Abstract
Transglutaminases (TGases) are Ca2+-dependent enzymes capable of catalysing transamidation of glutamine residues to form intermolecular isopeptide bonds. Nine distinct TGases have been described in mammals, and two of them (types 2 and 3) are regulated by GTP/ATP. TGase2 hydrolyses GTP and is therefore a bifunctional enzyme. In the present study, we report that TGase5 is also regulated by nucleotides. We have identified the putative TGase5 GTP-binding pocket by comparative amino acid sequence alignment and homology-derived three-dimensional modelling. GTP and ATP inhibit TGase5 cross-linking activity in vitro, and Ca2+ is capable of completely reversing this inhibition. In addition, TGase5 mRNA is not restricted to epidermal tissue, but is also present in different adult and foetal tissues, suggesting a role for TGase5 outside the epidermis. These results reveal the reciprocal actions of Ca2+ and nucleotides with respect to TGase5 activity. Taken together, these results indicate that TGases are a complex family of enzymes regulated by calcium, with at least three of them, namely TGase2, TGase3 and TGase5, also being regulated by ATP and GTP.
Keywords: apoptosis, GTP, GTPase activity, transglutaminase
Abbreviations: NHEK, normal human epidermal keratinocyte; TGase, transglutaminase
INTRODUCTION
Nine distinct, but closely related, TGases (transglutaminases; EC 2.3.2.13) have been identified in mammals; TGase1–TGase7, coagulation Factor XIIIα and band 4.2 [1,2]. TGases are Ca2+-dependent enzymes, which catalyse intermolecular isopeptide bond formation by transamidation of specific glutamine residues [3]. This reaction, which results in the post-translational modification of proteins by the formation of ε(γ-glutamyl)lysine isodipeptide and N,N-bis-(γ-glutamyl)polyamine linkages, renders the modified polypeptides highly resistant to mechanical and chemical damages.
Among the TGase family members, TGase2 and TGase3 have been shown to be regulated by guanine nucleotides. TGase2 is involved in both intracellular and extracellular biological processes such as receptor-mediated endocytosis, cell adhesion, apoptosis and wound healing [4–6]. Several reports indicate that TGase2 activation can be associated with apoptosis (although recent evidence would suggest that involvement of TGase2 in apoptosis is indirect [7,8]) or with anti-apoptotic effects mediated by Rb [9]. Furthermore, TGase2 knockout (−/−) mice do not show a clear apoptotic phenotype, but show only mild erythrocyte fragility [10], abnormal insulin secretion [11] and defects in phagocytosis [12]. TGase2 is multifunctional protein and a bifunctional enzyme with both protein-cross-linking and GTP-hydrolysing activities [2]. Moreover, TGase2 is also a signal-transduction GTPase protein (Gh), which is coupled with the α1-adrenergic receptor and mediates the activation of phospholipase C. The X-ray structure of human TGase2 bound with GDP has been solved, and the main GDP-binding residues have been identified [13].
Recent biochemical as well as crystallographic evidence demonstrates direct binding of GTP/GDP to the active TGase3 enzyme. Moreover, it has been shown that TGase3 is capable of hydrolysing GTP, and the structural results show that the GTP-binding pocket is conserved between TGase3 and TGase2 [14].
TGase5 is a novel member of the TGase family [16], and only limited characterization at functional and biochemical levels has been performed. Originally, TGase5 cDNA was identified in human foreskin keratinocytes and further studies confirmed its involvement in keratinocyte differentiation in vitro and in vivo [17,18]. In the present study, we report for the first time that TGase5 is negatively regulated by adenine–guanine nucleotides, and is capable of hydrolysing GTP.
EXPERIMENTAL
Enzymic assays
Recombinant purified TGase5 enzyme was obtained as described previously [18]. Human full-length TGase3 was expressed in the baculovirus system and purified as described previously [15]. The TGase3 proenzyme was activated using dispase as described previously [18]. Guinea-pig liver TGase2 was purchased from Sigma and purified in-house by expression in Escherichia coli. We dialysed guinea-pig TGase2 to remove contaminants such as Mg2+ and Ca2+, and compared its specific activity with baculovirus-expressed TGase2. Both the enzymes had comparable specific activity using N,N′-dimethylcasein as substrate (results not shown). However, we observed that guinea-pig TGase2 was more stable under our experimental conditions and, therefore, we have used the Sigma guinea-pig enzyme in these studies. TGase activity was determined by measuring the incorporation of [3H]putrescine into N,N′-dimethylcasein [3]. Each tube contained 7 nM TGase5, 10 nM TGase2 or TGase3; and nucleotides were added as indicated in the Results and discussion section. The mixtures were incubated for 30 min at 37 °C before performing the TGase assay in the presence of 100 μM or 1 mM CaCl2.
Measurements of the free-Ca2+ reduction in the reaction mixtures were performed using the low-affinity calcium indicator Fluo-5N, which is capable of detecting free-Ca2+ concentrations in the 1 μM to 1 mM range. Fluo-5N (10 μM) was incubated for 10 min at 37 °C with the TGase reaction mixture (1 mM putrescine/1 mM dithiothreitol/50 mM Tris/HCl, pH 8.5/10 nM TGase) with or without N,N′-dimethylcasein (3 mg/ml) at different Ca2+ concentrations. Fluorescence emission was measured at 520 nm. Calibration curves obtained using Ca2+/EGTA buffer confirmed that the free-Ca2+ concentrations in the reaction mixture were decreased by 80–85% with respect to the Ca2+ concentration, which is present in small amounts in the buffers.
GTPase–ATPase activities were measured as described previously [4]. The reactions were started by the addition of TGases and allowed to proceed at 37 °C for 15, 30 and 45 min.
Generation of the TGase5 three-dimensional model
A three-dimensional model of human TGase5 was generated using MODELLER [19]. The crystal structure of human TGase2 (PDB code 1KV3) was used as a starting point. Human TGase2 and TGase5 have 41% sequence identity, and their sequences were aligned using LALIGN v.2.0u6. The alignment was input into MODELLER to model TGase5 required to satisfy the spatial constraints. The model was optimized using molecular probability density function by conjugate gradient and simulated annealing. GDP was docked into TGase5 in the same position and orientation as observed in TGase2. GDP and side chains of residues interacting with GDP were refined using MacroModel. TGase5 has a 29-amino-acid insertion, relative to TGase2, between β-barrel 1 and the catalytic core, which could not be modelled.
Detection by dot-blot hybridization
The expression of TGase5 in different tissues was performed using the RNA Master Blot [Human poly(A)+ RNAs; ClonTech Laboratories, Palo Alto, CA, U.S.A.]. The PCR-amplified fragments TG5F2 (5′-CTCTGCAGAAGCTGAAGGCTAG-3′) and TG5R5(3) (5′-CGTCTGGCGCGTTGTTCCAG-3′), spanning bases 1377 to 2179 of the published TGase5 sequence (accession number AF035960), was used as a probe for both Northern and Master blots. The 802 bp fragment was labelled with Klenow enzyme and [32P]dCTP using standard procedures. To hybridize, we used ‘ExpressHyb’ (ClonTech Laboratories); prehybridization, hybridization, and washes were performed according to the manufacturer's instructions.
RESULTS AND DISCUSSION
Expression pattern of TGase5
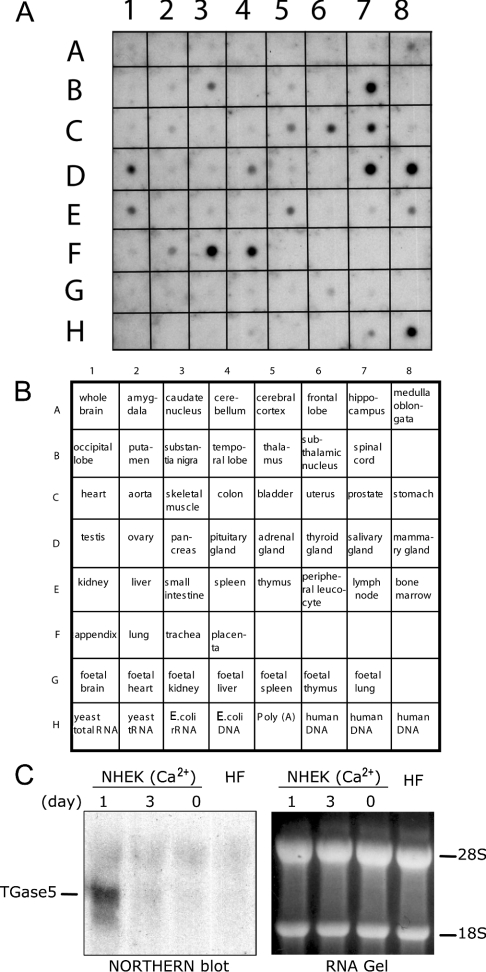
TGase5 is expressed in cultured keratinocytes induced to differentiate with calcium [17,18], but has also been detected in vitro in other cell lines [16]. To obtain a more detailed picture of its expression pattern, we performed a dot-blot Northern-blot analysis. The probe, comprising a C-terminal fragment of the cDNA encoding human TGase5, was hybridized to a commercially available poly(A+) (polyadenylated) RNA dot-blot (Figures 1A and 1B). To establish the specificity of the TGase5 probe, we performed Northern-blot analysis using mRNAs extracted from NHEKs (normal human epidermal keratinocytes) induced to differentiate with calcium, which express TGase1, TGase3 and TGase5 [18], and mRNA extracted from normal human fibroblasts, which express mainly TGase2 [7]. As already reported in our previous studies [18], we detected an accumulation of TGase5 mRNA within only 1 day of treatment of NHEK with Ca2+ (Figure 1C). Therefore the probe does not cross-hybridize with other TGases.
Figure 1. TGase5 expression pattern.
(A, B) A C-terminal cDNA fragment, encoding human TGase5, has been hybridized with a commercially available poly(A)+ RNA dot blot; see the Experimental section for details. One of the two identical blots is shown. (C) Northern blot is shown to demonstrate specificity of the probe used. RNA was extracted from NHEK after treatment with calcium for 1 and 3 days, and from normal human fibroblast (HF). Right panel: RNA gel as a loading control. Hybridization conditions are indicated in the Experimental section.
Although these results do not indicate the cell types within the tissue in which TGase5 is expressed, they do demonstrate that the human enzyme is not exclusively restricted to the epidermis (Figures 1A and 1B). The fact that TGase5 expression is not limited to the epidermis, as initially supposed, raises the possibility that it may also be involved in other biological functions.
Sequence alignments of TGases, predicted structural model of TGase5 and identification of the putative TGase5 GTP-binding pocket
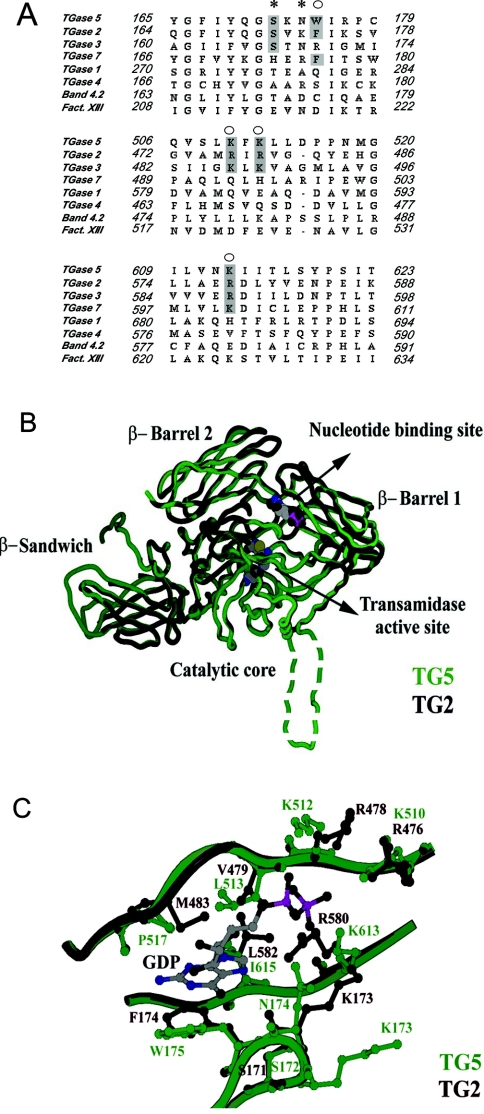
To explore whether TGase5 could also be regulated by GTP, we performed a sequence alignment of TGases in the region where the key residues for TGase2 interaction with GTP have been described (Figure 2A). These residues have been identified following a mutagenesis approach (indicated by the asterisk in Figure 2A) [20] and by X-ray studies of TGase2 complexed with GDP (indicated by the circles in Figure 2A) [13,14]. We observed that the key residues interacting with GTP in TGase2 (Ser-171, Phe-174 and Arg-476/478/578) are either identical or conservatively replaced in TGase5 (Ser-172, Trp-175 and Lys-510/512/613). These similarities are also seen on comparing TGase2 with TGase3. TGase3 has a GTP-binding site similar to TGase2, although the amino acid residues interacting with GTP are different and not always conservatively replaced (Ser-167, Arg-170, Lys-486/488 and Arg-588). On the basis of amino acid similarities, a computer-generated model of human TGase5 was built based on the three-dimensional structure of human TGase2. Not surprisingly, given the high degree of homology (41% identity), the overall predicted structure of TGase5 closely matches that of TGase2 (Figure 2B). In this model, GDP binds in the cleft between the catalytic core and β-barrel 1 in TGase2, and the close similarity of this region in the two proteins suggests that TGase5 can bind GDP in an analogous fashion. Trp-175, Leu-513, Pro-517 and Ile-615 (Figure 2C) form the hydrophobic pocket for the guanine in TGase5. Trp-175 may form a π–π stacking interaction with guanine, thereby playing the same role as Phe-174 in TGase2 (Figure 2C). In the latter enzyme, Lys-173, Arg-476, Arg-478 and Arg-580 interact with the phosphate group and facilitate GTP hydrolysis, whereas Asn-174, Lys-510, Lys-512 and Lys-613 essentially and conservatively replace these residues in TGase5. GTP hydrolysis in TGase2 may involve a nucleophilic water molecule bound to Lys-173 or Arg-476, as observed in the crystal structure. The counterparts of Lys-173 and Arg-476 are Asn-174 and Lys-510 in TGase5 (Figure 2B). From the computer-generated homology model of TGase5, it appears that Asn-175 could be hydrogen bonded to the β-phosphate and that Lys-613 or Lys-510 could be involved in the hydrolysis of the γ-phosphate (Figure 2C). Therefore we evaluated at the experimental level whether GTP regulated TGase5.
Figure 2. Identification of the TGase5 putative GTP-binding pocket.
(A) Sequence alignment of the GTP-binding region of TGase2 with the corresponding regions in other TGases. The key residues for TGase2 GTP interaction are indicated by * and ○. Those residues conservatively replaced or identical in TGase2, TGase3, TGase5 and TGase7 are shown in the shaded boxes. Sequences were aligned using ClustalX. (B) A computer-generated three-dimensional structural model of TGase5 (green) superimposed on the crystal structure of TGase2 (black: PDB code 1KV3). The GDP and the catalytic triad (Cys-278, His-337 and Asp-360) of TGase5 are shown as space-filling models at the nucleotide-binding site and the transamidase active site. The 29-amino-acid insertion between the catalytic core and the β-barrel 1 domains of TGase5 could not be modelled to any secondary structure and is shown as a dotted loop. (C) Comparison of the GDP-binding sites of TGase5 (green) and TGase2 (black).
Calcium activation of TGase2 and TGase5
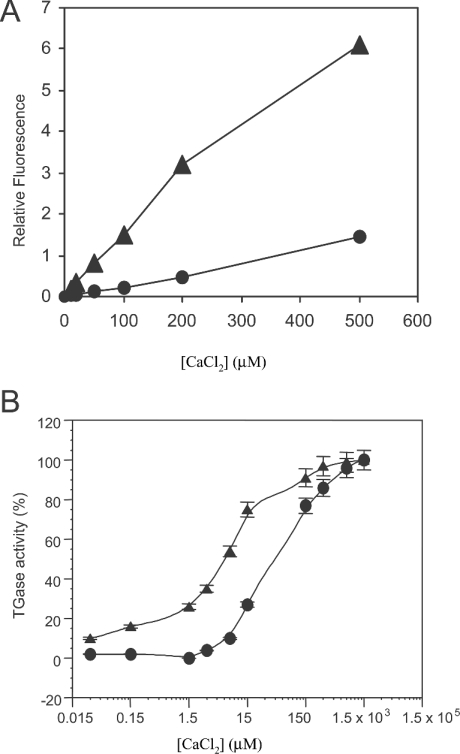
In the absence of Ca2+, TGase2 adopts an inactive conformation, which prevents the reactivity of Cys-277. Structural data show that the activation of TGase2 by Ca2+ [15,21] involves a displacement of protein domains, with increased reactivity and substrate accessibility to the active site. TGase2 activation by Ca2+ is probably counteracted by the inhibitor GTP. Therefore before investigating the role of guanine–adenine nucleotide in the regulation of TGase5 transamidating activity, we first studied the Ca2+ requirement for TGase5 activation. Figure 3(A) shows the reduction in free-Ca2+ under our experimental conditions due to the Ca2+-binding capacity of N,N′-dimethylcasein. Using a low-affinity calcium indicator (Fluo-5N), we found that N,N′-dimethylcasein binds 80–85% of the free Ca2+ contained in the reaction mixture. For clarity, we have indicated the effective free-Ca2+ concentrations measured in the reaction mixtures throughout the paper. Dose–response curves for Ca2+ activation of TGase2 and TGase5 are reported in Figure 3(B), showing that TGase2 is more sensitive to Ca2+ when compared with TGase5. Whereas TGase2 reaches half-maximal activation at 7.5 μM free-Ca2+ concentration, TGase5 activation at the same free-Ca2+ concentration is only 10%. TGase5 reaches half of its maximal activation at 45 μM free-Ca2+ concentration.
Figure 3. Calcium activation of TGase2 and TGase5.
(A) Measurement of the free-Ca2+ reduction in the reaction mixture due to the Ca2+-binding capacity of N, N′-dimethylcasein. Fluo-5N (10 μM) was incubated for 10 min at 37 °C with TGase reaction mixture with (•) or without (▴) N,N′-dimethylcasein (3 mg/ml) at different Ca2+ concentrations. Fluorescence was detected at 520 nm. Results presented are those calculated based on the amount of activity without nucleotides. The results are the mean for two independent experiments performed in duplicate. For clarity, Ca2+ concentrations shown in the text and on the graphs will be those effectively measured in the assay solutions (free-Ca2+ concentration). (B) Ca2+ activation of TGase2 (▴; 10 nM) and TGase5 (•; 10 nM) activity were assayed at ten different free-Ca2+ concentrations as described in the Experimental section; TGase5 activity was maximal (100%) at a free-Ca2+ concentration of 300 μM. Activities are reported as percentage of maximal TGase activity. Results are presented as means±S.E.M. for two independent experiments performed in triplicate.
These results show that, although TGase5 is also a Ca2+-dependent enzyme, its affinity for Ca2+ is lower in comparison with TGase2.
Guanine–adenine nucleotide inhibition of TGase2, TGase3 and TGase5 transamidating activity
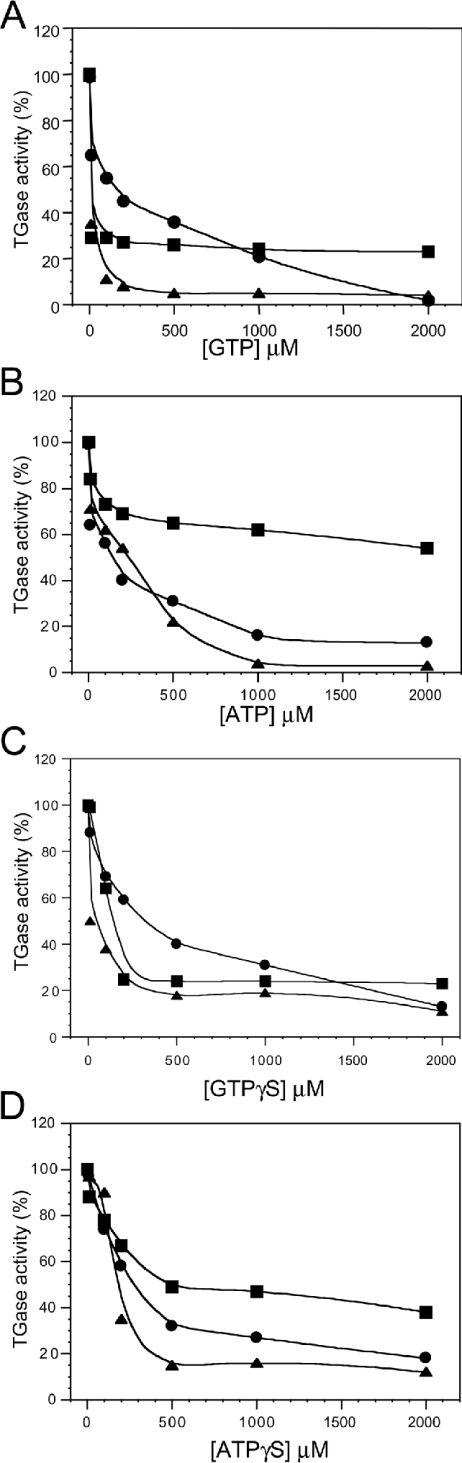
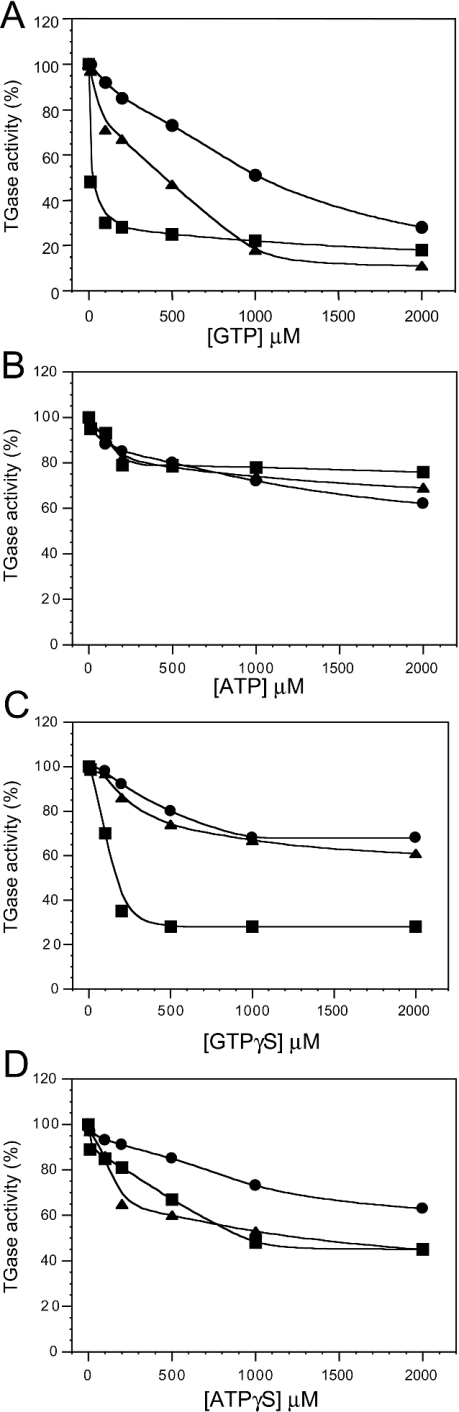
We evaluated the effects of various nucleotides on TGase5 in comparison with TGase2 and TGase3 at both 10 and 80% (Figure 3) of TGase5 Ca2+-stimulated activation (15 and 150 μM respectively; Figures 4 and 5). Both GTP and ATP were capable of inhibiting TGase5 transamidating activity as well as that of the TGase2 and TGase3 in a dose-dependent manner (Figures 4A and 4B). However, the degree of inhibition for the three enzymes is different depending on the Ca2+ concentration. At 15 μM Ca2+ (Figure 4A) and 100 μM GTP, an inhibition of the transamidating activity by 90, 70 and 45% of TGase2, TGase3 and TGase5 respectively was observed, whereas at a GTP concentration of 1 mM the inhibition for TGase2 was 95% and approx. 70% for TGase3 and TGase5. ATP also inhibited TGase5 but the difference was smaller at 15 μM Ca2+ (Figure 4) and 100 μM ATP. At 1 mM, ATP reduced the transamidating activity of TGase2, TGase5 and TGase3 by 95, 80 and 30% respectively. Similar results were obtained using GTPγS and ATPγS (Figures 4C and 4D). These experiments were repeated using 150 μM free-Ca2+ concentration. Again, we observed that both GTP and ATP were capable of inhibiting TGase5 as well as TGase2 and TGase3 (Figures 5A and 5B). The inhibition of TGase2 and TGase5 was observed only at a higher concentration of GTP in comparison with the above experiment. GTP, at the concentration of 1 mM, decreased the activity of TGase5, TGase2 and TGase3 by 45, 80 and 75% respectively (Figure 5A). Consistently, the ATP-mediated inhibition is lower at this Ca2+ concentration, being approx. 25–30% for all the three TGases (Figure 5B). Similar results were obtained using GTPγS and ATPγS (Figures 5C and 5D).
Figure 4. Guanine–adenine nucleotide inhibition of TGase2, TGase3 and TGase5 activity at 10% of Ca2+-stimulated TGase5 activity.
TGase5 (•), TGase2 (▴) and TGase3 (▪) (10 nM each) transamidating activities were performed using 15 μM free-Ca2+ concentration, in the presence of six different nucleotide concentrations. The activities are represented as a percentage of maximal TGase activity (100 % of the activity corresponds to 19.5×103 pmol·h−1·mg−1). Inhibitions by GTP (A), ATP (B), GTPγS (C) and ATPγS (D) are shown. The results are the means for three independent experiments performed in triplicate.
Figure 5. Guanine–adenine nucleotide inhibition of TGase2, TGase3 and TGase5 activity at 80% of Ca2+-stimulated TGase5 activity.
TGase5 (•), TGase2 (▪) and TGase3 (▴) (10 nM each) transamidating activities were performed using 150 μM Ca2+, in the presence of six different nucleotide concentrations (10, 100, 200, 500, 1000 and 2000 μM). The activities are represented as a percentage of maximal TGase activity (100% of the activity corresponds to 19.5×103 pmol·h−1·mg−1). Inhibitions by GTP (A), ATP (B), GTPγS (C) and ATPγS (D) are shown. The results are the means for three or four independent experiments performed in triplicate.
Together, these studies indicate that the GTP-mediated inhibition of TGase2, TGase3 and TGase5 is dependent on Ca2+ concentration (Figures 4A, 4B, 5A and 5B). Therefore Ca2+ and GTP concentrations appear to modulate the switch between the two activities of TGase2 and TGase5. The computer model of TGase5 structure also predicts a completely conserved catalytic pocket with respect to TGase2. The inactive conformation of TGase2 shows the nucleophilic Cys-277 hydrogen-bonded to Tyr-516, and the same interaction may well exist between Cys-278 and His-550 in TGase5. Liu et al. [13] have suggested that when Ca2+ binds to TGase2, the β-barrel 1 domain moves away from the catalytic core breaking this hydrogen bond and opening up the transamidation catalytic pocket. A similar mechanism may explain why Ca2+ binding in TGase5 down-regulates GTP hydrolysis.
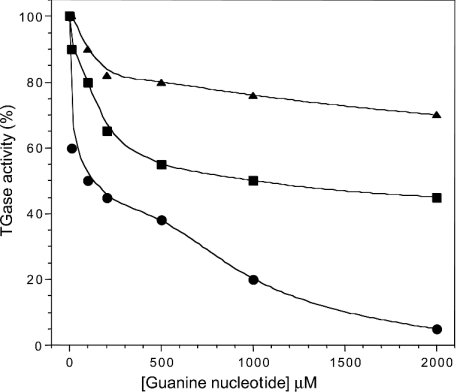
We also measured IC50 values for TGase2, TGase3 and TGase5 at 15 and 150 μM Ca2+. We observed that high calcium concentrations resulted in an increase in IC50 values (Table 1) which, for TGase5, increased from 205±54 μM (15 μM Ca2+) to 1165±354 μM (150 μM Ca2+) for GTP and from 168±47 to >2000 μM for ATP. Thus GTP and ATP inhibition of TGase5 activity are inversely proportional to Ca2+ activation and is highest under conditions where TGase activity is low. The results shown in Figure 6 indicate that the relative potency of inhibition of TGase5 by guanine nucleotides is GMP>GDP>GTP.
Table 1. IC50 measurements at 10 and 80% maximal TGase5 activity (respectively 15 and 150 μM Ca2+).
TGase activity was assayed as described in the Experimental section.
| IC50 (μM) | ||||||
|---|---|---|---|---|---|---|
| TGase5 | Tgase2 | Tgase3 | ||||
| 10% | 80% | 10% | 80% | 10% | 80% | |
| GTP | 205±54 | 1165±354 | 83±8 | 285±76 | 124±74 | 103±75 |
| GTPγS | 350±98 | >2000 | 78±27 | >2000 | 215±80 | 217±21 |
| ATP | 168±47 | >2000 | 202±27 | >2000 | >2000 | >2000 |
| ATPγS | 277±60 | >2000 | 166±85 | 772±222 | 489±152 | 847±217 |
Figure 6. Guanine nucleotide inhibition of TGase5 activity at 10% of Ca2+-stimulated TGase5 activity.
Increasing concentrations of the guanine nucleotides, GTP (•), GDP (▪) and GMP (▴), were preincubated for 30 min with purified recombinant TGase5. The remaining enzymic activities were calculated based on the activity in the absence of nucleotides. The results are the means for two or three independent experiments performed in triplicate.
TGase5 has GTPase activity
TGase2 had been shown to hydrolyse both GTP and ATP [20]. Therefore we tested TGase5 for both the activities in the presence of 1 mM Mg2+ and 10 μM Ca2+ (these bivalent cation concentrations were identified to be optimal based on the preliminary studies). We observed that TGase5 not only binds GTP and ATP, but is also capable of hydrolysing GTP. Thus incubation of purified recombinant human TGase5 with [γ-32P]GTP at 37 °C for 15, 30 and 45 min resulted in a linear release of [32P]PPi by the catalytic action of the enzyme. When spontaneous GTP hydrolysis was ignored, a net GTPase activity of 5.67±0.5 mmol/mol of TGase5 per min was calculated when compared with 8 mmol/mol for TGase2/min [20]. Unlike TGase2, but similarly to TGase3 [14], TGase5 showed no measurable ATPase activity.
Conclusion
For a long time, TGase2 was considered to be the only TGase family member with dual functions: a receptor signalling activity that requires GTP binding coupled with hydrolysis and a Ca2+-activated TGase activity, inhibited by GTP. In the present study, we show that TGase5 is also a dual-function enzyme with both a calcium-dependent transamidation activity as well as GTP binding and calcium-dependent GTP hydrolysis. Very recently [14], TGase3 has also been shown to bind GTP/ATP and also to undergo a GTPase cycle. Thus TGase2, TGase3 and TGase5 share common functional and regulatory controls. Moreover, GTP and ATP act as non-competitive inhibitors for TGase5 transamidating activity. Therefore, under physiological conditions, TGase5-transamidating activity appears to be regulated by local concentrations of calcium and nucleotides (GTP, GDP, GMP and ATP), as previously proposed for TGase2 [6].
The computer-simulated three-dimensional structure of TGase5, based on that of the TGase2, indicates that the putative GTP-binding pocket, transamidating catalytic site and the structural features involved in their reciprocal regulation are relatively conserved between the two enzymes. In both TGase2 and TGase5, the mechanism of inhibition by GTP probably involves both local and long-ranging events: the putative GTP pocket is located in a long β-structure far from the active-site Cys-271 in the primary structure, but spatially very close in the active conformation, so that a direct action of the nucleotide on the cysteine reactivity is feasible. Despite the high degree of structural homology between the two enzymes, TGase2 and TGase5 have different sensitivities to Ca2+ activation and GTP/ATP inhibition, suggesting that these two enzymes may have distinct functions.
In this study, we have shown that, at least in vitro, TGase5 is less responsive to Ca2+ activation than TGase2 and it is less sensitive to GTP inhibition. However, TGase5 may participate in different biological events. For instance, at physiological levels of calcium (10–15 μM), when TGase5 retains 25% of its maximal activity (75% for TGase2), a physiological level of GTP (100–200 μM) is able to inhibit significantly (45–55%) the TGase5 enzymic activity (90% for TGase2). Unfortunately, no biochemical data are available on substrate specificities for TGase2 and TGase5, except for skin-specific proteins; consequently, it is conceivable that in vivo these two enzymes cross-link different specific substrates with different kinetic constants at different time. In this contest, TGase5 enzymic activity and its regulation by nucleotides become physiologically relevant. Since we have shown a wider expression of TGase5, this regulation seems to be important also outside the skin.
In conclusion, the results reported in this study show that TGase5 has a complex set of biological functions suggesting additional physiological roles besides the cross-linking of proteins in the epidermis. However, the absence of sequence homology of TGase2, TGase3 and TGase5 with other GTP-binding proteins such as the monomeric and heterotrimeric G-proteins, elongation factors or dynamin, each of which contains four consensus GTP-binding motifs that fold into a structurally conserved GTP-binding site, suggests that TGase2 and TGase5 belong to a unique subfamily of GTPases.
Acknowledgments
We thank Dr M. Ranalli, Dr S. Rufini and Dr R.A. Knight for a stimulating discussion. E.C. was supported by Telethon (grant GGP02251) and G.M. by EU grant QLG1-1999-00739, Associazione Italiana di Ricerca contro il Cancro and Ministero Italiano Universita' e Ricerca.
References
- 1.Lorand L., Graham R. M. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 2003;4:140–156. doi: 10.1038/nrm1014. [DOI] [PubMed] [Google Scholar]
- 2.Griffin M., Casadio R., Bergamini C. Transglutaminases: nature's biological glues. Biochem. J. 2002;368:377–396. doi: 10.1042/BJ20021234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melino G., Candi E., Steinert P. M. Assays for transglutaminases in cell death. Methods Enzymol. 2000;322:433–472. doi: 10.1016/s0076-6879(00)22042-9. [DOI] [PubMed] [Google Scholar]
- 4.Davies P. J., Davies D. R., Levitzki A., Maxfield F. R., Milhaud P., Willingham M. C., Pastan I. H. Transglutaminase is essential in receptor-mediated endocytosis of α2-macroglobulin and polypeptide hormones. Nature (London) 1980;283:162–167. doi: 10.1038/283162a0. [DOI] [PubMed] [Google Scholar]
- 5.Melino G., Annicchiarico-Petruzzelli M., Piredda L., Candi E., Gentile V., Davies P. J., Piacentini M. Tissue transglutaminase and apoptosis: sense and antisense transfection studies with human neuroblastoma cells. Mol. Cell. Biol. 1994;14:6584–6596. doi: 10.1128/mcb.14.10.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haroon Z. A., Hettasch J. M., Lai T. S., Dewhirst M. W., Greenberg C. S. Tissue transglutaminase is expressed, active, and directly involved in rat dermal wound healing and angiogenesis. FASEB J. 1999;13:1787–1795. doi: 10.1096/fasebj.13.13.1787. [DOI] [PubMed] [Google Scholar]
- 7.De Laurenzi V., Melino G. Gene disruption of tissue transglutaminase. Mol. Cell. Biol. 2001;121:148–155. doi: 10.1128/MCB.21.1.148-155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nanda N., Iismaa S. E., Owens W. A., Husain A., Mackay F., Graham R. M. Targeted inactivation of Gh/tissue transglutaminase II. J. Biol. Chem. 2001;276:20673–20678. doi: 10.1074/jbc.M010846200. [DOI] [PubMed] [Google Scholar]
- 9.Boehm J. E., Singh U., Combs C., Antonyak M. A., Cerione R. A. Tissue transglutaminase protects against apoptosis by modifying the tumor suppressor protein p110 Rb. J. Biol. Chem. 2002;277:20127–20130. doi: 10.1074/jbc.C200147200. [DOI] [PubMed] [Google Scholar]
- 10.Bernassola F., Boumis G., Corazzari M., Bertini G., Citro G., Knight R. A., Amiconi G., Melino G. Osmotic resistance of high-density erythrocytes in transglutaminase 2-deficient mice. Biochem. Biophys. Res. Commun. 2002;291:1123–1127. doi: 10.1006/bbrc.2002.6558. [DOI] [PubMed] [Google Scholar]
- 11.Bernassola F., Federici M., Corazzari M., Terrinoni A., Hribal M. L., De Laurenzi V., Ranalli M., Massa O., Sesti G., McLean W. H., et al. Osmotic resistance of high-density erythrocytes in transglutaminase 2-deficient mice. FASEB J. 2002;16:1371–1378. doi: 10.1096/fj.01-0689com. [DOI] [PubMed] [Google Scholar]
- 12.Szondy Z., Sarang Z., Molnár P., Németh T., Piacentini M., Mastroberardino P. G., Falasca L., Aeschlimann D., Kovács J., Kiss I., et al. Transglutaminase 2−/− mice reveal a phagocytosis-associated crosstalk between macrophages and apoptotic cells. Proc. Natl. Acad. Sci. U.S.A. 2003;24:7812–7817. doi: 10.1073/pnas.0832466100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S., Cerione R. A., Clardy J. Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2743–2747. doi: 10.1073/pnas.042454899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahvazi B., Boeshans K. M., Idler W., Baxa U., Steinert P. M., Rastinejad F. Structural basis for the coordinated regulation of transglutaminase 3 by guanine nucleotides and calcium/magnesium. J. Biol. Chem. 2004;279:7180–7192. doi: 10.1074/jbc.M312310200. [DOI] [PubMed] [Google Scholar]
- 15.Ahvazi B., Kim H. C., Kee S. H., Nemes Z., Steinert P. M. Three-dimensional structure of the human transglutaminase 3 enzyme: binding of calcium ions changes structure for activation. EMBO J. 2002;21:2055–2067. doi: 10.1093/emboj/21.9.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aeschlimann D., Koeller M. K., Allen-Hoffmann B. L., Mosher D. F. Isolation of a cDNA encoding a novel member of the transglutaminase gene family from human keratinocytes. Detection and identification of transglutaminase gene products based on reverse transcription–polymerase chain reaction with degenerate primers. J. Biol. Chem. 1998;273:3452–3460. doi: 10.1074/jbc.273.6.3452. [DOI] [PubMed] [Google Scholar]
- 17.Candi E., Oddi S., Paradisi A., Terrinoni A., Ranalli M., Teofoli P., Citro G., Scarpato S., Puddu P., Melino G. Expression of transglutaminase 5 in normal and pathologic human epidermis. J. Invest. Dermatol. 2002;119:670–677. doi: 10.1046/j.1523-1747.2002.01853.x. [DOI] [PubMed] [Google Scholar]
- 18.Candi E., Oddi S., Terrinoni A., Paradisi A., Ranalli M., Finazzi-Agro A., Melino G. Transglutaminase 5 cross-links loricrin, involucrin, and small proline-rich proteins in vitro. J. Biol. Chem. 2001;276:35014–35023. doi: 10.1074/jbc.M010157200. [DOI] [PubMed] [Google Scholar]
- 19.Sali A., Blundell T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 20.Iismaa S. E., Wu M. J., Nanda N., Church W. B., Graham R. M. GTP binding and signaling by Gh/transglutaminase II involves distinct residues in a unique GTP-binding pocket. J. Biol. Chem. 2000;275:18259–18265. doi: 10.1074/jbc.M000583200. [DOI] [PubMed] [Google Scholar]
- 21.Mariani P., Carsughi F., Spinozzi F., Romanzetti S., Meier G., Casadio R., Bergamini C. Ligand-induced conformational changes in tissue transglutaminase: Monte Carlo analysis of small-angle scattering data. Biophys. J. 2000;78:3240–3251. doi: 10.1016/S0006-3495(00)76860-3. [DOI] [PMC free article] [PubMed] [Google Scholar]