Abstract
The GRU (glucocorticoid-response unit) within the distal enhancer of the gene encoding carbamoyl-phosphate synthase, which comprises REs (response elements) for the GR (glucocorticoid receptor) and the liver-enriched transcription factors FoxA (forkhead box A) and C/EBP (CCAAT/enhancer-binding protein), and a binding site for an unknown protein denoted P3, is one of the simplest GRUs described. In this study, we have established that the activity of this GRU depends strongly on the positioning and spacing of its REs. Mutation of the P3 site within the 25 bp FoxA–GR spacer eliminated GRU activity, but the requirement for P3 could be overcome by decreasing the length of this spacer to ≤12 bp, by optimizing the sequence of the REs in the GRU, and by replacing the P3 sequence with a C/EBPβ sequence. With spacers of ≤12 bp, the activity of the GRU depended on the helical orientation of the FoxA and GR REs, with highest activities observed at 2 and 12 bp respectively. Elimination of the 6 bp C/EBP–FoxA spacer also increased GRU activity 2-fold. Together, these results indicate that the spatial positioning of the transcription factors that bind to the GRU determines its activity and that the P3 complex, which binds to the DNA via a 75 kDa protein, functions to facilitate interaction between the FoxA and glucocorticoid response elements when the distance between these transcription factors means that they have difficulties contacting each other.
Keywords: architecture, carbamoyl-phosphate synthase, enhancer, glucocorticoid-response element, glucocorticoid-response unit
Abbreviations: C/EBP, CCAAT/enhancer binding protein; COUP-TF, chicken ovalbumin upstream promoter-transcription factor; CPS, carbamoyl-phosphate synthase I; DTT, dithiothreitol; EMSA, electrophoretic mobility-shift assay; FoxA, forkhead box A; GR, glucocorticoid receptor; GRE, glucocorticoid-response element; GRU, glucocorticoid-response unit; HNF, hepatocyte nuclear factor; PEPCK, phosphoenolpyruvate carboxykinase; RE, response element
INTRODUCTION
In liver, the enzymes of the urea cycle are responsible for the disposal of toxic ammonia originating from amino acid degradation into urea. The mitochondrial enzyme CPS (carbamoyl-phosphate synthase I; EC 6.3.4.16) is the rate-determining enzyme in this cycle [1]. Together with many genes encoding gluconeogenic and amino-acid-catabolizing enzymes, the genes encoding urea-cycle enzymes are expressed in the periportal region of the liver, and are activated by glucagon (via cAMP) and glucocorticoid hormones.
The hormonal regulation of the CPS gene is imposed by a 469 bp distal enhancer located 6.3 kb upstream of the transcription start site [2]. Within this distal enhancer, an 80 bp GRU (glucocorticoid-response unit) confers hormone responsiveness and tissue specificity upon the gene (Figure 1A) [3]. Hormone-response units are clusters of transcription factor-binding sites, comprising a hormone-responsive element and a number of cis-elements (accessory factors). Such a hormone-response unit allows the regulation of transcription of the gene in space and time by integrating multiple signal pathways [4]. We previously analysed the binding of transcription factors to the CPS GRU by in vitro footprinting [3]. At the 5′ end of the GRU is located a C/EBP (CCAAT/enhancer binding protein) RE (response element). C/EBP is a liver-enriched transcription factor that belongs to the basic zipper family of transcription factors and binds to its RE as a dimer [5]. Separated from the C/EBP-binding site by a 6 bp spacer is a binding site for FoxA/HNF3 (forkhead box A/hepatocyte nuclear factor 3). FoxA belongs to the winged-helix family of transcription factors, and is highly expressed in liver, stomach and intestine [6]. FoxA binds the DNA as a monomer and bends the DNA around itself, imposing a 13° angle upon the DNA [7]. At a distance of 25 bp downstream of the FoxA-binding site is located the GRE (glucocorticoid response element). The GR (glucocorticoid receptor) belongs to the steroid receptor family, and is expressed in most tissues. Upon ligand binding, the receptor dimerizes and binds to its RE. Within the 25 bp region between the FoxA- and GR-binding sites, a binding site for an unidentified protein denoted as P3 is present [3]. We showed previously that mutation of the GR, C/EBP or FoxA RE in the CPS GRU abolishes the response to glucocorticoids and, therefore, concluded that these elements are all necessary for a full glucocorticoid response [2].
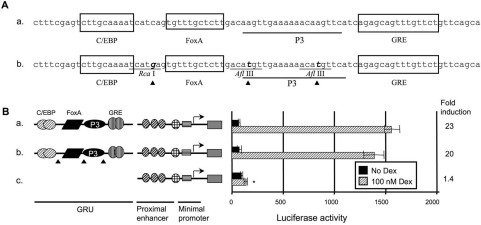
Figure 1. Introduction of restriction sites in the GRU does not affect GRU activity.
To facilitate cloning, restriction sites were introduced into the GRU. (A) Nucleotide sequence of the CPS GRU and the REs therein (construct a), and the modified GRU upon the introduction of restriction sites (construct b). The triangles indicate the positions of the point mutations. The GRU constructs were cloned upstream of the minimal promoter and proximal enhancer of the CPS gene. These reporter-gene constructs were transiently transfected into FTO-2B hepatoma cells and induced with glucocorticoids for 24 h. The reporter-gene activities, measured in the resulting lysates, are presented as means±S.E.M. luciferase values for at least four experiments and the fold induction. The asterisk indicates significantly different results relative to the wild-type construct a. The data show that the combination of proximal enhancer and minimal promoter does not result in a marked increase in activity when induced by glucocorticoids relative to basal activity (B, construct c). In contrast, both the parent and the modified GRUs exhibited a similarly high level of activity when cultured in presence of glucocorticoids (B, constructs a and b). Dex, dexamethasone.
To begin to address the question of how a GRU works, it is necessary to delineate the structural requirements with respect to positioning and spacing of the components of the unit. In the present study, we addressed these questions by constructing GRUs in which the sequences, relative distances apart and configurations of the REs were altered. These constructs were tested in transient transfection assays with FTO-2B hepatoma cells. We found that the sequences of the REs and their distance apart affected GRU activity greatly. As with the other participating REs, mutation of the P3 site within the FoxA–GR spacer eliminated GRU activity, but the effects of this mutation could be overcome by decreasing the length of this spacer to ≤12 bp, by optimizing the sequence of the REs, and by replacing the P3 sequence with a C/EBPβ sequence, suggesting that P3 functions to facilitate the binding of transcription factors to the GRU. These findings show that the spatial requirements of the relatively simple CPS GRU are strict.
MATERIALS AND METHODS
Tissue culture
The rat hepatoma cell line FTO-2B [8] and the monkey-kidney cell line COS-1 [9] were maintained in Dulbecco's modified Eagle's medium/F12 medium (Invitrogen Life Technologies, Carlsbad, CA, U.S.A.) supplemented with 10% (v/v) foetal calf serum at 37 °C and 5% CO2.
Transfection of reporter constructs
To determine the influence of the architectural composition of the CPS GRU on its activity, modifications were introduced into the GRU. All GRU constructs described were cloned into the BamHI and PstI sites of the pSPluc+ vector (Promega, Madison, WI, U.S.A.). For this purpose, the pSPluc+ reporter vector was modified by inserting the bovine growth hormone poly(A) tail into the XbaI–EcoRV sites downstream of the luciferase gene, and the minimal promoter containing the proximal enhancer (−161 to −38) of the CPS gene into the KpnI–HindIII sites of the polylinker upstream of the luciferase gene.
To test these constructs, 20 μg of the luciferase–reporter construct was co-transfected with 2 μg of pRL-CMV control DNA (Promega) into 1×107 FTO-2B hepatoma cells by electroporation [10]. Transfected cells were divided into two equal portions and cultured in two 9.6 cm2 wells. At 24 h post-transfection, the medium was replaced and the cells were cultured for another 24 h; in one of the two wells, this medium was supplemented with 100 nM dexamethasone (Centrafarm, Etten-Leur, The Netherlands). Luciferase activity was measured using the dual-luciferase reporter assay system (Promega) in an Autolumat plus (Berthold, Vilvoorde, Belgium). Luciferase values were corrected for differences in transfection efficiency and between-session variations. To analyse the differences between constructs, the nonparametric Kruskal–Wallis test was employed. Results were considered significantly different at P<0.05.
Preparation of nuclear extracts from COS-1 cells
Using poly(ethylene imine) as transfection agent [11], COS-1 cells were transfected with a C/EBPα expression vector and cultured for 2 days. Cells were harvested and resuspended in lysis buffer (10 mM Tris, pH 7.4, 15 mM NaCl, 60 mM KCl, 1 mM EDTA, 0.1 mM EGTA, 0.1% Triton X-100, 150 mM sucrose, 0.15 mM spermine and 0.5 mM spermidine) precooled to 4 °C. Nuclei were sedimented through a 1 M sucrose cushion for 20 min at 1400 g and resuspended in 1 vol. of low-salt buffer [20 mM Hepes, pH 7.9, 25% (v/v) glycerol, 1.5 mM MgCl2, 20 mM KCl, 0.2 mM EDTA, 0.2 mM PMSF and 0.5 mM DTT (dithiothreitol)]. Nuclear proteins were extracted for 20 min at 4 °C on a tilt board by dropwise addition of 1 vol. of high-salt buffer (same as low-salt buffer, except that the 20 mM KCl was substituted by 800 mM KCl). Following 2×2 h dialysis against buffer comprising 20 mM Hepes, pH 7.9, 20% (v/v) glycerol, 100 mM KCl, 0.2 mM EDTA, 0.2 mM PMSF and 0.5 mM DTT, proteins were frozen in liquid nitrogen and stored at −80 °C.
Nuclear extract preparation from rat liver
Nuclear extracts were prepared from rat livers according to the method of Sierra [12]. Briefly, nuclei were sedimented through a 2 M sucrose cushion. The nuclei were resuspended and lysed by addition of (NH4)2SO4. The resulting precipitate of chromatin was removed by centrifugation. Nuclear proteins were salted out by a further increase in the (NH4)2SO4 concentration.
To obtain a P3-enriched protein fraction, 20 mg of nuclear extract was loaded on a MonoS HR 5/5 (Amersham Biosciences, Little Chalfont, Bucks., U.K.) cationic exchanger column using an LCC 501-plus FPLC apparatus (Amersham Biosciences). Protein fractions of 1 ml were collected by eluting the column with buffer [10% (v/v) glycerol, 20 mM Hepes, pH 7.9, 1 mM EDTA, 1 mM DTT and 0.01% Nonidet P40] containing a gradient from 100 to 600 mM KCl. The resulting fractions were tested for the presence of P3 by EMSA (electrophoretic mobility-shift assay) analysis.
EMSA analysis
To study protein–DNA interactions, a double-stranded DNA probe was radiolabelled with [α-32P]dATP using Klenow polymerase and purified on a Sephadex G50 spin column (Amersham Biotechnologies). Each binding reaction contained 10 μg of nuclear extract, 20 mM Hepes, pH 7.9, 1 μg of poly(dI-dC)·poly(dI-dC), 10% (v/v) glycerol and 100 mM KCl in a final volume of 20 μl. Following a 10 min preincubation on ice, the probe was added (2×104 c.p.m.) and complexes were allowed to form for 20 min on ice. To perform competition experiments, unlabelled (non)specific oligonucleotides were added to the reaction mixture, whereas for supershift analysis 1 μl of antiserum was added 15 min after commencing the binding reaction. The resulting protein–DNA complexes were resolved on a 6% (w/v) polyacrylamide gel (acrylamide/bisacrylamide, 29:1) in 0.25×TBE buffer (1×TBE: 45 mM Tris, 45 mM boric acid, 1 mM EDTA) at room temperature. Before loading the samples, gels were pre-run for 1 h. Samples were loaded on to the gel without the use of dye; a Bromophenol Blue reference was loaded in a separate lane. Following electrophoresis at 10 V·cm−1 until the Bromophenol Blue reference had migrated two-thirds of the length of the gel, the latter was dried and exposed overnight to a phosphorus screen and analysed using a Storm 860 PhosphorImager apparatus (Molecular Dynamics, Sunnyvale, CA, U.S.A.).
Antibodies
Rabbit polyclonal antibodies against C/EBPα (cat. no. sc-61) and C/EBPβ (cat. no. sc-746) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.).
Southwestern blotting
Analysis of protein–DNA interactions by Southwestern blotting was performed according to the method of Labbé et al. [13]. A 25 μg portion of nuclear extract was mixed with 2×sample buffer (125 mM Tris/HCl, pH 6.8, 4% SDS, 0.1% Bromophenol Blue, 20% glycerol and 5% β-mercaptoethanol). The sample was loaded on an SDS/9%-PAGE minigel and electrophoresed in Laemmli buffer (50 mM Tris base, 384 mM glycine, 1% SDS, pH 8.3) at 50 V until the Bromophenol Blue had migrated off the gel. The gel was preincubated in blotting buffer (25 mM Tris base, 192 mM glycine, pH 8.3) for 1 h at 4 °C to remove SDS. The proteins were transferred to an Immobilon PVDF membrane (Millipore, Billerica, MA, U.S.A.) by electroblotting for 3 h at 50 V. After renaturing the proteins by incubation for 15 min in binding buffer (20 mM Hepes, pH 7.9, 5 mM MgCl2, 50 mM NaCl, 1 mM DTT), the membrane was incubated for 1 h in blocking buffer [5% (w/v) non-fat dried milk powder and 0.01% Tween-20 in binding buffer]. Binding was performed overnight by incubation in buffer [0.25% (w/v) non-fat dried milk powder and 0.01% Tween-20 in binding buffer] containing the radiolabelled P3 probe at a concentration of 1×106 c.p.m./ml. The membrane was washed for 2×20 min in binding buffer followed by 20 min in binding buffer. The blot was exposed to a phosphorus screen and analysed using a Storm 860 PhosphorImager apparatus (Molecular Dynamics).
RESULTS
Introduction of three restriction sites in the GRU does not influence activity
When reporter-gene constructs comprising different combinations of the minimal promoter, the proximal enhancer and the distal GRU enhancer were tested, we observed no glucocorticoid response when only the proximal enhancer and the promoter were present (Figure 1B, construct c). However, addition of the GRU rendered the construct highly glucocorticoid-responsive (construct a). To facilitate modification of this GRU, three restriction enzyme recognition sites were introduced by modifying only one nucleotide per restriction site (Figure 1A). Transfection experiments using FTO-2B hepatoma cells showed that these mutations did not significantly affect activity in response to glucocorticoids (Figure 1B, construct b).
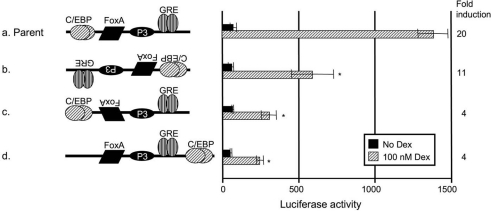
A specific arrangement of GRU elements is required for a glucocorticoid response
When we inverted the orientation of the GRU, we observed a 2.5-fold decrease in transcriptional activity (Figure 2, construct b). Because C/EBP and GR bind their REs as dimers [14,15], it is likely that their activity is independent of their orientation. In contrast, FoxA binds as a monomer. We therefore tested the possibility of a FoxA orientation-dependent glucocorticoid response. Inversion of the orientation of the FoxA site within the GRU indeed led to a 4.5-fold decrease in reporter-gene activity (Figure 2, construct c). We also tested whether the GRU REs are restricted to specific positions within the GRU by displacing the REs. Displacement of the C/EBP RE downstream of the GRE resulted in a markedly decreased activity in response to glucocorticoids (Figure 2, construct d). Together, these data indicate that the arrangement of the REs is crucial for GRU activity.
Figure 2. The specific arrangement of GRU elements is important for the glucocorticoid response.
The effects of different GRU arrangements on glucocorticoid-dependent transcription were tested by transient transfections into FTO-2B hepatoma cells. Modified GRUs were placed upstream of the proximal enhancer and minimal promoter. After 24 h of induction with or without glucocorticoids, cell lysates were prepared. Luciferase values are presented as means±S.E.M. for at least four experiments and the fold induction. The asterisk indicates significantly different results relative to parent construct a. The data show that inversion of the GRU (construct b) lowered the activity 2.5-fold, whereas inversion of the FoxA site only (construct c) decreased the activity 3-fold. Displacement of the C/EBP RE downstream of the FoxA- and GR-binding sites (construct d) decreased the activity 3-fold. Dex, dexamethasone.
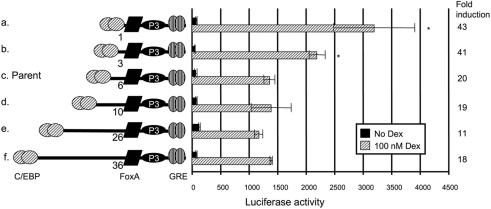
Elimination of the C/EBP–FoxA spacer increases GRU activity
The rotational phasing of a transcription factor on the DNA helix can affect its function [16,17]. Since a functional GRU is highly dependent on the presence of C/EBP [3] and its position relative to the FoxA and GR REs (Figure 2), we tested whether the distance between the GRU elements or their helical orientation affected the glucocorticoid response. When two or three helical turns (20 and 30 bp respectively) were added to the spacer between the C/EBP and FoxA REs (Figure 3, constructs e and f), activity was comparable with that of the parent construct (Figure 3, construct c). When half a helical turn was added to this region, GRU activity also remained unaffected (construct d). However, when the spacer between the C/EBP and FoxA elements was gradually eliminated, GRU activity increased 2-fold (constructs a and b).
Figure 3. Effects of variable spacer lengths between the C/EBP- and FoxA-binding sites in the GRU.
A series of GRU constructs was made with deletions or insertions between the C/EBP and FoxA REs. These modified GRUs were placed upstream of the proximal enhancer and minimal promoter of the CPS gene, and tested by transient transfection into FTO-2B hepatoma cells. After 24 h of treatment with or without glucocorticoids, cells were lysed and luciferase values were measured. The results are expressed as means±S.E.M. and fold induction from at least four experiments. Results that are significantly different from those with the parent construct c are indicated by asterisks. Shortening of the C/EBP–FoxA spacer (constructs a and b) significantly increased activity relative to the parent construct (c), whereas increasing the distance had no effect (constructs d–f). Dex, dexamethasone.
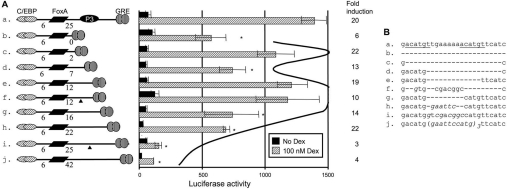
The FoxA–GRE distance is critical for GRU activity
As shown in Figure 1(A), the FoxA and GR REs are separated by a 25 bp region that harbours a binding site for an unknown protein, denoted P3 [3]. When this element was mutated, GRU activity was all but eliminated (Figure 4, construct i). The effects of this mutation could be overcome by decreasing the spacing between the FoxA and GR REs to 12 bp (Figure 4, constructs e–h). Interestingly, the length of the FoxA–GR spacer, rather than the sequence, was important once the FoxA–GR spacer was 12 bp (Figure 4, constructs e and f). When the spacer used in construct e was shortened further (Figure 4, constructs b–d), activity became dependent on the remaining length of the spacer. Removal of 0.5 or 1.2 helical turns (constructs d and b respectively) decreased activity to 40% and 50% respectively, but removal of one complete helical turn did not significantly decrease activity (construct c). These data show that an intact P3-binding site is only required if the distance between the two REs is 25 bp, and that the positions of FoxA and GR binding in the DNA helix do matter if less than two helical turns separate these sites.
Figure 4. Both the distance and the sequence of the region between the FoxA and GR REs influence the glucocorticoid response.
A series of GRU constructs was made with deletions or insertions between the FoxA and GR REs. Modified GRUs were tested in conjunction with the proximal enhancer and the minimal promoter by transfection into FTO-2B hepatoma cells (A). The results are given as means±S.E.M. for at least four experiments and the fold induction. The asterisks indicate that results are significantly different from those with construct e. The curved line indicates the helical expression profile with respect to the distance between the FoxA and GR REs. The triangles indicate the positions of the point mutations. (B) Sequences of the respective spacers. Italic characters represent nucleotides deviating from the parent sequence. Dex, dexamethasone.
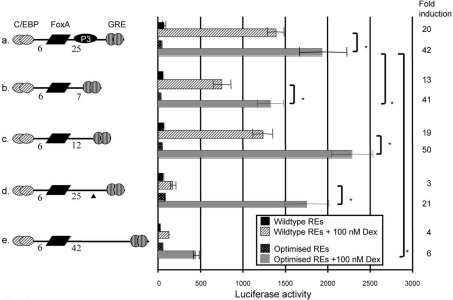
Optimization of the GRU REs increases activity and eliminates dependence on P3
When the CPS GRE was replaced by an optimized palindromic GRE, the C/EBP RE by a sequence selected for high-affinity binding [18] and the FoxA-binding site by a consensus sequence [19], glucocorticoid-induced GRU activity increased 1.4-fold (Figure 5, construct a). When the same optimizing modifications of the REs were introduced in the construct containing the inactivating P3 site mutation (Figure 5, construct d), GRU activity was increased 9-fold compared with the same construct carrying the native RE sequences, and became similar to that of the construct containing an intact P3 site and optimized REs (Figure 5, compare constructs a and d). Optimization of the REs can, therefore, also compensate for loss of P3. When the length of the FoxA–GR spacer exceeded 25 bp, optimization of the REs still increased GRU activity (Figure 5, construct e), but not to that of the P3-containing sequence. When we tested the effects of the optimized REs at FoxA–GR spacer lengths of 7 and 12 bp (Figure 5, constructs b and c), GRU activity was increased 1.8–1.9-fold with respect to the corresponding parent constructs, with preservation of the distance effects observed with the parent constructs. It thus seems that optimizing the GRU REs can compensate for loss of a functional P3-binding site, but leaves the activity of the GRU dependent on the helical positioning of the FoxA and GRE elements.
Figure 5. Optimization of the GRU REs increases the glucocorticoid response.
GRU variants, differing in the length of the FoxA–GRE spacer, were modified into GRUs with optimized REs. The GRE was replaced by a palindromic GRE (AGAACAnnnTGTTCT) that has a high affinity for GR [25]. The C/EBP REs were replaced by a sequence selected for optimal binding (ATTGCGTAAT) [18], while the FoxA RE was replaced by a consensus FoxA site (TATTGACTTAG) [19]. The triangle indicates the P3-inactivating mutation. The resulting GRUs were placed upstream of the proximal enhancer and minimal promoter of the CPS gene, and were transiently transfected into FTO-2B hepatoma cells. Following 24 h of induction with glucocorticoids, luciferase values were measured in the cell lysates. Shown are mean±S.E.M. luciferase values for at least four experiments and the fold induction. Asterisks indicate significantly different results between the indicated constructs. Dex, dexamethasone.
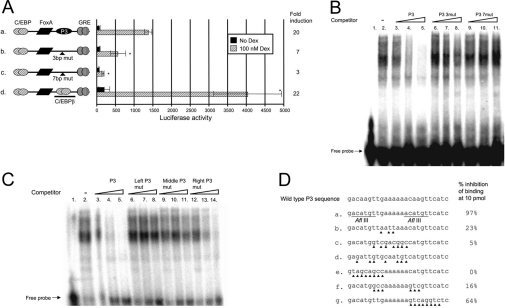
A C/EBPβ RE can functionally substitute for the P3 RE
Since a 7 bp substitution in the middle of the P3-binding region completely inactivated the GRU (Figures 6A and 6D, construct c), we modified fewer nucleotides. On substituting 3 bp in the P3-binding region (Figures 6A and 6D, construct b), hormone-induced GRU activity decreased to 40% of that of the parent construct (construct a). In accordance, these modified P3 elements could not compete with the wild-type P3 probe (Figure 6B) for protein binding. To define the P3-binding region further, we used oligonucleotides spanning the P3-binding region in which all nucleotides left of the hexameric A-stretch in the centre of the P3-binding region, the three nucleotides directly left and right of this A-stretch or all of the nucleotides right of the A-stretch were mutated (Figure 6D, construct e–g), and used them in competition experiments. Our results showed that while the left-sided P3 mutant could not compete with the radiolabelled P3 probe for protein binding (Figure 6C, lanes 6–8), the centrally (lanes 9–11) and right-sided (lanes 12–14) P3-mutated competitors functioned as moderate and strong competitors respectively. The core P3-binding region should, therefore, encompass the poly(A) stretch and the region left of it.
Figure 6. The P3 RE can be functionally substituted by a C/EBP RE.
Specific mutations (indicated by the triangles) were introduced in the P3-binding region of the GRU to give P3 mutants and a construct in which the P3 RE was altered to a consensus C/EBPβ RE (D). These GRUs were placed upstream of the proximal enhancer and minimal promoter of the CPS gene, and the constructs were transfected into FTO-2B hepatoma cells. After 24 h of induction with glucocorticoids, reporter-gene expression was measured in the cell lysates. Results are presented as mean±S.E.M. luciferase values from at least four experiments and the fold induction (A). Asterisks indicate results that are significantly different from those with the parent construct a. (B) To prove specificity, the P3 mutants were also used in a competition assay. A radiolabelled P3-binding region was incubated with 3 μg of rat liver nuclear extract and separated on a 6% (w/v) acrylamide gel (lane 2). Whereas addition of 0.1, 1 or 10 pmol of P3 oligonucleotide efficiently competed with the probe for protein binding (lanes 3–5), the same amounts of mutant P3 oligonucleotides could not (lanes 6–8 and 9–11). To define further the core P3-binding site, a similar competition assay was carried out (C): added to the reaction mixture was 0.1, 1 or 10 pmol of the parent P3 competitor (C, lanes 3–5; D, sequence a), or a competitor in which the left side (C, lanes 6–8; D, sequence e), middle (C, lanes 9–11; D, sequence f), or right side (C, lanes 12–14; D, sequence g) of the P3-binding region had been mutated. Dex, dexamethasone.
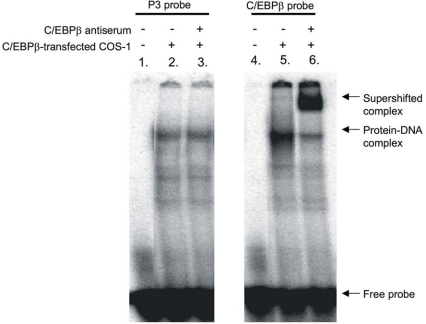
A transcription factor search showed similarity of this P3-binding region with the consensus C/EBPβ-binding site. When we altered the P3 element to a consensus C/EBPβ-binding site (Figure 6D, construct d), GRU activity increased 3-fold relative to that of the wild-type GRU (Figure 6A, construct d). The hypothesis that the enhanced activity was due to C/EBPβ binding to the modified P3 element was confirmed in an EMSA with C/EBPβ-enriched nuclear extracts of COS-1 cells (Figure 7). A specific protein–DNA complex was formed when the wild-type or C/EBPβ-modified P3 element was incubated with C/EBPβ-enriched COS-1 nuclear extract (Figure 7, lanes 2 and 5). However, upon addition of C/EBPβ-specific antibodies, a supershift was seen only with the C/EBP probe (lane 6), but not with the P3 probe (lane 3), demonstrating that the C/EBPβ consensus sequence does bind C/EBPβ protein, whereas the P3 RE does not. Altogether, these results indicate that the P3 site is not a C/EBPβ RE, but can be functionally replaced by a C/EBPβ RE.
Figure 7. The P3-binding region is not a C/EBPβ-binding site.
Supershift assays were performed using either the P3-binding region or the consensus C/EBPβ sequence as probe. The probes were incubated with a C/EBPβ-enriched COS-1 cell nuclear extract. To induce a supershift, C/EBPβ-specific antibodies were added to the samples. Both probes gave rise to a protein–DNA complex when incubated with nuclear extract (lanes 2 and 5). Addition of C/EBPβ-specific antibodies supershifted the C/EBPβ protein–DNA complex (lane 6), but not the P3 protein–DNA complex (lane 3).
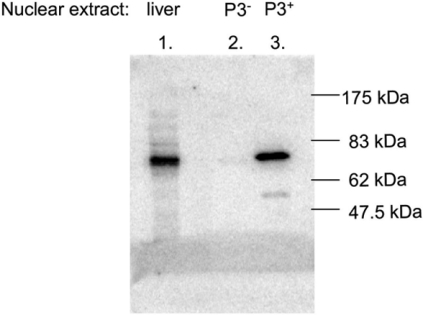
A 75 kDa protein interacts with the P3-binding sequence
A rat liver nuclear extract was fractionated on a MonoS cation-exchanger column and assayed with EMSAs for protein fractions that contained or were devoid of the P3 protein. Using these fractions, we performed Southwestern blotting to preliminarily identify the protein that interacts specifically with the P3 element. With a liver nuclear extract, a protein of approx. 75 kDa was found to interact with the radiolabelled P3-binding sequence (Figure 8, lane 1). The same result was found for a P3-containing protein fraction (lane 3), whereas a P3-negative fraction (lane 2) did not show any signal.
Figure 8. Identification of the protein that interacts with the P3 site by Southwestern blot analysis.
Crude and partially purified rat liver nuclear extracts were resolved on an SDS/9%-PAGE gel and transferred to a PVDF membrane. Following renaturation of the proteins, the blot was incubated with radiolabelled P3 probe. The positions of the molecular size markers are indicated. The liver nuclear extract contained a protein migrating at 75 kDa that was able to bind the P3 sequence (lane 1). A P3-positive MonoS fraction contained the same protein (lane 3), whereas a P3-negative MonoS protein fraction lacked this P3 protein (lane 2).
DISCUSSION
The present study shows that the functionally important elements of the CPS GRU are the binding sites for the GR and for the accessory proteins FoxA and C/EBP. The CPS GRU is, therefore, one of the simplest GRUs described. Although the experiments also showed that the P3 site was essential for GRU activity in its wild-type configuration, we identified three modifications that rendered the GRU independent of P3. This finding suggests that P3 is not necessary to make an efficient GRU, but serves to facilitate the formation of an active DNA–protein complex on the CPS GRU. The finding that the CPS GRU loses a large part of its transactivating activity when placed in the inverse orientation before the CPS promoter (Figure 2, construct b) is at first glance incompatible with the definition of enhancers as regulatory regions that can activate transcription from a promoter independent of their orientation [20]. However, we did not observe a dependence on orientation when a larger fragment comprising the GRU was tested [21], which suggests that this dependence of GRU activity on its orientation is caused by the close proximity of the GRU to the promoter.
The architectural composition of the GRU of the periportally expressed enzyme PEPCK (phosphoenolpyruvate carboxykinase) [22] has been studied in detail [23]. The components of the PEPCK GRU have to be in defined positions relative to each other, since swapping the HNF4/COUP-TF (chicken ovalbumin upstream promoter-transcription factor) (AF1) and FoxA (AF2) REs, or the FoxA (AF2) and COUP-TF (AF3) REs, resulted in a 2-fold decrease in GRU activity [24]. For CPS, the order of the REs in the GRU appears to be even more important, as displacement the C/EBP-binding site downstream of the FoxA and GR REs decreased the glucocorticoid-inducible activity 4.5-fold. In addition to the order of the respective REs, the distance between them may also be of importance, because it presumably affects the ability of the proteins occupying the elements to interact. Our data show that the activity of the GRU increases with decreasing distance between the C/EBP- and FoxA-binding sites. The increase in GRU activity that accompanies the increasing proximity of the C/EBP and FoxA REs suggests an interaction of C/EBP with one of the other transcription factors that bind to the GRU. We also tested the effects of spacing of the C/EBP and FoxA REs in the absence of a functional P3 RE (leaving 12 bp between the FoxA and GR REs), but the results were similar to the data obtained with an intact P3 site (results not shown). The effect of shortening of the C/EBP–FoxA spacer is, therefore, not dependent on a functional P3 site, implying that C/EBP does not interact directly with P3 or GR. Most probably, therefore, the increase in activity that accompanies the approximation of the C/EBP and FoxA sites results from an interaction between these two proteins. Because one helical turn is 10 bp and the lengths of the core C/EBP and FoxA REs are 10 and 11 bp respectively, the transcription factors will be located on the same side of the helix when the distance between the core elements is 1 bp (the most active configuration). It is thus conceivable that the observed increase in activity when these two elements are more closely apposed is the result of better alignment of the transcription factors on the DNA helix.
The spacing between the FoxA and GRE sites was also critical for GRU activity. In the absence of P3, the GRU was most active when the distance between the two binding sites was 2 or 12 bp, with the difference representing one helical turn of the DNA. This finding also implies that the positions of FoxA and GR on the DNA helix are important. At a spacing of 25 bp, however, P3 became necessary to ensure activity of the GRU. P3 was, however, redundant at this spacer length when the REs of the CPS GRU were modified into high-affinity elements, but this modification was no longer effective at greater spacer lengths. These findings suggest that P3 functions to facilitate interaction between the FoxA and GR REs at a distance where they have difficulty contacting each other. A strict requirement with respect to FoxA–GRE spacing is also observed in the PEPCK gene [24]. The distance of 18 bp that separates the FoxA-binding site (AF2) from GRE1 in this gene are critical for a full glucocorticoid response, with insertion of an additional helical turn decreasing expression levels.
Our results show that the CPS GRU is not maximally compact in the wild-type configuration. Instead, our data suggest that, by decreasing the C/EBP–FoxA spacer distance to 1 bp and the FoxA–GRE spacer distance to 2 bp (no P3), a unit would be generated that is very active, very responsive to glucocorticoids, very small and very simple, since it only comprises three REs. Clearly, a compact GRU does not need P3 for activity, since P3 is redundant at FoxA–GRE distances of 2 and 12 bp and when the other GRU REs are replaced by optimal REs. The question, therefore, emerges as to why P3 is integrated into this unit. Since the sequences of the GRU REs and the distance between the FoxA and GR REs have evolved in such a way that it allows the CPS GRU to be regulated by P3, it is probable that proper regulation of the wild-type GRU requires the interplay of all four transcription factors. To fully understand P3 function, it is necessary to establish its identity. DNA-sequence analysis suggested that P3 could be a C/EBPβ-binding site. When we tested this possibility by altering the P3 sequence into a consensus C/EBPβ site, we found a 3-fold increase in glucocorticoid-induced activity. However, using supershift assays, we found that C/EBPβ could not bind the wild-type P3 region. Although P3 has resisted the elucidation of its identity so far, our Southwestern analysis indicates that it is a 75 kDa protein.
In conclusion, our results provide new insight into how GRUs work. GRUs are not just clusters of REs that are end-points of regulatory pathways. Instead, the sequence, orientation and spacing of the participating REs are all crucial parameters in the effectiveness of a GRU.
References
- 1.Meijer A. J., Lamers W. H., Chamuleau R. A. Nitrogen metabolism and ornithine cycle function. Physiol. Rev. 1990;70:701–748. doi: 10.1152/physrev.1990.70.3.701. [DOI] [PubMed] [Google Scholar]
- 2.Christoffels V. M., van den Hoff M. J., Moorman A. F., Lamers W. H. The far-upstream enhancer of the carbamoyl-phosphate synthetase I gene is responsible for the tissue specificity and hormone inducibility of its expression. J. Biol. Chem. 1995;270:24932–24940. doi: 10.1074/jbc.270.42.24932. [DOI] [PubMed] [Google Scholar]
- 3.Christoffels V. M., Grange T., Kaestner K. H., Cole T. J., Darlington G. J., Croniger C. M., Lamers W. H. Glucocorticoid receptor, C/EBP, HNF3, and protein kinase A coordinately activate the glucocorticoid response unit of the carbamoylphosphate synthetase I gene. Mol. Cell. Biol. 1998;18:6305–6315. doi: 10.1128/mcb.18.11.6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell J., Noisin E., Hall R., O'Brien R., Imai E., Granner D. Integration of multiple signals through a complex hormone response unit in the phosphoenolpyruvate carboxykinase gene promoter. Mol. Endocrinol. 1994;8:585–594. doi: 10.1210/mend.8.5.8058068. [DOI] [PubMed] [Google Scholar]
- 5.Wedel A., Ziegler-Heitbrock H. W. The C/EBP family of transcription factors. Immunobiology. 1995;193:171–185. doi: 10.1016/s0171-2985(11)80541-3. [DOI] [PubMed] [Google Scholar]
- 6.Kaestner K. H., Hiemisch H., Luckow B., Schutz G. The HNF-3 gene family of transcription factors in mice: gene structure, cDNA sequence, and mRNA distribution. Genomics. 1994;20:377–385. doi: 10.1006/geno.1994.1191. [DOI] [PubMed] [Google Scholar]
- 7.Clark K. L., Halay E. D., Lai E., Burley S. K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature (London) 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 8.Killary A. M., Fournier R. E. A genetic analysis of extinction: trans-dominant loci regulate expression of liver-specific traits in hepatoma hybrid cells. Cell. 1984;38:523–534. doi: 10.1016/0092-8674(84)90507-5. [DOI] [PubMed] [Google Scholar]
- 9.Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 10.van den Hoff M. J., Christoffels V. M., Labruyere W. T., Moorman A. F., Lamers W. H. Electrotransfection with “intracellular” buffer. Methods Mol. Biol. 1995;48:185–197. doi: 10.1385/0-89603-304-X:185. [DOI] [PubMed] [Google Scholar]
- 11.Ogris M., Steinlein P., Kursa M., Mechtler K., Kircheis R., Wagner E. The size of DNA/transferrin-PEI complexes is an important factor for gene expression in cultured cells. Gene Ther. 1998;5:1425–1433. doi: 10.1038/sj.gt.3300745. [DOI] [PubMed] [Google Scholar]
- 12.Sierra F. Laboratory guide to in vitro transcription. In: Azzi A., Polak J. M., Saluz H. P., editors. Biomethods. Basel: Birkhauser Verlag; 1990. pp. 30–50. [Google Scholar]
- 13.Labbé S., Stewart G., LaRochelle O., Poirier G. G., Seguin C. Identification of sequence-specific DNA-binding proteins by southwestern blotting. Methods Mol. Biol. 2001;148:255–264. doi: 10.1385/1-59259-208-2:255. [DOI] [PubMed] [Google Scholar]
- 14.Lekstrom-Himes J., Xanthopoulos K. G. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 15.Reichardt H. M., Kaestner K. H., Tuckermann J., Kretz O., Wessely O., Bock R., Gass P., Schmid W., Herrlich P., Angel P., Schutz G. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- 16.Eisfeld K., Candau R., Truss M., Beato M. Binding of NF1 to the MMTV promoter in nucleosomes: influence of rotational phasing, translational positioning and histone H1. Nucleic Acids Res. 1997;25:3733–3742. doi: 10.1093/nar/25.18.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schule R., Muller M., Otsuka-Murakami H., Renkawitz R. Cooperativity of the glucocorticoid receptor and the CACCC-box binding factor. Nature (London) 1988;332:87–90. doi: 10.1038/332087a0. [DOI] [PubMed] [Google Scholar]
- 18.Osada S., Yamamoto H., Nishihara T., Imagawa M. DNA binding specificity of the CCAAT/enhancer-binding protein transcription factor family. J. Biol. Chem. 1996;271:3891–3896. doi: 10.1074/jbc.271.7.3891. [DOI] [PubMed] [Google Scholar]
- 19.Roux J., Pictet R., Grange T. Hepatocyte nuclear factor 3 determines the amplitude of the glucocorticoid response of the rat tyrosine aminotransferase gene. DNA Cell Biol. 1995;14:385–396. doi: 10.1089/dna.1995.14.385. [DOI] [PubMed] [Google Scholar]
- 20.Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 21.van den Hoff M. J., van de Zande L. P., Dingemanse M. A., Das A. T., Labruyere W., Moorman A. F., Charles R., Lamers W. H. Isolation and characterization of the rat gene for carbamoylphosphate synthetase I. Eur. J. Biochem. 1995;228:351–361. [PubMed] [Google Scholar]
- 22.Bartels H., Linnemann H., Jungermann K. Predominant localization of phosphoenolpyruvate carboxykinase mRNA in the periportal zone of rat liver parenchyma demonstrated by in situ hybridization. FEBS Lett. 1989;248:188–194. doi: 10.1016/0014-5793(89)80459-4. [DOI] [PubMed] [Google Scholar]
- 23.Scott D. K., Mitchell J. A., Granner D. K. The orphan receptor COUP-TF binds to a third glucocorticoid accessory factor element within the phosphoenolpyruvate carboxykinase gene promoter. J. Biol. Chem. 1996;271:31909–31914. doi: 10.1074/jbc.271.50.31909. [DOI] [PubMed] [Google Scholar]
- 24.Sugiyama T., Scott D. K., Wang J. C., Granner D. K. Structural requirements of the glucocorticoid and retinoic acid response units in the phosphoenolpyruvate carboxykinase gene promoter. Mol. Endocrinol. 1998;12:1487–1498. doi: 10.1210/mend.12.10.0187. [DOI] [PubMed] [Google Scholar]
- 25.Scott D. K., Stromstedt P. E., Wang J. C., Granner D. K. Further characterization of the glucocorticoid response unit in the phosphoenolpyruvate carboxykinase gene. The role of the glucocorticoid receptor-binding sites. Mol. Endocrinol. 1998;12:482–491. doi: 10.1210/mend.12.4.0090. [DOI] [PubMed] [Google Scholar]