oliveira V., Ariza M. E., Godzik A., Reed J. C. A novel PAAD-containing protein that modulates NF-κB induction by cytokines tumor necrosis factor-α and interleukin-1β. J. Biol. Chem. 2002;277:35333–35340. doi: 10.1074/jbc.M200446200. [DOI] [PubMed] [Google Scholar]
- 24.Inohara N., Koseki T., del Peso L., Hu Y., Yee C., Chen S., Carrio R., Merino J., Liu D., Ni J., Nunez G. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-κB. J. Biol. Chem. 1999;274:14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- 25.Hisamatsu T., Suzuki M., Reinecker H. C., Nadeau W. J., McCormick B. A., Podolsky D. K. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology. 2003;124:993–1000. doi: 10.1053/gast.2003.50153. [DOI] [PubMed] [Google Scholar]
- 26.Martinon F., Burns K., Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 27.Hu Y., Ding L., Spencer D. M., Nunez G. WD-40 repeat region regulates Apaf-1 self-association and procaspase-9 activation. J. Biol. Chem. 1998;273:33489–33494. doi: 10.1074/jbc.273.50.33489. [DOI] [PubMed] [Google Scholar]
- 28.Srinivasula S. M., Poyet J. L., Razmara M., Datta P., Zhang Z., Alnemri E. S. The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. J. Biol. Chem. 2002;277:21119–21122. doi: 10.1074/jbc.C200179200. [DOI] [PubMed] [Google Scholar]