Abstract
GFP (green fluorescent protein)-based FRET (fluorescence resonance energy transfer) technology has facilitated the exploration of the spatio-temporal patterns of cellular signalling. While most studies have used cyan- and yellow-emitting FPs (fluorescent proteins) as FRET donors and acceptors respectively, this pair of proteins suffers from problems of pH-sensitivity and bleeding between channels. In the present paper, we demonstrate the use of an alternative additional donor/acceptor pair. We have cloned two genes encoding FPs from stony corals. We isolated a cyan-emitting FP from Acropara sp., whose tentacles exhibit cyan coloration. Similar to GFP from Renilla reniformis, the cyan FP forms a tight dimeric complex. We also discovered an orange-emitting FP from Fungia concinna. As the orange FP exists in a complex oligomeric structure, we converted this protein into a monomeric form through the introduction of three amino acid substitutions, recently reported to be effective for converting DsRed into a monomer (Clontech). We used the cyan FP and monomeric orange FP as a donor/acceptor pair to monitor the activity of caspase 3 during apoptosis. Due to the close spectral overlap of the donor emission and acceptor absorption (a large Förster distance), substantial pH-resistance of the donor fluorescence quantum yield and the acceptor absorbance, as well as good separation of the donor and acceptor signals, the new pair can be used for more effective quantitative FRET imaging.
Keywords: donor/acceptor pair, fluorescence resonance energy transfer (FRET), green fluorescent protein (GFP), green fluorescent protein (GFP)-like protein
Abbreviations: CCD, charge-coupled device; CFP, cyan fluorescent protein; ECFP, enhanced CFP; FP, fluorescent protein; FRET, fluorescence resonance energy transfer; GFP, green fluorescent protein; KO, Kusabira-Orange; mKO, monomeric KO; MiCy, Midori-ishi cyan; mRFP1, monomeric red fluorescent protein; YFP, yellow fluorescent protein
INTRODUCTION
While spectral variant proteins with blue, cyan and yellow-green emissions have been generated from the bioluminescent jellyfish, Aequorea victoria [1], the discovery of GFP (green fluorescent protein)-like proteins from Anthozoa has significantly expanded the range of colours available for biotechnological applications. In 1999, Matz et al. [2] cloned six anthozoan naturally FPs (fluorescent proteins), all of which had 26–30% identity with Aequorea GFP. One of the novel FPs cloned from a red Discosoma species, drFP583, commercially known as DsRed, was demonstrated to show red-shifted excitation and emission spectra. The family of ‘GFP-like proteins’ continues to expand further [3,4]. To date, over 30 significantly different family members have been reported. In addition to sharing a modest degree of sequence identity, all GFP-like proteins probably share the same general β-can fold and possess intrinsic chromophores [3].
In the present paper, we present the molecular cloning and characterization of two FPs from stony coral animals, a CFP (cyan FP) derived from Acropara sp. and an orange FP from Fungia concinna. We utilized these two FPs as a novel donor/acceptor pair for FRET (fluorescence resonance energy transfer) measurements detecting caspase 3 activity during an apoptotic process. The performance of this donor/acceptor pair is highly effective, possibly proving to be as useful an experimental system as the common CFP/YFP (yellow FP) pair.
EXPERIMENTAL
cDNA cloning and gene construction
Samples of the Acropara and Fungia stony corals were acquired from the ocean near the Okinawa islands by Dr K. Iwao (Akajima Marine Science Laboratory, Okinawa, Japan). Total RNA was isolated from the corals by guanidine thiocyanate extraction [5]. Synthesis, amplification using degenerate primers, and generation of full-length cDNA were performed as previously described [2] using the following degenerate primers: 5′-GAAGGRTGYGTCAAYGGRCAY-3′ and 5′-ACVGGDCCATYDGVAAGAAARTT-3′. The cDNA encoding the protein-coding region was amplified using primers containing 5′ BamHI and 3′ EcoRI sites. The digested product was then cloned in-frame into the BamHI/EcoRI sites of pRSETB for bacterial expression. The 5′ end of the gene was modified by PCR to contain a Kozak consensus sequence (CCACCATG) after the BamHI site to promote efficient transcription. The BamHI/EcoRI fragment was then subcloned into the mammalian expression vector, pcDNA3 (Invitrogen).
Mutagenesis
Site-directed and semi-random mutations were introduced as described [6]. For semi-random mutants, multiple degenerative primers were used to mutate amino acid residues at multiple sites simultaneously.
Protein expression, in vitro spectroscopy and pH titrations
Proteins were expressed in Escherichia coli, purified, and characterized spectroscopically as described previously [7]. Fluorescence quantum yields were determined using fluorescein as a standard (0.91). For calculation of molar absorption coefficients, protein concentrations were measured using a Bradford assay kit (Bio-Rad) with BSA as the standard. pH titrations were performed as described [7].
Analytical ultracentrifugation
Sedimentation equilibrium experiments were performed using a Beckman An60Ti rotor on a Beckman XL-1 analytical ultracentrifuge at 25 °C. Absorbance was measured at the maximum wavelength as a function of radius at 25000 rev./min. Using this system, the tetramerization of DsRed [8] was verified.
Fluorescence imaging
HeLa cells were grown on 35-mm glass-bottom dishes in Dulbecco's modified Eagle's medium containing 10% (v/v) foetal bovine serum. At 2–4 days after transfection with Polyfect (Qiagen), cells in Hanks balanced salt solution buffer (Gibco) were treated with 500 ng/ml anti-Fas antibody: (CH-11; Medical & Biological Laboratories Co. Ltd), then subjected to imaging. FRET images were acquired on the Aquacosmos/Ashura system (Hamamatsu Photonics) using an IX-71 inverted microscope equipped with a Uapo 340 40×1.35 NA oil-immersion objective (Olympus), a 440AF21 excitation filter, a 455DRLP dichroic mirror, a 480 ALP emission filter and a three CCD (chargecoupled device) colour camera (C7780-22; Hamamatsu Photonics). Image acquisition and analysis were performed using Aquacosmos 2.5 software (Hamamatsu Photonics).
RESULTS AND DISCUSSION
Two fluorescent coral animals were collected in the Okinawa islands. Acropara sp. shows cyan coloration in the tips of its tentacle. Fungia concinna emits an orange fluorescence from its whole body. Degenerate primers served to amplify cDNAs from the samples. The primers represented variations of several regions whose amino acid sequences are conserved among GFP-like fluorescent proteins from Anthozoa species. The missing 5′ and 3′ ends of the cDNA fragments were amplified using RACE (rapid amplification of cDNA ends). cDNA clones #301 and #11 were obtained from Acropara sp. and F. concinna respectively.
A CFP, MiCy
Based on amino acid alignment (Figure 1), clone #301 appeared to encode the full-length protein. Transformation of the isolated cDNA into E. coli produced bright cyan colonies. The addition of a His6 tag at the N-terminus of the protein allowed purification using metal-affinity chromatography. As the protein, derived from the Acropara organism whose Japanese name is Midori-ishi, emitted cyan fluorescence, we named this novel FP MiCy. The closest orthologue is amFP486, a CFP cloned from Anemonia majano [2], which shares 48.2% identity. Similar to amFP486, MiCy possesses a tyrosine residue at the second amino acid of the chromophore-forming tripeptide, while ECFP (enhanced CFP) has a tryptophan residue at this position. The absorption spectrum of MiCy at pH 7.4 displayed two peaks, one at 472 nm (ε=27250 M−1·cm−1) and one at 380 nm (ε=15000 M−1·cm−1) (Figure 2A). With decreasing pH, the amplitude of the 472-nm peak decreased, while that of the 380-nm peak increased concomitantly, exhibiting an isosbestic point at 413 nm (results not shown). This pH-dependency indicates that the 380- and 472-nm peaks correspond to the neutral and ionized states of the phenolic hydroxy group of the chromophore respectively [1]. The protein had an apparent pKa of 6.6 (Figure 2D).
Figure 1. Amino acid sequence (single-letter code) alignment of DsRed, mRFP1, KO, mKO and MiCy.
In the sequence of MiCy, the β-sheet-forming regions are underlined. Residues whose side chains form the interior of the β-can [20] are shaded in grey. In the mRFP1 sequence, the substituted amino acids are shaded in yellow. In the sequence of mKO, the substituted amino acids that disrupt dimeric structure, increase folding efficiency and inhibit aggregation are shaded in blue, red and green respectively. In the sequences of both KO and mKO, the artificial regions are boxed in red. The residues responsible for chromophore synthesis are indicated by an asterisk.
Figure 2. Light-absorption properties of the new fluorescent proteins.
Absorption spectra of MiCy (A), KO (B) and mKO (C), and the pH-dependence of the absorption (Abs.) peak at the maximum (D).
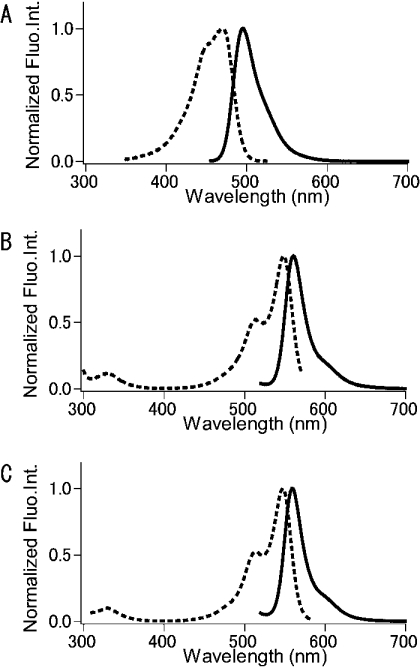
Excitation and emission spectra of MiCy are displayed in Figure 3(A). While the neutral form of the protein was non-fluorescent, the ionized form was highly fluorescent; the fluorescence quantum yield (Φ) was determined to be 0.90. The fluorescence spectra displayed the same pH dependency as the absorption spectrum, indicating that the Φ of MiCy was pH-resistant. Due to the high value and pH-resistance of the Φ, MiCy would probably be a more efficient and reliable donor for FRET than ECFP. In addition, fluorescence lifetime measurement revealed that MiCy fluorescence displayed as a single exponential decay with a time constant of 3.4 ns (Table 1). In contrast, ECFP exhibits a bi-exponential decay with 1.6 ns and 3.9 ns time constants. The simplicity of MiCy excited-state fluorescence decay makes it an ideal donor for fluorescence lifetime imaging experiments.
Figure 3. Normalized excitation (broken line) and emission (solid line) spectra of MiCy (A), KO (B) and mKO (C).
Fluo.Int., fluorescence intensity.
Table 1. Biochemical and spectral properties of ECFP, MiCy, KO and mKO.
| Protein | Excitation/emission maxima (nm) | Molar absorption coefficient (M−1·cm−1) | Fluorescence quantum yield | pH-sensitivity (pKa) | Fluorescence lifetime (ns) | Number of amino acids |
|---|---|---|---|---|---|---|
| ECFP | 435/478 | 28750 (435 nm) | 0.40 | 5.5 (Φ) | 1.60, 3.90 | 238 |
| MiCy | 472/495 | 27250 (472 nm) | 0.90 | 6.6 (ε) | 3.4 | 232 |
| KO | 548/561 | 109750 (548 nm) | 0.45 | <5.0 (ε) | 4.2 | 217 |
| mKO | 548/559 | 51600 (548 nm) | 0.60 | 5.0 (ε) | 4.1 | 217 |
We determined the absolute molecular mass of MiCy to be 59.7 kDa by analytical equilibrium ultracentrifugation analysis (Figure 4A). This value was twice as large as larger than the value (29.9 kDa) deduced from the primary structure of the protein, suggesting that MiCy forms a homodimeric complex similar to GFP from the bioluminescent sea pansy, Renilla reniformis [9]. Since all of the non-bioluminescent Anthozoan GFP-like proteins characterized so far form obligate tetramers [3], the dimer formation of MiCy may indicate structural divergence of the protein family.
Figure 4. The equilibrium radial absorbance profiles at 25000 rev./min by analytical ultracentrifugation analysis for MiCy (59.7 kDa) (A), KO (77.0 kDa) (B), and mKO (28.1 kDa) (C).
An orange fluorescent protein, KO (Kusabira-Orange)
The cDNA clone from F. concinna (#11) demonstrates 46.7% identity with amFP486. Upon transformation into E. coli, however, no fluorescent colonies were formed. Sequence alignment revealed that, in reference to other FPs, the encoded protein lacked approximately ten amino acids at the N-terminus. We therefore attached an artificial amino acid sequence, Met-Ser-Val-Ile-Lys-Pro-Glu, to the N-terminus (Figure 1). The resulting protein produced bright orange fluorescent bacterial colonies. As F. concinna is known in Japanese as Kusabira-Ishi, we named the protein Kusabira-Orange (KO).
We analysed the spectral properties of recombinant KO prepared from bacteria. At pH 7.4, KO displayed a major absorption wavelength maximum at 548 nm (ε=109750 M−1·cm−1) with a slight shoulder at 515 nm (Figure 2B). The large value of the molar absorption coefficient suggested the efficient maturation of the chromophore. A green-fluorescent intermediate appeared transiently in the course of chromophore maturation of KO, as seen for DsRed (results not shown). At significantly low pHs, an emission peak was also detected at approx. 423 nm. The relative intensities of these two absorption peaks changed with pH, indicating that the 423- and 548-nm peaks correspond to the neutral and ionized states of the phenolic hydroxy group of the chromophore respectively. The absorption spectrum of KO was pH-insensitive; the apparent pKa was determined to be below 5.0 (Figure 2D). Excitation and emission spectra of KO are shown in Figure 3(B). Following excitation at 500 nm, the protein emitted a bright orange fluorescence, peaking at 561 nm. The fluorescence intensity decreased slightly with increasingly acidic pH, exhibiting a similar apparent pKa (<5.0) as seen for the absorption spectrum. Thus the fluorescence quantum yield (Φ=0.45) is not pH-sensitive. We subjected the KO protein to analytical equilibrium ultracentrifugation analysis to calculate the absolute molecular mass of 77.0 kDa. This value was 2.75 times larger than that (28.0 kDa) deduced from the primary structure of the protein, suggesting that KO may exist in a heterogeneous oligomeric state, including dimers and tetramers.
Due to the development of new techniques allowing efficient discrimination of distinct fluorophores with overlapping emission spectra, such as the linear unmixing method [10], any small shift in the emission spectrum of novel FPs from established proteins would make the protein useful. Besides, there is a gap between green and red or between yellow and orange-red in the emission spectra of Aequorea GFP variants and GFP-like proteins from Anthozoa. Thus a true orange FP, such as KO, may greatly expand the repertoire of fluorescent dyes available for multicolour imaging.
Construction of a monomeric version of KO, mKO
A monomeric version of DsRed, called mRFP1, was previously generated by alteration of 33 amino acids [11]. KO has endogenous amino acid residues corresponding to six of the substitutions: Thr21→Ser, Asn42→Gln, Lys163→Met, Ser179→Thr, Ile180→Thr and Thr217→Ala (numbering is based on DsRed). Assuming that the KO oligomer exhibits a similar structure to the DsRed tetramer, we introduced three mutations (Phe102→Ser, Ala104→Ser and Val123→Thr) into the AB interface and four mutations (Cys151→Ser, Phe162→Tyr, Phe193→Tyr and Gly195→Ser) into the AC interface of KO. We then performed additional mutagenesis to obtain a brighter KO molecule. Twelve mutations (Lys11→Arg, Val25→Ile, Lys32→Arg, Ser55→Ala, Thr62→Val, Gln96→Glu, Glu117→Tyr, Val133→Ile, Ser139→Val, Thr150→Ala, Ala166→Glu and Gln190→Gly) and three mutations (Phe13→Tyr, Cys115→Thr and Cys217→Ser) were introduced to increase the folding efficiency and the solubility of the mutants respectively (Figure 1). The resulting protein retained the original bright orange fluorescence. The absolute molecular mass of 28.1 kDa (Figure 4C), determined by analytical equilibrium ultracentrifugation, is almost identical with the predicted size (28.0 kDa). This molecule was named mKO.
mKO exhibited similar fluorescence properties to KO (Figures 2C and 3C), but demonstrated a small decrease in light-absorbing ability (ε=51600 M−1·cm−1 at 548 nm) and a slight decrease in pH-resistance (apparent pKa=5.0) (Figure 2D). mKO also exhibits a higher fluorescence quantum yield than KO (0.60 compared with 0.45). Similar improvements in the fluorescence quantum yield were observed following oligomer into monomer conversion of a subset of Anthozoa fluorescent proteins, such as AG (Azami-Green) [12], although the production of mRFP1 was accompanied by decreased quantum yield [11].
Imaging of activity of caspase 3 through measurement of FRET between MiCy and mKO
The normalized excitation and emission spectra of MiCy and mKO (Figure 5A) demonstrate good overlap between the MiCy emission and the mKO excitation spectra, suggesting that the combination of MiCy and mKO may make a good donor/acceptor pair for FRET. Construction of the unimolecular fluorescent indicator, encoded by a single gene, principally requires that either the donor or acceptor FP be monomeric. Thus we did not disrupt the ability of MiCy to dimerize; we linked the C-terminus of MiCy and the N-terminus of mKO using a peptide containing the caspase 3 cleavage sequence, DEVD (Asp-Glu-Val-Asp) (Figure 5C) [13,14]. The recombinant protein, prepared in bacteria, was examined using an excitation wavelength of 440 nm. FRET was observed as a decrease in the emission peak at 495 nm and an increase in the peak at 559 nm. Incubation of the sample with an activated recombinant caspase 3 (3.7 μg/ml) at 30 °C for 120 min completely abolished the FRET signal. The change in colour before and after treatment with caspase 3 was easily recognizable by eye as seen using a common digital camera through a Wratten film passing light of longer than 480 nm with excitation at 440 nm (Figure 5D, inset). The emission ratio of 559/495 nm decreased 14-fold. Neither caspase 8 nor caspase 9 altered the emission spectrum (results not shown). Therefore the chimaeric MiCy–mKO protein functions as a specific sensor for caspase 3 activity, despite dimerization of MiCy. After expressing this caspase-3-sensor protein in HeLa cells, we detected the two signals from MiCy and mKO simultaneously using a colour CCD camera containing three CCD chips (Figure 6). Following a 1.5-h exposure to 500 ng/ml anti-Fas antibody (CH-11) and 10 μg/ml cycloheximide at 37 °C, the value of the 559/495 emission ratio dropped progressively from 1.2 to 0.2 just before the shrinking of cells characteristic of apoptosis.
Figure 5. FRET between MiCy and mKO.
(A) Normalized excitation (broken line) and emission (solid line) spectra of MiCy and mKO. (B) Normalized excitation (broken line) and emission (solid line) spectra of CFP and YFP. (C) Primary structure of the caspase-3-sensor protein. (D) Emission spectra of the caspase-3-sensor with excitation at 440 nm before (light grey) and after (dark grey) incubation with caspase 3. Inset, overt appearance of the sample tubes. Fluo.Int., fluorescence intensity.
Figure 6. FRET imaging of caspase 3 activity during an apoptotic process.
(A) FRET images of HeLa cells displayed in real colour and pseudo colour (ratios). Scale bar, 20 μM. (B) Time courses of the 559/495 nm emission ratios observed in the three cells indicated in (A).
For broader FRET applications, any oligomerization induced through the fusion of an oligomerizing donor or acceptor FP is problematic. The engineering of a completely monomeric MiCy variant is now under way.
pH-sensitivity of FRET pairs: MiCy/mKO compared with CFP/YFP
Multiple critical cellular organelles, such as lysosomes, have extreme pHs. Even within the cytosol, alkalinization and acidification may occur during metabolic stress or mitogenic stimulation. Thus FP resistance to pH fluctuation is often required for effective quantitative imaging. To use FRET in studies of environments with extreme pHs, both the donor quantum yield and the acceptor molar absorption coefficient must be indifferent to changes in pH [15]. While most GFP-based FRET experiments use CFP and YFP as FRET donors and acceptors, CFP has an acid-sensitive quantum yield and YFP has an acid-sensitive molar absorption coefficient [15]. When pH decreases, these two negative effects reinforce each other, resulting in loss of FRET. Below pH 6.5, FRET from CFP to YFP is significantly disturbed. Therefore development of alternative donor/acceptor pairs has been greatly desired for more effective quantitative FRET imaging.
The lower pH-sensitivity of the mKO molar absorption coefficient than pH-insensitive variants of YFP, such as EYFP.1 (enhanced YFP; apparent pKa=5.9) [15], citrine (apparent pKa=5.7) [16] and Venus (apparent pKa=6.0), makes this protein a desirable acceptor [17]. MiCy appears to be an ideal donor due to its pH-resistant quantum yield. While pH-sensitivity of the donor molar absorption coefficient is not important for pH-resistant FRET, a pH-resistant acceptor quantum yield would be preferable, because it facilitates the measurement of FRET efficiency by observing the ratio of donor to acceptor emissions. Again, mKO appears to make an ideal acceptor, as its quantum yield is completely indifferent to pH changes.
Optical factors for FRET measurements using MiCy/mKO compared with CFP/YFP
We compared the MiCy/mKO pair of excitation and emission spectra to that for CFP and YFP (Figure 5B). The MiCy/mKO pair appears to be free of the problems of the cross-excitation of acceptor and the cross-detection of the donor and FRET signals experienced with the CFP/YFP pair. Selective excitation of MiCy over mKO can be achieved using a 440AF21 (440±10.5 nm) bandpass filter. As emission spectra often rise steeply and fall gradually as a function of wavelength, the donor emission can often spill over into the FRET channel. Such cross-detection is prominent for the CFP/YFP pair [18]. In contrast, the emissions of MiCy and mKO should be easily separable using appropriate bandpass filters. Also, the good spectral overlap between MiCy emission and mKO absorption and the high fluorescence quantum yield of MiCy creates a Förster's distance for the MiCy/mKO pair as large as 5.3 nm, while the value for CFP/YFP is 4.9 nm [19]. Together, these advantages of the MiCy/mKO pair may be responsible for the relatively large changes in FRET signals observed as a measure of caspase 3 activity.
Acknowledgments
We thank Dr Kenji Iwao and Dr Saburo Hosaka at Akajima Marine Science Laboratory for acquiring stony coral animals, Dr Isogai for assistance with analytical centrifugation, and Dr H. Watanabe and Dr M. Watanabe for lifetime fluorescence measurements. This work was partly supported by grants from CREST of JST (Japan Science and Technology), the Special Coordination Fund for the promotion of Ministry of Education, Culture, Sports, Science and Technology, the Japanese Government, NEDO (the New Energy and Industrial Technology Development Organization) and HFSP (Human Frontier Science Program).
References
- 1.Tsien R. Y. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 2.Matz M. V., Fradokov A. F., Labas Y. A., Savitsky A. P., Zaraisky A. G., Markelov M. L., Lukyanov S. A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 3.Labas Y. A., Gurskaya N. G., Yanushevich Y. G., Fradkov A. F., Lukyanov K. A., Lukyanov S. A., Matz M. V. Diversity and evolution of the green fluorescent protein family. Proc. Natl. Acad. Sci. U.S.A. 2002;99:4256–4261. doi: 10.1073/pnas.062552299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyawaki A. Green fluorescent protein-like proteins in reef Anthozoa animals. Cell Struct. Funct. 2002;27:343–347. doi: 10.1247/csf.27.343. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Sawano A., Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 2000;28:E78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagai T., Sawano A., Park E. S., Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc. Natl. Acad. Sci. U.S.A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baird G. S., Zacharias D. A., Tsien R. Y. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. U.S.A. 2000;99:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ward W. W., Cormier M. J. An energy transfer protein in coelenterate bioluminescence: Characterization of the Renilla green-fluorescent protein (GFP) J. Biol. Chem. 1979;254:781–788. [PubMed] [Google Scholar]
- 10.Tsurui H., Nishimura H., Hattori S., Hirose S., Okumura K., Shirai T. Seven-color fluorescence imaging of tissue samples based on Fourier spectroscopy and singular value decomposition. J. Histochem. Cytochem. 2000;48:653–662. doi: 10.1177/002215540004800509. [DOI] [PubMed] [Google Scholar]
- 11.Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karasawa S., Araki T., Yamamoto-Hino M., Miyawaki A. A green-emitting fluorescent protein from Galaxeidae coral and its monomeric version for use in fluorescent labeling. J. Biol. Chem. 2003;278:34167–34171. doi: 10.1074/jbc.M304063200. [DOI] [PubMed] [Google Scholar]
- 13.Tyas L., Brophy V. A., Pope A., Rivett A. J., Tavaré J. M. Rapid caspase-3 activation during apoptosis revealed using fluorescence-resonance energy transfer. EMBO Rep. 2000;1:266–270. doi: 10.1093/embo-reports/kvd050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takemoto K., Nagai T., Miyawaki A., Miura M. Spatio-temporal activation of caspase revealed by indicator that is insensitive to environmental effects. J. Cell Biol. 2003;160:235–243. doi: 10.1083/jcb.200207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyawaki A., Tsien R. Y. Monitoring protein conformations and interactions by fluorescence resonance energy transfer between mutants of green fluorescent protein. Methods Enzymol. 2000;327:472–500. doi: 10.1016/s0076-6879(00)27297-2. [DOI] [PubMed] [Google Scholar]
- 16.Griesbeck O., Baird G. S., Campbell R. E., Zacharias D. A., Tsien R. Y. Reducing the environmental sensitivity of yellow fluorescent protein: mechanism and applications. J. Biol. Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 17.Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 18.Erickson M. G., Moon D. L., Yue D. T. DsRed as a potential FRET partner with CFP and GFP. Biophys. J. 2003;85:599–611. doi: 10.1016/S0006-3495(03)74504-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patterson G. H., Piston D. W., Barisas B. G. Förster distances between green fluorescent protein pairs. Anal. Biochem. 2000;284:438–440. doi: 10.1006/abio.2000.4708. [DOI] [PubMed] [Google Scholar]
- 20.Yang F., Moss L. G., Phillips G. N., Jr The molecular structure of green fluorescent protein. Nat. Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]