Abstract
The GHRPs (growth hormone-releasing peptides) are a class of small synthetic peptides known to stimulate GH release through binding of a G-protein-coupled receptor (designated GHS-R). We have found that hexarelin, a hexapeptide member of the GHRPs, binds to another protein identified as CD36, a scavenger receptor that is expressed in various tissues, including monocytes/macrophages and the endothelial microvasculature. CD36 is involved in the endocytosis of oxLDL (oxidized low-density lipoprotein) by macrophages, and in the modulation of angiogenesis elicited by thrombospondin-1 through binding to endothelial cells. To define the binding domain for hexarelin on CD36, covalent photolabelling of CD36 followed by enzymic and chemical degradation of the photoligand–receptor complex was performed. A 8 kDa photolabelled fragment corresponding to the CD36-(Asn132–Glu177) sequence has been identified as the hexarelin-binding site. Chemical cleavage of this fragment with CNBr resulted in the release of the free ligand, suggesting that Met169 is the contact point for the ligand within the receptor binding pocket. We conclude that the binding domain for hexarelin on CD36 overlaps with that for oxLDL, which corresponds to residues Gln155–Lys183 of CD36. Hence hexarelin might interfere with the CD36-mediated uptake of modified lipoproteins by macrophages. This may contribute, at least in part, to the anti-atherosclerotic effect of GHRPs in apolipoprotein E-deficient mice.
Keywords: atherosclerosis, CD36, growth hormone-releasing peptide, hexarelin, oxidized low-density lipoprotein, photoaffinity labelling
Abbreviations: apoE, apolipoprotein E; Bpa, p-benzoyl-L-phenylalanine; DMEM, Dulbecco's modified Eagle's medium; Endo F, N-glycosidase F; Endo Glu-C, endoproteinase Glu-C; GH, growth hormone; GHRP, growth hormone-releasing peptide; HEK293/hCD36 cells, human embryonic kidney 293 cells stably transfected with the human CD36 receptor; LDL, low-density lipoprotein; oxLDL, oxidized LDL; TSP-1, thrombospondin-1
INTRODUCTION
The GHRPs (growth hormone-releasing peptides) comprise a family of small synthetic peptides derived from Met-enkephalin that demonstrate potent and dose-dependent GH-releasing activity. This neuroendocrine activity of GHRPs was found to be mediated by a G-protein-coupled receptor, designated GHS-R1a, that is expressed predominantly in the hypothalamic–pituitary region. This receptor was identified further as the ghrelin receptor [1], the natural ligand of which is ghrelin, an endogenous GH secretagogue isolated initially from stomach [2].
In addition to neuroendocrine effects, hexarelin, a hexapeptide member of the GHRP family, was reported to demonstrate non-endocrine pleiotropic effects in the cardiovascular system [3,4] as well as in neoplastic tissues [5], raising questions about the nature of the receptor mediating these effects. We have reported previously that hexarelin binds to a novel class of binding sites in the mammalian heart that are distinct from the ghrelin receptor. This novel receptor was identified as CD36 [6], a multifunctional type B scavenger receptor that is expressed in various tissues, including monocytes/macrophages and the microvascular endothelium [7]. In monocytes/macrophages, CD36 signalling leads to the activation of pro-inflammatory genes by the endothelium and to the scavenging of oxLDL (oxidized low-density lipoprotein), thereby predisposing to the atherogenic formation of foam cells, the first step in the development of fatty streak lesions [8]. In agreement with this, we have recently demonstrated that long-term administration of GHRPs to apoE (apolipoprotein E)-deficient mice fed a high-fat, high-cholesterol diet significantly reduced atherosclerotic lesion development in a CD36-dependent manner [9].
Insight into the molecular basis of the binding of hexarelin to CD36 is of key importance for understanding the anti-atherosclerotic properties of this peptide. We report here a photoaffinity labelling approach using CD36 covalently modified with a hexarelin derivative containing the amino acid Bpa (p-benzoyl-L-phenylalanine) and limited enzymic and chemical proteolysis of the photoligand–receptor complex to identify the binding site for hexarelin on CD36.
MATERIALS AND METHODS
Animals
Sprague–Dawley rats were purchased from Charles Rivers (St-Constant, Que, Canada). The rats were fed normal chow and water ad libitum, and were used in accordance with the Institutional Animal Ethics Committee and the Canadian Council on Animal Care guidelines for the use of experimental animals.
Cell culture
HEK293/hCD36 cells (human embryonic kidney cells stably expressing human CD36) were maintained in DMEM (Dulbecco's modified Eagle's medium; Gibco/BRL, Burlington, Ont, Canada) supplemented with 10% (v/v) fetal bovine serum (Gibco/BRL) and 400 μg/ml geneticin (Gibco/BRL) in 5% CO2 at 37 °C.
Materials
Tyr-Bpa-Ala-hexarelin (see Figure 2A) was obtained by solid-phase synthesis according to Ong et al. [10]. Hexarelin and EP80317 were provided by Europeptides (Argenteuil, France). GHRP-2 and GHRP-6 were purchased from Peninsula Laboratories (Belmont, CA, U.S.A.). [D-Lys3]GHRP-6 was from Bachem Bioscience (King of Prussia, PA, U.S.A.). Ghrelin was from Neosystem (Strasbourg, France). Endo Glu-C (endoproteinase Glu-C) and Endo F (N-glycosidase F) were obtained from Roche Diagnostics (Indianapolis, IN, U.S.A.). BSA, bacitracin, leupeptin, Pefabloc, aprotinin, formic acid and CNBr were obtained from the Aldrich/Fluka/Sigma Group (St. Louis, MO, U.S.A.). PD-10 column was purchased from Amersham Pharmacia Biotech (San Francisco, CA, U.S.A.). Lipoproteins were from Intracel (Frederick, MD, U.S.A.). IODO-GEN®-precoated tubes were from Pierce Chemical Co. (Rockford, IL, U.S.A.). Platelets were donated from Hema-Quebec (St-Laurent, Que, Canada). All other reagents were of analytical grade unless specified.
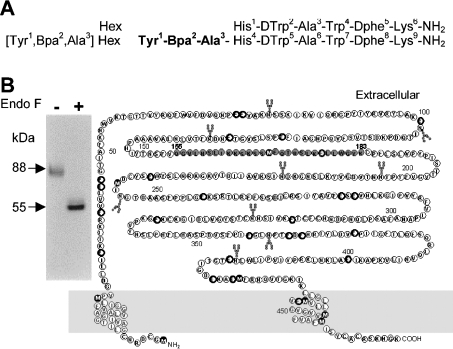
Figure 2. Photoaffinity cross-linking of [125I]Tyr-Bpa-Ala-hexarelin with CD36.
(A) Amino acid sequences of hexarelin and of its photoactivatable derivative used in photoaffinity labelling studies. (B) Primary amino acid sequence of the rat CD36 receptor. Bold circles indicate recognition residues for Endo Glu-C. Black filled circles indicate methionine residues, which are sensitive to CNBr treatment. The oxLDL-binding site is represented by grey filled circles. Putative glycosylation sites are indicated with Y-shaped pointers. Autoradiography (inset) of the labelled receptor was achieved by incubation of rat cardiac membranes as a source of CD36 (4 mg/ml) with 0.33 nmol/l [125I]Tyr-Bpa-Ala-hexarelin in Tris/HCl buffer at 22 °C for 1 h. The tubes were exposed to UV light (365 nm), treated or not with Endo F (50 m-units/μg of protein) and submitted to electrophoresis on a 7.5% (w/v) acrylamide gel. Arrows indicated the native radiolabelled receptor (88 kDa) and its deglycosylated form (55 kDa).
Membrane preparation
Following anaesthesia of the rats with sodium pentobarbital (CDMV, St-Hyacinthe, Que, Canada), the hearts were promptly removed and placed in ice-cold PBS for cardiac membrane preparation. Mice peritoneal macrophages were collected in saline containing 10 units/ml heparin. Cells were centrifuged and the pellet was kept at −80 °C until used. HEK293/hCD36 cells were incubated until detachment with 10 mM NaHCO3 containing 1 mM EDTA, and detaching cells were collected for membrane preparation. Human platelets were suspended in 0.05% EDTA and centrifuged twice (3 and 15 min at 2000 g), and the resulting pellet was then washed twice with 20 ml of 10 mM Tris/HCl buffer containing 96.5 mM NaCl, 85.7 mM dextrose, 1 mM EDTA and 5 mM EGTA, pH 7.4. All membranes were prepared according to Harigaya and Schwartz [11].
Lipoprotein labelling and oxidation
Radioiodination of LDL was performed by the method of Salacinski et al. [12] using IODO-GEN®-precoated tubes. Unbound iodine was removed by steric exclusion on a PD-10 column and by extensive dialysis against PBS. [125I]LDL and LDL (200 μg/ml) were oxidized with 5 μM CuSO4 for 12–16 h at 37 °C. Oxidation was terminated by addition of 100 μM EDTA. The degree of oxidation was monitored by measuring the amount of thiobarbituric acid-reactive substances, which was in the range of 35 malondialdehyde equivalents/mg of protein in oxLDL. The oxLDL was then concentrated with Centricon® Plus-20 (Millipore Co., Bedford, MA, U.S.A.) to approx. 1 mg/ml and stored at 4 °C until used (within 2 days). The final specific radioactivity of [125I]oxLDL ranged from 100 to 200 c.p.m./ng of oxLDL.
Western blot of CD36
Cell membranes were prepared from mouse and human hearts, mice peritoneal macrophages, human platelets and HEK293/hCD36 cells. Proteins (60 μg) were separated on SDS/7.5%-PAGE (200 V for 50 min) and transferred to nitrocellulose membranes. Blots were blocked for 2 h at room temperature with 50 mM Tris-buffered saline (2.9% NaCl), pH 7.4, containing 5% (w/v) non-fat milk and 0.1% (v/v) Tween-20, and incubated overnight at 4 °C with an affinity-purified rabbit polyclonal antibody to rat CD36 generated in our laboratory (1:200) [13] or with FA6-152 (1:500), a mouse monoclonal antibody to human CD36 (Immunotech, Marseille, France). After several washings, blots were incubated for 1 h with goat anti-rabbit (Pierce) or goat anti-mouse (Amersham Life Sciences Inc.) secondary antibody coupled to horseradish peroxidase (1:50000). Signals were visualized using SuperSignal chemiluminescence reagents (Pierce) with the FluorChem® imaging system. Protein bands were quantified by densitometry using AlphaEaseFC™ software (Alpha Innotech Corp., San Leandro, CA, U.S.A.). Results are expressed as the ratio of the density of the protein band to that of heart membranes. Heart membranes from CD36-null mice and HEK293 cells were used as negative controls for CD36 (results not shown).
Competition binding study
The photolabelling of CD36 with the photoactivatable derivative of hexarelin was carried out as described previously [10]. For competition studies, membranes were incubated with the photoactivatable radioligand in the presence of increasing concentrations (from 0.1 mM to 1 nM) of hexarelin, its analogues or ghrelin, or of oxLDL (from 0.14 to 142 μg/ml) or TSP-1 (thrombospondin-1; from 0.68 to 68 μg/ml) as endogenous CD36 ligands. The gels resulting from SDS/PAGE were fixed, dried, exposed to a storage phosphor screen (Amersham Biosciences), and analysed by using a Typhoon PhosphorImager (Amersham Biosciences) and ImageQuant 5.0 software. In some experiments, protein bands of 88 kDa were collected and the radioactivity counted.
Dissociation study
Experiments were performed using confluent HEK293/hCD36 cell monolayers in 12-well plates (Corning). For the binding of [125I]oxLDL, the cells were cooled on ice for 10 min and incubated with [125I]oxLDL (2.4 μg/ml) at 4 °C for 2 h in DMEM containing 0.5% (w/v) fatty acid-free BSA. After the incubation, cells were washed three times with ice-cold DMEM/BSA, and dissociation was initiated by addition of an excess concentration (300 μg/ml) of unlabelled oxLDL in the absence or presence of 10 μM hexarelin, or of 10 μM hexarelin alone, for up to 3 h at 4 °C. At each time point (20, 40, 60, 90, 120 and 180 min), the cell monolayers were washed once with cold PBS containing 0.5% BSA, followed by three washes with cold PBS. Washed cells were then solubilized in 0.1 M NaOH. Solubilized cells were counted for radioactivity and the protein content was measured. Non-specific binding was determined in the presence of a 100-fold excess of unlabelled oxLDL. Specific binding activities were defined by subtracting non-specific values from total binding activities.
Partial purification of the photoligand–receptor complex
The photolabelled receptor was purified by SDS/PAGE as described by Laemmli [14] using a 1.5 mm-thick 7.5% (w/v) acrylamide gel. Protein bands of 88 kDa were sliced into 1 mm3 fragments and extracted with 10 vol. of 100 mM NH4HCO3 containing 0.02% SDS for 5 h at 22 °C and for 2 days at 4 °C with constant shaking. The eluate was filtered through 0.22 μm-pore-size Minisart (Millipore) membranes, and concentrated with Centricon-10 (Amicon). Proteins were precipitated in 80% (v/v) acetone, dried and freeze-dried.
Enzymic and chemical digestion of the photoligand–receptor complex
For endoglycosidase-mediated digestion, the dry acetone precipitate of the photolabelled receptor was dissolved in 500 μl of 50 mM PBS containing 0.6% (v/v) octyl glucoside and 0.1% SDS. Endo F (35–100 units) was added and digestion was allowed to proceed for 18 h at 22 °C in the dark. For proteolytic digestion, partially purified native or deglycosylated CD36 receptors were dissolved in 100 mM NH4HCO3 buffer, pH 8.0, containing 0.1% SDS and Endo Glu-C (30–50 μg) for 72 h at 22 °C. Under these conditions, Endo Glu-C cleaves at the C-terminal side of glutamate residues. Subsequent digestions, where necessary, were performed using products of the first digestion that were run on SDS/PAGE; the band of interest was eluted in 100 mM NH4HCO3 containing 0.02% SDS, concentrated and freeze-dried. Prior to cleavage at methionine residues with CNBr, the receptor preparation was reduced by incubation with 100 mM β-mercaptoethanol for 30 min. CNBr hydrolysis of the labelled photoligand–receptor complex was performed in 50 μl of 70% (v/v) formic acid containing 100 mg/ml CNBr. Samples were incubated at 22 °C in the dark for 24 h under nitrogen. Reactions were terminated by addition of 10 vol. of water, and the reaction mixtures were freeze-dried.
Analysis of the products of enzymic and chemical cleavage
Fragments following proteolytic and chemical cleavage were separated by SDS/PAGE using 16.5% (w/v) polyacrylamide/Tris/Tricine gels and transferred on PVDF in 25 mM Tris buffer containing 192 nM glycine and 20% methanol [15]. The radiolabelled fragments were detected by autoradiography on X-ray films (Kodak BioMax® MS) with intensifying screens.
Data analysis
Calculation of the parameters for competition curves (IC50) and dissociation experiments (koff, t1/2) was performed by non-linear regression analysis using Graph Pad Prism software (Graph Pad Software Inc., San Diego CA, U.S.A.). The analysis of slope factors (Hill coefficient) of competition curves for hexarelin and oxLDL as competition ligands against the radiolabelled photoactivatable derivative of hexarelin were performed using ALLFIT for Windows software [16].
RESULTS
Covalent photolabelling of CD36 with [125I]Tyr-Bpa-Ala-hexarelin
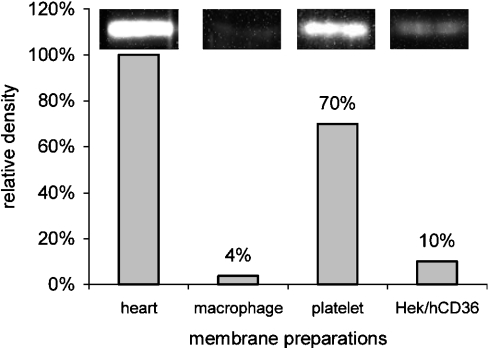
To determine the best source of CD36 for the mapping study, Western blots were carried out to assess the density of CD36 in various tissues, including monocytes/macrophages, platelets, HEK293/hCD36 cells and cardiac rat membrane preparations. As shown in Figure 1, CD36 was expressed at the highest density in cardiac membranes, and thus these were used as a source of CD36 for functional topology mapping.
Figure 1. Western blot analysis of CD36 expression.
Expression of the CD36 protein was assessed in membrane preparations from human and mouse hearts, mouse monocytes/macrophages, human platelets and HEK293/hCD36 cells. Total protein (60 μg) was separated by SDS/7.5%-PAGE and probed with specific anti-CD36 antibodies. Immunoblots for cardiac membranes, macrophages, platelets and HEK293/hCD36 cells are shown above the bars. Relative protein levels for CD36 are expressed as relative abundance (%) compared with that in the heart membrane preparations (set at 100%).
In order to identify the primary sequence of CD36 involved in hexarelin binding, photolabelling of the receptor was performed using [125I]Tyr-Bpa-Ala-hexarelin on rat cardiac membranes (Figure 2A). As shown in Figure 2B (lane 1), a single labelled protein of 88 kDa was obtained, as visualized by autoradiography after SDS/PAGE. Following treatment with Endo F, the receptor–ligand complex had an apparent molecular mass of 55 kDa (Figure 2, lane 2B), in agreement with the estimated molecular mass of the CD36 receptor (Figure 2B). Formation of the ligand–receptor conjugate was inhibited by an excess (10 μM) of unlabelled hexarelin (results not shown).
Among various biological functions, CD36 is known to mediate both the uptake of oxidatively modified lipoproteins by monocytes/macrophages [17] and the anti-angiogenic properties of TSP-1 [18]. The recognition of these endogenous ligands has been mapped to non-overlapping binding domains on this scavenger receptor. The region between amino acids Tyr93 and Ala120 has been shown to bind TSP-1 and to inhibit its anti-angiogenic activities. The binding domain for oxLDL is less definitively mapped. Monoclonal antibody and chimaeric protein studies suggest a critical role for the sequence between Gln155 and Lys183, while recombinant fusion proteins spanning the region Pro28–Arg93 have been shown to bind oxLDL directly [19,20].
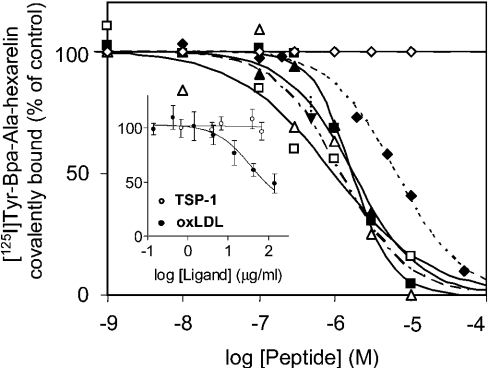
Our first studies were aimed at investigating whether hexarelin shares a binding domain with either of these two endogenous ligands of CD36. Competition curves for the binding of TSP-1 and oxLDL to CD36 were generated in the presence of [125I]Tyr-Bpa-Ala-hexarelin (Figure 3, inset). Analysis of the competition curves gave an IC50 value of 37 μg/ml for oxLDL; in contrast, TSP-1 did not inhibit the covalent photolabelling of the 88 kDa protein by the photoactivatable derivative of hexarelin. These results suggest that the hexarelin-binding site on CD36 might overlap with the binding domain for oxLDL. Furthermore, analysis of competition binding curves with GHRP analogues (Figure 3) gave IC50 values of 0.95±0.26 μM (mean±S.E.M.) for hexarelin, 1.79±0.47 μM for GHRP-2, 2.03±1.36 μM for GHRP-6, 1.7±0.14 μM for EP80317, 6.28±2.92 μM for [D-Lys3]GHRP-6 and >10 μM for ghrelin. Analysis of the competition binding curves of hexarelin and oxLDL with the photactivatable derivative of hexarelin by non-linear least-squares fit gave the same slope factor (−1), suggesting one class of binding site for both hexarelin and oxLDL on CD36.
Figure 3. Competition curves for endogenous CD36 ligands and GHRPs for binding of [125I]Tyr-Bpa-Ala-hexarelin to CD36.
A fixed concentration of [125I]Tyr-Bpa-Ala-hexarelin (0.33 nM) was incubated with rat cardiac membranes in the presence of increasing concentrations of GHRPs (1 nM to 0.1 mM) or of the endogenous ligands (inset) oxLDL (0.14–142 μg/ml) or TSP-1 (0.68–68 μg/ml) in a volume of 575 μl at 22 °C. Following UV irradiation, the covalently bound photoligand–CD36 complex was separated on SDS/PAGE, and bands corresponding to 88 kDa were counted for radioactivity. The IC50 for oxLDL as a competitive ligand was 37 μM. The GHRPs used were hexarelin (□), GHRP-2 (▴), GHRP-6 (▵), EP80317 (▪), [D-Lys3]GHRP-6 (♦) and ghrelin (⋄). Results are means±S.E.M. from at least two independent experiments.
Dissociation study with the [I125]oxLDL–CD36 complex
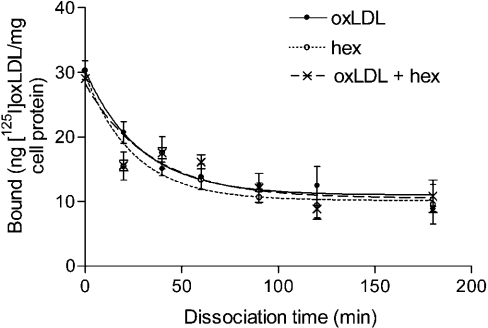
To determine whether hexarelin affects oxLDL binding to CD36 in an allosteric fashion, a dissociation study was conducted using [125I]oxLDL as radioligand and HEK293/hCD36 cells as binding sites. The time-dependent dissociation fitting curves for [I125]oxLDL in the presence of oxLDL (300 μg/ml), oxLDL plus 10 μM hexarelin and 10 μM hexarelin alone are shown in Figure 4, giving koff values (means±S.E.M.) of 0.035±0.008 min−1 (t1/2=19.8 min), 0.030±0.0095 min−1 (t1/2=23.1 min) and 0.040±0.012 min−1 (t1/2=17.4 min) respectively. These results suggest that hexarelin does not affect the binding of oxLDL to CD36 in an allosteric fashion.
Figure 4. Effect of hexarelin (hex) on the dissociation of [125I]oxLDL from HEK293/hCD36 cells.
Cells were incubated for 2 h at 4 °C with 2.4 μg/ml [125I]oxLDL. The cells were then washed and incubated with unlabelled oxLDL (300 μg/ml). At each time point (20, 40, 60, 90, 120 and 180 min), the cells were washed and cell-bound radioactivity was determined. The values are means±S.E.M. of triplicate determinations from one of two experiments giving similar results.
CNBr-mediated hydrolysis of the native and deglycosylated photoligand–receptor complex
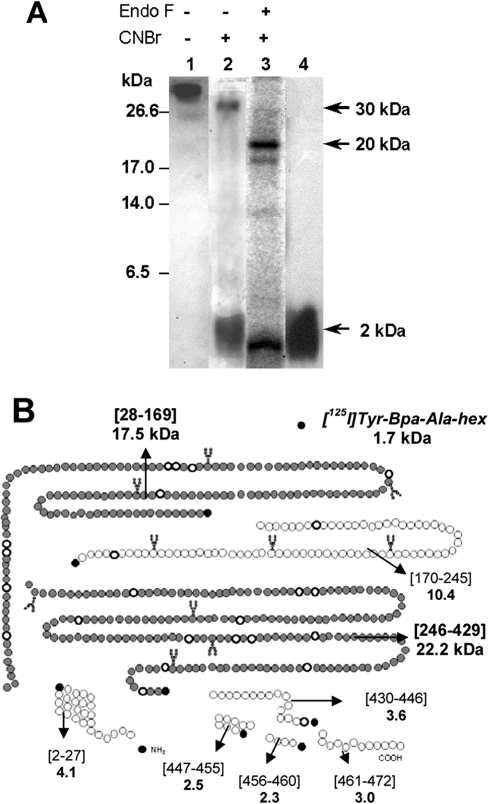
CD36 receptor photolabelled with [125I]Tyr-Bpa-Ala-hexarelin was isolated by electrophoresis and digested further with Endo F. CNBr cleavage of the native or deglycosylated photolabelled CD36 receptor yielded a similar major band of 2 kDa, as determined by SDS/PAGE (Figure 5A, lanes 2 and 3 respectively). This labelled fragment migrated with the same RF value as the radioiodinated photoactivatable peptide (Figure 5A, lane 4), suggesting that it might originate from the release of the ligand from the ε-methyl group on a methionine residue [21]. On the other hand, CNBr digestion of the native photolabelled CD36 receptor also provided a minor band with an apparent molecular mass of 30 kDa (Figure 5A, lane 2), whereas similar treatment on the deglycosylated photolabelled CD36 receptor yielded a fragment with an apparent mass of 20 kDa (Figure 5A, lane 3).
Figure 5. Electrophoretic properties of CNBr fragments of the [125I]Tyr-Bpa-Ala-hexarelin–CD36 conjugate.
(A) Rat heart CD36 was photolabelled with [125I]Tyr-Bpa-Ala-hexarelin as described in the Materials and methods section (lane 1). After electrophoresis, the covalent photolabelled CD36 receptor was extracted from the gel and submitted to degradation with CNBr (100 mg/ml), yielding fragments of 30 and 2 kDa (lane 2). The deglycosylated ligand–receptor conjugate treated with CNBr yielded two fragments of 20 and 2 kDa (lane 3). The free radiolabelled photoactivable hexarelin derivative was loaded alone as a reference (lane 4). Electrophoresis was performed in a 16.5% (w/v) acrylamide gel. (B) Potential CNBr fragmentation pattern of CD36. The positions of the proteolytic fragments are indicated in square brackets. The molecular masses of the predicted photoaffinity cross-linked unglycosylated fragments and ligand are also indicated (kDa). The putative contact domains of [125I]Tyr-Bpa-Ala-hexarelin, i.e. CD36-(Pro28–Met169) and CD36-(Ile246–Met429), are depicted with filled circles.
Endo Glu-C-mediated digestion of the photoligand–receptor complex
According to the theoretical CNBr cleavage map of the rat CD36 receptor (Figure 5B), only two fragments containing putative glycosylation site (Asn-Xaa-Ser or -Thr) could explain the molecular masses obtained experimentally: CD36-(Pro28–Met169) and CD36-(Ile246–Met429). Since release of the ligand seemed to be the main end-product of CNBr treatment, Met169 or Met429 was proposed to be the major point of attachment of [125I]Tyr-Bpa-Ala-hexarelin. Therefore we designed a protocol using enzymic cleavage with Endo Glu-C in order to overlap the derived methionine residue and to define more precisely which of Met169 and Met428 is cross-linked with the benzophenone.
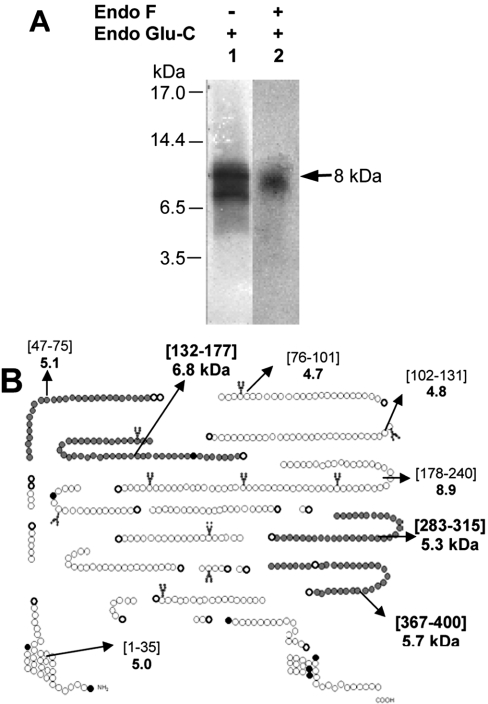
Treatment of the intact photolabelled CD36 receptor (88 kDa) with Endo Glu-C yielded a single radioactive band with an apparent molecular mass of ∼8 kDa (Figure 6A, lane 1). Subsequent treatment of this band with Endo F failed to alter its migration profile (results not shown). In a similar manner, treatment of the deglycosylated photolabelled CD36 receptor (55 kDa) with Endo Glu-C yielded a band of ∼8 kDa (Figure 6A, lane 2), supporting the conclusion that this fragment was not glycosylated, although we could not completely rule out the presence of a potential glycosylation site that was not effectively glycosylated.
Figure 6. Electrophoretic properties of Endo Glu-C fragments of the [125I]Tyr-Bpa-Ala-hexarelin–CD36 conjugate.
(A) The band corresponding to the photolabelled CD36 receptor was extracted from the electrophoresis gel and submitted or not to a deglycosylation step. The 88 kDa receptor conjugate (glycosylated form) and its 55 kDa deglycosylated form were treated with Endo Glu-C (30 units for 72 h at 22 °C) and reloaded on a 16.5% (w/v) acrylamide gel. In both cases, a unique band corresponding to an 8 kDa fragment was obtained. (B) Potential Endo Glu-C fragmentation pattern of CD36. The fragment (∼8 kDa) resulting from Endo Glu-C digestion of the photolabelled receptor conjugate may correspond to the following modified fragments: CD36-(Gly47–Glu75), CD36-(Asn132–Glu177), CD36-(Val283–Glu315) or CD36-(His367–Glu400) (depicted by filled circles).
By theoretical mapping of the Endo Glu-C cleavage of the CD36 receptor (Figure 6B), 22 possible fragments were identified. Five of them had a molecular mass within 3 kDa of the 8 kDa band found experimentally, namely fragments CD36-(Gly47–Glu75), CD36-(Asn132–Glu177), CD36-(Leu178–Glu240), CD36-(Val283–Glu315) and CD36-(His367–Glu400). The CD36-(Leu178–Glu240) fragment was not considered further, since as it did not overlap with the CNBr fragments identified [CD36-(Pro28–Met169) and CD36-(Ile246–Met429)]. The four remaining fragments had calculated molecular masses of 5.1, 6.8, 5.3 and 5.7 kDa respectively, which includes the mass of [125I]Tyr-Bpa-Ala-hexarelin.
[125I]Tyr-Bpa-Ala-hexarelin binds to Met169 on CD36
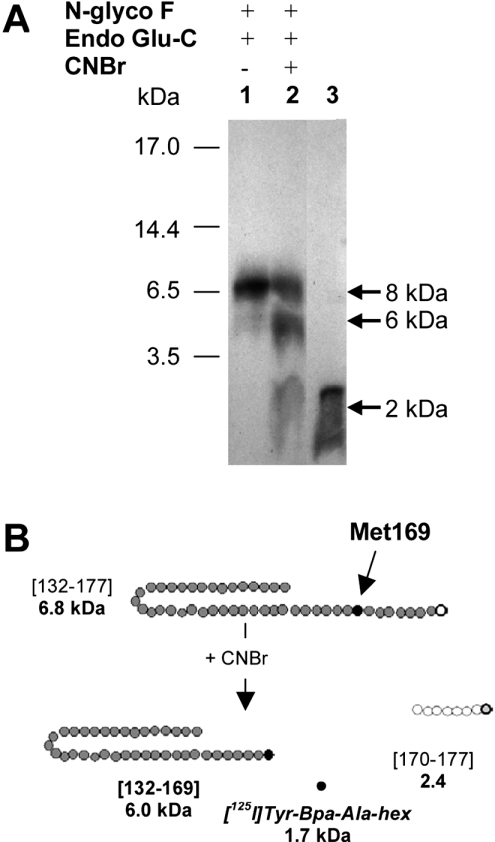
In order to identify the ∼8 kDa fragment from the Endo Glu-C digestion, we took advantage of the presence of a single methionine residue (Met169) within the CD36 domain comprising residues Asn132–Glu177. Further cleavage of the CD36-(Asn132–Glu177) fragment with CNBr was performed in order to release the [125I]Tyr-Bpa-Ala-hexarelin from Met169. As expected, a band of 2 kDa (Figure 7A, lane 2) was found that co-migrated with the free radioligand (Figure 7A, lane 3). Incomplete digestion, as indicated by the presence of the original band (8 kDa), was probably due to oxidation of the methionine during previous electrophoresis steps. Interestingly, an additional band of apparent molecular mass 6 kDa was also obtained (Figure 7B), which could have resulted from C-terminal cleavage after Met169, releasing the [125I]Tyr-Bpa-Ala-hexarelin-labelled fragment CD36-(Asn132–Met169).
Figure 7. CNBr cleavage of the partially purified Endo Glu-C- and Endo F-generated 8 kDa fragment of the photoligand–CD36 complex.
(A) Autoradiography of the extracted band of ∼8 kDa obtained after treatment with Endo Glu-C (lane 1). Following CNBr treatment, two radioactive species (6 and 2 kDa) were obtained (lane 2). The free radioligand of 2 kDa was run in lane 3. (B) The proteolysis fragment (∼8 kDa) of the ligand–receptor complex obtained after treatment with Endo Glu-C and Endo-F, i.e. CD36(Asn132–Glu177), was subsequently digested with CNBr. After treatment with CNBr, two fragments of ∼6 and ∼2 kDa were obtained, the latter corresponding to the molecular mass of [125I]Tyr-Bpa-Ala-hexarelin alone released from Met169 (black circle).
DISCUSSION
Our previous studies using a photolabelling approach identified CD36 as a receptor for hexarelin, used as a GHRP prototype [13]. As shown by competition binding studies, CD36 can also be considered as a binding site for other GHRP analogues. These derivatives bind to this scavenger receptor within the same affinity range as does hexarelin, which contrasts with the lower-affinity binding found for ghrelin, an endogenous GH secretagogue. CD36 is known to play major role in fatty acid translocation [22,23] as well as in the internalization of oxLDL, which points to a role in the pathological formation of macrophage foam cells as an early step in the development of atherosclerosis [8]. A wealth of evidence for an atherogenic role for oxLDL in the arterial wall [24] as well as for its circulating levels as a biomarker of atherosclerotic disease has been reported [25]. We have recently uncovered that long-term (12 weeks) treatment with GHRPs of apoE-deficient mice fed a high-fat, high-cholesterol diet significantly decreased atherosclerotic lesion formation [9].
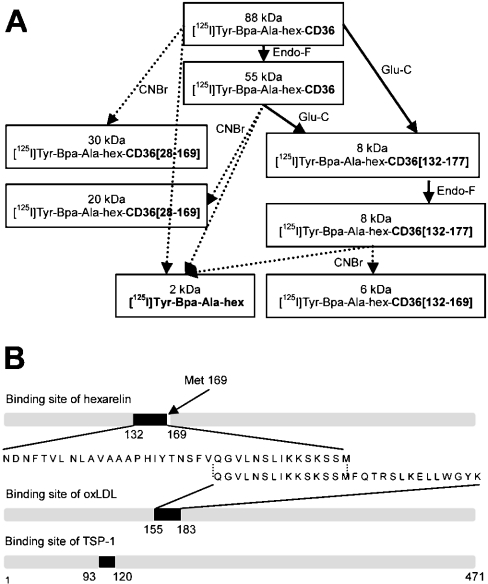
To document the molecular mechanisms involved in the beneficial anti-atherosclerotic effects of these peptides, we took advantage of a covalent photolabelling approach using a hexarelin derivative containing a Bpa residue, combined with limited enzymic and chemical proteolysis of the photoligand–receptor conjugate, to define the functional topology of this scavenger receptor (Figure 8A). Covalent photolabelling of ligands with benzophenone has been successfully applied to the functional mapping of G-protein-coupled receptors [26–28]. In the present study, photolabelling of the ligand–receptor complex led to the identification of residues Asn132–Met169 of CD36 as the binding domain for hexarelin. Interestingly, this binding domain was found to overlap with the CD36 multifunctional domain encompassing residues Gln155–Lys183, as shown in Figure 8(B). This immunodominant domain is involved in the binding and internalization of oxLDL by monocytes/macrophages, as demonstrated by the inhibition of the oxLDL–CD36 interaction using monoclonal antibodies directed against this domain [19]. This finding is in agreement with the competitive inhibition of the covalent photolabelling of CD36 with the photoactivatable derivative of hexarelin by oxLDL. Furthermore, the rate of dissociation of [125I]oxLDL from the [125I]oxLDL–CD36 complex was not affected by hexarelin, suggesting that GHRPs are unlikely to bind to a binding site distinct from that for oxLDL on CD36.
Figure 8. Location of the binding site for [125I]Tyr-Bpa-Ala-hexarelin on the primary sequence of CD36: comparison with the binding domains of CD36 for endogenous ligands.
(A) Schematic summary of the fragmentation scheme employed with the photolabelled ligand–receptor conjugate. Digestion with Endo F, Endo Glu-C and CNBr was carried out as described in the Materials and methods section. The molecular masses of the fragment are indicated in kDa, and represent the actual sizes of the digested fragments including the ligand [125I]Tyr-Bpa-Ala-hexarelin (hex). (B) Ligand-binding domains of CD36. The amino acid sequences of the binding domain for hexarelin, oxLDL and TSP-1 (endogenous ligands of CD36) are shown.
The hexarelin contact point within CD36 was identified as Met169, which was confirmed further by the release of the [125I]Tyr-Bpa-Ala-hexarelin thiocyanate adduct following CNBr treatment of the 8 kDa photoligand–receptor fragment. However, although this methionine residue is preferentially labelled with benzophenone, it may still be possible that an amino acid other than methionine is implicated, to a lower extent, in the binding of the photoactivatable hexarelin derivative, particularly in the photolabelling of the larger hydrolysis fragments observed. Alternatively, it may also be possible that the photoactivatable ligand was bound to the γ-carbon on the methionine, thereby impairing its release by CNBr hydrolysis. To resolve this issue, we attempted to replace Met169 with Ala by constructing an M169A CD36 mutant. However, transient transfection of M169A CD36 cDNA inserts in HEK293 cells was unable to generate enough material for photolabelling studies.
In addition, the predicted molecular mass of the photoligand–receptor complex of 6 kDa is in agreement with the apparent molecular mass of an additional 6 kDa band obtained by electrophoresis. The entity of 2 kDa obtained after CNBr cleavage of the native and deglycosylated CD36 receptors (Figure 5A, lanes 2 and 3) could correspond to cleavage of the methylthiocyanate adduct of [125I]-Tyr-Bpa-Ala-hexarelin, resulting from the insertion of the radioligand in the ε-methyl group of the methionine. On the other hand, the 6 kDa band obtained after CNBr treatment might indicate that a part of the radioligand was bound to another site on the CD36-(Asn132–Met169) fragment or simply bound to the γ-methylene group of the methionine. This result also confirms the non-glycosylated nature of the CD36-(Asn132–Glu177) domain, despite the presence of a potential glycosylation site at position 134 (Asn).
We may therefore hypothesize that the atheroprotective effect of GHRPs results from competition of GHRPs with oxLDL for binding to CD36, thereby decreasing the internalization of oxidized lipids within macrophages and hence the formation of atherosclerotic lesions. The specificity of GHRP binding to the oxLDL-binding site on CD36 is indicated by the absence of competition with TSP-1 binding [18]. In agreement with this, the TSP-1-binding domain encompasses residues Tyr93–Ala120 of CD36, as identified by competitive inhibition of TSP-1 binding using fusion proteins of CD36, and was found to be distinct from the hexarelin-binding site on CD36 [29].
In conclusion, our present study shows that the GHRP-binding domain on CD36 overlaps with that for oxLDL, which may interfere with the uptake of this pro-atherogenic ligand by monocytes/macrophages. This may explain, at least in part, the anti-atherosclerotic properties of GHRPs.
Acknowledgments
We are grateful to Eve-Marie Charbonneau and Petra Pohankova for expert technical assistance. This work was supported by grants from the Canadian Institute of Health Research (UOP 50059 and MOP 62837).
References
- 1.Howard A. D., Feighner S. D., Cully D. F., Arena J. P., Liberator P. A., Rosenblum C. I., Hamelin M., Hreniuk D. L., Palyha O. C., Anderson J., et al. A receptor in pituitary and hypothalamus that functions in growth hormone release. Science. 1996;273:974–977. doi: 10.1126/science.273.5277.974. [DOI] [PubMed] [Google Scholar]
- 2.Kojima M., Hosoda H., Date Y., Nakazato M., Matsuo H., Kangawa K. Ghrelin is a growth-hormone releasing acylated peptide from stomach. Nature (London) 1999;402:656–660. doi: 10.1038/45230. [DOI] [PubMed] [Google Scholar]
- 3.Muccioli G., Broglio F., Valetto M. R., Ghe C., Catapano F., Graziani A., Papotti M., Bisi G., Deghenghi R., Ghigo E. Growth hormone-releasing peptides and the cardiovascular system. Ann. Endocrinol. (Paris) 2000;61:27–31. [PubMed] [Google Scholar]
- 4.Ghigo E., Arvat E., Giordano R., Broglio F., Gianotti L., Maccario M., Bisi G., Graziani A., Papotti M., Muccioli G., et al. Biologic activities of growth hormone secretagogues in humans. Endocrine. 2001;14:87–93. doi: 10.1385/ENDO:14:1:087. [DOI] [PubMed] [Google Scholar]
- 5.Papotti M., Ghe C., Cassoni P., Catapano F., Deghenghi R., Ghigo D., Muccioli G. Growth hormone secretagogue binding sites in peripheral human tissues. J. Clin. Endocrinol. Metab. 2000;85:3803–3807. doi: 10.1210/jcem.85.10.6846. [DOI] [PubMed] [Google Scholar]
- 6.Bodart V., Bouchard J. F., McNicoll N., Escher E., Carriere P., Ghigo E., Sejlitz T., Sirois M. G., Lamontagne D., Ong H. Identification and characterization of a new growth hormone-releasing peptide receptor in the heart. Circ. Res. 1999;85:796–802. doi: 10.1161/01.res.85.9.796. [DOI] [PubMed] [Google Scholar]
- 7.Febbraio M., Hajjar D. P., Silverstein R. L. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J. Clin. Invest. 2001;108:785–791. doi: 10.1172/JCI14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Febbraio M., Podrez E. A., Smith J. D., Hajjar D. P., Hazen S. L., Hoff H. F., Sharma K., Silverstein R. L. Targeted disruption of the class B scavenger receptor CD36 protects against atherosclerotic lesion development in mice. J. Clin. Invest. 2000;105:1049–1056. doi: 10.1172/JCI9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ong H., Tremblay A., Febbraio M., Silverstein R. L., Avallone R., Iken K., Wang Y., Bujold K., Demers A., Sirois M. G., et al. Growth hormone-releasing peptides as inhibitors of fatty streaks formation: A new therapy for atherosclerosis. Program and Abstracts of the 85th Annual Meeting of The Endocrine Society, Philadelphia, 19–22 June 2003. 2003. p. 361. abstract P-223.
- 10.Ong H., McNicoll N., Escher E., Collu R., Deghenghi R., Locatelli V., Ghigo E., Muccioli G., Boghen M., Nilsson M. Identification of a pituitary growth hormone-releasing peptide (GHRP) receptor subtype by photoaffinity labeling. Endocrinology. 1998;139:432–435. doi: 10.1210/endo.139.1.5811. [DOI] [PubMed] [Google Scholar]
- 11.Harigaya S., Schwartz A. Rate of calcium binding and uptake in normal animal and failing human cardiac muscle. Membrane vesicles (relaxing system) and mitochondria. Circ. Res. 1969;25:781–794. doi: 10.1161/01.res.25.6.781. [DOI] [PubMed] [Google Scholar]
- 12.Salacinski P. R. P., McLean C., Sykes J. E. C., Clement-Jones V. V., Lowry P. J. Iodination of proteins, glycoproteins, and peptides using a solid-phase oxidizing agent, 1,3,4,6-tetrachloro-3α,6α-diphenyl glycoluril (Iodogen) Anal. Biochem. 1981;117:136–146. doi: 10.1016/0003-2697(81)90703-x. [DOI] [PubMed] [Google Scholar]
- 13.Bodart V., Febbraio M., Demers A., McNicoll N., Pohankova P., Perreault A., Sejlitz T., Escher E., Silverstein R. L., Lamontagne D., Ong H. CD36 mediates the cardiovascular action of growth hormone-releasing peptides in the heart. Circ. Res. 2002;90:844–849. doi: 10.1161/01.res.0000016164.02525.b4. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Schagger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 16.De Lean A., Munson P. J., Rodbard D. Simultaneous analysis of families of sigmoidal curves: application to bioassay, radioligand assay and physiological dose-response curves. Am. J. Physiol. 1978;235:E97–E102. doi: 10.1152/ajpendo.1978.235.2.E97. [DOI] [PubMed] [Google Scholar]
- 17.Han J., Hajjar D. P., Febbraio M., Nicholson A. C. Native and modified low density lipoproteins increase the functional expression of the macrophage class B scavenger receptor, CD36. J. Biol. Chem. 1997;272:21654–21659. doi: 10.1074/jbc.272.34.21654. [DOI] [PubMed] [Google Scholar]
- 18.Jimenez B., Volpert O. V., Crawford S. E., Febbraio M., Silverstein R. L., Bouck N. Signals leading to apoptosis-dependent inhibition of neovascularization by thrombospondin-1. Nat. Med. 2000;6:41–48. doi: 10.1038/71517. [DOI] [PubMed] [Google Scholar]
- 19.Puente Navazo M. D., Daviet L., Ninio E., McGregor J. L. Identification on human CD36 of a domain (155–183) implicated in binding oxidized low-density lipoproteins (Ox-LDL) Arterioscler. Thromb. Vasc. Biol. 1996;16:1033–1039. doi: 10.1161/01.atv.16.8.1033. [DOI] [PubMed] [Google Scholar]
- 20.Pearce S. F., Roy P., Nicholson A. C., Hajjar D. P., Febbraio M., Silverstein R. L. Recombinant glutathione S-transferase/CD36 fusion proteins define an oxidized low density lipoprotein-binding domain. J. Biol. Chem. 1998;273:34875–34881. doi: 10.1074/jbc.273.52.34875. [DOI] [PubMed] [Google Scholar]
- 21.Kage R., Leeman S. E., Krause J. E., Costello C. E., Boyd N. D. Identification of methionine as the site of covalent attachment of a p-Benzoyl-Phenylalanine-containing analogue of substance P on the substance P (NK) receptor. J. Biol. Chem. 1996;271:25797–25800. doi: 10.1074/jbc.271.42.25797. [DOI] [PubMed] [Google Scholar]
- 22.Abumrad N. A., el Maghrabi M. R., Amri E. Z., Lopez E., Grimaldi P. A. Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation. Homology with human CD36. J. Biol. Chem. 1993;268:17665–17668. [PubMed] [Google Scholar]
- 23.Brinkmann J. F. F., Abumrad N. A., Ibrahimi A., Van Der Vusse G. J., Glatz J. F. C. New insights into long-chain fatty acid uptake by heart muscle: a crucial role for fatty acid translocase/CD36. Biochem. J. 2002;367:561–570. doi: 10.1042/BJ20020747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boullier A., Bird D. A., Chang M. K., Dennis E. A., Friedman P., Gillotte-Taylor K., Horkko S., Palinski W., Quehenberger O., Shaw P., et al. Scavenger receptors, oxidised LDL, and atherosclerosis. Ann. N.Y. Acad. Sci. 2000:214–223. doi: 10.1111/j.1749-6632.2001.tb03943.x. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki T., Kohno H., Hasegawa A., Toshima S., Amaki T., Kurabayashi M., Nagai R., et al. on behalf of the Clinical Trial of Oxidised LDL (CTOL) investigators. Diagnostic implications of circulating oxidized low density lipoprotein levels as a biochemical risk marker of coronary artery disease Clin. Biochem. 2002;35:347–353. doi: 10.1016/s0009-9120(02)00326-0. [DOI] [PubMed] [Google Scholar]
- 26.Boucard A. A., Sauve S. S., Guillemette G., Escher E., Leduc R. Photolabelling the rat urotensin II/GPR14 identifies a ligand-binding site in the fourth transmembrane domain. Biochem. J. 2003;370:829–838. doi: 10.1042/BJ20021566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coulie B., Matsuura B., Dong M., Hadac E. M., Pinon D. I., Feighner S. D., Howard A. D., Miller L. J. Identification of peptide ligand-binding domains within the human motilin receptor using photoaffinity labeling. J. Biol. Chem. 2001;276:35518–35522. doi: 10.1074/jbc.M104489200. [DOI] [PubMed] [Google Scholar]
- 28.Behar V., Bisello A., Rosenblatt M., Chorev M. Direct indentification of two contact sites for parathyroid hormone (PTH) in the novel PTH-2 receptor using photoaffinity cross-linking. Endocrinology. 1999;140:4251–4261. doi: 10.1210/endo.140.9.6950. [DOI] [PubMed] [Google Scholar]
- 29.Pearce S. F., Wu J., Silverstein R. L. Recombinant GST/CD36 fusion proteins define a thrombospondin binding domain. J. Biol. Chem. 1995;270:2981–2986. doi: 10.1074/jbc.270.7.2981. [DOI] [PubMed] [Google Scholar]