Abstract
We have demonstrated previously that the EGFR (epidermal growth factor receptor) is a calmodulin (CaM)-binding protein. To establish whether or not the related receptor ErbB2/Neu/HER2 also binds CaM, we used human breast adenocarcinoma SK-BR-3 cells, because these cells overexpress this receptor thus facilitating the detection of this interaction. In the present paper, we show that ErbB2 could be pulled-down using CaM–agarose beads in a Ca2+-dependent manner, as detected by Western blot analysis using an anti-ErbB2 antibody. ErbB2 was also isolated by Ca2+-dependent CaM-affinity chromatography. We also demonstrate using an overlay technique with biotinylated CaM that CaM binds directly to the immunoprecipitated ErbB2. The binding of biotinylated CaM to ErbB2 depends strictly on the presence of Ca2+, since it was prevented by the presence of EGTA. Moreover, the addition of an excess of free CaM prevents the binding of its biotinylated form, demonstrating that this was a specific process. We excluded any interference with the EGFR, as SK-BR-3 cells express considerably lower levels of this receptor, and no detectable EGFR signal was observed by Western blot analysis in the immunoprecipitated ErbB2 preparations used to perform the overlay assays with biotinylated CaM. We also demonstrate that treating living cells with W7 [N-(6-aminohexyl)-5-chloro-1-naphthalenesulphonamide], a cell-permeant CaM antagonist, down-regulates ErbB2 phosphorylation, and show that W7 does not interfere non-specifically with the activity of ErbB tyrosine kinases. We also show that W7 inhibits the phosphorylation (activation) of both ERK1/2 (extracellular-signal-regulated kinases 1 and 2) and Akt/PKB (protein kinase B), in accordance with the inhibition observed in ErbB2 phosphorylation. In contrast, W7 treatment increased the phosphorylation (activation) of CREB (cAMP-response-element-binding protein) and ATF1 (activating transcription factor-1), two Ca2+-sensitive transcription factors that operate downstream of these ErbB2 signalling pathways, most likely because of the absence of calcineurin activity. We conclude that ErbB2 is a new CaM-binding protein, and that CaM plays a role in the regulation of this receptor and its downstream signalling pathways in vivo.
Keywords: calmodulin, calmodulin-binding protein, epidermal growth factor receptor, ErbB2/Neu/HER2 receptor
Abbreviations: ATF1, activating transcription factor-1; [Ca2+]cyt, cytosolic concentration of free calcium; CaM, calmodulin; CaM-BD, CaM-binding domain; CaMK-II, CaM-dependent protein kinase II; CaMK-IV, CaM-dependent protein kinase IV; CREB, cAMP-response-element-binding protein; DMEM, Dulbecco's modified Eagle's medium; ECL®, enhanced chemiluminescence; EGF, epidermal growth factor; EGFR, EGF receptor; ERK, extracellular-signal-regulated kinase; FBS, foetal bovine serum; HRGβ1, heregulin-β1; JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; NFAT, nuclear factor of activated T-cells; PI3K, phosphoinositide 3-kinase; PKB, protein kinase B; poly(L-Glu/L-Tyr), co-polymer of L-glutamic acid and L-tyrosine; W7, N-(6-aminohexyl)-5-chloro-1-naphthalenesulphonamide; W12, N-(4-aminobutyl)-1-naphthalenesulphonamide; W13, N-(4-aminobutyl)-5-chloro-1-naphthalenesulphonamide
INTRODUCTION
The ErbB2/Neu receptor, also named HER2, belongs to the EGFR (epidermal growth factor receptor)/ErbB1/HER1 tyrosine kinase subfamily, also formed by two additional members, ErbB3/HER3 and ErbB4/HER4 receptors [1,2]. ErbB2 plays an important role in the development of certain human cancers, particularly because it is overexpressed in adenocarcinomas of mammary and ovarian origin, among others [3,4]. Ligand-induced heterodimerization of ErbB2 with other members of the EGFR family appears to be the underlying mechanism for its activation, as a specific physiological soluble ligand for ErbB2 has not yet been identified. Thus heregulins induce the heterodimerization of ErbB2 with either ErbB3 or ErbB4, while EGF (epidermal growth factor) intervenes in the heterodimerization of ErbB2 with EGFR [5,6].
An early signal generated by the activation of the EGFR is a transient increase in [Ca2+]cyt (cytosolic concentration of free calcium) (see [7] for a review). This Ca2+ signal is also generated by EGFR–ErbB2 heterodimers, playing an important role in cell motility in this case because of the involvement in cytoskeletal reorganization [8]. Interestingly, the oscillations in [Ca2+]cyt generated by EGFR homodimers and EGFR–ErbB2 heterodimers present different patterns [9]. The calcium signal generated by ErbB receptors may have important functional implications, as the ubiquitous Ca2+-sensor calmodulin (CaM) becomes activated [10], and the Ca2+–CaM complex has been proposed to exert a regulatory action on these receptors [7,11].
In this context, we have demonstrated previously that the EGFR is a CaM-binding protein [12–16]. Although CaM is known to inactivate the EGFR via phosphorylation mediated by CaM-dependent protein kinase II (CaMK-II) [17–19], we have demonstrated that CaM also exerts a direct inhibitory action on the tyrosine kinase activity of the EGFR without the participation of this serine/threonine kinase [12,13]. It was also demonstrated that CaM binds directly to the EGFR in vitro [12,13,15] and in living cells [16], and that the CaM-binding domain (CaM-BD) of this receptor is located in its cytosolic juxtamembrane region [14,16,20]. Moreover, a reciprocal competitive interplay between CaM binding at this site and phosphorylation by protein kinase C of Thr654, also located in this segment, was demonstrated [14].
The CaM-BD of the EGFR is highly conserved among mammalian species, and presents high homology with similar regions in other ErbB receptors, including ErbB2 [11,14]. Therefore it is likely that CaM could interact with other members of the ErbB family. In the present paper, we demonstrate that ErbB2 is indeed a CaM-binding protein, and that CaM plays a role in the regulation of this receptor, and several ErbB2-initiated downstream signalling pathways, in intact cells.
EXPERIMENTAL
Reagents
Polyclonal anti-ErbB2 antibody (C-18) developed in rabbit against the C-terminus of the human receptor, mouse monoclonal anti-phospho-ERK (extracellular-signal-regulated kinase) antibody (E-4) against phosphorylated Tyr204 of human ERK1/2, rabbit polyclonal anti-Akt1/2 antibody (H-136) against a recombinant protein corresponding to amino acids 345–480 of human Akt1, rabbit polyclonal anti-phospho-CREB (cAMP-responseelement-binding protein) 1 antibody (Ser-133) [recognizing phosphorylated Ser133 of human CREB1, phosphorylated ATF1 (activating transcription factor-1)], rabbit polyclonal anti-CREB1 antibody (C-21) against a peptide mapping the C-terminus of human CREB1, mouse monoclonal anti-phospho-JNK (c-Jun N-terminal kinase) antibody (G-7) (against a peptide corresponding to human JNK1 containing phosphorylated Thr183 and Tyr185) and polyclonal rabbit anti-JNK1 antibody (C-17) (against a peptide mapping at the C-terminus of human JNK1) were from Santa Cruz Biotechnology. Monoclonal anti-EGFR antibody from clone 13 (recognizing the intracellular segment 996–1022 of the human receptor) developed in mouse, monoclonal anti-phosphotyrosine RC20 antibody conjugated to horseradish peroxidase and mouse monoclonal anti-NFAT1 antibody were obtained from BD Transduction Laboratories. Anti-phosphotyrosine 4G10 antibody was purchased from Upstate Biotechnology. Rabbit polyclonal anti-phospho-Akt (Thr308) antibody and rabbit polyclonal anti-ERK1/2 antibody were purchased from Cell Signaling Technology. Rabbit polyclonal anti-[phospho-p38 MAPK (mitogen-activated protein kinase)] antibody (against a peptide containing phosphorylated Thr180 and Tyr182 of human p38 MAPK) and rabbit polyclonal anti-(p38 MAPK) (against a peptide corresponding to residues 341–360 of human p38 MAPK) were purchased from Calbiochem. Anti-rabbit IgG (heavy and light chains) developed in goat and conjugated to horseradish peroxidase was purchased from Zymed Laboratories. Anti-mouse IgG (Fc-specific) developed in goat and conjugated to horseradish peroxidase, human recombinant HRGβ1 (heregulin-β1), CaM–agarose, deoxycholic acid (sodium salt), sodium orthovanadate, leupeptin, pepstatin A, aprotinin, PMSF, poly(L-Glu/L-Tyr) (co-polymer of L-glutamic acid and L-tyrosine; 4:1 stochiometric ratio) (20–50 kDa) and mouse monoclonal anti-α-tubulin antibody (clone DM 1A) were purchased from Sigma. Complete™ mini EDTA-free protease inhibitor tablets were obtained from Roche. Human EGF was obtained from PeproTech EC (London, U.K.), and pre-stained molecular mass standards for electrophoresis were from Bio-Rad. EZ-link™ NHS-LC-biotin and streptavidin conjugated to horseradish peroxidase were from Pierce. BioTrace™ PVDF membranes were purchased from Pall Gelman Laboratory (Mississauga, Ontario, Canada), and OptiPhase HiSafe 2 scintillation fluid was from Wallac. The ECL® (enhanced chemiluminescence) assay kit, [methyl-3H]thymidine (46 Ci/mmol) and [γ-32P]ATP (triethylammonium salt) (3000 Ci/mmol) were from Amersham Biosciences. Escherichia coli cultures expressing recombinant rat CaM [21] were a gift from Professor Nobuhiro Hayashi from Fujita Health University, Aichi, Japan. Other chemicals used in this work were of analytical grade.
Cell cultures
Human breast adenocarcinoma SK-BR-3 cells (A.T.C.C., Manassas, VA, U.S.A.) and human epidermoid carcinoma A431 cells were grown in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) FBS (foetal bovine serum), 2 mM L-glutamine and 40 μg/ml gentamicin in a humidified atmosphere of 5% (v/v) CO2 in air at 37 °C. The cells were maintained overnight in a FBS-free medium before performing the experiments.
[methyl-3H]Thymidine incorporation
Incorporation of [methyl-3H]thymidine into DNA was carried out in confluent cell cultures essentially as described in [22]. Cells grown to confluence in 24-well culture dishes and deprived of FBS overnight were washed twice with 130 mM NaCl, 2.7 mM KCl and 11.5 mM sodium/potassium phosphate (pH 7.4) (PBS) and incubated for 14–16 h in 0.5 ml of DMEM supplemented with 1.2 μM (2 μCi/ml) [methyl-3H]thymidine in the absence or presence of 10% (v/v) FBS and concentrations of EGF or HRGβ1 as indicated in the legends of the Figures. Thereafter, cells were treated with ice-cold 10% (w/v) trichloroacetic acid for 10 min, solubilized with 0.2 M NaOH for 24 h, and neutralized with 0.2 M HCl. The radioactivity incorporated into the acid-insoluble material was measured using a scintillation counter.
Purification of recombinant CaM and preparation of biotinylated CaM
Recombinant rat CaM was purified from E. coli essentially as described in [21], except that the soluble cell extract was heated at 95 °C for 5 min before the heat-resistant proteins of the supernatant were subjected to phenyl-Sepharose chromatography. The concentration of CaM was determined spectrophotometrically at 276 nm using a molar absorption coefficient of 3740 M−1·cm−1 [23]. Purified CaM was biotinylated using EZ-link™ NHS-LC-biotin as described in [24].
ErbB2 immunoprecipitation
Serum-starved confluent SK-BR-3 cells grown in dishes 15 cm in diameter were incubated in the absence and presence of 10 nM EGF or 100 ng/ml HRGβ1 for 2–5 min at room temperature (20–22 °C). Thereafter, the medium was removed and 1 ml of a medium containing 50 mM Tris/HCl (pH 8), 150 mM NaCl, 1 mM EGTA, 1% (w/v) Triton X-100, 0.5% (w/v) deoxycholic acid, 1 mM sodium orthovanadate, 1 mM PMSF, 10 μg/ml leupeptin, 10 μg/ml pepstatin A and 10 μg/ml aprotinin (lysis buffer) was added and incubation was continued for 30 min on ice. The cell extract was collected by centrifugation at 15600 g for 35 min, and 2–3 mg of solubilized proteins were incubated overnight with 4 μg of anti-ErbB2 antibody pre-coupled to 30 μl of a slurry of Protein A–agarose in 200 μl of a medium containing 50 mM Tris/HCl (pH 8), 150 mM NaCl, 1 mM sodium orthovanadate, 1 mM PMSF (TNOP buffer). The beads were collected by centrifugation at 3600 g for 40 s and washed six times in TNOP buffer. The samples were boiled for 5 min in Laemmli sample buffer, the beads were removed by centrifugation at 3600 g for 40 s, and the supernatant was processed by SDS/PAGE as described below.
ErbB2 pull-down with CaM–agarose
Serum-starved confluent SK-BR-3 cells were solubilized using the lysis buffer, the sample was centrifuged at 15600 g for 1 h at 4 °C, and 1 mM CaCl2 was added. The sample (1 ml) was incubated with a slurry of 30 μl of CaM–agarose equilibrated in a buffer containing 20 mM Hepes/NaOH (pH 7.5), 150 mM NaCl, 0.2 mM CaCl2 and 1 mM PMSF (Ca2+ buffer). Controls in the presence of 1 mM EGTA instead of CaCl2 were also performed (EGTA buffer). The CaM–agarose beads were washed six times with the Ca2+ buffer or with the EGTA buffer, as required, collecting the beads by centrifugation at 3600 g for 40 s. The material bound to the beads was separated by boiling for 5 min in Laemmli sample buffer, the beads were removed by centrifugation at 3600 g for 40 s, and the supernatant was subjected to SDS/PAGE and Western blot analysis as described below using an anti-ErbB2 antibody.
Isolation of ErbB2 by Ca2+-dependent CaM-affinity chromatography
A Triton X-100-solubilized cell extract from SK-BR-3 cells was processed through a 3 ml CaM-agarose column equilibrated with a buffer containing 25 mM Hepes/NaOH (pH 7.5), 135 mM NaCl and 0.2 mM CaCl2 (Ca2+ buffer). After an extensive wash (100 ml) with the Ca2+ buffer, CaM-binding proteins were eluted (1 ml fractions) with a similar buffer containing 2 mM EGTA instead of CaCl2 (EGTA buffer). Proteins in pooled fractions (10 ml) were precipitated with 10% (w/v) trichloroacetic acid, and subjected to SDS/PAGE and Western blot analysis as described below using an anti-ErbB2 antibody.
CaM overlay experiments
The whole cell extract or the immunoprecipitated ErbB2 receptor from SK-BR-3 cells from non-stimulated cultures, or from cultures treated with 10 nM EGF for 1 min, were prepared as described above, and subjected to SDS/PAGE as described below. The proteins were electrotransferred on to a PVDF membrane using a medium containing 48 mM Tris-base, 36.6 mM glycine, 20% (v/v) methanol and 0.04% (w/v) SDS (TGMS medium), and fixed with 0.2% (v/v) glutaraldehyde for 45 min in a medium containing 0.1% (w/v) Tween 20, 100 mM Tris/HCl (pH 8.8), 500 mM NaCl and 0.25 mM KCl (T-TBS medium). The PVDF membrane was blocked with 5% (w/v) BSA in T-TBS medium, and incubated with 0.45 μg/ml biotinylated CaM in the presence of 0.2 mM CaCl2 for 30 min at 37 °C, or overnight at 4 °C. Controls containing 1 mM EGTA instead of CaCl2 were also performed. After extensively washing the PVDF membrane in T-TBS medium, streptavidin conjugated to horseradish peroxidase at a 1/3000 dilution was added and incubated for 30 min at 37 °C or for 1 h at room temperature. The positive bands were developed using the ECL® method, following the manufacturer's instructions.
Western blot analysis
Serum-starved confluent SK-BR-3 or A431 cells grown in dishes 15 cm in diameter were incubated in the absence and presence of 10 nM EGF or 100 ng/ml HRGβ1 for 1 min at room temperature, and the whole cell extracts were processed by SDS/PAGE as indicated below. Proteins in the gel were electrotransferred on to a PVDF membrane in TGMS medium, and the proteins were fixed with 0.2% (v/v) glutaraldehyde for 45 min in T-TBS medium. The PVDF membrane was blocked with 5% (w/v) BSA or 10% (w/v) fat-free dried milk in T-TBS medium, and probed with the RC20 anti-phosphotyrosine antibody conjugated to horseradish peroxidase (1/2000 dilution), or alternatively with the 4G10 anti-phosphotyrosine antibody (1/2000 dilution), and an appropriate secondary antibody coupled to horseradish peroxidase (1/3000 dilution). The PVDF membrane was reprobed with the anti-EGFR antibody (1/2500 dilution) or the anti-ErbB2 antibody (1/1000 dilution) after stripping the membrane in a medium containing 100 mM 2-mercapthoethanol, 2% (w/v) SDS and 62.5 mM Tris/HCl (pH 6.7) at 50 °C for 30 min, or after overnight incubation at room temperature. The activation status of different signalling pathways in SK-BR-3 cells incubated in the absence and presence of 10 nM HRGβ1 was determined in whole cell extracts by probing the PVDF membranes with the following antibodies (1/1000 dilution): anti-phospho-ERK, anti-phospho-Akt/PKB (protein kinase B), anti-(phospho-p38 MAPK), antiphospho-JNK and anti-phospho-CREB/phospho-ATF1. Appropriate secondary antibodies coupled to horseradish peroxidase at a 1/3000 dilution were used as required. The positive bands were developed using the ECL® method following the manufacturer's instructions. Adequate loading was ascertained by Fast Green FCF protein staining of the PVDF membranes, or, alternatively, by stripping and reproving the membranes with any of the following antibodies (1/1000 dilution): anti-ErbB2, anti-α-tubulin, anti-ERK, anti-Akt/PKB, anti-(p38 MAPK), anti-JNK and anti-CREB.
Preparation of solubilized membrane fractions
Cells were washed twice with PBS and broken by six forced passages through a 25 G 5/8 needle in a buffer containing 50 mM Hepes/NaOH (pH 7.4), 150 mM NaCl, 10 mM EDTA, 2 mM sodium orthovanadate, and a cocktail of protease inhibitors (1 tablet for 10 ml) (buffer A). The sample was centrifuged at 600 g for 5 min, the pellet was discarded, the supernatant was resuspended in buffer A, and centrifuged at 160000 g for 1 h at 4 °C. The new pellet was collected and solubilized in buffer A containing 1% (w/v) Triton X-100 on ice for 45 min, centrifuged again at 160000 g for 1 h at 4 °C and the supernatant, denoted solubilized membrane fraction, was collected.
Isolation of EGFR and tyrosine kinase assay
In order to test possible non-specific interfering effects of W7 [N-(6-aminohexyl)-5-chloro-1-naphthalenesulphonamide] on ErbB receptor tyrosine kinase activity, rat liver EGFR was isolated by Ca2+-dependent CaM-affinity chromatography as described previously [12], and its auto(trans)phosphorylation and tyrosine kinase activity toward an exogenous substrate was assayed in the absence and presence of the CaM antagonist (15 μg/ml) using poly(L-Glu/L-Tyr), following described protocols [12].
Other analytical procedures
Slab gel electrophoreses were performed according to Laemmli [25] at 12 mA overnight in a 5–20% (w/v) linear gradient polyacrylamide gel in the presence of 0.1% (w/v) SDS at pH 8.3. Protein concentrations were determined using BCA (bicinchoninic acid; Sigma) and copper sulphate (BCA method) following the manufacturer's instructions and using BSA as a standard. Densitometric measurement of the different bands in the X-ray films were performed by a computer-assisted densitometer (Epson Perfection 1200U) using the Scion Image Release Beta 4.0.2 software program. All densitometric values given in the present work are related to the anti-phosphotyrosine/anti-ErbB2 signal ratio to correct for possible deviation due to loading errors. Little, if any, loading error, however, was observed by staining proteins in the PVDF membranes with Fast Green FCF.
RESULTS
ErbB2 activation in SK-BR-3 cells
We have demonstrated previously that CaM interacts and regulates the EGFR in vitro [12–15] and in living cells [16]. Therefore it was important to exclude the possibility that this receptor could interfere with the undertaken studies on the interaction between CaM and ErbB2, as both receptors have very close molecular masses (170 kDa for EGFR and 185 kDa for ErbB2). We chose SK-BR-3 cells for the present study, as this cell line is known to overexpress ErbB2 while expressing far lower levels of other ErbB receptors, including the EGFR [26].
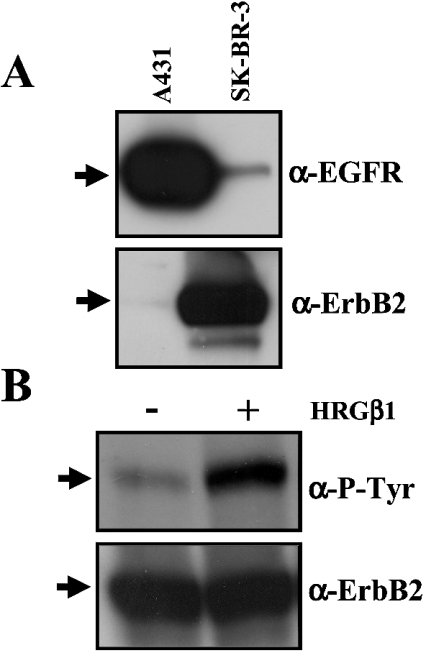
To confirm that this was the case in the clone of SK-BR-3 cells used, we performed Western blot analysis of whole cell extracts using anti-EGFR and anti-ErbB2 antibodies. Thus Figure 1(A) shows a weak EGFR signal on SK-BR-3 cells as compared with control A431 cells, which are known to overexpress this receptor [27]. In order to observe the weak EGFR signal in SK-BR-3 cells, the X-ray film was slightly overexposed. In contrast, a strong ErbB2 signal was detected on SK-BR-3 cells, while this signal was almost absent in A431 cells. This also demonstrates that the anti-ErbB2 antibody used in our experiments does not present cross-reactivity towards EGFR. To determine whether or not ErbB2 was able to undergo activation, we incubated SK-BR-3 cells in the absence and presence of HRGβ1 and performed Western blots using an anti-phosphotyrosine antibody. As shown in Figure 1(B), phosphorylation of tyrosine of ErbB2 was stimulated strongly upon addition of HRGβ1. The samples were reprobed with an anti-ErbB2 antibody to determine that they contain comparable amounts of receptor. Densitometric analysis showed that the mean±S.E.M. of HRGβ1-induced stimulation of ErbB2 phosphorylation increased to 641±179% (n=3) as measured in whole cell extracts, and to 346±84% (n=4) as measured in cell membrane fraction preparations.
Figure 1. HRGβ1 stimulates the phosphorylation of ErbB2 overexpressed in SK-BR-3 cells.
(A) Whole cell extracts (approx. 60 μg of protein) from A431 and SK-BR-3 cells were separately subjected to SDS/PAGE and Western blot analysis using anti-EGFR (α-EGFR) or anti-ErbB2 (α-ErbB2) antibodies as described in the Experimental section. The arrows point to EGFR or ErbB2. (B) Whole cell extracts (150 μg of protein) of SK-BR-3 cells from a non-treated culture (−) or a culture treated (+) with 100 ng/ml HRGβ1 for 5 min were subjected to SDS/PAGE and Western blot analysis as described in the Experimental section. The PVDF membrane was first probed with the anti-phosphotyrosine antibody (α-P-Tyr), and thereafter stripped and reprobed with the anti-ErbB2 antibody (α-ErbB2). The arrows point to ErbB2. Representative results from two (A) and three (B) separate experiments are presented.
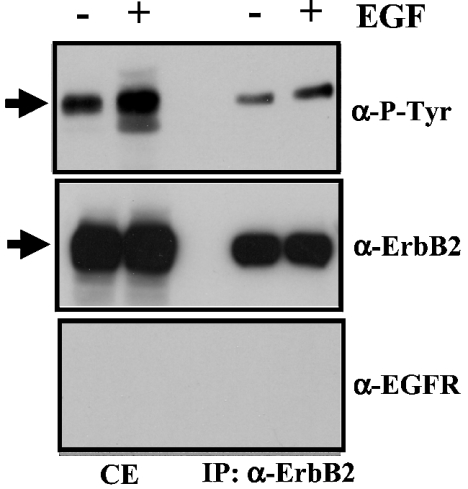
Figure 2 shows that ErbB2 can be activated by EGF, as observed by Western blot analysis using an anti-phosphotyrosine antibody. This was observed using both the whole cell extract and the immunoprecipitated ErbB2 from SK-BR-3 cells. The stimulation of tyrosine phosphorylation in ErbB2 induced by EGF was, however, less pronounced than the stimulation induced by HRGβ1, when the signals in the presence or absence of either of the two growth factors were compared (see Figure 1B and Figure 2, top panel). Densitometric analysis shows that the EGF-induced stimulation of ErbB2 phosphorylation increased to 179±23% (n=3) as measured in whole cell extracts, and to 128±3% (n=6) as measured using the immunoprecipitated receptor. As expected, ErbB2 was detected in both the whole cell extract and the immunoprecipitated material when probed with an anti-ErbB2 antibody (Figure 2, middle panel), while no EGFR signal was observed when samples where probed with an anti-EGFR antibody (Figure 2, bottom panel).
Figure 2. EGF activates ErbB2 while EGFR was undetected on immunoprecipitated ErbB2 preparations.
Whole cell extracts (CE) (120 μg of protein) or immunoprecipitated ErbB2 (IP) from 3.4 mg cell lysate proteins of SK-BR-3 cells of a non-treated culture (−) or a culture treated (+) with 10 nM EGF for 1 min were subjected to SDS/PAGE and Western blot analysis as described in the Experimental section. Separate samples were probed with anti-phosphotyrosine (α-P-Tyr), anti-ErbB2 (α-ErbB2) or anti-EGFR (α-EGFR) antibodies. The arrows point to ErbB2. Representative results from three (CE) and seven (IP) separate experiments are presented.
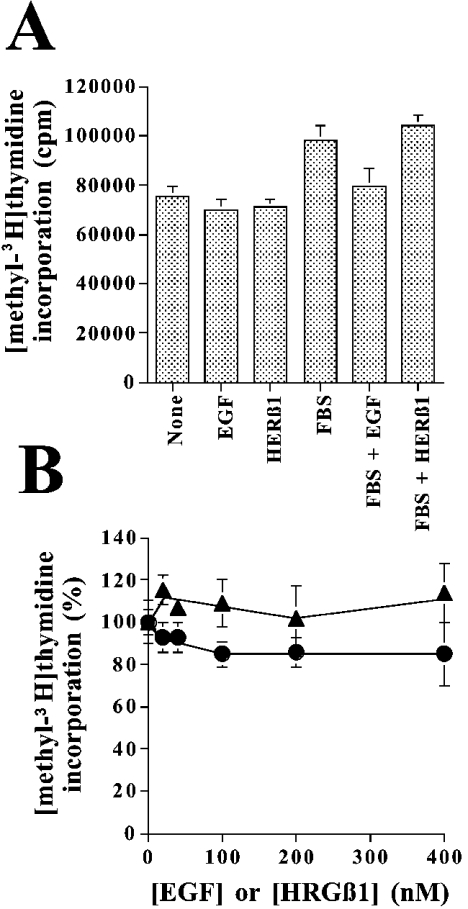
To characterize whether or not the activation of ErbB2 from SK-BR-3 cells generate a proliferative response upon addition of exogenous HRGβ1 or EGF, we measured the incorporation of [methyl-3H]thymidine into DNA under different experimental conditions. Figure 3(A) shows that SK-BR-3 cells incorporate [methyl-3H]thymidine with great efficiency in the absence of any added growth factor, and that the addition of 10 nM HRGβ1 or 10 nM EGF, in the absence or presence of 10% (v/v) FBS, does not stimulate DNA synthesis further. Moreover, increasing the concentrations of HRGβ1 or EGF also fails to enhance the incorporation of [methyl-3H]thymidine above its already high basal level, as observed in Figure 3(B).
Figure 3. Effect of EGF and HRGβ1 on SK-BR-3 cell proliferation.
Incorporation of [methyl-3H]thymidine into DNA was performed as described in the Experimental section. (A) The experiments where performed in the presence of 10 nM EGF, 10 nM HRGβ1 and/or 10% (v/v) FBS as indicated. Results are means±S.E.M. of eight determinations in two separate experiments. (B) The experiments were performed using the indicated concentrations of EGF (•) or HRGβ1 (▴). Results are means±S.E.M. of six determinations in two separate experiments. The 100% value corresponds to 70000 c.p.m.
ErbB2 binds CaM in a specific and Ca2+-dependent manner
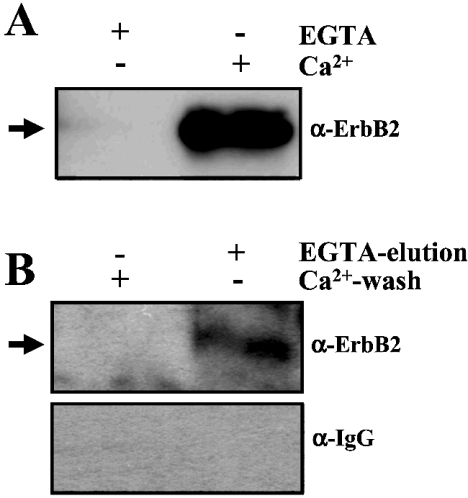
To determined whether or not ErbB2 could be isolated by CaM-affinity binding, we performed pull-down experiments using CaM-agarose. Figure 4(A) shows that ErbB2 could be detected by Western blot analysis using an anti-ErbB2 antibody in the material bound to the CaM–agarose beads when the experiments were performed in the presence of CaCl2, but not in its absence (in the presence of EGTA). Interestingly, in most preparations, the presence of the CaM inhibitor W7 during the incubation procedure increased the binding of ErbB2 to the CaM–agarose beads, most likely because the inactivation of endogenous CaM that binds and competes for ErbB2. Densitometric analysis shows that the binding of ErbB2 to CaM–agarose increased to 140±36% (n=4) in the presence of W7 as compared with controls in the absence of the CaM antagonist (results not shown). Furthermore, we isolated ErbB2 by Ca2+-dependent CaM-affinity chromatography. Thus Figure 4(B) shows the absence of ErbB2 in the CaM–agarose column effluent after an extensive wash with a buffer containing CaCl2, and the appearance of the receptor after elution with a buffer containing EGTA, as detected using an anti-ErbB2 antibody (Figure 4B, upper panel). A negative control using the secondary anti-IgG antibody alone is also presented (Figure 4B, lower panel). Overall, these experiments show that CaM binds to ErbB2 in a Ca2+-dependent manner.
Figure 4. ErbB2 binds to CaM–agarose.
(A) A solubilized whole cell extract (2.8 mg of protein) from non-stimulated SK-BR-3 cells was subjected to pull-down using CaM–agarose beads as described in the Experimental section in the presence (+) of 0.2 mM CaCl2 (Ca2+) or 1 mM EGTA as indicated. The material bound to the beads was subjected to SDS/PAGE and Western blot analysis using the anti-ErbB2 antibody (α-ErbB2). The arrow points to ErbB2. Representative results from two independent experiments are presented. (B) A solubilized whole cell extract (25 mg of protein) from non-stimulated SK-BR-3 cells was subjected to CaM-affinity chromatography as described in the Experimental section. Pooled fractions after Ca2+-wash and EGTA-elution were subjected to SDS/PAGE and Western blot analysis using an anti-ErbB2 antibody (α-ErbB2) (upper panel). A control using only the secondary antibody (α-IgG) is also presented (lower panel) The arrow points to ErbB2. Representative results from two separate experiments are presented.
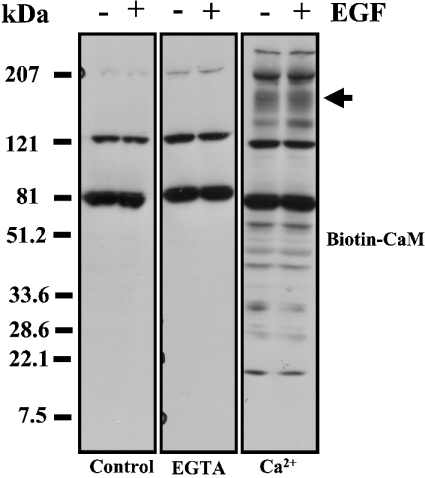
The detection of ErbB2 in the CaM–agarose beads in the presence of Ca2+, and its elution with EGTA, does not exclude, however, the possibility that this receptor could be isolated because of its indirect association with other CaM-binding protein(s). In fact, many different CaM-binding proteins can be detected in SK-BR-3 cells after overlaying the electrophoretically separated proteins with biotinylated CaM as shown in Figure 5. Hence, we detected CaM-binding proteins in the whole extracts from SK-BR-3 cells in experiments performed in the presence of CaCl2 (Figure 5, right-hand panel). In its absence (Figure 5, middle panel), however, the only observed bands were also detected in a mock experiment carried out in the absence of biotinylated CaM after development with streptavidin–peroxidase (Figure 5, left-hand panel), showing its non-specific binding. Little change, if any, in the binding of biotinylated CaM to the different CaM-binding proteins was observed between cell extracts from non-stimulated cells and cells stimulated with EGF. Significantly, a positive broad band of high molecular mass (190–160 kDa), the region of migration of ErbB receptors, was detected (Figure 5, arrow).
Figure 5. Ca2+-dependent CaM-binding proteins in SK-BR-3 cells.
A whole cell extract (100 μg of protein) from SK-BR-3 cells of a non-treated culture (−) or a culture treated (+) with 10 nM EGF for 1 min were subjected to SDS/PAGE, before electrotransfer of the proteins on to a PVDF membrane, and overlay with 0.45 μg/ml biotinylated CaM in the presence of 0.2 mM CaCl2 (Ca2+) and in its absence (EGTA) as described in the Experimental section. Samples were processed in the absence of biotinylated CaM (Control) to detect non-specific binding. The arrow indicates a broad high-molecular-mass (190–160 kDa) CaM-binding protein band. Representative results from two independent experiments are presented.
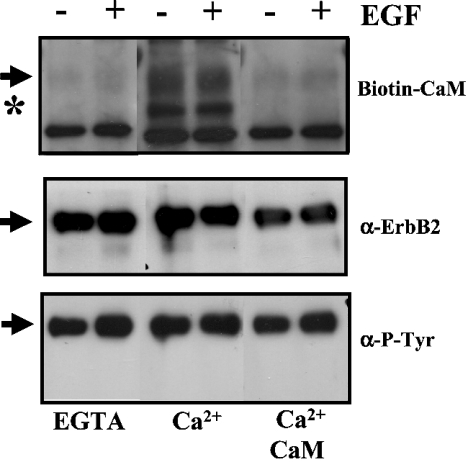
To determine whether ErbB2 could be responsible at least in part for the high-molecular-mass signal observed in overlay experiments with biotinylated CaM using whole cell extracts, we repeated the experiment using immunoprecipitated ErbB2. Figure 6 shows that biotinylated CaM binds to a 185 kDa band in the immunoprecipitated material in the presence of CaCl2 (Figure 6, Ca2+ lanes), but not in its absence (Figure 6, EGTA lanes). Moreover, the addition of an excess of free CaM chases the binding of its biotinylated form to this 185 kDa protein. No significant change was observed when non-stimulated cells or cells stimulated with EGF were used. Moreover, a prominent 160 kDa Ca2+-dependent CaM-binding protein (p160) co-immunoprecipitated with ErbB2 was detected (Figure 6, asterisk). An excess of free CaM also chases the binding of biotinylated CaM to p160. When the PVDF membrane was sequentially re-probed with anti-phosphotyrosine and anti-ErbB2 antibodies, we observed a perfect match between the biotinylated CaM signal, and the ErbB2 and tyrosine phosphorylation signals of the 185 kDa band. These experiments, therefore, show that CaM directly and specifically binds to ErbB2 in a Ca2+-dependent manner.
Figure 6. Direct and specific Ca2+-dependent binding of biotinylated CaM to immunoprecipitated ErbB2.
The immunoprecipitated ErbB2 from 2.6 mg of protein cell extract of SK-BR-3 cells of a non-stimulated culture (−) or a culture stimulated (+) with 10 nM EGF for 1 min, were subjected to overlay with 0.45 μg/ml biotinylated CaM in the presence of 0.2 mM CaCl2 (Ca2+) and 135 μg/ml free CaM as indicated, and described in the Experimental section. A control in the absence of CaCl2 and presence of 1 mM EGTA is also presented (EGTA). The PVDF membrane was sequentially stripped and reprobed with the anti-phosphotyrosine antibody (α-P-Tyr), and the anti-ErbB2 antibody (α-ErbB2). The arrows point to ErbB2, and the asterisk indicates p160. Representative results from two separate experiments are presented.
The CaM antagonist W7 inhibits HRGβ1-induced ErbB2 phosphorylation in intact cells
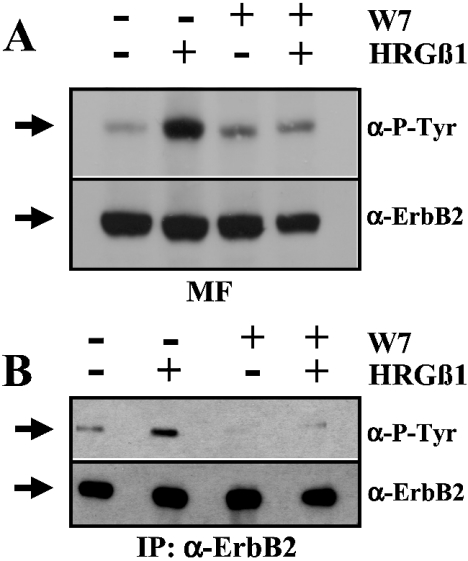
As mentioned above, the presence of the CaM antagonist W7 increases the binding of ErbB2 to CaM–agarose beads in vitro. We next decided to test whether or not this compound affects the activation of ErbB2 in intact cells. Figure 7(A) shows that the level of HRGβ1-induced phosphorylation of ErbB2 decreases in cells treated with W7 during 30 min before the addition of the ligand. The phosphorylation of the receptor was tested in isolated membrane fractions solubilized with Triton X-100 from HRGβ1-treated and untreated cells. Although the phosphorylation signal in the Western blot correlates perfectly with the ErbB2 signal, as detected using an anti-ErbB2 antibody, to ascertain further that the observed W7-induced decrease in phosphorylation took place in ErbB2 and not in any unrelated protein of similar molecular mass, we performed a similar experiment, but using the immunoprecipitated ErbB2 from HRGβ1-treated and untreated cells. Thus we observed again that W7 treatment strongly inhibits the HRGβ1-induced phosphorylation of ErbB2 (Figure 7B). In this instance, however, we also observed a significant W7-induced inhibition of the basal phosphorylation of ErbB2 in the absence of HRGβ1. Densitometric measurements show that treating living cells with 5 μg/ml W7 inhibited HRGβ1-dependent stimulation of ErbB2 phosphorylation to 81±12% (n=4) as measured in cell membrane fractions, and to 30±8% (n=2) as measured using immunoprecipitated ErbB2. In the absence of HRGβ1, W7 treatment inhibits ErbB2 phosphorylation to 16±1.5% (n=2) as measured using immunoprecipitated ErbB2. Furthermore, we observed that W7 treatment also inhibits HRGβ1-dependent tyrosine phosphorylation of four different phosphoproteins downstream of ErbB2, most likely protein substrates of its tyrosine kinase activity, denoted pp130, pp110, pp60 and pp51 (results not shown).
Figure 7. The CaM antagonist W7 inhibits ErbB2 phosphorylation in intact cells.
(A) Serum-deprived SK-BR-3 cells were treated with 5 μg/ml W7 for 30 min and then stimulated with 100 ng/ml HRGβ1 for 2 min as indicated. Thereafter, solubilized cell membrane extracts (100 μg of protein) were prepared and subjected to SDS/PAGE, before electrotransfer of the proteins on to a PVDF membrane, and Western blot analysis using an anti-phosphotyrosine antibody (α-P-Tyr) as described in the Experimental section. The PVDF membrane was subsequently stripped and reprobed with the anti-ErbB2 antibody (α-ErbB2). The arrows point to ErbB2. (B) Serum-deprived SK-BR-3 cells were treated with 5 μg/ml W7 for 30 min and then stimulated with 100 ng/ml HRGβ1 for 2 min as indicated. Thereafter, the immunoprecipitated ErbB2 from 1.3 mg of protein cell extract was subjected to SDS/PAGE, before electrotransfer of the protein on to a PVDF membrane, and Western blot analysis using an anti-phosphotyrosine antibody (α-P-Tyr) as described in the Experimental section. The PVDF membrane was subsequently stripped and reprobed with the anti-ErbB2 antibody (α-ErbB2). The arrows point to ErbB2. Representative results from five (A) and two (B) independent experiments are presented.
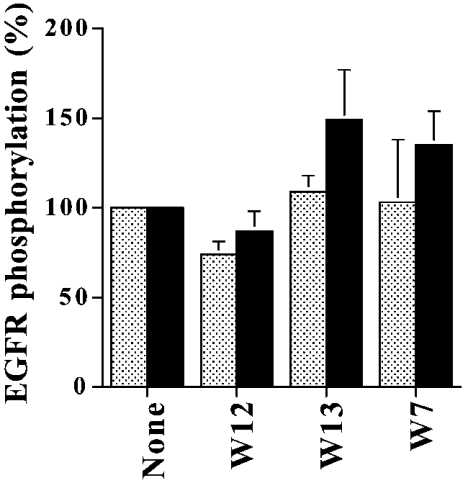
It has been reported that W7 at very high concentrations inhibits protein kinase C activity by two mechanisms: interacting with the phospholipid cofactor of the enzyme (IC50=260 μM), and acting as a competitive inhibitor of ATP (at even higher concentrations) [28]. Although the concentration of W7 used in our experiments with living cells was approx. 5–15-fold lower than the IC50 indicated above, we had to exclude nonetheless the possibility that W7 could be acting as a non-specific inhibitor of ErbB receptor tyrosine kinase activity. To this end, we directly assayed W7, and the related compounds W13 [N-(4-aminobutyl)-5-chloro-1-naphthalenesulphonamide] and W12 [N-(4-aminobutyl)-1-naphthalenesulphonamide], on the tyrosine kinase activity of an isolated preparation of EGFR, as ErbB2 cannot be assayed by itself in an isolated system. As shown in Figure 8, neither of these CaM antagonists, which have different inhibitory capacity for this protein (W7>W13≫W12), were able to inhibit either the auto(trans)phosphorylation of the receptor (Figure 8, grey bars), or its tyrosine kinase activity toward the exogenous substrate poly(L-Glu/L-Tyr) (Figure 8, black bars) at identical or 3-fold higher concentrations than the ones used in our experiments in living cells. On the contrary, W7 and W13, the most efficient CaM antagonists, significantly increased the tyrosine kinase activity of EGFR toward the substrate poly(L-Glu/L-Tyr). These results were expected, as these CaM antagonists could possibly inactivate endogenous CaM present in the EGFR preparations, which may partially inhibit the receptor tyrosine kinase activity as reported previously [12,13]. Furthermore, we have shown previously that treating living cells with these CaM antagonists also prevents the full activation of EGFR [16] in a similar fashion as observed with the activation of ErbB2 (this work).
Figure 8. Naphthalenesulphonamide-derivative CaM antagonists do not act as non-specific ErbB tyrosine kinase inhibitors.
Rat liver purified EGFR (40 μl) was incubated for 15 min on ice in the absence or presence of 15 μg/ml W12, 15 μg/ml W13 or 15 μg/ml W7 as indicated, and thereafter assayed for 2 min at 37 °C in 100 μl of a medium containing 15 mM Hepes/NaOH (pH 7.4), 5 mM MgCl2, 0.4 mM EGTA, 0.5 mM CaCl2 (0.1 mM free Ca2+), 0.1 mg/ml poly(L-Glu/L-Tyr), 1 μM EGF and 10 μM [γ-32P]ATP (2 μCi). The reaction was arrested upon addition of 10% (w/v) trichloroacetic acid, and the precipitated proteins were processed by SDS/PAGE and autoradiography as indicated in the Experimental section. Results are means±range of the densitometric intensities of the autophosphorylated 170 kDa EGFR band (grey bars) and the phosphorylated exogenous substrate poly(L-Glu/L-Tyr) (black bars) for two independent experiments, considering 100% to be the values obtained in the absence of CaM antagonists.
Diverse actions of W7 on signalling pathways downstream of ErbB2
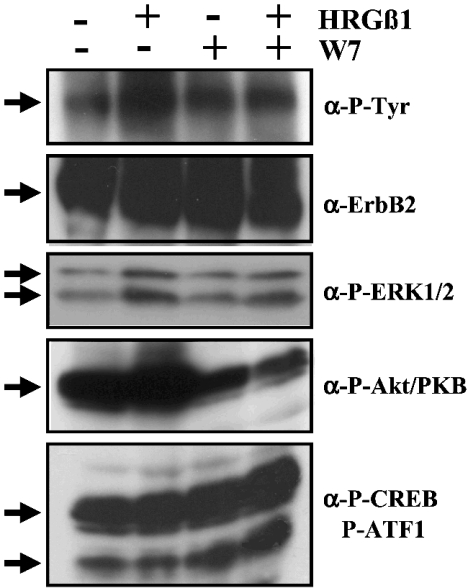
To test the possible involvement of CaM on signalling pathways downstream of ErbB2, we focused our attention on the activation status of different MAPK pathways (ERK, p38 MAPK and JNK), the PI3K (phosphoinositide 3-kinase)/Akt pathway and the activity of several ErbB2-activated Ca2+-dependent transcription factors [CREB, ATF1 and NFAT (nuclear factor of activated T-cells)] in the absence and presence of HRGβ1 and/or the CaM antagonist W7. Figure 9 shows that, concomitant with the inhibitory action of W7 on the HRGβ1-dependent tyrosine phosphorylation of ErbB2, (Figure 9, α-P-Tyr panel), the CaM antagonist also induces the down-regulation of the Ras/MAPK (ERK) and PI3K/Akt pathways, as shown determining the levels of phospho-ERK1/2 (Figure 9, α-P-ERK1/2 panel) and phospho-Akt/PKB (Figure 9, α-P-Akt/PKB panel) respectively. Little, if any, activation of the p38 MAPK and JNK pathways was observed upon W7 treatment, as determining the phosphorylated (active) forms of p38 MAPK and JNK (results not shown). In contrast, we observed that W7 induces a significant increase in the phosphorylation status of the Ca2+-dependent transcription factors CREB (Figure 9, α-P-CREB/P-ATF1 panel, upper band) and ATF1 (Figure 9, α-P-CREB/P-ATF1 panel, lower band) as determined using an anti-phospho-CREB/anti-phospho-ATF1 antibody. Significantly, the activation levels of both the Akt/PKB pathway, and the transcription factors CREB and ATF1, were already very high under basal conditions (absence of HRGβ1) in SK-BR-3 cells. Preliminary results show that the Ca2+-dependent transcription factor NFAT is localized in the nuclear fraction of non-stimulated (absence of HRGβ1) SK-BR-3 cells, which makes it difficult to evaluate the potential effect of W7 in the cytosol-to-nucleus translocation process (results not shown).
Figure 9. Effect of W7 on downstream ErbB2 signalling pathways.
Serum-deprived SK-BR-3 cells were incubated in the absence (−) and presence (+) of 15 μg/ml W7 for 30 min, and then stimulated with 10 nM HRGβ1 for 2 min as indicated. Thereafter, whole cell extracts (60 μg of protein) were prepared and subjected to SDS/PAGE. The proteins were electrotransferred on to a PVDF membrane, and different Western blot analyses were performed using anti-phosphotyrosine (α-P-Tyr), anti-ErbB2 (α-ErbB2), anti-phospho-ERK1/2 (α-P-ERK1/2), anti-phospho-Akt/PKB (α-P-Akt/PKB) and anti-phospho-CREB (α-P-CREB) (also recognizing phospho-ATF1, P-ATF1) antibodies as described in the Experimental section. The ErbB2 signal (α-ErbB2) constitutes one of the multiple loading controls performed (see the Experimental section) corresponding to the stripped and reprobed PVDF membrane presented in the α-P-Tyr panel. Representative results from six (α-P-Tyr), six (α-ErbB2), five (α-P-ERK1/2), five (α-P-Akt/PKB) and five (α-P-CREB) independent experiments performed in identical or similar conditions (5–20 μg/ml W7) are presented. Arrows point, respectively, to phosphorylated ErbB2 (α-P-Tyr), total ErbB2 (α-ErbB2), phosphorylated ERK1/2 (top and bottom arrows) (α-P-ERK1/2), phosphorylated Akt/PKB (α-P-Akt/PKB), and phosphorylated CREB (top arrow) and ATF1 (bottom arrow) (α-P-CREB/P-ATF1).
DISCUSSION
In the present report, we have extended our studies on the interaction of CaM with tyrosine kinase receptors of the EGFR family to ErbB2. Hence, we have shown that CaM binds specifically and directly to ErbB2 in a Ca2+-dependent manner. The functional activation of ErbB2 by HRGβ1 or EGF obligatorily requires its heterodimerization with another ErbB receptor partner. The human breast adenocarcinoma SK-BR-3 cells used in the present study overexpress ErbB2, and express limited amounts of EGFR, ErbB3 and ErbB4 [26], sufficient, however, to warrant the activation of ErbB2 upon ligand stimulation, either by HRGβ1 or EGF. Since we had demonstrated previously that EGFR interacts directly with CaM in a specific Ca2+-dependent manner [12–16], it was important to exclude the possibility that this receptor was interfering with the observed interaction between CaM and ErbB2. Our results clearly demonstrate that the low level of EGFR expression in this cell line, although it may be required for ErbB2 activation, was not interfering, as the immunoprecipitated ErbB2 preparations used in this study were free of EGFR. We also observed that the activation of ErbB2 does not appear to significantly modify the binding of CaM to this receptor, similarly to what we had previously shown with the binding of CaM to EGFR [15,16].
When we used immunoprecipitated ErbB2, in addition to demonstrating that the Ca2+–CaM complex binds directly and specifically to this receptor, we showed that these preparations contain an additional 160 kDa CaM-binding protein (p160), suggesting that this protein co-immunoprecipitated with ErbB2. Interestingly, we have shown previously that two CaM-binding proteins (p190 and p160) co-immunoprecipitated with EGFR from two different cell lines tested [15]. The nature and function of these CaM-dependent proteins are unknown, although it is tempting to speculate that the p160 that co-immunoprecipitated with both ErbB2 and EGFR is the same protein.
The high level of [methyl-3H]thymidine incorporation into DNA in these cells in the absence of added growth factors, and the absence of stimulation of this basal incorporation level upon addition of exogenous HRGβ1 or EGF, suggest the production by these cells of endogenous ErbB ligand(s) able to induce autocrine/paracrine stimulation, and/or that the activated ErbB2 may additionally intervene in non-proliferative pathways, for example in cell migration, as already demonstrated [29]. Therefore the potential functional role that CaM may have in modulating ErbB2 activity could encompass not only the regulation of cell proliferation, but perhaps additional functions mediated by this receptor such as the modulation of cell motility. Because ErbB2 cannot form functional homodimers [1,2,5,6], it was not possible to test the effect of CaM on the tyrosine kinase activity of an isolated preparation of this receptor directly, in contrast with the case of EGFR where we indeed demonstrated that CaM inhibits its tyrosine kinase activity [12,13]. Therefore indirect approaches should be employed to obtain some clues about the possible action of CaM on the functionality of ErbB2. A logical option was to use a cell-permeant CaM antagonist in intact living cells to determine its action on ErbB2 activation. To this end we used the CaM antagonist W7 [30].
When we tested W7 in vitro, we observed that this compound does not prevent the Ca2+-dependent association of ErbB2 to CaM–agarose. On the contrary, we observed an increase binding of ErbB2. This suggests that the region of CaM involved in the interaction with ErbB2 is not affected by the conformational change of CaM induced by this inhibitor, and that the inhibition of endogenous CaM, possibly present in the preparation, favours the binding of ErbB2 to the immobilized CaM. This does not exclude, however, that the CaM–W7 complex was incompetent in blocking a regulatory response of CaM on ErbB2. In fact, our results show that treating intact living cells with W7 significantly diminishes the HRGβ1-dependent phosphorylation of ErbB2. Similarly, we have observed in other cell lines that overexpress EGFR that W7, and a related CaM antagonist (W13), inhibit the EGF-dependent phosphorylation of EGFR in vivo [16]. Moreover, W7 has been shown to inhibit EGF-induced, but not EGF-independent, proliferation of a human hepatoma cell line [31].
The use of W7 in intact cells, where we observed the down-regulation of ErbB2 tyrosine phosphorylation, made it mandatory to test that this CaM antagonist does not act as a non-specific ATP-binding-site kinase inhibitor, as it has been shown for protein kinase C, albeit at extremely high concentrations [28]. We tested directly, not only W7, but also W13 and W12, other members of this family of CaM antagonists with different inhibitory potentials (W7>W13≫W12), on the tyrosine kinase activity of isolated EGFR. Our results show conclusively that none of these compounds inhibit the EGF-induced auto(trans)phosphorylation of the receptor nor the tyrosine kinase activity toward an exogenous substrate. Therefore we conclude that the observed effect of W7 on living cells on ErbB2 (in the present study) and EGFR [16] is not due to any artifactual inhibition of the receptor tyrosine kinase activity.
We also noticed that treatment of cells with W7 in the absence of HRGβ1 induced a decrease in the basal phosphorylation of ErbB2 as detected after immunoprecipitation of the receptor, but this inhibition was not detected after isolation of ErbB2 from detergent-solubilized membranes. The reason could be the existence of different pools of ErbB2 within different cell membranes, as we have detected a Triton X-100-insoluble membrane pool containing high levels of phosphorylated ErbB2, which appears not to be affected by W7 treatment (results not shown).
How the inhibition of CaM by W7 in intact cells mediates the inactivation of ErbB2 is not yet known. The indirect inhibition of CaM-dependent kinases, such as CaMK-II and CaMK-IV, was considered. This, however, is very unlikely, as the phosphorylation of ErbB2 at Thr1172 by CaMK-II has been documented, and the mutation of this phosphorylation site produces a defect in the desensitization of an ErbB2–EGFR chimaera and a more sustained EGF-induced phosphorylation of this receptor as compared with a non-mutated chimaeric receptor [32]. These observations, therefore, do not support the idea that failure in phosphorylation of ErbB2 by CaMK-II could be responsible for our findings because W7 treatment induced receptor down-regulation, not receptor up-regulation.
Consequently, some other possibilities were considered. Among these, several conceivable options should be mentioned: (i) that the inhibition of intracellular CaM could facilitate ErbB2 dephosphorylation by activation of a phosphatase down-regulated by CaM; (ii) that inhibition of CaM could increase the internalization of ErbB2, preventing in this manner access to extracellular HRGβ1; and (iii) that CaM inhibition could induce a redistribution of ErbB2 between different intracellular compartments, as we have noticed the presence of a conspicuous ErbB2 pool resistant to Triton X-100 solubilization in SK-BR-3 cells (results not shown). In this context, although several protein tyrosine phosphatases have been shown to dephosphorylate ErbB2 [33–35], we are not aware, however, that these dephosphorylation pathways are under the control of CaM. In contrast, with respect to ErbB2 internalization, there is the precedent that CaM has been shown to be involved in the intracellular trafficking of EGFR, a related receptor of the same family, as the treatment of living cells with the CaM antagonist W13 induces the sequestration of this receptor in endosomes [36]. Therefore it is likely that ErbB2 could behave in the same manner. Notwithstanding the uncertainties about the mechanism of action of W7 on the decreased phosphorylation of ErbB2, our results show clearly that CaM plays a direct or indirect role on the regulation of the activity or ErbB2 in intact cells.
Furthermore, we tested the effect of W7 on ErbB2-initiated downstream signalling pathways in living cells. As expected, the Ras/MAPK (ERK), and PI3K/Akt pathways were both inhibited concomitant with ErbB2 down-regulation. These inhibitory actions should not, however, be ascribed exclusively to a decrease in ErbB2 signalling capacity, as CaM is known to regulate several steps of the Ras/MAPK (ERK) pathway directly [37]. Thus W13, a related CaM antagonist, has been shown to inhibit the Ras/MAPK (ERK) pathway to the level of Raf-1, a kinase upstream of ERK1/2 [36]. Nevertheless, the action of CaM on the Ras/MAPK (ERK) pathway appears to be very complex, as in some cell types, it has been shown that CaM blockade by W13 induces a sustained activation of this signalling route at the level of Ras, Raf and MEK [38].
An increase in [Ca2+]cyt is known to control, with the intervention of CaM, the function of several Ca2+-responsive transcription factors such as CREB [39–42], ATF1 [43] and NFAT [44,45], among others. We have shown that the treatment of living cells with W7 elicits the up-regulation of CREB and ATF1. These results support the notion that W7 is indeed inactivating CaM, subsequently inducing a failure in calcineurin activity, and thereby an increase in the phosphorylation level of CREB and ATF1, as expected [40]. This is despite the fact that CREB appears to be in part phosphorylated by CaMK-II and CaMK-IV [39,41].
Overall, these results show that CaM is not only acting in living cells on the ErbB2 receptor itself, but also at different points of ErbB2-initiated downstream signalling pathways, such as the Ras/MAPK (ERK) and PI3K/Akt pathways, and their gene transcriptional activity mediated by downstream Ca2+-dependent transcription factors.
As mentioned above, the sequence similarity of the CaM-BD of EGFR with homologous regions of other ErbB receptors is very high. Thus we have postulated that ErbB2 could interact with CaM at its cytosolic juxtamembrane region, and that additional members of this family of tyrosine kinase receptors, ErbB3 and ErbB4, particularly the latter, could also be CaM-binding proteins [11,14]. Further work should be carried out to demonstrate if this is indeed the case.
Acknowledgments
We thank Amparo Jiménez for expert technical assistance, Dr Margarita Fernández and Dr Jorge Martín for gifts of anti-Akt/PKB antibodies, and Dr Miguel Quintanilla for the anti-α-tubulin antibody. This work was financed by grants (to A.V.) from the Comisión Interministerial de Ciencia y Tecnología (SAF2002-03258) and the Consejería de Educación de la Comunidad de Madrid (08.1/0027/2001-1). The generous support from the Instituto Carlos III, Fondo de Investigaciones Sanitarias (RTICCC C03/10) is also acknowledged. H.L. was supported by the Agencia Española de Cooperación Internacional (2002CN0013), A.dC. by a predoctoral fellowship from the Fundación Carolina and V.S. by a predoctoral fellowship from the Consejo de Desarrollo Científico y Humanístico de la Universidad Central de Venezuela.
References
- 1.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 2.Carraway K. L., III, Sweeney C. Localization and modulation of erbB receptor tyrosine kinases. Curr. Opin. Cell Biol. 2001;13:125–130. doi: 10.1016/s0955-0674(00)00188-5. [DOI] [PubMed] [Google Scholar]
- 3.Tzahar E., Yarden Y. The ErbB-2/HER2 oncogenic receptor of adenocarcinomas: from orphanhood to multiple stromal ligands. Biochim. Biophys. Acta. 1998;1377:M25–M37. doi: 10.1016/s0304-419x(97)00032-2. [DOI] [PubMed] [Google Scholar]
- 4.Rubin I., Yarden Y. The basic biology of HER2. Anal. Oncol. 2001;12:S3–S8. doi: 10.1093/annonc/12.suppl_1.s3. [DOI] [PubMed] [Google Scholar]
- 5.Alroy I., Yarden Y. The ErbB signaling network in embryogenesis and oncogenesis: signal diversification through combinatorial ligand–receptor interactions. FEBS Lett. 1997;410:83–86. doi: 10.1016/s0014-5793(97)00412-2. [DOI] [PubMed] [Google Scholar]
- 6.Olayioye M. A., Neve R. M., Lane H. A., Hynes N. E. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villalobo A., Ruano M. J., Palomo-Jiménez P. I., Li H., Martín-Nieto J. The epidermal growth factor receptor and the calcium signal. In: Pochet R., Donato R., Haiech J., Heizmann C., Gerke V., editors. Calcium: The Molecular Basis of Calcium Action in Biology and Medicine. Boston: Kluwer Academic Publishers; 2000. pp. 287–303. [Google Scholar]
- 8.Feldner J. C., Brandt B. H. Cancer cell motility – on the road from c-erbB-2 receptor steered signaling to actin reorganization. Exp. Cell Res. 2002;272:93–108. doi: 10.1006/excr.2001.5385. [DOI] [PubMed] [Google Scholar]
- 9.Dittmar T., Husemann A., Schewe Y., Nofer J.-R., Niggemann B., Zänker K. S., Brandt B. H. Induction of cancer cell migration by epidermal growth factor is initiated by specific phosphorylation of tyrosine 1248 of c-erbB-2 receptor via EGFR. FASEB J. 2002;16:1823–1825. doi: 10.1096/fj.02-0096fje. [DOI] [PubMed] [Google Scholar]
- 10.Persechini A., Cronk B. The relationship between the free concentrations of Ca2+ and Ca2+–calmodulin in intact cells. J. Biol. Chem. 1999;274:6827–6830. doi: 10.1074/jbc.274.11.6827. [DOI] [PubMed] [Google Scholar]
- 11.Martín-Nieto J., Cusidó-Hita D. M., Li H., Benguría A., Villalobo A. Regulation of ErbB receptors by calmodulin. In: Pandalai S. G., editor. Recent Research Developments in Biochemistry. Part I. Vol. 3. Trivandrum: Research Signpost; 2002. pp. 41–58. [Google Scholar]
- 12.San José E., Benguría A., Geller P., Villalobo A. Calmodulin inhibits the epidermal growth factor receptor tyrosine kinase. J. Biol. Chem. 1992;267:15237–15245. [PubMed] [Google Scholar]
- 13.Benguría A., Martín-Nieto J., Benaim G., Villalobo A. Regulatory interaction between calmodulin and the epidermal growth factor receptor. Ann. N.Y. Acad. Sci. 1995;766:472–476. doi: 10.1111/j.1749-6632.1995.tb26698.x. [DOI] [PubMed] [Google Scholar]
- 14.Martín-Nieto J., Villalobo A. The human epidermal growth factor receptor contains a juxtamembrane calmodulin-binding site. Biochemistry. 1998;37:227–236. doi: 10.1021/bi971765v. [DOI] [PubMed] [Google Scholar]
- 15.Li H., Villalobo A. Evidence for the direct interaction between calmodulin and the human epidermal growth factor receptor. Biochem. J. 2002;362:499–505. doi: 10.1042/bj3620499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., Ruano M. J., Villalobo A. Endogenous calmodulin interacts with the epidermal growth factor receptor in living cells. FEBS Lett. 2004;559:175–180. doi: 10.1016/S0014-5793(04)00067-5. [DOI] [PubMed] [Google Scholar]
- 17.Countaway J. L., Nairn A. C., Davis R. J. Mechanism of desensitization of the epidermal growth factor receptor protein-tyrosine kinase. J. Biol. Chem. 1992;267:1129–1140. [PubMed] [Google Scholar]
- 18.Theroux S. J., Latour D. A., Stanley K., Raden D. L., Davis R. J. Signal transduction by the epidermal growth factor receptor is attenuated by a COOH-terminal domain serine phosphorylation site. J. Biol. Chem. 1992;267:16620–16626. [PubMed] [Google Scholar]
- 19.Feinmesser R. L., Wicks S. J., Taverner C. J., Chantry A. Ca2+/calmodulindependent kinase II phosphorylates the epidermal growth factor receptor on multiple sites in the cytoplasmic tail and serine 744 within the kinase domain to regulate signal generation. J. Biol. Chem. 1999;274:16168–16173. doi: 10.1074/jbc.274.23.16168. [DOI] [PubMed] [Google Scholar]
- 20.Aifa S., Johansen K., Nilsson U. K., Liedberg B., Lundström I., Svensson S. P. S. Interactions between the juxtamembrane domain of the EGFR and calmodulin measured by surface plasmon resonance. Cell. Signal. 2002;14:1005–1013. doi: 10.1016/s0898-6568(02)00034-7. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi N., Matsubara M., Takasaki A., Titani K., Taniguchi H. An expression system of rat calmodulin using T17 phage promoter in Escherichia coli. Protein Expression Purif. 1998;12:25–28. doi: 10.1006/prep.1997.0807. [DOI] [PubMed] [Google Scholar]
- 22.Dicker P., Rozengurt E. Phorbol esters and vasopressin stimulate DNA synthesis by a common mechanism. Nature (London) 1980;287:607–612. doi: 10.1038/287607a0. [DOI] [PubMed] [Google Scholar]
- 23.Wolff D. J., Poirier P. G., Brostrom C. A., Brostrom M. A. Divalent cation binding properties of bovine brain Ca2+-dependent regulator protein. J. Biol. Chem. 1977;252:4108–4117. [PubMed] [Google Scholar]
- 24.Billingsley M. L., Pennypacker K. R., Hoover C. G., Brigati D. J., Kincaid R. L. A rapid and sensitive method for detection and quantification of calcineurin and calmodulin-binding proteins using biotinylated calmodulin. Proc. Natl. Acad. Sci. U.S.A. 1985;82:7585–7589. doi: 10.1073/pnas.82.22.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Kroll T., Odyvanova L., Clement J. H., Platzer C., Naumann A., Marr N., Höffken K., Wölfl S. Molecular characterization of breast cancer cell lines by expression profiling. J. Cancer Res. Clin. Oncol. 2002;128:125–134. doi: 10.1007/s00432-001-0308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ullrich A., Coussens L., Hayflick J. S., Dull T. J., Gray A., Tam A. W., Lee J., Yarden Y., Libermann T. A., Schlessinger J., et al. Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. Nature (London) 1984;309:418–425. doi: 10.1038/309418a0. [DOI] [PubMed] [Google Scholar]
- 28.O'Brian C. A., Ward N. E. Binding of protein kinase C to N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide through its ATP binding site. Biochem. Pharmacol. 1989;38:1737–1742. doi: 10.1016/0006-2952(89)90406-1. [DOI] [PubMed] [Google Scholar]
- 29.Verbeek B. S., Adriaansen-Slot S. S., Vroom T. M., Beckers T., Rijksen G. Overexpression of EGFR and c-erbB2 causes enhanced cell migration in human breast cancer cells and NIH3T3 fibroblasts. FEBS Lett. 1998;425:145–150. doi: 10.1016/s0014-5793(98)00224-5. [DOI] [PubMed] [Google Scholar]
- 30.Nishikawa M., Tanaka T., Hidaka H. Ca2+-calmodulin-dependent phosphorylation and platelet secretion. Nature (London) 1980;287:863–865. doi: 10.1038/287863a0. [DOI] [PubMed] [Google Scholar]
- 31.Wu B.-W., Wu Y., Wang J.-L., Lin J.-S., Yuan S.-Y., Li A., Cui W.-R. Study on the mechanism of epidermal growth factor-induced proliferation of hepatoma cells. World J. Gastroenterol. 2003;9:271–275. doi: 10.3748/wjg.v9.i2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feinmesser R. L., Gray K., Means A. R., Chantry A. HER-2/c-erbB2 is phosphorylated by calmodulin-dependent protein kinase II on a single site in the cytoplasmic tail at threonine-1172. Oncogene. 1996;12:2725–2730. [PubMed] [Google Scholar]
- 33.Hernández-Sotomayor S. M., Arteaga C. L., Soler C., Carpenter G. Epidermal growth factor stimulates substrate-selective protein-tyrosine-phosphatase activity. Proc. Natl. Acad. Sci. U.S.A. 1993;90:7691–7695. doi: 10.1073/pnas.90.16.7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhai Y., Wirth J., Kang S., Welsch C. W., Esselman W. J. LAR-PTPase cDNA transfection suppression of tumor growth of neu oncogene-transformed human breast carcinoma cells. Mol. Carcinog. 1995;14:103–110. doi: 10.1002/mc.2940140206. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X. B., Lee M. S., Zelivianski S., Lin M. F. Characterization of a prostate-specific tyrosine phosphatase by mutagenesis and expression in human prostate cancer cells. J. Biol. Chem. 2001;276:2544–2550. doi: 10.1074/jbc.M006661200. [DOI] [PubMed] [Google Scholar]
- 36.Tebar F., Villalonga P., Sorkina T., Agell N., Sorkin A., Enrich C. Calmodulin regulates intracellular trafficking of epidermal growth factor receptor and the MAPK signaling pathway. Mol. Biol. Cell. 2002;13:2057–2068. doi: 10.1091/mbc.01-12-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agell N., Bachs O., Rocamora N., Villalonga P. Modulation of the Ras/Raf/MEK/ERK pathway by Ca2+ and calmodulin. Cell. Signalling. 2002;14:649–654. doi: 10.1016/s0898-6568(02)00007-4. [DOI] [PubMed] [Google Scholar]
- 38.Bosch M., Gil J., Nachs O., Agell N. Calmodulin inhibitor W13 induces sustained activation of ERK2 and expression of p21cip1. J. Biol. Chem. 1998;273:22145–22150. doi: 10.1074/jbc.273.34.22145. [DOI] [PubMed] [Google Scholar]
- 39.Enslen H., Sun P., Brickey D., Soderling S. H., Klamo E., Soderling T. R. Characterization of Ca2+/calmodulin-dependent protein kinase IV: role in transcriptional regulation. J. Biol. Chem. 1994;269:15520–15527. [PubMed] [Google Scholar]
- 40.Schwaninger M., Blume R., Krüger M., Lux G., Oetjen E., Knepel W. Involvement of the Ca2+-dependent phosphatase calcineurin in gene transcription that is stimulated by cAMP through cAMP response elements. J. Biol. Chem. 1995;270:8860–8866. doi: 10.1074/jbc.270.15.8860. [DOI] [PubMed] [Google Scholar]
- 41.Wu G.-Y., Deisseroth K., Tsien R. W. Activity-dependent CREB phosphorylation: convergence of a fast, sensitive calmodulin kinase pathway and a slow, less sensitive mitogen-activated protein kinase pathway. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2808–2813. doi: 10.1073/pnas.051634198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mellström B., Naranjo J. R. Mechanisms of Ca2+-dependent transcription. Curr. Opin. Neurobiol. 2001;11:312–319. doi: 10.1016/s0959-4388(00)00213-0. [DOI] [PubMed] [Google Scholar]
- 43.Liu F., Thompson M. A., Wagner S., Greenberg M. E., Green M. R. Activating transcription factor-1 can mediate Ca2+- and cAMP-inducible transcriptional activation. J. Biol. Chem. 1993;268:6714–6720. [PubMed] [Google Scholar]
- 44.Dolmetsch R. E., Lewis R. S., Goodnow C. C., Healy J. I. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature (London) 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- 45.Holmberg C. I., Tran S. E. F., Eriksson J. E., Sistonen L. Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem. Sci. 2002;27:619–627. doi: 10.1016/s0968-0004(02)02207-7. [DOI] [PubMed] [Google Scholar]