Abstract
The social amoeba Dictyostelium discoideum exhibits high activities of phospholipase and lysophospholipase [Ferber, Munder, Fischer and Gerisch (1970) Eur. J. Biochem. 14, 253–257]. We assayed Dictyostelium lysates to demonstrate the presence of a highly active phospholipase B (PLB) enzyme that removed both fatty-acid chains from phosphatidylcholine and produced the water-soluble glycerophosphorylcholine. We purified the PLB activity from Dictyostelium cytosol using standard agarose media (size exclusion and ion exchange), and combined this with an affinity purification step using myristoylated ARF1 (ADP-ribosylation factor 1), a protein which has a single fatty acid at its N-terminus. Two proteins co-purified (48 kDa and 65 kDa), and the 48 kDa protein was digested with trypsin, peptide fragments were separated by reverse-phase chromatography, and the resultant peptides were sequenced by Edman degradation. From the peptide sequences obtained, database searches revealed a gene which encodes a protein of 65 kDa with unknown function. The 48 kDa protein therefore appears to be a fragment of the full-length 65 kDa product. Expression of the gene in Escherichia coli confirmed that it encodes a PLB. Characterization of its substrate specificity indicated that, in addition to phosphatidylcholine deacylation, the enzyme also hydrolysed phosphatidylinositol and phosphatidylethanolamine. The PLB identified in the present study is not related to existing PLBs found in bacteria, fungi or mammals. There are, however, genes similar to Dictyostelium PLB in mammals, flies, worms and Giardia, but not in yeast. We therefore have identified a novel family of intracellular PLBs.
Keywords: ADP-ribosylation factor (ARF), deacylation, Dictyostelium, glycerophosphorylcholine, non-esterified fatty acid (NEFA), phospholipase B
Abbreviations: ARF1, ADP-ribosylation factor 1; GPC, glycerophosphorylcholine; LPC, lysoPC; myrARF1, myristoylated ARF1; NEFA, non-esterified fatty acid; NHS, N-hydroxysuccinimide; PC, phosphatidylcholine; P-choline, phosphorylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PLB, phospholipase B; PLD, phospholipase D
INTRODUCTION
Phospholipase B (PLB) enzymes catalyse the hydrolytic cleavage of both acylester bonds of glycerophospholipids. Essentially these enzymes display phospholipase A/lysophospholipase activity, catalysing the total deacylation of phospholipids. Phosphatidylcholine (PC)-hydrolysing PLB activities have been described in bacteria [1], fungi [2], Dictyostelium discoideum [3] and in mammalian cells [4–11], and three distinct gene families have been identified from bacteria, fungi and mammals. In the micro-organism Moraxella bovis, the plb gene encodes a protein of 616 amino acids and the enzyme hydrolyses PC/LPC (lysoPC) to produce NEFAs (non-esterified fatty acids, also known as free fatty acids) and glycerophosphorylcholine (GPC) [1]. PLB enzymes have been cloned from different fungi, including Candida albicans [12], Saccharomyces cerevisae [13–15], Cryptococcus neoformans [16], Penicillium notatum and Torulaspora delbruekii, and are related by sequence [2], but not to M. bovis PLB. The fungal enzymes when examined indicate that PC is their main substrate. The fungal PLBs range 600–670 amino acids in length and have a potential role in virulence and in fungal pathogenesis [2]. In S. cerevisiae, the PLB enzyme, encoded by the SPO1 gene is the most divergent member and is required for meiosis [13,15]. PLB has also been cloned from mammalian cells, but is not sequence related to the fungal PLBs or to bacterial PLB. The enzyme was initially characterized from the intestine and is now known to be present in the external face of the brush-border membranes of mature enterocytes where it is involved in digestion of dietary lipids [4,7–11,17–19]. The enzyme is an ecto-phospholipase with a short C-terminal domain, inserted in the membrane by a hydrophobic segment connected to a larger, heavily glycosylated globular domain [8,19]. The latter contains four tandem repeat domains and several lipase and phospholipase consensus sequences, i.e. the GxSxG sequence similar to a classic lipase motif found in α/β hydrolases [20] and the GDSL sequence found in M. bovis [16,21]. Mutational analysis showed that the second repeat domain contains the essential serine residue required for catalysis and that it is located in the GDSL sequence rather than the GxSxG sequence [10]. The intestinal PLB is also expressed in the epidermis [11] and in the epididymis where it may be responsible for production of GPC, which is a main component of seminal plasma essential for sperm protection [9].
The social amoeba D. discoideum exhibits high activities of phospholipase A and lysophospholipase, suggesting the presence of a PLB activity [3]. Our knowledge of PLB genes in microorganisms, fungi and mammals indicated that unrelated sequences are responsible for PLB activity. We have therefore purified the PLB activity from Dictyostelium and obtained sufficient peptide sequence to isolate the corresponding gene that encodes for this enzyme. Dictyostelium PLB represents the first member of a new gene family of putative PLBs conserved in humans, mice, flies and worms, but not yeast.
EXPERIMENTAL
Growth of Dictyostelium
D. discoideum were grown axenically at 22 °C on an orbital shaker (New Brunswick Scientific INNOVA 4230; 180 rev./min). All experiments used vegetative cells from standard HL-5 medium (composition per litre: 14.3 g of Oxoid L37 bacteriological peptone, 7.15 g of Oxoid L21 yeast extract, 15.4 g of D-glucose, 0.51 g of disodium hydrogen phosphate, 0.49 g of potassium di-hydrogen phosphate, pH 6.4). Media was autoclaved following preparation. Dictyostelium can be grown on lawns of bacteria or in shaken culture rich in bacteria. However, to ensure there was no bacterial contamination they were grown wholly in shaken culture with synthetic media. Cells were grown to a density of approx. 2×107 cells/ml. For the initial experiments, 200 ml cultures were used, while for the purification, cells were grown in 2-litre flasks and 2–5 litres were used per purification. Cells were pelleted at low speed (700 g), and either frozen or filter lysed immediately [22]. Filter lysis (5 μM filter) was performed in 200 mM sucrose/50 mM NaCl in 50 mM phosphate buffer, pH 6.4, in the presence of protease inhibitors [1 mM PMSF and 200 μM tosyl-lysylchloromethane (‘TLCK’)].
Determination of choline and choline metabolites
The assay to determine PC-hydrolysing activity utilized radiolabelled PC presented in a mixture of lipids comprising PE (phosphatidylethanolamine), PtdIns(4,5)P2 and didecanoyl-PC in the molar ratios of 10:0.3:1. [This assay was originally designed to monitor ARF (ADP-ribosylation factor)-stimulated PLD (phospholipase D) activity.] The final concentration of PC used in the assay was 8.6 μM and the assay volume was 60 μl. [3H]Methyl-choline-labelled PC (C10:0/C10:0) was generally added to give 80000 d.p.m. per assay. Assays were performed at 37 °C for 60 min unless indicated otherwise. Assays were quenched with 250 μl of chloroform:methanol (1:1, v/v). [In initial experiments myrARF1 (myristoylated ARF1; 10 μM) and GTPγS (30 μM) were also included to examine whether PC could be metabolized via PLD, but no PLD activity was identified. Although myrARF or GTPγS had no effect on PLB activity, it was noted that in the presence of myrARF1 alone, GPC was metabolized to choline and this was due to a contaminant present in the myrARF1 preparation.] The phases were separated with the addition of 125 μl of water and centrifugation at 1500 g for 5 min at 4 °C. Choline was separated from phosphorylated metabolites [phosphorylcholine (P-choline) and GPC] using Bio-Rex 70 cation exchange resin as described previously [23]. In brief, the resin was prepared by addition of 3 bed volumes (bed volume, ∼1 ml) of 0.5 M NaOH, followed by 10 bed volumes of de-ionized water. The sample (200 μl) was applied to the pre-prepared resin, and phosphorylated metabolites removed by washing with 3 ml of water. Choline was eluted from the resin by the application of 1.5 ml of 0.05 M glycine/0.5 M NaCl, pH 3. The resultant radioactivity was counted in the presence of 4 ml of Ultima Gold XR scintillation fluid in the scintillation counter (Canberra Packard).
Chromatography of PC metabolites
Paper chromatography was performed using the solvent ethanol/water (17:3, v/v). Standards for choline, P-choline, and GPC were also run (50 mM, 10 μl per lane), and were seen to have RF values close to the reported values of 0.65, 0.24, and 0.38 respectively [23,24].
Separation of the lipid metabolites of PC was performed by TLC using chloroform/methanol/acetic acid/water (75:45:3:1, by vol.) as the solvent [25]. Standards (PC, fatty acids and diacylglycerol, 10 μg) were run alongside to identify the products.
Purification and amino acid sequence determination
Cytosol from 1–5 litre cultures of Dictyostelium (approx. 5×109 cells/litre) was precipitated with 20–45% saturated ammonium sulphate. The pellet was resuspended, dialysed and concentrated to a volume of 10 ml [20 mM Hepes (pH 7.0)/0.02% sodium azide]. Two identical separations of the concentrate (5 ml per run) were performed using a Pharmacia Superdex-200 column at a flow rate of 1.5 ml/min. Fractions of 10 ml were collected. Assays for activity were performed on 20 μl of each fraction using didecanoyl PC substrate. Incubations were performed for 60 min at 37 °C in the presence of 10 μM myrARF1 and 30 μM GTPγS. Water soluble metabolites were separated into choline and GPC. Peak fractions from the gel-filtration separation were combined (approx. 120 ml) and applied to a 5 ml Hi-Trap heparin column connected in-line to a 5 ml Hi-Trap Q-Sepharose column at 1 ml/min. The columns were separated, and each column was washed separately with 30 ml of buffer [20 mM Hepes (pH 7.0)/0.02% sodium azide]. Proteins were eluted with 2 M NaCl (3 ml fractions). Fractions were desalted and the activity of 20 μl of each fraction was determined in the presence of myrARF1. There was negligible activity in the flow-through and wash step. Peak fractions from the Q-Sepharose column were further fractionated using a pre-prepared myrARF1-bound NHS (N-hydroxysuccinimide)-affinity column. MyrARF1 (10 mg) was coupled to a 1 ml Hi-Trap NHS-affinity column according to the manufacturer's suggested conditions (Amersham Biosciences). (Hi-Trap NHS-activated Sepharose High Performance is a gel designed for coupling ligands containing primary amino groups and is based on highly cross-linked agarose beads with six-atom spacer arms attached to the matrix by epichlorohydrine and activated by N-hydroxysuccinimide.) Test runs were performed using a sample of this partially purified activity (0.5 ml) and applied to the 1 ml affinity column at 1 ml/min. The column was washed with 5 ml of buffer, and then the bound protein eluted with 2 M NaCl. Fractions (200 μl) were collected for the load, wash and elution stages. Activity in every second fraction was determined. Fractions (20 μl) were assayed using didecanoyl-PC substrate in the presence of myrARF1. For the final purification, only the peak activity was collected for 50 successive runs.
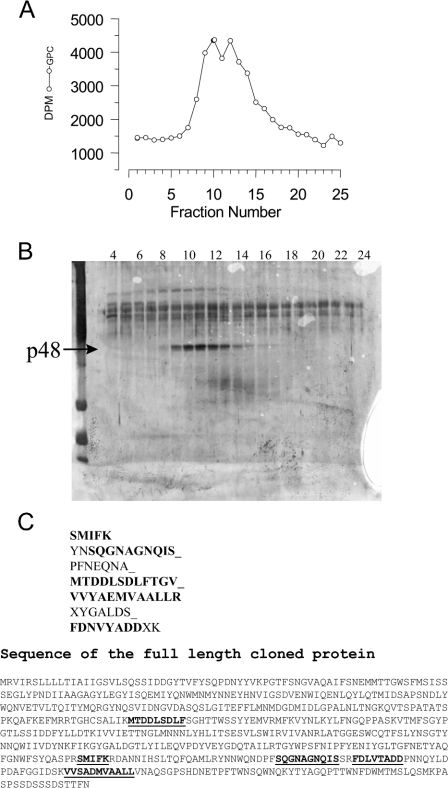
For sequence analysis, the purified material was additionally subjected to gel filtration (see Figure 3) and fractions containing PLB activity were concentrated and applied to SDS/PAGE for final purification, stained and digested with trypsin in the excised gel piece. Peptides were purified by HPLC as described previously [26,27].
Figure 3. Sequencing and alignment of peptide sequences and the predicted PLB protein.
PLB activity was purified from a scaled up preparation starting with a 5 litre Dictyostelium culture. The purification was taken through the steps identified in Figure 2. Following ARF affinity chromatography, the activities were again further subjected to chromatography on a Superose-12 column. Purified material was applied (2×100 μl aliquots) and 200 μl fractions collected using a flow rate of 0.5 ml/min in 20 mM Hepes (pH 7.0)/0.02% sodium azide. The individual fractions were assayed (A), subjected to SDS/PAGE and silver stained (B). The 48 kDa protein (p48) profile matched the activity profile. However, 65 kDa protein was masked by the presence of contaminants and sequence was only obtained from the 48 kDa band. The 48 kDa protein was digested with trypsin, and peptide fragments separated by reverse-phase chromatography. The resultant peptides were sequenced by Edman degradation, and the sequences generated are shown (C). The peptides shown in bold were identified in the fulllength cloned protein and are underlined. The accession number for the full-length cDNA is AF411829_1.
Molecular cloning and expression of Dictyostelium cDNA encoding PLB
E. coli (BL21, Amersham) was transformed with pGEX-5X-3 containing an in-frame Dictyostelium PLB clone or empty pGEX-5X-3. These bacterial expression plasmids produce GST (glutathione-S-transferase)-tagged fusion proteins. Cultures were grown overnight at 37 °C in Luria–Bertani broth (200 ml), and then expanded into 1-litre cultures and grown for a further 5 h at 37 °C. IPTG (isopropyl β-D-thiogalactoside; 0.5 mM) was then added for a further 18 h at 22 °C. Bacteria were harvested at 5000 g for 10 min at 4 °C. Pellets were resuspended in 30 ml of PBS/1 mg/ml lysozyme and incubated at 37 °C for 15 min. DNaseI (200 units) was added, and then the suspensions were sonicated with a large probe for three 5 s pulses on ice. The suspensions were allowed to stand at 18–25 °C for a further 10 min, and then re-sonicated for a further three 5 s pulses. Insoluble material was sedimented at 100000 g for 30 min at 4 °C. The clarified supernatants were then agitated with a 0.5 ml bed volume of glutathione–Sepharose (Amersham) for 30 min at 18–25 °C. Beads were sequentially washed with 5×20 ml PBS. Bound proteins were eluted with 3×1 ml of 50 mM Tris (pH 8.0)/10 mM GSH. The eluates were then concentrated in 10 kDa Centricons (Millipore) to a volume of 800 μl. SDS/PAGE and Western blotting were performed on the purified proteins using standard procedures. The anti-GST antibody was obtained from Abcam (goat polyclonal, number ab6613-1) and used at a 1:2000 dilution in PBS/Tween. PLB activity was assessed using the didecanoyl-PC substrate described above.
Determination of substrate specificity
Recombinant PLB (purified from a 2 litre E. coli culture) was concentrated to 600 μl, and 30 μl was used for assessing PC acyl-chain length selectivity and headgroup specificity. Since the amount of recombinant PLB obtained was limited, we also used partially purified PLB. PLB was partially purified from a 200 ml culture (2×108 cells) of axenically grown Dictyostelium. Briefly, the cells were lysed by sonication in 20 mM Hepes, pH 7.0, in the presence of protease inhibitors, and fractionated on a Superdex-200 gel-filtration column. Peak fractions were combined, and applied to a heparin–Sepharose column. The flow-through was then applied to a Mono-Q column, and the column washed extensively. PLB was eluted with 2 M NaCl, and the peak fractions desalted to 20 mM Hepes, pH 7.0, and concentrated. This resulted in the production of 800 μl of enzyme with a protein concentration of 1.5 mg/ml.
Labelled lipids were produced by growing HL60 cells in the presence of either [3H]choline, [3H]inositol or [3H]ethanolamine. HL60 cells were grown in M199 supplemented with 10% dialysed fetal calf serum with the relevant label at 10 μCi/ml for 48 h. Cells were washed extensively with PBS, and the lipids extracted using chloroform/methanol/water at a ratio of 1:1:0.9 (by vol.). The labelled lipids (present in the lower phase) were dried under vacuum, and separated using TLC. Lipids were resolved using solvent consisting of chloroform/methanol/acetic acid/water [75:45:12:2 for PC, and 75:45:3:1 for PI (phosphatidylinositol) and PE]. Regions containing the labelled lipids were scraped and extracted from the silica using chloroform/methanol/water in the ratio 1:1:0.9. Lower phases were dried, and the quantity of radioactivity determined. Vesicles were prepared using equimolar quantities of PE/PI/PC so that the final concentration of each lipid in the assay was 10 μM. When using commercially obtained PC (dipalmitoyl and didecanoyl) the chain length of the label was matched with the PC species being used. When the substrate contained one of the labelled lipids isolated from HL60 cells a comparable chain length match was used such that brain PE, brain PI and egg yolk PC were used. Radioactivity was added to give 100000 d.p.m. per reaction.
RESULTS
This work was initiated to examine the presence of PC-hydrolysing activity by PLD in Dictyostelium. Small samples of frozen, axenically-grown Dictyostelium cells were lysed by sonication in the presence of protease inhibitors. A post-nuclear supernatant was prepared and enzyme activity monitored using a well-established in vitro PLD assay [28,29]. Release of water-soluble (3H-labelled choline-containing) metabolites generated from PC were measured. These water-soluble metabolites were separated into phosphorylated (P-choline and GPC), and non-phosphorylated (choline) metabolites using BioRex 70 cation exchange resin. In Figure 1(A) a time course of the enzymic breakdown of PC by Dictyostelium lysates is shown. Principally, the products of this reaction were phosphorylated, and hence revealed that PLD activity was not a major enzyme activity in these crude lysates. We further characterized the enzyme activity present in these lysates, and were able to demonstrate that the activity was independent of ARF1, GTPγS and PtdIns(4,5)P2, further evidence that we were not measuring PLD activity (results not shown). We next looked for the cellular localization of the enzyme activity by further fractionating the lysates into cytosol and membrane fractions. As shown in Figure 1(B) the enzyme activity was apparently not partitioned specifically to either of these crude locations.
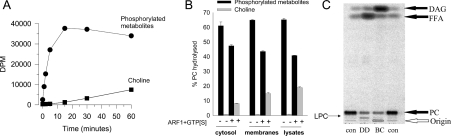
Figure 1. Identification of D. discoideum PLB enzyme activity.
(A) Dictyostelium lysate (10 μg) was incubated in the presence of GTPγS (30 μM) and myrARF1 (10 μM) at 37 °C for the indicated times. Water-soluble metabolites were separated into choline and phosphorylated metabolites. The results are expressed as d.p.m. of products liberated. Error bars represent range of two points (smaller than symbol in most instances). (B) Dictyostelium lysate was sedimented at 100000 g to separate membranes from cytosol. Lysate (40 μg), membranes (16 μg), and cytosol (18 μg) were assayed using didecanoyl-PC substrate (material used as cell equivalent). Incubations were performed for 60 min at 37 °C in the presence and absence of GTPγS and myrARF1. Water-soluble metabolites were separated into choline and phosphorylated metabolites. The results are expressed as the percentage of PC hydrolysed. Error bars represent range of two points. (C) Dictyostelium lysate (DD; 80 μg), and PLC from B. cereus (BC; 1 units/ml) were assayed using [14C]didecanoyl-PC substrate (labelled with 14C in both fatty acid chains). Counts were added to give 92000 d.p.m. per reaction. Incubations were performed for 60 min at 37 °C in the absence of any stimulus. The organic phase was dried, re-solubilized in chloroform, and separated using TLC. Radioactivity on the TLC plate was determined using a phosphorimager. DAG (diacylglycerol), FFA (NEFA) and LPC are indicated. con, substrate alone.
The results in Figure 1(A) show that we also generate small quantities of choline during these incubations. Further analysis demonstrated that this was as a consequence of myrARF1 being used in early experiments. In the absence of myrARF1, choline release did not occur (Figure 1B). Additional experiments showed that choline release was due to the breakdown of GPC by an activity that was present in the myrARF1 preparations (Figure 1B and results not shown).
In order to accurately describe the enzyme activity we used two further assays to examine more closely the products formed during the incubations. As indicated above the products released could either be P-choline or GPC which would be formed via a PLC or PLB respectively. Products from a 20 min incubation at 37 °C were subjected to paper chromatography to separate choline, P-choline and GPC. Products of the Dictyostelium reaction were excised in regions corresponding to the standards, and the radioactivity in each spot determined. The Dictyostelium enzyme activity liberated more than 95% GPC (results not shown). Additionally, another form of didecanoyl-PC substrate was used which was not labelled in the choline head-group as described above, but was labelled with 14C in each of the fatty acid chains. The products of these reactions were subjected to TLC and Figure 1(C) shows the major product produced by the lysate was NEFA. Only a small amount of LPC was also released. As a control, we used PLC from Bacillus cereus, which as expected produced diacylglycerol as its major product. Thus Dictyostelium lysates contain a very active PLB that forms GPC and NEFAs from PC.
Purification of the Dictyostelium PC-hydrolysing PLB activity
Since the activity was present in both the cytosol and membranes of lysed cells (Figure 1B), we used the cytosol for the purification, as most proteases are found in the membrane fraction. Small volumes of cytosol were tested on a selection of purification media. Chromatography media tested included gel filtration, Mono-Q, heparin, Mono-P, phenyl-Superose and affinity chromatography using myrARF1. The Dictyostelium enzyme activities were stable following chromatography on gel filtration, Mono-Q, heparin and a myrARF1 affinity column generated using Hi-Trap NHS-activated agarose beads (Amersham Biosciences). Attempts using Mono-P or phenyl-Superose proved to be unsuccessful. Additionally, ammonium sulphate precipitation was found to be useful for the first purification step. The cytosol was ammonium sulphate precipitated at 0–20%, 20–45% and 45–70% saturation. In the 20–45% pellet, 67% of the cytosolic protein, and 75% of the activity was recovered. (There was not a substantial enrichment of the enzyme activities during the process of ammonium sulphate precipitation, but it was a useful step for concentrating the cytosol.). Figures 2(A–C) illustrates the series of steps that we finally used for the complete purification.
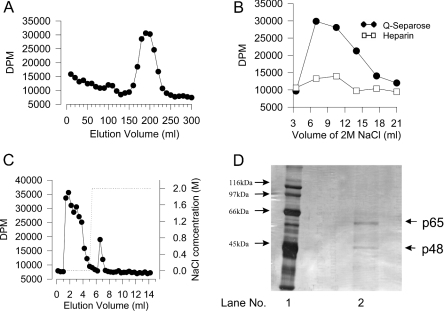
Figure 2. Purification of PLB activity.
(A) Superdex-200 fractionation. Cytosol from 5×109 cells (Dictyostelium) was precipitated with 20–45% saturated ammonium sulphate. The pellet was resuspended, dialysed, and concentrated to 10 ml [20 mM Hepes (pH 7.0)/0.02% sodium azide]. Two identical separations of the concentrate (5 ml per run) were performed using a Pharmacia Superdex-200 column at a flow rate of 1.5 ml/min. Fractions of 10 ml were collected. Assays for activity were performed on 20 μl of each fraction using didecanoyl-PC substrate. Incubations were performed for 60 min at 37 °C in the presence of 10 μM myrARF1 and 30 μM GTPγS. Water soluble metabolites were separated into choline and GPC. Only the results for GPC are shown. The results are expressed as d.p.m. released from the added 80000 d.p.m. The results shown are from one of the two identical separations. (B) heparin and Mono-Q fractionation of active Superdex-200 fractions. Peak fractions from the gel-filtration separation shown in (A) were combined. The combined material (approx. 120 ml) was applied to a 5 ml Hi-Trap heparin column connected in-line to a 5 ml Hi-Trap Q-Sepharose column at 1 ml/min. The columns were separated, and each column was washed separately with 30 ml of buffer. Proteins were eluted with 2 M NaCl (3.5 ml fractions). Fractions were desalted and the activity of 20 μl of each fraction was determined in the presence of myrARF1. The results are shown for proteins eluted from heparin and Q-Sepharose. There was negligible activity in the flow through and wash step (results not shown). (C) myrARF1 affinity purification. Peak fractions from the Q-Sepharose column shown in (B) were further fractionated using a pre-prepared myrARF1-bound NHS-affinity column. A sample of this partially purified activity (0.5 ml) was applied to the 1 ml affinity column at 1 ml/min. The column was washed with 5 ml of buffer, and then the bound protein eluted with 2 M NaCl. Fractions (200 μl) were collected for the load, wash and elution stages. Activity in every second fraction was determined. Fractions (20 μl) were assayed using didecanoyl-PC substrate in the presence of myrARF1. The results are expressed as d.p.m. released from the added 80000 d.p.m. (D) Silver-stained SDS/PAGE of purified enzyme after myrARF1 affinity purification (lane 2). Lane 1, molecular mass markers.
Cytosol was prepared by filter lysis using cells from a 1-litre Dictyostelium culture. The pellet from 20–45% ammonium sulphate saturation was solubilized, dialysed to reduce the concentration of ammonium sulphate, concentrated to 10 ml, and applied to a Pharmacia Superdex-200 column in two separate 5 ml runs. Fractions were then analysed for the Dictyostelium enzyme activity (Figure 2A). Calibration of the Superdex-200 column with the molecular mass markers, carbonic anhydrase (29 kDa), ovalbumin (45 kDa) and BSA (66 kDa) showed that the Dictyostelium activities migrated with a mass of 55–65 kDa. Peak fractions from Superdex-200 were passed through 5 ml columns of Hi-Trap heparin, and Q-Sepharose connected in-line (heparin→Q). The Dictyostelium enzyme was observed to pass through heparin resin, and bind to Q-Sepharose. The Q-column was disconnected from the combination, and the Dictyostelium enzyme activities were step eluted from the Q-Sepharose with 2 M NaCl. Figure 2(B) shows activities for fractions (following desalting) generated from 2 M NaCl elutions of both heparin and Q-Sepharose columns. It can clearly be seen from this data that the majority of the activity was eluted from the Q-column. No activity could be detected in the column flow-through (results not shown). The protein concentration of the Q-Sepharose eluate was undetectable using standard protein assays.
Desalted fraction 2 from Q-Sepharose (total volume 4 ml) was then applied (load 0.5 ml) to a pre-prepared myrARF1 affinity column. The affinity column consisted of an NHS-activated matrix which had been reacted with 10 mg of myrARF1. Protein was eluted with step elution using 2 M NaCl, and assayed (Figure 2C). The majority of the enzyme activity was found to pass through the column during the wash step (0 M NaCl), however, some activity bound to the column and was eluted with 2 M NaCl. If the unbound material was re-loaded on to the affinity column, another similar amount bound. This indicated that the affinity column had a finite binding capacity, and it was found that in order to achieve optimal recovery it was best to run the column several times with smaller load volumes. The amount of protein in both the affinity column load, and in the peak material eluted with 2 M NaCl, were below those detectable using Bradford or Pierce assays.
In order to maximize recovery, the ARF-affinity column load material was diluted to 50 ml, and the column was run 50 times with 1 ml injection volumes. Using a Pharmacia FPLC it was possible to program the controller to automatically deliver 1 ml load volumes, wash with 20 ml of buffer, elute with 5 ml of 2 M NaCl, and wash the column with 20 ml of buffer before re-loading. The fraction collector was set up to collect only the 5 ml of 2 M NaCl eluate. Following 50 repeated runs, the material was concentrated using an Amicon 10K pressure filtration system and 10K centricon to 1.5 ml. Protein was detectable in this material (150 μg total). SDS/PAGE of some of this material shows that the sample was relatively pure when stained with silver (Figure 2D). Two bands were found, one at 48 kDa and another at 65 kDa. The material obtained from this purification run was insufficient for sequence analysis and we therefore had to scale up using cells from 5 litres of Dictyostelium culture. Figure 3 illustrates the final run which allowed determination of sequence information from the 48 kDa band. The 65 kDa protein was obscured by contaminants and not sequenced. The 48 kDa band was subjected to trypsin digestion and micro-sequencing yielded data from 7 peptides of varying length (Figure 3C).
These peptide sequences were used to screen the Tsukuba Dictyostelium EST (expressed sequence tag) database. A partially sequenced cDNA for an unknown protein was identified. Five of the peptide sequences, including three of the longer peptides SQGNAGNQIS, MTDDLSDLF and VVYAEMVAALLR, could be identified in the predicted protein sequence and are marked in bold. The match between the peptide sequences and the cloned protein was from 75 to 100% identical. The sequences were found towards the C-terminus of the protein suggesting that the 48 kDa band was a cleaved product of the full-length 65 kDa protein from which the N-terminus had been removed. We searched the Dictyostelium genome project to find the complete sequence, which contained an open reading frame encoding a full-length protein of 65 kDa (Figure 3C). We also found a number of other related sequences, though none of them contained perfect matches to the sequenced peptides.
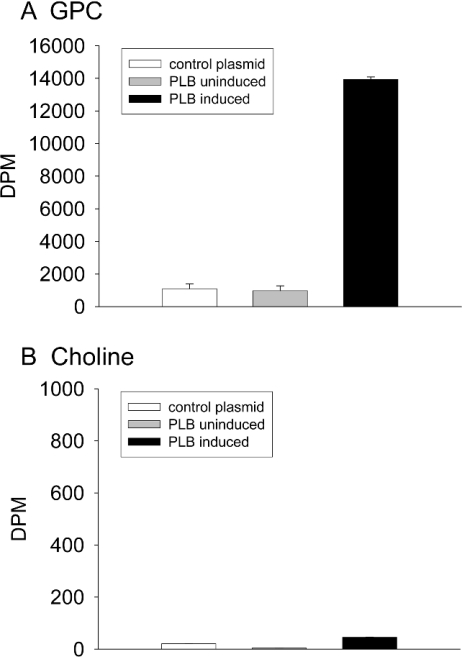
A full-length cDNA clone for PLB was obtained by reverse-transcriptase PCR, using mRNA extracted from vegetative cells and primers corresponding to the ends of the open reading frame. This was fully sequenced to verify the intron boundaries, then subcloned into the expression vector pGEX-5X-3. To assay activity we expressed the protein in the BL21 bacterial strain. Initial attempts to express the protein under standard conditions were unsuccessful. Large amounts of GST were produced, but no detectable PLB protein. We therefore induced the protein expression at 22 °C, a temperature more suited to Dictyostelium protein folding. Even at this temperature it was clear that the protein was not expressed in large quantities by the bacteria. We therefore assayed activity from bacteria transformed with an empty vector or with a PLB-containing plasmid (uninduced and induced cells in parallel) (Figure 4). These results show that IPTG-induced bacteria containing pGEX-PLB show a clear increase in PLB activity (Figure 4A), whereas uninduced bacteria or BL21 transformed with an empty pGEX-5X-3 vector do not. We measured choline levels in parallel and found no change under any conditions (Figure 4B). We were unable to detect GST–PLB by either SDS/PAGE or by Western blotting using an anti-GST antibody (results not shown).
Figure 4. Expression of Dictyostelium PLB in E. coli.
BL21 were transformed with control plasmid (pGEX-5X-3) and plasmid containing PLB (pGEX-5X-3-PLB). Three cultures were grown: control plasmid, PLB uninduced and PLB induced. The GST-fusion proteins were purified as described in the Experimental section, resulting in 800 μl of purified protein for each 1 litre culture. The proteins were assayed for PLB activity using the didecanoyl substrate. Briefly, 40 μl of each purified protein was incubated with 20 μl of vesicles, in a total volume of 120 μl for 60 min at 37 °C. myrARF1 and GTPγS were omitted from the assay. (A) shows GPC and (B) choline.
Analysis of PLB substrate specificity
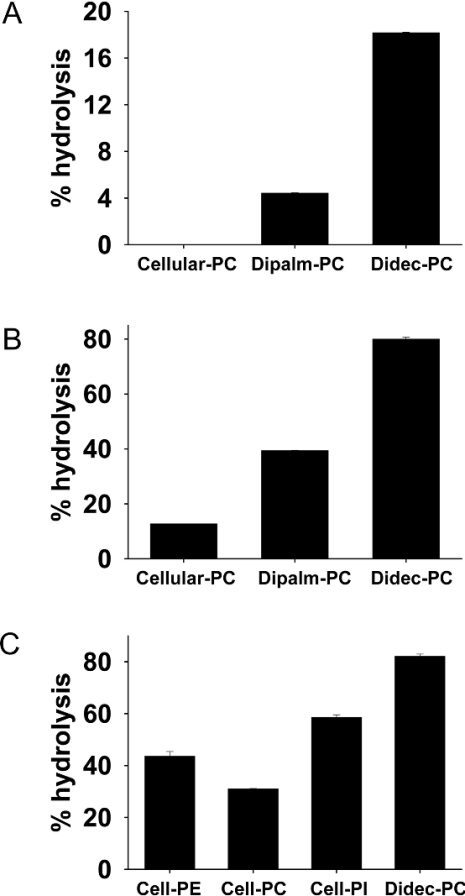
Following successful purification and cloning of Dictyostelium PLB, we next examined the substrate specificity of the enzyme with respect to the fatty acyl chain and head-group specificity. Initially, we examined the PLB enzyme for activity against PC species differing in their acyl chain length. In all experiments we have used synthetic PC (C10:0/C10:0) (didecanoyl-PC) as substrate and this was compared with PC (C16:0/C16:0) (dipalmitoyl-PC), which was also commercially available as a choline-labelled lipid. In addition, we used PC extracted from [3H]choline-labelled HL60 cells in order to provide the PLB enzyme with PC containing a mixture of acyl chain species. (The predominant acyl chain species in cellular PC are C16:0/C18:1>C16:0/C16:1>C18:1/C18:1>C16:1/C18:1 [30,31]). Data for the recombinant enzyme (Figure 5A), and for PLB isolated from Dictyostelium (Figure 5B) show that as the acyl chain length increases, the PLB activity decreases.
Figure 5. Acyl chain length specificity and headgroup specificity of PLB.
(A) Recombinant PLB was assayed using three different PC species: cellular PC extracted from [3H]choline-labelled HL60 cells and [3H]choline-labelled dipalmitoyl-PC (Dipalm-PC) and didecanoyl-PC (Didec-PC). Assays were performed for 60 min at 37 °C. (B) Partially purified PLB (purified on Superdex-200, Heparin, and Mono-Q) was assayed using the same substrates as in (A). Samples containing 7.5 μg of partially purified PLB were incubated for 20 min at 37 °C. (C) Partially purified PLB [as in (B)] was assayed for activity using appropriately-labelled lipids purified from HL60 cells (grown for 48 h with [3H]choline, [3H]ethanolamine or [3H]inositol). Samples containing 1.5 μg of partially purified PLB were incubated for 60 min at 37 °C. Water-soluble products were extracted, separated into phosphorylated and non-phosphorylated metabolites, and quantified. Assays were performed in duplicate and the results shown are representative of three independent experiments. Results are shown for percentage hydrolysis of substrate, of which >95% in each case was the deacylated lipid (glycerophosphorylcholine, glycerophosphorylinositol or glycerophosphorylethanolamine).
For the analysis of head-group specificity we were restricted with a choice of labelled substrates and thus we used purified phospholipids (PI, PC and PE) from appropriately labelled HL60 cells. The acyl chain species for PI and for PE are distinct, with PI containing mainly longer chain length and highly unsaturated with stearoyl arachidonyl (C18:0/C20:4) as a dominant species, whereas PE contains mainly C18:0/C18:1, C18:1/C18:1 and C18:0/C20:4 [30–32]. It can be clearly seen that the PLB enzyme was able to hydrolyse each of the lipids, with a preference for PI>PE>PC, despite the longer chain length found in PI (Figure 5C). From this information we cannot specify the relative activity against each of the phospholipids definitively, since the acyl chain composition is different for the three lipids. However, it is clear that both the head-group and the acyl chain length determine the activity of the enzyme.
DISCUSSION
Over 30 years ago, a very high phospholipase A/lysophospholipase activity against PC had been described in Dictyostelium lysates [3]. We identified the activity as an extremely active PLB enzyme which liberated GPC and NEFAs from PC. The protein responsible was purified using conventional column chromatography, and the in vitro release of GPC was monitored. The final step in the purification process was an affinity column using myristoylated ARF1 and this produced two bands on a gel at 48 kDa and 65 kDa. During the scale-up process, only sufficient protein was obtained from the 48 kDa band, and this was used for obtaining peptide sequence. It was fortuitous that the affinity chromatography using myrARF1 as bait was successful. It seems likely that the PLB was binding to the myristoyl group but was unable to cleave it.
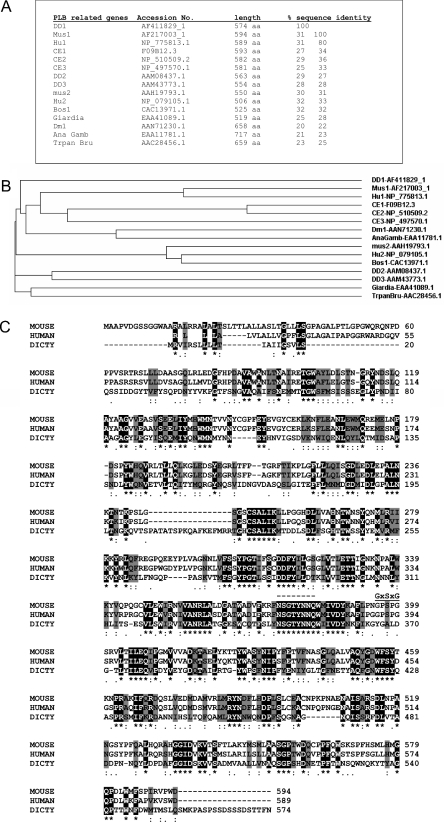
Although sequence information was only obtained from the 48 kDa band, this unambiguously identifies an approx. 65 kDa protein of previously unknown function in the Dictyostelium cDNA and genome databases. We assume that the 48 kDa protein is a degradation product of the 65 kDa full-length protein observed in the SDS gel (Figure 2D). Comparisons of the deduced amino acid sequence of the protein (574 amino acids long) against the non-redundant protein database using BLASTP program reveal a family of proteins of between 550–600 amino acids long (Figures 6A and 6B). The protein family is recorded as laminin A in CDART at NCBI (see also pfam04916.1). The one PubMed link refers to the identification of an evolutionary conserved gene, lamina ancestor (lama) expressed in the precursors of the Drosophila first optic ganglion, the lamina [33]. Lama was identified as a gene of unknown function that is expressed in both glial and neural progenitors during development of the first optic ganglion in the Drosophila eye. Although entered in the databases as laminin A family, this assignment appears to be incorrect, as these sequences are not discernibly related to laminins. The protein identified in Dictyostelium has PLB activity, and we suggest that the entire family should be renamed accordingly. BLAST searches reveal several related genes in D. discoideum. A single gene is detected in the flies Drosophila (where it is known as lama) and Anopheles gambiae. The parasites Trypanosoma brucei and Giardia lamblia also contain a single copy. Homologues of the 65 kDa protein are also identified in mammals (two human, one bovine and two mouse) and C. elegans (three homologues) (Figures 6A and 6B). No homologues were identified in S. cerevisiae, indicating the absence of this family of PLBs. All these genes have no clear biological roles that have hitherto been described. In Drosophila, loss-of-function mutants of the lama gene reveal no obvious phenotype [33], and in C. elegans small interfering RNA knockdowns are reported to be wild-type. However, it will be very interesting to re-examine this mutant in the light of the findings reported here.
Figure 6. Dictyostelium PLB-related sequences in mammals and other organsims.
(A) Sequences related to Dictyostelium PLB in several organisms. Accession numbers for the entries are used to identify the sequences. (B) Cladogram showing the relationship between the different sequences. DD, Dictyostelium discoideum; Mus, mouse; Hu, human; CE, C. elegans; Bos, Bos taurus; Dm, Drosophila; AnaGamb, Anopheles gambiae; Trpan Bru, Trypanosoma brucei. (C) A BLAST (NCBI) search was performed using the D. discoideum PLB sequence. Several sequences were retrieved with 30% identity or less. Alignment using Clustal (EBI) of mouse (Q8BHG8), human (Q8NHP8) and Dictyostelium (Q8MWQ0) sequences illustrates regions of identity (shaded black) and of similarity (shaded grey). The consensus lipase sequence GxSxG, and the region of strong PLB conservation NSGTYN(N/S)Q is indicated (broken line).
The PLB gene family identified here is not related to the bacterial PLB, secretory PLBs from fungi or to the mammalian intestinal PLB. Sequence-related fungal PLBs include Candida albicans (accession number U59710) [12], and related proteins from S. cerevisiae (accession number L23089) [14], P. notatum (accession number P39457) and T. Delbrueckii (accession number Q11121). Together with the PLB recently identified from the pathogenic fungus, Cryptococcus neoformans [16], these PLBs are secreted enzymes, are heavily glycosylated and are probably involved in virulence in some cases [2]. The PLBs in S. cerevisiae have been analysed and the first PLB enzyme (Spo1p) (accession number P53541) was identified as the product of the SPO1 gene which is required for meiosis and is only expressed during sporulation [13,15]. This PLB is localized to the nucleus and is required for spindle body duplication [15]. There are three other PLBs (plb1, plb2 and plb3) which are responsible for the PLB activity observed during vegetative growth. Deletion of all three PLBs displays no growth defects, thus the function of these enzymes remains to be identified. Sequence similarity between yeast PLBs and mammalian cytosolic PLA2 suggests that these enzymes may act by a common catalytic mechanism which comprises a Ser–Asp dyad [34,35], rather than the usual catalytic triad found in many lipases [20,36]. GxSxG together with aspartate and histidine form a catalytic triad conserved in many lipases, including monoglyceride lipase [37].
The first mammalian PLB cloned to date was the intestinal PLB, a glycosylated ecto-enzyme which is also unrelated to the PLB identified here [9,17]. The intestinal enzyme is also expressed in the epididymis and it may play a role in sperm maturation [9]. It is found mainly in membranes, and comprises a signal peptide, four tandem repeats of about 350 amino acids, two of them containing the lipase consensus sequence GxSxG, and a hydrophobic domain near the C-terminus [8,9,17,19]. However, the consensus sequence GxSxG was dispensable for activity and the nucleophile serine residue was located within the conserved GDSL sequence. Repeat 2 is fully equipped with the catalytic triad, but not the other repeats [10]. The secreted PLB enzyme from M. Bovis also has the GDSL motif as well [1]. Thus the catalytic triad can comprise the catalytic nucleophile serine (present in the motif GxSxG or GDSL), aspartate and histidine.
We have examined whether the PLB identified from D. discoideum may contain the catalytic triad found in many lipases. Figure 6(C) provides an alignment of the D. discoideum enzyme and the sequence-related human and mouse candidates. There are distinct regions of sequence conservation between the three proteins that are obvious. We also note that the mouse and human sequence do contain the GxSxG motif towards the C-terminus. However, this motif is not found in the Dictyostelium enzyme or in any other sequences identified in Figure 6, despite the obvious sequence identity between these proteins. This observation suggests the PLB is not a conventional lipase. The most conserved sequence found in the majority of the proteins related to Dictyostelium PLB is -NSGTYN(S/N)Q- which contains a serine residue that could potentially be involved in catalysis. The bacterial and fungal PLBs are mainly secreted enzymes, whereas the intestinal PLB are ecto-enzymes. Analysis of Dictyostelium PLB sequence (by PSORT and SignalP) indicates that it has a signal sequence with the most likely cleavage site between position 20 and 21: VLS–QS [38]. In summary, the present study identifies a novel PLB cDNA sequenced from D. discoideum and defines a new family of lipases. This family of lipases are not restricted to hydrolysing PC, as PI and PE are also substrates. When the enzyme was offered PC of different chain lengths, it had a preference for the shorter chain lengths. However, when the enzyme was given lipids with differing head-groups, a preference for PI was noted despite the presence of longer chain lengths. Based on this analysis, we would suggest the enzyme identified here is the same enzyme described in 1980 in extracts of D. discoideum on the basis of its PI-deacylating activity [39]. The ability of this PLB to hydrolyse PE is probably a major reason why expression of this protein in E. coli is compromised. It has also been previously reported that the salivary glands of adult blowflies (Calliphora erythrocephala) contain enzymes that deacylate PI, PC and PE, and were secreted in the saliva [40]. The presence of a signal sequence together with the knowledge that a similar enzyme is found in saliva would suggest that the enzyme is a secreted enzyme used for digestive purposes.
Acknowledgments
We are grateful to Dr Nick Totty (Ludwig Institute, University College London, U.K.) for sequencing the PLB. We thank Adrian Harwood for providing us with axenically grown D. discoideum. We thank Robin Irvine for bringing to our attention studies concerning the PI-deacylating activity identified in Dictyostelium and blowfly salivary gland. We thank the Medical Research Council for a studentship to C.P.M. (1994–1997) when this work was initiated and the Wellcome Trust for funding.
References
- 1.Farn J. L., Strugnell R. A., Hoyne P. A., Michalski W. P., Tennent J. M. Molecular characterization of a secreted enzyme with phospholipase B activity from Moraxella bovis. J. Bacteriol. 2001;183:6717–6720. doi: 10.1128/JB.183.22.6717-6720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghannoum M. A. Potential role of phospholipases in virulence and fungal pathogenesis. Clin. Microbiol. Rev. 2000;13:122–143. doi: 10.1128/cmr.13.1.122-143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferber E., Munder P. G., Fischer H., Gerisch G. High phospholipase activities in amoebae of Dictyostelium discoideum. Eur. J. Biochem. 1970;14:253–257. doi: 10.1111/j.1432-1033.1970.tb00284.x. [DOI] [PubMed] [Google Scholar]
- 4.Gassama-Diagne A., Rogalle P., Fauvel J., Willson M., Klaebe A., Chap H. Substrate specificity of phospholipase B from guinea pig intestine. A glycerol ester lipase with broad specificity. J. Biol. Chem. 1992;267:13418–13424. [PubMed] [Google Scholar]
- 5.Boll W., Schmid-Chanda T., Semenza G., Mantei N. Messenger RNAs expressed in intestine of adult but not baby rabbits. Isolation of cognate cDNAs and characterization of a novel brush border protein with esterase and phospholipase activity. J. Biol. Chem. 1993;268:12901–12911. [PubMed] [Google Scholar]
- 6.Delagebeaudeuf C., Gassama A., Collet X., Nauze M., Chap H. Guinea pig intestinal phospholipase B: protein expression during enterocyte maturation and effects of N-oligosaccharide removal on enzymatic activities and protein stability. Biochim. Biophys. Acta. 1996;1303:119–126. doi: 10.1016/0005-2760(96)00090-2. [DOI] [PubMed] [Google Scholar]
- 7.Tojo H., Ichida T., Okamoto M. Purification and characterization of a catalytic domain of rat intestinal phospholipase B/lipase associated with brush border membranes. J. Biol. Chem. 1998;273:2214–2221. doi: 10.1074/jbc.273.4.2214. [DOI] [PubMed] [Google Scholar]
- 8.Takemori H., Zolotaryov F. N., Ting L., Urbain T., Komatsubara T., Hatano O., Okamoto M., Tojo H. Identification of functional domains of rat intestinal phospholipase B/lipase. Its cDNA cloning, expression, and tissue distribution. J. Biol. Chem. 1998;273:2222–2231. doi: 10.1074/jbc.273.4.2222. [DOI] [PubMed] [Google Scholar]
- 9.Delagebeaudeuf C., Gassama-Diagne A., Nauze M., Ragab A., Li R. Y., Capdevielle J., Ferrara P., Fauvel J., Chap H. Ectopic epididymal expression of guinea pig intestinal phospholipase B. Possible role in sperm maturation and activation by limited proteolytic digestion. J. Biol. Chem. 1998;273:13407–13414. doi: 10.1074/jbc.273.22.13407. [DOI] [PubMed] [Google Scholar]
- 10.Lu T., Ito M., Tchoua U., Takemori H., Okamoto M., Tojo H. Identification of essential residues for catalysis of rat intestinal phospholipase B/lipase. Biochemistry. 2001;40:7133–7139. doi: 10.1021/bi010237n. [DOI] [PubMed] [Google Scholar]
- 11.Maury E., Prevost M. C., Nauze M., Redoules D., Tarroux R., Charveron M., Salles J. P., Perret B., Chap H., Gassama-Diagne A. Human epidermis is a novel site of phospholipase B expression. Biochem. Biophys. Res. Commun. 2002;295:362–369. doi: 10.1016/s0006-291x(02)00657-5. [DOI] [PubMed] [Google Scholar]
- 12.Leidich S. D., Ibrahim A. S., Fu Y., Koul A., Jessup C., Vitullo J., Fonzi W., Mirbod F., Nakashima S., Nozawa Y., Ghannoum M. A. Cloning and disruption of caPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans. J. Biol. Chem. 1998;273:26078–26086. doi: 10.1074/jbc.273.40.26078. [DOI] [PubMed] [Google Scholar]
- 13.Tevzadze G. G., Mushegian A. R., Esposito R. E. The SPO1 gene product required for meiosis in yeast has a high similarity to phospholipase B enzymes. Gene. 1996;177:253–255. doi: 10.1016/0378-1119(96)00261-2. [DOI] [PubMed] [Google Scholar]
- 14.Merkel O., Fido M., Mayr J. A., Pruger H., Raab F., Zandonella G., Kohlwein S. D., Paltauf F. Characterization and function in vivo of two novel phospholipases B/lysophospholipases from Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:28121–28127. doi: 10.1074/jbc.274.40.28121. [DOI] [PubMed] [Google Scholar]
- 15.Tevzadze G. G., Swift H., Esposito R. E. Spo1, a phospholipase B homolog, is required for spindle pole body duplication during meiosis in Saccharomyces cerevisiae. Chromosoma. 2000;109:72–85. doi: 10.1007/s004120050414. [DOI] [PubMed] [Google Scholar]
- 16.Chen S. C., Wright L. C., Golding J. C., Sorrell T. C. Purification and characterization of secretory phospholipase B, lysophospholipase and lysophospholipase/transacylase from a virulent strain of the pathogenic fungus Cryptococcus neoformans. Biochem. J. 2000;347:431–439. doi: 10.1042/0264-6021:3470431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delagebeaudeuf C., Gassama-Diagne A., Nauze M., Ragab A., Li R. Y., Capdevielle J., Ferrara P., Fauvel J., Chap H. Biochemical characterization and cloning of guinea pig intestinal phospholipase B. Ann. N.Y. Acad. Sci. 1998;859:192–193. doi: 10.1111/j.1749-6632.1998.tb11125.x. [DOI] [PubMed] [Google Scholar]
- 18.Tchoua U., Ito M., Okamoto M., Tojo H. Increased intestinal phospholipase A2 activity catalyzed by phospholipase B/lipase in WBN/Kob rats with pancreatic insufficiency. Biochim. Biophys. Acta. 2000;1487:255–267. doi: 10.1016/s1388-1981(00)00101-3. [DOI] [PubMed] [Google Scholar]
- 19.Nauze M., Gonin L., Chaminade B., Peres C., Hullin-Matsuda F., Perret B., Chap H., Gassama-Diagne A. Guinea pig phospholipase B, identification of the catalytic serine and the proregion involved in its processing and enzymatic activity. J. Biol. Chem. 2002;277:44093–44099. doi: 10.1074/jbc.M205761200. [DOI] [PubMed] [Google Scholar]
- 20.Schrag J. D., Cygler M. Lipases and alpha/beta hydrolase fold. Methods Enzymol. 1997;284:85–107. doi: 10.1016/s0076-6879(97)84006-2. [DOI] [PubMed] [Google Scholar]
- 21.Upton C., Buckley J. T. A new family of lipolytic enzymes? Trends Biochem. Sci. 1995;20:178–179. doi: 10.1016/s0968-0004(00)89002-7. [DOI] [PubMed] [Google Scholar]
- 22.Das O. P., Henderson E. J. A novel technique for cell lysis of eukaryotic cells: Isolation of plasma membranes from Dictyostelium discoideum. Biochim. Biophys. Acta. 1983;736:45–56. [Google Scholar]
- 23.Geny B., Fensome A., Cockcroft S. Rat brain cytosol contains a factor which reconstitutes G-protein-regulated phospholipase D activation in HL60 cells previously permeabilized with streptolysin O. Eur. J. Biochem. 1993;215:389–396. doi: 10.1111/j.1432-1033.1993.tb18045.x. [DOI] [PubMed] [Google Scholar]
- 24.Allan D., Quinn P. Resynthesis of sphingomyelin from plasma-membrane phosphatidylcholine in BHK cells treated with Staphyloccus aureus sphingomyelinase. Biochem. J. 1988;254:765–771. doi: 10.1042/bj2540765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allan D., Cockcroft S. A modified procedure for thin-layer chromatography of phospholipids. J. Lipid Res. 1982;23:1373–1374. [PubMed] [Google Scholar]
- 26.Totty N. F., Waterfield M. D., Hsuan J. J. Accelerated high sensitivity microsequencing of proteins and peptides using a miniature reaction cartridge. Protein Science. 1992;1:1215–1224. doi: 10.1002/pro.5560010914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas G. M. H., Cunningham E., Fensome A., Ball A., Totty N. F., Troung O., Hsuan J. J., Cockcroft S. An essential role for phosphatidylinositol transfer protein in phospholipase C-mediated inositol lipid signalling. Cell. 1993;74:919–928. doi: 10.1016/0092-8674(93)90471-2. [DOI] [PubMed] [Google Scholar]
- 28.Brown H. A., Gutowski S., Moomaw C. R., Slaughter C., Sternweis P. C. ADP-ribosylation factor, a small GTP-dependent regulatory protein, stimulates phospholipase D activity. Cell. 1993;75:1137–1144. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- 29.Vinggaard A. M., Jensen T., Morgan C. P., Cockcroft S., Hansen H. S. Didecanoylphosphatidylcholine is a superior substrate for assaying mammalian phospholipase D. Biochem. J. 1996;319:861–864. doi: 10.1042/bj3190861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunt A. N., Clark G. T., Attard G. S., Postle A. D. Highly saturated endonuclear phosphatidylcholine is synthesized in situ and colocated with CDP-choline pathway enzymes. J. Biol. Chem. 2001;276:8492–8499. doi: 10.1074/jbc.M009878200. [DOI] [PubMed] [Google Scholar]
- 31.Pettitt T. R., McDermott M., Saqib K. M., Shimwell N., Wakelam M. J. Phospholipase D1b and D2a generate structurally identical phosphatidic acid species in mammalian cells. Biochem. J. 2001;360:707–715. doi: 10.1042/0264-6021:3600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cockcroft S., Allan D. The fatty acid composition of phosphatidylinositol, phosphatidate and 1,2-diacylglycerol in stimulated human neutrophils. Biochem. J. 1984;222:557–559. doi: 10.1042/bj2220557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez S. E., Steller H. Molecular and genetic analyses of lama, an evolutionarily conserved gene expressed in the precursors of the Drosophila first optic ganglion. Mech. Dev. 1996;59:11–27. doi: 10.1016/0925-4773(96)00556-4. [DOI] [PubMed] [Google Scholar]
- 34.Dessen A. Structure and mechanism of human cytosolic phospholipase A2. Biochim. Biophys. Acta. 2000;1488:40–47. doi: 10.1016/s1388-1981(00)00108-6. [DOI] [PubMed] [Google Scholar]
- 35.Dessen A., Tang J., Schmidt H., Stahl M., Clark J. D., Seehra J., Somers W. S. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell. 1999;97:349–360. doi: 10.1016/s0092-8674(00)80744-8. [DOI] [PubMed] [Google Scholar]
- 36.Ollis D. L., Cheah E., Cygler M., Dijkstra B., Frolow F., Franken S. M., Harel M., Remington S. J., Silman I., Schrag J. The alpha/beta hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 37.Karlsson M., Contreras J. A., Hellman U., Tornqvist H., Holm C. cDNA cloning, tissue distribution, and identification of the catalytic triad of monoglyceride lipase. Evolutionary relationship to esterases, lysophospholipases, and haloperoxidases. J. Biol. Chem. 1997;272:27218–27223. doi: 10.1074/jbc.272.43.27218. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen H., Engelbrecht J., Brunak S., von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 39.Irvine R. F., Letcher A. J., Brophy P. J., North M. J. Phosphatidylinositol-degrading enzymes found in the cellular slime mould Dictyostelium discoideum. J. Gen. Microbiol. 1980;121:495–497. [Google Scholar]
- 40.Irvine R. F., Berridge M. J., Letcher A. J., Dawson R. M. Phosphatidylinositol-hydrolysing enzymes in blowfly salivary glands. Biochem. J. 1982;204:361–364. doi: 10.1042/bj2040361. [DOI] [PMC free article] [PubMed] [Google Scholar]