Abstract
Centromeres of fission yeast are arranged with a central core DNA sequence flanked by repeated sequences. The centromere-associated histone H3 variant Cnp1 (SpCENP-A) binds exclusively to central core DNA, while the heterochromatin proteins and cohesins bind the surrounding outer repeats. CHD (chromo-helicase/ATPase DNA binding) chromatin remodeling factors were recently shown to affect chromatin assembly in vitro. Here, we report that the CHD protein Hrp1 plays a key role at fission yeast centromeres. The hrp1Δ mutant disrupts silencing of the outer repeats and central core regions of the centromere and displays chromosome segregation defects characteristic for dysfunction of both regions. Importantly, Hrp1 is required to maintain high levels of Cnp1 and low levels of histone H3 and H4 acetylation at the central core region. Hrp1 interacts directly with the centromere in early S-phase when centromeres are replicated, suggesting that Hrp1 plays a direct role in chromatin assembly during DNA replication.
INTRODUCTION
Precise segregation of chromosomes during cell division is a fundamental and essential process for cell and organism viability. A successful transmission of genetic information to daughter cells is dependent on functional centromeres and kinetochores. Schizosaccharomyces pombe centromeres consist of 40–100 kb of repetitive DNA divided into the central core of the centromere (cnt/cc) and the flanking outer repetitive regions (otr) (1,2). These two regions form structurally and functionally distinct regions of the centromere (3,4). The heterochromatin protein 1 homolog, Swi6 in S.pombe together with Chp1, has been shown to be associated with the outer repeats but are excluded from the central core, whereas Mis6 and the H3 variant Cnp1 binds exclusively to the central core region (5–7). Mutations in genes that are important for the function of the central core and outer repeat of the centromere display distinct phenotypes, indicating functional differences between the domains. Mutants affecting the central core of the centromere, such as cnp1, mis6, mis12, mis14-18, mal2, nuf2 and ams2, typically display chromosome loss due to loss of bidirectionality likely caused by defective kinetochore-microtubuli attachments (6,8–12). In contrast, mutants defective in heterochromatin assembly in the flanking regions, such as rik1, swi6, clr4, csp1-12 and mutants in the RNAi-directed chromatin pathway, display a characteristic lagging chromosome phenotype, indicative of sister–centromere cohesion defects (13–18).
The centromere-specific histone H3 variant CENP-A is an essential and highly conserved protein, which is present in chromatin of inner centromere structures in all eukaryotes (6,19–21). CENP-A has been shown to be required for initial assembly of the kinetochore (22). Recently, three different protein complexes were shown to be required for proper Cnp1 localization to the central core region in S.pombe (9). Mis12 and Mis14 form one complex, Mis16 and Mis18 another and the third complex consist of Mis6, Mis15 and Mis17. In addition to these, the GATA-like factor Ams2 (12) and the coiled-oil protein Sim4 (23) are also required for Cnp1 localization.
Post-translational modifications of histones are also important in centromere function and structure. Studies in both S.pombe and human cells show that histone deacetylase (HDAC) activity is necessary for proper centromere function (24,25). If HDACs are inhibited by Trichostatin A (TSA), the elevated acetylation levels in the outer repeat region cause characteristic cohesion defects (24). In addition, histone methyltransferase activity is required in the outer repeat region to dimethylate histone H3 at lysine 9 (H3K9me2), thereby creating a binding site for Swi6 (26). Loss of H3K9me2 or Swi6 leads to loss of cohesin from the outer repeat region, which causes sister–centromere cohesion defects (16). Interestingly, high histone acetylation levels in the central core region correlate with loss of Cnp1 in mis16 and mis18 mutants, and Mis16 shows high similarity to the HDAC-associated protein RbAp48 (9).
Chromatin remodeling factors generally have a core ATPase/helicase domain and then additional domains, such as chromodomains [CHD (chromo-helicase/ATPase DNA binding)/Mi2 family] SANT domain (ISWI family) bromodomain (SWI/SNF family). These are ATP-dependent enzymes that can alter nucleosome position or structure and are involved in a broad range of cellular processes, such as DNA replication, repair, recombination, transcriptional regulation, elongation and termination (27,28). The SWI/SNF family member RSC of budding yeast and the human SNF-B chromatin-remodeling complex have previously been implicated in centromere and kinetochore function (29–31). Human ISWI (SNF2h) and budding yeast RSC have also been shown to be directly involved in establishing sister–chromatid cohesion along chromosome arms (32,33). However, RSC complexes are not needed for loading CENP-A (Cse4) to centromeres in budding yeast (31). The CHD, or Mi2 complex, has previously been copurified together with HDACs and the histone binding co-repressors RbAp46/48 in human and Xenopus, where the chromatin remodeling factors were suggested to facilitate histone deacetylation of chromatin (34–36). The human CHD/Mi2 complex co-localizes with Ikaros and the HP1 homolog M31 at centromeric heterochromatin (37,38), so although it remains to be demonstrated it is possible that Mi2 is required for centromere function. We have previously reported that the CHD/Mi2 family member Hrp1 (helicase-related protein in S.pombe) has a function in chromosome segregation (39). Overexpression and deletion of hrp1 resulted in chromosome segregation defects. Another fission yeast CHD family member, Hrp3, was shown to be directly required for silencing of the mating-type region mat2/3 but not at centromeres (40). In this paper, we investigate the role of Hrp1 in fission yeast centromere assembly and centromere function.
MATERIALS AND METHODS
S.pombe strains and media
The genotypes for the strains used in this study are listed in Table 1. Media and genetic techniques were prepared according to the standard procedures (41). G418 (Gibco) was used at a concentration of 200 mg/l. TSA (Sigma) was dissolved in dimethyl sulfoxide (DMSO) and used at 12.5–200 nM in the TSA growth assay and 100 nM was used in the lagging chromosome study. Thiabendazole (TBZ) (Sigma) was added from a stock solution containing 20 g/l TBZ dissolved in DMSO to a final concentration of 15 μg/ml in YEA plates. Low adenine indicator plates were supplemented with 7.5 mg/l of adenine. 5′-fluoroorotic acid (FOA) (US Biologicals) was used at a final concentration of 1.0 g/l. Endogenous epitope-tagged hrp1-HA and hrp1-myc gene fusions were constructed using a PCR-based approach, where endogenous genes were epitope-tagged after transformation and homologous recombination described in (42).
Table 1.
S.pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| FY367 | h+ leu1-32 ura4-D18 ade6-M210 | Ekwall et al. (24) |
| FY375 | h+ Ch16 LEU2+::ade6+-TEL leu1-32 ura4-DS/E ade6-M210 | Allshire et al. (47) |
| FY412 | h+ cc2(SphI)::ura4+ leu1-32, ura4-DS/E, ade6-M210 | Allshire et al. (47) |
| FY1180 | h+ otr1R(SphI)::ade6+ leu1-32 ura4-D18 ade6-M210 | Allshire et al. (47) |
| Hu15 | h90 mat3-M::ade6+ ura4-D18 ade6-210 | This study |
| Hu22 | h+ hrp1::ura4+ otr1R(SphI)::ade6+ leu1-32 ura4-D18 ade6-210 | This study |
| Hu26 | h+ hrp1::ura4+ Ch16 LEU2+::ade6+-TEL leu1-32 ura4-DS/E ade6-M210 | This study |
| Hu33 | h90 hrp1::ura4+ mat3::ade6+ ura4-D18 ade6-210 | This study |
| Hu61 | h+ hrp1::ura4+ otr1R(SphI)::ade6+ leu1-32 ura4-D18 ade6-DN/N | This study |
| Hu393 | h+ leu1/Ylp2.4pUCura4+-7 leu1-32 ura4-DS/E ade6-M216 | Bjerling et al. (55) |
| Hu465 | h− hrp1::leu2 leu1-32 ura4DS/E adeM210 | This study |
| Hu466 | h+ hrp1::LEU2+ leu1/YIp2.4 pUCura4+-7 leu1-32 ura4DS/E ade6-M210 | This study |
| Hu584 | h− hrp1::LEU2+ cc2(SphI)::ura4+ leu1-32, ura4-DS/E, ade6-M210 | This study |
| Hu602 | h+ hrp3::LEU2+ otr1R(SphI)::ade6+ leu1-32 ura4-D18 ade6-DN/N | This study |
| Hu615 | h+ hrp1-HA::kanMX6 leu1-32 ura4-D18 ade6-M210 | This study |
| Hu659 | h− dcr::kanMX6 leu1-32ura4-D18 ade6-M210 | Provost et al. (17) |
| Hu721 | h+ TM1::ade6 ade6 ΔNcoI mis6-302(ts) ura4DS/E | This study |
| Hu764 | h+ hrp1-myc::kanMX6 leu1-32ura4-D18 ade6-M210 | This study |
| Hu1121 | h+ hrp1::LEU2 dcr1::kanMX6 leu1-32ura4-D18 ade6-M210 | This study |
| Hu1176 | hrp1::leu2 ams2::ura4 leu1-32 ura4DS/E adeM210 | This study |
| Hu1181 | h− hrp1::leu2 mis6-302(ts) leu1-32 ura4DS/E adeM210 | This study |
| Sp45 | h− ams2::ura4 leu1 ura4 | K. Takahashi |
Immunofluorescence (IF), cell synchronization and fluorescence in situ hybridization (FISH)
The IF studies were performed as described previously (43) with some modifications. S.pombe cells were grown up to mid-log phase in YES media with or without 100 mg/l TSA. Cells walls were digested in PEMS with 1 mg/ml Zymolyase 100T (ICN Biomedicals) for 90 min at 37°C. We used the mouse α-tubulin antibody (Tat1) diluted 1:50 as primary antibodies (44). DNA was visualized by staining with 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma).
S.pombe cells were synchronized by cell size selection using d-Lactose gradients, in combination with hydroxyurea (HU) arrest. A total of 1 × 1010 mid-log phase cells were harvested and resuspended in 18 ml YES media. After the cells were shaken vigorously for 1 min, 3 ml of the cells were carefully applied to 50 ml d-Lactose gradients. The 20% d-Lactose gradients were created by freezing the solutions at −80°C overnight, followed by thawing at room temperature and equilibrated at 33°C for 1 h. Next, the cells were applied onto the gradient and centrifuged for 8 min, at 33°C in a swing out rotor. Immediately after centrifugation, 10 ml of small-sized cells were collected from the mid-zone of the gradient using a flat 10 ml pipette tip. The cells were pooled, resuspended in 3 ml, vortexed, loaded on a second gradient and the centrifugation step was repeated. The cells were pooled again, suspended into YES media containing 12 mM HU. Hundred and twenty minutes after the second gradient, the cells were collected and washed two times in phosphate-buffered saline and half of the cells were resuspended of YES media without HU and incubated for 1 h. The two batches of cells were subsequently used in a combined IF and FISH experiment. The FISH experiment was carried out as described previously (45). An aliquot of 1 μg of imr/otr probe DNA was incubated with a dNTP mixture containing 1 mM DIG-dUTP, 10 U of DNA polymerase, 1 μl DNase I diluted 1:50, for 90 min at 15°C according to the protocol. Cells were then pre-stained according to the protocol (see above). An aliquot of 4 μl of the probe in hybridization mixture was added to 1 × 107 cells. Fluorescein isothiocyanate-conjugated α-DIG antibody (Sheep) was used for visualization of the probe. The immunostained cells were visualized with a Zeiss Axioskop 2 imaging microscope and a Hamamatsu C4742-95 CCD camera. All images are deconvolved with the nearest-neighbor deconvolution algorithm and 0.2 μm in Z-steps, using Openlab 3 digital deconvolution software (Improvision).
Chromatin immunoprecipitation (ChIP)
ChIP assay was performed as described previously (24) with some modifications. The cell extract was resuspended and sonicated using a microtip in a Branson 200 sonicator on ice, for 1 min, 50% duty cycle, three times. An aliquot of 1 μl of antibody 9E10 α-myc (Sigma), 12CA5 α-HA (Sigma), α-acetylated isoforms of H4 and α-H3 (Upstate Biotechnology), and 2.5 μl of sheep α-Cnp1 (B. Mellone and R. Allshire, unpublished data) were used for precipitation of the respective crosslinked proteins. The washing of the protein-A beads was repeated two times with 140 mM NaCl lysis buffer, once with 500 mM NaCl lysis buffer [50 mM HEPES-KOH, pH 7.5, 1% Triton X-100, 0.1% (w/v) sodium deoxycholate, 1 mM EDTA and 500 mM NaCl], once with wash buffer [10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 0.5% (w/v) sodium deoxycholate, 0.5% NP-40 and 0.25 M LiCl] and once with TE buffer (1 mM EDTA, 10 mM Tris–HCl, pH 8.0). Finally, the ChIP DNA was quantified by duplex PCR, where two primer pairs, one primer pair for the locus of interest and one primer pair for a control locus, were used for every reaction. The Glutamyl tRNA synthetase gene was chosen as a control gene because it was expressed at moderate levels and is not affected by meiosis, cell cycle or environmental stress. Between 3 and 8 μl of precipitated DNA was amplified in a standard PCR for quantification, and input was diluted so that the PCR amplification was in the linear range.
32P labeled PCR products were separated on 4% polyacrylamide gels, visualized using Fujifilm PhosphoImager FLA3000 and quantified with Fujifilm Image gauge software.
RT–PCR
Total RNA was extracted using Qiagens RNeasy extraction kit (Qiagen). Purified RNA was reversed transcribed using SuperScript™ First-Strand Synthesis System for RT–PCR, following the manufacture's instructions (Invitrogen). cDNA was amplified with tRNA synthetase, dgIII or imrIII primers and PCR products were separated on 1.5% agarose gels and stained with ethidium bromide or SYBR green (Invitrogen). Quantification of the bands was performed using Fujifilm Image Gauge software.
Micrococcal nuclease (MNase) assay
MNase assay was performed as previously been described with some modifications (46). An aliquot of 1 mg/ml of Zymolyase 100-T was used for digestion of cell walls with a concentration of 2 × 108 cells/ml for 30 min with vigorous shaking at 37°C. MNase was used at 0, 25, 50, 100, 200, 300 and 500 U/ml to digest the DNA, for 5 min at 36°C. An aliquot of 3 μl of 10 mg/ml proteinase K was added to the digested samples and incubated overnight at 56°C. Next, the samples were phenol/chloroform purified and EtOH precipitated. The MNase-digested samples were separated on 1.5% agarose gels and analyzed using Southern blot.
RESULTS
The hrp1Δ mutant alleviates silencing at the centromere and the mating-type region
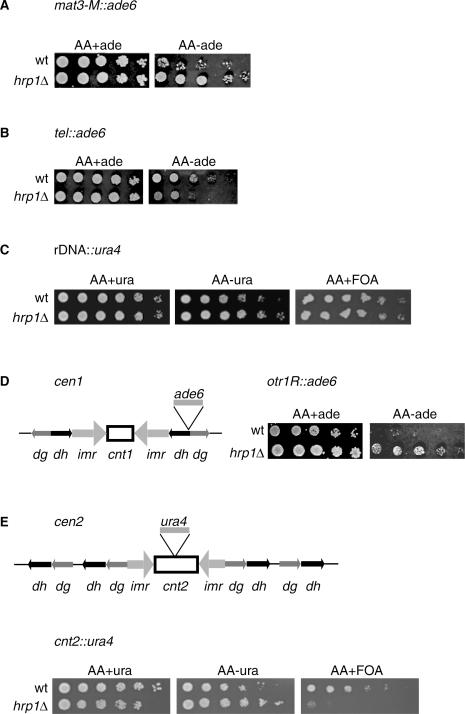
Previous studies have suggested that the hrp1 paralog in fission yeast, hrp3, was involved in mating-type silencing (40). These findings prompted us to investigate whether hrp1 has a role in transcriptional silencing. The hrp1Δ mutation was combined with marker genes integrated into the different heterochromatin regions in S.pombe, the mating-type region (mat3-M::ade6+), the telomere (tel::ade6+), the rDNA region (rDNA::ura4+) the outer repetitive domains of centromere 1 (otr1R::ade6+) and the central core of centromere 2 (cnt2::ura4+). Transcriptional silencing assays were carried out (Figure 1). In general, wild-type cells (wt) with marker genes inserted into heterochromatin regions grow poorly on selective plates, whereas mutants causing derepressing of heterochromatin grow better compared with wild type. It was clear that the hrp1Δ mutant alleviated mat3-M::ade6+ silencing, since the hrp1Δ colonies grew better than the wt colonies on −ade plates (Figure 1A). In contrast, telomeric silencing was increased in hrp1Δ as compared with wt, and rDNA silencing was unaffected by hrp1Δ (Figure 1B and C). The strongest silencing defects of hrp1Δ cells were observed at the centromeric insertions otr1R::ade6+ and cnt2::ura4+. hrp1Δ cells with the otr1R::ade6+ insertion grew well on −ade plates, whereas wt cells with the same insertion barely grew at all (Figure 1D). Growth of hrp1Δ cells with the cnt2::ura4+ insertion was completely inhibited by the FOA (ura4+ counter-selection), while wt cells grew well on FOA (Figure 1E). Thus, we concluded from these results that Hrp1 is required for transcriptional silencing of the central core and outer repeat regions of the centromere as well as the mating-type region.
Figure 1.
The hrp1Δ mutant alleviates silencing at the mating-type region, outer repeats and central core of the centromere. Cell suspensions of wt and hrp1Δ strains were 5-fold serially diluted and spotted on selective (−ura or −ade) or counter-selective plates (FOA) as indicated. The plates were incubated at 30°C for 3 days. (A) Silencing in the mating-type region (mat3-M::ade6+) was assayed in the Hu15 (wt) and Hu33 (hrp1Δ) strains. (B) Silencing at the telomeres (tel::ade6+) was assayed in the FY375 (wt) and Hu26 (hrp1Δ) strains. (C) Silencing at the rDNA (rDNA::ura4+) was assayed in the Hu393 (wt) and Hu466 (hrp1Δ) strains. (D) Left: schematic diagram of the otr1R::ade6+ insertion in centromere 1; right: silencing at the dg-dh repeats (otr1R::ade6+) was assayed in the FY1180 (wt) and Hu22 (hrp1Δ) strains. (E) Top: schematic diagram of the cen2::ura4+ insertion in centromere 2; bottom: silencing at the central core of the centromere (cen2::ura4+) was assayed in the FY412 (wt) and Hu584 (hrp1Δ) strains.
hrp mutants display chromosome segregation defects
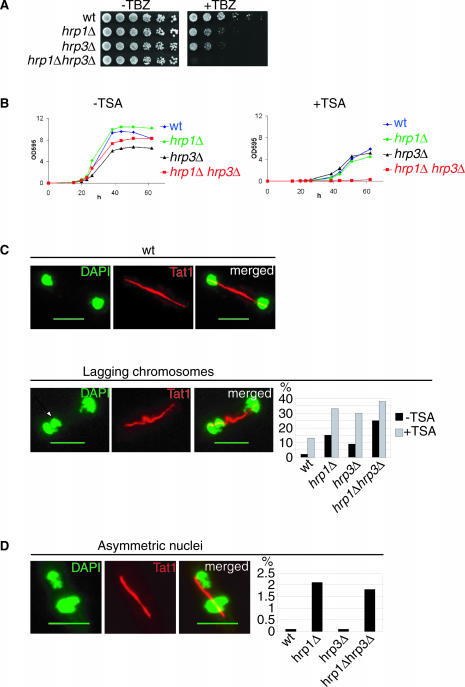
A common feature of mutants that alleviate centromeric silencing is the defects in chromosome segregation manifested as elevated chromosome loss and hypersensitivity to the microtubuli destabilizing drug TBZ (13,47). To investigate putative mitotic functions of Hrp1 and Hrp3, a TBZ sensitivity assay was performed. From this assay, it was clear that wt cells tolerated up to 15 μg/ml of TBZ (Figure 2A). In contrast, both of the single mutants were hypersensitive to this concentration of TBZ, showing a 5- to 25-fold growth reduction compared with wt. Interestingly, the hrp1Δ hrp3Δ double mutant displayed an additive increase in sensitivity to TBZ as compared with the single mutants, indicating that Hrp1 and Hrp3 have distinct functions in chromosome segregation.
Figure 2.
The hrp1Δ and hrp3Δ mutants are TBZ and TSA sensitive and display different chromosome segregation defects. (A) A spotting assay showing TBZ sensitivity of the hrp1Δ and hrp3Δ single and double mutants. Cells were 5-fold serially diluted and spotted on YEA plates with and without 15 μg/ml TBZ. The plates were incubated at 30°C for 3 days. Strains were FY1180 (wt) Hu61 (hrp1Δ), Hu602 (hrp3Δ) and Hu603 (hrp1Δ hrp3Δ). (B) Growth curves showing TSA sensitivity of wt (blue line), hrp1Δ (green line), hrp3Δ (black line) and hrp1Δ hrp3Δ double mutants (red line). Log phase cells grown in YEA were inoculated to a final concentration of 5 × 105 cells/ml in YES media without TSA (left panel) and with media containing 200 nM TSA (right panel). Growth was monitored measuring the optical density at 595 nm (OD595) for 64 h. Strains were Fy367 (wt) Hu61 (hrp1Δ), Hu602 (hrp3Δ) and Hu603 (hrp1Δ hrp3Δ). (C) Left, IF micrographs showing a representative cells in anaphase. The wt cell shows normal segregation and the hrp1Δ cell shows a lagging chromosome (arrow). Right: a bar diagram showing the percentages of wt and hrp mutant cells that displayed lagging chromosomes. Cells were grown at 18°C without TSA (black bars) and at 30°C in media containing 100 nM TSA (grey bars) prior to fixation. Strains were Fy367 (wt) Hu61 (hrp1Δ), Hu602 (hrp3Δ) and Hu603 (hrp1Δ hrp3Δ). (D) The hrp1Δ mutant and the hrp1Δ hrp3Δ double mutant display asymmetric segregation of nuclei. Left, an IF micrograph of an hrp1Δ cell with asymmetric nuclei. Right: a bar diagram that shows the percentages of wt and hrp mutant cells that display asymmetric segregation of nuclei. (C and D) The TAT1 antibody (red) was used for tubulin staining and DAPI (green) for staining DNA. Size bar = 5 µm.
It is known that other CHD family of proteins act in multi-protein complexes together with HDACs (34–36). To investigate whether Hrp1 and Hrp3 are involved in HDAC-dependent processes, hrp1Δ and hrp3Δ mutant strains were assayed sensitivity to the HDAC inhibitor TSA (Figure 2B). In cultures without TSA, the hrp1Δ cells grew slightly faster than wt cells as reported previously (48). However, hrp3Δ mutant cells showed a growth defect that was partially restored in the hrp1Δ hrp3Δ double mutant. In media containing 200 nM TSA, the growth of hrp single mutant cells were indistinguishable from wt cells. However, growth of the double mutant cells was completely inhibited by TSA. This indicated that both Hrp1 and Hrp3 are involved in HDAC-dependent processes via different pathways.
To further investigate the mitotic defects of hrp mutants and the relationship between Hrp1 and Hrp3 and HDACs, we used IF microscopy to analyze mitotic cells. Cells, wild-type, single and double mutants of hrp1Δ and hrp3Δ were grown at 30°C with and without TSA and subjected to IF using the α-tubulin antibody, Tat1, to visualize microtubules and DAPI to detect the chromosomal DNA. Late anaphase cells with long spindles (>5 μm) were analyzed for chromosome segregation defects (n = 160). The hrp1Δ cells displayed 15% lagging chromosomes (n = 270), hrp3Δ mutant 9% (n = 218) and the double mutants 25% (n = 162) of lagging chromosomes in cultures without TSA (Figure 2C, black bars). Both hrp1Δ and hrp3Δ single mutants displayed lagging chromosome frequencies significantly different from wt cells (Chi-square test; P < 0.05) (Figure 2C). The 25% lagging chromosomes detected in the double mutant relative to the single mutants was statistically significant (Chi-square test; P < 0.05) reinforcing the idea that Hrp1 and Hrp3 have distinct functions in the chromosome segregation process. To test whether the observed TSA sensitivity of hrp mutants was due to a cooperative function with HDACs in chromosome segregation, the effect of TSA on the lagging chromosome phenotypes was investigated. Cells in cultures grown in 100 nM TSA displayed a marked increase of defective anaphases. A total of 13% of the TSA-treated wt cells (n = 45), 33% of hrp1Δ cells (n = 212) and 30% of the hrp3Δ cells (n = 82) displayed lagging chromosomes (Figure 2C, gray bars). In the strain where both hrp1Δ and hrp3Δ were deleted, 35% lagging chromosomes were detected (n = 190). The elevated chromosome loss in TSA in the hrp mutants compared with wt was statistically significant (Chi-square test; P < 0.05). Hence, these results were consistent with the previous findings, suggesting an interplay between Hrp1, Hrp3 and HDACs in chromosome segregation.
Further examination of the IF samples revealed that hrp1Δ single and hrp1Δ hrp3Δ double mutants cells showed elevated numbers of asymmetric segregation (large and small nuclei) in late anaphase cells. The asymmetric segregation of nuclei is typical of central core/kinetochore defects and has been observed in several mis mutants (6,7,9). We found that <0.1% of the wt cells (n = 203) and hrp3Δ cells (n = 261) displayed asymmetric segregation of nuclei. In contrast, 2.1% of the hrp1Δ single mutants (n = 345) and 1.8% of the hrp1Δ hrp3Δ double mutant cells (n = 323) displayed statistically significant (Chi-square test; P < 0.05), unequal mitotic segregation events (Figure 2D). Importantly, hrp3Δ cells did not show asymmetric segregation of nuclei. This indicated that Hrp1 has a specialized role (not shared by Hrp3) in chromosome segregation, which is consistent with defects in the central core/kinetochore structure.
The hrp1Δ mutation causes elevated histone H4 acetylation levels and reduced SpCENP-A Cnp1 levels at the central core region
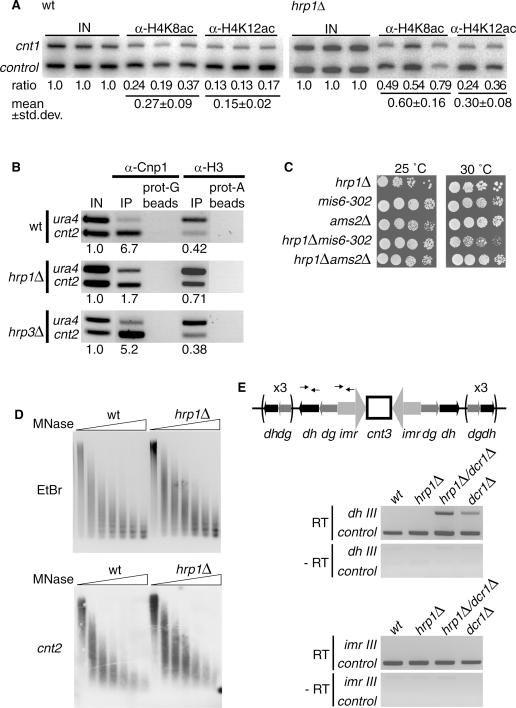
To expand on the specialized role of Hrp1 in the central core region, we investigated the chromatin modifications and histone variants in this chromosomal region by ChIP analysis. First, we used antibodies against histone H4 acetylated at lysine 8 (H4K8ac) or histone H4 lysine 12 (H4K12ac) for ChIP. The cnt1 locus was used for duplex PCR quantification of acetylated H4 histones together with the tRNA synthetase control. In wt cells, the acetylation levels of cnt1 were generally low: 0.27 for H4K8ac and 0.15 for H4K12ac (Figure 3A). In contrast, the hrp1Δ cells had significantly increased H4 acetylation levels at the central core of the centromere (paired t-test, P < 0.05). The acetylation levels in hrp1Δ were 2-fold increased (from 0.27 to 0.60) for H4K8ac and also for H4K12ac (from 0.15 to 0.30). Hence, these results suggested that that Hrp1 is important to prevent acetylation of histone H4 at the central core region of the centromere.
Figure 3.
The hrp1Δ mutant has elevated histone H4 acetylation levels, reduced levels of Cnp1 (SpCENP-A) in chromatin of the central core region, and it shows synthetic growth defects with mis6-302. (A) Quantitative PCR (32P images) analyses of ChIP samples. For each experiment, the normalized competitive PCR input ratios (IN), the mean ratio values and the SD is indicated below the lanes. The experiments were performed in duplicates or triplicates and both results were statistically significant (P < 0.05, independent sample t-test). α-H4K8ac and α-H4K12ac antibodies were used to measure H4K8 and H4K12 acetylation levels at cnt1 in wt and hrp1Δ cells with the glutamyl tRNA synthetase gene as a control. Strains were FY412 (wt) and Hu584 (hrp1Δ). (B) Quantitative PCR of ChIP samples. α-Cnp1 and α-H3 antibodies were used to determine Cnp1 and H3 levels, respectively, at cnt1 in wt and hrp1Δ cells. Strains were FY412 (wt) and Hu61 (hrp1Δ). (C) A spotting assay showing hrp1 genetically interacts with other genes important for Cnp1 loading. hrp1Δ, mis6-302, ams2Δ single and double mutant cells were serially diluted, spotted onto YES plates and incubated for 3 days at 25 or 30°C as indicated. Strains were Hu721 (mis6-302), Sp45 (ams2Δ), Hu465 (hrp1Δ), Hu1181 (hrp1Δ mis6-302) and Hu1176 (hrp1Δ ams2Δ). (D) Central core structure is not altered in the hrp1Δ mutant. MNase assay of wt and hrp1Δ chromatin extracts. Top: ethidium bromide (EtBr) stained gel with partially digested chromatin samples 0, 25, 50, 100, 200, 300 and 500 U MNase. Bottom: a Southern blot of the same gel hybridized with a [32P]labeled cnt2 probe. (E) Picture of RT–PCR products from cDNA samples to investigate a putative termination defect of hrp1Δ in dg-dh transcription. Top: schematic diagram of centromere 3. The position of the dhIII and imrIII primers used RT–PCR are indicated. Bottom: dhIII and imrIII RT–PCR analysis of dcr1Δ, hrp1Δ and hrp1Δ dcr1Δ mutant cells (as indicated). Primers for the tRNA synthetase gene were used as a control. Strains were Fy367 (wt), Hu61 (hrp1Δ), Hu659 (dcr1Δ) and Hu1121 (hrp1Δ dcr1Δ).
Next, we performed ChIP analysis of the histone H3 variant Cnp1 (SpCENP-A) in wt and hrp1Δ cells. Loss of CENP-A from the central core region in mis mutants has previously been shown to correlate well with an increase of histone H3 (9). This indicates that Cnp1 in wt cells replaces H3 in the central core region. Therefore, we used both α-Cnp1 and α-H3 specific antibodies for ChIP. Competitive PCR was used to determine the amount of Cnp1 or H3 binding to the central core in wt and hrp1Δ cells. The first primer was common for both reactions and anneals upstream of the ura4+ control gene. The other two primers were specific for the central core of the centromere (cnt2) or the ura4+ endogenous locus. The ChIP experiment showed 6.7-fold enrichment for Cnp1 at cnt2 as compared with ura4+ control gene (Figure 3B). There was a 4-fold reduction of Cnp1 at cnt2 in hrp1Δ cells, whereas Cnp1 levels in hrp3Δ remained relatively unchanged. Importantly, there was an increase in histone H3 at cnt2 in hrp1Δ cells (0.71) as compared with wt cells (0.42) correlating well with the decrease in Cnp1 in hrp1Δ cells. Again the H3 levels in hrp3Δ remained relatively unchanged. Hence, we concluded from this that Hrp1 has a specialized role (not shared with Hrp3) in promoting the stable association of Cnp1 and thereby reducing the histone H3 levels in nucleosomes of the central core region.
To determine whether hrp1 genetically interacted with other pathways involved in Cnp1 loading, the hrp1Δ mutation was combined with the temperature-sensitive mis6-302 mutation (7) and with the ams2Δ mutation (12). Growth of single and double mutants was assayed at 25°C and at 30°C for 4 days. The mis6-302 hrp1Δ double mutant had a reduced growth at 30°C as compared with the mis6-302 and hrp1Δ single mutants (Figure 3C). The ams2Δ hrp1Δ double mutant as well as the single mutants grew. Therefore, these results suggested that Hrp1 is involved in a pathway distinct from Mis6 and since no additive growth defects were seen in ams2Δ hrp1Δ it is likely that Hrp1 acts in the Ams2 pathway.
It is known that the central core region has a specialized chromatin structure that can be detected as a smear-like digestion pattern in MNase assays (49). In mutants where Cnp1 levels are low, the smear-like digestion pattern is generally changed into a regular digestion pattern consistent with a nucleosome ladder (6,7,9). Therefore, we carried out MNase assays on wild-type and hrp1Δ mutant cells (Figure 3D). Surprisingly, no extensive differences were observed in MNase digestion pattern at cnt2 in hrp1Δ mutant cells as compared with wt cells. Thus, this finding indicated that the residual Cnp1 levels in hrp1Δ mutant cells are sufficient to form the specialized chromatin structure at cnt2.
hrp1 has previously been isolated in a genetic screen for strains defective in transcriptional termination (50). It was shown that the hrp1 mutation causes read through of the ura4 transcriptional terminator. The centromeric dg-dh repeats are transcribed in both directions and the transcripts are processed by Dicer (Dcr1) and the RNAi machinery to direct heterochromatin formation over the dg-dh repeats (18). Therefore, it is possible that a putative defective termination of dg-dh transcripts caused by hrp1 mutation would lead to transcriptional read through into the central core region and thereby disturbing chromatin assembly. To test this possibility, a transcriptional termination assay was performed. Purified total RNA from wt, hrp1Δ, dcr1Δ and the hrp1Δ dcr1Δ double mutant cells was used in an RT–PCR assay to analyze transcripts in dg-dh repeats (dhIII) and the inner repeats (imrIII) of centromere 3. dhIII transcripts were detectable in hrp1Δ dcr1Δ and dcr1Δ cells, but not in the wild-type and hrp1Δ cells (Figure 4E). The dhIII transcripts were more abundant in hrp1Δ dcr1Δ cells than in dcr1Δ cells consistent with the reduced silencing observed at dg-dh in hrp1Δ (Figure 1E). If transcripts read through in hrp1Δ from dh-dg into the central core region, then they should be readily detectable in the intervening imrIII region. However, imrIII transcripts were not observed in hrp1Δ cells. From these results, we concluded that the hrp1Δ mutant does not cause read through of dg-dh transcripts into the central core region.
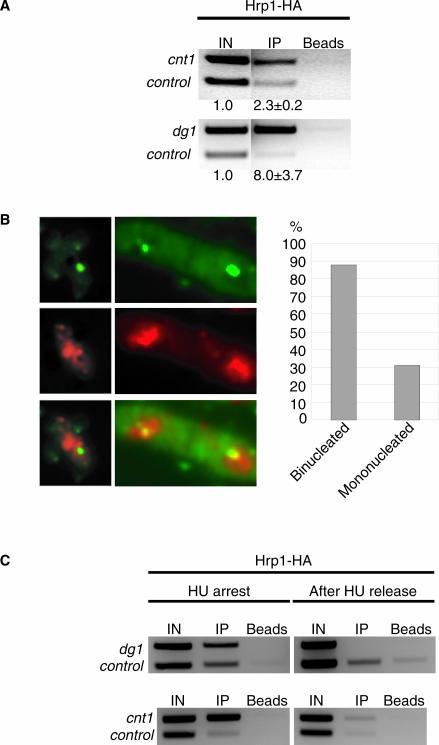
Figure 4.
Hrp1 directly binds to the centromere in a periodic, cell cycle-dependent manner. (A) An EtBr-stained gel with PCR products from ChIP with Hrp1-HA. The α-HA antibody was used for precipitating epitope-tagged Hrp1-HA in an asynchronous cell culture. cnt1, dg1 and tRNA synthetase (control) primers were used in the duplex PCR for quantification of Hrp1-HA binding. The strain was Hu615. (B) Left: an IF image with Hrp1-myc (red), combined with FISH using pRS140 centromere probe (green). Overlapping signals, indicating co-localization between Hrp1-myc and the cen-FISH signal, is visualized by a yellow signal. Right: a bar diagram showing the percentages of mononucleated and binucleated cells, in which Hrp1-myc signals co-localized with the cen-FISH signal. Size bar = 5 µm. The strain was Hu764. (C) An EtBr-stained gel of PCR quantified products from ChIP of synchronized cells. Half of the cells were collected immediately after HU arrest, the other half were washed and transferred to fresh media without HU and grown 1 h before harvest, and both cell batches were subjected to ChIP. The strain was Hu615 (Hrp1-HA).
Hrp1 is present at the centromere in a cell cycle-dependent manner
To determine whether Hrp1 was directly interacting with the centromere, a ChIP experiment was performed with cell cultures expressing the Hrp1-HA epitope-tagged protein. The ChIP samples were quantified using duplex PCR with primers specific for the central core 1 (cnt1), dg-dh repeats of the centromere 1 (dg1) and an internal control (Figure 4A). Hrp1-HA enriched both central core (2.3-fold) and the outer repetitive regions (8.0-fold). This demonstrated that Hrp1 directly interacts with both regions of the centromere.
To test whether Hrp1 was associated with centromeres at all stages of the cell cycle, cells expressing the Hrp1-myc epitope-tagged protein were processed for IF and then subjected to FISH using a centromere-specific probe (pRS140). Mononucleated and binucleated cells were analyzed for co-localization of Hrp1-myc to the centromere using deconvolution microscopy (Figure 4B). A high proportion (88%) of the binucleated cells (n = 100) displayed an (yellow) overlapping signal with Hrp1-myc (red) and the pRS140 probe (green). In contrast, only 31% of the mononucleated cells (n = 100) showed co-localization of Hrp1-myc and cen-FISH signals. The frequencies were statistically significantly different from wt cells if the co-localization was the same in binucleated and mononucleated cells (Chi-square, P < 0.001). Binucleated fission yeast cells are known to undergo G1/S stages of the cell cycle. Thus, this analysis suggested that Hrp1 periodically associates with the centromere during these stages.
To confirm the cell cycle-dependent localization of Hrp1 to the centromere, logarithmic growth phase cells expressing Hrp1-myc were synchronized using a two-step approach. In the first step, the small-sized cells (in early G2 phase) were collected in two consecutive lactose gradients. In the next step, the cells were transferred to media containing the DNA synthesis inhibitor HU to arrest the cells in G1/S-phase. Half of the cells were collected, and the other half was released from the HU arrest prior to ChIP for Hrp1-myc. The ChIP samples revealed a substantial enrichment for Hrp1-myc at outer repeats (dg1) and the central core (cnt1) in HU arrested cells, but this enrichment disappeared after 1 h release (Figure 4C). Hrp1-myc protein levels were unchanged at different stages of the cell cycle as judged by IF (Figure 4B), and Hrp1 mRNA levels are known to be constant during the cell cycle (51). The fission yeast central core region (cnt1) is known to replicate early in S-phase and to be replicated already at the HU arrest point (52). Thus, this result indicated that Hrp1 is associated with the central core region at the stage when DNA replication occurs.
DISCUSSION
Specialized and overlapping functions of Hrp1 and Hrp3
Our results demonstrate that the hrp1Δ deletion mutant causes a strong alleviation of transcriptional silencing in the central core region of the centromere and that Hrp1 is associated with this region of the centromere by ChIP. In contrast, the hrp3Δ mutant has no effect on central core silencing and was not associated with the central core region by ChIP (40). We show that hrp3Δ has no effect on Cnp1 (SpCENP-A) levels in the central core region, whereas hrp1Δ shows a strong reduction of Cnp1 levels. In addition, the chromosome segregation defects observed in hrp1Δ (unequal segregation) are consistent with a defect in the central core region. Such defects were not observed in hrp3Δ and were not increased in hrp1Δ hrp3Δ double mutant as compared with hrp1Δ single mutant. These findings indicate that out of the two closely related (52% identical) CHD paralogs, Hrp1 has a unique role in the central core region. In budding yeast, there is a single CHD protein with no role at centromeres (Chd1), and in humans there are four related CHD proteins, of which CHD3 and CHD4 are part of the Mi2 (35,53) complex known to be associated with centromeres (37,38). Phylogenetic analysis indicated that hrp1 and hrp3 genes were duplicated in fission yeast after the divergence of budding and fission yeasts (40). Duplicated loci often diverge both in sequence and function. Therefore, it seems logical that the two fission yeast hrp genes have evolved novel specialized functions, such as the role of Hrp1 in the central core region. We found that hrp1 and hrp3 mutants had additive effects on TBZ sensitivity and lagging chromosome phenotypes (Figure 2). Another shared function of Hrp1 and Hrp3 is silencing of mat2/3 (40). Thus, our findings are consistent with the idea that Hrp1 has evolved a specialized role at central core region of the centromere, but Hrp1 and Hrp3 still have some shared functions in sister–chromatid cohesion and mat2/3 silencing.
A link between CENP-A loading and histone deacetylation
Interestingly, the reduced Cnp1 levels and increased histone H3 levels observed in hrp1Δ correlated with increased histone H4 acetylation levels (Figure 3A and B). A similar increase in acetylation and loss of Cnp1 was also observed in mis16 and mis18 mutants, and Mis16 is 53% identical to RpAp48, a known component of HDAC complexes (9). The HDACs acting at central core could be Clr3 or Sir2 since both have been shown to be associated with DNA of the central core region in ChIP assays [(54), M. Wirén, I. Sinha, R.A. Silverstein, J. Walfridsson, H.-M. Lee, P. Laurenson, L. Pillus, D. Robyr, M. Grunstein and K. Ekwall, manuscript submitted] and Clr3-myc co-localizes with centromeres by FISH analysis (55). Furthermore, histone acetylation levels are high in sir2 and clr3 mutants (M. Wirén, I. Sinha, R.A. Silverstein, J. Walfridsson, H.-M. Lee, P. Laurenson, L. Pillus, D. Robyr, M. Grunstein and K. Ekwall, manuscript submitted). We speculate that Hrp1 could promote the activity of these HDACs. Another possibility is that Hrp1 counteracts the Mst1 histone H4 acetyl-transferase activity reported to act in the central core region (56). Our finding that chromosome segregation defects of hrp1 are additive with the HDAC inhibitor TSA (Figure 2) are consistent with such models, since it suggests that Hrp1 acts in parallel with HDACs. The Mi2 complex consists of CHD3 CHD4 remodeling factors, RpAp46, RpAp48, HDAC1/2 and other proteins (34–36). In experiments in vitro with the Mi2 complex and chromatin templates, it was shown that ATP-dependent remodeling by Mi2 was required for a subsequent histone deacetylation step. Therefore, it is conceivable that remodeling by Hrp1 has to precede histone deacetylation in the central core region. Whether Mis16 (like its homolog RpAp48) is part of an HDAC complex possibly including Clr3 or Sir2, which acts in the central core region, remains to be tested but we speculate that such activity, necessary for Cnp1 loading, could be aided by Hrp1.
Hrp1 localizes to the centromere at the G1/S stages of the cell cycle
Hrp1 is previously known to affect transcriptional termination (50). To test whether hrp1Δ indirectly affects centromere structure/function via transcriptional regulation, we carried out genome-wide cDNA expression profiling of hrp1Δ versus wild type. Only ∼20 genes were 2-fold affected by hrp1Δ and none of these is known to affect centromeres (J. Walfridsson, unpublished data). Another possibility is that Hrp1 has a direct role in the assembly of centromeric chromatin. Recently, the Drosophila CHD ortholog (CHD1) was shown to promote assembly of a regular nucleosome ladder in vitro from naked DNA and free histones (57). Hrp1 could act similarly during DNA replication to assemble centromeric chromatin containing Cnp1. Hrp1 could perhaps directly affect the exchange of histone H3 with Cnp1 during chromatin assembly. Previously, the SWI/SNF-related remodeling factor SWR1 was shown to catalyze histone variant exchange (58), so it is imaginable that CHD remodeling factors also are able to exchange histone variants. It is likely that the centromeric assembly function of Hrp1 is linked to DNA replication. The direct binding to the outer repeats and the central core of the centromere shown by ChIP (Figure 4) in HU-arrested cells argues for a direct functioning of Hrp1 at centromeres during early S-phase when centromeres are known to replicate (38). The co-localization of Hrp1 with centromere FISH signals supports a periodic localization to the centromere and likely represents a cell cycle-dependent centromere function. The Cnp1 loading factor Ams2 associates with chromatin only in binucleated cells (12), and Mis16 shows a similar periodic nuclear localization being most prominent after M-phase (i.e. in G1/S) (9). Therefore, it seems as though chromatin association of centromeric HDAC activities, Ams2 and Mis16 activities, all of which contribute to Cnp1 loading, is coordinated during S-phase. The fact that hrp1 and ams2 do not show additive growth defects (Figure 3C) indicates that they may act in the same pathway. Expression of Cnp1 mRNA peaks G1/S (6) and centromeres are replicated early in S-phase and during HU arrest (52). Altogether, these observations indicate that Hrp1 remodeling histone deacetylation and Ams2-mediated Cnp1 loading occurs during DNA replication in fission yeast.
Acknowledgments
The authors thank K. Takahashi and M. Yanagida for kindly providing them with strains, and B. Mellone and R. Allshire for sending them Cnp1 antibodies. The authors are also grateful for valuable advice on MNase assay from J. Cooper and on cell synchronization from B. Arcangioli. K.E. is a Royal Swedish Academy of Sciences Research Fellow supported by a grant from Knut and Alice Wallenberg Foundation. K.E is funded by Cancerfonden 4284-B02-04XBB and VR-M 31X-12562, VR-NT 621-2002-4311 grants. K.E is a ‘NET’ member funded by the ‘Epigenome’ EU network of excellence. Funding to pay the Open Access publication Charges for this article was provided by Cancerfonden.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chikashige Y., Kinoshita N., Nakaseko Y., Matsumoto T., Murakami S., Niwa O., Yanagida M. Composite motifs and repeat symmetry in S.pombe centromeres: direct analysis by integration of NotI restriction sites. Cell. 1989;57:739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- 2.Clarke L., Amstutz H., Fishel B., Carbon J. Analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA. 1986;83:8253–8257. doi: 10.1073/pnas.83.21.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kniola B., O'Toole E., McIntosh J.R., Mellone B., Allshire R., Mengarelli S., Hultenby K., Ekwall K. The domain structure of centromeres is conserved from fission yeast to humans. Mol. Biol. Cell. 2001;12:2767–2775. doi: 10.1091/mbc.12.9.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appelgren H., Kniola B., Ekwall K. Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J. Cell Sci. 2003;116:4035–4042. doi: 10.1242/jcs.00707. [DOI] [PubMed] [Google Scholar]
- 5.Partridge J.F., Borgstrom B., Allshire R.C. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi K., Chen E.S., Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- 7.Saitoh S., Takahashi K., Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- 8.Goshima G., Saitoh S., Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Jin Q.W., Pidoux A.L., Decker C., Allshire R.C., Fleig U. The mal2p protein is an essential component of the fission yeast centromere. Mol. Cell. Biol. 2002;22:7168–7183. doi: 10.1128/MCB.22.20.7168-7183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nabetani A., Koujin T., Tsutsumi C., Haraguchi T., Hiraoka Y. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma. 2001;110:322–334. doi: 10.1007/s004120100153. [DOI] [PubMed] [Google Scholar]
- 12.Chen E.S., Saitoh S., Yanagida M., Takahashi K. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol. Cell. 2003;11:175–187. doi: 10.1016/s1097-2765(03)00011-x. [DOI] [PubMed] [Google Scholar]
- 13.Ekwall K., Nimmo E.R., Javerzat J.P., Borgstrom B., Egel R., Cranston G., Allshire R. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell. Sci. 1996;109:2637–2648. doi: 10.1242/jcs.109.11.2637. [DOI] [PubMed] [Google Scholar]
- 14.Ekwall K., Cranston G., Allshire R.C. Fission yeast mutants that alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics. 1999;153:1153–1169. doi: 10.1093/genetics/153.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pidoux A.L., Uzawa S., Perry P.E., Cande W.Z., Allshire R.C. Live analysis of lagging chromosomes during anaphase and their effect on spindle elongation rate in fission yeast. J. Cell Sci. 2000;113:4177–4191. doi: 10.1242/jcs.113.23.4177. [DOI] [PubMed] [Google Scholar]
- 16.Bernard P., Maure J.F., Partridge J.F., Genier S., Javerzat J.P., Allshire R.C. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 17.Provost P., Silverstein R.A., Dishart D., Walfridsson J., Djupedal I., Kniola B., Wright A., Samuelsson B., Radmark O., Ekwall K. Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc. Natl Acad. Sci. USA. 2002;99:16648–16653. doi: 10.1073/pnas.212633199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volpe T., Schramke V., Hamilton G.L., White S.A., Teng G., Martienssen R.A., Allshire R.C. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11:137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan K.F., Hechenberger M., Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meluh P.B., Yang P., Glowczewski L., Koshland D., Smith M.M. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 21.Buchwitz B.J., Ahmad K., Moore L.L., Roth M.B., Henikoff S. A histone-H3-like protein in C.elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- 22.Van Hooser A.A., Ouspenski II, Gregson H.C., Starr D.A., Yen T.J., Goldberg M.L., Yokomori K., Earnshaw W.C., Sullivan K.F., Brinkley B.R. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- 23.Pidoux A.L., Richardson W., Allshire R.C. Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J. Cell Biol. 2003;161:295–307. doi: 10.1083/jcb.200212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ekwall K., Olsson T., Turner B.M., Cranston G., Allshire R.C. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 25.Taddei A., Maison C., Roche D., Almouzni G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nature Cell Biol. 2001;3:114–120. doi: 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- 26.Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 27.Muchardt C., Yaniv M. ATP-dependent chromatin remodelling: SWI/SNF and Co. are on the job. J. Mol. Biol. 1999;293:187–198. doi: 10.1006/jmbi.1999.2999. [DOI] [PubMed] [Google Scholar]
- 28.Varga-Weisz P. ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene. 2001;20:3076–3085. doi: 10.1038/sj.onc.1204332. [DOI] [PubMed] [Google Scholar]
- 29.Tsuchiya E., Hosotani T., Miyakawa T. A mutation in NPS1/STH1, an essential gene encoding a component of a novel chromatin-remodeling complex RSC, alters the chromatin structure of Saccharomyces cerevisiae centromeres. Nucleic Acids Res. 1998;26:3286–3292. doi: 10.1093/nar/26.13.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue Y., Canman J.C., Lee C.S., Nie Z., Yang D., Moreno G.T., Young M.K., Salmon E.D., Wang W. The human SWI/SNF-B chromatin-remodeling complex is related to yeast RSC and localizes at kinetochores of mitotic chromosomes. Proc. Natl Acad. Sci. USA. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu J.M., Huang J., Meluh P.B., Laurent B.C. The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol. Cell. Biol. 2003;23:3202–3215. doi: 10.1128/MCB.23.9.3202-3215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hakimi M.A., Bochar D.A., Schmiesing J.A., Dong Y., Barak O.G., Speicher D.W., Yokomori K., Shiekhattar R. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–998. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- 33.Huang J., Hsu J.M., Laurent B.C. The RSC nucleosome-remodeling complex is required for Cohesin's association with chromosome arms. Mol. Cell. 2004;13:739–750. doi: 10.1016/s1097-2765(04)00103-0. [DOI] [PubMed] [Google Scholar]
- 34.Tong J.K., Hassig C.A., Schnitzler G.R., Kingston R.E., Schreiber S.L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y., LeRoy G., Seelig H.P., Lane W.S., Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 36.Wade P.A., Jones P.L., Vermaak D., Wolffe A.P. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 37.Brown K.E., Guest S.S., Smale S.T., Hahm K., Merkenschlager M., Fisher A.G. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 38.Kim J., Sif S., Jones B., Jackson A., Koipally J., Heller E., Winandy S., Viel A., Sawyer A., Ikeda T., et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 39.Yoo E.J., Jin Y.H., Jang Y.K., Bjerling P., Tabish M., Hong S.H., Ekwall K., Park S.D. Fission yeast hrp1, a chromodomain ATPase, is required for proper chromosome segregation and its overexpression interferes with chromatin condensation. Nucleic Acids Res. 2000;28:2004–2011. doi: 10.1093/nar/28.9.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jae Yoo E., Kyu Jang Y., Ae Lee M., Bjerling P., Bum Kim J., Ekwall K., Hyun Seong R., Dai Park S. Hrp3, a chromodomain helicase/ATPase DNA binding protein, is required for heterochromatin silencing in fission yeast. Biochem. Biophys. Res. Commun. 2002;295:970–974. doi: 10.1016/s0006-291x(02)00797-0. [DOI] [PubMed] [Google Scholar]
- 41.Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 42.Bahler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 43.Hagan I.M., Hyams J.S. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- 44.Woods A., Sherwin T., Sasse R., MacRae T.H., Baines A.J., Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- 45.Ekwall K., Javerzat J.P., Lorentz A., Schmidt H., Cranston G., Allshire R. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- 46.Pidoux A., Mellone B., Allshire R. Analysis of chromatin in fission yeast. Methods. 2004;33:252–259. doi: 10.1016/j.ymeth.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 47.Allshire R.C., Nimmo E.R., Ekwall K., Javerzat J.P., Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 48.Jin Y.H., Yoo E.J., Jang Y.K., Kim S.H., Kim M.J., Shim Y.S., Lee J.S., Choi I.S., Seong R.H., Hong S.H., et al. Isolation and characterization of hrp1+, a new member of the SNF2/SWI2 gene family from the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 1998;257:319–329. doi: 10.1007/s004380050653. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi K., Murakami S., Chikashige Y., Funabiki H., Niwa O., Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alen C., Kent N.A., Jones H.S., O'Sullivan J., Aranda A., Proudfoot N.J. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell. 2002;10:1441–1452. doi: 10.1016/s1097-2765(02)00778-5. [DOI] [PubMed] [Google Scholar]
- 51.Rustici G., Mata J., Kivinen K., Lio P., Penkett C.J., Burns G., Hayles J., Brazma A., Nurse P., Bahler J. Periodic gene expression program of the fission yeast cell cycle. Nature Genet. 2004;36:809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- 52.Kim S.M., Dubey D.D., Huberman J.A. Early-replicating heterochromatin. Genes Dev. 2003;17:330–335. doi: 10.1101/gad.1046203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woodage T., Basrai M.A., Baxevanis A.D., Hieter P., Collins F.S. Characterization of the CHD family of proteins. Proc. Natl Acad. Sci. USA. 1997;94:11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Freeman-Cook L., Gomez E.B., Spedale E.J., Marlett J., Forsburg S.L., Pillus L., Laurenson P. Conserved locus-specific silencing functions of S.pombe sir2+ Genetics. 2004;169:1243–1260. doi: 10.1534/genetics.104.032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bjerling P., Silverstein R.A., Thon G., Caudy A., Grewal S., Ekwall K. Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol. Cell. Biol. 2002;22:2170–2181. doi: 10.1128/MCB.22.7.2170-2181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Minoda A., Saitoh S., Takahashi K., Toda T. BAF53/Arp4 homolog Alp5 in fission yeast is required for histone H4 acetylation, kinetochore-spindle attachment, and gene silencing at centromere. Mol. Biol. Cell. 2005;16:316–327. doi: 10.1091/mbc.E04-06-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lusser A., Urwin D.L., Kadonaga J.T. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nature Struct. Mol. Biol. 2005;12:160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 58.Mizuguchi G., Shen X., Landry J., Wu W.H., Sen S., Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]