Abstract
ISP-1 (myriocin) is a potent inhibitor of serine palmitoyltransferase, the primary enzyme of sphingolipid biosynthesis, and is a useful tool for studying the biological functions of sphingolipids in both mammals and yeast (Saccharomyces cerevisiae). In a previous study, we cloned yeast multicopy suppressor genes for ISP-1, and one of these, YPK1/SLI2, was shown to encode a serine/threonine kinase which is a yeast homologue of mammalian SGK1 (serum/glucocorticoid-regulated kinase 1). In the present study, another gene, termed SLI1 (YGR212W; GenBank accession number CAA97239.1), was characterized. Sli1p has weak similarity to Atf1p and Atf2p, which are alcohol acetyltransferases. Although a sli1-null strain grew normally, the IC50 of ISP-1 for the growth of this strain was markedly decreased compared with that for the parental strain, indicating that Sli1p is a major contributor to ISP-1 resistance in yeast. On a sli1-null background, the increase in resistance to ISP-1 induced by YPK1 gene transfection was almost abolished. These data indicate that Sli1p co-operates with Ypk1p in mediating resistance to ISP-1 in yeast. Sli1p was found to convert ISP-1 into N-acetyl-ISP-1 in vitro. Furthermore, N-acetyl-ISP-1 did not share the ability of ISP-1 to inhibit the growth of yeast cells, and the serine palmitoyltransferase inhibitory activity of N-acetyl-ISP-1 was much lower than that of ISP-1. These data suggest that Sli1p inactivates ISP-1 due to its N-acetyltransferase activity towards ISP-1.
Keywords: N-acetyltransferase, condensation domain, drug-resistance gene, ISP-1, myriocin, serine palmitoyltransferase
Abbreviations: EGFP, enhanced green fluorescent protein; MBP, maltose-binding protein; SGK1, serum/glucocorticoid-regulated kinase 1; SPT, serine palmitoyltransferase
INTRODUCTION
Sphingolipids are sphingoid-base-containing lipids that are found in both mammals and the yeast Saccharomyces cerevisiae. More than 300 derivatives of sphingolipids occur in mammals, and play crucial roles in adhesion, differentiation, growth and apoptosis. Although the molecular species of yeast sphingolipids are much simpler than in those of mammals, sphingolipids are known to be involved in several biological functions in yeast. Ceramide was shown to induce G1 arrest in yeast via a ceramide-activated protein phosphatase consisting of three protein phosphatase 2A components [1,2]. Dihydrosphingosine and phytosphingosine were shown to be transiently elevated and involved in cell cycle arrest during heat stress [3,4]. Phytosphingosine is postulated to activate ubiquitin-dependent proteolysis upon heat stress [5]. These sphingoid bases have also been shown to inhibit nutrient import [6,7]. The sphingolipid synthesis pathway was reported to be necessary for intracellular trafficking of glycosylphosphatidylinositol-anchored proteins [8,9] and for the internalization step of endocytosis [10]. However, most of the downstream pathway of sphingolipid-related signalling has not been well clarified in mammals and yeast.
Yeast is genetically tractable and is therefore a useful system for the study of sphingolipids. Indeed, most genes involved in the early stage of the sphingolipid biosynthetic pathway, which is shared by both mammals and yeast, were originally cloned in yeast and then their mammalian counterparts were studied. In a pioneering study using a sphingolipid auxotrophy yeast mutant, genes for the subunits of SPT (serine palmitoyltransferase; LCB1/2), which is the enzyme catalysing the first step in sphingolipid biosynthesis, were cloned [11,12]. Later, genes for the sphingosine kinases, sphingosine phosphate phosphatases and sphingosine phosphate lyase were also cloned in yeast before the corresponding mammalian genes [13–16]. More recently LAG1 and LAC1, which were known to be longevity genes in yeast, were identified as encoding ceramide synthases [17].
ISP-1 (myriocin) was originally isolated as an immunosuppressant from a fungus. Structural studies revealed that ISP-1 was identical to myriocin and thermozymocidin, which had been isolated previously as antibiotics [18,19]. ISP-1 exhibited an immunosuppressive potency 10–100-fold greater than that of cyclosporin A, a widely used immunosuppressant, when the activity was measured in a mouse allogenic mixed lymphocyte reaction [20]. Unlike cyclosporin A and FK506 (another commonly used immunosuppressant), ISP-1 did not interfere with interleukin-2 production in a mixed lymphocyte reaction [20], but instead suppressed alloreactive cytotoxic T lymphocyte generation in vivo and the interleukin-2-dependent growth of the mouse cytotoxic T cell line CTLL-2 [21]. FTY720 was developed as an analogue of ISP-1 and has little apparent toxicity in vivo [22], and therefore it has been tested in clinical immunosuppressive regimens for humans [23]. Both immunosuppressants are involved in sphingolipid function, but their direct targets are different, despite the structural similarity between the two. ISP-1 inhibits SPT in mammals, and therefore ISP-1 treatment caused sphingolipid depletion that led to apoptosis of the cytotoxic T cell line. On the other hand, FTY720 was phosphorylated in vivo, and the phosphorylated form of FTY720 bound agonistically to the sphingosine 1-phosphate receptors on the lymphocyte cell membrane, and the resulting signal induced the recruitment of lymphocytes from blood to secondary lymphocyte tissues [24,25].
In contrast with mammals, yeast does not have sphingosine 1-phosphate receptors. However, similarly to mammals, the initial step of sphingolipid synthesis in yeast is catalysed by SPT. In a previous study [28], we showed that ISP-1 inhibited SPT and induced a growth defect in yeast. ISP-1 is therefore a useful tool for studying sphingolipid function in yeast. YPK1/SLI2 was identified as a multicopy suppressor gene for ISP-1-induced growth defects in yeast. YPK1 encodes a serine/threonine protein kinase that is a yeast counterpart of SGK1 (serum/glucocorticoid-regulated kinase 1) in mammals. SGK1 was isolated as a kinase regulated by serum and glucocorticoid [26]. Previously, SGK1 has been reported to be a downstream kinase of PDK1 (phosphoinositide-dependent kinase 1) and to have similar function to protein kinase B/Akt [27]. In yeast, Ypk1p is thought to be involved in the pathway of sphingolipid synthesis [28]. Indeed, the in vitro kinase activity of Pkh1/2p, upstream kinases of Ypk1p, was shown to be regulated by sphingoid bases, and the overexpression of Pkh1/2p suppressed the sphingoid base synthesis requirement [29]. However, the pathway downstream of Ypk1p is not well understood.
In the present study, another multicopy suppressor gene for ISP-1, SLI1, was characterized, and Sli1p was shown to have N-acetyltransferase activity towards ISP-1.
EXPERIMENTAL
Strains and reagents
S. cerevisiae strains KMY1006a (MATa leu2-3 ura3-52 his3-200 trp1-901 lys2-802 ADE2), Δypk1 (MATa leu2-3 ura3-52 his3-200 trp1-901 lys2-802 ADE2 ypk1::KanMX), BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and Δsli1 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sli1::KanMX) were used. BY4741 and Δsli1 were obtained from the Yeast Genome Deletion Project (via Research Genetics Inc.). Δypk1 was obtained by disruption of the YPK1 locus.
N-Acetyl-ISP-1 lactone was synthesized as described previously [30]. N-Acetyl-ISP-1 was obtained by cleavage of N-acetyl-ISP-1 lactone with 0.1 M KOH at 60 °C for 1 h. N-acetyl-ISP-1 was then purified by HPLC on a C22 reverse-phase column (250 mm×4.6 mm internal diam.) with a 15-min gradient from water containing 25% (v/v) methanol to 100% (v/v) methanol. N-Acetyl-ISP-1 was identified by MS.
Disruption of the YPK1 locus
A replacement cassette with long flanking homology regions was used to disrupt the YPK1 gene in strain KMY1006. PCR amplification was performed with KOD-Plus-polymerase (TOYOBO) using pFA6a-kanMX6 containing the geneticin resistance gene with the oligonucleotide primers P1 (GTAATCAACACTATCGTCATACCAACAACAGTTGAGTTATTGCCAGCTGAAGCTTCGTAC) and P2 (CGCCCCATCGACACGGATCTCGGAAGGGTAAGAGAGGAAAGCCACTAGTGGATCTGATAT), and generated a DNA product corresponding to the kanMX6 marker gene, with extensions of 20 or 22 bp identical to YPK1. The fragment was used to transform KMY1006 cells. Correct integration at the YPK1 locus in geneticin-resistant cells was confirmed by whole-cell PCR as described previously [31].
Molecular cloning of SLI1
SLI1 was cloned as described previously [28]. The 2.3 kb BamHI–PvuII fragment containing the wild-type SLI1 gene from pSLI (a plasmid which carries the SLI gene) was subcloned into YEp351 (an episomal, multicopy yeast vector containing the LEU2 selectable marker).
Yeast culture conditions
SD medium (used for SD agar plates) consisted of yeast nitrogen base, essential amino acids and glucose. Colonies selected on agar plates were inoculated into SD medium and then incubated overnight at 30 °C. Exponentially growing cultures were diluted to the desired concentrations, aliquoted into sterile culture tubes, and then treated with ISP-1 or methanol as a control. Proliferation was determined as the A600.
Construction of plasmids
To construct a Sli1p–EGFP (enhanced green fluorescent protein) fusion protein, the stop codon of SLI1 was replaced with a leucine codon, and XhoI, NotI and HindIII sites were introduced after the leucine codon by long, accurate PCR using primers 5′-TGTCACGGATCCGCAAGAATGAATCTTAAACTT-3′ and 5′-TGGTAAGCTTGCGGCCGCAGCTCGAGGTATAAATTTAAGTAATCTT-3′. The BamHI–HindIII fragment of the PCR product was subcloned into YEp351 with the ADH1 promoter, yielding YEp351ADH1-Sli1deltaSTOP. The EGFP DNA fragment was isolated from pEGFP-N1 by XhoI and NotI digestion and then ligated into YEp351ADH1-Sli1deltaSTOP, yielding YEp351-ADH1-Sli1p-EGFP. To generate YEp351-Sli1p-EGFP, the StuI–HindIII fragment of YEp351ADH1-Sli1p-EGFP was swapped into wild-type SLI1. The BamHI–StuI fragment of YEp351ADH1-Sli1deltaSTOP was isolated and ligated into wild-type SLI1, yielding YEp351ADH1-Sli1. YEp351-Ypk1 was obtained as described by Sun et al. [28].
Preparation of antibodies specific for Sli1p
The 1.4 kb BamHI–HindIII fragment was introduced by long, accurate PCR using primers 5′-TGTCACGGATCCGCAAGAATGAATCTTAAACTT-3′ and 5′-TCAGAAGCTTCTAGTATAAATTTAAGTAAT-3′, and ligated in-frame to the 3′ end of the MBP (maltose-binding protein) gene in the cytoplasmic expression vector pMalc2 (New England BioLabs). The Sli1–MBP fusion protein was expressed in Escherichia coli BL21 following induction with isopropyl β-D-thiogalactopyranoside. After sonication of the cells, the fusion protein was recovered in the soluble fraction, purified using an amylose resin (New England BioLabs) column, and then eluted from an SDS/polyacrylamide gel. Immunization and IgG purification were carried out by Veritas Corp. (Tokyo, Japan).
Analysis of sphingolipid biosynthesis
Cultures of 2 ml were grown to 2×106 cells/ml in SD medium at 30 °C and then labelled with [3H]serine (20 μCi/ml) for the indicated times. The cultures were chilled on ice after the addition of 0.5 ml of unlabelled stationary-phase cells and then subjected to centrifugation at 2800 g for 10 min at 4 °C. The cells were washed twice with 5 ml of cold water and then treated with 5% (v/v) trichloroacetic acid at 4 °C for 20 min. Lipids were extracted twice with 1 ml of ethanol/water/diethyl ether/pyridine/NH4OH (15:15:5:1:0.018, by vol.) at 60 °C, as described elsewhere [32]. The [3H]serine-labelled extract was subjected to mild alkaline methanolysis with 0.1 M KOH. The lipid products were extracted with 3 ml of chloroform, 1 ml of methanol, and 4.5 ml of 0.5 M NH4OH, and the chloroform layer was washed three times with 5 ml of water. The lipid products were dried under N2, resuspended in 0.04 ml of chloroform/methanol/water (16:16:5, by vol.), applied to a silica gel TLC plate, and then resolved with chloroform/methanol/4.2 M NH4OH (9:7:2, by vol.). Radioactive bands were visualized with a BAS 3000 image analyser (Fuji Film Co.).
Preparation of yeast extracts and immunoprecipitation
Total protein extracts were prepared from cells grown to mid-exponential phase. The cells were harvested, resuspended in lysis buffer [20 mM Tris/HCl, pH 7.5, containing 1 mM EDTA, 5 mM MgCl2, 50 mM KCl, 5% (v/v) glycerol, 1 mM PMSF, 0.5 μg/ml leupeptin, 1 μg/ml pepstatin A, 50 mM NaF and 3 mM dithiothreitol] and vortexed for 10 min with 0.5 vol. of 0.3–0.5-mm-diam. glass beads to lyse the cells, as described by Kohno et al. [33]. Unbroken cells and debris were removed by centrifugation of the homogenates at 12000 g for 10 min. For immunoprecipitation, Triton X-100 was added to the resulting supernatant to make a 1% solution. Immunoprecipitates were prepared using anti-Sli1p antibodies (10 μl) preadsorbed to a 50:50 (w/v) slurry of Protein G–Sepharose (10 μl). The beads were washed three times with lysis buffer and then incubated with the lysate at 4 °C overnight. Immune complexes were washed three times with lysis buffer and then separated by SDS/PAGE. Sli1p–EGFP was detected using anti-GFP antibodies, and Ypk1p was detected using anti-Ypk1p antibodies. Blots were examined using a chemiluminescent reagent (Pierce) for the detection of overexpressed Sli1 protein. Protein concentrations were measured with a Bio-Rad protein assay kit.
Assay for growth inhibition by ISP-1 and N-acetyl-ISP-1
Cells were grown in SD medium. The culture was diluted with SD medium to A600=0.002. Aliquots of 100 μl of cells were diluted into a series of wells in a microtitre plate. Each well containing 100 μl of SD medium was supplemented with ISP-1 or N-acetyl-ISP-1. Final concentrations of ISP-1 from 10 μg/ml to 1.22 ng/ml and of N-acetyl-ISP-1 from 3.9 μg/ml to 1.9 ng/ml were prepared by serial 2-fold dilutions. The plates were incubated at 30 °C for 42–48 h. The contents of each well were mixed, and A595 was measured.
Enzymic acetylation of ISP-1
Enzymic acetylation of ISP-1 was carried out using a modification of the protocol for the enzymic acetylation of 12-D,L-hydroxystearic acid described by Malcorps and Dufour [34]. The reaction was carried out in a final volume of 250 μl, containing 100 mM KH2PO4, 30 mM n-octyl glucoside, 0.1 mM ISP-1, 200 μM [3H]acetyl-CoA (25 Ci/mol) and 125 μl of yeast cell lysate prepared with breaking buffer (20 mM Tris/HCl, 10 mM EDTA, 5 mM dithiothreitol and 1 mM PMSF) as enzyme source. After the indicated period of incubation at 30 °C, the reaction was stopped by chilling on ice. The reaction product was extracted two times with 125 μl of n-butanol. The butanol was dried under N2 and the residue resuspended in 50 μl of methanol, applied to a silica gel TLC plate, and then resolved with chloroform/methanol/4.2 M NH4OH (9:7:2, by vol.; solvent A) or chloroform/methanol/4 M acetic acid (9:7:2, by vol.; solvent B). Radioactive bands were visualized with a BAS 2000 image analyser (Fuji Film Co.). For alkaline treatment, the labelled compound(s) was dissolved in 0.1 M KOH in methanol and incubated for 1 h at 60 °C. After incubation, the sample was neutralized and extracted with n-butanol.
SPT assay
SPT assays were carried out by a modification of the method of Buede et al. [11]. For each assay, the following components were used, in a final volume of 0.2 ml: 0.1 M Hepes (pH 8.3), 5 mM dithiothreitol, 2.5 mM EDTA (pH 8.0), 50 μM pyridoxal phosphate, 200 μM palmitoyl-CoA, 5 mM L-serine, 5 μCi of L-[3H]serine, 100 nM ISP-1 or N-acetyl-ISP-1, and 0.5 mg of yeast total protein extract prepared with phosphate buffer (50 mM sodium phosphate, pH 7.0, containing 5 mM dithiothreitol and 1 mM PMSF). After incubation for 20 min at 30 °C, 0.5 ml of 0.5 M NH4OH containing 10 mM L-serine was added, and then the lipid products were extracted with chloroform. The chloroform layer was transferred to a scintillation vial and then dried down completely before the radioactivity was measured with a Beckman LS6000 liquid scintillation counter. The result for background without palmitoyl-CoA was subtracted from all the measurements to calculate the specific activity.
Fluorescence microscopy
For indirect immunofluorescence microscopy, cells were fixed with 3.7% (v/v) formaldehyde in 0.1 M potassium phosphate buffer (pH 6.5), collected by centrifugation, and washed with SP buffer (0.1 M potassium phosphate, pH 7.5, and 1 M sorbitol). The cells were then converted into spheroplasts by adding 1:500 vol. of 2-mercaptoethanol and 15 μg/ml Zymolyase 100T (Seikagaku Co.), and incubated for 30 min at 30 °C. The spheroplasts were centrifuged and permeabilized by adding 0.5 ml of SP with 0.1% Triton X-100 for 10 min, and blocked with PBT (1% BSA and 0.1% Triton X-100 in PBS) for 10 min. The cells were incubated with primary antibodies diluted in PBT overnight. Anti-Kar2p antiserum (a gift from Dr. K. Kohno, NAIST, Ikoma, Japan) was used at 1:500 dilution. After washing, the cells were incubated with secondary antibodies diluted in PBS for 2 h. For secondary staining, Alexa Fluor 546 goat anti-rabbit IgG (H+L) conjugate (10 μg/ml; Molecular Probes) was used. The cells were observed under a fluorescence confocal microscope (LSM510; Zeiss).
RESULTS
Sli1p is a major contributor to ISP-1 resistance in yeast cells
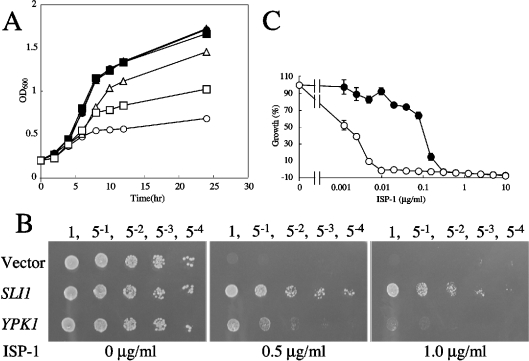
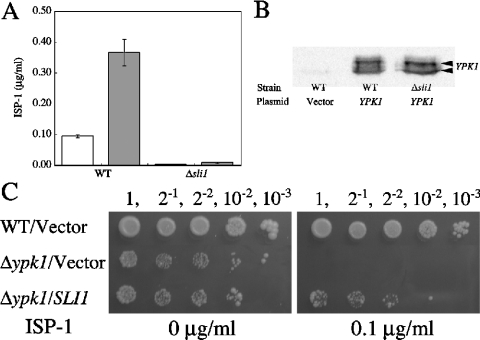
In our previous study, we showed that ISP-1 inhibited yeast cell growth, and characterized one of the suppressor genes for ISP-1 resistance, YPK1/SLI2 [28]. Another gene, termed SLI1 for sphingosine-like immunosuppressant resistant gene 1 (YGR212W; GenBank designation CAA97239.1), encodes a putative polypeptide (Sli1p) of 468 amino acids with a deduced molecular mass of 54.5 kDa (Figure 1). Sli1p-overexpressing yeast cells were much more resistant to ISP-1 than were Ypk1p-overexpressing cells (Figures 2A and 2B). To determine the phenotypic consequences of a complete loss of SLI1 function, we analysed the sli1-null mutant. The sli1-null mutant grew normally (results not shown). However, when the strain was cultured in the presence of ISP-1, cell growth was markedly reduced. The IC50 of ISP-1 for the growth decreased to approx. 0.01× that of the parental strain, suggesting that Sli1p is a major contributor to ISP-1 resistance in yeast (Figure 2C).
Figure 1. Amino acid sequence of Sli1p.
The amino acid sequence alignment of Sli1p, Atf1p and Atf2p was performed using CLUSTALW. The condensation domain-like region is underlined. Identical amino acids are marked with an asterisk (*), ‘strongly’ similar amino acids are marked with a colon (:), and ‘weakly’ similar amino acids are marked with a period (·).
Figure 2. ISP-1-resistance activity of Sli1p in yeast cells.
(A) An ISP-1-induced growth defect is blocked by overexpression of SLI1 and YPK1. KMY1006a cells containing YEp351 (•, ○, vector alone), YEp351-Sli1 (▴, ▵) or YEp351-Ypk1 (▪, □) were grown in medium containing methanol (•, ▴, ▪) or 1 μg/ml ISP-1 (○, ▵, □) for the indicated times. Proliferation was determined by measuring A600. (B) Resistance to ISP-1 of SLI1- and YPK1-overexpressing yeast cells on SD plates. Serially diluted KMY1006a cells containing YEp351 (Vector), YEp351-Sli1 or YEp351-Ypk1 were grown on SD plates containing methanol or 0.5 or 1.0 μg/ml ISP-1 for 2 days. Serial dilutions of each strain are indicated above the panels. (C) IC50 values of ISP-1 for the wild-type (•) and sli1-null (○) strains were measured as described in the Experimental section. A595 was measured after a 48 h incubation at 30 °C. Results are expressed as a percentage of the cell density achieved by the culture in the absence of ISP-1. Data are the means±S.D. from one experiment performed in triplicate.
The ISP-1-induced inhibition of sphingolipid biosynthesis is blocked by the overexpression of Sli1p
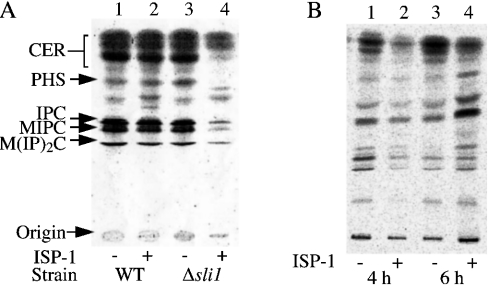
To clarify the mechanism underlying the hypersensitivity of the sli1-null mutant to ISP-1, the effect of ISP-1 on de novo sphingolipid biosynthesis was studied in sli1-null yeast cells, because the direct target of ISP-1 is SPT, the primary enzyme of sphingolipid biosynthesis. Sphingolipid biosynthesis was inhibited at a very low concentration of ISP-1 in the sli1-null cells, although biosynthesis in wild-type cells was not affected at this concentration (Figure 3A), indicating that the sphingolipid biosynthesis itself was hypersensitive to ISP-1, resulting in hypersensitivity of the null mutant to ISP-1.
Figure 3. Effects of the sli1 deletion and of Sli1p overexpression on sphingolipid biosynthesis in yeast cells.
Sphingolipids were labelled with [3H]serine and extracted, then separated on TLC plates as described in the Experimental section. The positions of standard lipids are indicated by arrows: CER, ceramide; PHS, phytosphingosine; IPC, inositol phosphoceramide; MIPC, mannose inositol phosphoceramide; M(IP)2C, mannose di-inositol diphosphoceramide. (A) Sphingolipid biosynthesis of sli1-null mutant cells (lanes 3 and 4) and wild-type (WT) cells (lanes 1 and 2) in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of 10 ng/ml ISP-1 for 2.5 h. [3H]Serine was added 0.5 h after the addition of ISP-1. (B) Sphingolipid biosynthesis of yeast cells (KMY1006a) containing YEp351-Sli1 (SLI1). Cells were treated with (lanes 2 and 4) or without (lanes 1 and 3) 0.5 μg/ml ISP-1 for 4 h (lanes 1 and 2) or 6 h (lanes 3 and 4). [3H]Serine was added 2 h (lanes 1 and 2) or 4 h (lanes 3 and 4) after the addition of ISP-1.
Two possibilities have been proposed to explain the mechanism of Sli1p-mediated ISP-1 resistance: Sli1p may decrease the ISP-1 concentration inside the cells, or Sli1p may increase the resistance of SPT to ISP-1. To test these possibilities, sphingolipid biosynthesis was monitored during ISP-1 treatment in Sli1p-overexpressing cells. At the beginning of ISP-1 treatment (Figure 3B, lanes 1 and 2), sphingolipid biosynthesis was decreased. However, during further incubation (Figure 3B, lanes 3 and 4), biosynthesis recovered to the level in untreated cells. These data indicate that Sli1p overexpression blocks the ISP-1-induced inhibition of sphingolipid biosynthesis. This is most probably due to a decrease in the ISP-1 concentration inside the cell, since the inhibition of biosynthesis was observed at the beginning of ISP-1 treatment.
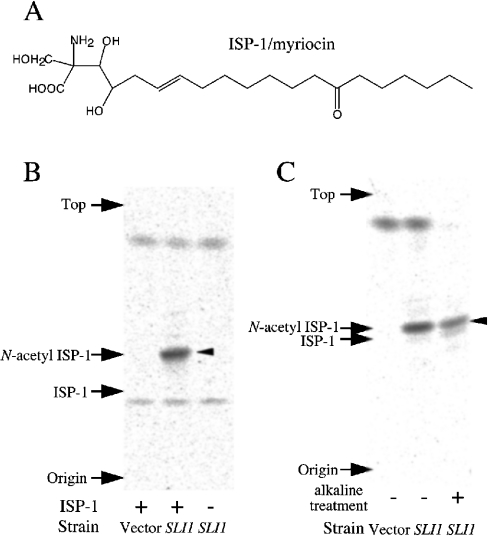
Sli1p acetylates ISP-1
Sli1p has weak similarity to Atf1p and Atf2p, which are yeast alcohol acetyltransferases [35,36] (Figure 1). Atf1p and Atf2p catalyse the synthesis of isoamyl acetate from acetyl-CoA and isoamyl alcohol. Based on the sequence similarity between Sli1p and Atf1/2p, we predicted that Sli1p may acetylate some hydroxy groups of ISP-1 (Figure 4A). We investigated whether Sli1p acetylates ISP-1 using 3H-labelled acetyl-CoA. One major band of putative acetylated ISP-1 was detected when ISP-1 and [3H]acetyl-CoA were incubated with yeast cell lysate containing Sli1p (Figure 4B). The band was not detected when the reaction was performed either with cell lysate that lacked Sli1p, or without ISP-1. Thus it is very likely that the labelled band represents acetyl-ISP-1 produced due to the acetyltransferase activity of Sli1p. Surprisingly, the radioactive compound co-migrated with N-acetyl-ISP-1 on a TLC plate in two different solvent systems (Figures 4B and 4C). Furthermore, the compound was stable after alkaline treatment (Figure 4C). These data suggest that the compound is not O-acetyl-ISP-1, but rather N-acetyl-ISP-1, because O-acetyl groups are known to be labile under alkaline treatment conditions. The acetyltransferase activity towards ISP-1 was dependent on the protein concentration of the enzyme source (results not shown). The specific activity of the crude cell lysate was calculated to be 7.69 nmol/min per mg of protein.
Figure 4. Acetylation activity towards ISP-1 of Sli1p-overexpressing cell lysate.
(A) Chemical structure of ISP-1. (B) ISP-1 acetyltransferase activity of Sli1p-overexpressing cells. Cell lysates prepared from sli1-null cells containing YEp351 (Vector) or YEp351ADH1-Sli1 (SLI1) were used as enzyme sources (0.5 μg/μl). Sli1p was overexpressed under the ADH1 promoter. Samples were incubated for 5 min at 30 °C with (left and middle lanes) or without (right lane) ISP-1. Products were extracted and separated on a TLC plate as described in the Experimental section using solvent A. A major putative band of acetylated ISP-1 is indicated by an arrowhead. Authentic standards are also indicated. (C) The radioactive acetyl-ISP-1 was treated under alkaline conditions as described in the Experimental section and separated on a TLC plate using solvent B. A major putative band of acetylated ISP-1 is indicated by an arrowhead. Authentic standards are also indicated.
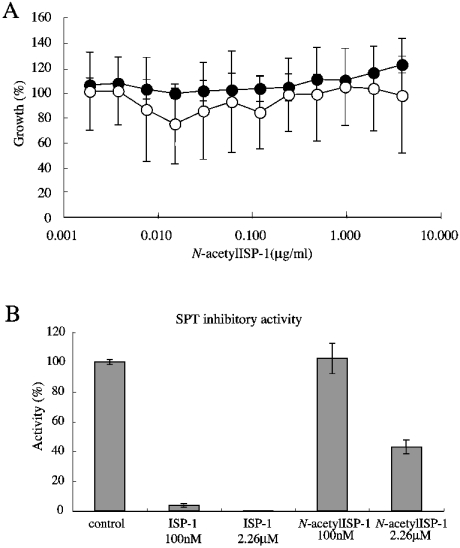
Acetylation activity towards ISP-1 was only detected when Sli1p was expressed under the ADH1 promoter, but not when expressed under its own promoter. The difficulty of detecting acetylation activity of the cell lysate may be due to a very low level of expression of Sli1p under its own promoter, even when overexpressed using a multicopy plasmid. Moreover, immunoprecipitated Sli1p did not show detectable acetyltransferase activity towards ISP-1. It is possible that Sli1p is a component of a multi-component complex enzyme. When wild-type yeast cells were treated with chemically synthesized N-acetyl-ISP-1, growth inhibition was not detected under the conditions used (Figure 5A). The sli1-null mutant was hypersensitive to ISP-1 (Figure 2C). However, the strain grew normally in the presence of N-acetyl-ISP-1. ISP-1 (100 nM) almost completely inhibited SPT activity in vitro, but N-acetyl-ISP-1 did not show any inhibitory activity against SPT at this concentration (Figure 5B). Furthermore, approx. 40% of wild-type SPT activity was still detected in the presence of a >20-fold molar excess of N-acetyl-ISP-1 (2.26 μM). These data suggest that Sli1p inactivates ISP-1 due to its N-acetyltransferase activity.
Figure 5. Effects of N-acetyl-ISP-1 on yeast cell growth and SPT activity.
(A) IC50 of N-acetyl-ISP-1 for the wild-type (•) and sli1-null (○) strains was measured as described in the Experimental section. A595 was measured after a 42 h incubation at 30 °C. Results are expressed as a percentage of the cell density achieved by the culture in the absence of ISP-1. Data are the means±S.D. from one experiment done in triplicate. (B) The effect of N-acetyl-ISP-1 on SPT activity was studied. SPT activity was measured using the wild-type cells as described in the Experimental section. Results are expressed as a percentage of control activity in the absence of reagents. Data are the means±S.D. from one experiment done in triplicate.
The ISP-1-resistance activity of Ypk1p is markedly decreased on a sli1-null background
YPK1/SLI2 has been reported to be a multicopy suppressor of ISP-1, and to encode a serine/threonine kinase [28]. Ypk1p is a functional homologue of the mammalian protein kinase SGK1, which was originally reported as a serine/threonine protein kinase that is transcriptionally regulated by serum and glucocorticoids in rat mammary tumour cells [37]. To clarify the relationship between Sli1p and Ypk1p, the effect of overexpression of Ypk1p on resistance to ISP-1 was examined on the sli1-null background. Comparison between Ypk1p-overexpressing cells and vector control cells on the wild-type background demonstrated that the IC50 of ISP-1 was increased by approx. 0.27 μg/ml in the Ypk1p-overexpressing cells (Figure 6A). Strikingly, the increase in IC50 due to Ypk1p expression was only 0.006 μg/ml on the sli1-null background. Under these conditions, the expression of Ypk1p was not significantly affected (Figure 6B), indicating that Sli1p is necessary for the ISP-1-resistance activity of Ypk1p. On the other hand, Sli1p-overexpressing ypk1-null cells showed ISP-1 resistance (Figure 6C). It has been shown that ypk1-null strains have a slow growth phenotype [38]. However, Sli1p overexpression did not recover the slow growth phenotype of the ypk1-null mutant in the absence of ISP-1 (Figure 6C).
Figure 6. Genetic interaction between YPK1 and SLI1 in mediating resistance to ISP-1.
(A) IC50 of ISP-1 in the wild-type (WT) strain and the sli1-null mutant strain. Cells containing YEp351 (empty bars) or YEp351-Ypk1 (shaded bars) were grown for 48 h at 30 °C. The IC50 of ISP-1 was measured as described in the Experimental section. Data are the means±S.D. from one experiment done in triplicate. (B) The expression level of Ypk1p was detected by immunoblotting. Cell-free extracts were prepared from the wild-type (WT) strain containing YEp351 (Vector) or YEp351-Ypk1 (YPK1), and the sli1-null mutant containing YEp351-Ypk1. Ypk1p was detected with anti-Ypk1p antibodies. (C) Resistance to ISP-1 of SLI1-overexpressing yeast cells on SD plates. Serially diluted wild-type (WT) and ypk1 null mutant strains containing YEp351 (Vector) or YEp351-Sli1 (SLI1) were grown on SD plates containing methanol or 0.1 μg/ml ISP-1 for 5 days. Serial dilutions of each strain are indicated above the panels.
Localization of Sli1p–EGFP fusion protein
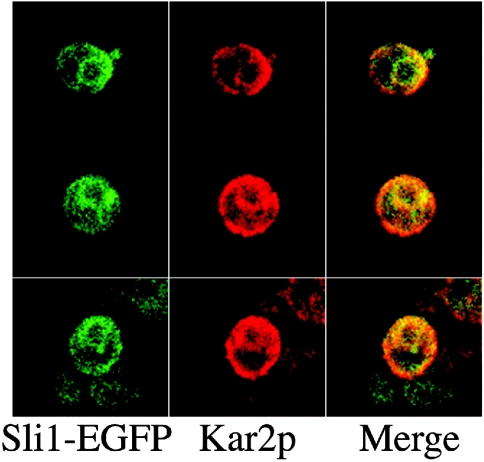
The intracellular localization of Sli1p was visualized by monitoring a Sli1p–EGFP fusion protein. The Sli1p–EGFP fusion protein was distributed in the area around the nucleus and co-localized with Kar2p, which was localized in both the endoplasmic reticulum and the plasma membrane (Figure 7), suggesting that the Sli1p–EGFP fusion protein localizes in the endoplasmic reticulum.
Figure 7. Intracellular localization of Sli1p.
Yeast cells containing YEp351ADH1-Sli1p-EGFP were fixed with formaldehyde, converted into spheroplasts, and permeabilized with 0.1% Triton X-100. They were then stained with anti-Kar2p antibodies. Green indicates the localization of Sli1p–EGFP (left panel) and red indicates the localization of Kar2p (middle panel). In the merged image (right panel), yellow indicates the co-localization of Sli1p–EGFP and Kar2p.
DISCUSSION
In the present study, one of the multicopy suppressor genes for ISP-1 was characterized. This gene, termed SLI1, showed a stronger ISP-1 resistance phenotype than YPK1/SLI2, which has already been characterized [28].
Sli1p is a major contributor to ISP-1 resistance in yeast cells. Cell lysates of Sli1p-overexpressing cells showed ISP-1 acetyltransferase activity (Figure 4). ISP-1 has three hydroxy groups and an amino group, which could potentially be acetylated (Figure 4A). Sli1p has weak similarity to the yeast alcohol acetyltransferases Atf1p and Atf2p, which catalyse the synthesis of short-chain and medium-chain aliphatic esters [34–36]. However, the labelled acetyl-ISP-1 was shown to co-migrate with N-acetyl-ISP-1 on a TLC plate using two different solvent systems. Furthermore, the labelled compound was stable under alkaline treatment conditions where O-acetyl groups are unstable. These data suggest that the amino group of ISP-1 is acetylated by Sli1p. This N-acetylation abolished the growth-inhibitory activity of ISP-1 in yeast cells (Figure 5A). In addition, N-acetylation of ISP-1 markedly reduced its inhibitory activity against SPT in vitro (Figure 5B).
The IC50 of ISP-1 in the sli1-null mutant was decreased to about 0.01 times that in the parental strain. The sphingolipid biosynthesis of the sli1-null mutant was inhibited at a low concentration of ISP-1 (10 ng/ml), although that of the parental strain was not. The concentration used was closer to the IC50 of ISP-1 for the inhibition of the in vitro activity of SPT in wild-type cells [28] than that for the inhibition of cell growth in wild-type cells (Figure 2C). These data suggest that the SPT activity is directly affected by ISP-1 in the sli1-null strain, as in vitro, but that inhibition of the enzyme activity by ISP-1 in wild-type cells is suppressed by the function of Sli1p to some extent. On the other hand, overexpression of Sli1p restores sphingolipid biosynthesis that was inhibited by ISP-1. Although sphingolipid biosynthesis was inhibited by ISP-1 at the beginning of the treatment period, the biosynthesis recovered to the same level as without ISP-1 during further incubation (Figure 3B). The ISP-1 concentration was probably high enough to inhibit SPT at early time points, but would be decreased due to the activity of Sli1p, and hence biosynthesis recovered at a later time point. This profile shows good accordance with the possibility that Sli1p inactivates ISP-1. Taken together, these data suggest that ISP-1 is converted into N-acetyl-ISP-1 and hence inactivated by Sli1p in vivo. Sli1p has a domain similar to the condensation domain (pfam 00668) (Figure 1). The condensation domain is usually involved in amide bond formation in non-ribosomal peptide biosynthesis [39]. Thus the condensation-like domain of Sli1p may be involved in amide bond formation in the N-acetylation reaction catalysed by Sli1p.
FTY720, an analogue of ISP-1, has been tested in clinical immunosuppressive regimens in humans [23]. In spite of the structural similarity between the two compounds, cell lysates prepared from Sli1p-overexpressing cells did not show any acetylation activity towards FTY720 (results not shown). The structure of ISP-1 is also similar to those of sphingosine and phytosphingosine, which are intermediates in the sphingolipid biosynthesis pathway. When yeast cells were treated with a high concentration of these compounds, a growth defect was also observed. Interestingly, Sli1p overexpression did not suppress the growth defect, despite the structural similarity. In accordance with these data, cell lysates prepared from Sli1p-overexpressing cells did not show any acetylation activity toward these long-chain bases (results not shown). Fumonisin B1 [40] and aureobasidin [41] also inhibit sphingolipid biosynthesis in a different manner to ISP-1: fumonisin B1 inhibits ceramide synthase [17,40] and aureobasidin inhibits inositol phosphoceramide synthase [41]. Treatment with these reagents also induces a growth defect in yeast cells. However, Sli1p did not overcome the growth defect caused by such treatment (results not shown). Taken together, these findings imply that the action of Sli1p seems to be specific for ISP-1.
YPK1/SLI2 is another multicopy suppressor gene for ISP-1, and encodes a serine/threonine kinase [28]. The multicopy suppressor activity of Ypk1p was almost abolished on the sli1-null background. On the other hand, Sli1p-overexpressing ypk1-null cells showed ISP-1 resistance. It is very difficult to compare the ISP-1 resistance activity of Sli1p in the ypk1-null mutant cells with that in wild-type cells, because ypk1-null strains show a slow growth phenotype [38]. These findings suggest that there is a genetic interaction between SLI1 and YPK1. However, we regard it likely that this interaction is not direct, because Sli1p was not phosphorylated in vitro by Ypk1p, while a peptide substrate was well phosphorylated by Ypk1p (results not shown), and the putative phosphorylation sequence for Ypk1p (RXRXXS/T) is not detected in Sli1p. These observations suggest that Sli1p is not a direct substrate of Ypk1p. Furthermore, the slow growth phenotype of ypk1-null strains was not compensated by the overexpression of Sli1p. Therefore the mechanism of interaction between SLI1 and YPK1 remains to be clarified.
To study the intracellular localization of Sli1p, a Sli1p–EGFP fusion protein was expressed. However, the fusion protein was not detected in yeast cells when expressed under its own promoter from a multicopy-base vector. The ADH1 promoter was therefore used to overcome this problem. The Sli1p–EGFP fusion protein was co-localized with the part of Kar2p known to localize in the endoplasmic reticulum [42], showing that Sli1p localizes in this organelle. SPT is also reported to localize in the endoplasmic reticulum. These data suggest that Sli1p may function in the endoplasmic reticulum and decrease the ISP-1 concentration in the vicinity of SPT.
ISP-1 was originally discovered as an immunosuppressant. This action of ISP-1 in mammalian cells is restricted to a few cell lines: CTLL-2 [21], Purkinje cells [43], and CHO cells [44,45]. However, the mechanism of action has not been clarified. Further studies on the function of Sli1p may facilitate elucidation of the detailed mechanism, although the mammalian homologue of Sli1p has yet to be identified.
Acknowledgments
This work was supported in part by Grants-in-Aid for Scientific Research (13470488 and 13877373 to Y.K.) from the Japan Society for the Promotion of Science, by a Grant-in-Aid for Scientific Research on Priority Areas (12140202 to Y.K.) from the Ministry of Education, Culture, Sports and Technology, and by RIKEN.
References
- 1.Fishbein J. D., Dobrowsky R. T., Bielawska A., Garrett S., Hannun Y. A. Ceramide-mediated growth inhibition and CAPP are conserved in Saccharomyces cerevisiae. J. Biol. Chem. 1993;268:9255–9261. [PubMed] [Google Scholar]
- 2.Nickels J. T., Broach J. R. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 1996;10:382–394. doi: 10.1101/gad.10.4.382. [DOI] [PubMed] [Google Scholar]
- 3.Jenkins G. M., Richards A., Wahl T., Mao C., Obeid L., Hannun Y. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J. Biol. Chem. 1997;272:32566–32572. doi: 10.1074/jbc.272.51.32566. [DOI] [PubMed] [Google Scholar]
- 4.Dickson R. C., Nagiec E. E., Skrzypek M., Tillman P., Wells G. B., Lester R. L. Sphingolipids are potential heat stress signals in Saccharomyces. J. Biol. Chem. 1997;272:30196–30200. doi: 10.1074/jbc.272.48.30196. [DOI] [PubMed] [Google Scholar]
- 5.Chung N., Jenkins G., Hannun Y. A., Heitman J., Obeid L. M. Sphingolipids signal heat stress-induced ubiquitin-dependent proteolysis. J. Biol. Chem. 2000;275:17229–17232. doi: 10.1074/jbc.C000229200. [DOI] [PubMed] [Google Scholar]
- 6.Skrzypek M. S., Nagiec M. M., Lester R. L., Dickson R. C. Inhibition of amino acid transport by sphingoid long chain bases in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:2829–2834. doi: 10.1074/jbc.273.5.2829. [DOI] [PubMed] [Google Scholar]
- 7.Chung N., Mao C., Heitman J., Hannun Y. A., Obeid L. M. Phytosphingosine as a specific inhibitor of growth and nutrient import in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:35614–35621. doi: 10.1074/jbc.M105653200. [DOI] [PubMed] [Google Scholar]
- 8.Sutterlin C., Doering T. L., Schimmoller F., Schroder S., Riezman H. Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J. Cell Sci. 1997;110:2703–2714. doi: 10.1242/jcs.110.21.2703. [DOI] [PubMed] [Google Scholar]
- 9.Zanolari B., Friant S., Funato K., Sutterlin C., Stevenson B. J., Riezman H. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 2000;19:2824–2833. doi: 10.1093/emboj/19.12.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath A., Sutterlin C., Manning-Krieg U., Movva N. R., Riezman H. Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. EMBO J. 1994;13:3687–3695. doi: 10.1002/j.1460-2075.1994.tb06678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buede R., Rinker-Schaffer C., Pinto W. J., Lester R. L., Dickson R. C. Cloning and characterization of LCB1, a Saccharomyces gene required for biosynthesis of the long-chain base component of sphingolipids. J. Bacteriol. 1991;173:4325–4332. doi: 10.1128/jb.173.14.4325-4332.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagiec M. M., Baltisberger J. A., Wells G. B., Lester R. L., Dickson R. C. The LCB2 gene of Saccharomyces and the related LCB1 gene encode subunits of serine palmitoyltransferase, the initial enzyme in sphingolipid synthesis. Proc. Natl. Acad. Sci. U.S.A. 1994;91:7899–7902. doi: 10.1073/pnas.91.17.7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagiec M. M., Skrzypek M., Nagiec E. E., Lester R. L., Dickson R. C. The LCB4 (YOR171c) and LCB5 (YLR260w) genes of Saccharomyces encode sphingoid long chain base kinases. J. Biol. Chem. 1998;273:19437–19442. doi: 10.1074/jbc.273.31.19437. [DOI] [PubMed] [Google Scholar]
- 14.Mao C., Wadleigh M., Jenkins G. M., Hannun Y. A., Obeid L. M. Identification and characterization of Saccharomyces cerevisiae dihydrosphingosine-1-phosphate phosphatase. J. Biol. Chem. 1997;272:28690–28694. doi: 10.1074/jbc.272.45.28690. [DOI] [PubMed] [Google Scholar]
- 15.Qie L., Nagiec M. M., Baltisberger J. A., Lester R. L., Dickson R. C. Identification of a Saccharomyces gene, LCB3, necessary for incorporation of exogenous long chain bases into sphingolipids. J. Biol. Chem. 1997;272:16110–16117. doi: 10.1074/jbc.272.26.16110. [DOI] [PubMed] [Google Scholar]
- 16.Mandala S. M., Thornton R., Tu Z., Kurtz M. B., Nickels J., Broach J., Menzeleev R., Spiegel S. Sphingoid base 1-phosphate phosphatase: a key regulator of sphingolipid metabolism and stress response. Proc. Natl. Acad. Sci. U.S.A. 1998;95:150–155. doi: 10.1073/pnas.95.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schorling S., Vallee B., Barz W. P., Riezman H., Oesterhelt D. Lag1p and Lac1p are essential for the Acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisae. Mol. Biol. Cell. 2001;12:3417–3427. doi: 10.1091/mbc.12.11.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kluepfel D., Bagli J., Baker H., Charest M. P., Kudelski A. Myriocin, a new antifungal antibiotic from Myriococcum albomyces. J. Antibiot. 1972;25:109–115. doi: 10.7164/antibiotics.25.109. [DOI] [PubMed] [Google Scholar]
- 19.Craveri R., Manachini P. L., Aragozzini F. Thermozymocidin new antifungal antibiotic from a thermophilic eumycete. Experientia. 1972;28:867–868. doi: 10.1007/BF01923181. [DOI] [PubMed] [Google Scholar]
- 20.Fujita T., Inoue K., Yamamoto S., Ikumoto T., Sasaki S., Toyama R., Chiba K., Hoshino Y., Okumoto T. Fungal metabolites. Part 11. A potent immunosuppressive activity found in Isaria sinclairii metabolite. J. Antibiot. 1994;47:208–215. doi: 10.7164/antibiotics.47.208. [DOI] [PubMed] [Google Scholar]
- 21.Miyake Y., Kozutsumi Y., Nakamura S., Fujita T., Kawasaki T. Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem. Biophys. Res. Commun. 1995;211:396–403. doi: 10.1006/bbrc.1995.1827. [DOI] [PubMed] [Google Scholar]
- 22.Chiba K., Yanagawa Y., Masubuchi Y., Kataoka H., Kawaguchi T., Ohtsuki M., Hoshino Y. FTY720, a novel immunosuppressant, induces sequestration of circulating mature lymphocytes by acceleration of lymphocyte homing in rats. I. FTY720 selectively decreases the number of circulating mature lymphocytes by acceleration of lymphocyte homing. J. Immunol. 1998;160:5037–5044. [PubMed] [Google Scholar]
- 23.Kahan B. D., Koch S. M. Current immunosuppressant regimens: considerations for critical care. Curr. Opin. Crit. Care. 2001;7:242–250. doi: 10.1097/00075198-200108000-00006. [DOI] [PubMed] [Google Scholar]
- 24.Mandala S., Hajdu R., Bergstrom J., Quackenbush E., Xie J., Milligan J., Thornton R., Shei G. J., Card D., Keohane C., et al. Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science. 2002;296:346–349. doi: 10.1126/science.1070238. [DOI] [PubMed] [Google Scholar]
- 25.Brinkmann V., Davis M. D., Heise C. E., Albert R., Cottens S., Hof R., Bruns C., Prieschl E., Baumruker T., Hiestand P., et al. The immune modulator FTY720 targets sphingosine 1-phosphate receptors. J. Biol. Chem. 2002;277:21453–21457. doi: 10.1074/jbc.C200176200. [DOI] [PubMed] [Google Scholar]
- 26.Webster M. K., Goya L., Ge Y., Maiyar A. C., Firestone G. L. Characterization of sgk, a novel member of the serine/threonine protein kinase gene family which is transcriptionally induced by glucocorticoids and serum. Mol. Cell. Biol. 1993;13:2031–2040. doi: 10.1128/mcb.13.4.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi T., Cohen P. Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem. J. 1999;339:319–328. [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y., Taniguchi R., Tanoue D., Yamaji T., Takematsu H., Mori K., Fujita T., Kawasaki T., Kozutsumi Y. Sli2 (Ypk1), a homologue of mammalian protein kinase SGK, is a downstream kinase in the sphingolipid-mediated signaling pathway of yeast. Mol. Cell. Biol. 2000;20:4411–4419. doi: 10.1128/mcb.20.12.4411-4419.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friant S., Lombardi R., Schmelzle T., Hall M. N., Riezman H. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 2001;20:6783–6792. doi: 10.1093/emboj/20.23.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujita T., Inoue K., Yamamoto S., Ikumoto T., Sasaki S., Toyama R., Yoneta M., Chiba K., Hoshino Y., Okumoto T. Fungal metabolites. Part 12. Potent immunosuppressant, 14-deoxomyriocin, (2S,3R,4R)-(E)-2-amino-3,4-dihydroxy-2-hydroxymethyleicos-6-enoic acid and structure–activity relationships of myriocin derivatives. J. Antibiot. 1994;47:216–224. doi: 10.7164/antibiotics.47.216. [DOI] [PubMed] [Google Scholar]
- 31.Roelants F. M., Torrance P. D., Bezman N., Thorner J. Pkh1 and pkh2 differentially phosphorylate and activate ypk1 and ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol. Biol. Cell. 2002;13:3005–3028. doi: 10.1091/mbc.E02-04-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson B. A., Lester R. L. The extraction of inositol-containing phospholipids and phosphatidylcholine from Saccharomyces cerevisiae and Neurospora crassa. J. Lipid Res. 1980;21:309–315. [PubMed] [Google Scholar]
- 33.Kohno K., Normington K., Sambrook J., Gething M. J., Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malcorps P., Dufour J. P. Short-chain and medium-chain aliphatic-ester synthesis in Saccharomyces cerevisiae. Eur. J. Biochem. 1992;210:1015–1022. doi: 10.1111/j.1432-1033.1992.tb17507.x. [DOI] [PubMed] [Google Scholar]
- 35.Fujii T., Nagasawa N., Iwamatsu A., Bogaki T., Tamai Y., Hamachi M. Molecular cloning, sequence analysis, and expression of the yeast alcohol acetyltransferase gene. Appl. Environ. Microbiol. 1994;60:2786–2792. doi: 10.1128/aem.60.8.2786-2792.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshimoto H., Fujiwara D., Momma T., Tanaka K., Sone H., Nagasawa N., Tamai Y. Isolation and characterization of the ATF2 gene encoding alcohol acetyltransferase II in the bottom fermenting yeast Saccharomyces pastorianus. Yeast. 1999;15:409–417. doi: 10.1002/(SICI)1097-0061(19990330)15:5<409::AID-YEA366>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 37.Casamayor A., Torrance P. D., Kobayashi T., Thorner J., Alessi D. R. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 1999;9:186–197. doi: 10.1016/s0960-9822(99)80088-8. [DOI] [PubMed] [Google Scholar]
- 38.Chen P., Lee K. S., Levin D. E. A pair of putative protein kinase genes (YPK1 and YPK2) is required for cell growth in Saccharomyces cerevisiae. Mol. Gen. Genet. 1993;236:443–447. doi: 10.1007/BF00277146. [DOI] [PubMed] [Google Scholar]
- 39.Marahiel M. A., Stachelhaus T., Mootz H. D. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 40.Wang E., Norred W. P., Bacon C. W., Riley R. T., Merrill A. H., Jr Inhibition of sphingolipid biosynthesis by fumonisins. Implications for diseases associated with Fusarium moniliforme. J. Biol. Chem. 1991;266:14486–14490. [PubMed] [Google Scholar]
- 41.Nagiec M. M., Nagiec E. E., Baltisberger J. A., Wells G. B., Lester R. L., Dickson R. C. Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J. Biol. Chem. 1997;272:9809–9817. doi: 10.1074/jbc.272.15.9809. [DOI] [PubMed] [Google Scholar]
- 42.Rose M. D., Misra L. M., Vogel J. P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- 43.Furuya S., Mitoma J., Makino A., Hirabayashi Y. Ceramide and its interconvertible metabolite sphingosine function as indispensable lipid factors involved in survival and dendritic differentiation of cerebellar Purkinje cells. J. Neurochem. 1998;71:366–377. doi: 10.1046/j.1471-4159.1998.71010366.x. [DOI] [PubMed] [Google Scholar]
- 44.Hanada K., Hara T., Nishijima M. Purification of the serine palmitoyltransferase complex responsible for sphingoid base synthesis by using affinity peptide chromatography techniques. J. Biol. Chem. 2000;275:8409–8415. doi: 10.1074/jbc.275.12.8409. [DOI] [PubMed] [Google Scholar]
- 45.Hanada K., Nishijima M., Fujita T., Kobayashi S. Specificity of inhibitors of serine palmitoyltransferase (SPT), a key enzyme in sphingolipid biosynthesis, in intact cells. A novel evaluation system using an SPT-defective mammalian cell mutant. Biochem. Pharmacol. 2000;59:1211–1216. doi: 10.1016/s0006-2952(00)00251-3. [DOI] [PubMed] [Google Scholar]