Abstract
The mechanisms of post-acute medical conditions and unexplained symptoms following SARS-CoV-2 infection (Long COVID; LC) are incompletely understood. There is growing evidence that viral persistence, immune dysregulation, and T cell dysfunction may play major roles. We performed whole-body positron emission tomography (PET) imaging in a well-characterized cohort of 24 participants at time points ranging from 27 to 910 days following acute SARS-CoV-2 infection using the radiopharmaceutical agent [18F]F-AraG, a selective tracer that allows for anatomical quantitation of activated T lymphocytes. Tracer uptake in the post-acute COVID-19 group, which included those with and without continuing symptoms, was higher compared to pre-pandemic controls in many regions, including the brain stem, spinal cord, bone marrow, nasopharyngeal and hilar lymphoid tissue, cardiopulmonary tissues, and gut wall. T cell activation in the spinal cord and gut wall was associated with the presence of LC symptoms. In addition, tracer uptake in lung tissue was higher in those with persistent pulmonary symptoms specifically. Increased T cell activation in these tissues was also observed in many individuals without LC. Given the high [18F]F-AraG uptake detected in the gut, we obtained colorectal tissue for in situ hybridization of SARS-CoV-2 RNA and immunohistochemical studies in a subset of five participants with LC symptoms. We identified intracellular SARS-CoV-2 single-stranded Spike protein-encoding RNA in rectosigmoid lamina propria tissue in all five participants and double-stranded Spike protein-encoding RNA in three participants up to 676 days following initial COVID-19, suggesting that tissue viral persistence could be associated with long-term immunologic perturbations.
One Sentence Summary:
Total-body PET imaging and microscopy analysis of rectosigmoid biopsies reveal prolonged T cell activation and SARS-CoV-2 RNA persistence following COVID-19.
INTRODUCTION
Some people do not return to their baseline health following Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2) infection (1, 2). Such individuals may experience an increased burden of new medical conditions including cardiovascular disease or diabetes (3). They may also experience Long COVID (LC): unexplained symptoms following SARS-CoV-2 infection not attributable to an alternative diagnosis (2). According to the U.S. Centers for Disease Control and Prevention (CDC), approximately 15% of American adults have ever experienced LC and 6% are currently experiencing the condition (4); 18 million adults in the U.S. alone might be affected (5). Despite the scale of the problem, there are no accepted treatments.
Acute coronavirus disease 2019 (COVID-19) is highly inflammatory (6, 7). In the post-acute phase, inflammation, immune activation, and dysregulation of virus-specific immune responses have consistently been identified in blood (8–19). These responses are associated with factors including clotting dysfunction (20–24), reactivation of Epstein Barr Virus (EBV) (8, 25, 26), and autoimmunity (8, 27–31). Our group recently demonstrated that compared to complete symptomatic recovery, LC is associated with increased frequencies of tissue-migrating CD4+ T cells and exhausted SARS-CoV-2-specific CD8+ T cells up to 8 months after COVID-19 (18). There is growing evidence that persistent SARS-CoV-2 antigens can be detected in various tissues for months following acute infection (32–40). Tissue viral persistence may explain ongoing aberrant immune responses, especially T cell dysfunction, inflammation, and symptomatology (18, 41).
Data regarding SARS-CoV-2 persistence or aberrant immune responses in non-blood tissues are sparse. Most studies have been limited to samples from convenience cohorts (33, 36), with many individuals requiring hospitalization or without detailed post-acute data. Clinical studies of tissue pathology in living participants have assessed limited quantities of tissue obtained through minimally invasive biopsies. Furthermore, many potential anatomic sites of SARS-CoV-2 persistence, such as the brain, spinal cord, and heart, cannot be sampled in living individuals (42, 43). As a result, characterization of immune responses in these locations has been limited. When it has been attempted, it has utilized non-specific tracers, such as [18F]Fluorodeoxyglucose (FDG), or limited follow-up of clinical symptoms (44–51). There is an urgent need to develop non-invasive techniques to identify persistent T cell immune responses in highly characterized cohorts to better understand the tissue-level biology that might drive findings observed in peripheral blood.
In this study, we performed whole-body positron emission tomography (PET) imaging of 24 highly characterized participants ranging from 27 to 910 days following COVID-19 onset. Given the potential importance of dysregulated T cell responses in the pathogenesis of LC and the potential for tissue-based persistence of SARS-CoV-2 eliciting such responses, we used a radiopharmaceutical agent, [18F]F-AraG (Fluorine-18 labeled arabinofuranosyl guanine), a selective and sensitive tracer that allows for anatomical localization of activated CD8+ and CD4+ T lymphocytes (52–54), and characterized viral persistence in gut tissue from a subset of participants with LC.
RESULTS
Study cohort
[18F]F-AraG PET/CT imaging was performed on 24 participants from the UCSF-based Long-term Impact of Infection with Novel Coronavirus (LIINC) study (NCT04362150) (55). We enrolled two groups: (1) those in the early post-acute phase (<90 days from COVID-19 onset with and without complete recovery (e.g., resolution of all COVID-19-attributed symptoms by the time of the visit; n=3 and n=6, respectively), and (2) those in the later post-acute phase (>90 days from COVID-19 onset) with and without complete recovery (n=3 with complete recovery and n=12 with LC symptoms (Table 1)). Images from 6 participants who underwent [18F]F-AraG PET imaging (3 females, 3 males) before 2020 served as pre-pandemic controls. Although we did not have access to detailed demographic information or in-depth medical histories for these control participants, we elected to use pre-pandemic rather than contemporaneous comparators given the high rate of subclinical or undiagnosed SARS-CoV-2 infection during the study period (4). Control participants were excluded, however, if they had systemic comorbidities (such as malignancy or immunosuppression) or recent infection that would influence T cell responses.
Table 1.
Participant Demographics, Clinical Factors, and Long COVID History
| Pt # | Variant Wave at Time of Infection | Hosp. During Acute Infection | Days from Infection to PET Scan | Days From Vaccine to PETa | Sex | Age b | LC Symptom Countc | Presence of LC Symptom Phenotype at Time of Imaging | Anti-N Ab at PETd | Days from Infection to Gut Biopsye | % Gut Cells with Spike RNA (ss, ds) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||||||
| Fatigue | Pulm | Cardiac | URI | GI | Neuro | |||||||||||
|
| ||||||||||||||||
| 1 | Pre-Omicron | N | 27 | 135 | F | 26 | 2 | Y | Y | N | N | N | N | Y | - | |
| 2 | Pre-Omicron | N | 29 | 113 | M | 37 | 1 | N | Y | N | N | N | N | Y | - | |
| 3 | Pre-Omicron | N | 42 | 151 | M | 30 | 4 | N | Y | Y | N | N | N | Y | - | |
| 4 | Pre-Omicron | N | 44 | 149 | M | 64 | 0 | N | N | N | N | N | N | Y | - | |
| 5 | Pre-Omicron | N | 48 | 199 | M | 29 | 5 | Y | N | Y | N | Y | Y | Y | - | |
| 6 | Pre-Omicron | N | 50 | N/Af | F | 32 | 4 | Y | Y | N | N | N | Y | N | - | |
| 7 | Omicron | N | 64 | 6 | M | 42 | 14 | Y | Y | N | N | Y | Y | N | - | |
| 8 | Omicron | N | 65 | 386 | F | 64 | 0 | N | N | N | N | N | N | N | - | |
| 9 | Omicron | N | 83 | 183 | F | 59 | 0 | N | N | N | N | N | N | Y | - | |
| 10 | Omicron | N | 95 | 246 | M | 47 | 0 | N | N | N | N | N | N | Y | 158 | 2.56, 0.09 |
| 11 | Omicron | N | 114 | 286 | F | 30 | 8 | Y | N | Y | Y | Y | N | Y | - | |
| 12 | Pre-Omicron | N | 193 | 137 | F | 28 | 13 | Y | Y | N | N | Y | Y | N | - | |
| 13 | Omicron | N | 205 | 122 | F | 47 | 6 | Y | Y | N | N | N | Y | N | - | |
| 14 | Omicron | N | 231 | 63 | M | 35 | 4 | Y | N | Y | N | Y | Y | Y | - | |
| 15 | Omicron | N | 246 | 302 | M | 65 | 6 | Y | N | N | Y | Y | Y | N | 213 | 1.57, 0 |
| 16 | Pre-Omicron | Yg | 260 | 118 | M | 54 | 8 | Y | Y | N | Y | N | Y | N | 442 | 1.83, 0.7 |
| 17 | Pre-Omicron h | N | 406 | 219 | F | 33 | 0 | N | N | N | N | N | N | Y | - | |
| 18 | Pre-Omicron | N | 617 | 320 | F | 32 | 7 | Y | N | Y | N | N | Y | N | 645 | 0.38, 0 |
| 19 | Pre-Omicron | N | 625 | 122 | M | 46 | 14 | Y | Y | N | N | Y | Y | Y | - | |
| 20 | Pre-Omicron | N | 641 | 321 | M | 28 | 11 | Y | N | Y | N | Y | Y | Y | - | |
| 21 | Pre-Omicron | N | 654 | 425 | F | 33 | 15 | Y | Y | Y | N | Y | Y | N | - | |
| 22 | Pre-Omicron | N | 663 | 248 | M | 52 | 6 | Y | Y | N | Y | Y | Y | Y | 676 | 1.55,1.22 |
| 23 | Pre-Omicron | N | 890 | 211 | M | 52 | 0 | N | N | N | N | N | N | Y | - | |
| 24 | Pre-Omicron | Yi | 910 | 155 | F | 59 | 6 | Y | Y | N | N | Y | Y | N | - | |
Pt, participant; Hosp., hospitalized; Pulm, pulmonary; URI, upper respiratory; GI, gastrointestinal; Neuro, neurocognitive; LC, Long COVID; Y, Yes, N, No; F, female; M, male; ss, single-stranded SARS-CoV-2 Spike RNA, ds, double-stranded SARS-CoV-2 Spike RNA
Days from last COVID-19 vaccine dose to PET imaging
Age in years
Number of participant reported symptoms at the time of PET imaging (out of 32 total)
Anti-Nucleocapsid antibody detected at time of PET
Days from COVID-19 infection to biopsy; all participants had a least one Long COVID symptoms at the time of sigmoidoscopy and rectosigmoid tissue collection
Participant was not vaccinated prior to PET imaging
Participant did not require intensive care but received supplemental oxygen during hospitalization
Participant initially infected during pre-Omicron wave but had two documented re-infections. Days from infection to PET scan calculated from last Omicron reinfection
Participant did not require intensive care or supplemental oxygen during hospitalization
Demographics, clinical factors, and LC symptoms at the time of imaging were recorded (Table 1, Fig. 1). The median age was 39.5 years (range 26 to 65), and those with LC were younger than those without LC (median 33 versus 55.5 years, respectively; P=0.026). Eleven participants were female, and most were infected before the emergence of Omicron variants in the U.S. (table S1) (56). Only two were hospitalized for acute COVID-19; one had required supplemental oxygen, but not intensive care and the other had not required supplemental oxygen or intensive care. The median number of LC symptoms was 5.5 (range 0 to 15), and the median number of days between initial COVID-19 onset and PET imaging was 199 (range 27 to 910). No differences in the number of days between the last documented SARS-CoV-2 infection and PET imaging were observed between groups. All but one participant had received at least one COVID-19 vaccination prior to PET imaging (median number of days from most recent vaccine to tracer injection was 183). Among those with LC, the most common symptoms were fatigue (n=16) and neurocognitive complaints (n=14). Six participants did not report any LC symptoms.
Figure 1. COVID-19 PET imaging cohort demographics and clinical characteristics.
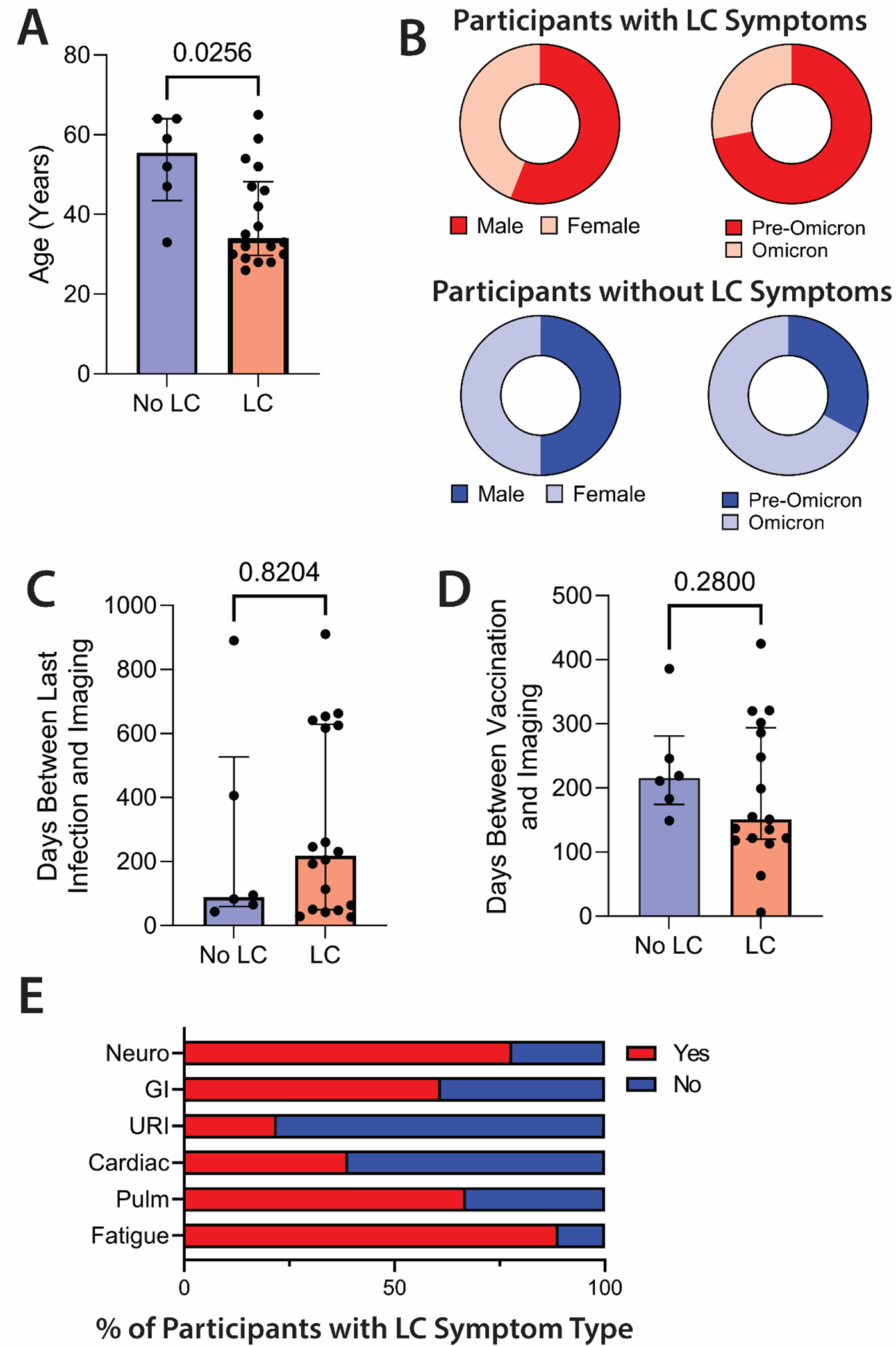
(A and B) Comparisons in age (A), sex assigned at birth, and variant wave (B) are shown in those with (n=18) and without LC (n=6). (C and D) The days between last documented SARS-CoV-2 infection and PET imaging (C) and last COVID-19 vaccine dose and PET imaging (D) are shown for participants with and without LC. (E) Shown are the percent of participants with and without specific LC symptoms phenotype. LC, Long COVID; GI, gastrointestinal; URI, upper respiratory infection symptoms; Pulm, pulmonary symptoms. Bars represent median values with all individual data points and interquartile ranges shown. P values were calculated by two-sided non-parametric Mann-Whitney tests.
To minimize the impact of vaccination on T cell activation, PET imaging was performed greater than 4 weeks from any vaccine dose (SARS-CoV-2 or otherwise), but one participant received a SARS-CoV-2 booster vaccine dose 6 days prior to imaging without notifying the study. There was no difference in the time between vaccination and PET imaging between those with and without LC (Fig. 1). One participant (participant 17) was initially infected during the ancestral wave but experienced two documented re-infections with presumed Omicron variants prior to imaging. Except for participant 17, no one reported acute symptoms suggestive of reinfection with SARS-CoV-2 between the initial COVID-19 episode and PET imaging; during the study period none had subsequent positive COVID-19 tests beyond their initial confirmatory test.
Clinical review of chest CTs showed little relationship between pulmonary LC symptoms and abnormal findings
On clinical review of chest CTs, four participants had apical scarring or reticulation suggesting mild pulmonary fibrosis; one had a bulla in the right lower lobe. The remainder of the scans were normal except for incidental findings likely not attributable to COVID-19, such as calcified granulomas (table S2). Most lung CT findings were determined by the radiologist to be without clinical importance. Furthermore, there was no association between the presence of pulmonary LC symptoms and abnormal clinical CT findings. This observation is consistent with prior studies demonstrating a relatively low rate of clinically meaningful fibrotic lung disease detectable by CT in patients with recent COVID-19 (57).
[18F]F-AraG PET/CT imaging was safe and identified tissue regions of T cell activation
[18F]F-AraG is a PET imaging agent developed to assess T cell activation and cycling. It is an analog of arabinosyl guanine (AraG), an FDA-approved chemotherapy agent (nelarabine) used to treat refractory T cell malignancies (58). [18F]F-AraG can be phosphorylated by cytoplasmic deoxycytidine kinase (dCK) and deoxyguanosine kinase (dGK), enzymes upregulated in activated T cells, which traps [18F]F-AraG intracellularly (59). [18F]F-AraG is highly selective for activated T cells in humans and selective activated T cell uptake has been confirmed using murine models and first-in-human studies of T cell responses following PD-1 therapy for malignancy (52, 60, 61). [18F]F-AraG is safe with no major adverse events in healthy volunteers, patients with metastatic cancer, or individuals with immune deficiencies (52–54). Of note, non-specific [18F]F-AraG uptake has been observed in the thyroid, choroid plexus, left ventricle wall, and some glandular tissue, and accumulates in the liver and kidneys during metabolism and excretion.
[18F]F-AraG was prepared in a two-step method using a modified previously reported procedure (58). [18F]F-AraG was administered intravenously (166.5–185 MBq) followed by PET/CT (Siemens Biograph Vision) for post-acute COVID-19 participants and (299.7–329.3 MBq) followed by PET/MR (GE SIGN) for pre-pandemic controls. Whole-body imaging was performed over a median interval of 50 (IQR 47–55) minutes post-injection, covering vertex to mid-thigh. Regions of interest (ROIs) were drawn around various tissues using isometric or 3-dimensional ball tools depending on the anatomical structure. Maximum and mean standardized uptake values (SUVmax and SUVmean) were determined. SUVs are a function of the concentration of radioactivity within a ROI, the administered activity, participant weight as a surrogate for volume, timing of injection, and scan time (62), allowing cross-participant comparisons. Overall, the tracer was well-tolerated, with no serious adverse events during or following tracer injection. CT was chosen for anatomical localization and PET attenuation correction to provide information on lung parenchymal and structural pathology following COVID-19.
Increased [18F]F-AraG tissue uptake was observed in post-acute COVID-19 participants compared to pre-pandemic controls
Higher SUVmax and SUVmean values were observed across a variety of anatomic regions (such as lymphoid tissues, glandular tissue, vascular tissue, and spinal cord) in post-acute COVID-19 participants compared with uninfected controls (Fig. 2 and 3). Maximum Intensity Projections (MIP) of PET data from all post-acute COVID-19 and pre-pandemic control participants were generated (fig. S1). Although [18F]F-AraG uptake was low overall in the brain and spinal cord (SUVmax and SUVmean <1), significantly higher SUVmax (p=0.003, p<0.001) and SUVmean (p=0.009, p<0.001) were identified in the thoracic cord and cauda equina (at the level of the fourth lumbar vertebra) and higher SUVmean (p=0.002) was identified in the brain stem (pons) (Fig. 3). The CNS choroid plexus is known to highly express angiotensin converting enzyme 2 (ACE2), the receptor for SARS-CoV-2, but this region had high background uptake and there were no differences between post-acute COVID-19 cases and pre-pandemic controls. It is possible that cauda equina ROIs included some overlap with extra-tissue cerebrospinal fluid, but similar differences in thoracic spinal cord SUVs were observed where ROIs were exclusively in tissue parenchyma. Significantly higher [18F]F-AraG uptake (SUVmax and SUVmean) was also observed in the aortic arch wall (p=0.007 and p=0.004), pulmonary artery wall (p=0.019 SUVmax only), lower lung lobes (p=0.039 and p=0.04) and right ventricle wall (p<0.001 and p=0.001) compared with pre-pandemic controls. Significant increases in [18F]F-AraG uptake were observed in nasal turbinates (SUVmax p=0.004 and SUVmean p=0.022), hilar lymph node regions (right-sided; SUVmax, p=0.008), proximal colon wall (SUVmax p=0.019), rectal wall (SUVmax p=0.004 and SUVmean p=0.015), lumbar (SUVmax p<0.001 and SUVmean p<0.001) and iliac crest (SUVmax p=0.023) bone marrow, and pharyngeal tonsils (SUVmax p=0.037). Uptake in the liver (a metabolic and excretory organ for [18F]F-AraG), abdominal adipose tissue, and quadriceps muscles were similar across participants (all p>0.05). No differences in SUV were observed in testes, penile tissue, prostate, or uterine tissue, although sample size was limited for these comparisons (fig. S2). [18F]F-AraG uptake was also compared between male and female participants. Although statistical power was limited in these four-way comparisons, male participants had significantly higher SUVmax (p=0.002) and SUVmean (p=0.041) uptake in right hilar ROIs compared to female participants following COVID-19 (fig. S3).
Figure 2. Increased [18F]F-AraG uptake in participants following COVID-19 compared to pre-pandemic control volunteers.
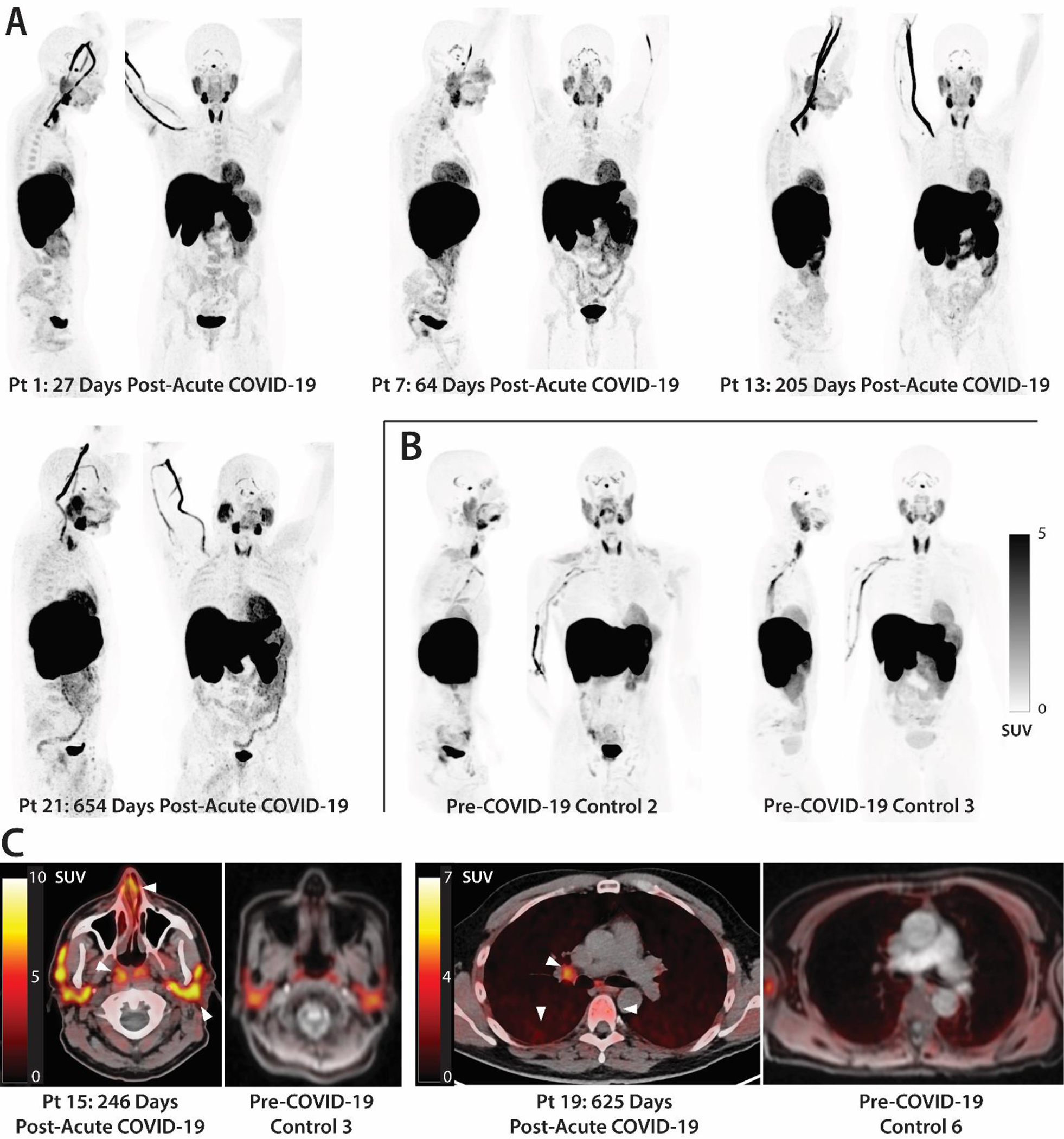
(A and B) Maximum intensity projections (MIP; coronal and sagittal views of 3-dimesional reconstructions) are shown for four representative participants at various times following SARS-CoV-2 infection (A) and male and female uninfected controls (B). (C) Axial PET/CT overlay images show signal in nasal turbinates, parotid glands, tonsillar tissue, hilar lymph node, lung parenchyma, and lumbar bone marrow in representative post-acute COVID-19 and control participants (white arrows). MIPs for all participants are shown in fig. S1. Pt, patient. SUV, standardized uptake value.
Figure 3. Increased [18F]F-AraG in many tissues from post-acute COVID-19 cases compared to pre-pandemic control volunteers.
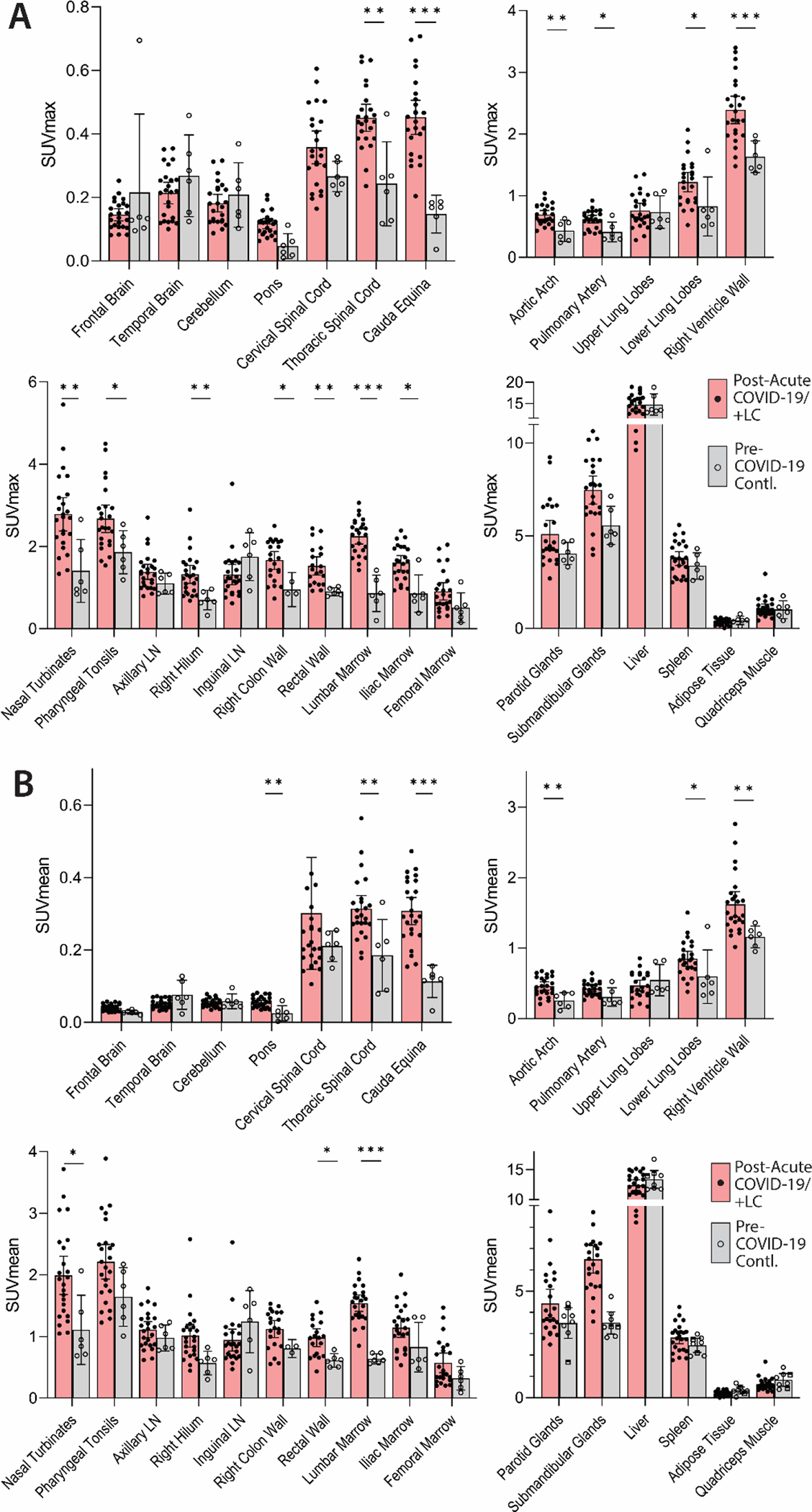
(A and B) Maximum standardized uptake values (SUVmax) (A) and mean SUV (SUVmean) (B) for various anatomical regions of interest (ROI) are shown for post-acute COVID-19 participants, including those with any number of Long COVID symptoms (Post-acute COVID-19/+LC; n=18), as well as pre-pandemic controls (n=6). Bars represent mean SUVmax or SUVmean and error bars represent 95% confidence interval. Adjusted P values <0.05, <0.01 and <0.001 represented by *, **, and *** respectively from two-sided non-parametric Kruskal–Wallis tests using a Benjamini-Hochberg adjustment for false discovery rates across multiple comparisons (q value = adjusted P value). All data points are shown on the graph. ROI determination was not possible in 3 and 5 post-acute COVID-19 participants for proximal colon and rectal wall ROIs, respectively, and 3 pre-pandemic control participants for proximal colon. LN, lymph node.
[18F]F-AraG uptake remained elevated throughout the post-acute phase of COVID-19 in most tissues
We performed imaging over a span of nearly 2.5 years following COVID-19 onset to determine the duration of T cell activation. [18F]F-AraG SUVmax for tissues of interest was stratified by timing of PET imaging before or after 90 days following initial COVID-19 onset (Fig. 4A; fig. S4). Although there is variability in case definitions for LC (63), the World Health Organization recommends that symptoms are present at least 90 days following COVID-19 (64). Our group has consistently used this definition in prior work, and sensitivity analyses for this project demonstrated near-maximal separation between PET imaging uptake measures between groups stratified by this time point. We observed modestly decreased uptake in thoracic and lumbar spinal cord and colon/rectal wall ROIs in participants imaged beyond 90 days following COVID-19 onset, but SUVs in these later-imaged individuals remained significantly elevated compared to pre-pandemic controls (thoracic spinal cord: SUVmax p=0.025 only; cauda equina: SUVmax p<0.001, SUVmean p<0.001; rectal wall: SUVmax only p=0.038), except for the right colon wall. We observed significant inverse correlations between [18F]F-AraG SUVmax uptake in colon/rectal wall ROIs and the number of days from initial infection to PET imaging (P=0.004 and P=0.035, respectively by two-tailed Spearman tests; fig. S5) but no other significant correlations between time from infection to imaging and tracer uptake in other tissue regions.
Figure 4. Differential [18F]F-AraG uptake in post-acute COVID-19 cases and control participants grouped by time from initial COVID-19 symptom onset to PET imaging and by Long COVID symptoms.
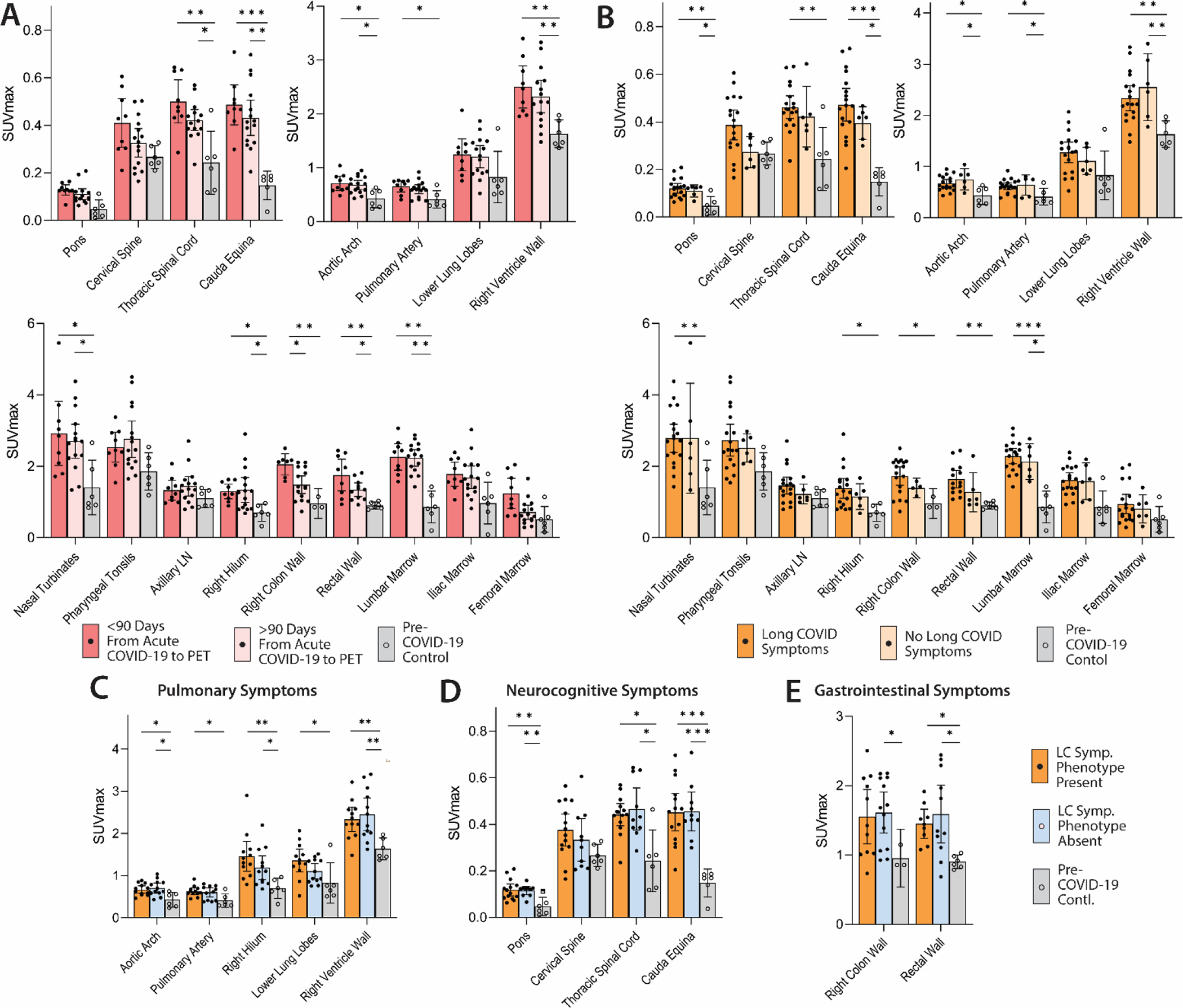
(A) SUVmax values in tissue ROIs in post-acute COVID-19 participants imaged <90 days or >90 days from acute infection onset and control volunteers are shown. (B) SUVmax values in tissue ROIs in post-acute COVID-19 participants with or without Long COVID symptoms reported at the time of imaging and control volunteers are shown in. (C to E) SUVmax values in tissue ROIs in post-acute COVID-19 participants with or without pulmonary symptoms (C), neurocognitive symptoms (D) and gastrointestinal symptoms (E) are also shown. Bars represent mean SUVmax and error bars represent 95% confidence interval. Adjusted P values <0.05, <0.01 and <0.001 represented by *, **, and *** respectively from two-sided non-parametric Kruskal–Wallis tests using a Benjamini-Hochberg adjustment for false discovery rates across multiple comparisons (q value = adjusted P). All data points are shown. SUVmean values are shown in fig. S4.
Long COVID symptoms were associated with higher [18F]F-AraG uptake in some tissues
Associations between T cell activation and LC symptoms were assessed between post-acute COVID-19 participants with (N=18) and without (N=6) LC symptoms at the time of imaging. Participants with LC symptoms were generally highly symptomatic, with a median of 5.5 symptoms. Whereas tracer uptake in many tissue regions was significantly higher in post-acute COVID-19 participants with and without LC symptoms compared with pre-pandemic controls (pons, cauda equina, aortic arch, pulmonary artery, right ventricle wall, lumbar bone marrow; all p<0.05), uptake in the right hilum (SUVmax p=0.004 SUVmean p=0.047), colon wall (SUVmax only p=0.034) and rectal wall (SUVmax p=0.004, SUVmean p=0.013) were only significantly higher than pre-pandemic controls in those that experienced LC symptoms (Fig. 4B; fig. S4). However, no statistically significant correlations were observed between the number of LC symptoms and PET uptake in any ROI, (all P>0.05 by two-sided Spearman tests).
Increased [18F]F-AraG uptake in some tissues was associated with Long COVID symptom phenotypes
We investigated whether specific LC symptom phenotypes correlated with SUVmax and SUVmean uptake in tissue ROIs. Significantly higher uptake in the lower lung parenchyma (SUVmax p=0.032, SUVmean p=0.042) and pulmonary artery wall (SUVmax only p=0.047) was only observed in participants with pulmonary LC symptoms (cough, shortness of breath, dyspnea) present at time of imaging compared to pre-pandemic controls, whereas uptake in the aortic arch wall, right hilum and right ventricle wall in those with and without pulmonary symptoms was significantly higher in post-acute COVID-19 participants (all p<0.05) compared to pre-pandemic controls (Fig. 4C and fig. S4). Direct relationships between other phenotypes (neurocognitive, gastrointestinal) and tissue uptake in related tissues (CNS tissues, gut wall) were not observed (Fig. 4D and E). Of note, all but two participants in the LC group reported fatigue and all persons with fatigue also reported neurological symptoms. Of those with GI symptoms, only 3 reported constipation, and none reported other lower tract symptoms such as diarrhea.
SARS-CoV-2 vaccination had little effect on the biodistribution of [18F]F-AraG
We analyzed [18F]F-AraG uptake in participants grouped by receipt of a SARS-CoV-2 vaccine greater than or less than 180 days prior to PET imaging. Timing from most recent vaccination to imaging appeared to have little effect on [18F]F-AraG uptake across most tissues, with the exception of modestly lower colon (SUVmax only p=0.034) and rectal (SUVmax p=0.003, SUVmean p=0.009) wall tracer uptake in those whose last dose of SARS-CoV-2 vaccine was >180 days prior to imaging (fig. S6). Although our protocol did not enroll those who planned to receive any vaccine within 4 weeks of imaging, as noted above one participant received a SARS-CoV-2 mRNA booster 6 days prior to imaging without informing the study team. Post-hoc investigation of this participant’s PET/CT images revealed similar [18F]F-AraG uptake across all tissue ROIs to other post-acute COVID-19 participants, with values in the middle range of the observed SUVmax values, and without marked uptake in the deltoid muscle injection site. In addition, no significant correlations were observed between the time from vaccine to imaging and PET uptake in any ROI (all P>0.05 by two-sided Spearman tests).
Detectable SARS-CoV-2 nucleocapsid antibody responses were not with [18F]F-AraG uptake
SARS-CoV-2 nucleocapsid IgG titers were measured from plasma obtained at the time of PET imaging to provide information about potential recent reinfection that may have influenced our findings. Ten of 24 post-acute COVID-19 participants had no detectable nucleocapsid antibody detection at the time of PET imaging (signal to cutoff [SC] ratio <1; Table 1). Overall, among the post-acute COVID-19 group, the presence of a detectable nucleocapsid IgG response did not have a major influence on [18F]F-AraG uptake across ROIs (fig. S7).
Modules of circulating markers of inflammation and immune activation were associated with [18F]F-AraG PET uptake in some tissues
To identify associations between [18F]F-AraG PET imaging and systemic immune activation, we assessed circulating protein mediators in plasma just prior to PET imaging using the Olink EXPLORE 384 Inflammation panel. Proteomic data were available for 19 of 24 participants. Differentially expressed proteins were observed in participants grouped by time since initial COVID-19 onset, LC symptom count (proteomic data were only available on two cases without LC symptoms), and higher PET tracer uptake in various ROIs of interest; they did not achieve individual significance after conservative adjustments for multiple comparisons. Clustered heatmaps of differentially expressed genes (DEGs), however, yielded modules of related proteins defined by non-hierarchical k-means clustering by time from initial COVID-19 onset to PET imaging > or <90 days, the presence of >5 or <5 LC symptoms at the time of imaging, and high or low tracer [18F]F-AraG uptake across various anatomical ROIs (Fig. 5, fig. S8). High and low [18F]F-AraG uptake was defined as participants with ROI SUVmax values one to three standard deviations above the mean SUVmax value of the pre-pandemic controls (standard deviations cutoffs were based on overall variation within case and control SUVmax values to define clusters of individuals with [18F]F-AraG uptake higher than the control population). Whereas individual proteins were not increased in adjusted analyses, these modules of proteins had shared directional expression suggesting biological plausibility and warrant further study in larger cohorts.
Figure 5. Modules of circulating markers of inflammation and immune activation are associated with reported Long COVID symptom number and [18F]F-AraG PET uptake in representative tissues.
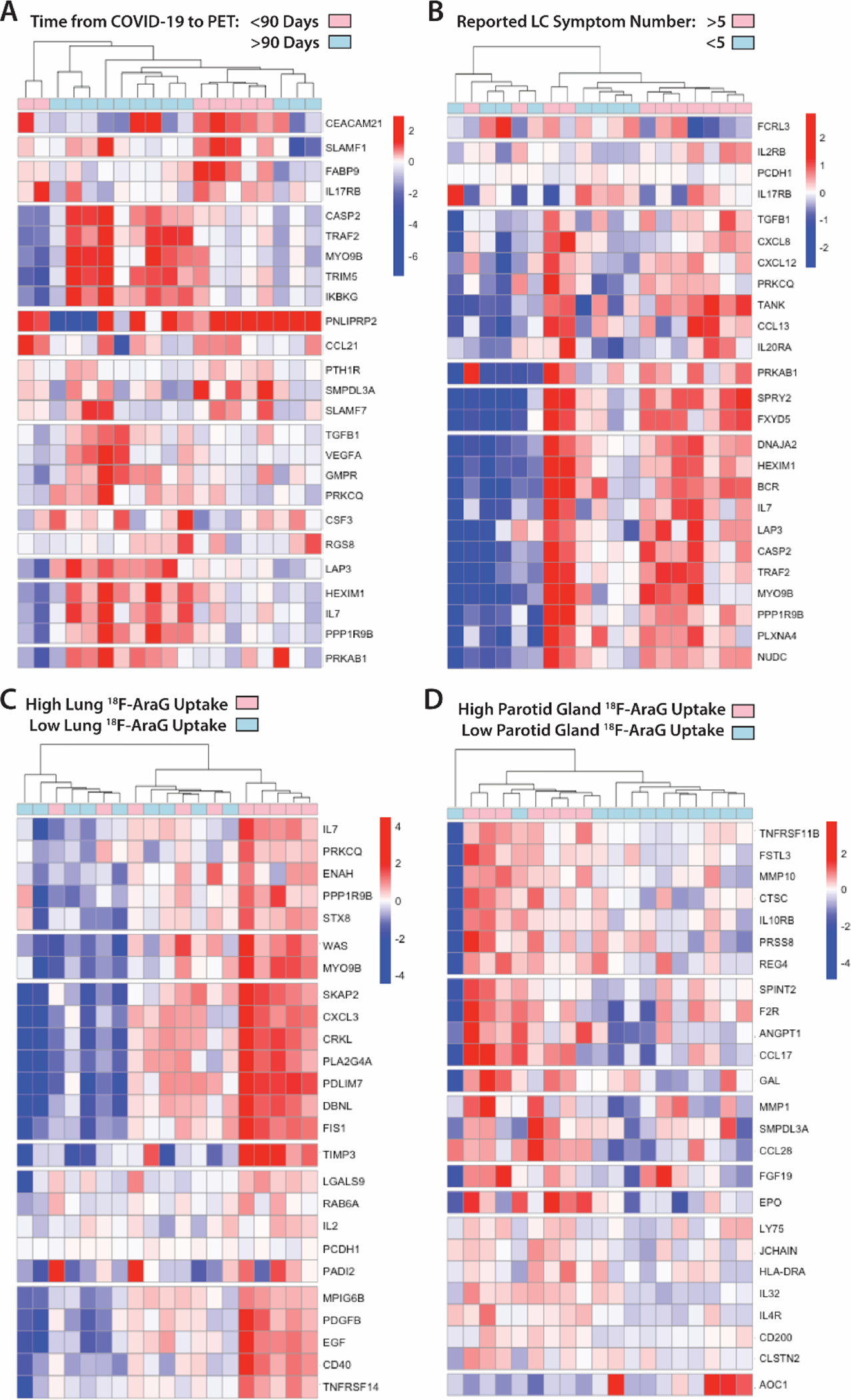
(A to D) Clustered heat maps of the top 25 differentially expressed plasma proteins from Olink Proximity Extension Assay EXPLORE 384 panel based on unadjusted P values with markers grouped into k-clusters based on similarity are shown for participants imaged early (<90 days) or later (>90 days) after symptom onset (A), those reporting >5 or ≤5 Long COVID symptoms (out of a total of 32 surveyed across multiple organ systems) at the time of PET imaging (B), and in those with high lower lung lobe [18F]F-AraG uptake (C; defined as SUVmax >2 standard deviations [SD] above the average SUVmax value measured in Pre-pandemic control volunteers), and parotid gland tissue [18F]F-AraG uptake (D; defined as SUVmax >1 SD above the average SUVmax value measured in pre-pandemic control participants). Heat maps clustered by high uptake in other tissues ROIs are shown in fig. S8. Scale bars represent mean subtracted normalized log2 protein expression values.
High-dimensional flow cytometry of peripheral blood and gut-derived mononuclear cell phenotypes did not reveal significant relationships
A multi-dimensional spectral flow cytometry panel that characterized CD4+ and CD8+ T cell, and B cell phenotypes, including markers of activation, naïve/memory phenotypes, regulatory function, and immune checkpoint/exhaustion was performed on peripheral blood mononuclear cells (PBMCs) from 16 participants who had sufficient specimens around the time of PET imaging and from gut tissue from 5 participants who underwent colorectal biopsies. Overall, we observed higher frequencies of effector memory CD8+ and CD4+ T cells in gut tissue versus peripheral blood, but similar frequencies of CD8+ and CD4+ T cell subsets, CD8+ and CD4+ lymphocytes expressing the activation markers CD38 and HLA-DR, T cells expressing PD-1, and class-switched (IgD-) and unswitched (IgD+) memory B cells in participants grouped by time from infection, having greater than 5 LC symptoms, or time from last COVID-19 vaccine dose to imaging and vaccine (fig. S9 and S10). In addition, there were no significant correlations between PET tissue tracer uptake, LC symptom counts, timing of infection or vaccination, or specific LC symptom phenotypes with CD4+T cell, CD8+ T cell, and B cell phenotypes (all p>0.05).
Intestinal biopsies in a subset of post-acute COVID-19 participants showed evidence of persistent SARS-CoV-2 Spike protein-encoding RNA in rectal tissue
Prior studies suggest that SARS-CoV-2 RNA or proteins may be detected in the gut or shed in stool for several months following COVID-19 (32). Given the higher [18F]F-AraG uptake in proximal colon and rectal wall across many post-acute COVID-19 participants, it is possible that viral persistence may be driving increased activated T cell migration to gastrointestinal tissues. We explored this by collecting rectosigmoid tissue by flexible sigmoidoscopy in 5 participants who had undergone PET imaging ranging from 158 to 676 days following infection (Table 1). Four of 5 participants had biopsies within 63 days of PET imaging; one was biopsied 182 days after imaging. All participants reported at least two LC symptoms at the time of biopsy. None had received a SARS-CoV-2 vaccine dose in the prior month. None reported any history, symptoms, or testing suggestive of SARS-CoV-2 reinfection, and three of the five participants (participants 15, 16 and 18) had no detectable SARS-CoV-2 nucleocapsid IgG detected around the time of tissue collection (SC ratios <1 at time of PET imaging). No clinical evidence of reinfection was observed in those who continued to have detectable N IgG responses.
Single-stranded SARS-CoV-2 RNA was detected in multiple cells in all three tissue regions surveyed from all but one individual (participant 18) who had SARS-CoV-2 RNA+ cells detected in only one of three gut tissue regions sampled 645 days following initial infection. The percentage of gut cells that harbored SARS-CoV-2 Spike protein-encoding RNA for each participant was determined (Table 1). Nearly all RNA was detected in cells in the lamina propria, without an epithelial signal (Fig. 6). A small percentage of RNA+ cells expressed CD68, a macrophage and monocyte lineage marker, but many SARS-CoV-2 RNA+ cells did not express CD68 and none expressed CD3. Nonetheless, we observed relatively high numbers of CD68+ and CD3+ positive cells within the vicinity of SARS-CoV-2 RNA+ cells (Fig. 6). [18F]F-AraG SUVmax values in proximal colon and rectal tissue in the participants who underwent biopsy were at least 3 standard deviations above the mean SUVmax of pre-pandemic controls (1.86 vs 0.95 and 1.5 vs 0.9, respectively).
Figure 6. SARS-CoV-2 single-stranded Spike protein-encoding RNA was identified in recto-sigmoid tissue of individuals with LC months to years following acute infection.
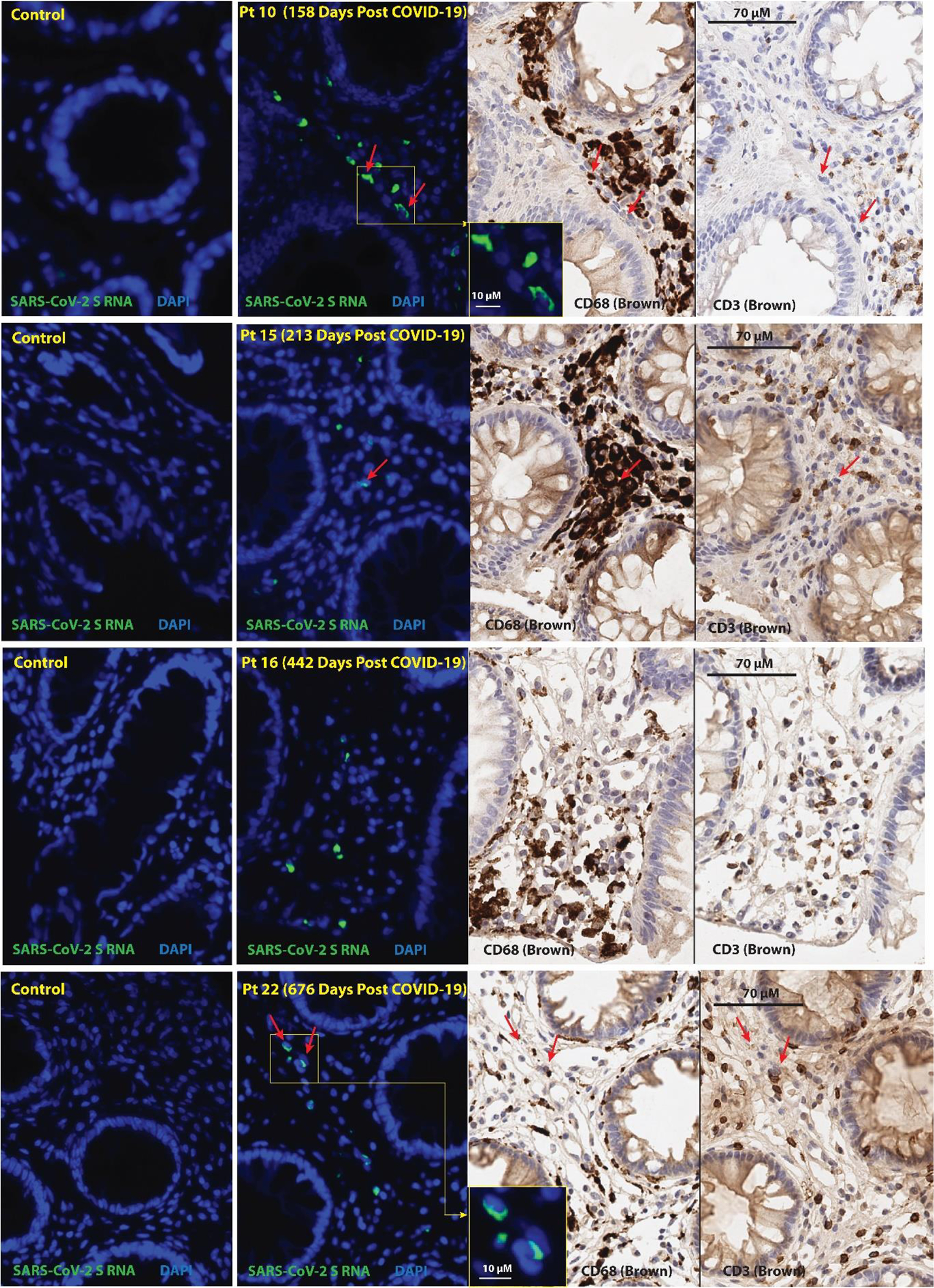
Panels represent from left to right: SARS-CoV-2 spike protein-encoding RNA staining by ISH in pre-pandemic tissue, SARS-CoV-2 spike protein-encoding RNA staining by ISH in post-acute COVID-19 participant sample, CD68 immunostaining, and CD3 immunostaining. Red arrows denote representative areas of RNA detection across images for each sample (not all RNA detection is marked). Spike single-stranded (ss)RNA was detected in all five of the post-acute COVID-19 participants that underwent biopsy from 158 to 676 days following initial COVID-19 symptom onset and signal was primarily observed in cells located within the lamina propria. Four participants with detectable ssRNA in three distinct gut regions are shown, a fifth participant had rare Spike ssRNA detected in only one of three regions imaged. A minority of SARS-CoV-2 ssRNA signal was localized in CD68+ cells and very rarely in CD3+ cells. No viral ssRNA was detected in control tissue from a pre-pandemic participant.
Double-stranded SARS-CoV-2 RNA was also identified in gut lamina propria in 3 of the 4 participants with detectable single-stranded RNA in all three gut regions surveyed (Table 1). dsRNA is only produced during active viral transcription and translation activity or during replication. We observed intracellular dsRNA in relatively discreet clusters and not distributed widely across all regions, which may represent either local replication with limited viral spread or lack of ability to achieve immune clearance of actively infected cells (Fig. 7). Polymerase Chain Reaction (PCR) was also performed on RNA isolated from bulk rectal tissue lysates (separate biopsy from the FFPE tissue used above) targeting the Spike, N1, N2, Envelope (E) and RNA-dependent RNA polymerase (RdRp) protein-encoding regions of SARS-CoV-2. No RNA was detected using this method in any participant.
Figure 7. SARS-CoV-2 Spike protein-encoding double-stranded (ds)RNA was observed in recto-sigmoid tissue from individuals with LC months to years following acute infection.
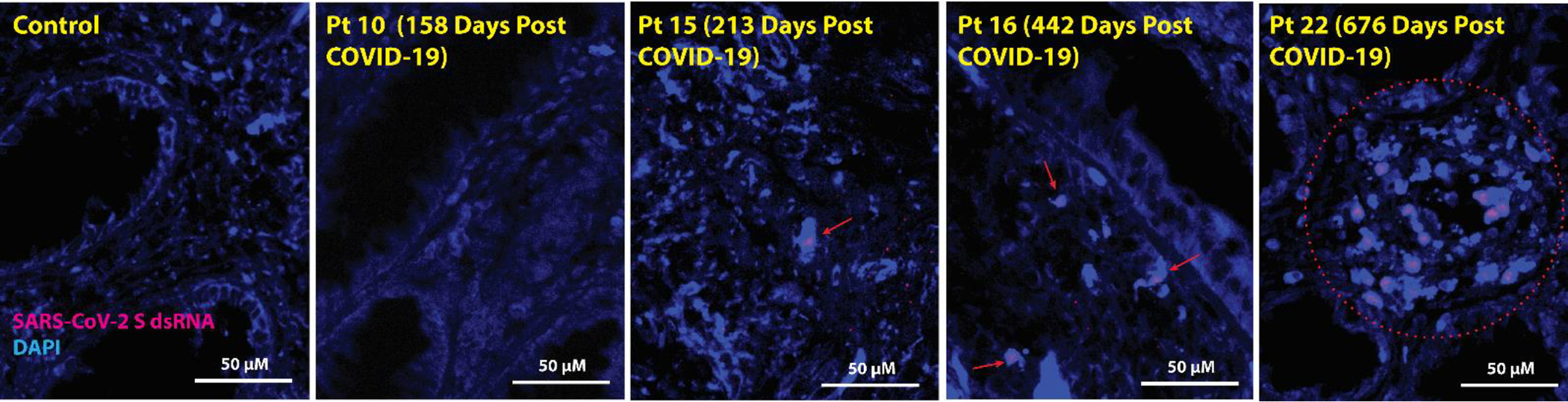
Red arrows and the red circle denote representative areas of dsRNA detection in cells across images for each sample. Spike protein-encoding dsRNA was detected in three of the post-acute COVID-19 participants who underwent biopsy from 213 to 676 days following initial COVID-19 symptom onset, and the signal was primarily observed exclusively in cells located within the lamina propria. Four representative participants are shown. No viral dsRNA was detected in the control tissue from a pre-pandemic participant.
DISCUSSION
In this first-in-human T cell activation PET imaging study of individuals following SARS-CoV-2 infection, we found evidence of persistent T cell activation in a variety of tissues. In some individuals, this activity may persist for years following initial COVID-19 onset and be associated with systemic changes in immune activation as well as the presence of LC symptoms. Finally, we found evidence of SARS-CoV-2 persistence in gut tissue including potential ongoing viral transcriptional activity. Taken together, these observations suggest that even clinically mild SARS-CoV-2 infection could have long-term consequences on tissue-based immune homeostasis and potentially result in an active viral reservoir in deeper tissues. Although correlative, our findings provide additional evidence to support the role of tissue-based immune activation and viral persistence as contributors to post-acute sequelae of SARS-CoV-2 infection, including LC.
This study adds to our knowledge of tissue-based immune responses following SARS-CoV-2 infection. Whereas traditional PET-based imaging using radio-labeled glucose (FDG) as a marker of tissue inflammation has been applied to the study of acute COVID-19 and, to a lesser extent, the post-acute phase (44–50), this method is non-specific because FDG is taken up by any metabolically active tissue. One pilot PET study using a CD8-specific minibody radiotracer provides some evidence that T cells traffic to tissues such as bone marrow following acute infection and remain up to 4 months thereafter (51), but the relationship between T cell activation state, clinical symptoms, and viral persistence in the post-acute phase was not addressed. Dysregulated T cell responses in peripheral blood have previously been associated with LC (18) and methods that identify activated T cell responses in tissues have the potential to provide insights into the breadth of immune response and dysfunction across the whole body.
We found that [18F]F-AraG uptake was higher in post-acute COVID-19 participants compared to pre-pandemic controls in many anatomical regions. These observations were identified up to 2.5 years following initial COVID-19 symptom onset, in the absence of confirmed or suspected re-infection. Although [18F]F-AraG uptake in some tissues (spinal cord, colon/rectal wall) appeared to decline with time, the degree of uptake often remained elevated above those measured in pre-pandemic healthy controls. These data extend prior observations of a durable and dysfunctional cellular immune response to SARS-CoV-2 (11, 18, 19, 70) and suggest that SARS-CoV-2 infection could result in a new immunologic steady state in the years following COVID-19.
In this study, T cell activation in some tissues (spinal cord and gut wall) was higher in participants reporting LC symptoms of any type compared to both pre-pandemic controls and those with complete recovery. Increased lung and pulmonary artery wall [18F]F-AraG uptake was only identified in those with persistent pulmonary symptoms, and not those without these symptoms, compared to pre-pandemic controls, suggesting a potential link between ongoing aberrant tissue immune responses and long-term clinical symptoms. Larger studies involving well-curated clinical cohorts will be needed to further characterize relationships between whole-body T cell activation and LC symptoms.
This study provides evidence for ongoing immune responses in tissues, a potential source of inflammation observed in peripheral blood (8–18, 71). Furthermore, as LC is increasingly framed as having potential neurological underpinnings, it is possible that spinal cord and brainstem [18F]F-AraG uptake observed in our study may represent T cell trafficking to CNS tissues with residual viral components. This is consistent with an autopsy study that identified SARS-CoV-2 spike RNA and protein in the spinal cord and basal ganglia in two individuals who died 65 and 230 days following COVID-19 (36). We note, however, that the increase in PET signal in the lumbar cauda equina regions may also be due, in part, to accumulation of tracer in the CSF through altered blood-brain barrier permeability, as well as activated T cell accumulation.
We identified cellular single-stranded SARS-CoV-2 RNA in rectosigmoid lamina propria tissue in situ in participants studied up to two years following COVID-19. This observation extends prior reports limited to 4 to 6 months post-COVID-19 (32, 33, 36). Importantly, we also observed clusters of lamina propria cells that harbored double-stranded RNA in several participants. Double-stranded RNA is produced only during replication, active infection, or ongoing transcriptional activity. Therefore, our findings suggest that there may be ongoing replication or active SARS-CoV-2 “reservoirs” that persist and evade immune clearance through mechanisms yet to be identified (40). Further tissue-based study of viral persistence and virus-immune-host responses in these microenvironments is needed to determine if viral persistence has a direct association with LC symptoms.
We employed several measures to guarantee virus-specific RNA in situ hybridization, including use of pre-pandemic control tissue on each slide to minimize technique differences or batch testing effects, and repeated RNA staining in contiguous slices to verify consistency of location of the viral RNA signal. We also employed different RNA hybridization probes to minimize confounding by autofluorescence. PCR from bulk tissue lysates was unsuccessful (and hence viral sequencing was not possible). SARS-CoV-2 RNAscope methods have been shown to be more sensitive in gut tissues in prior analyses (33). As [18F]F-AraG uptake was observed in this anatomical region in nearly all post-acute COVID-19 participants, this finding suggests that virus persistence might contribute to the sustained T cell activation in our cohort. Because all participants who underwent gut biopsy met LC criteria, it is difficult to draw concrete conclusions regarding the impact of gut viral persistence on LC symptoms or [18F]F-AraG uptake. Further investigation, including assessment of larger numbers of fully recovered comparators, will be needed to definitively determine whether viral persistence is associated with LC.
A key question for the field is in which cell type(s) SARS-CoV-2 might persist. We observed spike protein-encoding RNA in regions of CD68+ and CD3+ immune cell infiltration, although most infected cells were not macrophages or T cells. ACE2 expression in the lamina propria and on myeloid immune cells has been shown to be low, and we observed little-to-no RNA in the gut epithelium, where ACE2 expression is higher (32, 72). These data suggest that macrophages or other immune cells may be acquiring virus or viral contents either through phagocytosis of other infected cells or through viral-immune complexes, as previously proposed (73). Viral transcriptional activity, which is suggested by the observed presence of double-stranded RNA within cells, may lead to innate immune sensing and downstream, tissue-based inflammation that could lead to infiltration of other immune cells (e.g., T cells), tissue damage and systemic inflammation even without replication or de novo infection. For example, residual HIV-1 transcriptional activity in the setting of otherwise suppressive antiretroviral therapy without replication has been observed (74, 75).
Collectively, our data are consistent with a model in which persistence of SARS-CoV-2 results in chronic tissue-based inflammation, T cell activation, and perhaps LC. However, we acknowledge that this story of persistent immune activation and viral antigen in LC is still developing, and no causal relationships have been verified. The lack of observed associations between many LC symptoms and T cell activation may be due to a smaller cohort or the well-described challenges in measuring LC (55). Larger studies will be needed to further define the relationships between immune responses at the tissue level and specific LC endotypes.
This study has several limitations, many inherent to PET imaging protocols. First, whereas the relatively small sample size limited power in correlative studies, we were powered to examine the primary imaging endpoints. Second, because of the rapidly evolving pandemic, including intermittent variant waves, the recognition of LC as a relevant clinical entity, and the rapid but inconsistent rollout of SARS-CoV-2 vaccines, we were required to adapt the protocol over time. This resulted in a shift from imaging participants closer to initial SARS-CoV-2 symptom onset to prioritizing those with LC symptoms months to years after COVID-19. As a result, those enrolled earlier in the pandemic tended to be imaged closer to the time of acute infection and were not selectively enrolled based upon the presence of LC symptoms. Third, we relied mainly on pre-pandemic controls who were imaged in [18F]F-AraG protocols using similar PET acquisition strategies and timing from tracer injection to image acquisition. Pre-pandemic control volunteers were imaged using PET/MRI and, on average, received a higher dose of [18F]F-AraG, but SUV were used as comparison which take into account tracer injection dose, participant size and isotope decay rates. Sensitivity of the PET/CT and PET/MRI scanners is similar. We would expect any confounding from this higher tracer dose to lead to higher uptake in the pre-pandemic controls compared to post-acute COVID-19 participants. Nonetheless, the use of historical controls may introduce further bias, but we note that it has become difficult to enroll contemporary, never-infected control participants in research studies now that most of the population has been infected with SARS-CoV-2 (sometimes subclinically) (4). Individuals who participate in LC research also tend to be highly vaccinated, so at this point it is not feasible to identify individuals who never received a SARS-CoV-2 vaccination. Given the limited clinical information available on the pre-pandemic control participants, there may be differences in the timing of PET with regard to other infectious exposures (e.g., seasonal respiratory viruses) between the pre-pandemic and post-acute COVID-19 cohorts. During the early stages of the pandemic when we recruited many of our post-acute COVID-19 participants, there were reductions in other circulating respiratory viruses. We followed each of our post-acute COVID-19 participants closely with ongoing assessment for infections, which were uncommon in the post-acute COVID-19 cohort during the study period. As a result, it is probable that the pre-COVID-19 controls, recruited in an era without the public health restrictions brought on by the pandemic, had more frequent exposure to other respiratory infections around the time of imaging. If this were the case, it would be expected to bias our results towards higher tracer uptake in the control participants, and thus decrease our ability to identify differences between pre-pandemic and post-acute COVID-19 participants. Finally, although mitigated by frequent assessments in LIINC, it remains possible that some participants may have had asymptomatic or undetected reinfections between initial COVID-19 onset and their rectosigmoidoscopy or imaging; although this was less common during the study period, it is now quite common and will be an important consideration for future research.
In summary, our results provide evidence of long-term immune system activation in several tissues following SARS-CoV-2 infection, including in those experiencing LC. We also show that SARS-CoV-2 RNA may persist in gut tissue for nearly 2 years. Overall, these observations challenge the paradigm that COVID-19 is a transient acute infection, building upon recent observations in blood (76). These data provide evidence for T cell activation and viral persistence in tissues well beyond the initial illness.
METHODS
Study design.
Adults with SARS-CoV-2 infection confirmed on nucleic acid-based or antigen-based testing were enrolled 14 days or longer following initial COVID-19 symptom onset and assessed prospectively approximately every 4 months following initial symptom onset. At each visit, participants completed interviewer-administered questionnaires that assessed demographics, medical history, and SARS-CoV-2 infection, vaccination, and treatment history, as well as Long COVID symptoms. Participants with and without concurrent LC symptoms then underwent single [18F]F-AraG PET CT imaging, and a subset of 5 individuals with LC additionally provided written informed consent to undergo colorectal biopsies weeks to months following completion of imaging. Participants were excluded from the imaging protocol if they had a history of excessive radiation, underwent a prior research study involving radiation within 1 year of enrollment, were pregnant or breastfeeding, had screening absolute neutrophil count <1000 cells/mm3, platelet count <75,000 cells/mm3, hemoglobin <8 g/dL, estimated creatinine clearance <60 mL/min, aspartate aminotransferase >3x ULN units/L, alanine aminotransferase >3x ULN units/L, had recent use of medication including guanosine or cysteine analogs, had known SARS-CoV-2 nasopharyngeal shedding within 5 days of scan, had a known SARS-CoV-2 vaccine within 4 weeks of scan, or had a prior history of immunoproliferative or autoimmune disease. We also did not image potential participants if they reported upper respiratory symptoms or other viral symptoms in the 4 weeks prior to PET imaging.
Participants.
Study volunteers were participants in the UCSF-based LIINC study (NCT04362150) (55) and opted into the imaging protocol (NCT04815096). Participants provided written informed consent for participation in both the LIINC cohort and the imaging protocol. The study was approved by the UCSF Institutional Review Board and the UCSF Radiation Safety Committee. ClinicalTirals.gov numbers: NCT04362150 and NCT04815096. Procedures for LIINC have been described in detail elsewhere (55).
Questionnaire-based measurements.
At the first LIINC visit, participants completed an interviewer-administered questionnaire that assessed demographics, medical history, and SARS-CoV-2 infection, vaccination, and treatment history. At all subsequent visits, participants completed additional interviewer-administered questionnaires that queried the presence and severity of any symptoms that were new or worsened since the initial SARS-CoV-2 diagnosis, quality of life, interim medical diagnoses, and interim treatments and vaccinations. Symptoms that predated SARS-CoV-2 infection and were unchanged were not considered to represent LC. The questionnaire included up to 32 symptoms which we grouped into distinct LC phenotypes based on our prior published work showing biological differences between these groups as follows (26, 55): [1] fever/chills (fever, subjective fever, chills); [2] fatigue; [3] pulmonary (cough, shortness of breath); [4] cardiac (chest pain, abnormal heart rate); [5] upper respiratory (runny nose, sore throat); [6] muscle pain; [7] gastrointestinal (loss of appetite, nausea, vomiting, abdominal pain, constipation, diarrhea); [8] rash; [9] changes in taste and smell; [10] neurological/cognitive (concentration problems (such as “brain fog”), headache, vision changes, dizziness, feeling faint, balance problems, numbness/tingling); [11] back pain; [12] sleep disturbances. Concurrent with PET imaging, detailed histories were obtained including symptoms, prior or subsequent SARS-CoV-2 PCR or antigen testing, and clinical symptoms potentially related to SARS-CoV-2 re-infection.
[18F]F-AraG imaging.
Participants were intravenously administered [18F]F-AraG (166.5–185 MBq) and PET/CT whole-body imaging was carried out for 20 minutes at approximately 50 minutes post injection. [18F]F-AraG was prepared as documented elsewhere (58). Images were taken from the top of head to mid thighs. Routine urinalysis was performed 7 to 14 days after imaging to ensure proper excretion of [18F]F-AraG.
PET and CT image analysis.
Standardized uptake values (SUV) in various tissue regions of interest (ROI) from PET/CT data were determined using the OsiriX DICOM viewer software package (Pixmeo). ROI determination was performed in complex structures such as brain sections, heart wall, spleen, and gut wall using two-dimensional isometric ROIs (fig. S11). For simpler structures such as the spinal cord, bone marrow, and whole lymph nodes, three-dimensional spherical VOIs were used). For axillary and inguinal lymph node ROI determination, the most prominent nodes on both the right and left side were included and SUV values averaged. ROI were placed independently by two individuals blinded to the study group following ROI determination upon a subcohort and comparison to ensure consistency across reviewers. Two- and three-dimensional PET or PET/CT images were generated in OsiriX keeping window levels consistent between participants. SUVs from participants reporting LC symptoms were compared with six pre-pandemic PET/MRI controls. ROI determination on bowel tissue was challenging as we observed intermittent intraluminal collection of tracer, which was highly anatomically variable across all participants. As intraluminal signal does not represent specific tissue tracer uptake, regions in gut wall tissue were only drawn in areas without clear contiguous intraluminal signal. As a result, ROI placement was not possible in 3 and 5 post-acute COVID-19 participants for proximal colon and rectal wall ROIs, respectively, and 3 pre-pandemic control participants for proximal colon. All PET images and CT images of the chest were further reviewed independently by two dual board-certified radiologists and nuclear medicine physicians (R.R.F. and Y.W.). Qualitative abnormalities were tabulated. To constrain the large number of potential comparisons in this study, we focused SUV comparisons in ROIs that demonstrated differences between pre-COVID-19 and post-acute COVID-19 participants for focused, three-way analyses.
Circulating Markers of Inflammation.
A Protein Extension Assay (PEA) using the Olink EXPLORE Inflammation panel from plasma samples was performed in post-acute COVID-19 imaging participants to characterize 365 unique plasma proteins associated with inflammation and immune signaling. Protein Extension Assay (PEA) involves dual-recognition of two matched antibodies labeled with unique DNA oligonucleotides that simultaneously bind to specific target proteins. The simultaneous antibody binding leads to hybridization of unique DNA oligonucleotides that serve as templates for polymerase-dependent extension (DNA barcoding) followed by PCR amplification and NovaSeq (Illumina) DNA sequencing. Protein expression values were log-transformed and centered using the average expression value for each protein. clustered heatmaps were generated by the UCSF Gladstone Bioinformatics Core using the R package HOPACH to find the best cluster number. Gene product expression values were log-transformed and centered using the average expression value for each protein. Gene products were then clustered by running the Kmeans algorithm using the best cluster number K found, and the results were plotted using the pheatmap package as modules. Standard deviation (SD) cutoff values between those with higher and lower [18F]F-AraG uptake for each tissue were decided based on variance of SUVmax values of uninfected controls, with higher variance allowing for one or more SDs above the mean to qualify as “higher” uptake.
qPCR and In situ hybridization of SARS-CoV-2 Spike protein-encoding RNA in rectal biopsies.
Rectal wall tissue samples were obtained by flexible sigmoidoscopy with tissue being fixed in fresh paraformaldehyde (PFA) followed by Paraffin embedding approximately 48 hours after fixation or were cryopreserved at −180°C in Fetal Bovine Serum (FBS) and 20% Dimethyl sulfoxide (DMSO) (77). A minimum of three formalin-fixed paraffin embedded (FFPE) rectosigmoid tissue biopsies from each participant were used for RNAscope experiments, along with comparative uninfected tissue, were mounted on the same slide to control for batch effects from processing, staining, microscopy, and image analysis. Experiments were performed at least twice on contiguous sections to verify signal over non-specific staining or autofluorescence. Contiguous sections underwent hematoxylin and eosin staining and immunohistochemical visualization of CD3 and CD68 expression to localize viral RNA signals with anatomical tissue regions and immune cell types of interest. Quantitative PCR assays were performed using Integrated DNA Technology’s (IDT) SARS-CoV-2 RUO qPCR Primer & Probe Kits for N1, N2, E, and RDRP detection (catalog no. 10006713, 10006804, 10006805, and 10006806). Positive controls consisted of fragments of human RPP30 and SARS-CoV-2 isolate Wuhan-Hu-1 (GenBank: NC_045512.2) provided in each IDT kit. Quantitative PCR was performed with TaqPath 1-Step RT-qPCR Master Mix with the following conditions: 95 °C 2 min, 95 °C 3 sec, 55 °C 30 sec using the StepOnePlus Real-Time PCR System. SARS-Cov-2 RNA was considered detectable for cycle threshold (ct) values <40. Positive and non-template controls were run for all samples tested. Each sample was run in duplicate wells. Spike qPCR was also performed on RNA extracted from cryopreserved rectal tissue pieces using Thermo Fisher’s single-tube TaqMan Microbe Detection SARS-CoV-2 S gene Assay (catalogue no. A50137, Assay ID Vi07918636_s1) to detect the S1 domain of the spike protein. Thermo Fisher’s Comprehensive Microbiota Control (catalogue no. A50832), a multi-target plasmid pool containing the sequences for the S gene-specific assay, was used as a positive control. Quantitative PCR was run using TaqPath 1-Step RT-qPCR Master Mix with the following conditions: 50°C 2 min, 95°C 20 sec, 95°C 1 sec, 60°C 20 sec using the StepOnePlus Real-Time PCR System. SARS-Cov-2 RNA was considered detectable for cycle threshold (ct) values <40. Positive controls were run for all samples tested.
The manual RNAscope 2.5 HD assay (Advanced Cell Diagnostics; catalog no. 322310) was used to identify SARS-CoV-2 Spike protein-encoding RNA in situ. Paraffin-embedded tissue blocks were sectioned at 5 μm, mounted onto SuperFrost Plus slides, and stored at 4°C prior to staining. Slides were baked in a dry-air oven at 60°C for 1 h, then deparaffinized in 100% Xylene (5 min) twice and washed in 95% ethanol (3 min) twice, 80% ethanol (3 min) once, and dH2O (1 min) twice, all at room temperature. To prevent drying, 3 to 4 drops of TBS were placed on each tissue section. A hydrophobic barrier was then drawn around each tissue section and allowed to dry for 10 min. To block endogenous peroxidase, slides were then pretreated with hydrogen peroxide for 10 mins at room temperature followed by washing with dH2O. Next, heat-induced epitope retrieval was performed with Target Retrieval Reagent (ACDBio) and incubated at 100˚C for 15 minutes followed by rinsing with dH2O. Protease digestion was accomplished by treatment with Protease Plus solution (ACDBio) for 30 minutes at 40˚C followed by dH2O wash. Hybridization was performed with RNAscope probe-V-nCoV2019-S (catalog no. 848561-C3, Advanced Cell Diagnostics) at 40°C for 2 h at a 1:50 dilution. Following hybridization, 3 amplification steps were carried out as indicated in the original protocol. Slides were then incubated with HRP-C3 at room temperature for 15 min, TSA Vivid Fluorophore 520 for 40°C for 30 min, and HRP blocker at 40°C for 15 min. Finally, slides were counterstained with DAPI, washed in PBS, and cover-slipped using Prolong Diamond Mounting Media. Sense Spike RNAscope was also performed on each tissue as above [Probe- V-nCoV2019-S-sense (catalogue no. 845701-C3)] to determine presence of double-stranded RNA.
Images were captured using the Zeiss AxioObserver Z1 (RNAscope) or the Leica Aperio VERSA (chromogenic staining on contiguous tissue sections). FFPE rectal tissue from a pre-pandemic control participant was mounted on each participant slide to control for any staining or imaging technique differences. Only linear brightness and contrast adjustments were made to image files and all adjustments were applied identically for each image.
Spectral Flow Cytometry.
PBMCs or cells obtained from gut tissue following a previously published collagenase disaggregation protocol (78) were stained with the Cytek 25-Color Immunoprofiling Assay 25-plex kit (Cat# SKU R7–40002) and with the BioLegend 7 Color Immunoprofiling kit antibodies (Cat#900004160); Zombie UV dye (Cat#423108) was used for live-dead staining. 3e6 cells were stained with 5μl of antibodies for 30 min at 25°C in a total volume of 130μl, including the addition of Brilliant stain buffer plus (cat#566385) according to kit manufacturer’s guidelines. Cells were washed twice with FACS buffer (PBS+10%FBS+1mM EDTA) before fixation with 1% PFA in PBS. Cells were acquired on the 5L Cytek Aurora the day after. Spectraflow beads were used to verify laser alignment and power consistency. The forward and side scatter profiles were established using human PBMCs, similar to the test samples. Single- stained cellular reference controls were used to unmix the data according to the 25-plex Cytek acquisition protocol, with unstained cells as an autofluorescence control. Unmixing errors were corrected by spillover correction using OMIP-069 supplementary information as a guide. The Cytek analysis template was used to replicate the gating strategy in FlowJo 10 (fig. S12 and S13). Data was exported, and further analysis performed on GraphPad Prism.
SARS-CoV-2 Nucleocapsid Antibody Testing.
Nucleocapsid IgG antibodies from post-acute COVID-19 participants from plasma tested at the time of PET imaging were measured using the Abbott Architect i2000 two step Chemiluminescent microparticle immunoassay (CMIA). Signal to cutoff (S/C) ratios were determined and samples were considered positive if the S/C ratio was above the assay-defined threshold (S/C >1 were considered positive).
Statistical analysis.
Individual-level data are presented in data file S1. We used two-tailed, non-parametric Kruskal-Wallis tests using a Benjamini-Hochberg adjustment for false discovery rates from multiple comparisons within specific tissue regions (lymphoid tissues, glandular tissue, vascular, spinal cord, etc.) to compare ROI SUV data and flow cytometric data across participants (adjusted P values being analogous to the q value obtained from FPR adjustmentsNonparametric tests were used given the assumption that SUV data may not be normally distributed across comparator groups. Two-sided Welch Two Sample t-tests were used to compare protein expression obtained from Olink assays. Two-sided Spearman rank tests were performed to determine correlations between continuous variables. Sensitivity analyses were performed to determine cutoff values for grouping SUV PET results by time from infection and time from infection to imaging. An alpha level of 0.05 was used to define statistically significant results.
Supplementary Material
Acknowledgements
We are grateful to the study participants and their medical providers. We acknowledge current and former LIINC clinical study team members Tamara Abualhsan, Andrea Alvarez, Grace Anderson, Khamal Anglin, Urania Argueta, Mireya Arreguin, Tyrine Bailey, Alexus Clark, Emily Conway, Nicole DelCastillo, Avery Eun, Marin Ewing, Emily Fehrman, Tony Figueroa, Diana Flores, Halle Grebe, Heather Hartig, Yanel Hernandez, Beatrice Huang, Marian Kerbleski, Raushun Kirtikar, Megan Lew, James Lombardo, Monica Lopez, Michael Luna, Lynn Ngo, Enrique Martinez Ortiz, Jesus Pineda-Ramirez, Justin Romero, Ruth Diaz Sanchez, Matthew So, Celina Chang Song, Alex Tang, Cassandra Thanh, Fatima Ticas, Leonel Torres, Brandon Tran, Julian Uy, Daisy Valdivieso, Deepshika Verma, Meghann Williams, and Andhy Zamora; and LIINC laboratory team members Belen Altamirano-Poblano, Joanna Donatelli, Lilian Grimbert, Jill Hakim, Nikita Iyer, Owen Janson, and Keirstinne Turcios. We thank Jessica Chen, Aidan Donovan, Carrie Forman, Rania Ibrahim, Sean Thomas, and Badri Viswanathan for assistance with data entry and review. We thank the UCSF AIDS Specimen Bank for processing specimens and maintaining the LIINC biospecimen repository. We are grateful to Elnaz Eilkhani for regulatory support. We are also grateful for the contributions of additional LIINC leadership team members: Bryan Greenhouse, Isabelle Rodriguez-Barraquer, and Rachel Rutishauser. We would like to dedicate this work to Aruna Gambhir who passed away at the age of 61 on November 29, 2023, after a long battle with breast cancer. Her obituary can be found at https://jnm.snmjournals.org/content/early/2024/02/08/jnumed.124.267375.
Funding
PET-imaging and peripheral blood immune testing was supported by a Merck Investigator Studies Program Grant (to TJH). The LIINC clinical core, PET imaging program, and gut biopsy collection and testing was supported by grants from the PolyBio Research Foundation (to TJH, HV, SGD, and MJP). This work was also supported by NIH/National Institute of Allergy and Infectious Diseases grants (3R01AI141003–03S1 to TJH, K23AI157875 to MJP, and K24AI145806 to TJH), NIH/National Institute of Neurological Disorders and Stroke (R01NS136197 to MJP) and the Zuckerberg San Francisco General Hospital Department of Medicine and Division of HIV, Infectious Diseases, and Global Medicine.
Footnotes
Conflicts of Interest
MJP reports consulting fees for Gilead Sciences and AstraZeneca and research support from Aerium Therapeutics, outside the submitted work. SGD reports consulting for Enanta Pharmaceuticals and Pfizer and research support from Aerium Therapeutics, outside the submitted work. TJH reports consulting fees for Roche and Regeneron outside the submitted work. JL is employed by CellSight Technologies, holds CellSight stock options, and holds patents relating to [18F]F-AraG (Methods and Materials for Making PET Radiotracers, US20220370648A1; Methods and Materials for Using [18F]F-AraG in Cardiac Imaging, US20230309833A1). AG was employed by CellSight Technologies.
Data Availability:
All data associated with this study are in the paper or supplementary materials. Source data without participant-level identifying information for analyses and figures will be made available upon request of the communicating authors.
REFERENCES
- 1.Nalbandian A, Sehgal K, Gupta A, Madhavan MV, McGroder C, Stevens JS, Cook JR, Nordvig AS, Shalev D, Sehrawat TS, Ahluwalia N, Bikdeli B, Dietz D, Der-Nigoghossian C, Liyanage-Don N, Rosner GF, Bernstein EJ, Mohan S, Beckley AA, Seres DS, Choueiri TK, Uriel N, Ausiello JC, Accili D, Freedberg DE, Baldwin M, Schwartz A, Brodie D, Garcia CK, Elkind MSV, Connors JM, Bilezikian JP, Landry DW, Wan EY, Post-acute COVID-19 syndrome. Nat Med 27, 601–615 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis HE, McCorkell L, Vogel JM, Topol EJ, Long COVID: major findings, mechanisms and recommendations. Nat Rev Microbiol 21, 133–146 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Aly Z, Xie Y, Bowe B, High-dimensional characterization of post-acute sequelae of COVID-19. Nature 594, 259–264 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention COVID Data Tracker. US Department of Health and Human Services, CDC. (https://covid.cdc.gov/covid-data-tracker). [Google Scholar]
- 5.Robertson MM, Qasmieh SA, Kulkarni SG, Teasdale CA, Jones HE, McNairy M, Borrell LN, Nash D, The Epidemiology of Long Coronavirus Disease in US Adults. Clin Infect Dis 76, 1636–1645 (2023). [DOI] [PubMed] [Google Scholar]
- 6.Del Valle DM, Kim-Schulze S, Huang HH, Beckmann ND, Nirenberg S, Wang B, Lavin Y, Swartz TH, Madduri D, Stock A, Marron TU, Xie H, Patel M, Tuballes K, Van Oekelen O, Rahman A, Kovatch P, Aberg JA, Schadt E, Jagannath S, Mazumdar M, Charney AW, Firpo-Betancourt A, Mendu DR, Jhang J, Reich D, Sigel K, Cordon-Cardo C, Feldmann M, Parekh S, Merad M, Gnjatic S, An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med 26, 1636–1643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, Takahashi T, Tokuyama M, Lu P, Venkataraman A, Park A, Mohanty S, Wang H, Wyllie AL, Vogels CBF, Earnest R, Lapidus S, Ott IM, Moore AJ, Muenker MC, Fournier JB, Campbell M, Odio CD, Casanovas-Massana A, Yale IT, Herbst R, Shaw AC, Medzhitov R, Schulz WL, Grubaugh ND, Dela Cruz C, Farhadian S, Ko AI, Omer SB, Iwasaki A, Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584, 463–469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su Y, Yuan D, Chen DG, Ng RH, Wang K, Choi J, Li S, Hong S, Zhang R, Xie J, Kornilov SA, Scherler K, Pavlovitch-Bedzyk AJ, Dong S, Lausted C, Lee I, Fallen S, Dai CL, Baloni P, Smith B, Duvvuri VR, Anderson KG, Li J, Yang F, Duncombe CJ, McCulloch DJ, Rostomily C, Troisch P, Zhou J, Mackay S, DeGottardi Q, May DH, Taniguchi R, Gittelman RM, Klinger M, Snyder TM, Roper R, Wojciechowska G, Murray K, Edmark R, Evans S, Jones L, Zhou Y, Rowen L, Liu R, Chour W, Algren HA, Berrington WR, Wallick JA, Cochran RA, Micikas ME, Unit IS-SC-B, Wrin T, Petropoulos CJ, Cole HR, Fischer TD, Wei W, Hoon DSB, Price ND, Subramanian N, Hill JA, Hadlock J, Magis AT, Ribas A, Lanier LL, Boyd SD, Bluestone JA, Chu H, Hood L, Gottardo R, Greenberg PD, Davis MM, Goldman JD, Heath JR, Multiple early factors anticipate post-acute COVID-19 sequelae. Cell 185, 881–895 e820 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durstenfeld MS, Peluso MJ, Kelly JD, Win S, Swaminathan S, Li D, Arechiga VM, Zepeda V, Sun K, Shao S, Hill C, Arreguin MI, Lu S, Hoh R, Tai V, Chenna A, Yee BC, Winslow JW, Petropoulos CJ, Kornak J, Henrich TJ, Martin JN, Deeks SG, Hsue PY, Role of antibodies, inflammatory markers, and echocardiographic findings in postacute cardiopulmonary symptoms after SARS-CoV-2 infection. JCI Insight 7, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giron LB, Peluso MJ, Ding J, Kenny G, Zilberstein NF, Koshy J, Hong KY, Rasmussen H, Miller GE, Bishehsari F, Balk RA, Moy JN, Hoh R, Lu S, Goldman AR, Tang HY, Yee BC, Chenna A, Winslow JW, Petropoulos CJ, Kelly JD, Wasse H, Martin JN, Liu Q, Keshavarzian A, Landay A, Deeks SG, Henrich TJ, Abdel-Mohsen M, Markers of fungal translocation are elevated during post-acute sequelae of SARS-CoV-2 and induce NF-kappaB signaling. JCI Insight 7, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peluso MJ, Deitchman AN, Torres L, Iyer NS, Munter SE, Nixon CC, Donatelli J, Thanh C, Takahashi S, Hakim J, Turcios K, Janson O, Hoh R, Tai V, Hernandez Y, Fehrman EA, Spinelli MA, Gandhi M, Trinh L, Wrin T, Petropoulos CJ, Aweeka FT, Rodriguez-Barraquer I, Kelly JD, Martin JN, Deeks SG, Greenhouse B, Rutishauser RL, Henrich TJ, Long-term SARS-CoV-2-specific immune and inflammatory responses in individuals recovering from COVID-19 with and without post-acute symptoms. Cell Rep 36, 109518 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peluso MJ, Lu S, Tang AF, Durstenfeld MS, Ho HE, Goldberg SA, Forman CA, Munter SE, Hoh R, Tai V, Chenna A, Yee BC, Winslow JW, Petropoulos CJ, Greenhouse B, Hunt PW, Hsue PY, Martin JN, Daniel Kelly J, Glidden DV, Deeks SG, Henrich TJ, Markers of Immune Activation and Inflammation in Individuals With Postacute Sequelae of Severe Acute Respiratory Syndrome Coronavirus 2 Infection. J Infect Dis 224, 1839–1848 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peluso MJ, Sans HM, Forman CA, Nylander AN, Ho HE, Lu S, Goldberg SA, Hoh R, Tai V, Munter SE, Chenna A, Yee BC, Winslow JW, Petropoulos CJ, Martin JN, Kelly JD, Durstenfeld MS, Hsue PY, Hunt PW, Greene M, Chow FC, Hellmuth J, Henrich TJ, Glidden DV, Deeks SG, Plasma Markers of Neurologic Injury and Inflammation in People With Self-Reported Neurologic Postacute Sequelae of SARS-CoV-2 Infection. Neurol Neuroimmunol Neuroinflamm 9, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein J, Wood J, Jaycox J, Lu P, Dhodapkar RM, Gehlhausen JR, Tabachnikova A, Tabacof L, Malik AA, Kamath K, Greene K, Monteiro VS, Pena-Hernandez M, Mao T, Bhattacharjee B, Takahashi T, Lucas C, Silva J, McCarthy D, Breyman E, Tosto-Mancuso J, Dai Y, Perotti E, Akduman K, Tzeng TJ, Xu L, Yildirim I, Krumholz HM, Shon J, Medzhitov R, Omer SB, van Dijk D, Ring AM, Putrino D, Iwasaki A, Distinguishing features of Long COVID identified through immune profiling. medRxiv, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ong SWX, Fong SW, Young BE, Chan YH, Lee B, Amrun SN, Chee RS, Yeo NK, Tambyah P, Pada S, Tan SY, Ding Y, Renia L, Leo YS, Ng LFP, Lye DC, Persistent Symptoms and Association With Inflammatory Cytokine Signatures in Recovered Coronavirus Disease 2019 Patients. Open Forum Infect Dis 8, ofab156 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultheiss C, Willscher E, Paschold L, Gottschick C, Klee B, Henkes SS, Bosurgi L, Dutzmann J, Sedding D, Frese T, Girndt M, Holl JI, Gekle M, Mikolajczyk R, Binder M, The IL-1beta, IL-6, and TNF cytokine triad is associated with post-acute sequelae of COVID-19. Cell Rep Med 3, 100663 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phetsouphanh C, Darley DR, Wilson DB, Howe A, Munier CML, Patel SK, Juno JA, Burrell LM, Kent SJ, Dore GJ, Kelleher AD, Matthews GV, Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat Immunol 23, 210–216 (2022). [DOI] [PubMed] [Google Scholar]
- 18.Yin K, Peluso MJ, Luo X, Thomas R, Shin MG, Neidleman J, Andrew A, Young KC, Ma T, Hoh R, Anglin K, Huang B, Argueta U, Lopez M, Valdivieso D, Asare K, Deveau TM, Munter SE, Ibrahim R, Standker L, Lu S, Goldberg SA, Lee SA, Lynch KL, Kelly JD, Martin JN, Munch J, Deeks SG, Henrich TJ, Roan NR, Long COVID manifests with T cell dysregulation, inflammation and an uncoordinated adaptive immune response to SARS-CoV-2. Nat Immunol, (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodruff MC, Bonham KS, Anam FA, Walker TA, Faliti CE, Ishii Y, Kaminski CY, Ruunstrom MC, Cooper KR, Truong AD, Dixit AN, Han JE, Ramonell RP, Haddad NS, Rudolph ME, Yalavarthi S, Betin V, Natoli T, Navaz S, Jenks SA, Zuo Y, Knight JS, Khosroshahi A, Lee FE, Sanz I, Chronic inflammation, neutrophil activity, and autoreactivity splits long COVID. Nat Commun 14, 4201 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruger A, Vlok M, Turner S, Venter C, Laubscher GJ, Kell DB, Pretorius E, Proteomics of fibrin amyloid microclots in long COVID/post-acute sequelae of COVID-19 (PASC) shows many entrapped pro-inflammatory molecules that may also contribute to a failed fibrinolytic system. Cardiovasc Diabetol 21, 190 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pretorius E, Venter C, Laubscher GJ, Kotze MJ, Oladejo SO, Watson LR, Rajaratnam K, Watson BW, Kell DB, Prevalence of symptoms, comorbidities, fibrin amyloid microclots and platelet pathology in individuals with Long COVID/Post-Acute Sequelae of COVID-19 (PASC). Cardiovasc Diabetol 21, 148 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pretorius E, Venter C, Laubscher GJ, Lourens PJ, Steenkamp J, Kell DB, Prevalence of readily detected amyloid blood clots in ‘unclotted’ Type 2 Diabetes Mellitus and COVID-19 plasma: a preliminary report. Cardiovasc Diabetol 19, 193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pretorius E, Vlok M, Venter C, Bezuidenhout JA, Laubscher GJ, Steenkamp J, Kell DB, Persistent clotting protein pathology in Long COVID/Post-Acute Sequelae of COVID-19 (PASC) is accompanied by increased levels of antiplasmin. Cardiovasc Diabetol 20, 172 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cervia-Hasler C, Bruningk SC, Hoch T, Fan B, Muzio G, Thompson RC, Ceglarek L, Meledin R, Westermann P, Emmenegger M, Taeschler P, Zurbuchen Y, Pons M, Menges D, Ballouz T, Cervia-Hasler S, Adamo S, Merad M, Charney AW, Puhan M, Brodin P, Nilsson J, Aguzzi A, Raeber ME, Messner CB, Beckmann ND, Borgwardt K, Boyman O, Persistent complement dysregulation with signs of thromboinflammation in active Long Covid. Science 383, eadg7942 (2024). [DOI] [PubMed] [Google Scholar]
- 25.Gold JE, Okyay RA, Licht WE, Hurley DJ, Investigation of Long COVID Prevalence and Its Relationship to Epstein-Barr Virus Reactivation. Pathogens 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peluso MJ, Deveau TM, Munter SE, Ryder D, Buck A, Beck-Engeser G, Chan F, Lu S, Goldberg SA, Hoh R, Tai V, Torres L, Iyer NS, Deswal M, Ngo LH, Buitrago M, Rodriguez A, Chen JY, Yee BC, Chenna A, Winslow JW, Petropoulos CJ, Deitchman AN, Hellmuth J, Spinelli MA, Durstenfeld MS, Hsue PY, Kelly JD, Martin JN, Deeks SG, Hunt PW, Henrich TJ, Chronic viral coinfections differentially affect the likelihood of developing long COVID. J Clin Invest 133, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bodansky A, Wang CY, Saxena A, Mitchell A, Takahashi S, Anglin K, Huang B, Hoh R, Lu S, Goldberg SA, Romero J, Tran B, Kirtikar R, Grebe H, So M, Greenhouse B, Durstenfeld MS, Hsue PY, Hellmuth J, Kelly JD, Martin JN, Anderson MS, Deeks SG, Henrich TJ, DeRisi JL, Peluso MJ, Autoantigen profiling reveals a shared post-COVID signature in fully recovered and Long COVID patients. medRxiv, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woodruff MC, Ramonell RP, Haddad NS, Anam FA, Rudolph ME, Walker TA, Truong AD, Dixit AN, Han JE, Cabrera-Mora M, Runnstrom MC, Bugrovsky R, Hom J, Connolly EC, Albizua I, Javia V, Cashman KS, Nguyen DC, Kyu S, Singh Saini A, Piazza M, Tipton CM, Khosroshahi A, Gibson G, Martin GS, Maier CL, Esper A, Jenks SA, Lee FE, Sanz I, Dysregulated naive B cells and de novo autoreactivity in severe COVID-19. Nature 611, 139–147 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Son K, Jamil R, Chowdhury A, Mukherjee M, Venegas C, Miyasaki K, Zhang K, Patel Z, Salter B, Yuen ACY, Lau KS, Cowbrough B, Radford K, Huang C, Kjarsgaard M, Dvorkin-Gheva A, Smith J, Li QZ, Waserman S, Ryerson CJ, Nair P, Ho T, Balakrishnan N, Nazy I, Bowdish DME, Svenningsen S, Carlsten C, Mukherjee M, Circulating anti-nuclear autoantibodies in COVID-19 survivors predict long COVID symptoms. Eur Respir J 61, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tesch F, Ehm F, Vivirito A, Wende D, Batram M, Loser F, Menzer S, Jacob J, Roessler M, Seifert M, Kind B, Konig C, Schulte C, Buschmann T, Hertle D, Ballesteros P, Bassler S, Bertele B, Bitterer T, Riederer C, Sobik F, Reitzle L, Scheidt-Nave C, Schmitt J, Incident autoimmune diseases in association with SARS-CoV-2 infection: a matched cohort study. Clin Rheumatol, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang R, Yen-Ting Chen T, Wang SI, Hung YM, Chen HY, Wei CJ, Risk of autoimmune diseases in patients with COVID-19: A retrospective cohort study. EClinicalMedicine 56, 101783 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaebler C, Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Tokuyama M, Cho A, Jankovic M, Schaefer-Babajew D, Oliveira TY, Cipolla M, Viant C, Barnes CO, Bram Y, Breton G, Hagglof T, Mendoza P, Hurley A, Turroja M, Gordon K, Millard KG, Ramos V, Schmidt F, Weisblum Y, Jha D, Tankelevich M, Martinez-Delgado G, Yee J, Patel R, Dizon J, Unson-O’Brien C, Shimeliovich I, Robbiani DF, Zhao Z, Gazumyan A, Schwartz RE, Hatziioannou T, Bjorkman PJ, Mehandru S, Bieniasz PD, Caskey M, Nussenzweig MC, Evolution of antibody immunity to SARS-CoV-2. Nature 591, 639–644 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zollner A, Koch R, Jukic A, Pfister A, Meyer M, Rossler A, Kimpel J, Adolph TE, Tilg H, Postacute COVID-19 is Characterized by Gut Viral Antigen Persistence in Inflammatory Bowel Diseases. Gastroenterology 163, 495–506 e498 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Natarajan A, Zlitni S, Brooks EF, Vance SE, Dahlen A, Hedlin H, Park RM, Han A, Schmidtke DT, Verma R, Jacobson KB, Parsonnet J, Bonilla HF, Singh U, Pinsky BA, Andrews JR, Jagannathan P, Bhatt AS, Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA suggest prolonged gastrointestinal infection. Med (N Y) 3, 371–387 e379 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swank Z, Senussi Y, Manickas-Hill Z, Yu XG, Li JZ, Alter G, Walt DR, Persistent Circulating Severe Acute Respiratory Syndrome Coronavirus 2 Spike Is Associated With Post-acute Coronavirus Disease 2019 Sequelae. Clin Infect Dis 76, e487–e490 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stein SR, Ramelli SC, Grazioli A, Chung JY, Singh M, Yinda CK, Winkler CW, Sun J, Dickey JM, Ylaya K, Ko SH, Platt AP, Burbelo PD, Quezado M, Pittaluga S, Purcell M, Munster VJ, Belinky F, Ramos-Benitez MJ, Boritz EA, Lach IA, Herr DL, Rabin J, Saharia KK, Madathil RJ, Tabatabai A, Soherwardi S, McCurdy MT, Consortium NC-A, Peterson KE, Cohen JI, de Wit E, Vannella KM, Hewitt SM, Kleiner DE, Chertow DS, SARS-CoV-2 infection and persistence in the human body and brain at autopsy. Nature 612, 758–763 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheung CCL, Goh D, Lim X, Tien TZ, Lim JCT, Lee JN, Tan B, Tay ZEA, Wan WY, Chen EX, Nerurkar SN, Loong S, Cheow PC, Chan CY, Koh YX, Tan TT, Kalimuddin S, Tai WMD, Ng JL, Low JG, Yeong J, Lim KH, Residual SARS-CoV-2 viral antigens detected in GI and hepatic tissues from five recovered patients with COVID-19. Gut 71, 226–229 (2022). [DOI] [PubMed] [Google Scholar]
- 38.Craddock V, Mahajan A, Spikes L, Krishnamachary B, Ram AK, Kumar A, Chen L, Chalise P, Dhillon NK, Persistent circulation of soluble and extracellular vesicle-linked Spike protein in individuals with postacute sequelae of COVID-19. J Med Virol 95, e28568 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peluso MJ, Deeks SG, Mustapic M, Kapogiannis D, Henrich TJ, Lu S, Goldberg SA, Hoh R, Chen JY, Martinez EO, Kelly JD, Martin JN, Goetzl EJ, SARS-CoV-2 and Mitochondrial Proteins in Neural-Derived Exosomes of COVID-19. Ann Neurol 91, 772–781 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Proal AD, VanElzakker MB, Aleman S, Bach K, Boribong BP, Buggert M, Cherry S, Chertow DS, Davies HE, Dupont CL, Deeks SG, Eimer W, Ely EW, Fasano A, Freire M, Geng LN, Griffin DE, Henrich TJ, Iwasaki A, Izquierdo-Garcia D, Locci M, Mehandru S, Painter MM, Peluso MJ, Pretorius E, Price DA, Putrino D, Scheuermann RH, Tan GS, Tanzi RE, VanBrocklin HF, Yonker LM, Wherry EJ, SARS-CoV-2 reservoir in post-acute sequelae of COVID-19 (PASC). Nat Immunol 24, 1616–1627 (2023). [DOI] [PubMed] [Google Scholar]
- 41.Sherif ZA, Gomez CR, Connors TJ, Henrich TJ, Reeves WB, Force RMPT, Pathogenic mechanisms of post-acute sequelae of SARS-CoV-2 infection (PASC). Elife 12, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henrich TJ, Hsue PY, VanBrocklin H, Seeing Is Believing: Nuclear Imaging of HIV Persistence. Front Immunol 10, 2077 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henrich TJ, Jones T, Beckford-Vera D, Price PM, VanBrocklin HF, Total-Body PET Imaging in Infectious Diseases. PET Clin 16, 89–97 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verger A, Barthel H, Tolboom N, Fraioli F, Cecchin D, Albert NL, van Berckel B, Boellaard R, Brendel M, Ekmekcioglu O, Semah F, Traub-Weidinger T, van de Weehaeghe D, Morbelli S, Guedj E, 2-[(18)F]-FDG PET for imaging brain involvement in patients with long COVID: perspective of the EANM Neuroimaging Committee. Eur J Nucl Med Mol Imaging 49, 3599–3606 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Debs P, Khalili N, Solnes L, Al-Zaghal A, Sair HI, Yedavalli V, Luna LP, Post-COVID-19 Brain [(18)F] FDG-PET Findings: A Retrospective Single-Center Study in the United States. AJNR Am J Neuroradiol 44, 517–522 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferrucci R, Cuffaro L, Capozza A, Rosci C, Maiorana N, Groppo E, Reitano MR, Poletti B, Ticozzi N, Tagliabue L, Silani V, Priori A, Brain positron emission tomography (PET) and cognitive abnormalities one year after COVID-19. J Neurol 270, 1823–1834 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martini AL, Carli G, Kiferle L, Piersanti P, Palumbo P, Morbelli S, Calcagni ML, Perani D, Sestini S, Time-dependent recovery of brain hypometabolism in neuro-COVID-19 patients. Eur J Nucl Med Mol Imaging 50, 90–102 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen LL, van de Burgt A, Smit F, Audhoe RS, de Boer SM, van Velden FHP, de Geus-Oei LF, Investigating the potential added value of [ 18 F]FDG-PET/CT in long COVID patients with persistent symptoms: a proof of concept study. Nucl Med Commun 44, 495–501 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goehringer F, Bruyere A, Doyen M, Bevilacqua S, Charmillon A, Heyer S, Verger A, Brain (18)F-FDG PET imaging in outpatients with post-COVID-19 conditions: findings and associations with clinical characteristics. Eur J Nucl Med Mol Imaging 50, 1084–1089 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Nardo L, Spencer BA, Abdelhafez YG, Li EJ, Omidvari N, Chaudhari AJ, Badawi RD, Jones T, Cherry SR, Wang G, Total-Body Multiparametric PET Quantification of (18) F-FDG Delivery and Metabolism in the Study of COVID-19 Recovery. medRxiv, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Omidvari N, Jones T, Price PM, Ferre AL, Lu J, Abdelhafez YG, Sen F, Cohen SH, Schmiedehausen K, Badawi RD, Shacklett BL, Wilson I, Cherry SR, First-in-human immunoPET imaging of COVID-19 convalescent patients using dynamic total-body PET and a CD8-targeted minibody. medRxiv, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levi J, Lam T, Goth SR, Yaghoubi S, Bates J, Ren G, Jivan S, Huynh TL, Blecha JE, Khattri R, Schmidt KF, Jennings D, VanBrocklin H, Imaging of Activated T Cells as an Early Predictor of Immune Response to Anti-PD-1 Therapy. Cancer Res 79, 3455–3465 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ronald JA, Kim BS, Gowrishankar G, Namavari M, Alam IS, D’Souza A, Nishikii H, Chuang HY, Ilovich O, Lin CF, Reeves R, Shuhendler A, Hoehne A, Chan CT, Baker J, Yaghoubi SS, VanBrocklin HF, Hawkins R, Franc BL, Jivan S, Slater JB, Verdin EF, Gao KT, Benjamin J, Negrin R, Gambhir SS, A PET Imaging Strategy to Visualize Activated T Cells in Acute Graft-versus-Host Disease Elicited by Allogenic Hematopoietic Cell Transplant. Cancer Res 77, 2893–2902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levi J, Duan H, Yaghoubi S, Packiasamy J, Huynh L, Lam T, Shaikh F, Behera D, Song H, Blecha J, Jivan S, Seo Y, VanBrocklin HF, Biodistribution of a Mitochondrial Metabolic Tracer, [(18)F]F-AraG, in Healthy Volunteers. Mol Imaging 2022, 3667417 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peluso MJ, Kelly JD, Lu S, Goldberg SA, Davidson MC, Mathur S, Durstenfeld MS, Spinelli MA, Hoh R, Tai V, Fehrman EA, Torres L, Hernandez Y, Williams MC, Arreguin MI, Ngo LH, Deswal M, Munter SE, Martinez EO, Anglin KA, Romero MD, Tavs J, Rugart PR, Chen JY, Sans HM, Murray VW, Ellis PK, Donohue KC, Massachi JA, Weiss JO, Mehdi I, Pineda-Ramirez J, Tang AF, Wenger MA, Assenzio MT, Yuan Y, Krone MR, Rutishauser RL, Rodriguez-Barraquer I, Greenhouse B, Sauceda JA, Gandhi M, Scheffler AW, Hsue PY, Henrich TJ, Deeks SG, Martin JN, Persistence, Magnitude, and Patterns of Postacute Symptoms and Quality of Life Following Onset of SARS-CoV-2 Infection: Cohort Description and Approaches for Measurement. Open Forum Infect Dis 9, ofab640 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hodcroft EB. 2021. “CoVariants: SARS-CoV-2 Mutations and Variants of Interest.” https://covariants.org/.
- 57.Lee JH, Yim JJ, Park J, Pulmonary function and chest computed tomography abnormalities 6–12 months after recovery from COVID-19: a systematic review and meta-analysis. Respir Res 23, 233 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Namavari M, Chang YF, Kusler B, Yaghoubi S, Mitchell BS, Gambhir SS, Synthesis of 2’-deoxy-2’-[18F]fluoro-9-beta-D-arabinofuranosylguanine: a novel agent for imaging T-cell activation with PET. Mol Imaging Biol 13, 812–818 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Rodriguez CO Jr., Mitchell BS, Ayres M, Eriksson S, Gandhi V, Arabinosylguanine is phosphorylated by both cytoplasmic deoxycytidine kinase and mitochondrial deoxyguanosine kinase. Cancer Res 62, 3100–3105 (2002). [PubMed] [Google Scholar]
- 60.Guglielmetti C, Levi J, Huynh TL, Tiret B, Blecha J, Tang R, VanBrocklin H, Chaumeil MM, Longitudinal Imaging of T Cells and Inflammatory Demyelination in a Preclinical Model of Multiple Sclerosis Using (18)F-FAraG PET and MRI. J Nucl Med 63, 140–146 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levi J, Goth S, Huynh L, Lam T, Huynh TL, Schulte B, Packiasamy JA, (18)F-AraG PET for CD8 Profiling of Tumors and Assessment of Immunomodulation by Chemotherapy. J Nucl Med 62, 802–807 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kinahan PE, Fletcher JW, Positron emission tomography-computed tomography standardized uptake values in clinical practice and assessing response to therapy. Semin Ultrasound CT MR 31, 496–505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCorkell L, Peluso MJ, Long COVID research risks losing momentum - we need a moonshot. Nature 622, 457–460 (2023). [DOI] [PubMed] [Google Scholar]
- 64.Soriano JB, Murthy S, Marshall JC, Relan P, Diaz JV, WHO Clinical Case Definition Working Group on Post-COVID-19 Condition, A clinical case definition of post-COVID-19 condition by a Delphi consensus. Lancet Infect Dis 22, e102–e107 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bennet AM, Reynolds CA, Eriksson UK, Hong MG, Blennow K, Gatz M, Alexeyenko A, Pedersen NL, Prince JA, Genetic association of sequence variants near AGER/NOTCH4 and dementia. J Alzheimers Dis 24, 475–484 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang LC, Almazan G, Cdon, a cell surface protein, mediates oligodendrocyte differentiation and myelination. Glia 64, 1021–1033 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Vargas-Franco D, Kalra R, Draper I, Pacak CA, Asakura A, Kang PB, The Notch signaling pathway in skeletal muscle health and disease. Muscle Nerve 66, 530–544 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ogasawara M, Nishino I, A review of core myopathy: central core disease, multiminicore disease, dusty core disease, and core-rod myopathy. Neuromuscul Disord 31, 968–977 (2021). [DOI] [PubMed] [Google Scholar]
- 69.Jakovcevski M, Akbarian S, Di Benedetto B, Pharmacological modulation of astrocytes and the role of cell type-specific histone modifications for the treatment of mood disorders. Curr Opin Pharmacol 26, 61–66 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ryan FJ, Hope CM, Masavuli MG, Lynn MA, Mekonnen ZA, Yeow AEL, Garcia-Valtanen P, Al-Delfi Z, Gummow J, Ferguson C, O’Connor S, Reddi BAJ, Hissaria P, Shaw D, Kok-Lim C, Gleadle JM, Beard MR, Barry SC, Grubor-Bauk B, Lynn DJ, Long-term perturbation of the peripheral immune system months after SARS-CoV-2 infection. BMC Med 20, 26 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peluso MJ, Donatelli J, Henrich TJ, Long-term immunologic effects of SARS-CoV-2 infection: leveraging translational research methodology to address emerging questions. Transl Res, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo Y, Wang B, Gao H, Gao L, Hua R, Xu JD, ACE2 in the Gut: The Center of the 2019-nCoV Infected Pathology. Front Mol Biosci 8, 708336 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patterson BK, Francisco EB, Yogendra R, Long E, Pise A, Rodrigues H, Hall E, Herrera M, Parikh P, Guevara-Coto J, Triche TJ, Scott P, Hekmati S, Maglinte D, Chang X, Mora-Rodriguez RA, Mora J, Persistence of SARS CoV-2 S1 Protein in CD16+ Monocytes in Post-Acute Sequelae of COVID-19 (PASC) up to 15 Months Post-Infection. Front Immunol 12, 746021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Singh K, Natarajan V, Dewar R, Rupert A, Badralmaa Y, Zhai T, Winchester N, Scrimieri F, Smith M, Davis I, Lallemand P, Giglietti A, Hensien J, Buerkert T, Goshu B, Rehm CA, Hu Z, Lane HC, Imamichi H, Long-term persistence of transcriptionally active ‘defective’ HIV-1 proviruses: implications for persistent immune activation during antiretroviral therapy. AIDS 37, 2119–2130 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dube M, Tastet O, Dufour C, Sannier G, Brassard N, Delgado GG, Pagliuzza A, Richard C, Nayrac M, Routy JP, Prat A, Estes JD, Fromentin R, Chomont N, Kaufmann DE, Spontaneous HIV expression during suppressive ART is associated with the magnitude and function of HIV-specific CD4(+) and CD8(+) T cells. Cell Host Microbe 31, 1507–1522 e1505 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peluso MJ, Swank ZN, Goldberg SA, Lu S, Dalhuisen T, Borberg E, Senussi Y, Luna MA, Chang Song C, Clark A, Zamora A, Lew M, Viswanathan B, Huang B, Anglin K, Hoh R, Hsue PY, Durstenfeld MS, Spinelli MA, Glidden DV, Henrich TJ, Kelly JD, Deeks SG, Walt DR, Martin JN, Plasma-based antigen persistence in the post-acute phase of COVID-19. Lancet Infect. Dis. (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vasquez JJ, Aguilar-Rodriguez BL, Rodriguez L, Hogan LE, Somsouk M, McCune JM, Deeks SG, Laszik ZG, Hunt PW, Henrich TJ, CD32-RNA Co-localizes with HIV-RNA in CD3+ Cells Found within Gut Tissues from Viremic and ART-Suppressed Individuals. Pathog Immun 4, 147–160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Park LM, Lannigan J, Jaimes MC, OMIP-069: Forty-color full spectrum flow cytometry panel for deep immunophenotyping of major cell subsets in human peripheral blood. Cytometry A 97, 1044–1051 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are in the paper or supplementary materials. Source data without participant-level identifying information for analyses and figures will be made available upon request of the communicating authors.


