Abstract
Carnivorous plants are known to secrete acid proteinases to digest prey, mainly insects, for nitrogen uptake. In the present study, we have purified, for the first time, to homogeneity two acid proteinases (nepenthesins I and II) from the pitcher fluid of Nepenthes distillatoria (a pitcher-plant known locally as badura) and investigated their enzymic and structural characteristics. Both enzymes were optimally active at pH approx. 2.6 towards acid-denatured haemoglobin; the specificity of nepenthesin I towards oxidized insulin B chain appears to be similar, but slightly wider than those of other APs (aspartic proteinases). Among the enzymic properties, however, the most notable is their unusual stability: both enzymes were remarkably stable at or below 50 °C, especially nepenthesin I was extremely stable over a wide range of pH from 3 to 10 for over 30 days. This suggests an evolutionary adaptation of the enzymes to their specific habitat. We have also cloned the cDNAs and deduced the complete amino acid sequences of the precursors of nepenthesins I and II (437 and 438 residues respectively) from the pitcher tissue of N. gracilis. Although the corresponding mature enzymes (each 359 residues) are homologous with ordinary pepsin-type APs, both enzymes had a high content of cysteine residues (12 residues/molecule), which are assumed to form six unique disulphide bonds as suggested by computer modelling and are supposed to contribute towards the remarkable stability of nepenthesins. Moreover, the amino acid sequence identity of nepenthesins with ordinary APs, including plant vacuolar APs, is remarkably low (approx. 20%), and phylogenetic comparison shows that nepenthesins are distantly related to them to form a novel subfamily of APs with a high content of cysteine residues and a characteristic insertion, named ‘the nepenthesin-type AP-specific insertion’, that includes a large number of novel, orthologous plant APs emerging in the gene/protein databases.
Keywords: aspartic proteinase, carnivorous plant, characterization, Nepenthes, nepenthesin, plant proteinase
Abbreviations: AP, aspartic proteinase; CND41, chloroplast nucleoid DNA-binding protein 41; Cya, cysteic acid; DAN, diazoacetyl-D,L-norleucine methyl ester; NAP, nepenthesin-type AP
INTRODUCTION
There are several carnivorous plants of different genera in nature, which catch preys, mainly insects, and digest their proteins primarily by their endogenous proteinase(s) and absorb the digestion products as the nitrogen source [1,2]. Hooker [3] was the first to document that Nepenthes is carnivorous, inspired directly by Charles Darwin. Since then, it has been the object of studies for nearly 130 years to understand how Nepenthes accomplishes this process [4]. Nepenthesin [5,6] is an acid proteinase secreted in the pitcher of Nepenthes species. So far, the acid proteinases from Nepenthes and Drosera species were only partially purified and poorly characterized [7–12]. Although they were shown to be members of APs (aspartic proteinases) [9,11], none of the enzymes secreted from carnivorous plants were purified to homogeneity, mainly due to the difficulty to obtain a sufficient amount of their digestive fluids.
APs are widely distributed in living organisms and extensive studies have been performed on mammalian, microbial and viral APs [13,14]. They are also distributed widely in the plant kingdom, and are also present in seeds, leaves and flowers in various plants [15] as well as in the digestive fluids of carnivorous species. Plant APs, such as those from barley [16,17] and rice [18] and cyprosins (or cardosins) [19–23] have been purified and well characterized. All these plant proteinases have a plant-specific insertion sequence in the middle of the molecule, and are supposed to be intracellular vacuolar enzymes. In contrast, APs from the digestive fluids of the carnivorous plants are the only known extracellular proteinases of plant origin. Therefore these enzymes are interesting from various points of view, such as physiological roles, structure–function relationships and molecular evolution.
In the present study, we have for the first time purified to homogeneity carnivorous plant APs in the pitcher fluid from Nepenthes distillatoria (i.e. nepenthesins I and II), a pitcher-plant known locally as badura, and elucidated their molecular and enzymic characteristics, including their remarkable stability in a wide range of pH over a long period of incubation time. This stability seems to indicate that they have evolutionally well adapted to their original habitat. We have also cloned cDNAs for the enzymes from the pitcher tissue of N. gracilis to deduce the complete amino acid sequences. These results (slender pitcher-plant) have revealed that they are unique enzymes belonging to a novel subfamily of APs with a high content of cysteine residues, which presumably form disulphide bonds to stabilize the enzymes. This family appears to include a large number of new plant orthologues distantly related to already known APs.
EXPERIMENTAL
Materials
Nepenthes pitcher fluid was collected from the plant N. distillatoria in the Singharaja forest (Sri Lanka). N. gracillis was obtained from the Taishoen plantation (Numazu, Shizuoka, Japan). In the present study, N. distillatoria was used for studies at the protein level and N. gracilis for those at the DNA level. This is because N. distillatoria was available only in Sri Lanka, whereas fresh Nepenthes tissue was required for cDNA cloning that had to be performed in Japan. DEAE-cellulose (DE-52) was a product of Whatman (Kent, U.K.). Sephacryl S-200 and a Mono Q HR5/5 column were purchased from Amersham Biosciences (Uppsala, Sweden) and pepstatin–Sepharose, DAN (diazoacetyl-D,L-norleucine methyl ester), the B chain of oxidized bovine insulin and porcine pepsin A were obtained from Sigma (St. Louis, MO, U.S.A.). Pepstatin A was from Peptide Institute (Osaka, Japan). Reagents for automated amino acid analysis and sequencing were obtained from Applied Biosystems (Foster City, CA, U.S.A.). Other reagents used were of the highest grade available.
Determination of enzyme activity
Proteolytic activity of the enzyme was determined essentially as described in [24]. In the standard assay, the reaction was performed in a mixture containing 200 μl of the enzyme solution and 400 μl of 2% acid-denatured haemoglobin in 0.1 M formate buffer (pH 3.0) as a substrate at 37 °C for 2 h, and then stopped by the addition of 800 μl of 5% (w/v) trichloroacetic acid. After centrifugation at 10000 g for 10 min, the absorbance at 280 nm of the resulting supernatant was measured against a blank sample. One unit of activity was defined as the increase of one absorbance unit per hour.
Protein determination
Protein was determined by measuring the absorbance at 280 nm of the sample solution or by the method of Smith et al. [25] using the bicinchoninic acid reagent.
Purification procedures
All purification procedures were performed at 4 °C. The collected fluid (30 litres) from the open pitchers was filtered to remove the insoluble materials and dialysed against 0.02 M sodium phosphate buffer, pH 7.5 (0.02 M phosphate; Buffer A). The proteins in the dialysed fluid were adsorbed on to wet DEAE cellulose (approx. 2 litres) equilibrated with the same buffer by batch-wise treatment and the cellulose was placed into a glass column (6.0 cm×70 cm). The column was washed with Buffer A and the protein was eluted with 0.5 M NaCl. The fractions with proteolytic activity were pooled and applied to a second DEAE-cellulose column (3.0 cm×35 cm) equilibrated with Buffer A. The column was washed with the same buffer and the protein was eluted with a linear gradient of 0–0.5 M NaCl in 2 litres of the same buffer. The active fractions in each activity peak were pooled separately and concentrated by using a small DEAE-cellulose column. Each concentrated sample was applied to a Sephacryl S-200 column (3.1 cm×114 cm) equilibrated with Buffer A and 0.2 M NaCl. The active fractions were pooled and dialysed against 0.02 M sodium acetate buffer (pH 4.0) and applied to a pepstatin–Sepharose column (1.3 cm×1.6 cm) equilibrated with the same buffer. The column was washed with the same buffer and the protein was eluted with 0.05 M Tris/HCl buffer (pH 8.0) containing 1 M NaCl, and then with 0.05 M sodium borate buffer (pH 10.0) containing 1 M NaCl. The fraction eluted at pH 10.0 was immediately adjusted to pH 8.0 by the addition of 1 M Tris/HCl buffer (pH 8.0). The active fractions were pooled and dialysed against 0.02 M Tris/HCl buffer (pH 7.8) and applied to a Mono Q column. The protein was eluted with a linear gradient of 0–1 M NaCl in 0.02 M Tris/HCl buffer (pH 7.8) and the active fractions were pooled. The Mono Q chromatography of the major enzyme fraction was performed three times with one-third of the sample at a time. The digestive fluid collected from the unopened pitchers was submitted to essentially the same purification procedures.
Purity check and molecular-mass determination
SDS/PAGE [26] and gel filtration on a column (3.1 cm×114 cm) of Sephacryl S-200 were used for purity check and molecular-mass determination of the purified enzymes. In SDS/PAGE, the protein was stained with Coomassie Brilliant Blue and the carbohydrate with Schiff's reagent after periodate oxidation [27].
Determination of the N-terminal and partial internal amino acid sequences
The N-terminal amino acid sequence of each protein was determined by using an Applied Biosystems pulse-liquid protein sequencer model 477A. To determine the partial internal sequences of the enzyme, the protein was reduced and carboxymethylated as described by Crestfield et al. [28]. The protein (approx. 150 μg) was then digested at 37 °C with endoproteinase Asp-N (1 μg) for 12 h in 300 μl of 0.1 M ammonium bicarbonate (pH 7.8), with trypsin (3 μg) for 4 h in 300 μl of 0.1 M ammonium bicarbonate (pH 8.1) or with Staphylococcus aureus V8 protease (1 μg) for 4 h in 300 μl of 0.1 M ammonium bicarbonate (pH 8.1). The resulting peptides were separated by HPLC using a Hitachi (Tokyo, Japan) 655A-11 system on a column (0.46 cm×25 cm) of TSKgel ODS-120T (Tosoh, Tokyo, Japan). The peptides were eluted with a linear gradient of acetonitrile (0–50%) in 0.1% trifluoroacetic acid at a flow rate of 0.8 ml/min. The effluent was monitored by measuring the absorbance at 215 nm and the peptide peak fractions were collected and freeze-dried. An aliquot of each peptide fraction dissolved in water was submitted to automated amino acid sequencing.
Digestion of oxidized insulin B chain and analysis of the cleavage sites
The B chain of oxidized insulin (150 nmol) was digested with the purified nepenthesin I (0.3 nmol) in 600 μl of 0.1 M formate buffer (pH 3.0) at 37 °C for 3 h. The resulting peptides were separated by HPLC and analysed in the same manner as described above.
Reaction of DAN
Each purified enzyme (50 μg) was treated with DAN (200 μg) in 3.0 ml of 0.05 M sodium acetate buffer (pH 5.0) at 14 °C in the presence or absence of 2 mM cupric sulphate. Aliquots were withdrawn at appropriate intervals and the remaining activity was determined. Porcine pepsin was treated with DAN under the same conditions for comparison.
Stability studies
To investigate the effect of pH on the stability, each enzyme (50 μg/ml of buffer) and the crude pitcher fluid were incubated at different pH values at 37 °C, and after 7 and 30 days the remaining activity was determined at pH 3.0. The buffers used were 0.05 M sodium formate buffer (pH 3.0), 0.05 M sodium acetate buffers (pH 4.0 and 5.0), 0.05 M sodium phosphate buffers (pH 6.0 and 7.0), 0.05 M Tris/HCl buffers (pH 8.0 and 9.0) and 0.05 M sodium borate buffer (pH 10.0). Porcine pepsin A (50 μg/ml) in the respective buffers was incubated in the same manner for comparison.
To investigate the effect of temperature on the stability, each enzyme (50 μg/ml) in 0.02 M sodium formate buffer (pH 3.0) as well as the crude pitcher fluid (pH 3.0) was incubated at different temperatures for 2 h, and the remaining activity was determined at 37 °C. Furthermore, each enzyme sample was incubated at different temperatures (4, 25, 37 and 50 °C), and after 7 and 30 days the remaining activity was determined at 37 °C. Porcine pepsin A solution (50 μg/ml of 0.02 M sodium formate buffer, pH 3.0) was incubated in the same manner for comparison.
Preparation of antibody and immunohistochemical staining
The purified nepenthesin I mixed with the complete Freund's adjuvant was injected into rabbits (1.0 mg for the primary injection and 0.5 mg for each of the three booster injections) and IgG was purified from the antiserum by 40% (w/v) ammonium sulphate precipitation followed by Protein A–Sepharose affinity chromatography. Longitudinal sections of fresh N. distillatoria tissues prepared by using a microtome were fixed by dipping in 3% (w/v) formaldehyde solution for 1 h. They were successively incubated in PBS with 1% BSA for 2 h, in a 1:8000 diluted purified primary antibody preparation for 2 h, in a 1:2000 diluted secondary antibody with a VECTASTAIN ABC immunochemical staining kit (Vector Laboratories, Burlingame, CA, U.S.A.) for 2 h and in the ABC solution for 1 h to be stained with an alkaline phosphatase substrate as described in the manufacturer's manual. As a control experiment, the primary antibody preparation preincubated with an excess of nepenthesin I immobilized by PVDF membrane was used.
cDNA cloning
Total RNA was isolated from the N. gracilis pitchers using the lower, approx. one-third, part of each pitcher, possessing the digestive glands, according to the modified hot borate method [29], and used for the isolation of poly(A)+ (polyadenylated) RNA using an Oligotex™-dT30 Super mRNA Purification kit (Takara, Kyoto, Japan). Single-stranded cDNA was prepared using a cDNA synthesis kit (Takara) and an oligo(dT) primer.
On the basis of the partial amino acid sequences determined at the protein level, degenerate oligonucleotide primers were synthesized as follows (see also Table 1): Nep1s, YAGDGEY; Maj3a, PTFVMHF; Maj2s, IWTQCQP; Maj1a, QIPTFVM; Min1a, TCFGEPS; Min2s, IWTQCEP; Min4a, FGCGQTV. PCR was performed using Ex Taq DNA polymerase (Takara) and a thermal cycler GeneAmp2400 (PerkinElmer Biosystems, Norwalk, CT, U.S.A.). The PCR analysis was typically based on 30 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min and completed by a 7 min extension at 72 °C, and then the block temperature was held at 4 °C.
Table 1. Primers used for cDNA cloning of nepenthesins I and II.
D, H, N, R, S, W and Y indicate degenerate sites (D, A/G/T; H, A/C/T; N, A/C/G/T; R, A/G; S, C/G; W, A/T; and Y, C/T).
| Primer | Orientation | Sequence (5′→3′) | Target | Location |
|---|---|---|---|---|
| Nep1s | Forward | TAYGCNGGNGAYGGNGARTA | I | 265–284 |
| II | 268–287 | |||
| Maj3a | Reverse | AARTGCATNACRAANGTNGG | I | 1099–1118 |
| Maj2s | Forward | ATHTGGACNCARTGYCARCC | I | 355–374 |
| Maj1a | Reverse | CATNACRAANGTNGGDATYTG | I | 1093–1113 |
| Maj7a | Reverse | TGATCCCAATAGAAGAGTGC | I | 749–768 |
| Maj7s | Forward | ATGACCCCCATTGGTAGC | I | 721–738 |
| ASP16-RT-P | Reverse | AAGGRAGGGTRGAGAAGGA | I | 430–448 |
| II | 433–451 | |||
| Maj12s | Forward | TGGATACCGGTAGCGATCTT | I | 335–354 |
| Maj13a | Reverse | TTGCGGAGAAAGGTTGTGCT | I | 312–331 |
| Maj14a | Reverse | GTTCCAATCGATAAGTTCATC | I | 288–308 |
| Min1a | Reverse | SWNGGYTCNCCRAARCANGT | II | 1057–1076* |
| Min2s | Forward | ATHTGGACNCARTGYGARCC | II | 358–377 |
| Min4a | Reverse | ACNGTYTGNCCRCANCCRAA | II | 604–623* |
| Min8a | Reverse | GTGAAGGTCTCGGTTGCC | II | 555–572 |
| Min11s | Forward | AAACCTGCAATAATAATGAA | II | 485–504 |
| Min12s | Forward | TGGATACCGGCAGTGATCTC | II | 338–357 |
| Min13a | Reverse | TGGCCGAGAAAGAACTATCC | II | 315–334 |
| Min14a | Reverse | GGAGTACCAATTGCTACGTTC | II | 294–314 |
* There are some differences in sequence between the designed primer and the obtained cDNA.
The single-stranded cDNA was used as a template for PCR amplification of partial cDNAs using primers Nep1s and Maj3a for nepenthesin I and Nep1s and Min1a for nepenthesin II. The amplified fragments were further subjected to nested PCR using primers Maj2s and Maj1a for nepenthesin I and Min2s and Min4a for nepenthesin II. Sequencing of the fragments obtained was performed using the terminator cycle sequencing method using BigDye Terminator v3.0 Cycle Sequencing Ready Reaction kit and a DNA sequencer ABI PRISM 3100 (Applied Biosystems).
The complete 3′-end of the cDNA was amplified using a specific forward primer Maj7s (Table 1) for nepenthesin I or Min11s for nepenthesin II and oligo(dT) primer as a reverse primer. DNA fragments amplified using primers Nep1s and Maj7a for nepenthesin I and primers Nep1s and Min8a for nepenthesin II were sequenced. 5′-RACE (rapid amplification of cDNA ends) was performed using 5′-Full RACE Core Set (Takara) according to the manufacturer's instructions. Single-stranded cDNA was prepared using phosphorylated primer Nep16-RT-P, and was cyclized or concatemerized by ligation. Using the obtained DNA as a template, PCRs were performed with primers Maj12s and Maj13a for nepenthesin I and with Min12s and Min13a for nepenthesin II. Then nested PCRs were performed with primers Maj2s and Maj14a for nepenthesin I and with Min2s and Min14a for nepenthesin II. The obtained DNAs were sequenced. The sequence between the N- and C-termini was completed by PCR using the cDNA and the upstream and the downstream primers.
Molecular modelling
The tertiary structures of nepenthesins I and II were predicted by the homology modelling method with the program MODELLER [30], using the crystal structure of porcine pepsin A (PDB ID 5PEP) [31] as a template. The alignment of the amino acid sequences of the target enzyme (nepentheshin I or II) and the template (porcine pepsin) was performed by the program ClustalW [32] to conserve the active-site architecture [33]. The modelling was performed by setting all the parameters to their default values and assuming there was no information on disulphide bonds. Using the resultant structures, the disulphide bonds were assigned and energy minimization calculation was performed. All the procedures for performing the energy minimization and manipulating the atomic co-ordinates of the cysteine residues were performed by the SYBYL molecular modelling system (Tripos, St. Louis, MO, U.S.A.). The energy calculations were performed using a cut-off distance of 10 Å (1 Å=0.1 nm), a dielectric constant of 4r and default values for the other parameters. The graphic image was produced using the MidasPlus program [34].
Constuction of a phylogenetic tree
The amino acid sequences of nepenthesins and related homologues were aligned and a phylogenetic tree was constructed by the program ClustalW [32]. The amino acid sequences of the homologues of nepenthesins were obtained by FASTA/BLASTP searches of the gene/protein databases.
RESULTS
Purification of acid proteinases from the pitcher fluid
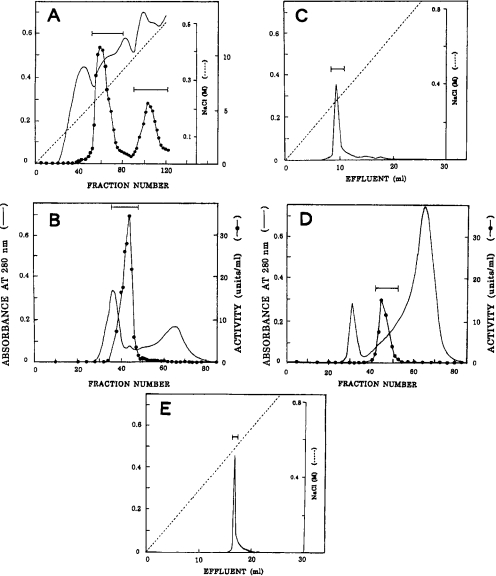
After DEAE-cellulose chromatography of the digestive fluid from the open pitchers of N. distillatoria, two acid proteinase activity peaks were obtained (Figure 1A); the major peak was eluted with 0.25 M NaCl and the minor peak with 0.42 M NaCl. These enzyme fractions were separately purified by successive steps of chromatography on columns of Sephacryl S-200 (Figures 1B and 1D), pepstatin–Sepharose and Mono Q (Figures 1C and 1E). After pepstatin–Sepharose chromatography, the major enzyme was completely eluted with 0.05 M Tris/HCl and 1 M NaCl. On the other hand, the minor enzyme was eluted only partially (approx. 28%) with this buffer and mostly (approx. 72%) with 0.05 M sodium borate buffer (pH 10) and 1 M NaCl; these fractions were combined for further purification since they were apparently indistinguishable in other properties. Thus approx. 1.8 mg of the purified major enzyme (200-fold purification in a yield of 21%) and approx. 0.46 mg of the purified minor enzyme (186-fold purification in a yield of 4.8%) were obtained (Table 2). Each of the purified enzymes gave a single protein band on SDS/PAGE (Figure 2). Similar results were obtained with the digestive fluid from the unopened pitchers (results not shown). The major and minor enzymes were designated nepenthesins I and II respectively.
Figure 1. Purification of Nepenthes acid proteinases.
(A) DEAE-cellulose chromatography. The enzyme fraction eluted from the first DEAE-cellulose column was rechromatographed; fraction size, 10 ml. (B) Sephacryl S-200 chromatography of the major enzyme from the second DEAE-cellulose column; fraction size, 11 ml. (C) Mono Q chromatography of the major enzyme. The enzyme fraction obtained by successive chromatography on Sephacryl S-200 and pepstain–Sepharose was chromatographed. The Figure shows the result obtained with one-third of the enzyme sample. (D) Sephacryl S-200 chromatography of the minor enzyme. (E) Mono Q chromatography of the minor enzyme. The conditions were the same as in (C) except that the whole sample was used. In each chromatography, the fractions under the bar were pooled. The purified major and minor enzymes were designated nepenthesins I and II respectively.
Table 2. Purification of acid proteinases from Nepenthes pitcher fluid.
| Step | Enzyme | Total protein (mg) | Total activity (units) | Specific activity (units/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| Crude fluid | 1770 | 7710 | 4.36 | 100 | 1 | |
| 1st DE-52 | 500 | 5580 | 11.2 | 72.0 | 2.6 | |
| 2nd DE-52 | I | 99.0 | 2600 | 26.3 | 34.0 | 6.0 |
| II | 18.0 | 1140 | 63.3 | 14.8 | 14.5 | |
| Sephacryl S-200 | I | 13.6 | 2080 | 153 | 27.0 | 35.1 |
| II | 3.70 | 790 | 214 | 9.1 | 49.1 | |
| Pepstatin–Sepharose | I | 2.52 | 1810 | 718 | 23.5 | 165 |
| II | 0.98 | 500 | 510 | 6.5 | 117 | |
| FPLC (Mono Q) | I | 1.83 | 1600 | 874 | 21.0 | 200 |
| II | 0.46 | 372 | 809 | 4.8 | 186 |
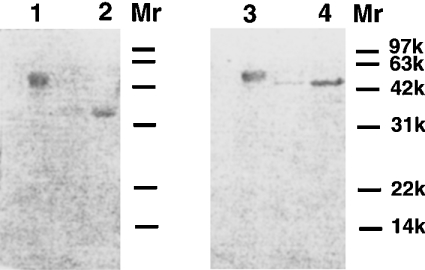
Figure 2. SDS/PAGE of the purified nepenthesins I and II.
Electrophoresis was performed under non-reducing (lanes 1 and 2) and reducing (lanes 3 and 4) conditions. The protein was stained with Coomassie Brilliant Blue. Lanes 1 and 3, nepenthesin I; lanes 2 and 4, nepenthesin II.
Molecular masses of nepenthesins
The molecular mass of nepenthesin I was estimated to be approx. 45 and 51 kDa by SDS/PAGE under non-reducing and reducing conditions respectively (Figure 2, lanes 1 and 3) and 58 kDa by gel filtration. The protein band obtained by SDS/PAGE was stained positively with the periodic acid–Schiff's base reagent. On the other hand, the molecular mass of nepenthesin II was estimated to be approx. 35 and 45 kDa by SDS/PAGE under non-reducing and reducing conditions respectively (Figure 2, lanes 2 and 4). The protein band on SDS/PAGE was negative to the periodic acid–Schiff's base reagent.
The N-terminal and partial internal amino acid sequences of nepenthesins
The N-terminal 24- and 19-residue sequences of nepenthesins I and II respectively were determined as follows. Nepenthesin I: IGPSGVETTVYAGDGEYLMXLSIG and II: QTVQVEPPYYAGDGEYLMV. In addition, partial internal sequences, including 134 and 158 residues for nepenthesins I and II respectively were determined at the protein level, which will be shown later.
Inhibition of nepenthesins by pepstatin and DAN
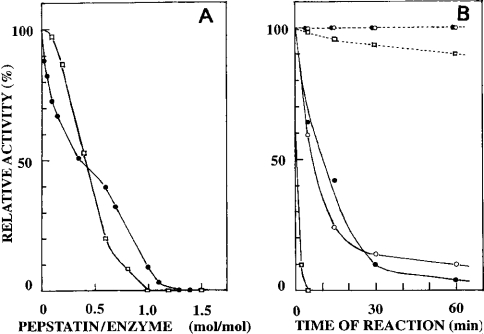
Nepenthesin I was strongly inhibited by pepstatin under acidic conditions similar to porcine pepsin A and complete inhibition was obtained at 0.1 mM pepstatin. The acid proteinase activity in the crude pitcher fluid was also inhibited completely by pepstatin under similar conditions. As can be seen from Figure 3(A), pepstatin appeared to bind to nepenthesin I in a 1:1 stoichiometry similar to porcine pepsin. Nepenthesin II was also shown to be inhibited completely with 0.1 mM pepstatin (results not shown).
Figure 3. Inhibition of nepenthesins by pepstatin and DAN.
(A) Pepstatin inhibition of nepenthesin I (•) at pH 3.0. Inhibition of porcine pepsin A (□) by pepstatin was also examined under the same conditions. (B) DAN inhibition of nepenthesin I in the presence (–•–) and absence (--•--) of cupric ions and of nepenthesin II in the presence (–○–) and absence (--○--) of cupric ions. Porcine pepsin A was treated with DAN in the same manner in the presence (–□–) and absence (--□--) of cupric ions.
Both enzymes were inhibited strongly by DAN in the presence of cupric ions (Figure 3B) and nearly complete inhibition was obtained after 3 h. They were inactivated at similar rates, but much more slowly than porcine pepsin A. In the absence of cupric ions, none of them were inactivated.
Cleavage specificity of nepenthesin towards oxidized insulin B chain
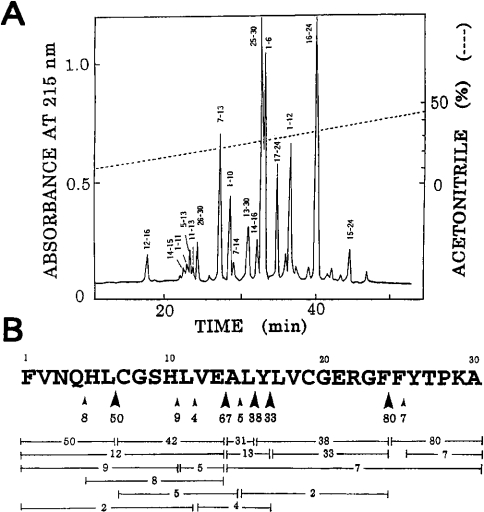
An HPLC pattern of a 3 h digest of oxidized insulin B chain at pH 3.0 by nepenthesin I is shown in Figure 4(A). Several peptide bonds were cleaved and especially the peptide bonds, Phe24-Phe25, Glu13-Ala14, Leu6-Cya7 (where Cya stands for cysteic acid), Leu15-Tyr16 and Tyr16-Leu17, were cleaved significantly (Figure 4B). The extents of cleavage of these bonds were estimated to be 80, 67, 50, 38 and 33% respectively under the conditions used.
Figure 4. Action of nepenthesin I on oxidized insulin B chain.
(A) HPLC of a 3 h digest of oxidized insulin B chain with the purified enzyme at pH 3.0 and 37 °C. The numbers on each peak indicate the residue positions. (B) Cleavage sites and yields of peptides. The number on each horizontal bar indicates the yield (%) of the corresponding peptide. Large and small arrowheads show the major and minor cleavage sites respectively. The number under each arrowhead indicates an estimated extent (%) of cleavage.
Effects of pH on the activity and stability of nepenthesins
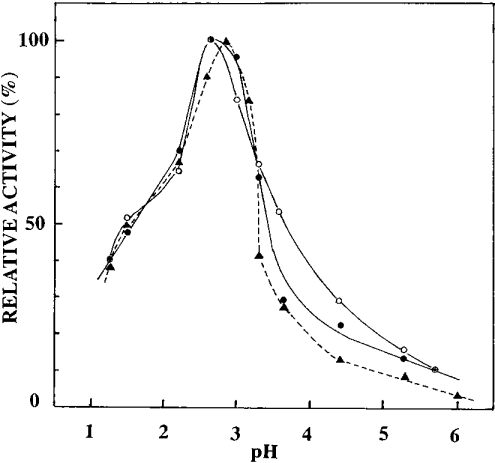
The pH–activity profiles are shown in Figure 5. The optimal activity of each enzyme towards acid-denatured haemoglobin was observed at pH∼2.6; the crude pitcher fluid showed a similar pH–activity profile with an optimum pH at 2.8. It is notable that the purified enzymes as well as the crude fluid possess some activity at pH∼6.0.
Figure 5. pH dependence of the activity of nepenthesins towards haemoglobin.
The activity was measured at various pH values at 37 °C towards acid-denatured haemoglobin. –•–, nepenthesin I; –○–, nepenthesin II; --▴--, crude pitcher fluid.
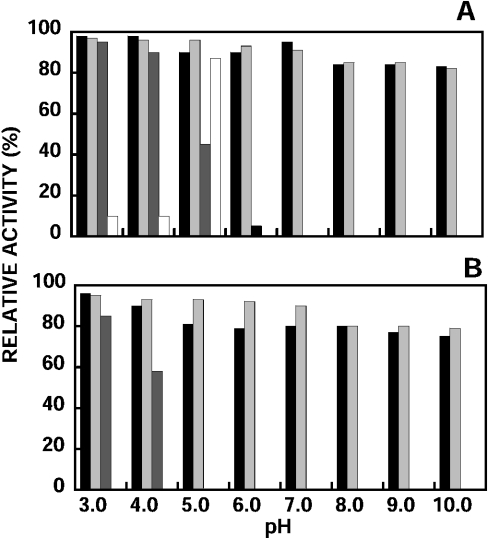
The results obtained when the enzymes were incubated at 37 °C at various pH values (3.0–10.0) are shown in Figures 6(A) and 6(B). Nepenthesin I was most stable at pH 3.0 and 95% of the original activity was retained after incubation for 30 days. Under the same conditions, the enzyme was considerably stable even at pH 10.0, where it retained 79% of the original activity after incubation for 30 days. The results obtained with the crude pitcher fluid were similar to those obtained with nepenthesin I. On the other hand, nepenthesin II was somewhat less stable. It was most stable at pH 3.0 and retained 85% of the original activity after 30 days, whereas the activity was completely lost at pH 5.0 and above within 30 days. Under the same conditions, porcine pepsin A was extremely unstable; it rapidly lost the activity over a wide range of pH. Porcine pepsin A was only stable at pH 5.0 for 7 days, where the enzyme is known to be most stable.
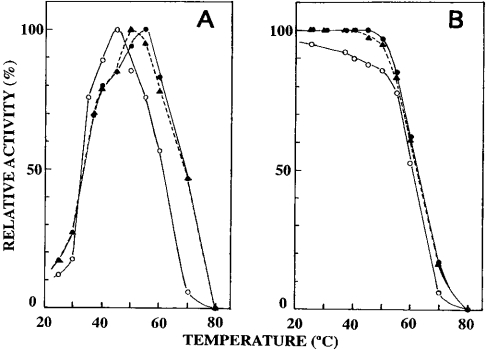
Figure 6. Effects of temperature on the activity and stability of nepenthesins.
(A) The activity was measured at various temperatures at pH 3.0. (B) The activity was measured at 37 °C after incubation of each enzyme at various temperatures at pH 3.0 for 2 h. –•–, nepenthesin I; –○–, nepenthesin II; --▴--, crude pitcher fluid.
Effects of temperature on the activity and stability of nepenthesins
The temperature–activity profiles are shown in Figure 6(A). The optimal temperature of nepenthesin I was 55 °C and above this temperature the activity gradually decreased and was lost completely at 80 °C. On the other hand, the optimal temperature of nepenthesin II was 45 °C and the activity was largely lost at 70 °C. The temperature–activity profile of the crude pitcher fluid was rather similar to that of nepenthesin I with an optimum at 50 °C. When the enzymes were incubated at pH 3.0 and at different temperatures for 1 h and then assayed at 37 °C, the results shown in Figure 7(B) were obtained. Nepenthesin I as well as the activity in the crude fluid was stable up to 50 °C, and above this temperature it became unstable, whereas under the same conditions nepenthesin II was less stable. These results are in good agreement with those shown in Figure 7(A).
Figure 7. Stability of nepenthesins at different pH values.
The activity was measured after incubation at 37 °C for 7 days (A) and 30 days (B) at different pH values: closed bar, crude pitcher fluid; light bar, nepenthesin I; dark bar, nepenthesin II; open bar, porcine pepsin A.
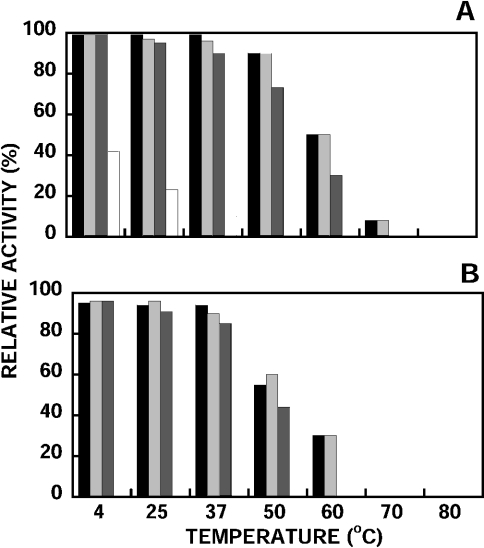
The results obtained when the enzymes were incubated at different temperatures at pH 3.0 for 7 and 30 days, and then assayed for the remaining activity, are shown in Figures 8(A) and 8(B). Nepenthesin I was very stable up to 30 days even at 50 °C. After 30 days at 50 °C, the enzyme retained 60% of the original activity. Similar results were obtained with the crude pitcher fluid. Nepenthesin II was also stable at various temperatures; at 50 °C it retained 44% of the original activity after 30 days. Under the same conditions, porcine pepsin A was significantly unstable; it retained only 10% of the original activity after 7 days at 37 °C, whereas nepenthesins I and II retained 96 and 90% respectively of the original activity.
Figure 8. Stability of nepenthesins at different temperatures.
The activity was measured after incubation at pH 3.0 for 7 days (A) and 30 days (B) at different temperatures. The same notations as in Figure 7 are used.
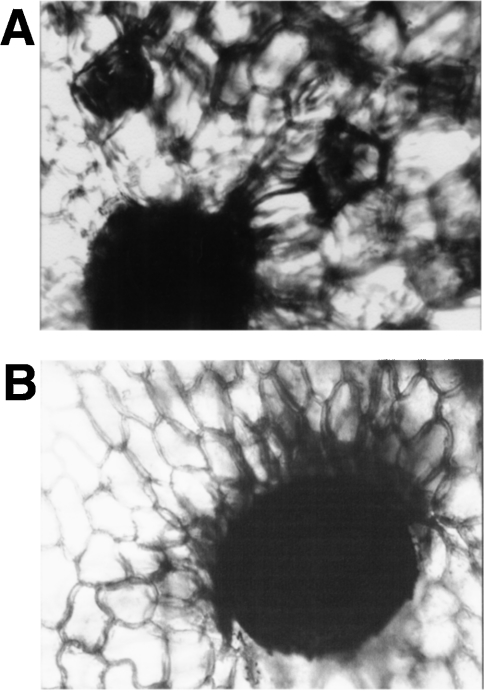
Cellular localization of nepenthesin
The results of immunohistochemical staining of nepenthesin I are shown in Figure 9. Parenchymal cells surrounding the secretory glands were stained purple with the antibody (Figure 9A), which was not observed with the control sample without the primary antibody (Figure 9B). Positive staining was not observed for the upper part of the pitcher and the bine portion connecting the pitcher with the leaf, both of which lack the secretory glands (results not shown). Under the conditions used, the secretory glands appeared as black dots and it was difficult to see how much they were positively stained.
Figure 9. Immunohistochemical staining of Nepenthes pitcher tissue with an anti-nepenthesin I antibody.
Longitudinal sections of the lower one-third part of the Nepenthes pitcher tissue were stained with the specific antibody against nepenthesin I (A) and without the specific antibody (control) (B). Magnification,×400.
Amino acid sequences of nepenthesins I and II
The complete amino acid sequences of the prepro forms of nepenthesins I and II were deduced by cloning and nucleotide sequencing of the cDNA clones obtained from N. gracilis as shown in Figures 10(A) and 10(B). The N-terminal and internal sequences determined at the protein level with the enzymes from N. distillatoria are also shown in Figure 10. The N-terminus of each mature enzyme was deduced by comparison of the deduced amino acid sequence with the N-terminal sequences of the N. distillatoria enzymes. Thus prepro-nepenthesin I is composed of 437 residues, including a 24-residue putative signal sequence, a 56-residue putative propeptide and a 359-residue mature enzyme (nepenthesin I), and prepro-nepenthesin II is composed of 438 residues, including a 24-residue putative signal sequence, a 55-residue putative propeptide and a 359-residue mature enzyme (nepenthesin II). Nepenthesin I was found to have two variants: one (nepenthesin Ia) having Asp, Asn and Gly and the other (nepenthesin Ib) having Val, Thr and Glu at positions 233, 251 and 392 respectively (the numbering of the prepro-nepenthesin I is used throughout the text unless otherwise specified).
Figure 10. The nucleotide and the deduced amino acid sequences of nepenthesin Ia (A) and nepenthesin II (B) from N. gracilis.
The putative active-site aspartic acids and the flap tyrosine are shown by an asterisk. The potential N-glycosylation site motifs are underlined. Under the sequences are also shown the partial amino acid sequences of peptides obtained from the N. distillatoria enzymes determined at the protein level. Peptide numbers are shown in parenthesis before or after each sequence. N-Term, N-terminal sequence determined with the whole protein; DN, endopeptidase Asp-N; T, trypsin; V, Staphylococcus aureus V8 protease.
The calculated molecular masses and pI values of the mature enzymes are respectively 37.476 kDa and 3.94 for nepenthesin Ia, 37.519 kDa and 3.94 for nepenthesin Ib and 37.511 kDa and 3.09 for nepenthesin II. Nepenthesin I contains six potential N-glycosylation sites whereas nepenthesin II has none. Each enzyme contains the active-site sequence motifs, Asp-Thr-Gly and Asp-Ser-Gly, the so-called flap tyrosine residue (residue 174 corresponding to Tyr75 in the porcine pepsin numbering) and notably 12 cysteine residues per molecule. Each lacks the plant-specific insertion, typical of plant vacuolar APs at positions between Asn340 and Leu341, but appears to have an approx. 22-residue insertion (tentatively assigned to residues 148–169) preceding the flap tyrosine residue. This insertion contains four cysteine residues and was named ‘the nepenthesin-type AP (NAP)-specific insertion.’
The partial amino acid sequences determined at the protein level for nepenthesin I (total 157 residues) and nephenthesin II (total 177 residues) from N. distillatoria were different at eight and eleven positions respectively from the sequences deduced from N. gracilis; thus the sequence identity of each enzyme from the two species was 94%. In addition, sequence variations, Ser/Tyr and Thr/Asp, were observed at position 109 in nepenthesin I and at position 148 in nepenthesin II respectively from N. distillatoria.
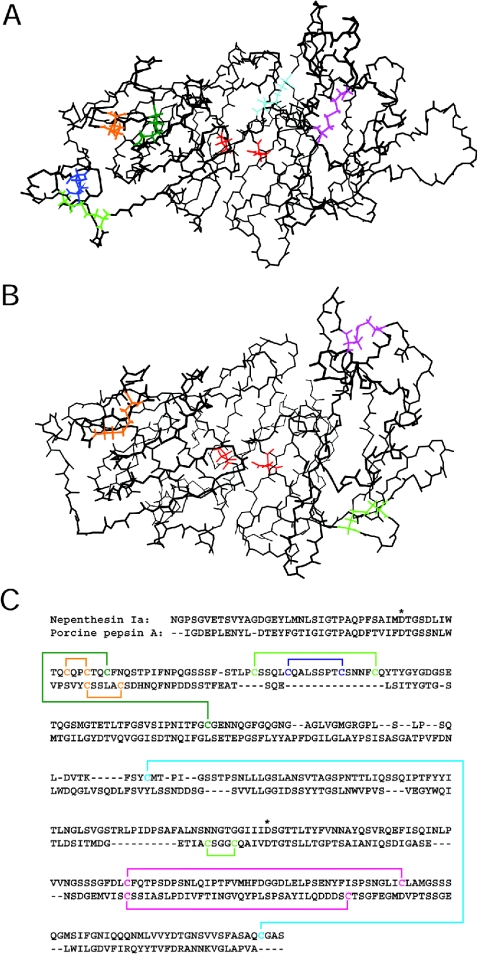
Tertiary structures of nepenthesins predicted by molecular modelling
Figure 11 shows the tertiary structure of nepenthesin Ia predicted by molecular modelling. The predicted backbone structures of nepenthesins Ib and II (results not shown) were essentially the same as that of nepenthesin Ia. In the predicted structures based on homology modelling, the two cysteine residues in each of the two pairs, Cys45/Cys48 and Cys162/Cys356, were located close enough to form disulphide bonds (the cysteine residue numbering is based on the sequence of the mature enzyme). Therefore the disulphide bonds were created in the above pairs. The pairing of the four cysteine residues, Cys72, Cys77, Cys85 and Cys90, which are closely located in the NAP-specific insertion, could not be predicted since there was very little structural information on this region. Therefore the energy minimization calculation was performed with the two disulphide bonds formed, but with the four cysteine residues unconnected. In the resultant structure, the locations of the remaining four cysteine residues (Cys51, Cys125, Cys276 and Cys317) were still somewhat distant from each other, but the Cys276/Cys317 pair was assumed to form a disulphide bond since porcine pepsin has a disulphide bond (Cys249-Cys282) at a similar location. Thus the disulphide bonds were introduced between Cys276 and Cys317 and between Cys51 and Cys125. After the energy minimization, the location of the two disulphide bonds was shown to be reasonable, and there were no steric or energetic hindrances in the whole molecule. For the cysteine residues in the NAP-specific insertion, the most plausible pairings would be Cys72–Cys90 and Cys77–Cys85 if the insertion sequence were looped out from the rest of the enzyme molecule. Figure 11 also shows the predicted disulphide bonds including the tentative pairs within the insertion sequence.
Figure 11. The backbone structure and disulphide bond pairing of nepenthesin Ia predicted by molecular modelling and comparison with porcine pepsin A.
The side chains of the cysteine residues and the putative catalytic aspartic acid residues are presented in (A, B). (A) The backbone structure of N. gracilis nepenthesin Ia. All the cysteine residues are assumed to form disulphide bonds. Cys45–Cys48 is coloured in orange, Cys51–Cys125 in green, Cys72–Cys90 in light green, Cys77–Cys85 in blue, Cys162–Cys356 in cyan and Cys276–Cys317 in magenta. The catalytic residues, Asp35 (left) and Asp237 (right), are coloured in red. (The numbering used is for the mature nepenthesin Ia.) (B) The backbone structure of porcine pepsin A. Cys45–Cys50, Cys206–Cys210 and Cys249–Cys282 (pepsin numbering) are shown in orange, light green and magenta respectively and the catalytic residues, Asp32 (left) and Asp215 (right), are shown in red. (C) Predicted arrangements of the disulphide bonds in nepenthesin Ia (upper row) and porcine pepsin A (lower row). The cysteine residues and the disulphide bonds are shown in the same colours as in (A, B). The catalytic aspartic acid residues are shown by asterisks.
Figure 11(B) shows the tertiary structure of porcine pepsin A [33] for comparison, and Figure 11(C) shows the disulphide bond arrangements in the primary structures of nepenthesin I and porcine pepsin A. When compared with pepsin A, it appears that three additional disulphide bonds (Cys51–Cys125, Cys72–Cys90 and Cyc77–Cys85) are introduced into the N-terminal lobe of nepenthesin, and one additional disulphide bond (Cys162–Cys356) between the N- and C-terminal lobes and that one disulphide bond (Cys206–Cys210) in the C-terminal lobe of pepsin A is lost from nepenthesin.
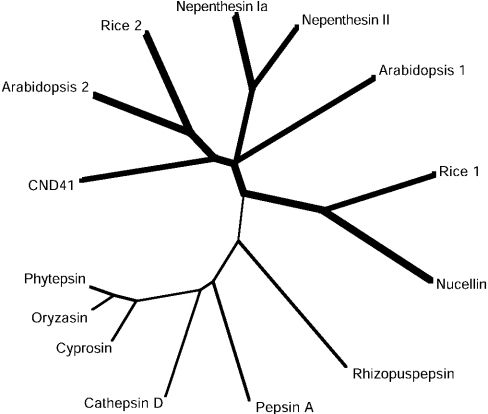
Molecular phylogeny
The amino acid sequences of the prepro forms of nepenthesins and some of their typical homologues were aligned with those of some typical homologues to compare them and to construct a phylogenetic tree. For this comparison, we included eight NAPs with 12 conserved cysteine residues at similar positions [nepenthesins and orthologue enzymes from Arabidopsis thaliana, barley (nucellin), rice and tobacco (CND41, chloroplast nucleoid DNA-binding protein 41)], which have been identified at the protein and/or cDNA level. As for the Arabidopsis and rice enzymes, two typical ones with much different pI values were selected. We also included six of the already known typical pepsin-type APs with six or four conserved cysteine residues at similar positions (pepsin A, cathepsin D, rhizopuspepsin and the plant vacuolar enzyme cyprosin, oryzasin and phytepsin). All these enzymes appeared to be roughly similar in size except that the three vacuolar APs had an additional sequence of approx. 100 residues, called a plant-specific insertion. The two active-site aspartic acid residues in the Asp-Thr-Gly and Asp-Ser/Thr/Cys-Gly motifs are conserved among all the enzymes. The tyrosine residue on the flap of porcine pepsin also appears to be conserved among them, although we have to consider the presence of the NAP-specific insertion for the nepenthesin-type enzymes, which contains two putative disulphide bonds. The sequence identities were calculated for the mature enzymes using the sequences corresponding to those of residue 17 to the C-terminal cysteine of nepenthesins after removing the NAP-specific insertion from the nepenthesins and their orthologues and the plant-specific insertion from the three plant vacuolar APs. The identities thus obtained are 67% between nepenthesins I and II, 23–38% (average, 30%) between nepenthesins and the rest of the nepenthesin-type enzymes and 12–22% (average, 18%) between nepenthesins and the ordinary pepsin-type enzymes. It is also notable that the potential N-glycosylation sites are rich in nepenthesin I (six sites), one of the Arabidopsis enzymes (accession no. AY088536) (five sites) and one of the rice enzymes (accession no. AK106097) (five sites). On the basis of the sequence information, a phylogenetic tree was constructed as shown in Figure 12.
Figure 12. A phylogenetic tree for nepenthesins and related typical APs.
Bold line, the branches of the NAPs; thin line, the ordinary pepsin-type APs. The phylogenetic tree was produced by ClustalW using the sequence of residues 95–434 of prepro-nepenthesin Ia and the corresponding sequences of the other APs, after removing the NAP-specific insertion and the plant-specific insertion. The sequences of the following 12 APs are compared: (1) nepenthesin Ia, N. gracilis AP Ia (AB114914); (2) nepenthesin II, N. gracilis AP II (AB114915); (3) Arabidopsis 1, A. thaliana AP (AF37029) (Ip, 4.3); (4) Arabidopsis 2, A. thaliana AP (AY088536) (Ip, 9.5); (5) nucellin, H. vulgare (barley) AP (U87148); (6) rice 1, O. sativa AP (AK068348) (Ip, 8.7); (7) rice 2, O. sativa AP (AK165097) (Ip, 5.3); (8) CND41, N. tabacum (tobacco) AP (D26015); (9) cyprosin, Cynara caldunclus AP (X81984); (10) oryzasin, O. sative AP (D32114); (11) phytepsin, H. vulgare (barley) AP (X56136); (12) cathepsin D, porcine cathepsin D (M11233); (13) pepsin A, porcine pepsin A (M20920); and (14) Rhisopuspepsin, Rhizopus chinensis AP (P06026). The number in parenthesis is the accession number.
DISCUSSION
Two acid proteinases, nepenthesins I and II, were purified to homogeneity as examined by SDS/PAGE and N-terminal amino acid sequencing from the pitcher fluid of N. distillatoria. Moreover, the primary structures of the two enzymes were deduced by cloning and sequencing the corresponding cDNAs from N. gracilis. This is the first case of complete purification and primary structure determination of extracellular proteinases in the digestive fluid of a carnivorous plant. Recently, Ann et al. [35] have reported the cloning of APs from the pitcher tissue of N. alata (winged pitcher-plant). However, these enzymes clearly belong to vacuolar APs since they share a so-called plant-specific insersion and lack the primary structure features characteristic of nepenthesins. In the present study, we used N. distillatoria for studies at the protein level and N. gracilis for those at the DNA level. Nepenthesins I and II from the former are supposed to be essentially the same enzymes as the corresponding enzymes from the latter; the partial amino acid sequences of nepenthesins I and II from N. distillatoria determined at the protein level were 93–94% identical with the corresponding sequences of nepenthesins I and II from N. gracilis respectively.
Nepenthesins I and II from N. distillatoria are significantly different from each other in properties as judged from the differences in chromatographic behaviour, especially on DEAE-cellulose, molecular mass, N-terminal and partial internal amino acid sequence and effects of temperature and pH on the activity and stability. These results are consistent with the amino acid sequences deduced from the cDNAs for the enzymes from N. gracilis. The amino acid sequence identity between nepenthesins I and II from N. gracilis is 66.6%. Since nepenthesin I was eluted from the DEAE-cellulose column before nepenthesin II, the former should be less acidic than the latter. Indeed, the pI values calculated from the amino acid composition of each enzyme are 3.94 and 3.09 for N. gracilis nepenthesins I and II respectively. The approximate molecular masses of N. distillatoria nepenthesins I and II were estimated from SDS/PAGE to be 45 and 35 kDa respectively under non-reducing conditions and 51 and 45 kDa respectively under reducing conditions. On the other hand, the calculated molecular masses of N. gracilis nepenthesins I and II were both 37.5 kDa. This discrepancy is supposed to be due to the presence of carbohydrate in nepenthesin I, but not in nepenthesin II. This is consistent with the fact that six potential N-glycosylation sites are present in N. gracilis nepenthesin I, but none in N. gracilis nepenthesin II (see Figure 10). The difference in molecular mass of 6–10 kDa between N. distillatoria nepenthesins I and II indicates that the carbohydrate content in nepenthesin I should be more than 10% by weight. In the partial amino acid sequencing of N. distillatoria nepenthesin I, the asparagine residues in the first and second potential N-glycosylation sites (residues 98 and 131) could not be positively identified at the protein level, whereas the asparagine residues at the third and fourth sites (residues 98 and 167) were identified, although no information has been obtained as yet for the remaining three sites at the protein level. Therefore these results seem to suggest that at least the first and second sites are glycosylated and that the third and fourth sites are not.
As shown by immunohistochemical staining (Figure 9), the enzymes appear to be synthesized in the parenchymal cells surrounding the secretory glands. However, the pitcher tissue was not so strongly stained. Furthermore, the extract of the pitcher tissues failed to show a significant level of acid proteinase activity. It is unlikely that these results are due to the occurrence of the enzymes in the zymogen form in the tissue, since they are supposed to react with the polyclonal antibody similar to the mature enzyme and to be rapidly activated under the acidic assay conditions. Therefore these results seem to indicate that the zymogens, which are supposed to be synthesized in the pitcher tissue are rapidly secreted into the pitcher fluid and activated without accumulation in the tissue, although other possibilities cannot be completely excluded.
The AP-specific inhibitor pepstatin and DAN were shown to inhibit the purified enzymes strongly similar to porcine pepsin A. Previously, we have shown that the acid proteinase activity in the crude pitcher fluid of N. ampullaria was strongly inhibited by pepstatin and DAN [11]. Thus the previous finding was confirmed with the purified enzymes, indicating that the enzymes belong to the typical pepstatin-sensitive AP family. On pepstatin–Sepharose chromatography, nepenthesin II was more firmly bound to pepstatin–Sepharose when compared with nepenthesin I. This suggests that nepenthesin II should be more sensitive to pepstatin when compared with nepenthesin I.
The specificity of nepenthesin I towards an oxidized insulin B chain appears to be similar, but somewhat wider when compared with those of other APs such as pepsin A and cathepsin D; human pepsin A cleaved mainly the Leu11–Val12, Leu15–Tyr16 and Phe25–Tyr26 bonds and rat cathepsin D cleared the Leu15–Tyr16, Phe24–Phe25 and Phe25–Tyr26 bonds [36]. Most peptide bonds susceptible to these enzymes were more or less cleaved by nepenthesin I. Interestingly, the Leu6–Cya7 bond was found to be one of the major sites of cleavage by nepenthesin I; the cleavage of this bond by other APs has not been reported. Leucine residue seems to be one of the preferred P1 site of the enzyme since three of the four Leu–Xaa bonds in oxidized insulin B chain were more or less cleaved.
At or below 50 °C, nepenthesin I was found to be extraordinarily stable in a wide range of pH for a long period. Nepenthesin II was also fairly stable, but less stable than nepenthesin I. Such an unusual stability, especially of nepenthesin I, has never, to our knowledge, been reported for other proteinases. The cDNA sequencing revealed that both nepenthesins have a high content of cysteine residues, 12 residues per molecule of protein, which would form six disulphide bonds. APs with such a high disulphide bond content have not been known before except for plant vacuolar APs that have three additional disulphide bonds within the plant-specific insersion [37], which is absent in nepenthesins. The high content and specific pairing of the disulphide bonds should contribute greatly to the stability of both nepenthesins. Indeed, both nepenthesins are predicted to have three disulphide bonds, each linking two cysteine residues distantly located in the primary structure, i.e. one in the N-terminal lobe, the second between the N- and C-terminal lobes and the third in the C-terminal lobe (Figure 11). On the other hand, most of the ordinary pepsin-type APs have only one such disulphide bond in the C-terminal lobe. In nepenthesins, one disulphide bond present in the C-terminal lobe of porcine pepsin A is absent; this would not affect the stability significantly since this disulphide bond is formed between two nearby cysteine residues and is not always conserved in ordinary pepsin-type APs, such as rhizopuspepsin and penicillopepsin.
The presence of carbohydrate and a higher pI value of nepenthesin I are supposed to contribute to render nepenthesin I much more stable than nepenthesin II. The carbohydrate moieties may help increase the stability by reducing the possibility of autolysis and/or denaturation. On the other hand, nepenthesin I (pI, 3.94) has 20 acidic residues (12 Asp and 8 Glu) and five basic residues (1 Lys, 1 His and 3 Arg), whereas nepenthesin II (pI, 3.09) has 33 acidic residues (18 Asp and 15 Glu) and one basic residue (1 Lys). Therefore the charge repulsion among the dissociating carboxylate groups, which will lead to denaturation, should be less pronounced in nepenthesin I than in nepenthesin II as the pH is raised. Thus, these enzymes, especially nepenthesin I, appear to be so designed that they are capable of remaining stable in a wide range of pH at relatively high temperatures to work for a long period in the pitcher fluid of the Nepenthes plants in a tropical habitat, indicating an evolutionary adaptation of the enzymes to their specific environments. To extend further the structure–function studies, it is absolutely necessary to elucidate the three-dimensional structure of the enzymes.
As can be seen from the phylogenetic tree (Figure 12), nepenthesins clearly belong to a novel subfamily of APs. Previously, Chen and Foolad [38] have reported that the barley gene encoded an AP-like protein with no plant-specific insertion, named it nucellin and suggested that it may be involved in nucellar cell death, although no studies were performed at the protein level. On the other hand, Nakano et al. [39] isolated a similar 41 kDa AP-like protein with DNA-binding activity from the chloroplast nucleoids of tobacco cells, named it CND41 and suggested its possible function as a negative regulator of chloroplast gene expression. Later, this protein was purified and shown to have proteolytic activity optimally at acidic pH, which, however, was reported to be only weakly inhibited by pepstatin [40]. The elucidation of the primary structures of nepenthesins in this study has shown that nepenthesins, nucellin and CND41 belong to the same type of APs, and facilitated us to make a further search for the same types of enzymes. FASTA and BLASTP homology searches for nepenthesins in various databases revealed that at the genomic DNA level, there are nearly 90 and 30 orthologues in A. thaliana and Oryza sativa respectively, two in Nicotiana tabacum, one in Zea mays and one in Hordeum vulgar. So far the cDNAs have been obtained for 16 orthologues in A. thaliana, six in O. sativa, one in N. tabacum (CND41) and one in H. vulgar (barley nucellin).
These NAPs have the NAP-specific insertion, but no plant-specific insertion, and many of them have 12 cysteine residues at the positions corresponding to those in nepenthesins, whereas some others lack one or few of these cysteine residues. The NAP-specific insertion mostly contains four (and occasionally fewer) cysteine residues, and those of the six nepenthesin-type enzymes (Figure 12) are shown below. Among them, there are significant variations in sequence and it is tempting to assume that they may play some specific role(s) in intracellular targeting or functional regulation of the enzymes.
(1) Nepenthesin Ia: LPCSSQLCQALSSPTCSNNFCQ
(2) Nepenthesin II: LPCESQYCQDLPSETCNNNECQ
(3) Arabidopsis 1: LTCSAPQCSLLETSACRSNKCL
(4) Arabidopsis 2: VFCNSSTCQDLVAATSNSGPCGGNNGVVKTPCE
(5) Nucellin: VVCGSPLCVAVRRDVPGIPECSRNDPHRCH
(6) Rice 1: VPCANSICTALHSGSSPNKKCTTQQQCD
(7) Rice 2: VPCNASACEASLKAATGVPGSCATVGGGGGGGKSERCY
(8) CND41: ISCTSAACSSLKSATGNSPGCSSSNCV.
It is also interesting to note that the putative mature forms of these enzymes can be classified into two groups with different pI values as for nepenthesins, although their pI values are generally higher than those of nepenthesins: one group is weakly acidic with pI values of approx. 4–6 and the other group is basic with pI values of approx. 8–10. These differences might be correlated with their physiological roles.
Construction of a phylogenetic tree including all of these over 100 orthologues (K. Takahashi, unpublished work) has shown that they all belong to the NAPs distinct from already known ordinary pepsin-type APs. Thus nepenthesins and those orthologues constitute a novel subfamily of APs, suggesting their ubiquitous distribution and multiple roles in the plant kingdom. With the identification of more and more gene sequences, more APs of this type will be found in other plants as well. From the sequences examined so far, an orthologous enzyme appears to be present even in Chlamydomanas, although not in Cyanobacteria. Among these enzymes, nepenthesins are the only known extracellular enzymes, and the others seem to be intracellular enzymes. Therefore nepenthesins are supposed to have adapted specifically to become extracellular digestive enzymes during the course of molecular evolution. The physiological roles of the intracellular NAPs remain to be elucidated.
Acknowledgments
We are grateful to Dr M. Tanji (Department of Biophysics and Biochemistry, Graduate School of Science, University of Tokyo, Tokyo, Japan) for his help in the preliminary studies on carnivorous plant acid proteinases, and to Professor N.S. Andreeva (Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow, Russia) and Professor C. Faro (Department of Biochemistry, University of Coimbra, Coimbra, Portugal) for helpful discussions on the sequence alignment regarding the flap tyrosine. This study was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science, Culture and Sports of Japan and the Japan Society for Promotion of Science and by a grant (NSF/BT/97/03) from National Science Foundation (Sri Lanka).
References
- 1.Darwin C. London: John Murray; 1875. Insectivorous Plants. [Google Scholar]
- 2.Lloyd F. E. New York: Ronald Press; 1942. The Carnivorous Plants. [Google Scholar]
- 3.Hooker J. D. The carnivorous habits of plants. Nature (London) 1874;10:366–372. [Google Scholar]
- 4.Frazer C. K. The enduring controversies concerning the process of protein digestion in Nepenthes (Nepenthaceae) Carniv. Pl. Newslett. 2000;29:56–62. [Google Scholar]
- 5.Woessner J. F. Nepenthesin. In: Barrett A. J., Rawlings N. D., Woessner J. F., editors. Handbook of Proteolytic Enzymes. San Diego, CA: Academic Press; 1998. pp. 846–848. [Google Scholar]
- 6.Nakayama S., Amagase S. Acid protease in Nepenthes: partial purification and properties of the enzyme. Proc. Jpn. Acad. 1968;44:358–362. [Google Scholar]
- 7.Amagase S., Nakayama S., Tsugita A. Acid protease in Nepenthes. II. Study on the specificity of nepenthesin. J. Biochem. (Tokyo) 1969;66:431–439. doi: 10.1093/oxfordjournals.jbchem.a129166. [DOI] [PubMed] [Google Scholar]
- 8.Amagase S. Digestive enzymes in the insectivorous plants. III. Acid proteases in the genus Nepenthes and Drosera peltata. J. Biochem. (Tokyo) 1972;72:73–81. doi: 10.1093/oxfordjournals.jbchem.a129899. [DOI] [PubMed] [Google Scholar]
- 9.Lobareva L. S., Rudenskaya G. N., Stepanov V. M. Pepsin-like protease from the insectivorous plant Nepenthes. Biokhimiya. 1973;38:640–642. [PubMed] [Google Scholar]
- 10.Jenttsch J. Isolation of the peotease nepthacin. FEBS Lett. 1972;21:273–276. doi: 10.1016/0014-5793(72)80181-9. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K., Chang W.-J., Ko J.-S. Specific inhibition of acid proteases from brain, kidney, skeletal muscle and insectivorous plants by diazoacetyl-D,L-norleucine methyl ester and by pepstatin. J. Biochem. (Tokyo) 1974;76:897–899. [PubMed] [Google Scholar]
- 12.Tökés Z. A., Woon W. C., Chambers S. M. Digestive enzymes secreted by the carnivorous plant Nepenthes macfarianei. Planta. 1974;119:39–46. doi: 10.1007/BF00390820. [DOI] [PubMed] [Google Scholar]
- 13.Rawlings N. D., Barrett A. J. Families of aspartic peptidases, and those of unknown catalytic mechanism. Methods Enzymol. 1995;248:105–120. doi: 10.1016/0076-6879(95)48009-9. [DOI] [PubMed] [Google Scholar]
- 14.Barrett A. J., Rawlings N. D., Woessner J. F., editors. Handbook of Proteolytic Enzymes. San Diego, CA: Academic Press; 1998. Aspartic peptidases; pp. 799–986. [Google Scholar]
- 15.Kervinen J., Törmäkangas K., Runeberg-Roos P., Guruprasad K., Blundell T., Teeri T. H. Structure and possible function of aspartic proteinases in barley and other plants. In: Takahashi K., editor. Aspartic Proteinases: Structure, Function, Biology, and Biomedical Implications. New York: Prenum Press; 1995. pp. 241–254. [DOI] [PubMed] [Google Scholar]
- 16.Kervinen J. Phytepsin. In: Barrett A. J., Rawlings N. D., Woessner J. F., editors. Handbook of Proteolytic Enzymes. San Diego, CA: Academic Press; 1998. pp. 836–839. [Google Scholar]
- 17.Runeberg-Roos P., Törmäkangas K., Ostman A. Primary structure of a barley-grain aspartic proteinase. A plant aspartic proteinase resembling mammalian cathepsin D. Eur. J. Biochem. 1991;202:1021–1027. doi: 10.1111/j.1432-1033.1991.tb16465.x. [DOI] [PubMed] [Google Scholar]
- 18.Asakura T., Watanabe H., Abe K., Arai S. Rice aspartic proteinase, oryzasin, expressed during seed ripening and germination, has a gene organization distinct from those of animal and microbial aspartic proteinases. Eur. J. Biochem. 1995;232:77–83. doi: 10.1111/j.1432-1033.1995.tb20783.x. [DOI] [PubMed] [Google Scholar]
- 19.Cordeiro C. M., Brodelius P. E., Pais S. M. Cyprosin. In: Barrett A. J., Rawlings N. D., Woessner J. F., editors. Handbook of Proteolytic Enzymes. San Diego, CA: Academic Press; 1998. pp. 839–842. [Google Scholar]
- 20.Cordeiro M., Xue Z.-T., Pietrzak M., Pais M. S., Brodelius P. Plant aspartic proteinases from Cynara cardunculus spp. flavescens cv. Cardoon: nucleotide sequence of a cDNA encoding cyprosin and its organ-specific expression. In: Takahashi K., editor. Aspartic Proteinases: Structure, Function, Biology and Biomedical Implications. New York: Plenum Pess; 1995. pp. 367–372. [DOI] [PubMed] [Google Scholar]
- 21.Pires E. M. V. Cardosin A. In: Barrett A. J., Rawlings N. D., Woessner J. F., editors. Handbook of Proteolytic Enzymes. San Diego, CA: Academic Press; 1998. pp. 843–844. [Google Scholar]
- 22.Pires E. M. V. Cardosin B. In: Barrett A. J., Rawlings N. D., Woessner J. F., editors. Handbook of Proteolytic Enzymes. San Diego, CA: Academic Press; 1998. pp. 844–846. [Google Scholar]
- 23.Faro C., Verissimo P., Lin Y., Tang J., Pires E. Cardosin A and B, aspartic proteinases from the flowers of cardoon. In: Takahashi K., editor. Aspartic Proteinases: Structure, Function, Biology and Biomedical Implications. New York: Plenum Pess; 1995. pp. 374–377. [Google Scholar]
- 24.Athauda S. B. P., Tanji M., Kageyama T., Takahashi K. A comparative study on the NH2-terminal amino acid sequences and some other properties of six isozymic forms of human pepsinogens and pepsins. J. Biochem. (Tokyo) 1989;106:920–927. doi: 10.1093/oxfordjournals.jbchem.a122952. [DOI] [PubMed] [Google Scholar]
- 25.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Zacharius R. M., Zell T. E., Morrison J. H., Woodlock J. J. Glycoprotein staining following electrophoresis on acrylamide gels. Anal. Biochem. 1969;30:148–152. doi: 10.1016/0003-2697(69)90383-2. [DOI] [PubMed] [Google Scholar]
- 28.Crestfield A. M., Moore S., Stein W. H. The preparation and enzymatic hydrolysis of reduced and S-carboxymethylated proteins. J. Biol. Chem. 1963;238:622–627. [PubMed] [Google Scholar]
- 29.Wan C.-Y., Wilkins T. A. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.) Anal. Biochem. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- 30.Sali A., Blundell T. L. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 31.Cooper J. B., Khan G., Taylor G., Tickle I. J., Blundell T. L. X-ray analyses of aspartic proteinases. II. Three-dimensional structure of the hexagonal crystal form of porcine pepsin at 2.3 Å resolution. J. Mol. Biol. 1990;214:199–222. doi: 10.1016/0022-2836(90)90156-G. [DOI] [PubMed] [Google Scholar]
- 32.Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andreeva N. S., Rumsh L. D. Analysis of crystal structures of aspartic proteinases: on the role of amino acid residues adjacent to the catalytic site of pepsin-like enzymes. Protein Sci. 2001;10:2439–2450. doi: 10.1110/ps.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrin T. E., Huang C. C., Jarvis L. E., Langridge R. The MIDAS display system. J. Mol. Graph. 1988;6:13–27. [Google Scholar]
- 35.Ann C.-I., Fukusaki E., Kobayashi A. Aspartic proteinases are expressed in pitchers of the carnivorous plant Nepenthes alata Blanco. Planta. 2000;214:661–667. doi: 10.1007/s004250100665. [DOI] [PubMed] [Google Scholar]
- 36.Matsuzaki O., Takahashi K. Improved purification of slow moving protease from human gastric mucosa and its action on the B chain of oxidized bovine insulin. Biomed. Res. 1988;9:515–523. [Google Scholar]
- 37.Kervinen J., Tobin G. J., Costa J., Waugh D. S., Wlodawer A., Zdanov A. Crystal structure of plant aspartic proteinase prophytepsin: inactivation and vacuolar targeting. EMBO J. 1999;18:3947–3955. doi: 10.1093/emboj/18.14.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen F., Foolad M. R. Molecular organization of a gene in barley which encodes a protein similar to aspartic protease and its specific expression in nucellar cells during degeneration. Plant Mol. Biol. 1997;35:821–831. doi: 10.1023/a:1005833207707. [DOI] [PubMed] [Google Scholar]
- 39.Nakano T., Murakami S., Shoji T., Yoshida S., Yamada Y., Sato F. A novel protein with DNA binding activity from tobacco chloroplast nucleoids. Plant Cell. 1997;9:1673–1682. doi: 10.1105/tpc.9.9.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murakami S., Kondo Y., Nakano T., Sato F. Protease activity of CND41, a chloroplast nucleoid DNA-binding protein, isolated from cultured tobacco cells. FEBS Lett. 2000;468:15–18. doi: 10.1016/s0014-5793(00)01186-8. [DOI] [PubMed] [Google Scholar]