Abstract
Two members of the NF-κB (nuclear factor κB)/Rel transcription factor family, NF-κB1 and NF-κB2, are produced as precursor proteins, NF-κB1 p105 and NF-κB2 p100 respectively. These are proteolytically processed by the proteasome to produce the mature transcription factors NF-κB1 p50 and NF-κB2 p52. p105 and p100 are known to function additionally as IκBs (inhibitors of NF-κB), which retain associated NF-κB subunits in the cytoplasm of unstimulated cells. The present review focuses on the latest advances in research on the function of NF-κB1 and NF-κB2 in immune cells. NF-κB2 p100 processing has recently been shown to be stimulated by a subset of NF-κB inducers, including lymphotoxin-β, B-cell activating factor and CD40 ligand, via a novel signalling pathway. This promotes the nuclear translocation of p52-containing NF-κB dimers, which regulate peripheral lymphoid organogenesis and B-lymphocyte differentiation. Increased p100 processing also contributes to the malignant phenotype of certain T- and B-cell lymphomas. NF-κB1 has a distinct function from NF-κB2, and is important in controlling lymphocyte and macrophage function in immune and inflammatory responses. In contrast with p100, p105 is constitutively processed to p50. However, after stimulation with agonists, such as tumour necrosis factor-α and lipopolysaccharide, p105 is completely degraded by the proteasome. This releases associated p50, which translocates into the nucleus to modulate target gene expression. p105 degradation also liberates the p105-associated MAP kinase (mitogen-activated protein kinase) kinase kinase TPL-2 (tumour progression locus-2), which can then activate the ERK (extracellular-signal-regulated kinase)/MAP kinase cascade. Thus, in addition to its role in NF-κB activation, p105 functions as a regulator of MAP kinase signalling.
Keywords: IκB kinase (IKK), nuclear factor κB (NF-κB), p100, p105, Toll-like receptor (TLR), tumour progression locus-2 (TPL-2)
Abbreviations: ABIN, A20-binding inhibitor of nuclear factor κB; BAFF, B-cell activating factor; BMDM, bone-marrow-derived macrophage; βTrCP, β-transducin repeat-containing protein; COX-2, cyclo-oxygenase-2; DC, dendritic cell; DD, death domain; Dif, Dorsal-related immunity factor; EBNA1, EBV nuclear antigen 1; EBV, Epstein–Barr virus; ERK, extracellular-signal-regulated kinase; Fn14, fibroblast-growth-factor-inducible 14; GC, germinal centre; GM-CSF, granulocyte–macrophage colony-stimulating factor; GRR, glycine-rich region; GSK, glycogen synthase kinase; HTLV-1, human T-cell leukaemia virus type 1; IFNβ, interferon-β; IκB, inhibitor of nuclear factor κB; IKK, IκB kinase; IL, interleukin; Imd, immune deficiency; JNK, c-Jun N-terminal kinase; LMP1, latent membrane protein 1; LPS, lipopolysaccharide; LTβR, lymphotoxin-β receptor; MAP kinase, mitogen-activated protein kinase; MAP 3-kinase, MAP kinase kinase kinase; MEF, mouse embryo fibroblast; MEK, MAP kinase/ERK kinase; MIP, macrophage inflammatory protein; NEMO, nuclear factor κB essential modulator; NF-κB, nuclear factor κB; NIK, NF-κB-inducing kinase; PEST region, polypeptide sequence enriched in proline (P), glutamic acid (E), serine (S) and threonine (T); PGRP-LC, peptidoglycan recognition protein LC; RANKL, receptor activator of NF-κB ligand; RHD, Rel homology domain; SCF, Skp1/Cul1/F-box; TH1, T-helper 1; TH2, T-helper 2; TLR, Toll-like receptor; TNF, tumour necrosis factor; TPL-2, tumour progression locus-2; TRAF, TNF-receptor-associated factor; TWEAK, TNF-like weak inducer of apoptosis
INTRODUCTION
NF-κB (nuclear factor κB) transcription factors play an important role in the regulation of immune and inflammatory responses. Upon infection, pathogenic micro-organisms activate NF-κB transcription factors via triggering of TLRs (Toll-like receptors), which are expressed on cells of the innate immune system, including macrophages, DCs (dendritic cells) and mucosal epithelial cells [1]. TLRs recognize invariant microbial molecules, including components of the bacterial cell wall such as LPS (lipopolysaccharide) and microbial nucleic acids [2]. NF-κB induction is essential for the expression of a wide variety of immune-response genes. These include pro-inflammatory cytokines [e.g. TNFα (tumour necrosis factor-α), IL-1 and -6 (interleukin-1 and -6)], chemokines [e.g. MIP-1α (macrophage inflammatory protein-1α) and RANTES (regulated upon activation, normal T-cell expressed and secreted)] and adhesion molecules [e.g. E-selectin and VCAM-1 (vascular cell adhesion molecule-1)], which collectively regulate the recruitment of immune cells to sites of infection [3]. TNFα and IL-1 stimulation of their respective receptors in turn strongly activates NF-κB, which plays an important role in amplifying and extending the duration of the innate immune response [4,5]. Effector molecules which have direct microbicidal activity (e.g. defensins) and enzymes which generate reactive intermediates (e.g. inducible nitric oxide synthase) are also transcriptionally induced by NF-κB [3]. NF-κB facilitates T-cell activation via up-regulation of MHC proteins and CD80/86 on antigen-presenting cells, and therefore forms a molecular link between activation of the innate and adaptive immune systems [6]. Subsequently, during the adaptive phase of an immune response, NF-κB is involved in the activation of T- and B-lymphocytes by their antigen and co-stimulatory receptors [6]. In addition, NF-κB is required for LTβR (lymphotoxin-β receptor) regulation of peripheral lymphoid organogenesis [7] and stimulation of B-cell differentiation and survival by the receptor for B-cell activating factor (BAFF) [8].
Dysregulation of NF-κB can lead to the constitutive overproduction of pro-inflammatory cytokines, which is associated with a number of chronic inflammatory disorders, including rheumatoid arthritis and Crohn's disease [9,10]. NF-κB also plays an important role in regulating the expression of anti-apoptotic proteins (e.g. c-IAP-1/2, AI, Bcl-2 and Bcl-XL) and the cell-cycle regulator cyclin D1, which increase cellular survival and proliferation respectively [11,12]. Consequently NF-κB has been implicated in cell transformation [12], and persistent NF-κB activation may explain the known causative link between chronic inflammation and certain types of cancer [13,14]. A glossary of key terms used in this review is given in Table 1.
Table 1. Glossary of key terms used in this review.
| Protein | Function | Reference |
|---|---|---|
| TLRs | Recognition of invariant microbial molecules (e.g. TLR4/LPS). | [2] |
| Induction of innate immune response. | ||
| TNF receptor family (e.g. TNFR1, CD40, LTβR) | Regulation of development and function of immune system | [189] |
| TRAF proteins | Intracellular adapter proteins | [71] |
| MAP kinase pathway [MAP 3-K (e.g. TPL-2); MAP 2-kinase (e.g. MEK); MAP-kinase (e.g. ERK)] | Intracellular signalling cascade | [190] |
| IKK complex (IKK1/IKK2/NEMO) | Multisubunit protein complex; regulation of IκB proteolysis | [24] |
| IκB proteins (IκBα, IκBβ, IκBε, NF-κB1 p105, NF-κB2 p100) | NF-κB inhibitors | [15] |
| NF-κB (Rel) proteins (RelA, RelB, c-Rel, NF-κB1 p50, NF-κB2 p52) | Dimeric transcription factors | [15] |
| Ubiquitination | Covalent protein modification; regulates target protein stability or function | [27] |
| E3 ligase (e.g. SCFβTrCP) | Substrate recognition and ubiquitin transfer | [27] |
| Proteasome | Multisubunit protease; proteolysis of ubiquitinated proteins (e.g. IκBs) | [28] |
NF-κB AND IκB PROTEINS
NF-κB is composed of homo- and hetero-dimeric complexes of Rel family polypeptides, which are characterized by an N-terminal RHD (Rel homology domain), which mediates DNA binding, nuclear localization and subunit dimerization [15]. Mammals express five NF-κB proteins, namely RelA (p65), RelB, c-Rel, NF-κB1 p50 and NF-κB2 p52 (Figure 1A). NF-κB1 and NF-κB2 are synthesized as large precursors of 105 (p105) and 100 kDa (p100) respectively. These are partially proteolysed by the proteasome (termed processing), which removes their C-terminal halves, to produce the active NF-κB1 p50 and NF-κB2 p52 subunits. RelA and c-Rel both have C-terminal transactivation domains in their RHDs and can strongly activate transcription from NF-κB binding sites in target genes. RelB also contains a transactivation domain in its RHD and can function as an NF-κB activator when complexed with p50 or p52. However, RelB/RelA heterodimeric complexes are inhibitory, since they cannot bind DNA [16]. NF-κB1 p50 and NF-κB2 p52 lack a transactivation domain, and can only promote transcription when heterodimerized with a transactivating Rel subunit [6]. Homodimers of p50 and p52 function as transcriptional repressors, but can stimulate transcription when bound to the IκB-like nuclear protein BCL-3 [17–19].
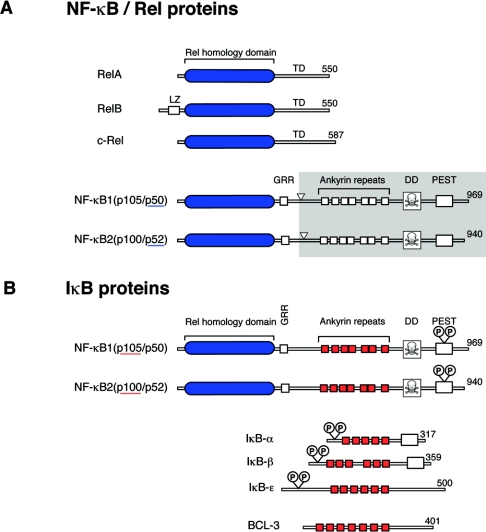
Figure 1. Schematic representation of members of the Rel/NF-κB (A) and IκB (B) families of proteins.
Rel/NF-κB proteins are characterized by their conserved N-terminal RHDs (blue) which mediate DNA binding, nuclear localization and subunit dimerization. IκB proteins each contain ankyrin repeat regions (red), which interact with RHDs of NF-κB subunits to prevent nuclear translocation. The numbers of amino acids in each protein are shown on the right. In (A), arrowheads point to the C-terminal residues of p50 and p52, which are produced by proteolytic processing of the C-terminal halves p105 and p100 respectively (grey shading). In (B) the positions of the IKK phosphorylation sites, which regulate the inducible proteolysis of IκB proteins, are shown. LZ, leucine zipper; TD, transactivation domain.
In unstimulated cells, NF-κB dimers are inactive, since they are sequestered in the cytoplasm by interaction with inhibitory proteins termed IκBs (inhibitors of NF-κB) [15]. In response to stimulation with agonists, IκBs are proteolytically degraded by the proteasome, releasing associated NF-κB dimers to translocate into the nucleus and modulate gene expression (Figure 2). The transcriptional activity of certain NF-κB dimers is additionally regulated by phosphorylation, providing an extra level of control of NF-κB activation [20].
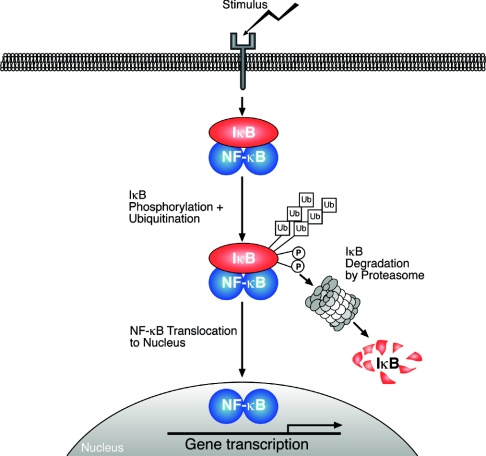
Figure 2. Model of the generic NF-κB activation pathway.
Various stimuli induce the phosphorylation ( ) and subsequent polyubiquitination (
) and subsequent polyubiquitination ( ) of IκBs, which are then targeted for degradation by the 26 S proteasome. Associated NF-κB dimers are thereby released to translocate into the nucleus, where they bind to the promoter regions of NF-κB-responsive genes to modulate their expression. The transactivating capacity of nuclear NF-κB dimers can also be regulated by phosphorylation.
) of IκBs, which are then targeted for degradation by the 26 S proteasome. Associated NF-κB dimers are thereby released to translocate into the nucleus, where they bind to the promoter regions of NF-κB-responsive genes to modulate their expression. The transactivating capacity of nuclear NF-κB dimers can also be regulated by phosphorylation.
IκBs comprise a family of structurally related proteins, each of which contain multiple ankyrin repeats that interact with the nuclear localizing signals of RHDs to prevent nuclear translocation of Rel subunits (Figure 1B). This family includes IκBα, IκBβ and IκBε, which are composed of a central ankyrin repeat region and an N-terminal regulatory domain, which controls their inducible degradation. The precursor NF-κB proteins, NF-κB1 p105 and NF-κB2 p100, also function as IκBs as a result of ankyrin repeat regions in their C-terminal halves. Alternate promoter usage of the NF-κB1 gene can generate an mRNA encoding only the C-terminal IκB-like portion of p105, termed IκBγ, but this has so far only been reported in murine cells [21,22].
REGULATION OF THE CANONICAL NF-κB PATHWAY BY THE IκB KINASE COMPLEX
The signalling pathway by which cytokines regulate the degradation of IκBα to release p50/RelA and p50/c-Rel heterodimers has been characterized in greatest detail (Figure 3, left) and is known as the ‘canonical’ NF-κB pathway [23]. A critical event in this pathway is the rapid agonist-induced phosphorylation of IκBα by the classical IKK (IκB kinase) complex. This comprises two kinase subunits, IKK1 (also known as IKKα) and IKK2 (IKKβ), and a structural subunit NEMO (NF-κB essential modulator; IKKγ), which is required to couple the IKK complex to upstream signals [24]. Of the two catalytic subunits, only IKK2 is essential for IκBα phosphorylation triggered by TNFα and IL-1. Activation of IKK2 by these stimuli has been shown to require the MAP 3-Ks (MAP kinase kinase kinases), MEKK3 [MAP kinase/ERK (extracellular-signal-regulated kinase) kinase kinase kinase kinase] and TAK1 (transforming-growth-factor-β-activated kinase), which are thought to directly phosphorylate the IKK2 activation loop [25,26]. IκBα is phosphorylated by IKK on two N-terminal serine residues, which creates a binding site for βTrCP (β-transducin repeat-containing protein), the receptor subunit of an SCF (Skp1/Cul1/F-box) ubiquitin E3 ligase complex [27]. This catalyses the rapid polyubiquitination of IκBα on two adjacent lysine residues, targeting it for degradation by the 26 S proteasome. This releases associated NF-κB dimers to translocate into the nucleus and modulate gene transcription [28]. Agonist-induced degradation of IκBβ and IκBε is similarly triggered by their N-terminal phosphorylation by the classical IKK complex, which promotes their βTrCP-mediated ubiquitination and subsequent proteolysis by the proteasome [6].
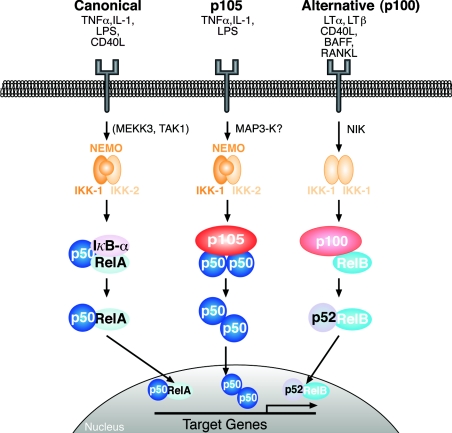
Figure 3. Model of different NF-κB signalling pathways.
The canonical pathway (left) is activated by a large number of agonists, a few of which are listed. Activation of this pathway depends on the IKK complex (IKK1–IKK2–NEMO), which phosphorylates IκBα to induce its rapid degradation. (IκBβ and IκBε are similarly regulated by IKK.) This pathway is essential for immune responses, inflammation and promoting cell survival. The alternative NF-κB pathway (right) is activated a limited number of agonists and is important for secondary lymphoid organogenesis, maturation of B-cells and adaptive humoral immunity. This pathway requires NIK and IKK1 and induces the slow processing of p100 to p52, resulting in nuclear translocation of p52/RelB heterodimers. The p105 pathway (centre) is specifically involved in immune and inflammatory responses. Agonist activation of this pathway induces phosphorylation of the p105 PEST region by the classical IKK complex. This triggers p105 polyubiquitination and subsequent degradation, releasing p50 homodimers which undergo nuclear translocation and positively or negatively regulate gene transcription.
DROSOPHILA HAS TWO DISTINCT NF-κB PATHWAYS
The NF-κB/IκB pathway is conserved in evolution, and innate immune defence in the fruitfly Drosophila is regulated by the NF-κB-related transcription factors Dorsal, Dif (Dorsal-related immunity factor) and Relish [29]. NF-κB activation in Drosophila is controlled by two distinct signalling pathways (Figure 4): the Toll pathway, which regulates Dif and Dorsal, and the Imd (immune deficiency) pathway, which regulates Relish [30]. These pathways are induced by specific microbial stimuli and predominantly regulate distinct target genes.
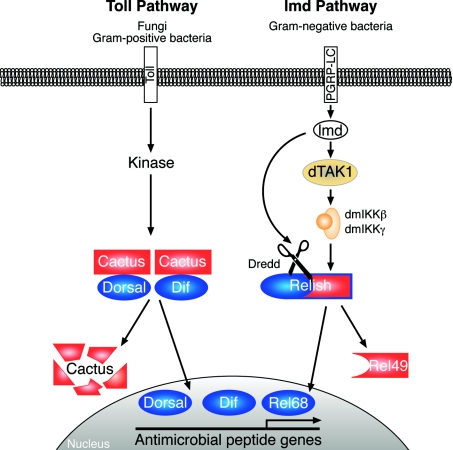
Figure 4. Drosophila Toll and Imd NF-κB signalling pathways.
Stimulation of the Toll pathway by fungi and Gram-positive bacteria induces Cactus phosphorylation by an unknown kinase, triggering Cactus degradation by the proteasome. This releases Dif and Dorsal NF-κB-like transcription factor to translocate into the nucleus and induce the expression of antimicrobial peptides directed against Gram-positive bacteria and fungi. The Imd pathway is activated by Gram-negative bacteria, which trigger a cell-membrane-bound peptidoglycan recognition protein, PGRP-LC. Downstream of this receptor, the Imd signalling pathway bifurcates. One branch triggers the activation of the Drosophila melanogaster (dm) IKK complex, via the MAP 3-kinase dTAK1, which then directly phosphorylates Relish. The other branch regulates the activation of Dredd caspase. This directly cleaves phosphorylated Relish, generating the N-terminal transcription factor fragment (Rel-68) that translocates into the nucleus and induces the expression of antimicrobial peptides directed against Gram-negative bacteria. The IκB-like C-terminal fragment, Rel-49, remains in the cytoplasm, but does not prevent nuclear translocation of Rel-68. Imd is an adapter protein, containing a DD, which is required for activation of both the IKK and Dredd branches. ANK, ankyrin repeat region.
In unstimulated cells, Dorsal and Dif are retained in the cytoplasm by their interaction with the IκB protein Cactus, which is homologous with mammalian IκBα. Stimulation of the Toll membrane receptor by fungi and Gram-positive bacteria induces phosphorylation of the N-terminus of Cactus by an unknown kinase, triggering Cactus degradation by the proteasome [31]. This releases Dif and Dorsal to translocate into the nucleus and induce expression of anti-microbial peptides active against these micro-organisms (e.g. drosomycin [30]). While only Dif is required for antifungal immunity in the adult fruitfly, either Dif or Dorsal can mediate immune responses in larvae. Dorsal is additionally required in early embryogenesis for the Toll-dependent patterning of the dorsoventral axis [32].
Relish has intrinsic IκB function, similar to NF-κB1 p105 and NF-κB2 p100, and is proteolytically processed to produce the active transcription factor Rel-68 [31]. The Imd pathway is activated by Gram-negative bacteria, which trigger cell-membrane-bound PGRP-LC (peptidoglycan recognition protein LC). PGRP-LC stimulation rapidly activates a Drosophila IKK complex (comprising an IKK2-related kinase, dIKKβ, and a structural subunit homologous with NEMO, dIKKγ) which directly phosphorylates Relish [33]. This facilitates Relish cleavage by the caspase Dredd to generate Rel-68, which translocates into the nucleus and induces the expression of antimicrobial peptides against Gram-negative bacteria (e.g. diptericin and attacins) [31,34,35]. The residual C-terminal fragment, Rel-49, remains in the cytoplasm, but does not prevent nuclear translocation of Rel-68. Given the conservation of NF-κB in evolution, it is not surprising that there are similarities in the regulation of Relish and the homologous mammalian proteins p100 and p105. These are highlighted in the remainder of the present review.
Recently, significant advances have been made in the understanding of the signalling pathways controlling proteolysis of the mammalian precursor proteins NF-κB1 p105 and NF-κB2 p100 and their specific functions. Our current knowledge and outstanding questions in this field are discussed in the following sections.
NF-κB2 p100/p52
Processing of p100 generates p52, which can translocate to the nucleus in association with other Rel subunits. An important role for p100 in the regulation of p52 has been inferred from analysis of mutant mice (nfkb2ΔC/ΔC) which lack full-length p100, but still express a functional p52 protein (Table 2; [36]). Such mice have a dramatic induction of p52-containing NF-κB binding activity in the nucleus and display increased lymphocyte proliferation, enlarged lymph nodes and gastric mucosal hyperplasia. However, one caveat with this mouse model is that all of the transcribed nfkb2 mRNA makes p52. This results in constitutively elevated p52 levels compared with wild-type mice, in which p52 is produced very inefficiently from p100. Therefore it is unclear whether the detected phenotype in nfkb2ΔC/ΔC mice results from lack of p100, overproduction of p52, or both.
Table 2. Phenotypes of gene knockout mice important for the delineation of the alternative NF-κB signalling pathway.
Abbreviations and symbols used: ↑, increase; ↓, decrease; =, unaffected; Ab, antibody; Ag, antigen; BM, bone marrow; ConA, concanavalin A; DC, dendritic cells; GC, germinal center; LN, lymph node; MEF, mouse embryo fibroblast; MZ, marginal zone; PP, Peyer's patches. Tissues and stimuli are emboldened for clarity.
| Peripheral lymphoid organs | T-cells | B-cells | Other comments | Reference(s) | |
|---|---|---|---|---|---|
| nf-κb2−/− (no p100/p52) | Spleen: architecture disrupted | αCD3, ConA: IL-2 ↑ | LPS, αCD40, αIgD: | LPS/macrophage: | [42,43,191] |
| MZ: absent | Splenic numbers ↑ | proliferation ↓ | IL-10 ↓ | ||
| LN: architecture disrupted | Numbers ↓ Ag-specific Ab ↓ | IL-12 ↑ | |||
| GC: absent | TNFα, IL6, NO= | ||||
| PP: absent | |||||
| nf-κb2ΔC/ΔC (no p100, high p52) | Spleen: atrophy | αCD3+αCD28: proliferation ↑ | LPS: proliferation ↑ | Gastric hyperplasia | [36] |
| LN: enlarged | IL-2, IL-4, IL-10, GM-CSF, TNFα ↑ | Die post-natally | |||
| relb−/− | Spleen: enlarged | Thymic atrophy | LPS, αCD40, αIg: proliferation ↓ | Multi-organ inflammation (T-cell-dependent) | |
| MZ: disrupted | |||||
| GC: absent | |||||
| LN: absent | |||||
| PP: absent | |||||
| bcl3−/− | Spleen: architecture disrupted | αCD3: proliferation= | Ag-specific Ab ↓ | Myeloid hyperplasia | [41,192–196] |
| LN: architecture disrupted | IFN-γ= | Splenic/thymic DC ↓ | Defective Ag priming of T-cells | [18,197,198] | |
| PP: size/numbers ↓ | Numbers ↓ Ag-specific Ab ↓ | ||||
| GC: absent | |||||
| nik−/− | Spleen: architecture disrupted | Ag-specific Ab ↓ | LTβR/MEF: | [48] | |
| LN: absent | NF-κB transactivation ↓ | ||||
| PP: absent | |||||
| aly/aly (NIKG855R, low p52, normal p100) | Spleen: architecture disrupted | αCD3: proliferation ↓ | BAFF, αCD40: p100 processing ↓ | LTβR/MEF: p100 processing ↓ | [8,45,47,49,57,58,61,199] |
| MZ: absent | IL-2 ↓ | ||||
| GC: absent | LPS, αCD40, αIgM: Proliferation ↓ | ||||
| LN: absent | |||||
| PP: absent | Maturation impaired Ag-specific Ab ↓ | ||||
| ikk1−/− (BM chimaeras) | Spleen: architecture disrupted | αCD3, ConA: proliferation= | LPS, αIgM: Proliferation ↓ | LTβR/MEF: p100 processing ↓ | [44,50,57] |
| GC: absent | |||||
| Mature B-cells ↓ | |||||
| Apoptosis ↑ | |||||
| p100 processing ↓ (basal) | |||||
| ikk1aa (IKK1SS176/180AA, kinase inactive) | GC: absent | Mature B-cells ↓ | LTβR/MEF: p100 processing ↓ | ||
| PP: absent | p100 processing ↓ (basal) | [44,57] | |||
| ltβr−/− | Spleen: architecture disrupted, | Ab affinity maturation ↓ | Lymphocyte infiltration (muliple organs); similar phenotype in ltα−/− and ltβ−/− mice | [200] | |
| MZ: absent | |||||
| GC: absent | |||||
| LN: absent | |||||
| PP: absent |
Early studies indicated that NF-κB2 p100 can retain p50, RelA and c-Rel in the cytoplasm [37,38]. However, more recently it has been shown that p100 is the sole IκB involved in the cytoplasmic retention of RelB and regulation of its transcriptional activity [39,40]. Consistently, nfkb2−/− and relb−/− mice have partially overlapping phenotypes (Table 2), with both mutant mice lacking lymph nodes, Peyer's patches and splenic GCs (germinal centres) [7,41–43]. Thus the regulation of RelB by NF-κB2 p100 appears to be important in controlling peripheral lymphoid organogenesis and GC formation.
THE ALTERNATIVE NF-κB PATHWAY
Research over the last 3 years has characterized a novel signalling pathway, activated by a subset of NF-κB agonists, which stimulates processing of p100 to p52 (Figure 3, right). This is distinct from the canonical NF-κB pathway, which regulates IκBα degradation, and has become known as the ‘alternative’ or ‘non-canonical’ NF-κB pathway [23].
The first clue that p100 processing might be regulated was reported by the laboratory of Shao-Cong Sun in 2001 [44], which demonstrated that p100 processing is stimulated by ectopic expression of NIK (NF-κB binding kinase). NIK has homology within its kinase domain with MAP kinase kinase kinases (MAP 3-Ks). Furthermore, splenocytes from the alymphoplasia (aly/aly) mutant mouse strain, which contains a mutation in NIK that blocks its ability to induce p100 processing in transfected cells, were shown to have dramatically reduced steady-state levels of p52, although p100 levels were normal [45,46]. Consistent with the hypothesis that NIK and p100 might be components of the same signalling pathway, aly/aly and nik−/− mice [47–49] have similar phenotypes to nfkb2−/− mice [42,43], each displaying a systemic absence of lymph nodes and Peyer's patches as well as disorganized splenic and thymic architectures (Table 2).
Induction of p100 processing to p52 by overexpressed NIK in fibroblasts requires IKK1 expression [44]. Thus IKK1 acts downstream of NIK in a signalling pathway that controls p100 proteolysis. In accordance with this hypothesis, peripheral lymphoid organogenesis is disrupted in IKK1−/− radiation chimaeras (Table 2; [44,50]). Furthermore, IKK1 was identified as a NIK-interacting protein in a yeast two-hybrid screen [51], and NIK can phosphorylate the activation loop of IKK1, stimulating its kinase activity [52]. In vitro experiments suggest that IKK1 directly phosphorylates p100 to induce its processing [44]. IKK2 and NEMO are not required for NIK-induced p100 processing [44,53], raising the possibility that the pool of IKK1 regulating p100 processing may not be a component of the IKK1–IKK2–NEMO complex that controls the degradation of IκBα. Indeed, it has been demonstrated that a fraction of IKK1 is not associated with either IKK2 or NEMO in murine fibroblasts [54]. It is not known whether this pool of IKK1 is complexed with other regulatory proteins.
RECEPTOR ACTIVATION OF THE ALTERNATIVE NF-κB PATHWAY
Analysis of LTβR knockout mice (Table 2) has indicated that LTβRs on non-haematopoietic and myeloid lineage cells are essential for the normal development of lymph nodes and Peyer's patches [55]. The absence of these peripheral lymphoid organs in aly/aly, nik−/−, ikk1−/− chimaeric, nfκb2−/− and relb−/− mice [7] suggests that NIK, IKK1, NF-κB2 and RelB act in a common signalling pathway downstream of LTβRs in stromal cells, which regulates peripheral lymphoid tissue organogenesis. Consistent with this hypothesis, anti-LTβR antibody induces processing of p100 to p52 in mouse embryonic fibroblasts (MEFs), resulting in nuclear translocation of p52/RelB heterodimers [41,56,57]. Furthermore, analysis of knockout MEFs indicates that NIK and IKK1 are required for LTβR induction of p100 processing, whereas IKK2 and NEMO are dispensable [41,56,57]. Interestingly, nuclear translocation of p50/RelB heterodimers in MEFs by anti-LTβR antibody also requires p100 processing, suggesting that p52/p50 exchange may occur after p100 processing [54].
Two studies have described cell autonomous defects in aly/aly B-cell function [45,58]. In addition, radiation chimaeras generated with ikk1−/− bone marrow or bone marrow from mice expressing a non-activatable IKK1 mutant (IKK1AA) have cell autonomous defects in B-cell maturation and p100 processing to p52 [44]. Since B-cells do not express LTβR [59], this raises the possibility that distinct receptors regulate p100 processing via NIK and IKK1 in B-cells. Two of these have been identified, namely BAFF-R [8,60] and CD40 [61], which trigger p100 processing via a NIK-dependent pathway. BAFF-induced p100 processing is important for promoting both survival and maturation of transition-1-stage splenic B-cells [8,62]. On the basis of a comparison of nfkb2−/− and wild-type splenic B-cells, it has recently been suggested that activation of the alternative pathway by CD40 is important for optimal promotion of cell survival and homotypic aggregation [63]. However, it is also possible that these phenotypes reflect functional differences in nfkb2−/− B-cells present prior to an encounter with CD40 ligand, rather than being due solely to the acute failure of CD40 to induce p100 processing.
RANKL (receptor activator of NF-κB ligand), which controls the differentiation of osteoclasts [64], induces processing of p100 to p52 in osteoclast precursors via a NIK-dependent pathway [65]. Furthermore, induction of osteoclastogenesis by RANKL is blocked in nik−/− cells and also by expresssion of a non-processable p100 mutant [65]. Thus the alternative NF-κB pathways also appears to play a role in bone formation. TWEAK (TNF-like weak inducer of apoptosis) [66] and LPS [67,68] also induce p100 processing, but the physiological significance of activation of the alternative pathway for these agonists is not known.
Several studies have shown TNFα stimulation does not induce p100 processing, although it strongly induces IκBα degradation [54,56,57,61]. However, TNFα stimulation does increase the expression of both p100 and RelB, which are transcriptional targets of the canonical NF-κB signalling pathway [41,54]. Thus the canonical NF-κB pathway is indirectly linked to the alternative NF-κB pathway and may influence the amplitude and duration of its activation.
KINETICS OF THE ALTERNATIVE AND CANONICAL NF-κB PATHWAYS
Time-course experiments have indicated that receptor induction of p100 processing is very slow, taking place over several hours. However, each of the identified receptors which trigger the alternative NF-κB pathway also induce IκBα degradation within minutes, although to different extents.
The kinetics of NF-κB dimer binding to specific target promoters has been analysed in human DCs [67]. For example, LPS stimulation of DCs has been found to rapidly increase binding of p50/RelA heterodimers released from IκBα to the IL-12p40 promoter. These are then slowly replaced by p52/RelB dimers resulting from LPS-induced p100 processing [67]. Since these heterodimers have different transcriptional activity, it has been suggested that sequential activation of the two NF-κB signalling pathways may allow fine tuning of IL-12 gene transcription over time. However, analysis of knockout MEFs has indicated that LTβR activation of the canonical NF-κB pathway induces a set of genes (encoding VCAM-1 and MIP-2) not overlapping with that activated by the alternative pathway {encoding EBI-1 [EBV (Epstein–Barr-virus)-induced gene 1]-ligand chemokine, B-lymphocyte chemoattractant and BAFF} [54,57]. Thus it appears that the canonical and alternative NF-κB pathways can regulate distinct or overlapping genes, which may depend on factors such as cell type, stimulus and target gene involved. In an analogous fashion, the Relish NF-κB pathway in Drosophila induces antibiotic peptides targeting Gram-negative bacteria (e.g. diptericin), whereas Cactus is mainly involved in the up-regulation of peptides against Gram-positive bacteria and fungi (e.g. drosomycin) [29]. However, cecropin can be activated by either pathway and is thought to have activity towards both Gram-positive and Gram-negative bacteria, as well as fungi [31,69].
In mammalian cells, activation of p100 processing by LTβR, BAFF-R and CD40 is blocked by inhibition of protein synthesis with cycloheximide [8,56,61]. It is possible that this reflects a requirement for ongoing p100 synthesis, since it has been proposed that LTβR induces p52 by a co-translational mechanism [68]. However, as induction of p52 by these receptors correlates with a corresponding reduction in p100 levels [8,57,60,61], it is more likely that p52 is produced by post-translational proteolysis of p100. Thus signal-induced p100 processing may require the induced or continued synthesis of a protein or proteins. LTβ induction of the p100 kinase activity of IKK1 also occurs over several hours [57], suggesting that de novo protein synthesis may be required upstream of IKK1 activation.
TRAF REGULATION OF THE ALTERNATIVE NF-κB PATHWAY
An important outstanding question is how receptor ligation stimulates NIK to induce p100 processing. An initial clue was provided by the identification of NIK as a binding partner for TRAF2 (TNF-receptor-associated factor-2) in a two-hybrid screen [70]. TRAF2 is a member of a family of intracellular proteins required for transducing signals from receptors of the TNF receptor and IL-1 receptor/TLR families [71].
A binding site for TRAF2 in the CD40 cytoplasmic tail is essential for induction of p100 processing by CD40 [61,72]. Similarly, TWEAK induction of p100 processing via its receptor, Fn14 (fibroblast-growth-factor-inducible 14), is blocked by mutation of the TRAF-binding site in the Fn14 cytoplasmic tail [73]. Furthermore, TWEAK fails to induce p100 processing in fibroblasts deficient for TRAF2 and TRAF5, and this defect can be rescued by reconstitution of cells with TRAF2 [73]. It is possible that TRAFs may directly link receptors to the activation of NIK by recruitment of the kinase to plasma membrane [70]. However, membrane recruitment of TRAF2 and TRAF5 is unlikely to be sufficient, as TNFα stimulation does not induce p100 processing [41,54,57,61], although it activates the canonical NF-κB pathway via TRAF2 and TRAF5 [74]. Thus these TRAFs probably co-operate with other signalling proteins to activate the alternative NF-κB signalling pathway.
TRAF2 and TRAF6 both function as E3 ligases which, in conjunction with the heterodimeric E2, Ubc13/Uev1A, promote addition of Lys63-linked polyubiquitin chains to target proteins [75]. Lys63-linked polyubiquitination does not target proteins for degradation by the proteasome, but rather is a regulatory modification which controls an essential, but undefined, step in the activation of the classical IKK complex [76]. TRAF2/6 and NEMO have been shown to undergo this modification in TNFα-stimulated cells [75,77,78], although it is unclear how this regulates IKK signalling function. It will clearly be important in the future to determine whether Lys63 ubiquitination is also involved in receptor activation of p100 processing via TRAF-2/5 and IKK1.
A recent study has demonstrated that TRAF3 negatively regulates the alternative pathway via its association with a novel-sequence motif at the N-terminus of NIK [79]. TRAF3 binding targets NIK for ubiquitination and degradation by the proteasome, thereby blocking p100 processing induced by transfected NIK. Although it was not investigated, TRAF3 presumably promotes addition of Lys48-linked polyubiquitin chains to NIK, since this modification targets proteins to the proteasome [27]. Consistent with these data, stimulation of M12 B-cells with either anti-CD40 antibody or BAFF, which both activate the alternative NF-κB pathway in these cells, induces degradation of endogenous TRAF3, with concomitant enhancement of endogenous NIK protein levels [79]. Thus an essential step in receptor activation of the alternative pathway is the rescue of NIK from TRAF3-induced proteolysis. These data suggest that that de novo synthesis of NIK is required for agonist activation of the alternative pathway, perhaps explaining the inhibitory effects of cycloheximide on receptor-induced p100 processing (see above).
MECHANISM OF p100 PROCESSING
Signal-induced p100 processing involves increased polyubiquitination of p100 (again presumably via Lys48-linked ubiquitin) and is mediated by the 26 S proteasome [46,53,61,80]. RNA-interference experiments have clearly indicated that ubiquitination and processing of p100 induced by overexpressed NIK requires βTrCP, the recognition component of the same SCF E3 ubiquitin ligase that catalyses IκBα ubiquitination [28,81]. It has been proposed that βTrCP binds to a phosphorylated sequence in the p100 PEST region (Asp-phosphoSer866-Ala-Tyr-Gly-phosphoSer870) which is related to the βTrCP binding site on IκBα and is thought to be directly phosphorylated by IKK1 [44] [PEST regions are polypeptide sequences enriched in proline (P), glutamic acid (E), serine (S) and threonine (T)]. Lys855, N-terminal to the βTrCP binding site, serves as the major ubiquitin-anchoring residue on p100 [82]. NIK-induced ubiquitination of p100 may trigger its recruitment to the proteasome by promoting interaction of the p100 DD (death domain) with S9, a non-ATPase subunit in the 19 S ‘lid’ of the 26 S proteasome [83].
Constitutive p100 processing has been detected in C-terminal deletion mutants of p100 in which a processing inhibitory domain that encompasses the p100 DD has been removed [46]. However, it remains to be demonstrated that this is relevant for physiological production of the low levels of p52 that are present in unstimulated cells. Nevertheless, the mechanisms for signal-induced and constitutive p100 processing appear to be different. Thus βTrCP is not required for constitutive processing of p100 deletion mutants [81]. Although it has not been directly shown that such p100 mutants are ubiquitinated, this raises the possibility that another E3 ligase is involved in constitutive p100 processing. Also, whereas NIK-induced p100 processing is clearly a post-translational process [44], it is possible that constitutive p100 processing occurs co-translationally [83,84]. The subcellular localization of signal-induced and constitutive p100 processing is also distinct. Constitutive p100 processing requires its nuclear/cytoplasmic shuttling, while NIK-induced processing is independent of nuclear localization [85].
ACTIVATION OF p100 PROCESSING IN CANCER
Chromosomal translocations and deletions affecting the 3′ region of the nfkb2 gene are found in a small percentage of B-cell non-Hodgkin lymphomas, chronic lymphocytic leukaemias and myelomas [86]. Nfκb2 rearrangements are also frequently detected in cutaneous T-cell lymphomas. Genetic alterations of p100 invariably result in deletion of the DD and part of the ankyrin repeat region, with the N-terminal RHD remaining intact. Such p100 deletion mutants transform fibroblasts, confirming their oncogenic potential [87].
Early studies suggested that C-terminal truncation of p100 in lymphomas allowed direct nuclear translocation of the mutant p100 molecule, which could then directly activate transcription, without requiring processing to p52 [88]. However, more recently it has been shown that oncogenic p100 deletion mutants are constitutively processed to p52 in the nucleus due to removal of the processing inhibitory p100 DD [46,85]. Thus transcriptional activity of mutant p100 molecules may result from receptor-independent production of p52. In addition, some truncated p100 molecules lose their IκB activity and fail to inhibit RelA-induced NF-κB transcription [89]. This loss of function appears to require fusion of extra amino acids at the C-terminus of the truncated p100 molecule [89]. The lymphoid hyperplasia that develops in mice which overproduce p52 in the absence of p100 [36] supports the hypothesis that increased p52 and loss of p100 IκB function play an important role in promoting proliferation and preventing apoptosis of lymphoid cells. In addition, p52–BCL-3 complexes up-regulate the transcription of cyclin D1 and Bcl-2 genes, which promote proliferation and block apoptosis respectively [90,91].
Overproduction of p52 is also associated with T-cell transformation induced by HTLV-1 (human T-cell leukaemia virus type 1) [92]. HTLV-1 activation of p52 production is mediated by its transforming protein, Tax, which stimulates p100 processing by a NIK-independent mechanism. This involves recruitment of the classical IKK complex (IKK1–IKK2–NEMO) to p100 via NEMO and contrasts with receptor-induced activation of the alternative pathway which does not require NEMO [53]. One of the transforming proteins of EBV, LMP1 (latent membrane protein 1), also induces p100 processing [73,80,93,94]. LMP1 activation of p100 processing is independent of NEMO and requires a TRAF binding site in its C-terminal cytoplasmic tail and NIK, similar to CD40 [61]. The fact that two transforming viral proteins have evolved to activate p100 processing via distinct mechanisms suggests an important role for this NF-κB signalling pathway in viral propagation. Both Tax and LMP1 also activate NF-κB by inducing IκBα degradation [95–97], indicating that the canonical and alternative NF-κB pathways may function together to promote cell transformation.
A novel pro-apoptotic function for p100 has been proposed which involves activation of caspase 8 and requires the p100 DD [98]. This suggests that C-terminal truncation of p100 would also abrogate its pro-apoptotic function, thereby facilitating cell transformation. Further experimentation is required to determine the validity of this model [99].
NF-κB1 p105/p50
Cellular levels of p105 are regulated by two distinct proteolytic mechanisms which are mediated by the proteasome. Processing of p105 to generate p50 occurs constitutively, resulting in similar levels of p105 and p50 in most cell types [24]. By contrast, agonist stimulation predominantly triggers complete degradation of p105, which releases associated Rel subunits to translocate into the nucleus. p105 can retain RelA, c-Rel and p50, but not RelB, in the cytoplasm [38,40,100]. However, genetic studies in mice suggest that p105 is only essential for cytoplasmic retention of p50 [101]. It is unclear whether p105 binds directly to p50 homodimers or p50 monomers which subsequently dimerize upon release. IκBα binds inefficiently to p50 homodimers [102], but can bind to p50, which is heterodimerized with other Rel subunits, such as RelA.
PROCESSING OF NF-κB1 p105 TO p50
The proteasome normally mediates the complete degradation of ubiquitinated proteins [103], and the partial proteolysis of p105 and p100 by the proteasome to generate p50 and p52 respectively is exceptional [104]. The mechanism of p105 processing has been the subject of intensive research (Figure 5).
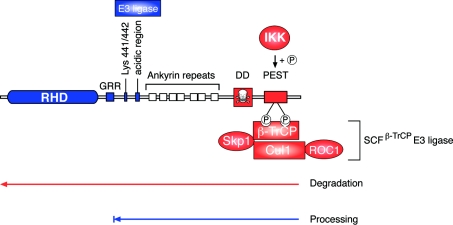
Figure 5. Proteolysis of NF-κB1 p105.
NF-κB1 p50 is constitutively produced from p105 by proteolytic removal of the C-terminal half of p105 by the proteasome (a mechanism termed processing). The GRR acts as a stop signal, preventing entry of the RHD into the proteasome, which is thought to degrade p105 from the C-terminus. The stability of the folded RHD is important in preventing its entry into the proteasome and proteolysis. The identity of the E3 ligase involved in p105 processing is not known, but has been proposed to bind to an acidic region adjacent to the ankyrin repeats. Following agonist stimulation, p105 is phosphorylated by the IKK complex on Ser927 and Ser932 in the p105 PEST region. These phosphorylated residues act a binding site for βTrCP, the recognition subunit of a multisubunit SCF-type E3 ubiquitin ligase, which catalyses the polyubiquitination of phospho-p105 on multiple lysine residues, triggering its subsequent complete degradation by the proteasome. ROC1 is a RING finger protein. Elements involved in processing are shown in blue. Elements involved in degradation are shown in red.
A glycine-rich region (GRR), in the central part of p105 between the RHD and ankyrin repeats (residues 376–404 of human p105), is essential for constitutive processing to p50 [105]. A homologous GRR in p100 is similarly required for both its constitutive and signal-induced processing to p52 [61,80,84,106]. The GRR in p105 is not required for p105 ubiquitination, and it has been proposed to function as a processing stop signal, preventing entry of the RHD of p105 into the proteasome [107]. Similarly, a related glycine-alanine repeat domain of EBNA1 (EBV nuclear antigen 1) blocks proteolysis of EBNA1 by the proteasome [108]. Indeed, insertion of this glycine-alanine repeat into different positions within IκBα prevents its signal-induced degradation, although phosphorylation and ubiquitination are not affected [109]. Thus GRRs may have a more general role in protecting proteins from proteasome-mediated proteolysis.
An alternative role for the p105 GRR in p105 processing has also been proposed [105]. Generation of chimaeric proteins in which the flexible p105 GRR is placed between two stably folded domains allows the proteasome to cut in the middle of the fusion protein [105]. In addition, the proteasome has been shown to catalyse endoproteolytic cleavage of polypeptide bonds [110]. Together, these data suggest that the GRR may promote the endoproteolytic cleavage of p105 to produce p50, while the resulting C-terminal fragment is degraded by the proteasome. However, C-terminal p105 fragments have not been detected in cells, and it is unclear whether p105 actually undergoes endoproteolytic cleavage.
Several residues N-terminal to the p105 GRR, which are required for correct folding of p50 RHD, are also essential for p105 processing [111]. The stability of the RHD appears to be important in protecting it from proteolysis, probably by inhibiting unfolding and entry into the proteasome. Thus the p105 GRR is necessary, but may not be sufficient, to prevent proteolysis of the RHD by the proteasome. Processing of p105 is also negatively regulated by its association with p50, which may interfere with entry of p105 into the proteasome [112,113]. Similarly, RelB binding to p100 has an inhibitory effect on its processing to p52 [114].
Experiments in which the p105 GRR has been transferred within p105 or inserted into homologous or heterologous proteins have indicated that the GRR is not usually sufficient to promote processing [107]. An acidic region (residues 446 and 454 of human p105) has been shown to be additionally required [107]. This has been proposed to function as a recognition site for an uncharacterized ubiquitin E3 ligase which targets p105 Lys441 and Lys442 for ubiquitination [107].
It has been suggested that p105 is processed co-translationally to produce p50 and that synthesis of the complete p105 molecules is not required for p50 generation [115]. Consistent with this hypothesis, a truncated form of p105, p60, can be processed into p50 [116]. It has also been proposed that the p50 RHD undergoes co-translational dimerization and that this is essential for both p50 homodimer and p105 generation [117]. However, several investigators have demonstrated a clear precursor–product relationship between p105 and p50 in pulse–chase experiments [38,116,118,119]. The physiological importance of co-translational p105 processing, therefore, remains uncertain.
STIMULUS-INDUCED PROTEOLYSIS OF NF-κB1 p105
Following cellular stimulation with agonists, such as TNFα, IL-1 and LPS, p105 is phosphorylated and proteolysed more rapidly by the proteasome [118,120,121]. The PEST region in the p105 C-terminus contains a conserved motif (Asp-Ser927-Gly-Val-Gly-Thr-Ser932) homologous with the IKK target sequence in IκBα (Figure 5). Ser927 and Ser932 within this motif are rapidly phosphorylated by the classical IKK complex after TNFα stimulation with kinetics similar to the phosphorylation of IκBα [122,123]. This induces the recruitment of the SCFβTrCP E3 ligase to p105, which then catalyses p105 ubiquitination, promoting subsequent p105 proteolysis. Genetic studies have confirmed that stimulus-induced p105 proteolysis requires the expression of a functional IKK complex and is blocked in cells lacking both IKK1 and IKK2 or NEMO [122–124]. Furthermore, IKK1 and IKK2 must both interact with p105 via its DD to mediate phosphorylation of the p105 PEST region [125,126]. In contrast with the signal-induced proteolysis of p100, TNFα promotes the proteolysis of p105 independently of IKK1 [122]. Together these data suggest that the classical IKK complex, which triggers IκBα degradation, also regulates p105 proteolysis. However, it is not known whether specific TRAFs or MAP 3-kinases function upstream of the IKK complex in this signalling pathway.
Proteolysis of p105 stimulated by co-transfected IKK2 in COS-7 cells increases production of p50 [127]. However, similar experiments in HEK-293 cells demonstrate that overexpressed IKK2 promotes a decrease in p105 without affecting p50 generation, suggesting that IKK-induced p105 proteolysis in these cells results in p105 degradation [46,124]. TNFα causes a modest increase in production of p50 in an osteoclastoma cell line, although this is small relative to the corresponding decrease in p105 [121]. In contrast, LPS or plasmin stimulation of primary human monocytes or LPS stimulation of THP-1 cells strongly increases proteolysis of p105 without affecting p50 levels [113,128]. Together these data suggest that the major result of signal-induced proteolysis is the complete degradation of p105. This continues over several hours [118,122] and is thought to result in the slow translocation of p50-containing NF-κB dimers into the nucleus, which can then complex with the IκB-like protein BCL-3 [126]. The NF-κB1 gene is itself up-regulated by NF-κB, which may be important for negative-feedback regulation of signalling pathways dependent on induced p105 proteolysis [129].
RNA-interference experiments have demonstrated that, similarly to IκBα and p100, βTrCP is required for TNFα stimulation of p105 ubiquitination and proteolysis [123]. Gene knockdown experiments have indicated that p50 production, on the other hand, is independent of βTrCP [130], consistent with the hypothesis that a distinct E3 ligase is involved in constitutive p105 processing and signal-induced p105 degradation [107,127]. SCFβTrCP-mediated ubiquitination of phosphorylated p105 targets multiple lysine residues for ubiquitination [130], whereas only two lysine residues appear to be important for p105 processing [107]. It is possible that the ubiquitination of multiple lysine residues by SCFβTrCP overcomes the barriers of the GRR and RHD to proteasome-mediated proteolysis, facilitating complete p105 degradation. In accordance with this idea, signal-induced proteolysis of p100, which results in p52 production rather than complete degadation of p100, predominantly involves ubiquitination of a single lysine residue [82].
The β-propeller of βTrCP is known to interact with phosphorylated motifs containing the amino acid sequence Asp-phospho-Ser-Gly-Φaa-Xaa-phosphoSer, where Φaa is a hydrophobic amino acid [131]. The Asp-phosphoSer927-Gly-Val-Gly-Thr-phosphoSer932 phosphorylated motif in the PEST domain of p105 differs from this consensus sequence, owing to the introduction of an extra residue between the two phosphorylated serine residues, indicating that there is some flexibility in binding of βTrCP to phosphorylated target sequences [123]. However, in vitro binding experiments suggest that βTrCP has a significantly lower affinity for phospho-p105 than for phospho-IκBα [123], which may reduce the efficiency of βTrCP-mediated ubiquitination of p105 compared with that of IκBα. This may explain why TNFα stimulation induces partial proteolysis of p105, in contrast with IκBα, which is completely degraded [122].
p105 is phosphorylated on Ser903 and Ser907 by glycogen synthase kinase (GSK)-3β, with which it forms a complex [132,133]. Furthermore, TNFα-induced proteolysis of p105 is blocked in GSK-3β-deficient fibroblasts or by the introduction of p105 S903A (Ser903→Ala) or S907A point mutations [132]. It is currently unclear whether phosphorylation of p105 by GSK-3β affects IKK-mediated phosphorylation of p105 Ser927 and Ser932, and subsequent recruitment of the SCFβTrCP E3 ligase. However, induction of p105 ubiquitination by TNFα is blocked in GSK-3β-deficient fibroblasts [132], suggesting that either of these events may require p105 phosphorylation by GSK-3β.
TPL-2 A p105-REGULATED MAP 3-KINASE
In addition to NF-κB activation, LPS induction of many immune-response genes involves stimulation of each of the major MAP kinase subtypes [ERK-1/2, JNK (c-Jun N-terminal kinase) and p38] [2,134]. Activation of ERK-1/2 MAP kinases by LPS in macrophages requires the serine/threonine kinase TPL-2 (tumour progression locus-2) [135], which is also known as COT [136]. TPL-2 functions as a MAP 3-kinase that phosphorylates and activates the ERK-1/2 kinase MEK-1/2 [137] (Figure 6 and Table 3). LPS up-regulation of TNFα and COX-2 (cyclooxygenase-2) is dramatically reduced in TPL-2-deficient macrophages, owing to defective ERK-1/2 activation [138,139]. Consequently, TPL-2 knockout mice are resistant to endotoxin shock induced by LPS/D-galactosamine [138]. TPL-2 is also required for TNFα and CD40 ligand stimulation of the ERK/MAP kinase cascade [140], suggesting an important role for TPL-2 in both innate and adaptive immune responses.
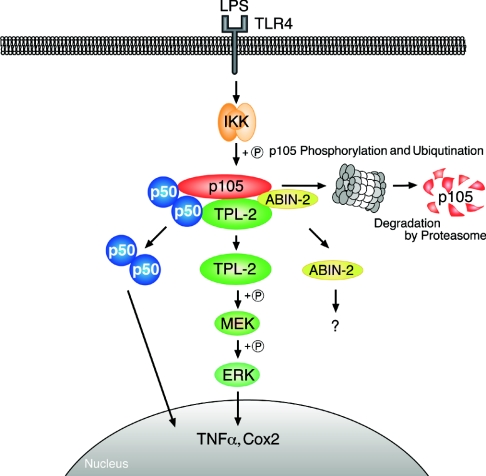
Figure 6. Diagram illustrating the TLR4–TPL-2–ERK–MAP kinase cascade.
Stimulation of TLR4 by LPS activates the IKK complex which phosphorylates Ser927 and Ser932 in the p105 PEST region. This triggers p105 ubiquitination and subsequent degradation, liberating TPL-2. p105-free TPL-2 then phosphorylates and activates MEK, which in turn phosphorylates and activates ERK. In unstimulated cells, TPL-2 and p105 form a ternary complex with ABIN-2. ABIN-2 is released from p105 after LPS stimulation, but its function is currently unknown. IKK-induced p105 proteolysis also liberates associated p50 (and other Rel subunits) to translocate into the nucleus and modulate target gene expression.
Table 3. Summary of phenotypes of NF-κB1 and TPL-2 knockout mice.
Abbreviations and symbols used: ↑, increase; ↓, decrease; =, unaffected; Ab, antibody; Ag, antigen; sCD40L, soluble CD40 ligand; mCD40L, membrane-bound CD40 ligand; D-Gal, D-galactosamine; LCMV, lymphocytic choriomeningitis virus; LN, lymph node.
| Macrophages | T-cells | B-cells | Other comments | Reference(s) | |
|---|---|---|---|---|---|
| nf-κb1−/− (no p105/p50, TPL-2/ABIN-2 ↓) | LPS: | αCD3+αCD28: | LPS, sCD40L: | Morphology/development= | [141–143,158,161,165] |
| MEK/ERK activation ↓ | Proliferation ↓ | Proliferation ↓ | |||
| COX-2, IL-6, IL-12, MCP1 ↓ | IL-2R ↓ | αIgM, mCD40L: | |||
| Proliferation= | |||||
| TH2 differentiation ↓ | Ab isotype switch ↓ | ||||
| GATA-3 ↓ | Ag-specific Abs ↓ (T-dependent) | ||||
| nf-κb1ΔC/ΔC (no p105, high p50) (TPL-2, ABIN-2 ↓?) | LPS: | αCD3+αCD28: | LPS, αIgM: | Enlarged LN, splenomegaly, lymphoid hyperplasia | [101] |
| IL-1β, IL-6, IL-10, TNFα ↓ | Proliferation ↓ | Proliferation ↑ | |||
| IL-2, IL-4, TNFα ↓ | |||||
| Basal Ig ↑ | |||||
| Ag-specific Abs ↑ (T-dependent) | Die post-natally | ||||
| tpl-2−/− | LPS: | αCD3+αCD28: | αCD40: | Morphology/development= | [138–140,188] |
| MEK/ERK activation ↓ | Proliferation= | MEK/ERK activation ↓ | |||
| TNFα, IL-1, COX-2 ↓ | IL-2, TNFα, IFN-γ= | Proliferation= | |||
| TNFα, αCD40: | CD54, CD69, CD44, CD95= | Septic shock ↓ (LPS/D-Gal) | |||
| MEK/ERK activation ↓ | Cytotoxic response (LCMV)= | Ag-specific Ab= | Inflammatory bowel disease ↓ |
The C-terminal half of NF-κB1 p105 forms a high-affinity stoichiometric association with TPL-2 via two distinct interactions [119,141]. The TPL-2 C-terminus (residues 398–467) binds to a region N-terminal to the p105 ankyrin repeat region (human p105 residues 497–534), whereas the TPL-2 kinase domain interacts with the p105 DD [141]. In unstimulated macrophages, all detectable TPL-2 is associated with p105 [119,142]. Initial overexpression experiments indicated that TPL-2 might act upstream of p105, regulating its inducible proteolysis and activating NF-κB [119]. This hypothesis is not apparently supported by the normal LPS induction of NF-κB in TPL-2-deficient macrophages [138]. However, since only a small fraction of p105 is actually associated with TPL-2 [142], it remains possible that the proteolysis of this pool of p105 is regulated by TPL-2. This may contribute to only a small fraction of total NF-κB activity in LPS-stimulated macrophages.
Although it is unclear whether TPL-2 regulates p105 proteolysis, recent research has revealed that p105 is essential for the regulation of two aspects of TPL-2 function. First, binding to p105 maintains TPL-2 protein stability [141,143]. Consequently, steady-state levels of TPL-2 are very low in p105-deficient cells [141,143], and LPS activation of MEK is severely reduced in p105-deficient macrophages [143]. Secondly, binding to p105 inhibits TPL-2 MEK kinase activity by preventing access to MEK [141,143]. Interaction of both the TPL-2 kinase domain and the C-terminus is required for inhibition of TPL-2 by p105 in vivo and, consequently, the catalytic activity of a C-terminally truncated TPL-2 mutant is unaffected by p105 [141]. Significantly, activation of the oncogenic potential of TPL-2 requires deletion of its C-terminus [144], and it is therefore likely that insensitivity to p105 negative regulation contributes to the ability of C-terminally truncated TPL-2 mutants to transform cells.
In unstimulated macrophages, TPL-2 MEK kinase activity is blocked as a result of its association with p105 [142,143]. LPS stimulates TPL-2 MEK kinase activity by promoting its release from its inhibitor, p105 [143], which has recently been shown to occur as a consequence of IKK-induced p105 proteolysis (S. Beinke, M. Robinson, M. Huginin and S. C. Ley, unpublished work; S.-C. Sun, personal communication). The IKK complex, therefore, regulates both NF-κB and ERK activation via p105 in LPS-stimulated macrophages (Figure 6). This mechanism of TPL-2 activation raises the question of the physiological advantage of linking LPS activation of ERK and NF-κB via p105. One possibility is that this facilitates more efficient induction by LPS of genes which are regulated both by ERK and NF-κB. TNFα [138,145] and COX-2 [139,146,147] are examples of such genes.
Interestingly, Relish-mediated transcription has recently been shown to negatively regulate JNK activation by controlling the proteasomal degradation of TAK1, the upstream MAP 3-kinase required for JNK activation [148]. NF-κB also negatively regulates JNK activation in mammals [149]. However, it is not yet known whether this also involves transcription driven by either of the precursor IκBs.
OTHER p105-INTERACTING PROTEINS
To fully understand the regulation and function of p105, it is essential to identify any additional proteins with which it associates. Two potential p105-interacting proteins have been found in separate yeast two-hybrid screens; first, the basic helix–loop–helix transcription factor LYL1 (lymphoblastic leukaemia derived sequence 1) [150], chromosomal gene rearrangements of which are associated with T-cell acute leukaemia [151] and, secondly, Casper/c-FLIP (FLICE-inhibitory protein) [152], a caspase 8-related protein that regulates death-receptor-induced apoptosis [153]. Although these interactions with p105 also occur in co-transfected mammalian cells, they have yet to be confirmed with the endogenous proteins. Overexpression of either LYL1 or Casper/c-FLIP modestly decreases the p50/p105 ratio, suggesting that p105 proteolysis is inhibited [150,152]. However, currently there is no genetic evidence (gene knockout or knockdown) to confirm that these proteins actually regulate p105 processing under more physiological conditions.
Two groups independently used a proteomic approach to identify novel p105-associated proteins [142,154]. In each case, tagged versions of p105 were expressed in mammalian cells, isolated by affinity purification, and associated proteins were identified by MS. Both studies identified ABIN (A20-binding inhibitor of NF-κB)-2 [155] as a protein that co-purifies with p105. Indeed, the majority of endogenous ABIN-2 in BMDMs (bone-marrow-derived macrophages) is associated with p105 and TPL-2 in a ternary complex [142]. RNA interference experiments indicate that ABIN-2 is required to maintain the metabolic stability of TPL-2 protein, while analysis of nf-κb1−/− cells has revealed that p105 is required to stabilize ABIN-2 as well as TPL-2 [141–143]. Thus, in unstimulated cells, ABIN-2 and TPL-2 are confined to a ternary complex with p105.
ABIN-2 was originally identified in a two-hybrid screen using the zinc-finger protein A20 as a bait [155]. A20 is encoded by a primary response gene which is induced by a number of stimuli which activate NF-κB, including TNFα and LPS [156]. A20 is required for termination of TNFα-induced NF-κB activation and also for blockade of TNFα-triggered apoptosis [157]. ABIN-2 overexpression blocks TNFα activation of NF-κB, and consequently it has been suggested that ABIN-2 may contribute to the NF-κB inhibitory functions of A20 [155]. The association of ABIN-2 with the TPL-2–p105 complex suggests that ABIN-2 may also be involved in the regulation of the TPL-2–MEK–ERK signalling pathway (Figure 6).
NF-κB1 FUNCTION IN IMMUNE AND INFLAMMATORY RESPONSES
The physiological function of NF-κB1 in immunity was initially investigated by generation and analysis of nfkb1−/− mice, which do not produce either p105 or p50 (Table 3). These mice display no histopathological changes, but have multifocal defects in immune system function [158].
The survival of mature quiescent splenic B-cells requires p105/p50 [159] and the proliferative response of splenic nfkb1−/− B-cells to LPS and soluble CD40 is impaired, owing to increased mitogen-induced apoptotis and G1-phase blockade [159–161]. These defects in B-cell responsiveness are selective for these stimuli, and antigen–receptor ligation or stimulation with membrane-bound CD40 induce normal-level proliferation of nfkb1−/− B-cells [161]. Consistent with NF-κB1 p105/p50 playing an important function in B-cells, nfkb1−/− mice fail to mount a normal humoral response to the T-cell-dependent antigen NP15-CG [160].
Thymopoiesis occurs normally in the absence of NF-κB1 p105/p50 [160]. However, CD4+ T-cells from nf-kb1−/− mice have an intrinsic defect in antigen–receptor-driven proliferation, which is associated with reduced IL-2-receptor expression [162,163]. Furthermore, functional maturation of CD4+ T cells to both TH1 (T-helper 1) and TH2 (T helper 2) subsets requires NF-κB1 p105/p50 expression [162–165]. Macrophages lacking NF-κB1 p105/p50 have defects in the production of IL-6 and IL-12 in response to LPS stimulation [160,166,167]. In contrast, up-regulation of TNFα and IL-1 by LPS is unaffected. Interestingly, one study found that, although LPS up-regulation of TNFα is normal in nf-kb1−/− macrophages, it is substantially reduced in nf-kb1−/− DCs [168]. Thus the contribution of NF-κB1 to cytokine induction by the same agonist may be cell-type-specific.
Consistent with the identified effects of NF-κB1 deficiency on haematopoeitic-cell function, the immune response to certain infectious organisms is impaired in nf-kb1−/− mice. For example, nf-kb1−/− mice are susceptible to the intracellular protozoan parasite Leishmania major owing to a defective TH1 response, developing chronic unresolving lesions associated with persistent parasitaemia [163]. Control of the replication of an extracellular Gram-positive bacterium, Streptococcus pneumoniae, is also compromised in nf-kb1−/− mice [160]. However, the immune response of nf-kb1−/− mice to two Gram-negative bacteria, Haemophilus influenzae and Escherichia coli K1, controlled infection effectively [160]. Surprisingly, nf-kb1−/− mice are actually more resistant to infection with encephalomyocarditis virus than wild-type mice, possibly due to increased production of IFNβ (interferon-β) by the knockout mice [160]. This suggests that NF-κB1 may negatively regulate IFNβ transcription.
Positive and negative effects of NF-κB1 have also been observed with respect to inflammatory responses. nf-kb1−/− mice are resistant to both collagen-induced (chronic) arthritis and methylated BSA/IL-1-induced (acute) arthritis [169]. Similarly, allergic airway inflammation requires NF-κB1 p105/p50 expression [170]. In contrast, colitis induced by intragastric infection with Helicobacter hepaticus is more severe in nf-kb1−/− mice, owing to overproduction of pro-inflammatory cytokines in the gut compared with wild-type animals [171]. Consistent with these data, a functional polymorphism of the promoter of the human NF-κB1 gene, which decreases NF-κB1 transcription, is associated with increased incidence of ulcerative colitis [172].
The function of NF-κB1 has also been investigated by generation of a different mutant mouse strain, nfkb1ΔC/ΔC (Table 3), which lacks the exons encoding the C-terminal half of p105 and therefore expresses p50 in the absence of p105 [101]. Such mice develop splenomegaly, enlarged lymph nodes and have an inflammatory phenotype composed of lymphocytic infiltration in the lungs and liver. These phenotypes require the production of active p50 homodimers, which are found at high levels in the nuclei of cells from nfkb1ΔC/ΔC mice [101,173]. p50 homodimers may function as transcriptional activators or repessors, depending on cell type, which may explain the positive and negative roles for NF-κB1 in immune responses and inflammation. For example, mRNA levels of GM-CSF (granulocyte–macrophage colony-stimulating factor), IL-2, M-CSF (macrophage colony-stimulating factor) and TNFβ are increased in thymocytes in the absence of p105, whereas in macrophages the mRNA expression of GM-CSF, TNFα and IL-6 are dramatically reduced. In addition, the lack of p105 increases the proliferation of splenic B-cells to both anti-Ig and LPS in vitro, while decreasing the proliferative response of T-cells induced by antigen–receptor cross-linking.
nfkb1 MUTANT MICE: A COMPOUND PHENOTYPE?
Much of what is known about the in vivo function of NF-κB1 has been deduced from the analysis of nfκb1−/− mice (see above). However, the interpretation of the molecular phenotype of these knockout mice is complicated by secondary effects on the stability of the proteins with which p105 associates. Thus steady-state levels of both TPL-2 and ABIN-2 are severely reduced in nfkb1−/− macrophages, owing to lack of p105 [141–143]. Some of the phenotypes of the mutant mouse strains therefore probably result from combined defects in the activation of NF-κB and the TPL-2–MEK–ERK pathway.
NF-κB1 AND CANCER
Unlike its homologue nfkb2, alterations in the structure and expression of the nfkb1 gene in leukaemias and lymphomas are rare [86]. nfkb1 gene rearrangements have been reported in certain acute lymphoblastic leukaemias [174], and overexpression of p50 has been demonstrated in a large percentage of non-small cell carcinomas [175,176]. However, it has not been demonstrated that expression of such mutated p105 alleles or overexpression of the wild-type p105 actually transforms cells.
NF-κB1 p50 may play a role in oncogenic transformation when complexed with BCL-3. Chromosomal translocation of the gene encoding BCL-3 is a rare, but recurrent, event in B-cell chronic lymphocytic leukaemia, resulting in overexpression of an intact BCL-3 protein [177], which can interact with homodimers of p50 and p52 to promote NF-κB-dependent transcription [178]. Consistent with BCL-3 acting as an oncogene in B-cells, Eμ-bcl-3 transgenic mice develop a lymphoproliferative disorder characterized by the accumulation of mature B-cells [179]. p50–BCL-3 complexes may also be important in the development of an epithelial malignancy nasopharyngeal carcinoma [180].
Activation of NF-κB by HTLV-1 Tax plays an essential role in its transforming function [181] and may be mediated in part by its interaction with p105 [182,183]. Tax has been proposed to activate p105-mediated NF-κB activation either by inducing the dissociation of p50 from p105 without affecting p105 proteolysis [184] or by increasing processing of p105 to p50 by the proteasome [185]. The functional importance of p105-mediated NF-κB activation for transformation by Tax has not been established.
THERAPEUTIC PERSPECTIVES
Components of the canonical NF-κB signalling pathway have been regarded as good potential drug targets for therapeutic intervention in inflammatory and autoimmune diseases [186]. However, chronic blockade of this pathway may have many unwanted side effects, since it is activated by multiple stimuli and has a broad physiological role [12,24]. Nevertheless, acute blockade of the canonical pathway may be tolerable in the treatment of cancers [186]. The p100 and p105 NF-κB pathways offer an alternative source of drug targets to the canonical pathway.
Pharmacological blockade of the alternative NF-κB pathway might be possible through the development of small-molecule inhibitors that target the kinases NIK or IKK1. This would be expected to have profound general immunosuppressive effects, since B-cell survival depends on BAFF-induced p100 processing [8,62], probably ruling out the use of such drugs for treatment of chronic inflammatory diseases. However, increased p100 processing often occurs in lymphoid cancers [187] and may be generally important for malignant transformation of this cell type. It is therefore possible that pharmacological blockade of NIK or IKK1 might be useful for specific treatment of lymphoid cancers in which p100 is constitutively processed as a result of increased activation of the alternative NF-κB signalling pathway. Potential examples of this are EBV-positive Hodgkin's lymphomas and HTLV-1-transformed T-cell lymphomas [53,80,93]. Such an approach, however, would not be appropriate for B-cell lymphomas in which increased p52 results from nfκb2 gene translocation, since this is probably independent of NIK and IKK1 [86].
Targeting proteins that control the NF-κB signalling pathway that regulates the proteolysis of p105 may be useful for treatment of inflammatory diseases, since it has a more restricted role in immune responses than the canonical pathway. Therefore this might allow the development of more specific anti-inflammatory drugs. One possible approach would be to develop drugs to block the function of upstream signalling components which are unique to the p105 pathway. In this respect, future research may identify a MAP 3-kinase that specifically regulates signal-induced p105 proteolysis and would be a very attractive target for small-molecule inhibitors.
The p105-associated kinase TPL-2 may be a good target for the development of small-molecule inhibitors to block activation of the ERK/MAP kinase cascade in immune responses. It might be possible to generate inhibitors which are very specific for TPL-2, since its kinase domain is not closely homologous with others in the database. Studies with TPL-2 knockout mice suggest that such TPL-2 inhibitors would be useful for treatment of septic shock and Crohn's disease [138,188].
CONCLUSIONS AND FUTURE RESEARCH DIRECTIONS
Although NF-κB1 p105 and NF-κB2 p100 have similar overall structures (Figure 1), current data indicate that their functions are distinct, as is the control of their proteolysis. It has been established that p100 processing to p52 is critical in the organogenesis of peripheral lymphoid tissues and for B-cell development. Induction of p100 processing is tightly regulated and induced by only a subset of ligands which activate NF-κB. In contrast, processing of p105 to p50 is constitutive, and receptor stimulation induces its complete degradation. Genetic studies have revealed an important role for NF-κB1 in immune and inflammatory responses.
Several key questions about p100 and p105 remain to be answered:
Are there specific receptors that induce p105 processing to p50, rather than p105 degradation, similar to p100?
What is the contribution of signal-induced p105 proteolysis to the amplitude and kinetics NF-κB activation?
Is there an equivalent MAP 3-kinase to NIK which is required for signal-induced p105 proteolysis?
What are the roles of ABIN-2 and other p105-associated proteins in the regulation of NF-κB and ERK/MAP kinase signalling pathways?
Is the activity of NIK regulated by its interaction with the terminal half of p100 [143], similar to the regulation of TPL-2 by p105?
Does p100 directly or indirectly regulate the activation of any of the MAP kinase cascades?
It is likely that many of these issues will be resolved in the near future in this fast-moving area of research.
Acknowledgments
We thank Dr Peter Atkinson, Dr Anne O'Garra, Dr Lee Johnston and Dr Matt Robinson, all of this Institute, and Dr Hamish Allen (Abbott Bioresearch Center, Worcester, MA, U.S.A.) for their helpful comments during the preparation of this review, Mr Frank Johnson (NIMR Photographics) for producing the illustrations, and Professor Shao-Cong Sun (Pennsylvania State University College of Medicine, Hershey, PA, U.S.A.) for communicating unpublished data to us. The advice of Professor Aaron Ciechanover (Technion-Israel Institute of Technology, Haifa, Israel) on models for p105 processing and Dr Ulrich Siebenlist (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, U.S.A.) on the role of NF-κB2 in B-cell development is also gratefully acknowledged. We apologize to any investigators whose work, owing to space limitations, has not been discussed. S.B. and S.C.L. are funded by the U.K. Medical Research Council.
References
- 1.Medzhitov R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 2.Takeda K., Kaisho T., Akira S. Toll-like receptors. Annu. Rev. Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 3.Zhang G., Ghosh S. Toll-like receptor-mediated NF-κB activation: a phylogenetically conserved paradigm in innate immunity. J. Clin. Invest. 2001;107:13–19. doi: 10.1172/JCI11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Neill L. A. J., Dinarello C. A. The IL-1 receptor/Toll-like receptor superfamily: crucial receptors for inflammation and host defense. Immunol. Today. 2000;21:206–209. doi: 10.1016/s0167-5699(00)01611-x. [DOI] [PubMed] [Google Scholar]
- 5.Wallach D., Varfolomeev E. E., Malinin N. L., Goltsev Y. V., Kovalenko A. V., Boldin M. P. Tumor necrosis factor receptor and Fas signaling mechanisms. Annu. Rev. Immunol. 1999;17:331–367. doi: 10.1146/annurev.immunol.17.1.331. [DOI] [PubMed] [Google Scholar]
- 6.Li Q., Verma I. M. NF-κB regulation in the immune system. Nat. Rev. Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 7.Weih F., Caamano J. Regulation of secondary lymphoid organ development by the nuclear factor-κB signal transduction pathway. Immunol. Rev. 2003;195:91–105. doi: 10.1034/j.1600-065x.2003.00064.x. [DOI] [PubMed] [Google Scholar]
- 8.Claudio E., Brown K., Park S., Wang H., Siebenlist U. BAFF-induced NEMO-independent processing of NF-κB2 in maturing B cells. Nat. Immunol. 2002;3:958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- 9.Girardin S. E., Hugot J.-P., Sansonetti P. J. Lessons from Nod2 studies: towards a link between Crohn's disease and bacterial sensing. Trends Immunol. 2003;24:652–658. doi: 10.1016/j.it.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Feldmann M., Maini R. N. Anti-TNFα therapy of rheumatoid arthritis: what have we learned? Annu. Rev. Immunol. 2001;19:163–196. doi: 10.1146/annurev.immunol.19.1.163. [DOI] [PubMed] [Google Scholar]
- 11.Karin M., Lin A. NF-κB at the crossroads of life and death. Nat. Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 12.Karin M., Cao Y., Greten F. R., Zhi-Wei L. NF-κB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 13.Farrow B., Evers B. M. Inflammation and the development of pancreatic cancer. Surg. Oncol. 2002;10:153–169. doi: 10.1016/s0960-7404(02)00015-4. [DOI] [PubMed] [Google Scholar]
- 14.Normark S., Nilsson C., Normark B. H., Hornef M. W. Persistent infection with Helicobacter pylori and the development of gastric cancer. Adv. Cancer Res. 2003;90:63–89. doi: 10.1016/s0065-230x(03)90002-9. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh S., May M. J., Kopp E. B. NF-κB and Rel proteins: evolutionary conserved mediators of immune responses. Annu. Rev. Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 16.Marienfeld R., May M. J., Berberich I., Serfling E., Ghosh G., Neumann M. RelB forms transcriptionally inactive complexes with RelA/p65. J. Biol. Chem. 2003;278:19852–19860. doi: 10.1074/jbc.M301945200. [DOI] [PubMed] [Google Scholar]
- 17.Bours V., Franzoso G., Azarenko V., Park S., Kanno T., Brown K., Siebenlist U. The oncoprotein Bcl-3 directly transactivates through κB motifs via association with DNA-binding p50B homodimers. Cell. 1993;72:729–739. doi: 10.1016/0092-8674(93)90401-b. [DOI] [PubMed] [Google Scholar]
- 18.Franzoso G., Bours V., Park S., Tomita-Yamaguchi M., Kelly K., Siebenlist U. The candidate oncoprotein Bcl-3 is an antagonist of p50/NF-κB mediated inhibition. Nature (London) 1992;359:339–342. doi: 10.1038/359339a0. [DOI] [PubMed] [Google Scholar]
- 19.Fujita T., Nolan G. P., Liou H.-C., Scott M. L., Baltimore D. The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through p50 homodimers. Genes Dev. 1993;7:1354–1363. doi: 10.1101/gad.7.7b.1354. [DOI] [PubMed] [Google Scholar]
- 20.Chen L.-F., Greene W. C. Shaping the nuclear action of NF-κB. Nat. Immunol. Rev. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 21.Gerondakis S., Morrice N., Richardson I. B., Wettenhall R., Fecondo J., Grumont R. J. The activity of a 70kDa IκB molecule identical to the carboxyl terminus of the p105 NF-κB precursor is modulated by protein kinase A. Cell Growth Differ. 1993;4:617–627. [PubMed] [Google Scholar]
- 22.Inoue J., Kerr L. D., Kakizuka A., Verma I. M. IκBγ, a 70 kDa protein identical to the C-terminal half of p100 NF-κB: a new member of the IκB family. Cell. 1992;68:1109–1120. doi: 10.1016/0092-8674(92)90082-n. [DOI] [PubMed] [Google Scholar]
- 23.Pomerantz J. L., Baltimore D. Two pathways to NF-κB. Mol. Cell. 2002;10:693–701. doi: 10.1016/s1097-2765(02)00697-4. [DOI] [PubMed] [Google Scholar]
- 24.Karin M., Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 25.Yang J., Lin Y., Guo Z., Cheng J., Huang J., Deng L., Liao W., Chen Z., Liu Z.-G., Su B. The essential role of MEKK3 in TNF-induced NF-κB activation. Nat. Immunol. 2001;2:620–624. doi: 10.1038/89769. [DOI] [PubMed] [Google Scholar]
- 26.Takaesu G., Surabhi R. M., Park K.-J., Ninomiya-Tsuji J., Matsumoto K., Gaynor R. B. TAK1 is critical for IκB kinase-mediated activation of the NF-κB pathway. J. Mol. Biol. 2003;326:105–115. doi: 10.1016/s0022-2836(02)01404-3. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y.-C. Ubiquitin ligases and the immune response. Annu. Rev. Immunol. 2004;22:81–127. doi: 10.1146/annurev.immunol.22.012703.104813. [DOI] [PubMed] [Google Scholar]
- 28.Ben-Neriah Y. Regulatory functions of ubiquitination in the immune system. Nat. Immunol. 2002;3:20–26. doi: 10.1038/ni0102-20. [DOI] [PubMed] [Google Scholar]
- 29.Silverman N., Maniatis T. NF-κB signaling pathways in mammalian and insect innate immunity. Genes Dev. 2001;15:2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann J. A. The immune response of Drosophila. Nature (London) 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 31.Hultmark D. Drosophila immunity: paths and patterns. Curr. Opin. Immunol. 2003;15:12–19. doi: 10.1016/s0952-7915(02)00005-5. [DOI] [PubMed] [Google Scholar]
- 32.Morisato D., Anderson K. V. Signaling pathways that establish the dorsal–ventral pattern of the Drosophila embryo. Annu. Rev. Genet. 1995;29:371–399. doi: 10.1146/annurev.ge.29.120195.002103. [DOI] [PubMed] [Google Scholar]
- 33.Silverman N., Zhou R., Stoven S., Pandey N., Hultmark D., Maniatis T. A Drosophila IκB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stoven S., Ando I., Kadalayil L., Engstrom Y., Hultmark D. Activation of the Drosophila NF-κB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 2000;1:347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoven S., Silverman N., Junell A., Hedengren-Olcott M., Erturk D., Engstrom Y., Maniatis T., Hultmark D. Caspase-mediated processing of the Drosophila NF-κB factor Relish. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5991–5996. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishikawa H., Carrasco D., Claudio E., Ryseck R.-P., Bravo R. Gastric hyperplasia and increased proliferative responses of lymphocytes in mice lacking the COOH-terminal ankyrin domain of NF-κB2. J. Exp. Med. 1997;186:999–1014. doi: 10.1084/jem.186.7.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheinman R. I., Beg A. A., Baldwin A. S. NF-κB p100 (Lyt-10) is a component of H2TF1 and can function as an IκB-like molecule. Mol. Cell. Biol. 1993;13:6089–6101. doi: 10.1128/mcb.13.10.6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mercurio F., DiDonato J. A., Rosette C., Karin M. p105 and p98 precursor proteins play an active role in NF-κB-mediated signal transduction. Genes Dev. 1993;7:705–718. doi: 10.1101/gad.7.4.705. [DOI] [PubMed] [Google Scholar]
- 39.Dobrzanski P., Ryseck R. P., Bravo R. Specific inhibition of RelB/p52 transcriptional activity by the C-terminal domain of p100. Oncogene. 1995;10:1003–1007. [PubMed] [Google Scholar]
- 40.Solan N. J., Miyoshi H., Bren G. D., Paya C. V. RelB cellular regulation and transcriptional activity are regulated by p100. J. Biol. Chem. 2001;277:1405–1418. doi: 10.1074/jbc.M109619200. [DOI] [PubMed] [Google Scholar]
- 41.Yilmaz Z. B., Weih D., Sivakumar V., Weih F. RelB is required for Peyer's patch development: differential regulation of p52–RelB by lymphotoxin and TNF. EMBO J. 2003;22:121–130. doi: 10.1093/emboj/cdg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caamano J. H., Rizzo C. A., Durham S. K., Barton D. S., Raventos-Suarez C., Snapper C. M., Bravo R. Nuclear Factor (NF)-κB2 (p100/p52) is required for normal splenic microarchitecture and B cell-mediated immune responses. J. Exp. Med. 1998;187:185–196. doi: 10.1084/jem.187.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franzoso G., Carlson L., Poljak L., Shores E. W., Epstein S., Leonardi A., Grinberg A., Tran T., Scharton-Kersten T., Anver M., et al. Mice deficient in nuclear factor (NF)-κB/p52 present with defects in humoral responses, germinal center reactions, and splenic microarchitecture. J. Exp. Med. 1998;187:147–159. doi: 10.1084/jem.187.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Senftleben U., Cao Y., Xiao G., Greten F. R., Krahn G., Bonizzi G., Chen Y., Hu Y., Fong A., Sun S.-C., Karin M. Activation by IKKα of a second evolutionary conserved, NF-κB signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 45.Yamada T., Mitani T., Yorita K., Uchida D., Matsushima A., Iwamasa K., Fujita S., Matsumoto M. Abnormal immune function of hematopoietic cells from alymphoplasia (aly) mice, a natural strain with mutant NF-κB-inducing kinase. J. Immunol. 2000;165:804–812. doi: 10.4049/jimmunol.165.2.804. [DOI] [PubMed] [Google Scholar]
- 46.Xiao G., Harhaj E. W., Sun S.-C. NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol. Cell. 2001;7:401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- 47.Koike R., Nishimura T., Yasumizu R., Tanaka H., Hataba Y., Watanabe T., Miyawaki S., Miyasaka M. The splenic marginal zone is absent in alymphoplastic aly mutant mice. Eur. J. Immunol. 1996;26:669–675. doi: 10.1002/eji.1830260324. [DOI] [PubMed] [Google Scholar]
- 48.Yin L., Wu L., Wesche H., Arthur C. D., White J. M., Goeddel D. V., Schreiber R. D. Defective lymphotoxin-β receptor-induced NF-κB transcriptional activity in NIK-deficient mice. Science. 2001;291:2162–2165. doi: 10.1126/science.1058453. [DOI] [PubMed] [Google Scholar]
- 49.Miyawaki S., Nakamura Y., Suzuka H., Koba M., Ikehara S., Shibata Y. A new mutation, aly, that induces a generalized lack of lymph nodes accompanied by immunodeficiency in mice. Eur. J. Immunol. 1994;24:429–434. doi: 10.1002/eji.1830240224. [DOI] [PubMed] [Google Scholar]
- 50.Kaisho T., Takeda K., Tsujimura T., Kawai T., Nomura F., Terada N., Akira S. IκB kinase α is essential for mature B cell development and function. J. Exp. Med. 2001;193:417–426. doi: 10.1084/jem.193.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Regnier C. H., Song H. Y., Gao X., Goeddel D. V., Cao Z., Rothe M. Identification and characterization of an IκB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 52.Ling L., Cao Z., Goeddel D. V. NF-κB-inducing kinase activates IKK-α by phosphorylation of Ser-176. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3792–3797. doi: 10.1073/pnas.95.7.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao G., Cvijic M. E., Fong A., Harhaj E. W., Uhlik M. T., Waterfield M., Sun S.-C. Retroviral oncoprotein Tax induces processing of NF-κB2/p100 in T cells: evidence for the involvement of IKKα. EMBO J. 2001;20:6805–6815. doi: 10.1093/emboj/20.23.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Derudder E., Dejardin E., Pritchard L. L., Green D. R., Korner M., Baud V. RelB/p50 dimers are differentially regulated by tumor necrosis factor-α and lymphotoxin-β receptor activation. J. Biol. Chem. 2003;278:23278–23284. doi: 10.1074/jbc.M300106200. [DOI] [PubMed] [Google Scholar]
- 55.Gommerman J. L., Browning J. L. Lymphotoxin/LIGHT, lymphoid microenviroments and autoimmunedisease. Nat. Immunol. Rev. 2003;3:642–655. doi: 10.1038/nri1151. [DOI] [PubMed] [Google Scholar]
- 56.Muller J. R., Siebenlist U. Lymphotoxin β receptor induces sequential activation of distinct NF-κB factors via separate signaling pathways. J. Biol. Chem. 2003;278:12006–12012. doi: 10.1074/jbc.M210768200. [DOI] [PubMed] [Google Scholar]
- 57.Dejardin E., Droin N. M., Delhase M., Haas E., Cao Y., Makris C., Li Z.-W., Karin M., Ware C. F., Green D. R. The lymphotoxin-β receptor induces different pathways of gene expression via two NF-κB pathways. Immunity. 2002;17:525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- 58.Karrer U., Althage A., Odermatt B., Hengartner H., Zinkernagel R. M. Immunodeficiency of alymphoplasia mice (aly/aly) in vivo: structural defect in secondary lymphoid organs and functional B cell defect. Eur. J. Immunol. 2000;30:2799–2807. doi: 10.1002/1521-4141(200010)30:10<2799::AID-IMMU2799>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 59.Ware C. F., VanArsdale T. L., Crowe P. D., Browning J. L. The ligands and receptors of the lymphotoxin system. Curr. Top. Microbiol. Immunol. 1995;198:175–218. doi: 10.1007/978-3-642-79414-8_11. [DOI] [PubMed] [Google Scholar]
- 60.Kayagaki N., Yan M., Seshasayee D., Wang H., Lee W., French D. M., Grewal I. S., Cochran A. G., Gordon N. C., Yin J., et al. BAFF/BLys receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-κB2. Immunity. 2002;10:515–524. doi: 10.1016/s1074-7613(02)00425-9. [DOI] [PubMed] [Google Scholar]
- 61.Coope H. J., Atkinson P. G. P., Huhse B., Belich M. P., Janzen J., Holman M., Klaus G. G. B., Johnston L. H., Ley S. C. CD40 regulates the processing of NF-κB2 p100 to p52. EMBO J. 2002;21:5375–5385. doi: 10.1093/emboj/cdf542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mackay F., Schneider P., Rennert P., Browning J. BAFF and APRIL: a tutorial on B cell survival. Annu. Rev. Immunol. 2003;21:231–264. doi: 10.1146/annurev.immunol.21.120601.141152. [DOI] [PubMed] [Google Scholar]
- 63.Zarnegar B., He J. Q., Oganesyan G., Hoffman A., Baltimore D., Cheng G. Unique CD40-mediated biological program in B cell activation requires both type 1 and type 2 NF-κB activation pathways. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8108–8113. doi: 10.1073/pnas.0402629101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teitelbaum S. L. Bone resorption by osteoclasts. Science. 2000;289:1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 65.Novack D. V., Yin L., Hagen-Stapleton A., Schreiber R. D., Goeddel D. V., Ross F. P., Teitelbaum S. L. The IκB function of NF-κB2 p100 controls stimulated osteoclastogenesis. J. Exp. Med. 2003;198:771–781. doi: 10.1084/jem.20030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saitoh T., Nakayama M., Nakano H., Yagita H., Yamamoto N., Yamaoka S. TWEAK induces NF-κB2 p100 processing and long-lasting NF-κB activation. J. Biol. Chem. 2003;278:36005–36012. doi: 10.1074/jbc.M304266200. [DOI] [PubMed] [Google Scholar]
- 67.Saccani S., Pantano S., Natoli G. Modulation of NF-κB activity by exchange of dimers. Mol. Cell. 2003;11:1563–1574. doi: 10.1016/s1097-2765(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 68.Mordmuller B., Krappmann D., Esen M., Wegener E., Scheidereit C. Lymphotoxin and lipopolysaccharide induce NF-κB p52 generation by a co-translational mechanism. EMBO Rep. 2003;4:82–87. doi: 10.1038/sj.embor.embor710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ekengren S., Hultmark D. Drosophila cecropin as an antifungal agent. Insect Biochem. Mol. Biol. 1999;29:965–972. doi: 10.1016/s0965-1748(99)00071-5. [DOI] [PubMed] [Google Scholar]
- 70.Malinin N. L., Boldin M. P., Kovalenko A. V., Wallach D. MAP 3K-related kinase involved in NF-κB induction by TNF, CD95 and IL-1. Nature (London) 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 71.Wajant H., Henkler F., Scheurich P. The TNF-receptor-associated factor family: scaffold molecules for cytokine receptors, kinases and their regulators. Cell. Signalling. 2001;13:389–400. doi: 10.1016/s0898-6568(01)00160-7. [DOI] [PubMed] [Google Scholar]
- 72.Grammer A. C., Lipsky P. E. CD40-mediated regulation of immune responses by TRAF-dependent and TRAF-independent signaling mechanisms. Adv. Immunol. 2001;76:61–178. doi: 10.1016/s0065-2776(01)76019-1. [DOI] [PubMed] [Google Scholar]
- 73.Saito N., Courtois G., Chiba A., Yamamoto N., Nitta T., Hironaka N., Rowe M., Yamamoto N., Yamaoka S. Two carboxy-terminal activation regions of Epstein–Barr virus latent membrane protein 1 activate NF-κB through distinct signaling pathways in fibroblast cell lines. J. Biol. Chem. 2003;278:46565–46575. doi: 10.1074/jbc.M302549200. [DOI] [PubMed] [Google Scholar]
- 74.Tada K., Okazaki T., Sakon S., Kobarai T., Kurosawa K., Yamaoka S., Hashimoto H., Mak T. W., Yagita H., Okumura K., et al. Critical roles of TRAF2 and TRAF5 in TNF-induced NF-κB activaiton and protection from cell death. J. Biol. Chem. 2001;276:36530–36534. doi: 10.1074/jbc.M104837200. [DOI] [PubMed] [Google Scholar]
- 75.Wang C., Deng L., Hong M., Akkaraju G. R., Inoue J.-I., Chen Z. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature (London) 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 76.Sun L., Chen Z. J. The novel functions of ubiquitination in signaling. Curr. Opin. Cell Biol. 2004;16:1–8. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 77.Kovalenko A., Chable-Bessia C., Cantarella G., Israel A., Wallach D., Courtois G. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature (London) 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 78.Trompouki E., Hatzivassiliou E., Tsichritzis T., Farmer H., Ashworth A., Mosalios G. CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activation by TNFR family members. Nature (London) 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 79.Liao G., Zhang M., Harhaj E. W., Sun S.-C. Regulation of the NF-κB inducing kinase by TRAF3-induced degradation. J. Biol. Chem. 2004;279:26243–26250. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- 80.Atkinson P. G. P., Coope H. J., Rowe M., Ley S. C. Latent membrane protein 1 of Epstein–Barr virus stimulates processing of NF-κB p100 to p52. J. Biol. Chem. 2004;279:51134–51142. doi: 10.1074/jbc.M304771200. [DOI] [PubMed] [Google Scholar]
- 81.Fong A., Sun S.-C. Genetic evidence for the essential role of β-transducin repeat-containing protein in the inducible processing of NF-κB2/p100. J. Biol. Chem. 2002;277:22111–22114. doi: 10.1074/jbc.C200151200. [DOI] [PubMed] [Google Scholar]
- 82.Amir R. E., Haecker H., Karin M., Ciechanover A. Mechanism of processing of the NF-κB2 p100 precursor: identification of the specific polyubiquitin chain-anchoring lysine residue and analysis of the role of NEDD8-modification on the SCFβ-TrCP ubiquitin ligase. Oncogene. 2004;23:2540–2547. doi: 10.1038/sj.onc.1207366. [DOI] [PubMed] [Google Scholar]
- 83.Fong A., Zhang M., Neely J., Sun S.-C. S9, a 19 S proteasome subunit interacting with ubiquitinated NF-κB2/p100. J. Biol. Chem. 2002;277:40697–40702. doi: 10.1074/jbc.M205330200. [DOI] [PubMed] [Google Scholar]
- 84.Heusch M., Lin L., Geleiunas R., Greene W. C. The generation of nfkb2 p52: mechanism and efficiency. Oncogene. 1999;18:6201–6208. doi: 10.1038/sj.onc.1203022. [DOI] [PubMed] [Google Scholar]
- 85.Liao G., Sun S.-C. Regulation of NF-κB2/p100 processing by its nuclear shuttling. Oncogene. 2003;22:4868–4874. doi: 10.1038/sj.onc.1206761. [DOI] [PubMed] [Google Scholar]
- 86.Rayet B., Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 87.Ciana P., Neri A., Cappellini C., Cavallo F., Pomati M., Chang C.-C., Maiolo A. T., Lombardi L. Constitutive expression of lymphoma-associated NF-κB/Lyt-10 proteins is tumorigenic in murine fibroblasts. Oncogene. 1997;14:1805–1810. doi: 10.1038/sj.onc.1201015. [DOI] [PubMed] [Google Scholar]
- 88.Chang C.-C., Zhang J., Lombardi L., Neri A., Dalla-Favera R. Rearranged NFKB-2 genes in lymphoid neoplasms code for constitutively active nuclear transactivators. Mol. Cell. Biol. 1995;15:5180–5187. doi: 10.1128/mcb.15.9.5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim K.-E., Gu C., Thakur S., Vieira E., Lin J. C., Rabson A. B. Transcriptional regulatory effects of lymphoma-associated NFKB2/lyt10 protooncogenes. Oncogene. 2000;19:1334–1345. doi: 10.1038/sj.onc.1203432. [DOI] [PubMed] [Google Scholar]
- 90.Westerheide S. D., Mayo M. W., Anest V., Hanson J. L., Baldwin A. S. The putative oncoprotein Bcl-3 induces cyclin D1 to stimulate G1 transition. Mol. Cell. Biol. 2001;21:8428–8436. doi: 10.1128/MCB.21.24.8428-8436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Viatour P., Bentires-Alj M., Chariot A., Deregowski V., de Leval L., Merville M.-P., Bours V. NF-κB2/p100 induces Bcl-2 expression. Leukemia. 2003;17:1349–1356. doi: 10.1038/sj.leu.2402982. [DOI] [PubMed] [Google Scholar]
- 92.Lanoix J., Lacoste J., Pepin N., Rice N., Hiscott J. Overproduction of NF-κB2 (lyt-10) and c-Rel: a mechanism for HTLV-1 Tax-mediated trans-activation via the NF-κB signaling pathway. Oncogene. 1994;9:841–852. [PubMed] [Google Scholar]
- 93.Eliopoulos A. G., Caamano J., Flavell J., Reynolds G. M., Murray P. G., Poyet J.-L., Young L. S. Epstein–Barr virus-encoded latent infection membrane protein 1 regulates the processing of p100 NF-κB2 to p52 via an IKKγ/NEMO-independent signalling pathway. Oncogene. 2003;22:7557–7569. doi: 10.1038/sj.onc.1207120. [DOI] [PubMed] [Google Scholar]
- 94.Luftig M. A., Yasui T., Soni V., Kang M.-S., Jacobson N., Cahir-McFarland E., Seed B., Kieff E. Epstein–Barr virus latent infection membrane protein 1 TRAF-binding site induces NIK/IKKα-dependent non-canonical NF-κB activation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:141–146. doi: 10.1073/pnas.2237183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Geleziunas R., Ferrell S., Lin X., Mu Y., Cunningham E. T. J., Grant M., Connelly M. A., Hambor J. E., Marcu K. B., Greene W. C. Human T cell leukemia virus type I Tax induction of NF-κB involves activation of the IκB kinase α (IKKα) and IKKβ cellular kinases. Mol. Cell. Biol. 1998;18:5157–5165. doi: 10.1128/mcb.18.9.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chu Z.-L., DiDonato J. A., Hawiger J., Ballard D. W. The Tax oncoprotein of human T cell leukemia virus type I associates with and persistently activates IκB kinases containing IKKα and IKKβ. J. Biol. Chem. 1998;273:15891–15894. doi: 10.1074/jbc.273.26.15891. [DOI] [PubMed] [Google Scholar]
- 97.Hatzivassiliou E., Mosalios G. Cellular signaling pathways engaged by the Epstein–Barr virus transforming protein LMP1. Front. Biosci. 2002;7:319–329. doi: 10.2741/hatziva. [DOI] [PubMed] [Google Scholar]
- 98.Wang Y., Cui H., Schroering A., Ding J. L., Lane W. S., McGill G., Fisher D. E., Ding H.-F. NF-κB2 p100 is a pro-apoptotic protein with anti-oncogenic function. Nat. Cell Biol. 2002;4:888–893. doi: 10.1038/ncb872. [DOI] [PubMed] [Google Scholar]
- 99.Hacker H., Karin M. Is NF-κB2/p100 a direct activator of programmed cell death? Cancer Cell. 2002;2:431–433. doi: 10.1016/s1535-6108(02)00213-1. [DOI] [PubMed] [Google Scholar]
- 100.Rice N. R., MacKichan M. L., Israel A. The precursor of NF-κB p50 has IκB-like functions. Cell. 1992;71:243–253. doi: 10.1016/0092-8674(92)90353-e. [DOI] [PubMed] [Google Scholar]
- 101.Ishikawa H., Claudio E., Dambach D., Raventos-Suarez C., Ryan C., Bravo R. Chronic inflammation and susceptibility to bacterial infections in mice lacking the polypeptide (p) 105 precursor (NF-κB1) but expressing p50. J. Exp. Med. 1998;187:985–996. doi: 10.1084/jem.187.7.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liou H.-C., Nolan G. P., Ghosh S., Fujita T., Baltimore D. The NF-κB p50 precursor, p105, contains an internal IκB-like inhibitor that preferentially inhibits p50. EMBO J. 1992;11:3003–3009. doi: 10.1002/j.1460-2075.1992.tb05370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ciechanover A., Orian A., Schwartz A. L. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 104.Rape M., Jentsch S. Taking a bite: proteasomal protein processing. Nat. Cell Biol. 2002;4:113–116. doi: 10.1038/ncb0502-e113. [DOI] [PubMed] [Google Scholar]
- 105.Lin L., Ghosh S. A glycine-rich region in NF-κB p105 functions as a processing signal for the generation of the p50 subunit. Mol. Cell. Biol. 1996;16:2248–2254. doi: 10.1128/mcb.16.5.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Betts J. C., Nabel G. J. Differential regulation of NF-κB2 (p100) processing and control by amino-terminal sequences. Mol. Cell. Biol. 1996;16:6363–6371. doi: 10.1128/mcb.16.11.6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Orian A., Schwartz A. L., Israel A., Whiteside S., Kahana C., Ciechanover A. Structural motifs involves in ubiquitin-mediated processing of the NF-κB precursor p105: roles of the glycine-rich region and a downstream ubiquitination domain. Mol. Cell. Biol. 1999;19:3664–3673. doi: 10.1128/mcb.19.5.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Levitskaya J., Shapiro A., Leonchiks A., Ciechanover A., Masucci M. G. Inhibition of ubiquitination/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein–Barr virus nuclear antigen 1. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12616–12621. doi: 10.1073/pnas.94.23.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sharipo A., Imreh M., Leonchiks A., Imreh S., Masucci M. G. A minimal glycine-alanine repeat prevents the interaction of ubiquitinated IκBα with the proteasome: a new mechanism for selective inhibition of proteolysis. Nat. Med. 1998;4:939–944. doi: 10.1038/nm0898-939. [DOI] [PubMed] [Google Scholar]
- 110.Liu C. W., Corboy M. J., DeMartino G. N., Thomas P. J. Endoproteolytic activity of the proteasome. Science. 2003;299:408–411. doi: 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee C., Schwartz M. P., Prakash S., Iwakura M., Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them for the degradation signal. Mol. Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 112.Cohen S., Orian A., Ciechanover A. Processing of p105 is inhibited by docking of p50 active subunits to teh ankyrin repeat domain, and inhibition is alleviated by signaling via the carboxyl-terminal phosphorylation/ubiquitin-ligase binding domain. J. Biol. Chem. 2001;276:26769–26776. doi: 10.1074/jbc.M102448200. [DOI] [PubMed] [Google Scholar]
- 113.Harhaj E. W., Maggirwar S. B., Sun S. C. Inhibition of p105 processing by NF-κB proteins in transiently transfected cells. Oncogene. 1996;12:2385–2392. [PubMed] [Google Scholar]
- 114.Maier H. J., Marienfeld R., Wirth T., Baumann B. Critical role of RelB serine 368 for dimerization and p100 stabilization. J. Biol. Chem. 2003;278:39242–39250. doi: 10.1074/jbc.M301521200. [DOI] [PubMed] [Google Scholar]
- 115.Lin L., DeMartino G. N., Greene W. C. Cotranslational biogenesis of NF-κB p50 by the 26 S proteasome. Cell. 1998;92:819–828. doi: 10.1016/s0092-8674(00)81409-9. [DOI] [PubMed] [Google Scholar]
- 116.Fan C.-M., Maniatis T. Generation of p50 subunit of NF-κB by processing of p105 through an ATP-dependent pathway. Nature (London) 1991;354:395–398. doi: 10.1038/354395a0. [DOI] [PubMed] [Google Scholar]
- 117.Li L., DeMartino G. N., Greene W. C. Cotranslational dimerization of the Rel homology domain of NF-κB1 generates p50–p105 heterodimers and is required for effective p50 dimerization. EMBO J. 2000;19:4712–4722. doi: 10.1093/emboj/19.17.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Donald R., Ballard D. W., Hawiger J. Proteolytic processing of NF-κB/IκB in human monocytes. J. Biol. Chem. 1995;270:9–12. doi: 10.1074/jbc.270.1.9. [DOI] [PubMed] [Google Scholar]
- 119.Belich M. P., Salmeron A., Johnston L. H., Ley S. C. TPL-2 kinase regulates the proteolysis of the NF-κB inhibitory protein NF-κB1 p105. Nature (London) 1999;397:363–368. doi: 10.1038/16946. [DOI] [PubMed] [Google Scholar]
- 120.MacKichan M. L., Logeat F., Israel A. Phosphorylation of p105 PEST sequences via a redox-insensitive pathway up-regulates processing to p50 NF-κB. J. Biol. Chem. 1996;271:6084–6091. doi: 10.1074/jbc.271.11.6084. [DOI] [PubMed] [Google Scholar]
- 121.Mellits K. H., Hay R. T., Goodbourn S. Proteolytic degradation of MAD3 (IκBα) and enhanced processing of the NF-κB precursor p105 are obligatory steps in the activation of of NF-κB. Nucleic Acids Res. 1993;21:5059–5066. doi: 10.1093/nar/21.22.5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Salmeron A., Janzen J., Soneji Y., Bump N., Kamens J., Allen H., Ley S. C. Direct phosphorylation of NF-κB p105 by the IκB kinase complex on serine 927 is essential for signal-induced p105 proteolysis. J. Biol. Chem. 2001;276:22215–22222. doi: 10.1074/jbc.M101754200. [DOI] [PubMed] [Google Scholar]
- 123.Lang V., Janzen J., Fischer G. Z., Soneji Y., Beinke S., Salmeron A., Allen H., Hay R. T., Ben-Neriah Y., Ley S. C. βTrCP-mediated proteolysis of NF-κB1 p105 requires phosphorylation of p105 serines 927 and 932. Mol. Cell. Biol. 2003;23:402–413. doi: 10.1128/MCB.23.1.402-413.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Heissmeyer V., Krappmann D., Hatada E. N., Scheidereit C. Shared pathways of IκB kinase-induced SCFβTrCP-mediated ubiquitination and degradation for the NF-κB precursor p105 and IκBα. Mol. Cell. Biol. 2001;21:1024–1035. doi: 10.1128/MCB.21.4.1024-1035.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Beinke S., Belich M. P., Ley S. C. The death domain of NF-κB1 p105 is essential for signal-induced p105 proteolysis. J. Biol. Chem. 2002;277:24162–24168. doi: 10.1074/jbc.M201576200. [DOI] [PubMed] [Google Scholar]
- 126.Heissmeyer V., Krappmann D., Wulczyn F. G., Scheidereit C. NF-κB p105 is a target on IkB kinases and controls signal induction of BCL-3–p50 complexes. EMBO J. 1999;18:4766–4788. doi: 10.1093/emboj/18.17.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Orian A., Gonen H., Bercovich B., Fajerman I., Eytan E., Israel A., Mercurio F., Iwai K., Schwartz A. L., Ciechanover A. SCFβTrCP ubiquitin ligase-mediated processing of NF-κB p105 requires phosphorylation of its C-terminus by IκB kinase. EMBO J. 2000;19:2580–2591. doi: 10.1093/emboj/19.11.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Syrovets T., Jendrach M., Rohwedder A., Schule A., Simmet T. Plasmin-induced expression of cytokines and tissue factor in human monocytes involves AP-1 and IKK-β-mediated NF-κB activation. Blood. 2001;97:3941–3950. doi: 10.1182/blood.v97.12.3941. [DOI] [PubMed] [Google Scholar]
- 129.Ten R. M., Paya C. V., Israel N., Le Bail O., Mattei M. G., Virelizier J. L., Kourilsky P., Israel A. The characterization of the promoter of the gene encoding the p50 subunit of NF-κB indicates that it participates in its own regulation. EMBO. J. 1992;11:195–203. doi: 10.1002/j.1460-2075.1992.tb05042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cohen S., Achbert-Weiner H., Ciechanover A. Dual effects of IκB kinase β-mediated phosphorylation on p105 fate: SCFβ-TrCP-dependent degradation and SCFβ-TrCP-independent processing. Mol. Cell. Biol. 2004;24:475–486. doi: 10.1128/MCB.24.1.475-486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wu G., Xu G., Schulman B. A., Jeffrey P. D., Harper J. W., Pavletich N. P. Structure of a β-TrCP1–Skp1–β-catenin complex: destruction motif binding and lysine specificity of the SCFβTrCP1 ubiquitin ligase. Genes Dev. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 132.Demarchi F., Bertoli C., Sandy P., Schneider C. Glycogen synthase-3β regulates NF-κB1/p105 stability. J. Biol. Chem. 2003;278:39583–39590. doi: 10.1074/jbc.M305676200. [DOI] [PubMed] [Google Scholar]
- 133.Demarchi F., Verardo R., Varnum B., Brancolini C., Schneider C. Gas6 anti-apoptotic signaling requires NF-κB activation. J. Biol. Chem. 2001;276:31738–31744. doi: 10.1074/jbc.M104457200. [DOI] [PubMed] [Google Scholar]
- 134.Takeda K., Akira S. TLR signaling pathways. Semin. Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 135.Patriotis C., Makris A., Bear S. E., Tsichlis P. N. Tumor progression locus 2 (Tpl-2) encodes a protein kinase involved in the progression of rodent T cell lymphomas and in T cell activation Proc. Natl. Acad. Sci. U.S.A. 1993;90:2251–2255. doi: 10.1073/pnas.90.6.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Miyoshi J., Higashi T., Mukai H., Ohuchi T., Kakunaga T. Structure and transforming potential of the human cot oncogene encoding a putative protein kinase. Mol. Cell. Biol. 1991;11:4088–4096. doi: 10.1128/mcb.11.8.4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Salmeron A., Ahmad T. B., Carlile G. W., Pappin D., Narsimhan R. P., Ley S. C. Activation of MEK-1 and SEK-1 by Tpl-2 proto-oncoprotein, a novel MAP kinase kinase kinase. EMBO J. 1996;15:817–826. [PMC free article] [PubMed] [Google Scholar]
- 138.Dumitru C. D., Ceci J. D., Tsatsanis C., Kontoyiannis D., Stamatakis K., Lin J.-H., Patriotis C., Jenkins N. A., Copeland N. G., Kollias G., Tsichlis P. N. TNFα induction by LPS is regulated post-transcriptionally via a TPL2/ERK-dependent pathway. Cell. 2000;103:1071–1083. doi: 10.1016/s0092-8674(00)00210-5. [DOI] [PubMed] [Google Scholar]
- 139.Eliopoulos A. G., Dumitru C. D., Wang C.-C., Cho J., Tsichlis P. N. Induction of COX-2 by LPS in macrophages is regulated by TPL2-dependent CREB activation signals. EMBO J. 2002;21:4831–4840. doi: 10.1093/emboj/cdf478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Eliopoulos A. G., Wang C.-C., Dumitru C. D., Tsichlis P. N. TPL-2 transduces CD40 and TNF signals that activate ERK and regulates IgE induction by CD40. EMBO J. 2003;22:3855–3864. doi: 10.1093/emboj/cdg386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Beinke S., Deka J., Lang V., Belich M. P., Walker P. A., Howell S., Smerdon S. J., Gamblin S. J., Ley S. C. NF-κB p105 negatively regulates TPL-2 MEK kinase activity. Mol. Cell. Biol. 2003;23:4739–4752. doi: 10.1128/MCB.23.14.4739-4752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lang V., Symons A., Watton S. J., Janzen J., Soneji Y., Beinke S., Howell S., Ley S. C. ABIN2 forms a ternary complex with TPL-2 and NF-κB1 p105 and is essential for TPL-2 protein stability. Mol. Cell. Biol. 2004;24:5235–5248. doi: 10.1128/MCB.24.12.5235-5248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Waterfield M. R., Zhang M., Norman L. P., Sun S.-C. NF-κB1/p105 regulates lipopolysaccharide-stimulated MAP kinase signaling by governing the stability and function of the TPL-2 kinase. Mol. Cell. 2003;11:685–694. doi: 10.1016/s1097-2765(03)00070-4. [DOI] [PubMed] [Google Scholar]
- 144.Ceci J. D., Patriotis C. P., Tsatsanis C., Makris A. M., Kovatch R., Swing D. A., Jenkins N. A., Tsichlis P. N., Copeland N. G. TPL-2 is an oncogenic kinase that is activated by carboxy-terminal truncation. Genes Dev. 1997;11:688–700. doi: 10.1101/gad.11.6.688. [DOI] [PubMed] [Google Scholar]
- 145.Swantek J. L., Christerson L., Cobb M. H. Lipopolysaccaride-induced tumor necrosis factor-α promoter activity is inhibitor of nuclear factor-κB kinase dependent. J. Biol. Chem. 1999;274:11667–11671. doi: 10.1074/jbc.274.17.11667. [DOI] [PubMed] [Google Scholar]
- 146.Chen C.-C., Sun Y.-T., Chen J.-J., Chang Y.-J. Tumor necrosis factor-α-induced cyclooxygenase-2 expression via sequential activation of ceramide-dependent mitogen-activated protein kinases, and IκB kinase 1/2 in human alveolar epithelial cells. Mol. Pharmacol. 2001;59:493–500. doi: 10.1124/mol.59.3.493. [DOI] [PubMed] [Google Scholar]
- 147.Caivano M., Gorgoni B., Cohen P., Poli V. The induction of cyclooxygenase-2 mRNA in macrophages is biphasic and requires both CCAAT enhancer-binding protein β (C/EBP β) and C/EBPδ transcript. J. Biol. Chem. 2001;276:48693–48701. doi: 10.1074/jbc.M108282200. [DOI] [PubMed] [Google Scholar]
- 148.Park J. M., Brady H., Ruocco M. G., Sun H., Williams D., Lee S. J., Kato T., Richards N., Chan K., Mercurio F., et al. Targeting of TAK1 by the NF-κB protein Relish regulates the JNK-mediated immune response in Drosophila. Genes Dev. 2004;18:584–594. doi: 10.1101/gad.1168104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Varfolomeev E. E., Ashkenazi A. Tumor necrosis factor: an apoptosis JuNKie? Cell. 2004;116:491–497. doi: 10.1016/s0092-8674(04)00166-7. [DOI] [PubMed] [Google Scholar]
- 150.Ferrier R., Nougarede R., Doucet S., Kahn-Perles B., Imbert J., Mathieu-Mahul D. Physical interaction of the bHLH LYL1 protein and NF-κB1 p105. Oncogene. 1999;18:995–1005. doi: 10.1038/sj.onc.1202374. [DOI] [PubMed] [Google Scholar]
- 151.Baer R. TAL1, TAL2 and LYL1: a family of basic helix–loop–helix proteins implicated in T cell acute leukaemia. Semin. Cancer Biol. 1993;4:341–347. [PubMed] [Google Scholar]
- 152.Li Z., Zhang J., Chen D., Shu H.-B. Casper/c-FLIP is physically and functionally associated with NF-κB1 p105. Biochem. Biophys. Res. Commun. 2003;309:980–985. doi: 10.1016/j.bbrc.2003.08.104. [DOI] [PubMed] [Google Scholar]
- 153.Krueger A., Baumann S., Krammer P. H., Kirchhoff S. FLICE-inhibitory proteins: regulators of death receptor-mediated apoptosis. Mol. Cell. Biol. 2001;21:8247–8254. doi: 10.1128/MCB.21.24.8247-8254.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bouwmeester T., Bauch A., Ruffner H., Angrand P.-O., Bergamini G., Croughton K., Cruciat C., Eberhard D., Gagneur J., Ghidelli S. A physical and functional map of the human TNFα/NF-κB signal transduction pathway. Nat. Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 155.van Huffel S., Delaei F., Heyninck K., de Valck D., Beyaert R. Identification of a novel A20-binding inhibitor of nuclear factor-κB activation termed ABIN-2. J. Biol. Chem. 2001;276:30216–30223. doi: 10.1074/jbc.M100048200. [DOI] [PubMed] [Google Scholar]
- 156.Beyaert R., Heyninck K., van Huffel S. A20 and A20-binding proteins as cellular inhibitors of nuclear factor-κB-dependent gene expression and apoptosis. Biochem. Pharmacol. 2000;60:1143–1151. doi: 10.1016/s0006-2952(00)00404-4. [DOI] [PubMed] [Google Scholar]
- 157.Lee E. G., Boone D. L., Chai S., Libby S. L., Chien M., Lodolce J. P., Ma A. Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sha W. C., Liou H.-C., Tuomanen E. I., Baltimore D. Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 159.Grumont R. J., Rourke I. J., O'Reilly L. A., Strasser A., Miyake K., Sha W., Gerondakis S. B lymphocytes differentially use the Rel and nuclear factor κB1 (NF-κB1) transcription factors to regulate cell cycle progression and apoptosis in quiescent and mitogen-activated cells. J. Exp. Med. 1998;187:663–674. doi: 10.1084/jem.187.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sha W. C., Lious H.-C., Tuomanen E. I., Baltimore D. Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 161.Snapper C., Zelazowski P., Rosas F. R., Kehry M. R., Tian M., Baltimore D., Sha W. C. B cells from p50/NF-κB knockout mice have selective defects in proliferation, differentiation, germ-line CH transcription, and Ig class switching. J. Immunol. 1996;156:183–191. [PubMed] [Google Scholar]
- 162.Zheng Y., Ouaaz F., Bruzzo P., Singh V., Gerondakis S., Beg A. A. NF-κB RelA (p65) is essential for TNF-α-induced Fas expression but dispensable for both TCR-induced expression and activation-induced cell death. J. Immunol. 2001;166:4949–4957. doi: 10.4049/jimmunol.166.8.4949. [DOI] [PubMed] [Google Scholar]
- 163.Artis D., Speirs K., Joyce K., Goldschmidt M., Caamano J., Hunter C. A., Scott P. NF-κB1 is required for optimal CD4+ Th1 cell development and resistance to Leishmania major. J. Immunol. 2003;170:1995–2003. doi: 10.4049/jimmunol.170.4.1995. [DOI] [PubMed] [Google Scholar]
- 164.Artis D., Shapira S., Mason N., Speirs K. M., Goldschmidt M., Caamano J., Liou H. C., Hunter C. A., Scott P. Differential requirement for NF-κB family members in control of helminth infection and intestinal inflammation. J. Immunol. 2002;169:4481–4487. doi: 10.4049/jimmunol.169.8.4481. [DOI] [PubMed] [Google Scholar]
- 165.Das J., Chen C. H., Yang L., Cohn L., Ray P., Ray A. A critical role for NF-κB in GATA3 expression and TH2 differentiation in allergic airway inflammation. Nat. Immunol. 2001;2:45–50. doi: 10.1038/83158. [DOI] [PubMed] [Google Scholar]
- 166.Kanters E., Gijbels M. J., van der Made I., Vergouwe M. N., Heeringa P., Kraal G., Hofker M. H., de Winther M. P. Hematopoietic NF-κB1 deficiency results in small atherosclerotic lesions with an inflammatory phenotype. Blood. 2004;103:934–940. doi: 10.1182/blood-2003-05-1450. [DOI] [PubMed] [Google Scholar]
- 167.Bohuslav J., Kravchenko V. V., Parry G. C. N., Erlich J. H., Gerondakis S., Mackman N., Ulevitch R. J. Regulation of an essential innate immune response by the p50 subunit of NF-κB. J. Clin. Invest. 1998;102:1645–1652. doi: 10.1172/JCI3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lamhamedi-Cherradi S.-E., Zheng S., Hilliard B. A., Xu L., Sun J., Alsheadat S., Liou H.-C., Chen Y. H. Transcriptional regulation of type I diabetes by NF-κB. J. Immunol. 2003;171:4886–4892. doi: 10.4049/jimmunol.171.9.4886. [DOI] [PubMed] [Google Scholar]
- 169.Campbell I. K., Gerondakis S., O'Donnell K., Wicks I. P. Distinct roles for the NF-κB1 (p50) and c-Rel transcription factors in inflammatory arthritis. J. Clin. Invest. 2000;105:1799–1806. doi: 10.1172/JCI8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yang L., Cohn L., Zhang D. H., Homer R., Ray A., Ray P. Essential role of nuclear factor κB in the induction of eosinophilia in allergic airway inflammation. J. Exp. Med. 1998;188:1739–1750. doi: 10.1084/jem.188.9.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Erdman S. E., Fox J. G., Dangler C. A., Feldman D., Horwitz B. H. Typhlocolitis in NF-κB-deficient mice. J. Immunol. 2001;166:1443–1447. doi: 10.4049/jimmunol.166.3.1443. [DOI] [PubMed] [Google Scholar]
- 172.Karban A. S., Okazaki T., Panhuysen C. I., Gallegos T., Potter J. J., Bailey-Wilson J. E., Silverberg M. S., Duerr R. H., Cho J. H., Gregersen P. K., et al. Functional annotation of a novel NFKB1 promoter polymorphism that increases risk for ulcerative colitis. Hum. Mol. Genet. 2004;13:35–45. doi: 10.1093/hmg/ddh008. [DOI] [PubMed] [Google Scholar]
- 173.Ishikawa H., Ryseck R. P., Bravo R. Charaterization of ES cells deficient for the p105 precursor (NF-κB1): role of p50 NLS. Oncogene. 1996;13:255–263. [PubMed] [Google Scholar]
- 174.Liptay S., Schmid R. M., Perkins N. D., Meltzer P., Altherr M. R., McPherson J. D., Walsmuth J. J., Nabel G. J. Related subunits of NF-κB map to two distinct loci associated with translations in leukemia, NF-κB1 and NF-κB2. Genomics. 1992;13:287–292. doi: 10.1016/0888-7543(92)90244-m. [DOI] [PubMed] [Google Scholar]
- 175.Bours V., Dejardin E., Goujon-Letawe F., Merville M.-P., Castronovo V. The NF-κB transcription factor and cancer: high expression of NF-κB. Biochem. Pharmacol. 1994;47:145–149. doi: 10.1016/0006-2952(94)90448-0. [DOI] [PubMed] [Google Scholar]
- 176.Mukhopadhyay T., Roth J. A., Maxwell S. A. Altered expression of the p50 subunit of the NF-κB transcription factor complex in non-small cell lung carcinoma. Oncogene. 1995;11:999–1003. [PubMed] [Google Scholar]
- 177.Michaux L., Mecucci C., Stul M., Wlodarska I., Hernandez J. M., Meeus P., Michaux J. L., Scheiff M. M., Noel H., Louwagie A., et al. BCL3 rearrangement and t(14;19) (q32;q13) in lymphoproliferative disorders. Genes Chromosomes Cancer. 1996;15:38–47. doi: 10.1002/(SICI)1098-2264(199601)15:1<38::AID-GCC6>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 178.Lenardo M., Siebenlist U. Bcl-3-mediated nuclear regulation of the NF-κB transactivating factor. Immunol. Today. 1994;15:145–147. doi: 10.1016/0167-5699(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 179.Ong S. T., Hackbarth M. L., Degenstein L. C., Baunoch D. A., Anastasi J., McKeithan T. W. Lymphadenopathy, splenomegaly and altered immunoglobulin production in BCL-3 transgenic mice. Oncogene. 1998;16:2333–2343. doi: 10.1038/sj.onc.1201771. [DOI] [PubMed] [Google Scholar]
- 180.Thornburg N. J., Pathmanathan R., Raab-Traub N. Activation of nuclear factor-κB p50 homodimer/BCL-3 complexes in nasopharyngeal carcinoma. Cancer Res. 2003;63:8293–8301. [PubMed] [Google Scholar]
- 181.Sun S.-C., Ballard D. W. Persistent activation of NF-κB by the Tax transforming protein of HTLV-1: hijacking cellular IκB kinases. Oncogene. 1999;18:6948–6958. doi: 10.1038/sj.onc.1203220. [DOI] [PubMed] [Google Scholar]
- 182.Watanabe M., Muramatsu M. A., Hirai H., Suzuki T., Fujisawa J., Yoshida M., Arai K. I., Arai N. HTLV-1 encoded Tax in association with NF-κB precursor p105 enhances nuclear localization of NF-κB p50 and p65 in transfected cells. Oncogene. 1993;8:2949–2958. [PubMed] [Google Scholar]
- 183.Hirai H., Fujisawa J., Suzuki T., Ueda K., Muramatsu M., Tsuboi A., Arai N., Yoshida M. Transcriptional activator Tax of HTLV-1 binds to the NF-κB precursor p105. Oncogene. 1992;7:1737–1742. [PubMed] [Google Scholar]
- 184.Munoz E., Courtois G., Veschambre P., Jalinot P., Israel A. Tax induces nuclear translocation of NF-κB through dissociation of cytoplasmic complexes containing p105 or p100 but does not induce degradation of IκBα/MAD3. J. Virol. 1994;68:8035–8044. doi: 10.1128/jvi.68.12.8035-8044.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rousset R., Desbois C., Bantignies F., Jalinot P. Effects on NF-κB1/p105 processing of the interaction between the HTLV-1 transactivator Tax and the proteasome. Nature (London) 1996;381:328–331. doi: 10.1038/381328a0. [DOI] [PubMed] [Google Scholar]
- 186.Karin M., Yamamoto Y., Wang Q. M. The IKK NF-κB system: a treasure trove for drug development. Nat. Rev. Drug Discov. 2004;3:17–26. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 187.Sun S.-C., Xiao G. Deregulation of NF-κB and its upstream kinases in cancer. Cancer Metastasis Rev. 2003;22:405–422. doi: 10.1023/a:1023733231406. [DOI] [PubMed] [Google Scholar]
- 188.Kontoyiannis D., Boulougouris G., Manoloukos M., Armaka M., Apostolaki M., Pizarro T. T., Kotlyarov A., Forster I., Flavell R. A., Gaestel M., et al. Genetic dissection of the cellular pathways and signaling mechanisms in modeled tumor necrosis factor-induced Crohn's-like inflammatory bowel disease. J. Exp. Med. 2002;196:1563–1574. doi: 10.1084/jem.20020281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Locksley R. M., Killeen N., Lenardo M. J. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 190.Marshall C. J. MAP kinase kinase kinase, MAP kinase kinase and MAP kinase. Curr. Opin. Gene. Dev. 1994;4:82–89. doi: 10.1016/0959-437x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 191.Caamano J., Tato C., Cai G., Villegas E. N., Speirs K., Craig L., Alexander J., Hunter C. A. Identification of a role for NF-κB2 in the regulation of apoptosis and in maintainance of T cell-mediated immunity to Toxoplasma gondii. J. Immunol. 2000;165:5720–5728. doi: 10.4049/jimmunol.165.10.5720. [DOI] [PubMed] [Google Scholar]
- 192.Weih F., Warr G., Yang H., Bravo R. Multifocal defects in immune responses in RelB-deficient mice. J. Immunol. 1997;158:5211–5218. [PubMed] [Google Scholar]
- 193.Weih D. S., Yilmaz Z. B., Weih F. Essential role of RelB in germinal center and marginal zone formation and proper expression of homing chemokines. J. Immunol. 2001;167:1909–1919. doi: 10.4049/jimmunol.167.4.1909. [DOI] [PubMed] [Google Scholar]
- 194.Weih F., Carrasco D., Durham S. K., Barton D. S., Rizzo C. A., Ryseck R.-P., Lira S. A., Bravo R. Multiorgan inflammation and hematopoietic abnormalities in mice with targeted disruption of RelB, a member of the NF-κB/Rel family. Cell. 1995;80:331–340. doi: 10.1016/0092-8674(95)90416-6. [DOI] [PubMed] [Google Scholar]
- 195.Snapper C. M., Rosas F. R., Zelazowski P., Moorman M. A., Kehry M. R., Bravo R., Weih F. B cells lacking RelB are defective in proliferative responses, but undergo normal B cell maturation to Ig secretion and Ig class switching. J. Exp. Med. 1996;184:1537–1541. doi: 10.1084/jem.184.4.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Burkly L., Hession C., Ogata L., Reilly C., Marconi L. A., Olson D., Tizard R., Cate R., Lo D. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature (London) 1995;373:531–536. doi: 10.1038/373531a0. [DOI] [PubMed] [Google Scholar]
- 197.Paxian S., Merkle H., Riemann M., Wilda M., Adler G., Hameister H., Liptay S., Pfeffer K., Schmid R. M. Abnormal organogenesis of Peyer's patches in mice deficient for NF-κB1, NF-κB2, and Bcl-3. Gastroenterology. 2002;122:1853–1868. doi: 10.1053/gast.2002.33651. [DOI] [PubMed] [Google Scholar]
- 198.Schwarz E. M., Krimpenfort P., Berns A., Verma I. M. Immunological defects in mice with a targeted disruption in Bcl-3. Genes Dev. 1997;11:187–197. doi: 10.1101/gad.11.2.187. [DOI] [PubMed] [Google Scholar]
- 199.Matsumoto M., Yamada T., Yoshinaga S. K., Boone T., Horan T., Fujita S., Li Y., Mitani T. Essential role of NF-κB-inducing kinase in T cell activation through the TCR/CD3 pathway. J. Immunol. 2002;169:1151–1158. doi: 10.4049/jimmunol.169.3.1151. [DOI] [PubMed] [Google Scholar]
- 200.Futterer A., Mink K., Luz A., Kosco-Vilbois M. H., Pfeffer K. The lymphotoxin β receptor controls organogenesis and affinity maturation in peripheral lymphoid tissue. Immunity. 1998;9:59–70. doi: 10.1016/s1074-7613(00)80588-9. [DOI] [PubMed] [Google Scholar]