Abstract
BACKGROUND:
Clonal hematopoiesis of indeterminate potential (CHIP) occurs due to acquired mutations in bone marrow progenitor cells. CHIP confers a 2-fold risk of atherosclerotic cardiovascular disease. However, there are limited data regarding specific cardiovascular phenotypes. The purpose of this study was to define the coronary artery disease phenotype of the CHIP population-based on coronary angiography.
METHODS:
We recruited 1142 patients from the Vanderbilt University Medical Center cardiac catheterization laboratory and performed DNA sequencing to determine CHIP status. Multivariable logistic regression models and proportional odds models were used to assess the association between CHIP status and angiography phenotypes.
RESULTS:
We found that 18.4% of patients undergoing coronary angiography had a CHIP mutation. Those with CHIP had a higher risk of having obstructive left main (odds ratio, 2.44 [95% CI, 1.40–4.27]; P=0.0018) and left anterior descending (odds ratio, 1.59 [1.12–2.24]; P=0.0092) coronary artery disease compared with non-CHIP carriers. We additionally found that a specific CHIP mutation, ten eleven translocase 2 (TET2), has a larger effect size on left main stenosis compared with other CHIP mutations.
CONCLUSIONS:
This is the first invasive assessment of coronary artery disease in CHIP and offers a description of a specific atherosclerotic phenotype in CHIP wherein there is an increased risk of obstructive left main and left anterior descending artery stenosis, especially among TET2 mutation carriers. This serves as a basis for understanding enhanced morbidity and mortality in CHIP.
Keywords: atherosclerosis, genetics, heart failure, mutation, observational cohort
Clonal hematopoiesis of indeterminate potential (CHIP) results from acquired mutations in bone marrow progenitor cells. More than 10% of individuals older than 65 years old have CHIP, and the presence of a CHIP mutation is associated with a 50% increase in mortality across numerous studies.1–4 CHIP leads to a 2-fold risk of atherosclerotic cardiovascular disease (ASCVD), which accounts for a large fraction of the increased mortality.1,5,6 However, there are limited data detailing patient-level CVD manifestations in humans.
Mutations in DNA methyltransferase 3A (DNMT3A) or ten eleven translocase 2 (TET2) together represent >70% of all CHIP cases.3,4 TET2 is the second most common CHIP mutation and has been consistently linked to the development of atherosclerotic disease in animal models, human studies, and population-based biobanks.1,7,8 Mechanistic studies suggest these effects are mediated through an altered inflammatory axis involving myeloid lineage cells.7,9,10 For example, a myeloid lineage-specific TET2-knockout mice, and specifically TET2-knockout macrophages, promote accelerated atherosclerosis development, dependent on an enhanced expression of chemokines and secretion of IL (interleukin)-1β and IL-6.7 Conversely, DNMT3A has been more closely linked to heart failure.11–14 Patients with heart failure with DNMT3A mutations are more likely to have progressive disease with higher rates of mortality, possibly due to enhanced fibroblast activation and subsequent cardiac fibrosis.15–17
To date, evaluation of specific coronary phenotypes has been limited. Using coronary computed tomography calcium quantification, Jaiswal et al found that CHIP status was associated with an elevated Agatston score, suggesting more severe calcific coronary artery disease (CAD) in the setting of CHIP. This study also identified an association with increasing variant allele frequency size. In another study, retrospective analysis of CHIP in patients from the CULPRIT-SHOCK trial (Culprit Lesion Only PCI versus Multivessel PCI in Cardiogenic Shock) revealed that among patients in cardiogenic shock after myocardial infarction, those with CHIP suffered increased morbidity and mortality when compared with non-CHIP patients.18 Secondary analysis found that there were neither differences in the rates of ST-segment elevation myocardial infarction (STEMI) nor the frequency of triple vessel disease within the CULPRIT-SHOCK cohort. Finally, a study evaluating patients with STEMI found that while the presence of either DNMT3A or TET2 CHIP led to poorer outcomes, it did not influence the rates of 1-, 2-, or 3-vessel coronary disease.19
Despite the availability of ample population-based biobank epidemiology and several retrospective analyses, there have been no prospective studies of CHIP in the cardiovascular setting, and the current literature lacks deep characterization of cardiovascular phenotypes, including evaluation with coronary angiography.
METHODS
Full methods are available in the Supplemental Material. Patients were enrolled under the Vanderbilt University Medical Center. Institutional Review Board approval no. 090828 in accordance with the treaty of Helsinki. The data repository will be posted to an open-source repository such as Open Science Framework as a spreadsheet file without any identifiable information. The spreadsheet will be granted without restriction upon publication of this and ongoing work using this data. Statistical analyses were performed using the statistical programming language, R, version 4.3.1, with packages Hmisc and rms.20,21
RESULTS
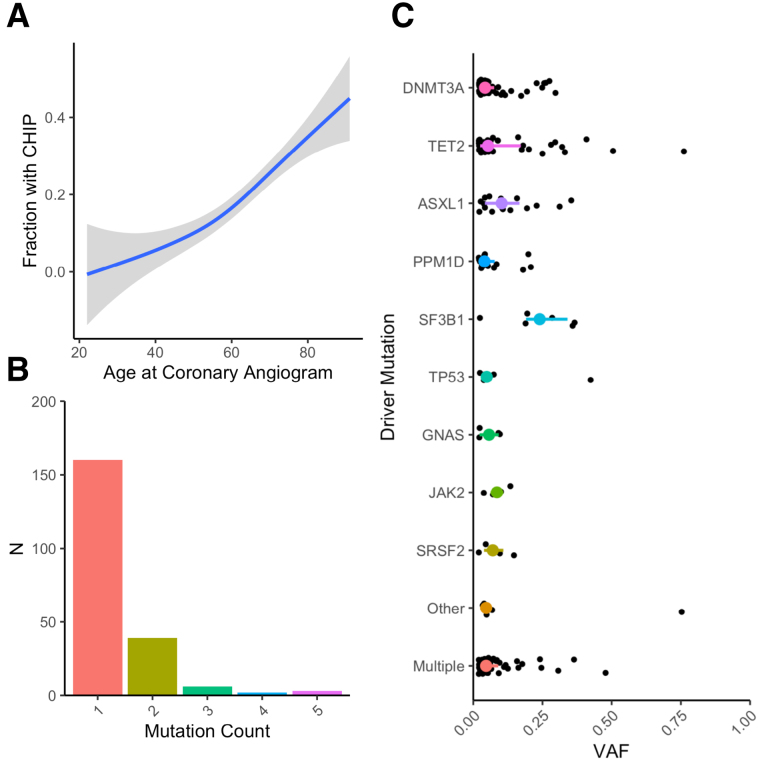
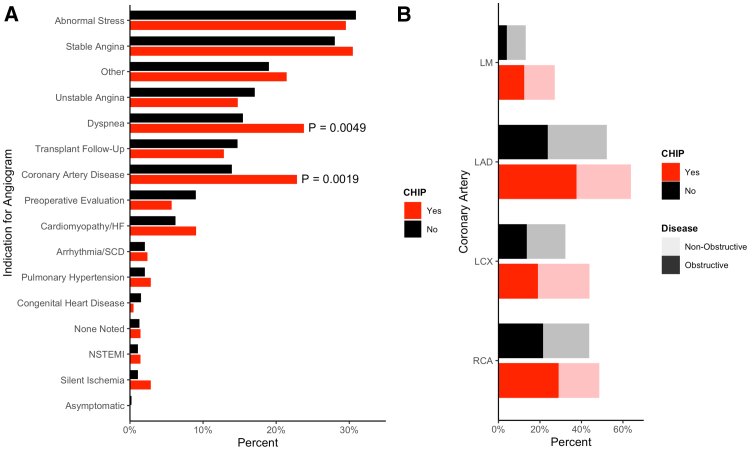
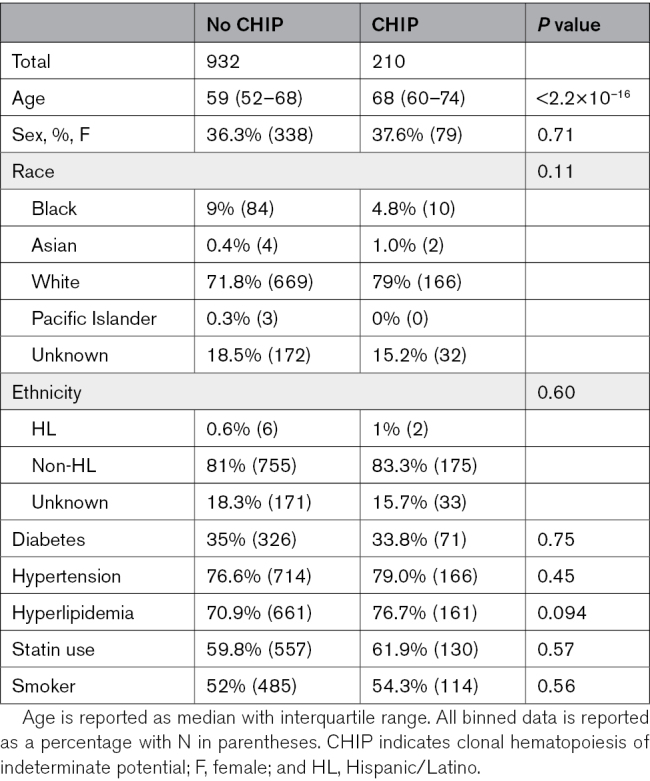
We sequenced 1469 patients, of whom 1142 had complete data and did not have a STEMI at the time of catheterization. Of these, 210 (18.4%) had CHIP mutations with a variant allele frequency of >2%. CHIP mutations spanned the expected spectrum of common mutations (Figure 1). CHIP patients were significantly older (median, 68 [interquartile range, 60–74] versus 59 [52–68] years; P<2.2×10−16; Table; Figure 1A) and more likely to be taking dual antiplatelet therapy, beta blockers, and calcium channel blockers (Table S3). CAD and dyspnea were more commonly listed as indications for angiogram among patients with CHIP (Figure 2A). When fully adjusted, CHIP remains significantly associated with the CAD indication alone (odds ratio [OR], 1.56 [1.03–2.34]; P=0.034; dyspnea OR, 1.45 [0.99–2.14]; P=0.057). Across all vascular territories, patients with CHIP more often had both any plaque and obstructive plaque (Figure 2B).
Figure 1.
Clonal hematopoiesis of indeterminate potential (CHIP) is highly prevalent in a catheterization laboratory population. CHIP prevalence by age (A) and mutation count (B), and variant allele fraction (VAF) distribution by driver gene (C).
Figure 2.
Referral patterns and plaque burden in patients with clonal hematopoiesis of indeterminate potential (CHIP). Documented referral indication for coronary angiogram provided by the referring physician (A). Proportion of patients that has coronary atherosclerosis in specific coronary arteries stratified by CHIP status (B). LAD indicates left anterior descending; LCX, left circumflex; LM, left main; NSTEMI, non-ST elevation myocardial infarction; RCA, right coronary artery; and SCD, sudden cardiac death.
Table.
Baseline Cohort Demographics Stratified by CHIP Status
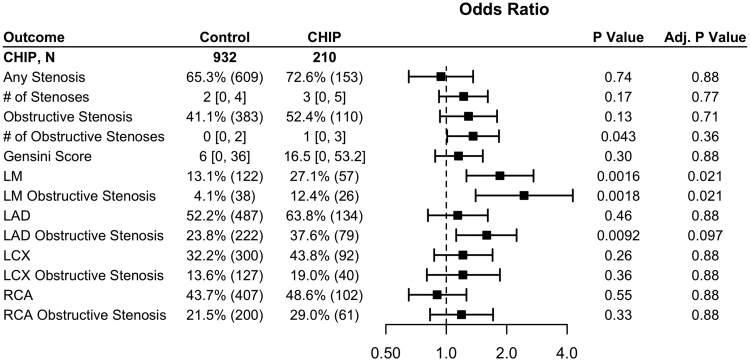
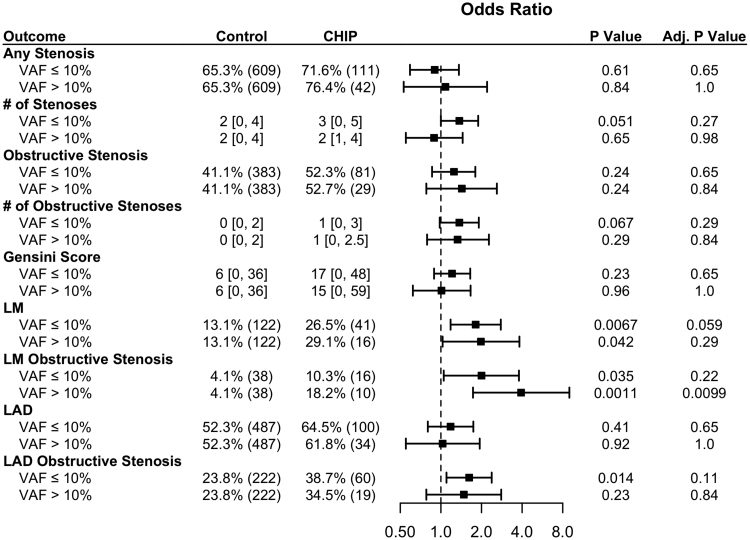
In multivariable-adjusted analysis, CHIP carrier status was significantly correlated with any stenosis and obstructive stenosis in the left main (LM) coronary artery (OR, 1.85 [1.26–2.72]; P=0.021 and OR, 2.44 [1.40–4.27]; P=0.021; Figure 3) and nonsignificantly correlated with obstructive stenosis in the left anterior descending coronary artery (OR, 1.59 [1.12–2.24]; P=0.097; Figure 3). There were no associations with other vascular territories.
Figure 3.
Patients with clonal hematopoiesis of indeterminate potential (CHIP) have higher rates of left main (LM) stenosis. Adjusted odds ratios for angiography outcomes stratified by CHIP. Numeric data is displayed as percentage (N) for binary outcomes, and median (interquartile range) for continuous data. LAD indicates left anterior descending; LCX, left circumflex; and RCA, right coronary artery.
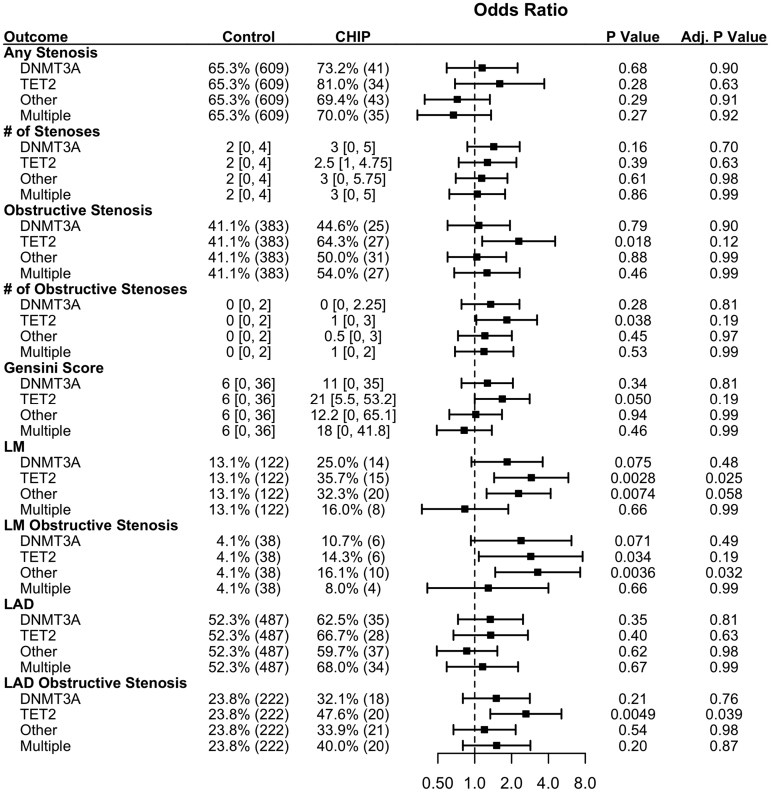
When stratified by driver genes, TET2 mutants and other non-DNMT3A mutants were associated with LM stenosis and obstructive LM stenosis (Figure 4; Table S4). TET2 was also associated with obstructive left anterior descending stenosis (OR, 2.62 [1.34–5.11]; P=0.0049). When all patients with CHIP were stratified by variant allele frequency, there was a association of obstructive LM disease only among those with larger variant allele frequency (Figure 5), but there were no other significant associations across variant allele frequency.
Figure 4.
TET2 (ten eleven translocase 2) clonal hematopoiesis of indeterminate potential (CHIP) demonstrates more severe coronary artery disease (CAD). Fifty percent of patients with multiple CHIP mutations have a TET2 mutation. Adjusted odds ratios for angiography outcomes stratified by CHIP gene mutation. Numeric data are displayed as percentage (N) for binary outcomes, and median (interquartile range) for continuous data. LAD indicates left anterior descending; and LM, left main.
Figure 5.
Left main (LM) stenosis is associated with higher variant allele frequency (VAF). Adjusted odds ratios for angiography outcomes stratified by clonal hematopoiesis of indeterminate potential (CHIP) VAF. Numeric data are displayed as percentage (N) for binary outcomes, and median (interquartile range) for continuous data. LAD indicates left anterior descending.
DISCUSSION
CHIP is a known risk factor for the development of ASCVD; however, there have been no cohort-based studies evaluating specific cardiovascular phenotypes, leaving a critical gap in the current literature. In a single-center, prospective, observational cohort study of patients undergoing cardiac catheterization, we report nearly 1 in 5 had a detectable CHIP mutation. This is a population highly enriched for CHIP carriers in the cardiac catheterization laboratory. The expected rate of CHIP is between 5% to 7% given the median age of this cohort when compared with large biobank data.1,3,5 However, our study population is similarly enriched for CHIP mutations when compared with other targeted studies of patients with cardiovascular disease. For example, 3 separate groups evaluating CHIP in heart failure cohorts reported frequencies ranging from 18.4% to 38.7%.15,16,22 This observation affirms the notion that cardiovascular cohorts are enriched for patients with CHIP and represents an opportunity for future studies focused on primary prevention in CHIP-related ASCVD.
Invasive analysis with coronary angiography revealed a specific risk pattern for those with CHIP in our study. In fully adjusted analyses, we find a significant association between CHIP status and LM stenosis, LM obstructive stenosis, and left anterior descending obstructive stenosis by coronary angiography. Together, this represents a novel finding and potentially explains the enhanced morbidity and mortality associated with cardiogenic shock and ASCVD in the setting of CHIP.18,19,23 To the best of our knowledge, this is the first documentation of a specific coronary atherosclerotic phenotype in patients with CHIP.
A larger effect size for obstructive lesions in any vessel was seen for TET2 CHIP. This finding corroborates a growing set of data suggesting this subgroup exhibits more severe cardiovascular phenotypes, likely driven by inflammatory signaling. For example, in a subgroup analysis from the CANTOS (Canakinumab Anti-inflammatory Thrombosis Outcome Study), TET2 but not DNMT3A derived benefit from IL-1B inhibition on cardiovascular disease outcomes.9 Furthermore, in an analysis of the most recent release of ≈450K whole exomes from the UK Biobank, Vlasschaert et al5 showed that a genetic polymorphism in the IL-6 receptor gene (p.Asp358Ala) is protective against TET2-mediated incident CAD. Our findings in TET2 CHIP patients support the notion that precision approaches may be an effective means for identifying high-risk individuals for intensive primary or secondary prevention strategies.
Our study has several important limitations. Sample size limits our ability to interrogate differences, particularly between driver genes and clone size. Additionally, the indications for catheterization vary across CHIP and non-CHIP patients, highlighting the heterogeneity of the referred population. Interestingly, although patients with CHIP were more likely to have CAD as an indication for angiography, CHIP does not seem to be associated with the presence of CAD alone in this population. Instead, there is an increased risk of a high-risk lesion. Patients with STEMI were excluded in the current analysis for several reasons. STEMI represents a fundamentally divergent pathophysiology when compared with stable coronary disease. The STEMI subgroup was small in our analysis (n=8), and their exclusion did not alter the significance of any results reported. Furthermore, there has been 1 retrospective analysis of STEMI in the presence of either TET2 or DNMT3A CHIP, finding that patients who had a CHIP mutation predicted worse outcomes including major adverse cardiac events and death.19 This may reflect an increased prevalence of high-risk obstructive lesions in these patients, similar to the pattern described herein. Given the study design, these data are also not revealing of causality. Although the association between CHIP and ASCVD has been extensively examined, our understanding of how CHIP results in cardiovascular pathology is being refined as additional studies are published,4,24,25 and this approach adds to that body of literature.
The current study underscores the enormous potential of precision approaches in cardiovascular medicine. Here, we identify a genetic biomarker of coronary disease in ≈20% of the patients in a cardiac catheterization laboratory using commonplace and increasingly affordable methods. Our cost to perform this assay was <$5 USD per patient. Through invasive coronary angiography, we define a specific CAD risk profile for patients with CHIP, noting that those with a TET2 driver mutation are at enhanced risk. In our cohort, 65% of patients carrying an isolated TET2 mutation had obstructive stenosis in a coronary vessel. Given these findings, early identification of CHIP mutations in patients could significantly alter current practice recommendations and primary prevention strategies.
ARTICLE INFORMATION
Sources of Funding
The National Institutes of Health Early Independence Award grant DP5 OD029586 and a Burroughs Wellcome Fund Career Award for Medical Scientists was given to Dr Bick.
Disclosures
Dr Bick reports personal fees and serves on the scientific advisory board of TenSixteen Bio unrelated to the present work. The other authors report no conflicts.
Supplemental Material
Supplemental Methods
Tables S1–S4
Supplementary Material
Nonstandard Abbreviations and Acronyms
- ASCVD
- atherosclerotic cardiovascular disease
- CAD
- coronary artery disease
- CANTOS
- Canakinumab Anti-inflammatory Thrombosis Outcome Study
- CHIP
- clonal hematopoiesis of indeterminate potential
- DNMT3A
- DNA methyltransferase 3A
- IL
- interleukin
- LM
- left main
- OR
- odds ratio
- STEMI
- ST segment elevation myocardial infarction
- VAF
- variant allele frequency
J.B. Heimlich and M.A. Raddatz contributed equally as first authors to this work.
For Sources of Funding and Disclosures, see page 344.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCGEN.123.004415.
Contributor Information
J. Brett Heimlich, Email: brett.heimlich@gmail.com.
John Wells, Email: jawells4@gmail.com.
Caitlyn Vlasschaert, Email: caitlyn.vlasschaert@queensu.ca.
Sydney Olson, Email: sydney.olson@outlook.com.
Marcus Threadcraft, Email: marcus.threadcraft@vumc.org.
Kristoff Foster, Email: kristoff.foster@vumc.org.
Emmanuel Boateng, Email: e.k.boateng2020@gmail.com.
Dan M. Roden, Email: Dan.Roden@VUMC.org.
Colin M. Barker, Email: colin.m.barker@vumc.org.
REFERENCES
- 1.Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, McConkey M, Gupta N, Gabriel S, Ardissino D, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377:111–121. doi: 10.1056/NEJMoa1701719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bick AG, Weinstock JS, Nandakumar SK, Fulco CP, Bao EL, Zekavat SM, Szeto MD, Liao X, Leventhal MJ, Nasser J, et al. ; NHLBI Trans-Omics for Precision Medicine Consortium. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature. 2020;586:763–768. doi: 10.1038/s41586-020-2819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kessler MD, Damask A, O’Keeffe S, Banerjee N, Li D, Watanabe K, Marketta A, Van Meter M, Semrau S, Horowitz J, et al. Common and rare variant associations with clonal haematopoiesis phenotypes. Nature. 2022;612:301–309. doi: 10.1038/s41586-022-05448-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlasschaert C, Heimlich JB, Rauh MJ, Natarajan P, Bick AG. Interleukin-6 receptor polymorphism attenuates clonal hematopoiesis-mediated coronary artery disease risk among 451 180 individuals in the UK biobank. Circulation. 2023;147:358–360. doi: 10.1161/CIRCULATIONAHA.122.062126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gumuser ED, Schuermans A, Cho SMJ, Sporn ZA, Uddin MM, Paruchuri K, Nakao T, Yu Z, Haidermota S, Hornsby W, et al. Clonal hematopoiesis of indeterminate potential predicts adverse outcomes in patients with atherosclerotic cardiovascular disease. J Am Coll Cardiol. 2023;81:1996–2009. doi: 10.1016/j.jacc.2023.03.401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, Wu C-L, Sano S, Muralidharan S, Rius C, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 2017;355:842–847. doi: 10.1126/science.aag1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bick AG, Pirruccello JP, Griffin GK, Gupta N, Gabriel S, Saleheen D, Libby P, Kathiresan S, Natarajan P. Genetic interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation. 2020;141:124–131. doi: 10.1161/CIRCULATIONAHA.119.044362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Svensson EC, Madar A, Campbell CD, He Y, Sultan M, Healey ML, Xu H, D’Aco K, Fernandez A, Wache-Mainier C, et al. TET2-driven clonal hematopoiesis and response to canakinumab: an exploratory analysis of the CANTOS randomized clinical trial. JAMA Cardiol. 2022;7:521–528. doi: 10.1001/jamacardio.2022.0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heimlich JB, Bhat P, Parker A, Jenkins MT, Vlasschaert C, Ulloa J, Van Amburg J, Potts CR, Olson S, Silver AJ, et al. Multiomic profiling of human clonal hematopoiesis reveals genotype and cell-specific inflammatory pathway activation. Blood Advances. 2024:bloodadvances.2023011445. doi: 10.1182/bloodadvances.2023011445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abplanalp WT, Cremer S, John D, Hoffmann J, Schuhmacher B, Merten M, Rieger MA, Vasa-Nicotera M, Zeiher AM, Dimmeler S. Clonal hematopoiesis–driver DNMT3A mutations alter immune cells in heart failure. Circ Res. 2021;128:216–228. doi: 10.1161/CIRCRESAHA.120.317104 [DOI] [PubMed] [Google Scholar]
- 12.Abplanalp WT, Mas-Peiro S, Cremer S, John D, Dimmeler S, Zeiher AM. Association of clonal hematopoiesis of indeterminate potential with inflammatory gene expression in patients with severe degenerative aortic valve stenosis or chronic postischemic heart failure. JAMA Cardiol. 2020;5:1170–1175. doi: 10.1001/jamacardio.2020.2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abplanalp W, Cremer C, Cremer S, John D, John D, Hoffmann J, Hoffmann J, Rieger MA, Rieger MA, Vasa-Nicotera M, et al. Clonal hematopoiesis-driver DNMT3A mutations alter immune cells in heart failure. Eur Heart J. 2021;128:216–228. doi: 10.1161/CIRCRESAHA.120.317104 [DOI] [PubMed] [Google Scholar]
- 14.Dorsheimer L, Assmus B, Rasper T, Ortmann CA, Abou-El-Ardat K, Kiefer KC, Hoffmann J, Seeger F, Bonig H, Dimmeler S, et al. Hematopoietic alterations in chronic heart failure patients by somatic mutations leading to clonal hematopoiesis. Haematologica. 2020;105:e328–e332. doi: 10.3324/haematol.2019.224402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pascual-Figal DA, Bayes-Genis A, Díez-Díez M, Hernández-Vicente A, Vázquez-Andrés D, de la Barrera J, Vazquez E, Quintas A, Zuriaga MA, Asensio-López MC, et al. Clonal hematopoiesis and risk of progression of heart failure with reduced left ventricular ejection fraction. J Am Coll Cardiol. 2021;77:1747–1759. doi: 10.1016/j.jacc.2021.02.028 [DOI] [PubMed] [Google Scholar]
- 16.Dorsheimer L, Assmus B, Rasper T, Ortmann CA, Ecke A, Abou-El-Ardat K, Schmid T, Brüne B, Wagner S, Serve H, et al. Association of mutations contributing to clonal hematopoiesis with prognosis in chronic ischemic heart failure. JAMA Cardiol. 2019;4:25–33. doi: 10.1001/jamacardio.2018.3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shumliakivska M, Luxán G, Hemmerling I, Abplanalp WT, Li X, Scheller M, Müller-Tidow C, Leuschner F, Schuhmacher B, Debes A, et al. DNMT3A clonal hematopoiesis-driver mutations induce cardiac fibrosis by paracrine activation of fibroblasts. Physiology. 2023;15:606. doi: 10.1101/2023.01.07.521766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Böhme M, Desch S, Rosolowski M, Scholz M, Krohn K, Büttner P, Cross M, Kirchberg J, Rommel K-P, Pöss J, et al. Impact of clonal hematopoiesis in patients with cardiogenic shock complicating acute myocardial infarction. J Am Coll Cardiol. 2022;80:1545–1556. doi: 10.1016/j.jacc.2022.08.740 [DOI] [PubMed] [Google Scholar]
- 19.Wang S, Hu S, Luo X, Bao X, Li J, Liu M, Lv Y, Zhao C, Zeng M, Chen X, et al. Prevalence and prognostic significance of DNMT3A- and TET2- clonal haematopoiesis-driver mutations in patients presenting with ST-segment elevation myocardial infarction. EBioMedicine. 2022;78:103964. doi: 10.1016/j.ebiom.2022.103964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrell F. Hmisc: harrell miscellaneous. Accessed December 21, 2023. Available from: https://hbiostat.org/R/Hmisc/
- 21.Harrell FE. Regression modeling strategies: with applications to linear models, logistic and ordinal regression, and survival analysis. Second edition. Cham Heidelberg New York: Springer; 2015 [Google Scholar]
- 22.Shi C, Aboumsallem JP, Suthahar N, De Graaf AO, Jansen JH, Van Zeventer IA, Bracun V, De Wit S, Screever EM, Van Den Berg PF, et al. Clonal haematopoiesis of indeterminate potential: associations with heart failure incidence, clinical parameters and biomarkers. Eur J Heart Fail. 2023;25:4–13. doi: 10.1002/ejhf.2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scolari FL, Abelson S, Brahmbhatt DH, Medeiros JJF, Fan CS, Fung NL, Mihajlovic V, Anker MS, Otsuki M, Lawler PR, et al. Clonal haematopoiesis is associated with higher mortality in patients with cardiogenic shock. Eur J Heart Fail. 2022;24:1573–1582. doi: 10.1002/ejhf.2588 [DOI] [PubMed] [Google Scholar]
- 24.Kar SP, Quiros PM, Gu M, Jiang T, Mitchell J, Langdon R, Iyer V, Barcena C, Vijayabaskar MS, Fabre MA, et al. Genome-wide analyses of 200,453 individuals yield new insights into the causes and consequences of clonal hematopoiesis. Nat Genet. 2022;54:1155–1166. doi: 10.1038/s41588-022-01121-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakao T, Bick AG, Taub MA, Zekavat SM, Uddin MM, Niroula A, Carty CL, Lane J, Honigberg MC, Weinstock JS, et al. ; Samoan Obesity, Lifestyle and Genetic Adaptations Study (OLaGA) Group. Mendelian randomization supports bidirectional causality between telomere length and clonal hematopoiesis of indeterminate potential. Sci Adv. 2022;8:eabl6579. doi: 10.1126/sciadv.abl6579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller PG, Fell GG, Foy BH, Scherer AK, Gibson CJ, Sperling AS, Burugula BB, Nakao T, Uddin MM, Warren H, et al. Clonal hematopoiesis of indeterminate potential and risk of death from COVID-19. Blood. 2022;140:1993–1997. doi: 10.1182/blood.2022018052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vlasschaert C, Mack T, Heimlich JB, Niroula A, Uddin MM, Weinstock JS, Sharber B, Silver AJ, Xu Y, Savona MR, et al. A practical approach to curate clonal hematopoiesis of indeterminate potential in human genetic datasets. Blood. 2023;141:2214. doi: 10.1182/blood.2022018825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–e164. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, Bejar R, Berti E, Busque L, Chan JKC, et al. The 5th edition of the World Health organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36:1703–1719. doi: 10.1038/s41375-022-01613-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rampidis GP, Benetos G, Benz DC, Giannopoulos AA, Buechel RR. A guide for gensini score calculation. Atherosclerosis. 2019;287:181–183. doi: 10.1016/j.atherosclerosis.2019.05.012 [DOI] [PubMed] [Google Scholar]
- 32.Guo W, Romano J. A generalized Sidak-Holm procedure and control of generalized error rates under independence. Stat Appl Genet Mol Biol. 2007;6:Article3. doi: 10.2202/1544-6115.1247 [DOI] [PubMed] [Google Scholar]