ABSTRACT
Shigella bacteria utilize the type III secretion system (T3SS) to invade host cells and establish local infection. Invasion plasmid antigen D (IpaD), a component of Shigella T3SS, has garnered extensive interest as a vaccine target, primarily due to its pivotal role in the Shigella invasion, immunogenic property, and a high degree of conservation across Shigella species and serotypes. Currently, we are developing an epitope- and structure-based multivalent vaccine against shigellosis and require functional epitope antigens of key Shigella virulence determinants including IpaD. However, individual IpaD B-cell epitopes, their contributions to the overall immunogenicity, and functional activities attributing to bacteria invasion have not been fully characterized. In this study, we predicted continuous B-cell epitopes in silico and fused each epitope to a carrier protein. Then, we immunized mice intramuscularly with each epitope fusion protein, examined the IpaD-specific antibody responses, and measured antibodies from each epitope fusion for the activity against Shigella invasion in vitro. Data showed that all epitope fusion proteins induced similar levels of anti-IpaD IgG antibodies in mice, and differences were noted for antibody inhibition activity against Shigella invasion. IpaD epitope 1 (SPGGNDGNSV), IpaD epitope 2 (LGGNGEVVLDNA), and IpaD epitope 5 (SPNNTNGSSTET) induced antibodies significantly better in inhibiting invasion from Shigella flexneri 2a, and epitopes 1 and 5 elicited antibodies more effectively at preventing invasion of Shigella sonnei. These results suggest that IpaD epitopes 1 and 5 can be the IpaD representative antigens for epitope-based polyvalent protein construction and protein-based cross-protective Shigella vaccine development.
IMPORTANCE
Shigella is a leading cause of diarrhea in children younger than 5 years in developing countries (children’s diarrhea) and continues to be a major threat to public health. No licensed vaccines are currently available against the heterogeneous Shigella species and serotype strains. Aiming to develop a cross-protective multivalent vaccine against shigellosis and dysentery, we applied novel multiepitope fusion antigen (MEFA) technology to construct a broadly immunogenic polyvalent protein antigen, by presenting functional epitopes of multiple Shigella virulence determinants on a backbone protein. The functional IpaD epitopes identified from this study will essentially allow us to construct an optimal polyvalent Shigella immunogen, leading to the development of a cross-protective vaccine against shigellosis (and dysentery) and the improvement of global health. In addition, identifying functional epitopes from heterogeneous virulence determinants and using them as antigenic representatives for the development of cross-protective multivalent vaccines can be applied generally in vaccine development.
KEYWORDS: Shigella spp., epitope mapping, vaccine, invasion plasmid antigen, IpaD, type III secretion system (T3SS)
INTRODUCTION
Shigella is one of the major causes of diarrheal mortality worldwide and is estimated to lead to 212,000 deaths in 2016 (1). Shigellosis predominantly affects children under the age of 5 years, and accounts for approximately 54,900 deaths of young children annually (2). In addition, Shigella infections are reported to result in linear growth faltering (3, 4) and a significant impact of environmental enteric dysfunction (EED) (5), leading to poor longer-term outcomes including impaired cognitive development, academic underachievement at school, and limited economic opportunities (6–8).
Shigellosis could be treated and/or prevented theoretically. Oral rehydration therapy and antibiotic drugs are the typical treatments for severe infection but that brings heavy pressure and financial burdens to families and local healthcare systems in developing countries or regions (9). Indeed, many rural areas cannot access these facilities and resources. In addition, the increased and repeated usage of antibiotics led to the rapid acquisition of antimicrobial resistance among Shigella and other bacteria (10). Improved water, sanitation, and hygiene (WASH) could effectively prevent shigellosis and other enteric diseases, but financial constraints faced by developing countries, particularly countries in Southeast Asia and sub-Saharan Africa, prevent this program from being implemented for the foreseeable future. Vaccines would be a more cost-effective and practical prevention countermeasure against shigellosis. Unfortunately, there are no licensed vaccines for Shigella infections.
A safe, cost-effective, cross-protective, and ideally cold chain-free vaccine against heterogeneous Shigella species and serotypes is urgently needed. Such a vaccine would reduce morbidity and mortality in children under 5 years and also curb the threat of antibiotic resistance. However, developing vaccines against Shigella has been proven to be challenging. Immunological heterogeneity among Shigella serotypes and unique disease mechanisms are among the key difficulties in Shigella vaccine development. There are 4 species and over 50 serotypes in the genus of Shigella. These bacteria elaborate on a unique pathogenesis strategy to establish local infection, including adhesion, invasion, immune evasion, replication, and intercellular mobility (11, 12). Particularly, the type III secretion system (T3SS), a needle-like structure with an associated tip complex plays a major role in the disease mechanism (13). In T3SS, invasion plasmid antigens including invasion plasmid antigen D (IpaD) provide a physical platform and are responsible for sensing the host environment, leading to the activation of the T3SS upon contact with the host epithelial cells (11, 14). In addition, Shigella can impair host-adapted immunity development via IpaD-mediated B lymphocyte apoptosis (15). Targeting IpaD was indicated to be a promising approach against Shigella infections. Indeed, anti-IpaD antibodies were found to neutralize Shigella virulence properties, including invasion (16, 17) and hemolysis (18, 19). Furthermore, IpaD was identified as a broadly protective vaccine immunogen (20, 21).
Recently, based on a novel vaccinology platform, multiepitope fusion antigen (MEFA) (22), we designed an epitope- and structure-based polyvalent protein antigen called Shigella MEFA. This Shigella MEFA was constructed by incorporating epitopes from several virulence factors (invasion plasmid antigen B - IpaB, IpaD, VirG, GuaB, and Shiga toxins 1 and 2) and demonstrated to induce functional antibodies against Shigella invasion and Shiga toxin cytotoxicity and cross-protect mice from lethal pulmonary infection by S. sonnei or S. flexneri serotype 2a, 3a, or 6 (23). However, the epitopes included in that Shigella MEFA protein were predicted based on antigenicity in silico but not functional immunity empirically. Knowing that an immunodominant epitope may not be able to induce functional immunity, this in-silico-based Shigella MEFA may not be optimal functionally. To optimize the Shigella MEFA protein design and maximize functional immunity, we need to first identify functional epitopes from each target virulence determinant. We have mapped the IpaB epitopes and identified the top-ranked ones for this particular virulence (24), but protective epitopes from the other key virulence factors are yet to be determined.
In this study, we mapped the functional B-cell epitopes for virulence determinant IpaD. We first in silico predicted the immunodominant B-cell epitopes, then genetically fused each epitope to a carrier for an epitope fusion protein, subsequently immunized mice with each epitope fusion protein, and measured the functional activity of epitope fusion-induced antibodies against Shigella bacteria invasion to identify functional epitopes for IpaD. Top-ranked IpaD epitopes can then be the antigenic representatives for this key virulence determinant and used to optimize the Shigella MEFA protein and potentially enhance vaccine efficacy against Shigella infections.
RESULTS
IpaD epitopes were in silico predicted and genetically fused to carrier protein CsaB
Eight continuous immunodominant B-cell epitopes were identified based on in silico antigenicity scores, using the BepiPred 2.0 epitope prediction software (25). They were EP1 (SPGGNDGNSV), EP2 (LGGNGEVVLDNA), EP3 (SAEDETMKNN), EP4 (EYPINKDARE), EP5 (SPNNTNGSSTET), EP6 (TLTNSISTSS), EP7 (VQKYSNANSIFD), and EP8 (KWLTELGGTIG) (Fig. 1A). All epitopes were conserved among S. dysenteriae Sd197, S. boydii, S. sonnei, and S. flexneri 2a, and protein modeling suggested these epitopes were surface exposed on the IpaD protein (Fig. 1A and B; each epitope in a different color). Nucleotides encoding each epitope were genetically fused to carrier protein CsaB, a major subunit protein of the enterotoxigenic Escherichia coli (ETEC) adhesin CS4, leading to eight epitope fusion genes (Fig. 2A). Each epitope fusion gene was cloned into vector pET28a and confirmed with DNA sequencing. Expressed in E. coli BL21 (DE3), eight recombinant epitope fusion proteins, at the size of around 18 kDa, were generated (Fig. 2B). Recombinant epitope fusion proteins were characterized with anti-IpaBD fusion mouse antiserum in Western blot (Fig. 2C).
Fig 1.
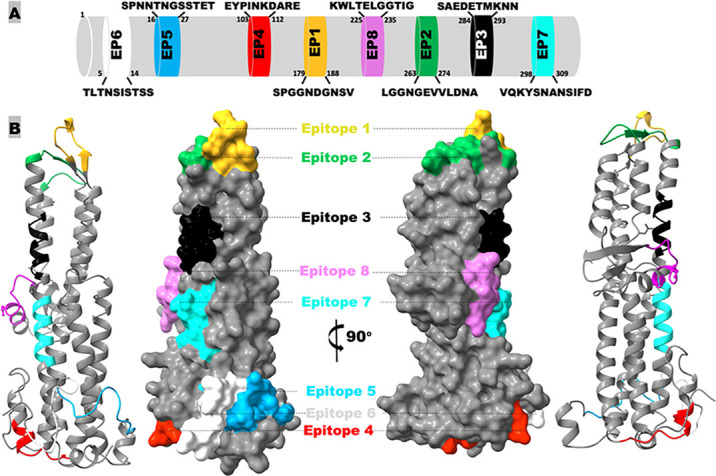
In silico identification of continuous B-cell epitopes of Shigella IpaD protein. (A) The epitope positions and amino acid sequences. (B) Illustration of IpaD epitope surface exposure and location on protein modeling, generated by Phyre 2 based on the S. flexneri 2a sequence (protein no. NP_858259) and visualized by ChimeraX-1.6.1. This model is archived in the Model archive database with the identifier “ma-rf0iy.”
Fig 2.
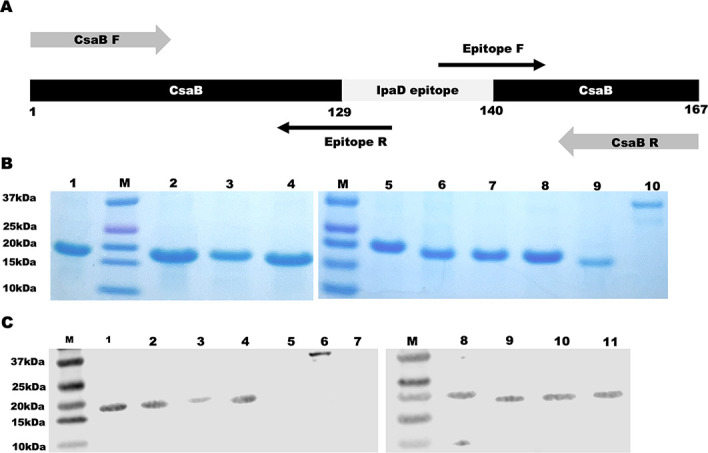
Construction, extraction, and characterization of IpaD epitope fusion proteins. (A) Illustration of IpaD epitope fusion construction, with two overlapping PCR amplifications using primers CsaB-F/epitope-R and epitope-F/CsaB-R to substitute a backbone CsaB epitope (amino acids 130–139) with each IpaD epitope. (B) IpaD epitope fusion protein in SDS-PAGE with Coomassie blue staining; epitope fusion protein from EP1 to EP8 in lanes 1 through 8, backbone protein CsaB in lane 9, and recombinant IpaD protein in lane 10. (C) IpaD epitope fusion protein characterization in Western blot with anti-DB fusion (IpaD and IpaB fusion protein) mouse serum (diluted 1:6,000); EP1 to EP4 in lanes 1 to 4, EP5 to EP8 in lanes 8 to 11, CsaB in lane 5, and IpaD in lane 6, and host cell E. coli BL21 proteins in lane 7.
IpaD epitope fusion proteins induced anti-IpaD antibody response in mice
Mice intramuscularly (i.m.) immunized with each IpaD epitope fusion protein (CsaB-IpaD epitope fusion) developed robust antibody responses to IpaD protein (Fig. 3). Anti-IpaD IgG titers (log10) in serum samples of the mice immunized with EP1, EP2, EP3, EP4, EP5, EP6, EP7, or EP8 epitope fusion protein were detected at 3.68 ± 0.21, 3.74 ± 0.22, 3.42 ± 0.06, 3.73 ± 0.26, 3.81 ± 0.36, 3.84 ± 0.18, 3.67 ± 0.24, 3.71 ± 0.19, respectively. Mice i.m. immunized with recombinant protein IpaD, which was used as a positive control, had anti-IpaD IgG detected at 4.63 ± 0.19 (log10). Serum samples from the groups of control mice, which were administrated with phosphate-buffered saline (PBS) or carrier protein CsaB (mouse sera from the other studies in the laboratory), showed no detectable anti-IpaD IgG response.
Fig 3.
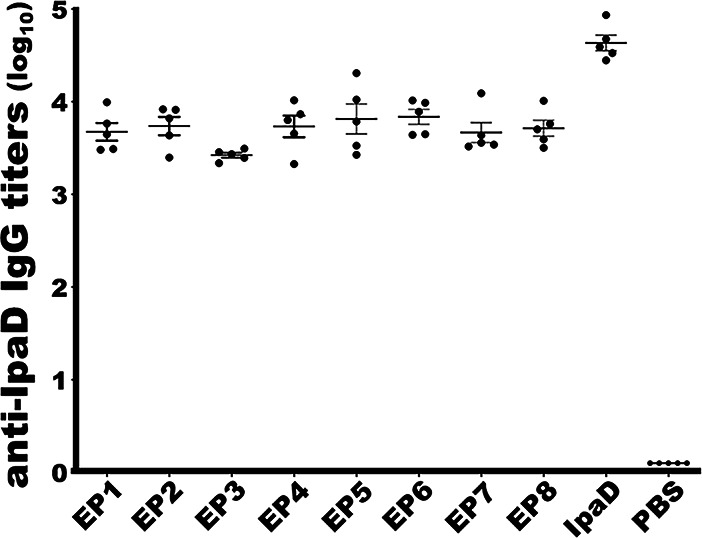
Mouse serum anti-IpaD IgG titers (in log10) from each immunization group or the control group. EP1 to EP8, IpaD, or PBS represents mouse sera from the group i.m. immunized with IpaD epitope fusion 1, 2, 3, 4, 5, 6, 7, or epitope fusion 8, recombinant IpaD protein, or PBS the control. 40 µg epitope fusion protein (EP1 to EP8) or the IpaD recombinant protein, or 40 µl PBS, adjuvanted with 0.2 µg of dmLT, were i.m. injected into each mouse in the group. Bars indicated IgG titer means and standard deviations.
Antibodies derived from EP1, 2, or 5 epitope fusion protein significantly inhibited Shigella bacteria invasion in vitro
Antibody invasion inhibition assays showed that the number of S. sonnei bacteria (CFUs) invaded HeLa cells was reduced by 39.5% ± 16.62%, 25.2% ± 14.89%, 14.7% ± 21.30%, 17.6% ± 16.35%, 47.2% ± 6.73%, 17.5% ± 17.42%, 1% ± 17.24%, 22.2% ± 11.66%, and 64.2% ± 10.45%, respectively, after S. sonnei bacteria being incubated with heat-inactivated sera from the group immunized with EP1, EP2, EP3, EP4, EP5, EP6, DP7, EP8 epitope fusion protein, or the IpaD recombinant protein, with numbers of the invaded bacteria treated with the control serum were referred to as 100% (Fig. 4). Statistical analyses showed that the reduction from incubation with sera of the mice immunized with epitope fusion protein EP1 or EP5 was significantly different than the PBS control group (P < 0.0001).
Fig 4.
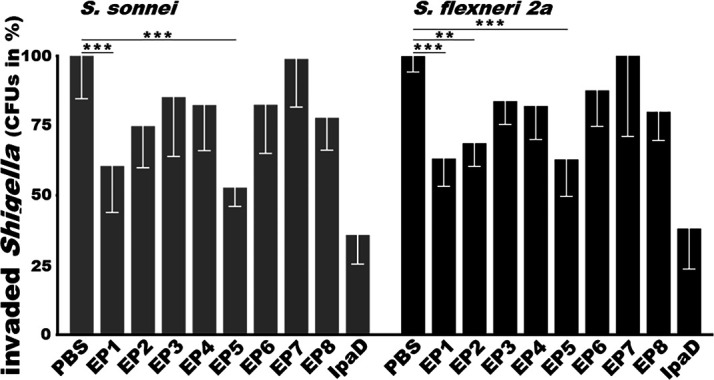
In-vitro antibody invasion inhibition against Shigella sonnei or S. flexneri 2a. Heat-inactivated mouse sera from the group i.m. immunized with PBS (control), IpaD epitope fusion 1, 2, 3, 4, 5, 6, 7, or epitope fusion 8, or IpaD recombinant protein were mixed with S. sonnei or S. flexneri 2a bacteria, and then transferred to confluent monolayered Hela cells. After eliminating extracellular Shigella bacteria, cells were lysed; the invaded Shigella bacteria were collected, cultured overnight, and counted (CFUs). CFUs were converted to percentages, referring to the CFUs from treatment with control mouse sera as 100%. Boxes and bars indicated means and standard deviations. ** and *** indicated P < 0.01 and P < 0.001, respectively.
When S. flexneri 2a bacteria were examined, the in vitro invasion was reduced by 36.9% ± 9.93%, 31.4% ± 8.29%, 16.2% ± 8.35%, 18.0% ± 11.99%, 37.2% ± 13.24%, 12.4% ± 12.87%, -8.6% ± 37.52%, 20.1% ± 10.28%, and 62.0% ± 14.43% respectively, after incubation with heat-inactivated sera from the group immunized with EP1, EP2, EP3, EP4, EP5, EP6, DP7, EP8 epitope fusion protein, or the IpaD recombinant protein. Statistical analyses indicated the reduction from the sera of the group immunized with epitope fusion EP1 (P = 0.0002), EP2 (P = 0.0041), or EP5 (P = 0.0002) was significantly different than the control (Fig. 4).
EP1 and EP5 were the top-ranked functional epitopes of Shigella IpaD
Serum antibodies from the mice immunized with each IpaD epitope fusion protein reduced S. sonnei or S. flexneri 2a invasion in vitro, except for the sera from epitope fusion EP7. However, sera from the mice immunized with EP1epitope fusion or EP5 epitope fusion protein were shown more effective against invasion of S. sonnei and S. flexneri 2a. The antibody invasion inhibition levels from EP1 or EP5 epitope fusion protein were not significantly different compared to those from the positive control IpaD protein. For invasion inhibition against S. sonnei, 39.5% of EP1 was not statistically different from 64.2% of IpaD (P = 0.08), and 47.2% of EP5 was not significantly different than 64.2% of IpaD (P = 0.999). For invasion inhibition against S. flexneri 2a, 36.9% from EP1 compared to 62% from IpaD (P = 0.07), or 37.2% from EP5 than 62% from IpaD (P = 0.08), was not significantly different.
DISCUSSION
One of the major challenges in developing vaccines against shigellosis is the heterogeneity among Shigella species and serotypes. Differed from the LPS-based vaccines that target specific serotypes and thus are unable to cover different Shigella species and heterogeneous serotypes, a protein-based multivalent vaccine to target protein antigens conserved across Shigella spp. and serotypes can potentially be cross-protective. Such a multivalent vaccine can be produced conventionally, by mixing a few protein immunogens into a cocktail product or genetically fusing two or more proteins as a fusion protein, but with the disadvantage of vaccine formulation and manufacturing complexity (26). Since not all protein segments or domains elicit host immune responses and only functional peptide domains or epitopes induce protective immunity, an alternative vaccine antigen approach has been developed. Instead of an entire protein, functional peptides or epitopes, if they are conserved, can be used as vaccine antigens to induce broadly functional immunity against heterogeneous pathogenic strains or isolates (27). To develop an epitope-based multivalent protein vaccine against heterogeneous Shigella spp. and serotypes, we applied an epitope- and structure-based vaccine technology, MEFA (22), and constructed a polyvalent protein antigen “Shigella MEFA” by presenting epitopes from IpaB, VirG, GuaB, Shiga toxin A and B subunits on backbone protein IpaD (28). We further demonstrated that the resultant polyvalent Shigella MEFA induced broadly functional antibodies and the antibodies inhibited invasion of all Shigella species and the important serotype strains and neutralized cytotoxicity of Shiga toxins; more importantly, this Shigella MEFA protein protected mice against pulmonary lethal challenge from Shigella sonnei or Shigella flexneri 2a, 3a, or 6 strain (28).
While the epitopes of that Shigella MEFA protein functioned collectively, it is unknown if each epitope was functional individually or if an individual epitope was the better one representing the target virulence factor even if it was functional. We believe that the use of top-ranked functional epitopes from each target virulence factor (IpaB, IpaD, VirG, GuaB, and Shiga toxins) will improve Shigella MEFA in eliciting protective immunity, thus becoming a better antigen for a more protective Shigella vaccine. In addition, since both IpaD and IpaB are involved in Shigella bacterial invasion into host cells, epitopes of these two virulence factors can function synergistically; therefore, we do not know whether the representative IpaB or IpaD epitope in the previous Shigella MEFA protein is functional individually. With the functional epitopes of IpaB identified empirically in a recent study (24), we focused on functional epitope mapping for IpaD in the current study. Moreover, because IpaD was designed as the backbone of the Shigella MEFA immunogen, identifying IpaD functional epitopes will allow us to retain the top-ranked IpaD epitope on the backbone for maximum IpaD protective immunity and improve future design of an optimal Shigella MEFA immunogen by substituting the less or not functional IpaD epitopes with the top-ranked functional epitopes from the other target virulence factors.
Data from this study indicated that each IpaD-epitope fusion protein elicited robust anti-IpaD antibody responses, but sera from the mice immunized with EP3, EP4, EP6, EP7, or EP8 epitope fusion were significantly less effective in inhibiting in vitro invasion of S. sonnei and S. flexneri 2a. By contrast, the fusion proteins with EP1 (IpaD 179–188), EP2 (IpaD 263–274), or EP5 (IpaD 16–27) were as effective as the IpaD recombinant protein in inducing functional antibodies against S. flexneri 2a invasion. Similarly, sera from the mice immunized with EP1 fusion, EP5 fusion, or recombinant IpaD showed similar inhibitory activities against in vitro invasion of S. sonnei. These results suggest that IpaD epitope 1 and epitope 5 can serve as surrogate antigens of IpaD to induce functional antibodies against this key virulence factor.
Early studies suggested that peptides nearby at or overlapping with EP1 or EP2 are important against Shigella invasion. Monoclonal antibody “IpaD-318” was demonstrated to directly bind and neutralize S. flexneri 2a invasion into HeLa cells (16). The epitope associated with MAb IpaD-318 was suggested to be within residues 273–278 and the area between 175 and 191 of IpaD (16). Interestingly, the EP1 (IpaD 179–188) and EP2 (IpaD 263–274) share overlapping regions with the reported binding areas of MAb IpaD-318. In addition, early site-directed mutagenesis studies showed that a deletion of IpaD peptide 161–180 (29), 171–180 (30), or 181–190 (30), or substitution of IpaD N186Y significantly impaired Shigella invasion (31). These peptide deletions or the substitution occurred within the region of EP1 (179-188), supporting the important role of epitope EP1 in Shigella invasion and the selection of this epitope as a vaccine antigen for immunity against Shigella invasion.
The current study found that EP2 (IpaD 263–274) induced functional antibodies against S. flexneri 2a invasion. The importance of peptides in this site was also suggested by early studies. It was demonstrated that mutations, regardless they were deletions or substitutions, including deletion of IpaD peptide 241–280 (29), 261–270, or 271–280 (30), or alterations of a residue at positions N273I and Q277L (31), consistently led to an impairment in bacteria invasion. Peptide IpaD 192–267, which overlaps with EP2 (263-274), was also suggested to be the binding site for IpaB since the removal of this domain eliminated the interaction of IpaD with IpaB, a requirement for T3SS function and Shigella bacteria invasion (32).
Results from the current study, while they agree with some early studies, also challenge other observations from previous studies. Specifically, earlier studies showed that deletions of IpaD peptide 1–20 (29), 221–230 (30), 231–240 (30), 281–290 (30), or 281–320 (29) significantly altered Shigella invasion. In this study, we found that antibodies derived from the fusion with EP3 (IpaD 284–293, overlaps with 281–290 and 281–320), EP6 (IpaD5-14, overlaps with 1–20), EP7 (IpaD298-309, overlaps with 281–320), or EP8 (IpaD 225–235, overlaps with 221–230 and 231–240) did not significantly affect in vitro invasion of either Shigella strain tested (S. sonnei, S. flexneri 2a). On the other hand, our data support other early studies, which indicated residue substitution at the N292Y (within EP3) (31), Y301A and N305A (within EP7) (33), W226A (within EP8) (33) not significant in affecting Shigella invasion. We found that EP3 (IpaD 284–293, including N292), EP7 (IpaD 298–309, including Y301 and N305), and EP8 (IpaD225-235, including W226) play a less important role in Shigella sonnei or Shigella flexneri 2a invasion. While the exact causes of these discrepancies among these early studies remain unclear to us, one possible reason could be the structural or antigenic alteration resulting from residue deletion in the region, since a peptide deletion could more drastically alter the antigenicity when compared to a substitution mutation (34).
Data from this study showed that EP5 (IpaD16-27) plays a role in inducing functional antibodies against Shigella bacteria invasion. EP5 is located in the N-terminal which is poorly understood. An early study showed that, unlike deletions in the C-terminus, truncation of the N terminus resulted in a less severe attenuation in Shigella invasion ability (29). However, other studies demonstrated that IpaD N-terminus induced protective immunity against Shigella infection in the Sereny test and suggested that antibodies derived from IpaD N terminus could potentially block Shigella invasion (35–37).
In conclusion, data from this study indicated that antibodies derived from fusion proteins with epitope EP1 (179-188), EP2 (263-274), or EP5 (IpaD16-27) inhibited Shigella sonnei or flexneri 2a invasion in vitro. We need to point out that the binding structure of IpaD to the functional antibodies is yet to be determined, future monoclonal antibody production and mapping studies, or better in vivo studies such as immunizing mice with the epitope fusion antigen and then challenging mice with Shigella bacteria in a pulmonary model, should help us to further validate IpaD functional or protective epitopes. Also, we focused on B-cell epitopes in the current study, T-cell epitopes which play a role in lasting immunity will need to be examined in future studies. In addition, we tested the two most important species in this study, whether antibodies from these epitopes also protect against invasion from other Shigella spp. or other serotypes is yet to be determined. Nevertheless, results from this study could be significant for understanding the antigenicity of IpaD, and the functional epitopes identified from this study will guide us to optimize the Shigella MEFA immunogen and develop a better protective vaccine against shigellosis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers
Bacteria and plasmids used in this study are listed in Table 1. E. coli DH5α (Promega; Madison, WI) was used to host the plasmid carrying each IpaD epitope fusion gene, and BL21(DE3) (Agilent Technologies, Santa Clara, CA) was used for epitope fusion protein expression. S. sonnei 53g (9904) and S. flexneri 2a 2457T (9905) provided by Dr. David A. Sack at the Johns Hopkins University and Dr. Eileen Barry at the University of Maryland School of Medicine, respectively, were used in antibody invasion inhibition assays. Plasmid 9765, pET28α vector with ETEC adhesin CS4 major subunit csaB gene (38), was used as the template to construct IpaD epitope fusion genes. All IpaD epitope fusion genes were cloned into vector pET28a.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source/reference |
|---|---|---|
| E. coli strains | ||
| DH5α | fhuA2 Δ(argF-lacZ) U169 phoA glnV44 φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | Promega |
| BL21(DE3) | huA2 Δ(argF-lacZ) U169 phoA glnV44 φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | Agilent |
| 9765 | CS4 major subunit (CsaB) in pET28α/ DH5α, Kan+ | (38) |
| 9736 | IpaD in pET28α/BL21, Kan+ | This study |
| 9933 | IpaD-epitope-1-CsaB in pET28α/BL21 | This study |
| 9934 | IpaD-epitope-2-CsaB in pET28α/BL21 | This study |
| 9935 | IpaD-epitope-3-CsaB in pET28α/BL21 | This study |
| 9936 | IpaD-epitope-4-CsaB in pET28α/BL21 | This study |
| 9939 | IpaD-epitope-5-CsaB in pET28α/BL21 | This study |
| 9940 | IpaD-epitope-6-CsaB in pET28α/BL21 | This study |
| 9941 | IpaD-epitope-7-CsaB in pET28α/BL21 | This study |
| 9942 | IpaD-epitope-8-CsaB in pET28α/BL21 | This study |
| Shigella strains | ||
| 9904 | Shigella sonnei 53G | Johns Hopkins University |
| 9905 | Shigella flexneri 2457T | University of Maryland |
| Plasmid | ||
| pET28α | Protein expression vector | Novagen |
In silico prediction of IpaD B-cell immunodominant epitopes and construction of epitope fusion genes
IpaD protein continuous B-cell immunodominant epitopes were in silico predicated first based on IpaD amino acid sequence (accession: NP_858259), using BepiPred-2.0 software (25). As previously described (22, 24), amino acid sequences of the predicted epitopes were examined and confirmed to be conserved among S. dysenteriae Sd197, S. boydii, S. sonnei, and S. flexneri 2a. Epitopes were ranked based on antigenicity scores and the top eight epitopes were selected and designed as epitopes 1 through 8 (EP1 to EP8). IpaD protein structure was visualized with UCSF ChimeraX-1.6.1 (39), and each epitope was verified for surface exposure and localized on the IpaD protein model (ID ma-rf0iy) using the Protein Homology/AnalogY Recognition Engine V 2.0 (Phyre 2) (40).
Plasmid 9765 with the csaB gene cloned in pET28a was used as the DNA template for IpaD epitope fusion gene construction. Nucleotides coding each of the eight selected IpaD epitopes were inserted into the csaB gene using splicing overlap extension (SOE) PCR as previously described (24), with specific PCR primers (Table 2). Each CsaB-IpaD epitope fusion gene was cloned into pET28α, at NheI and EagI restriction enzyme sites for tag-less recombinant proteins, and the resultant plasmid was hosted by E. coli DH5a or BL21 (DE3) strain.
TABLE 2.
PCR primers used to insert each IpaD epitope into E. coli CS4 adhesin major subunit gene csaB in the study
| Primer | Sequence (5′−3′) | Amplified region |
|---|---|---|
| IpaD-F | CGGGCTAGCATGAATATAACAACTCTGACTAA | Upstream of ipaD, with NheI site |
| IpaD-R | CGGCCGTCAGAAATGGAGAAAAAGTT | Downstream of ipaD, with EagI site |
| Ep1F | CGGAGGTAACGACGGAAACTCCGTG ttagtgattggtgcgactac | Paired with CsaB-R to insert IpaD-ep1 into CsaB |
| Ep1R | AGTTTCCGTCGTTACCTCCGGGAGA tgacgttccaaaatttaatt | Paired with CsaB-F to insert IpaD-ep1 into CsaB |
| Ep2F | AAATGGCGAGGTTGTGCTAGATAATGCA ttagtgattggtgcgactac | Paired with CsaB-R to insert IpaD-ep2 into CsaB |
| Ep2R | CTAGCACAACCTCGCCATTTCCACCTAG tgacgttccaaaatttaatt | Paired with CsaB-F to insert IpaD-ep2 into CsaB |
| Ep3F | CGAAGATGAAACAATGAAAAATAAT ttagtgattggtgcgactac | Paired with CsaB-R to insert IpaD-ep3 into CsaB |
| Ep3R | TTTTCATTGTTTCATCTTCGGCAGA tgacgttccaaaatttaatt | Paired with CsaB-F to insert IpaD-ep3 into CsaB |
| Ep4F | TCCAATTAATAAAGACGCAAGAGAA ttagtgattggtgcgactac | Paired with CsaB-R to insert IpaD-ep4 into CsaB |
| Ep4R | TTGCGTCTTTATTAATTGGATATTC tgacgttccaaaatttaatt | Paired with CsaB-F to insert IpaD-ep4 into CsaB |
| Ep5F | CAATACCAACGGTTCATCAACCGAAACA ttagtgattggtgcgactac | Paired with CsaB-R to insert IpaD-ep5 into CsaB |
| Ep5R | TTGATGAACCGTTGGTATTGTTTGGACT tgacgttccaaaatttaatt | Paired with CsaB-F to insert IpaD-ep5 into CsaB |
| Ep6F | GACTAATAGTATTTCCACCTCATCA ttagtgattggtgcgactac | Paired with CsaB-R to insert IpaD-ep6 into CsaB |
| Ep6R | AGGTGGAAATACTATTAGTCAGAGT tgacgttccaaaatttaatt | Paired with CsaB-F to insert IpaD-ep6 into CsaB |
| Ep7F | ATACAGTAATGCCAATAGTATTTTTGAT ttagtgattggtgcgactac | Paired with CsaB-R to insert IpaD-ep7 into CsaB |
| Ep7R | TACTATTGGCATTACTGTATTTTTGAAC tgacgttccaaaatttaatt | Paired with CsaB-F to insert IpaD-ep7 into CsaB |
| Ep8F | CTTACAGAATTAGGTGGAACAATCGGC ttagtgattggtgcgactac | Paired with CsaB-R to insert IpaD-ep8 into CsaB |
| Ep8R | TGTTCCACCTAATTCTGTAAGCCATTT tgacgttccaaaatttaatt | Paired with CsaB-F to insert IpaD-ep8 into CsaB |
| CsaB-F | CTAGCTAGCgtagagaaaaatatcactgtaa | Forward primer to amplify csaB gene |
| CsaB-R | TCACGGCCGTTA ttatgatgctaaggtcattaa | Reverse primer to amplify csaB gene |
IpaD epitope fusion protein expression, purification, and characterization
Tag-less epitope fusion proteins were expressed, extracted, and refolded as we described previously (23, 24, 41). Briefly, a single overnight-grown colony of IpaD epitope fusion strain (in E. coli BL21 DE3) was cultured overnight in Luria-Bertani (LB) broth supplemented with kanamycin (30 µg/mL), then sub-cultured in fresh 2xYT broth (kanamycin 30 µg/mL) and grown to the log phase (OD600 ranging from 0.5 to 0.7) in a shaking incubator (37°C, 200 rpm). Bacteria were induced with 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG; Sigma, MO) for approximately 4 h, then harvested with centrifugation (13,000 × g, for 15 min at 4°C), and stored in a −80°C freezer overnight. Bacteria pellets were thawed on ice, suspended with 10 mL bacterial protein extraction reagent (B-PER, in phosphate buffer, pH 7.5; Thermo Fisher Scientific, Rochester, NY), further lysed with lysozyme (200 µg/mL), and sonication (130W, 20 kHz for 15 min by Ultrasonic processor GEX130; Cole Parmer, Vernon Hills, IL). Lysates were collected for extraction of inclusion body proteins by following the manufacturer’s protocol (Thermo Fisher Scientific).
Inclusion body proteins were solubilized in 50 mM CAPS [3-(cyclohexylamine) propane sulfonic acid, pH11.0; Sigma] supplemented with 0.3% N-lauroylsarcosine and 1 mM dithiothreitol (DTT). Solubilized proteins in Spectrum membrane tubing (6–8 kDa; Fisher Scientific) were refolded with dialysis buffer 1M Tris-HCl (pH 8.5) in a cold room, with 2× exchanges of the buffer (8 h per exchange). Refolded proteins were collected, aliquoted, examined in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) Coomassie blue staining, and characterized in Western blotting using anti-IpaBD fusion antibodies (42).
Mouse intramuscular immunization with IpaD epitope fusion proteins
A total of 10 groups of 8-week-old BALB/c female mice (Charles River Laboratories, Wilmington, MA), 5 mice per group, were included in the immunization study. Eight groups were administrated intramuscularly the eight IpaD epitope fusion proteins respectively, 40 µg fusion protein per mouse. One group was immunized with 40 µg IpaD recombinant IpaD as the positive control, and another group with 1× PBS as the control. Double mutant heat-labile toxin (dmLT, LTR192G/L211A; a gift from Dr. Jacob Bitoun at Tulane University), at a dose of 0.2 µg, was included in all groups. One primary was followed by two boosters, at an interval of 2 weeks. Two weeks after the final booster, all mice were sacrificed; blood samples were collected from each mouse, and sera were stored at −80°C until use. The mouse study (protocol #22088) was approved by the IACUC at the University of Illinois at Urbana-Champaign.
Mouse serum anti-IpaD IgG titration
Serum samples from each mouse were examined for antibody response to IpaD in enzyme-linked immunosorbent assay (ELISA) as described previously (43). Briefly, 100 ng IpaD recombinant protein, in 100 µL bicarbonate/carbonate antigen coating buffer (50 mM), was coated to each well of 96-well Immulon 2-HB plates (Thermo Fisher Scientific). After incubation at 37°C for 1 h and 4°C overnight, wells were blocked with 10% skim milk (in PBST), and then incubated with twofold serum dilutions (1:400 to 1:25,600), in triplicates. Wells were washed with PBST and incubated with horse radish peroxidase (HRP)-conjugated goat anti-mouse IgG secondary antibody (Sigma, St. Louis, MO) at 37°C for 1 h. After washes (3×) with PBST, wells were added with 3,3,5,5-tetramethylbenzidine (TMB) substrate (KPL, Gaithersburg, MD) and incubated at room temperature for 20 min. OD was read at a wavelength of 650 nm with a SPECTROstarNano spectrometer (BMG LABTECH, Germany). OD650 readings at the highest serum dilution (>0.3 after subtraction of background readings) were converted to antibody titers and expressed in log10 as previously described (43).
Mouse serum antibody invasion inhibition assay to measure antibody function against Shigella bacteria invasion
Shigella bacteria invade Hela cells; functional antibodies derived from IpaD or IpaD epitopes block Shigella invasion in vitro. As described previously (24), mouse sera from each group, after inactivation at 56°C for 30 min, were mixed with S. sonnei or S. flexneri 2a (~ 2×107 CFUs, colony forming units). After 25 min at room temperature, each serum and bacteria mixture was transferred to 95%–100% confluent monolayered HeLa cells, a gift from Dr. Dongwan Yoo at the University of Illinois Urbana-Champaign and cultured in wells of a 24-well tissue culture plate. After 2 h of incubation, cells were rinsed with PBS and then treated with DMEM containing gentamicin (300 µg/mL, Sigma) to eliminate extracellular bacteria. Cells were then rinsed and lysed with 0.5% Triton X-100 solution to release the invaded intracellular Shigella bacteria. Lysates were suspended with PBS, serially diluted, and plated on LB agar plates. After overnight culture at 37°C, CFUs were counted and converted to a percentage by referring the CFUs from bacteria treated with the control mouse sera as 100%.
Statistical analyses
Statistical analyses were performed using the GraphPad Prism version 7.0.0 (GraphPad Software, San Diego, California, USA). Two-way analysis of variance (ANOVA) was performed, and Bonferroni’s multiple comparisons test was used to examine the statistical difference in antibody invasion inhibition across treatment groups. A P < 0.05 indicated a difference statistically significant. Error bars represent standard deviations from the means.
ACKNOWLEDGMENTS
The authors thank Drs. David A. Sack (Johns Hopkins University) and Eileen M. Barry (University of Maryland) for providing us with Shigella sonnei or S. flexneri 2a, Jacob P. Bitoun (Tulane University) for adjuvant dmLT, Wendy Picking (University of Kansas) for anti-IpaDB mouse sera, and Dongwan Yoo (University of Illinois at Urbana-Champaign) for Hela cells. Chongyang Zhang and Shafiullah MD Parvej provided technical assistance.
Financial support for this study was provided by NIH R01 AI175214-01.
Contributor Information
Weiping Zhang, Email: wpzhang@illinois.edu.
Christopher A. Elkins, Centers for Disease Control and Prevention, Atlanta, Georgia, USA
REFERENCES
- 1. Organization WH . 2021. WHO preferred product characteristics for vaccines against Shigella. Available from: https://www.who.int/publications/i/item/9789240036741
- 2. Hosangadi D, Smith PG, Kaslow DC, Giersing BK, Armah G, Black R, Chilengi R, Guerrant DL, Houpt E, Khalil I, Kirkwood C, Kotloff K, Lamberti L, Mok W, Muhib F, Silva CP, Rheingans R, Riddle M, Walker R, Cons WESV . 2019. WHO consultation on ETEC and Shigella burden of disease, Geneva, 6-7th April 2017: meeting report. Vaccine 37:7381–7390. doi: 10.1016/j.vaccine.2017.10.011 [DOI] [PubMed] [Google Scholar]
- 3. Rogawski ET, Liu J, Platts-Mills JA, Kabir F, Lertsethtakarn P, Siguas M, Khan SS, Praharaj I, Murei A, Nshama R, et al. 2018. Use of quantitative molecular diagnostic methods to investigate the effect of enteropathogen infections on linear growth in children in low-resource settings: longitudinal analysis of results from the MAL-ED cohort study. Lancet Glob Health 6:e1319–e1328. doi: 10.1016/S2214-109X(18)30351-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kasumba IN, Pulford CV, Perez-Sepulveda BM, Sen S, Sayed N, Permala-Booth J, Livio S, Heavens D, Low R, Hall N, et al. 2021. Characteristics of Salmonella recovered from stools of children enrolled in the global enteric multicenter study. Clin Infect Dis 73:631–641. doi: 10.1093/cid/ciab051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rogawski McQuade ET, Platts-Mills JA, Gratz J, Zhang J, Moulton LH, Mutasa K, Majo FD, Tavengwa N, Ntozini R, Prendergast AJ, Humphrey JH, Liu J, Houpt ER. 2020. Impact of water quality, sanitation, handwashing, and nutritional interventions on enteric infections in rural Zimbabwe: the sanitation hygiene infant nutrition efficacy (SHINE) trial. J Infect Dis 221:1379–1386. doi: 10.1093/infdis/jiz179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lorntz B, Soares AM, Moore SR, Pinkerton R, Gansneder B, Bovbjerg VE, Guyatt H, Lima AM, Guerrant RL. 2006. Early childhood diarrhea predicts impaired school performance. Pediatr Infect Dis J 25:513–520. doi: 10.1097/01.inf.0000219524.64448.90 [DOI] [PubMed] [Google Scholar]
- 7. Soni A, Fahey N, Bhutta ZA, Li W, Frazier JA, Moore Simas T, Nimbalkar SM, Allison JJ. 2021. Early childhood undernutrition, preadolescent physical growth, and cognitive achievement in India: a population-based cohort study. PLoS Med 18:e1003838. doi: 10.1371/journal.pmed.1003838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Libby TE, Delawalla MLM, Al-Shimari F, MacLennan CA, Vannice KS, Pavlinac PB. 2023. Consequences of Shigella infection in young children: a systematic review. Int J Infect Dis 129:78–95. doi: 10.1016/j.ijid.2023.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Riewpaiboon A, Youngkong S, Sreshthaputra N, Stewart JF, Samosornsuk S, Chaicumpa W, von Seidlein L, Clemens JD. 2008. A cost function analysis of shigellosis in Thailand. Value Health 11 Suppl 1:S75–83. doi: 10.1111/j.1524-4733.2008.00370.x [DOI] [PubMed] [Google Scholar]
- 10. Anderson JD, Bagamian KH, Muhib F, Amaya MP, Laytner LA, Wierzba T, Rheingans R. 2019. Burden of enterotoxigenic Escherichia coli and shigella non-fatal diarrhoeal infections in 79 low-income and lower middle-income countries: a modelling analysis. Lancet Glob Health 7:e321–e330. doi: 10.1016/S2214-109X(18)30483-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mattock E, Blocker AJ. 2017. How do the virulence factors of Shigella work together to cause disease? Front Cell Infect Microbiol 7:64. doi: 10.3389/fcimb.2017.00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brunner K, Samassa F, Sansonetti PJ, Phalipon A. 2019. Shigella-mediated immunosuppression in the human gut: subversion extends from innate to adaptive immune responses. Hum Vaccin Immunother 15:1317–1325. doi: 10.1080/21645515.2019.1594132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu B, Morado DR, Margolin W, Rohde JR, Arizmendi O, Picking WL, Picking WD, Liu J. 2015. Visualization of the type III secretion sorting platform of Shigella flexneri. Proc Natl Acad Sci USA 112:1047–1052. doi: 10.1073/pnas.1411610112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Muthuramalingam M, Whittier SK, Picking WL, Picking WD. 2021. The Shigella type III secretion system: an overview from top to bottom. Microorganisms 9:451. doi: 10.3390/microorganisms9020451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nothelfer K, Arena ET, Pinaud L, Neunlist M, Mozeleski B, Belotserkovsky I, Parsot C, Dinadayala P, Burger-Kentischer A, Raqib R, Sansonetti PJ, Phalipon A. 2014. B lymphocytes undergo TLR2-dependent apoptosis upon Shigella infection. J Exp Med 211:1215–1229. doi: 10.1084/jem.20130914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sierocki R, Jneid B, Orsini Delgado ML, Plaisance M, Maillère B, Nozach H, Simon S. 2021. An antibody targeting type III secretion system induces broad protection against Salmonella and Shigella infections. PLoS Negl Trop Dis 15:e0009231. doi: 10.1371/journal.pntd.0009231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Espina M, Olive AJ, Kenjale R, Moore DS, Ausar SF, Kaminski RW, Oaks EV, Middaugh CR, Picking WD, Picking WL. 2006. IpaD localizes to the tip of the type III secretion system needle of Shigella flexneri. Infect Immun 74:4391–4400. doi: 10.1128/IAI.00440-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barta ML, Shearer JP, Arizmendi O, Tremblay JM, Mehzabeen N, Zheng Q, Battaile KP, Lovell S, Tzipori S, Picking WD, Shoemaker CB, Picking WL. 2017. Single-domain antibodies pinpoint potential targets within Shigella invasion plasmid antigen D of the needle tip complex for inhibition of type III secretion. J Biol Chem 292:16677–16687. doi: 10.1074/jbc.M117.802231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Espina M, Olive AJ, Kenjale R, Moore DS, Ausar SF, Kaminski RW, Oaks EV, Middaugh CR, Picking WD, Picking WL. 2006b. IpaD localizes to the tip of the type III secretion system needle of Shigella flexneri. Infect Immun 74:4391–4400. doi: 10.1128/IAI.00440-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jneid B, Rouaix A, Féraudet-Tarisse C, Simon S. 2020. SipD and IpaD induce a cross-protection against Shigella and Salmonella infections. PLoS Negl Trop Dis 14:e0008326. doi: 10.1371/journal.pntd.0008326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martinez-Becerra FJ, Kissmann JM, Diaz-McNair J, Choudhari SP, Quick AM, Mellado-Sanchez G, Clements JD, Pasetti MF, Picking WL. 2012. Broadly protective Shigella vaccine based on type III secretion apparatus proteins. Infect Immun 80:1222–1231. doi: 10.1128/IAI.06174-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li S, Lee KH, Zhang W. 2022. Methods Mol Biol, p 151–169. In Bidmos Fadil (ed), Multiepitope fusion antigen: MEFA, an Epitope- and structure-based vaccinology platform for multivalent vaccine development. Vol. 2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li S, Anvari S, Ptacek G, Upadhyay I, Kaminski RW, Sack DA, Zhang W. 2023. A broadly immunogenic polyvalent Shigella multiepitope fusion antigen protein protects against Shigella sonnei and Shigella flexneri lethal pulmonary challenges in mice. Infect Immun 91:e0031623. doi: 10.1128/iai.00316-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li S, Han X, Upadhyay I, Zhang W. 2022. Characterization of functional B-cell epitopes at the amino terminus of Shigella invasion plasmid antigen B (IpaB). Appl Environ Microbiol 88:e0038422. doi: 10.1128/aem.00384-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jespersen MC, Peters B, Nielsen M, Marcatili P. 2017. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 45:W24–W29. doi: 10.1093/nar/gkx346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schlingmann B, Castiglia KR, Stobart CC, Moore ML. 2018. Polyvalent vaccines: high-maintenance heroes. PLoS Pathog 14:e1006904. doi: 10.1371/journal.ppat.1006904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Okuno Y, Isegawa Y, Sasao F, Ueda S. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 67:2552–2558. doi: 10.1128/JVI.67.5.2552-2558.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li S, Seo H, Upadhyay I, Zhang W. 2023. A polyvalent adhesin-toxoid multiepitope-fusion-antigen-induced functional antibodies against five enterotoxigenic Escherichia coli adhesins (CS7, CS12, CS14, CS17, and CS21) but not enterotoxins (LT and STa). Microorganisms 11:2473. doi: 10.3390/microorganisms11102473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Picking WL, Nishioka H, Hearn PD, Baxter MA, Harrington AT, Blocker A, Picking WD. 2005. IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect Immun 73:1432–1440. doi: 10.1128/IAI.73.3.1432-1440.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schiavolin L, Meghraoui A, Cherradi Y, Biskri L, Botteaux A, Allaoui A. 2013. Functional insights into the Shigella type III needle tip IpaD in secretion control and cell contact. Mol Microbiol 88:268–282. doi: 10.1111/mmi.12185 [DOI] [PubMed] [Google Scholar]
- 31. Roehrich AD, Guillossou E, Blocker AJ, Martinez-Argudo I. 2013. Shigella IpaD has a dual role: signal transduction from the type III secretion system needle tip and intracellular secretion regulation. Mol Microbiol 87:690–706. doi: 10.1111/mmi.12124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson S, Roversi P, Espina M, Olive A, Deane JE, Birket S, Field T, Picking WD, Blocker AJ, Galyov EE, Picking WL, Lea SM. 2007. Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J Biol Chem 282:4035–4044. doi: 10.1074/jbc.M607945200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meghraoui A, Schiavolin L, Allaoui A. 2014. Single amino acid substitutions on the needle tip protein IpaD increased Shigella virulence. Microbes Infect 16:532–539. doi: 10.1016/j.micinf.2014.03.010 [DOI] [PubMed] [Google Scholar]
- 34. Studer RA, Dessailly BH, Orengo CA. 2013. Residue mutations and their impact on protein structure and function: detecting beneficial and pathogenic changes. Biochem J 449:581–594. doi: 10.1042/BJ20121221 [DOI] [PubMed] [Google Scholar]
- 35. Jahantigh D, Saadati M, Fasihi Ramandi M, Mousavi M, Zand AM. 2014. Novel intranasal vaccine delivery system by chitosan nanofibrous membrane containing N-terminal region of IpaD antigen as a nasal Shigellosis vaccine, studies in Guinea pigs. Journal of Drug Delivery Science and Technology 24:33–39. doi: 10.1016/S1773-2247(14)50005-6 [DOI] [Google Scholar]
- 36. Arianzad SA, Zeinoddini M, Haddadi A, Nazarian S, Sajedi RH. 2020. In Silico design of chimeric and immunogenic protein-containing IpaB and IpaD as a vaccine candidate against Shigella dysenteriae. Current Proteomics 17:333–341. doi: 10.2174/1570164617666190906145843 [DOI] [Google Scholar]
- 37. Hesaraki M, Saadati M, Honari H, Olad G, Heiat M, Malaei F, Ranjbar R. 2013. Molecular cloning and biologically active production of IpaD N-terminal region. Biologicals 41:269–274. doi: 10.1016/j.biologicals.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 38. Ruan X, Knudsen DE, Wollenberg KM, Sack DA, Zhang W. 2014. Multiepitope fusion antigen induces broadly protective antibodies that prevent adherence of Escherichia coli strains expressing colonization factor antigen I (CFA/I), CFA/II, and CFA/IV. Clin Vaccine Immunol 21:243–249. doi: 10.1128/CVI.00652-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pettersen EF, Goddard TD, Huang CRC, Meng EEC, Couch GS, Croll TI, Morris JH, Ferrin TE. 2021. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30:70–82. doi: 10.1002/pro.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seo H, Garcia C, Ruan X, Duan Q, Sack DA, Zhang W. 2021. Preclinical characterization of immunogenicity and efficacy against diarrhea from MecVax, a multivalent enterotoxigenic E. coli vaccine candidate. Infect Immun 89:e0010621. doi: 10.1128/IAI.00106-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Martinez-Becerra FJ, Chen X, Dickenson NE, Choudhari SP, Harrison K, Clements JD, Picking WD, Van De Verg LL, Walker RI, Picking WL. 2013. Characterization of a novel fusion protein from IpaB and IpaD of Shigella spp. and its potential as a pan-Shigella vaccine. Infect Immun 81:4470–4477. doi: 10.1128/IAI.00859-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lu T, Moxley RA, Zhang W. 2019. Mapping the neutralizing epitopes of enterotoxigenic Escherichia coli K88 (F4) fimbrial adhesin and major subunit FaeG. Appl Environ Microbiol 85:e00329-19. doi: 10.1128/AEM.00329-19 [DOI] [PMC free article] [PubMed] [Google Scholar]


