Abstract
Studies on the biochemical properties of very-large-size eukaryotic DNA polymerases have been limited by the difficulty in obtaining sufficient purified forms of each enzyme. Our aim was to determine and elucidate the biochemical properties of one such polymerase, pol ζ (DNA polymerase ζ) from Drosophila melanogaster (Dmpol ζ). Using an REV1 (UV-revertible gene 1) protein-affinity column, we have isolated the enzyme directly from Drosophila embryos. Completely purified Dmpol ζ was found to have a molecular mass of approx. 240 kDa, and to be sensitive to aphidicolin and resistant to ddTTP (2′,3′-dideoxythymidine-5-triphosphate) and N-ethylmaleimide. The enzyme has a preference for poly(dA)/oligo(dT)10:1 as a template primer and has high processivity for DNA synthesis. Moreover, Dmpol ζ showed significantly higher fidelity compared with Rattus norvegicus DNA polymerase, an error-prone DNA polymerase, in an M13 forward mutation assay. The activities of bypassing pyrimidine dimers and (6-4) photoproducts and extending from mismatched primer-template termini in (6-4) photoproduct by Dmpol ζ were not detected. Drosophila REV7 interacted with Dmpol ζ in vitro, but did not influence the DNA synthesis activity of Dmpol ζ. The present study is the first report about characterization of purified pol ζ from multicellular organisms, and the second concerning the characterization of yeast pol ζ.
Keywords: DNA polymerase ζ (pol ζ), Drosophila melanogaster, fidelity, processivity, REV1 protein-affinity column chromatography, REV7
Abbreviations: CPD, cis-syn-cyclobutane pyrimidine dimer; ddTTP, 2′,3′-dideoxythymidine-5-triphosphate; Dmpol ζ, pol ζ from Drosophila melanogaster; DmRPA, replication protein A from Drosophila melanogaster; DTT, dithiothreitol; GST, glutathione S-transferase; GST–Dmpol ζ 858–1217aa, GST-tagged Dmpol ζ protein amino acid residues 858–1217; NEM, N-ethylmaleimide; NP40, Nonidet P40; PCNA, proliferating-cell nuclear antigen; (6-4)PP, (6-4) photoproducts; pol ζ, DNA polymerase ζ; REV1, UV-revertible gene 1; Rnpol β, Rattus norvegicus pol β; RPA, replication protein A
INTRODUCTION
Studies of recombinant enzymes are beset by difficulties, one of the most serious problems being generation of recombinant proteins with high molecular mass by conventional genetic engineering methods. Moreover, some recombinant proteins, even though active, may not accurately reflect the native forms. For example, the biochemical properties of endogenous pol λ (DNA polymerase λ) [1] were found to differ from those of recombinant pol λ in several respects [2]. To characterize large-size enzymes accurately, they must be isolated directly from the tissues or cells, as in the pre-genomic era, and then compared with recombinant enzymes.
Our aim was to study the functions of Dmpol ζ (pol ζ from Drosophila melanogaster). The exact in vitro and in vivo functions of pol ζ remain mostly unclear. To establish firmly the general properties of pol ζ, the enzyme should be obtained from many organisms and investigated. So far, a clear understanding of the physiological role of Dmpol ζ has been hampered by the lack of knowledge of its biochemical properties, mainly due to the fact that, although the gene has been identified at the genetic level, recombinant products have never been obtained as intact and active enzymes [3]. To isolate quickly a large amount of endogenous Dmpol ζ, we developed the REV1 (UV-revertible gene 1) protein-affinity column chromatography discussed below in detail. This method works extremely well for the isolation of Dmpol ζ and, hence, we succeeded in purifying a sufficient amount of enzyme to near-homogeneity.
As reviewed by Lawrence [4] recently, most of the spontaneous and DNA damage-induced mutations in eukaryotes result from replication processes in which pol ζ plays major roles; an important motive for studying pol ζ is the expectation that information about its activity will increase our understanding of the origins of cancer and development of diversity in the immune system, in both of which mutagenesis is an important feature. In this connection, when compared with yeast and mammals, D. melanogaster is a valuable model system for biochemical, genetic and developmental investigations [5–7], and transgenic animals can be prepared rather easily. Furthermore, this animal features various kinds of cell cycles: DNA replication and nuclear division without cell division in the early embryos, DNA amplification without M-phase in various larval tissues, including the salivary glands, and usual cell cycles consisting of G1, S-, G2 and M-phases in sites such as neural tissues and imaginal discs. Using Drosophila, it should be possible to assess the function of Dmpol ζ in these different cell cycles. In the present study, we report the biochemical properties of the native Dmpol ζ purified directly from embryos using a REV1 protein-affinity isolation method.
EXPERIMENTAL
Materials
The nucleotides, calf thymus and chemically synthetic templates or primers such as poly(dA)/oligo(dT)10:1, [3H]dTTP (4800 c.p.m./mmol), [α-32P]dTTP and [γ-32P]ATP (∼3000 Ci/mmol) were purchased from Amersham Biosciences. The DNA polymerase inhibitor ddTTP (2,3-dideoxythymidine-5-triphosphate) was purchased from Amersham Biosciences, whereas aphidicolin, NEM (N-ethylmaleimide) and heparin were obtained from Sigma (St. Louis, MO, U.S.A.). DE81 filters and P11-cellulose phosphate were purchased from Whatman Biosystems (Maidstone, Kent, U.K.). DEAE-Sepharose, Sephadex G-75 and SP-Sepharose were purchased from Amersham Biosciences. Bradford reagents and molecular-mass standards for SDS/PAGE were from Bio-Rad (Hercules, CA, U.S.A.). Calf thymus DNA used as a template primer was activated with pancreatic DNase I by the method described in [8]. PCNA (proliferating-cell nuclear antigen) cDNA of Drosophila was a gift from Dr M. Yamaguchi (Division of Biotechnology, Faculty of Textile Sciences, Kyoto Institute of Technology, Japan). RPA (replication protein A) from Drosophila (DmRPA) was isolated in our laboratory based on the method described previously [9]. Monoclonal antibody against Dmpol α (5E12) was a gift from Dr K. Kuroda (Mitsubishi Chemical Institute of Life Science, Japan). All other reagents were of analytical grade and were purchased from Sigma, Amersham Biosciences or Wako Chemicals (Osaka, Japan).
Buffers
TEMG buffer contained 50 mM Tris/HCl (pH 7.5), 1 mM EDTA, 5 mM 2-mercaptoethanol, 10% (v/v) glycerol, 0.01% (v/v) NP40 (Nonidet P40) and three protease inhibitors (1 μg/ml pepstatin A, 1 μg/ml leupeptin and 1 mM PMSF). PEMG buffer contained 137 mM NaCl, 8.1 mM Na2HPO4, 1.47 mM KH2PO4, 2.68 mM KCl, 1 mM EDTA, 5 mM 2-mercaptoethanol, 10% glycerol, 0.01% NP40 and the three protease inhibitors mentioned above. The glutathione buffer contained 30 mg/ml GSH, 50 mM Tris/HCl (pH 8.0), 1 mM EDTA, 5 mM 2-mercaptoethanol, 10% glycerol, 0.01% NP40 and the three protease inhibitors.
Collection of Drosophila melanogaster embryos
The basic procedure for collection of Drosophila embryos has been described previously [10]. D. melanogaster (Oregon R and M316) at 0–24 h were collected from large population cages.
Production of polyclonal antibody against Dmpol ζ
PCR was performed using the EST clone, SD07383, with the partial Dmpol ζ coding sequence. Primers were selected based on information from the Berkeley Drosophila Genome Project (http://www.fruitfly.org/index.html). The primers synthesized were 5′-GAATTCCGATGCTATCGAACACAGTTTG-3′ and 5′-CCGCTCGAGCGGGTAGCCATAGGTAACATTGG-3′. The resultant truncated sequence (residues 1329–1728) of the Dmpol ζ coding region was cloned into the pET28b(+) expression vector and transformed into Escherichia coli BL21(DE3) for protein induction. Extracts prepared from cells induced for 3 h were shown to contain a His6-tagged N-terminal Dmpol ζ fusion peptide. This was purified by His-bind resin column chromatography and SDS/PAGE and used for the immunization of rabbits.
Expression and purification of the GST–DmREV1-C (GST-tagged truncated DmREV1 protein)
PCR was performed using the EST clone, GH11153, containing the entire Drosophila REV1 (DmREV1) coding sequence. The primers synthesized were 5′-CGCGGATCCGCGCACGGAGTTTGCGATATA-3′ and 5′-CCGGAATTCCGGGGAGCACTTTATGCAACG-3′. The 1110 bp BamHI–EcoRI DNA fragment, which contains the C-terminal 370 residues, was cloned into the pGEX 6P-1 expression vector and transformed into E. coli BL21(DE3). One colony was incubated in 100 ml of Luria–Bertani medium, containing 40 mM glucose and 50 μg/ml ampicillin at 30 °C. The culture was grown overnight and separated into small batches for transferring to fresh medium. After 2 h of growth, 6 ml of 100 mM isopropyl β-D-thiogalactoside was added. After 3 h of growth, each culture was centrifuged at 3000 g for 10 min at 4 °C in a Hitachi 10A rotor. The pellet was resuspended in PEMG buffer, sonicated five times for 30 s and centrifuged at 30000 g for 20 min at 4 °C. The supernatant was loaded on to a glutathione–Sepharose 4B column pre-equilibrated with PEMG buffer, extensively washed with the same buffer and eluted with glutathione buffer. The purified GST (glutathione S-transferase)-tagged truncated DmREV1 protein (GST–DmREV1-C) was dialysed against TEMG buffer (containing 50% glycerol) and stored at −20 °C until use.
Expression and purification of Drosophila REV7
Drosophila REV7 (DmREV7) cDNA fragments were isolated by reverse transcriptase–PCR from Kc cell using primers based on the Berkeley Drosophila Genome Project. The primers synthesized were 5′-CCGGAATTCCGGATGCAGGCGGAGATTAAG-3′ and 5′-CCCTCGAGGGATTAACGATGAGGACATCCA-3′. The full-length sequence of the DmREV7 coding region was cloned into the pET28a(+) expression vector and transformed into E. coli BL21(DE3) for protein induction. Extracts from the cells induced for 3 h at 30 °C were purified by His-bind resin.
Expression and purification of the GST–Dmpol ζ 858–1217aa (GST-tagged truncated Dmpol ζ protein amino acid residues 858–1217)
PCR was performed using the cDNA library from Kc cells. The primers synthesized were 5′-AACGGAATTCTTACAACGTGAGCTAATTCCACAGG-3′ and 5′-ATAGCTCGAGTTAGAGCCTAGTTCCGCGCTTAA-3′. The truncated sequence (residues 858–1217) of the Dmpol ζ coding region was cloned into the pGEX 6P-1 expression vector and transformed into E. coli BL21(DE3). Extracts from the cells induced for 3 h at 30 °C were purified by glutathione–Sepharose 4B and SP–Sepharose column chromatographies.
Purification of Dmpol ζ
Dmpol ζ was purified from D. melanogaster embryos by the method described below. All the steps were performed below 4 °C.
Step 1. The purified GST–DmREV1-C protein was diluted three times in TEMG buffer containing 0.25 M KCl and loaded on to a glutathione–Sepharose 4B column pre-equilibrated with TEMG buffer containing 0.2 M KCl. The column was washed extensively with the same buffer (REV1 protein-affinity column).
Step 2. Frozen Drosophila embryos (20 g) were crushed in a Waring-type blender (maximum speed) and suspended in 50 ml of TEMG buffer containing 0.2 M KCl and 3 mM MgCl2. The embryo suspension was then homogenized with a homogenizer (LH-21; Yamato, Tokyo, Japan) and the homogenate was filtered through two layers of nylon mesh and then centrifuged (40000 g, 60 min). The final supernatant, designated as the S100 fraction, was used for the following purification step.
Step 3. The S100 fraction was mixed with 3 vol. of TEMG buffer and loaded on to a P11-cellulose phosphate column pre-equilibrated with TEMG buffer containing 50 mM KCl. The column was then washed with TEMG buffer containing 150 mM KCl and elution was performed with a linear gradient from 150 mM to 0.8 M KCl in TEMG buffer, and the fractions were tested for activity. The fractions of interest were pooled (fraction a).
Step 4. Fraction a was diluted 9-fold in the TEMG buffer and loaded on to a DEAE-Sepharose column pre-equilibrated with TEMG buffer containing 50 mM KCl. After washing with the same buffer, elution was performed with a linear gradient from 50 mM to 0.2 M KCl in TEMG buffer. The fractions were tested for activity, and those of interest were collected (fraction b).
Step 5. Fraction b was loaded on to a REV1 protein-affinity column pre-equilibrated with TEMG buffer containing 0.2 M KCl. The column was extensively washed with the same buffer and eluted with TEMG buffer containing 0.8 M KCl. The fractions were tested for activity, and those of interest were collected (fraction c).
Step 6. Fraction c was fractionated on a Sephadex G-75 column pre-equilibrated with TEMG buffer containing 0.5 M KCl. After testing for activity, the fractions of interest were collected (fraction d).
Step 7. Fraction d was diluted 9-fold in TEMG buffer and loaded on to an SP–Sepharose column pre-equilibrated with TEMG buffer containing 50 mM KCl. The column was washed with the same buffer and eluted with a linear gradient of 50 mM to 0.8 M KCl in the TEMG buffer. The fractions were tested for activity, and those of interest were collected (fraction e). Fraction e was combined, dialysed against TEMG buffer (containing 50% glycerol) and stored at −20 °C.
Other enzymes and proteins
Dmpol α was purified as described previously [11]. Recombinant Dmpol η, Drosophila PCNA (DmPCNA) and recombinant Rnpol β (Rattus norvegicus pol β) were purified as described in [12–14]. Klenow fragment was obtained from Toyobo (Osaka, Japan). Dmpol α, Dmpol η, Rnpol β and Klenow fragment had specific activities of 75000, 6700, 200000 and 7100000 units/mg respectively.
DNA polymerase assay
The reaction mixture contained 50 mM Tris/HCl (pH 7.5), 1 mM DTT (dithiothreitol), 5 mM MgCl2, 15% glycerol, 100 μg/ml poly(dA)/oligo(dT)10:1, 5 μM [3H]dTTP (4800 c.p.m./pmol) and the enzyme fraction. The enzyme sample was generally diluted with TEMG buffer to reduce the carried-over salt. When activated DNA (250 μg/ml) was used as a template primer, the [3H]dTTP concentration was reduced to 0.5 μM and 20 μM of dATP, dGTP and dCTP were added. The reaction was allowed to progress by incubation at 37 °C for 30 min, and then stopped by cooling on ice. The mixture was then spotted on to a Whatman DE81 ion-exchange filter (2 cm×2 cm) and air-dried and then washed three times with 5% (w/v) Na2HPO4 (5 ml/filter), three times with distilled water and twice with ethanol. The radioactivity of each dried filter was finally measured in a toluene-based scintillator. One unit of polymerase activity was defined as the amount catalysing the incorporation of 1 pmol of [3H]dTMP into activated DNA in 60 min.
To examine the effects of DmREV7, DmPCNA or DmRPA on the DNA synthetic activity of Dmpol ζ, activated DNA was used as a template primer. Dmpol ζ (0.2 pmol) was preincubated with 10 pmol of DmPCNA (homotrimer), DmRPA (heterotrimer), DmREV7 and BSA (control protein) for 30 min on ice, and the activity was assayed under the standard conditions.
Gel filtration
The stored fraction (50 units) was dialysed against TEMG buffer containing 0.5 M KCl and concentrated to a volume of 0.4 ml; 200 μl was loaded on to a gel-filtration column (Superose 6 HR 10/30; Amersham Biosciences) and pre-equilibrated with TEMG buffer containing 0.5 M KCl. Fractions (200 μl) were collected and assayed for DNA polymerase activity using activated DNA as a template. The molecular mass was estimated from a calibration curve using chymotrypsinogen A (25 kDa), BSA (66 kDa), catalase (232 kDa) and ferritin (440 kDa).
Translesion synthesis and mispair extension of DNA lesions
The 67 nt oligomer template (5′-ATGTTCGCGGATCCACCATGCCTGCACCAATTAAGCATCGTAATCATGGTCATGAATTCCGGTTATG-3′) was used as the ‘non-damaged template’. The same 67 nt oligomers containing the CPD (cis-syn-cyclobutane pyrimidine dimer) and the TT (6-4)PP (6-4 photoproduct) at the underlined sites were synthesized as described in [15,16]. A 35 nt oligomer with a sequence complementary to the 67 nt template was synthesized and used as the primer. The reaction mixtures (20 μl) used for testing the bypass of CPD contained 50 mM Tris/HCl (pH 7.5), 1 mM DTT, 5 mM MgCl2, 10% glycerol, 100 μM each of dATP, dCTP, dGTP and dTTP, 5 nM of 5′-labelled primer template and 10 fmol of Klenow fragment, 40 fmol of Dmpol η or 300 fmol of Dmpol ζ. The reaction mixtures (20 μl) used for testing the bypass of (6-4)PP contained 50 mM Tris/HCl (pH 7.5), 1 mM DTT, 5 mM MgCl2, 10% glycerol, 100 μM each of dATP, dCTP, dGTP and dTTP, 5 nM 5′-labelled primer template and 30 fmol of Klenow fragment, 300 fmol of Dmpol η or 300 fmol of Dmpol ζ. After incubation at 37 °C for 1, 5 or 10 min, reactions were terminated with 20 μl of 20 mM EDTA and 95% (v/v) formamide, and the products were subjected to 8 M urea in 10% acrylamide gel, and the resulting products were subjected to 8 M urea in 10% acrylamide gel, and the resulting products were then dried and exposed to BioMax MS-1 (Kodak, Rochester, NY, U.S.A.).
To test the ability of extension from mispair template-primer termini by Dmpol ζ, the 36 nt oligomers in which an A or a G is paired with the 3′-T of (6-4)PP or a non-damaged T residue were used as the primers. Dmpol ζ (300 fmol) was incubated with the matched or mismatched primer template at 37 °C for 1, 5 or 10 min in the presence 100 μM each of the four dNTPs (2′-deoxynucleotide triphosphates). The products were subjected to 8 M urea in 10% acrylamide gel and then dried and exposed as mentioned above.
Processivity assay
To compare the primer extension reactions of Dmpol ζ with those of Klenow fragment (Figure 3A), the 5′-labelled 30 nt oligomer primer annealed to the 67 nt ‘non-damaged template’ (as shown previously) was used as the template DNA. The reaction mixture (20 μl) contained 50 mM Tris/HCl (pH 7.5), 1 mM DTT, 5 mM MgCl2, 10% glycerol, 100 μM each of dATP, dCTP, dGTP and dTTP, 5 nM of 5′-labelled primer template and 0.5 unit of Klenow fragment (0.2 nM) or Dmpol ζ (20 nM). After incubation at 37 °C for 10 min, reactions were terminated with 20 μl of 20 mM EDTA and 95% formamide, and the reaction products were subjected to 8 M urea in 10% acrylamide gel, then dried and exposed to BioMax MS-1 (Kodak).
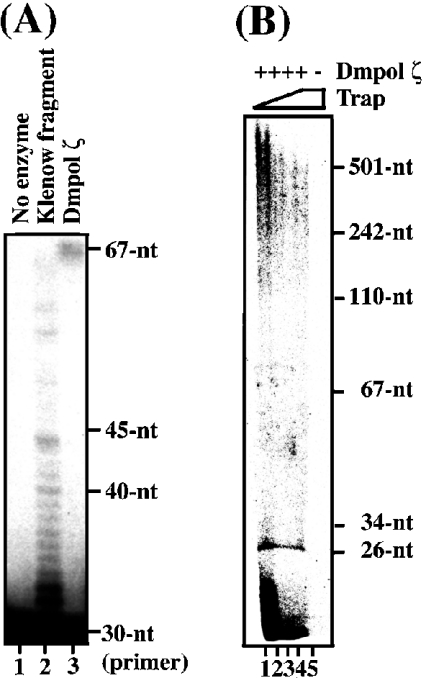
Figure 3. Processivity of Dmpol ζ.
(A) 5′-labelled 30 nt oligomer primer (5 nM) annealed to the 67 nt oligomer template was incubated with 0.5 unit of Klenow fragment (0.2 nM) or Dmpol ζ (20 nM) respectively. The reaction products were separated on a 10% acrylamide/8 M urea gel. Control fraction with no enzyme is shown in lane 1. (B) 5 μg/ml poly(dA)/oligo(dT)10:1 (100 nM primer molecule) and Dmpol ζ (12.5 nM) were incubated with four different amounts of heparin as trapping reagent (100, 500, 2500 and 10000 nM in lanes 1–4 respectively). The products were separated on a 10% acrylamide/8 M urea gel. Control reaction with no enzyme and 10000 nM heparin is shown in lane 5.
To examine the processivity of Dmpol ζ (Figure 3B), the homopolymer, poly(dA)/oligo(dT)10:1, was used as template DNA. The reaction mixture (100 μl) contained 50 mM Tris/HCl (pH 7.5), 1 mM DTT, 5 mM MgCl2, 10% glycerol, 0.5 μM dTTP, 0.05 μM [α-32P]dTTP, 5 μg/ml poly(dA)/oligo(dT)10:1 (100 nM primer molecule), 12.5 nM Dmpol ζ and four different amounts (100, 500, 2500 and 10000 nM) of heparin as the trapping reagent. After incubation at 37 °C for 5 min, samples were precipitated twice with ethanol, and resuspended in loading buffer containing 10 mM Tris/HCl (pH 8.0), 1 mM EDTA, 95% formamide and 0.1% (w/v) Bromophenol Blue. The reaction products were separated on a 10% acrylamide/8 M urea gel and then dried and exposed as mentioned above.
In vitro protein–protein interaction assay between DmREV7 and GST–Dmpol ζ 858–1217aa
GST–Dmpol ζ 858–1217aa (1 nmol) or 1 nmol of GST protein (alone) was incubated with 1 nmol of DmREV7 on ice for 30 min in 900 μl of T-TEG buffer [50 mM Tris/HCl (pH 7.4), 1 mM EDTA, 10% glycerol and 0.01% Tween 20] containing 150 mM NaCl, and mixed with 50 μl of glutathione–Sepharose 4B in 100 μl of the same buffer. After incubation at 4 °C for 1 h, the beads were washed five times with the same buffer, and eluted with 200 μl of T-TEG buffer containing 0.8 M NaCl. The bound protein (T7–DmREV7) was detected by Western-blot analysis with an anti-T7-tagged antibody (Novagen).
Proofreading assay
The assay was performed as described in our previous report [17]. A synthetic oligonucleotide (19-mer; 5′-CACGACGTTGTAAAACGAT-3′) was labelled with [γ-32P]ATP by T4 polynucleotide kinase and then hybridized to M13mp18 DNA to create a targeted GTEMPLATE-TPRIMER mispair at the 3′-OH end. Electrophoretic analysis was performed on a 20% polyacrylamide/8 M urea gel, dried and then exposed to BioMax MS-1 (Kodak).
Fidelity assay
DNA replication fidelity was assayed by the method described previously [18] using Bacteriophage M13mp18 and E. coli JM109 strain. Replicative form M13mp18 was digested with the restriction endonuclease PvuII, and the reaction product, an approx. 6800 bp DNA fragment, was subjected to agarose-gel electrophoresis and excised. After the DNA electroeluted from this gel fragment was purified, gapped circular M13mp18 DNA was prepared by mixing the restriction enzyme fragments and single-stranded circular M13mp18 DNA in 30 mM NaCl and 300 mM sodium citrate. The mixture was heated at 95 °C for 10 min, cooled in an ice bath and then incubated at 65 °C for 60 min. The gapped circular M13mp18 DNA was then purified by agarose-gel electrophoresis. Replication reactions for a forward mutation assay were carried out for 1 h at 37 °C in 20 μl containing 50 mM Tris/HCl (pH 7.5), 1 mM DTT, 5 mM MgCl2, 10% glycerol, 100 ng of gapped circular M13mp18 DNA, 100 μM each of dATP, dCTP, dGTP and dTTP and 1 unit each of Dmpol α, Dmpol ζ and Rnpol β. Under this condition, approx. 350 nt were incorporated per DNA template, although it was necessary to fill 270 nt of 420 gapped for calculation of the error rate.
RESULTS AND DISCUSSION
Purification of Dmpol ζ from Drosophila embryos by REV1 protein-affinity column chromatography
In Drosophila, the cells contain several DNA polymerases (pol α, γ, δ, ε, ζ, η, θ, ι and σ) and REV1 [19]. From our past 18 years experience [6,17,20,21], we concluded that the best approach to obtain large amounts of purified large-size DNA polymerases from D. melanogaster is direct isolation from embryos by a few chromatographic steps. Here, a purified form of the recombinant protein of GST–DmREV1-C (GST-tagged, truncated Drosophila REV1 protein containing the C-terminal 370 residues) was conjugated with a glutathione–Sepharose 4B column (see the Experimental section). In higher eukaryotes, the C-terminal domain of REV1 protein reportedly interacts with the REV7–REV3 complex, and REV3 corresponds to the catalytic subunit of pol ζ [22]. We therefore hypothesized that the Dmpol ζ complex selectively binds to Drosophila REV1 protein. The GST–DmREV1-C protein never dissociated from glutathione–Sepharose 4B, unless eluted with GSH. By using this old-fashioned pre-genomic era method, we succeeded in obtaining enough purified Dmpol ζ to allow biochemical properties to be elucidated.
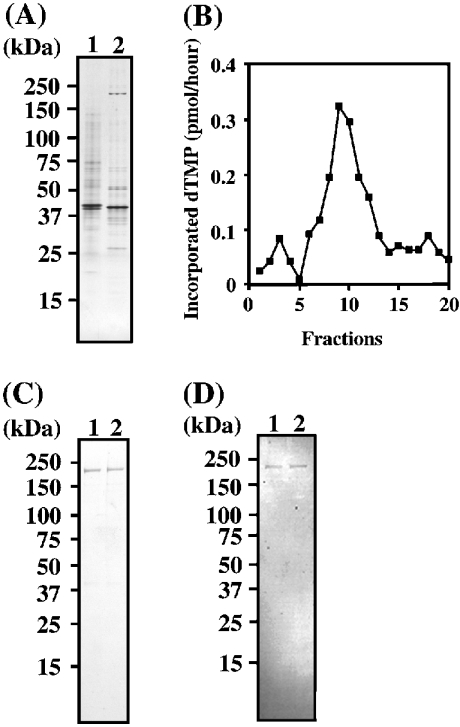
As described in the Experimental section, the crude extract of the embryos was chromatographed through P11-cellulose phosphate, DEAE-Sepharose and REV1 protein-affinity columns. In Figure 1A (lanes 1 and 2), we show fraction b after passing through the DEAE-Sepharose column and fraction c after REV1 protein-affinity purification respectively (see the Experimental section). With advancing purification, a band with a molecular mass of approx. 240 kDa appeared (Figure 1A, lane 2). Fraction c was purified finally on an SP-Sepharose column after Sephadex G-75 column chromatography. Figure 1(B) shows the elution pattern with SP-Sepharose column chromatography, and Figure 1(C) indicates SDS/PAGE of fractions in the peak area (fractions 9 and 10) in Figure 1(B). The resulting peak fractions in Figure 1B (lanes 1 and 2) showed a single band of 240 kDa. The same fractions in Figure 1(B) were analysed by immunoblotting for a variety of known DNA polymerases from Drosophila such as Dmpol α, δ and ε. The fractions in Figure 1D (lanes 1 and 2) were identical with those in Figure 1C (lanes 1 and 2) respectively. No bands were observed with antibodies against Dmpol α, δ and ε (results not shown). However, a clear signal was detected with a polyclonal antibody against Dmpol ζ, shown at a molecular mass of 240 kDa (Figure 1D), indicating that a significant amount of the purified Dmpol ζ was obtained only through use of five columns.
Figure 1. Purification of Dmpol ζ.
(A) SDS/PAGE analysis of the fractions (20 ng) after passing through DEAE-Sepharose (fraction b) and REV1 protein-affinity column (fraction c; see the Experimental section). Fractions b and c (lanes 1 and 2 respectively) were analysed by SDS/PAGE (5–20% gel). The gel was stained with silver. (B) Elution pattern with SP-Sepharose column chromatography. Fractions were tested under the standard assay conditions (see the Experimental section) using activated DNA as the template primer. (C, D) SDS/PAGE and immunoblot analyses of the SP-Sepharose fractions (fraction e); 100 ng of the peak fractions (fraction 9, lane 1; fraction 10, lane 2) were analysed by SDS/PAGE (5–20% gel). The gel was stained with silver (C) and immunoblotted with a polyclonal antibody against Dmpol ζ (D).
Biochemical properties of Dmpol ζ
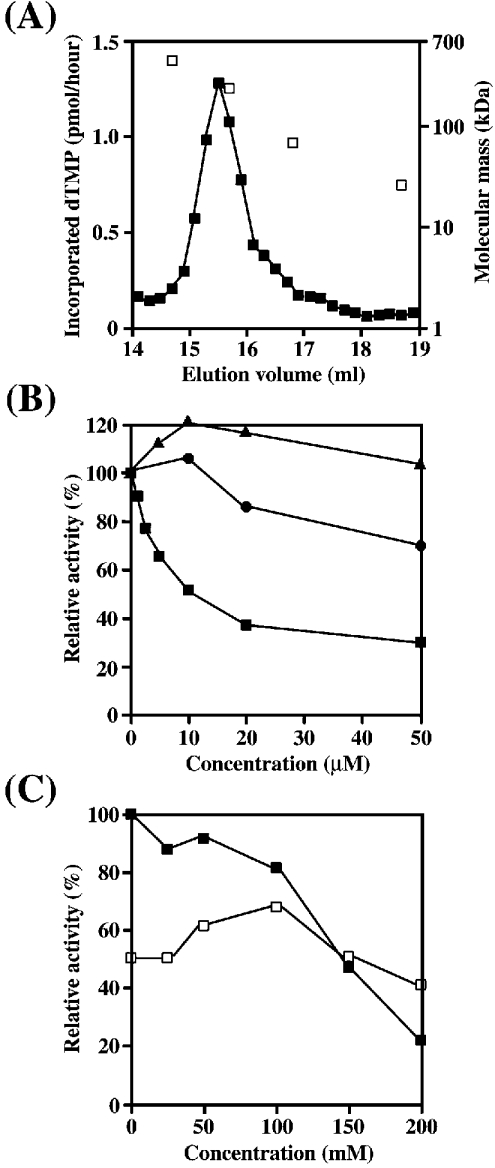
Next, to test whether this 240 kDa protein was responsible for the DNA polymerase activity, the SP-Sepharose fraction (fraction e) was concentrated and analysed by Superose 6 gel filtration. The single peak of the DNA polymerase activity appeared at a point, calculated to be 240 kDa (Figure 2A). Optimum reaction conditions for Dmpol ζ were determined using the homopolymer poly(dA)/oligo(dT)10:1 as template DNA. The optimum pH was 7.5 for the 50 mM Tris/HCl buffer, with about half the optimum activity at either pH 6.5 or 8.5 (results not shown). The optimum pH of Dmpol ζ was similar to those of Dmpol δ and yeast pol ζ as reported previously [17,23], and was distinct from those of other DNA polymerases, such as Dmpol α and Dmpol γ, which require a higher pH range [24,25]. Dmpol ζ was found to require bivalent cations, and the optimum concentration was 5 mM Mg2+ ions. Mn2+ ions at 2.5 mM provided half the optimum activity (results not shown). Figure 2(B) shows results on the sensitivity of the activity to typical DNA polymerase inhibitors, such as aphidicolin, ddTTP and NEM. Unlike yeast pol ζ, the enzyme was sensitive to aphidicolin (IC50=10 μM), although it was insensitive to ddTTP and NEM [23]. As shown in Figure 2(C), high KCl concentrations (150–200 mM) suppressed the activity, whereas Dmpol γ required much higher salt concentrations for DNA polymerization to be optimized [25]. Additionally, the response of the DNA polymerase activity was found to be dependent on the template-primer species; the activity was very sensitive to KCl on the poly(dA)/oligo(dT)10:1: the optimum dose was 0 mM KCl, whereas 100 mM KCl was optimal for activated DNA. Three DNA polymerases from Drosophila, Dmpol ε, ζ and θ, have expected molecular masses of 230–250 kDa on the basis of their cDNA sequences. Dmpol θ is insensitive to aphidicolin [6], and Dmpol ε is optimal at 50 mM KCl on poly(dA)/oligo(dT)10:1 [21]. Therefore these results also suggested that the purified DNA polymerase contained in the SP-Sepharose fraction was Dmpol ζ and not any other DNA polymerase. From this purification, the isolated activity in the SP-Sepharose fraction had a specific activity of 4500 units/mg (Table 1). The specific activity of purified Dmpol ζ was lower than those of other pols used in our experiments (see the Experimental section). The activity of Dmpol ζ alone, as well as that of yeast REV3, might be unstable [23].
Figure 2. Biochemical properties of Dmpol ζ.
(A) The gel-filtration pattern of the SP-Sepharose fraction (25 units) in Superose 6 HR 10/30. DNA polymerase activity (▪) and molecular-mass standards (□). The molecular mass was estimated from a calibration curve with chymotrypsinogen A (25 kDa), BSA (66 kDa), catalase (232 kDa) and ferritin (440 kDa). (B) Effects of DNA polymerase inhibitors. Significant sensitivity of Dmpol ζ to aphidicolin (▪), but not to ddTTP (•) or NEM (▾), was detected using poly(dA)/oligo(dT)10:1 as the template primer. (C) Effects of the ionic strength of KCl. The response of the DNA polymerase activity could be changed with template-primer species: the activity was very sensitive to KCl on poly(dA)/oligo(dT)10:1 (▪), whereas 100 mM KCl was optimal for activated DNA (□).
Table 1. Purification of Dmpol ζ from Drosophila embryos.
The total activity and specific activity in the S100 fraction, P11-cellulose phosphate and DEAE-Sepharose column samples were not calculated, since there were a few DNA polymerases in the samples.
| Fraction step | Total protein (mg) | Total activity (units) | Specific activity (units/mg) |
|---|---|---|---|
| S100 fraction | 942 | – | – |
| P11-cellulose phosphate column | 227 | – | – |
| DEAE-Sepharose column | 124 | – | – |
| REV1 protein-affinity column | 0.195 | 993 | 5080 |
| Sephadex G-75 column | 0.116 | 426 | 3670 |
| SP-Sepharose column | 0.0196 | 88.0 | 4500 |
Processivity of Dmpol ζ
To examine the processivity of Dmpol ζ, we first compared primer extension reactions on the 5′-labelled 30 nt oligomer primer annealed to the 67 nt oligomer template (see the Experimental section) between Dmpol ζ and Klenow fragment. Klenow fragment is known to be a middle-processive enzyme [26]. We performed this test with enzymes that elongate few primers. Under this condition, the pattern of Dmpol ζ was more processive than that of Klenow fragment (Figure 3A). Whereas most of the reaction products were extended up to approx. 15 nt by Klenow fragment (Figure 3A, lane 2), they were extended to the end of the 67 nt oligomer template by Dmpol ζ (Figure 3A, lane 3). This result suggests that Dmpol ζ may be a higher-processive polymerase.
Next, we measured the processivity of Dmpol ζ. For the analysis of products resulting from a single binding event of Dmpol ζ to the substrate, the assay was performed in the presence of various amounts of heparin. In this test, the homopolymer poly(dA)/oligo(dT)10:1 was used as template DNA. The products from Dmpol ζ were extended up to approx. 500 nt in the presence of an excess of heparin as trapping reagent (Figure 3B, lanes 3 and 4). This result indicates that Dmpol ζ has a high processivity.
Fidelity of Dmpol ζ
First, we tested for the presence of 3′-5′ exonuclease activity using a 5′-labelled template primer containing a 3′-terminal mismatch. Reportedly, yeast pol ζ has no 3′-5′ proofreading exonuclease activity [23]. To determine whether Dmpol ζ has such a potential, its terminal mismatch nucleotide excision ability was tested. After the nuclease reaction using 5′-labelled substrates, the products were analysed by denaturing PAGE. Dmpol ζ demonstrated no proofreading exonuclease activity. It lacked the 3′-5′ exonuclease function that can proofread replication errors (results not shown).
The phenotype of the Drosophila mus205, whose gene encodes the catalytic subunit of Dmpol ζ, is strikingly different from that of the yeast rev3 mutant [3]. The mus205 mutant shows no effect on methyl methanesulphonate-induced mutations and recombinations. Thus we examined the fidelity of Dmpol ζ using M13mp18, containing a gap in the lacZ gene region, on non-damaged DNA in vitro (see the Experimental section). The mutation frequency for Dmpol ζ was determined in a forward mutation assay. Dmpol ζ showed significantly higher fidelity than Rnpol β (seven times higher) and almost the same level as Dmpol α (Table 2). It is known that eukaryotic pol α misincorporates nucleotides with a frequency of approx. 10−4, and eukaryotic pol β has an error frequency of approx. 10−3 [27,28]. Thus Dmpol ζ incorporates correct nucleotides with a high frequency at least on the non-damaged template, suggesting that Dmpol ζ is not an error-prone DNA polymerase.
Table 2. Mutation frequencies of Rnpol β, Dmpol α and Dmpol ζ in a forward mutation assay.
This assay was performed as described in the Experimental section using M13mp18 containing a gap in the lacZα gene region. ‘No enzyme’ indicates the background of this assay.
| DNA polymerase | Total plaques | Mutant plaques | Mutation frequency (×10−4) |
|---|---|---|---|
| No enzyme | 6652 | 3 | 4.5 |
| Rnpol β | 2383 | 53 | 222.4 |
| Dmpol α | 3696 | 12 | 32.5 |
| Dmpol ζ | 3352 | 11 | 32.8 |
Translesion synthesis and mispair extension of DNA lesions by Dmpol ζ
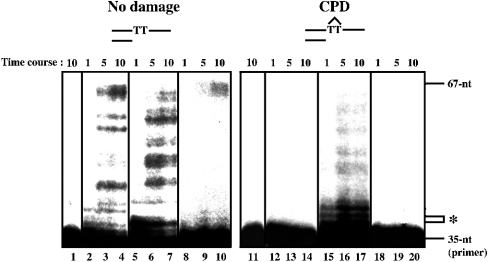
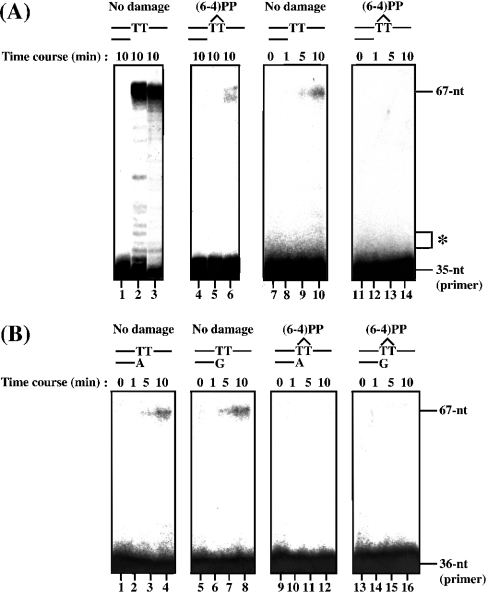
We were also interested in the behaviour of Dmpol ζ on damaged DNA templates. In previous reports, it was found that yeast pol ζ bypassed CPD very inefficiently, but not (6-4)PP [23,29]. Although the polymerase species were different, Ishikawa et al. [12] found that both Dmpol η and ι are capable of bypassing CPD, and Dmpol η is also capable of bypassing (6-4)PP. These characteristics are strikingly different from those of pol η and pol ι from humans and yeast [29–31]. Therefore we examined whether Dmpol ζ is capable of bypassing CPD and (6-4)PP. In the translesion synthesis assay, the thymidine residues of nt 36 and 37 form the CPD or the (6-4)PP. Dmpol η was capable of bypassing CPD efficiently under our assay conditions, but the bypass of CPD by Dmpol ζ was not efficient (Figure 4). In addition, bypass of (6-4)PP by Dmpol ζ was not detected (Figure 5A).
Figure 4. Translesion synthesis of CPD by Dmpol ζ.
DNA lesions on the templates are CPD (lanes 11–20). The positions of CPD (36 and 37 nt) are marked with an asterisk on the right side of the panel. No damage controls were included for CPD (lanes 1–10). The reaction mixtures contained 10 fmol of Klenow fragment (lanes 2–4 and 12–14), 40 fmol of Dmpol η (lanes 5–7 and 15–17) and 300 fmol of Dmpol ζ (lanes 8–10 and 18–20). After incubation at 37 °C for 1, 5 or 10 min, the products were separated on a 10% polyacrylamide/8 M urea gel. Control reactions with no enzyme are shown in lanes 1 and 11.
Figure 5. Translesion synthesis and mispair extension of (6-4)PP by Dmpol ζ.
(A) Translesion synthesis of (6-4)PP. The DNA lesions on the templates are (6-4)PP (lanes 4–6 and 11–14). The positions of (6-4)PP (36 and 37 nt) are marked with an asterisk on the right side of the panel. No-damage controls for (6-4)PP were included (lanes 1–3 and 7–10). Control reactions with no enzyme are shown in lanes 1 and 4. The reaction mixtures contained 30 fmol of Klenow fragment (lanes 2 and 5), 300 fmol of Dmpol η (lanes 3 and 6) and 300 fmol of Dmpol ζ (lanes 7–14). After incubation at 37 °C, the reaction products were separated on a 10% polyacrylamide/8 M urea gel. (B) Extension from matched and mismatched primer-template termini by Dmpol ζ. Dmpol ζ (300 fmol) was incubated with a primer in which an A (lanes 9–12) or a G (lanes 13–16) is paired with the 3′-T of (6-4)PP in the template. No-damage controls for (6-4)PP were included (lanes 1–8). The products were separated on a 10% polyacrylamide/8 M urea gel.
Next, we tested whether Dmpol ζ could extend from mismatched primer-template termini on (6-4)PP. Since pol η preferentially inserts a G residue opposite to the 3′-T of (6-4)PP [12,31,32], we examined the ability of Dmpol ζ to extend from a primer in which an A or a G is paired with the 3′-T of the lesion. Yeast pol ζ has the ability to extend from a mismatched base opposite to the 3′-T of (6-4)PP, and it is more efficient at extending from the G opposite to the 3′-T of (6-4)PP than from the G paired with the T on non-damaged template [29,32]. Extension by Dmpol ζ from both matched and mismatched primer-template termini on non-damaged template was clearly detectable (Figure 5B, lanes 1–8), but it was not detected on (6-4)PP (Figure 5B, lanes 9–16). This result indicates that the ability of Dmpol ζ to extend from the G opposite to the 3′-T of (6-4)PP is different from that of yeast pol ζ. Dmpol η is capable of bypassing CPD efficiently [12], and Drosophila cultured Kc cells reportedly have a 6-4 photolyase that human cells lack, repairing (6-4)PP in a light-dependent manner [33]. Therefore Dmpol ζ might not be essential for the bypass of such lesions.
Drosophila REV7 directly interacts with Dmpol ζ, but does not influence the DNA synthetic activity of Dmpol ζ
In performing this experiment, we also found a sequence termed Drosophila REV7 (DmREV7) that was homologous with human REV7 (GenBank® accession no. AF15782) using the database of the Berkeley Drosophila. In humans and yeast, REV7 is supposed to be a subunit of pol ζ [23,34], interacting with the enzyme to increase its activity, at least in the case of yeast [23]. Therefore we tried to examine the interaction between DmREV7 and Dmpol ζ and the effect of DmREV7 on the DNA synthetic activity of Dmpol ζ.
First, we isolated the open reading frame region of DmREV7 by reverse transcriptase–PCR using specific primers from a cDNA library of Kc cells. The amino acid sequence of the gene product shared 27% identity and 47% similarity with human REV7. The nucleotide sequence data have been submitted to the DDBJ, EMBL, GenBank® and GSDB Nucleotide Sequence Databases with accession number AB115908.
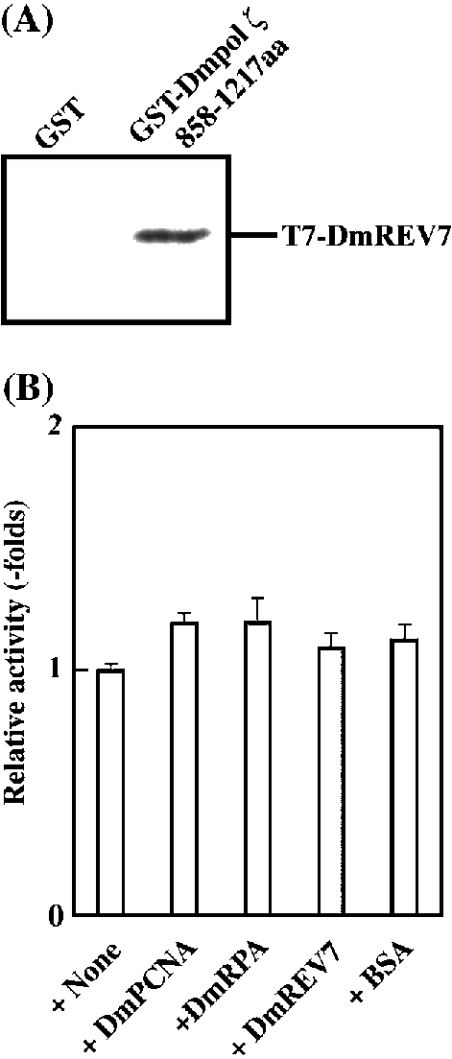
To examine where DmREV7 directly interacts with Dmpol ζ, we performed an in vitro binding assay. In humans, REV7 interacts with the middle region of REV3 protein [22]. Although the middle region of human REV3, except for a small 50–60 amino acid region, is not related to that of Dmpol ζ, we tried to examine where DmREV7 interacted with the middle region of Dmpol ζ. A GST fusion protein containing Dmpol ζ amino acid residues 858–1217 (GST–Dmpol ζ 858–1217aa) and T7-tagged DmREV7 (T7–DmREV7) were expressed in bacteria and purified. T7–DmREV7 was tested for binding to GST–Dmpol ζ 858–1217aa compared with a control containing GST alone by using glutathione–Sepharose beads. As shown in Figure 6(A), T7–DmREV7 bound to GST–Dmpol ζ 858–1217aa, but not to GST alone. This result indicates that DmREV7 directly interacts with Dmpol ζ similar to the yeast and human REV7.
Figure 6. DmREV7 directly interacts with Dmpol ζ, but does not influence the activity of Dmpol ζ.
(A) Interaction between DmREV7 and Dmpol ζ in vitro. GST alone (1 nmol) and 1 nmol of GST fusion protein containing Dmpol ζ amino acid residues 858–1217 (GST–Dmpol ζ 858–1217aa) immobilized on glutathione–Sepharose beads were incubated with 1 nmol of T7-tagged DmREV7 (T7–DmREV7). The bound protein (T7–DmREV7) was detected by Western-blot analysis with an anti-T7-tagged antibody. (B) Effects of DmPCNA, DmRPA or DmREV7 on the DNA synthetic activity of Dmpol ζ. Dmpol ζ (0.2 pmol) was preincubated with 10 pmol of DmPCNA (homotrimer), DmPRA (heterotrimer), DmREV7 or BSA (control protein) for 30 min on ice and the activity was assayed as described in the Experimental section. Results shown are representative of three independent experiments. The error bars indicate S.D.
Next, we tested the effect of DmREV7 on the DNA synthetic activity of Dmpol ζ. At the same time, we also tested the effects of PCNA (DmPCNA) and RPA (DmRPA) isolated from D. melanogaster. Figure 6(B) shows the effects of DmREV7, DmPCNA, DmRPA and BSA (control). Although this assay was performed in the presence of a significant amount of DmREV7, DmPCNA, DmRPA or BSA (the molar ratio of Dmpol ζ to these proteins was 1:50), all of them did not influence the DNA synthetic activity of Dmpol ζ (Figure 6B). Recently, Murakumo et al. [34] showed that human REV7 interacts with the spindle assembly checkpoint protein MAD2 (mitotic arrest-deficient 2), and suggested that REV7 may act as an adapter between DNA repair and the spindle assembly checkpoint. Furthermore, Masuda et al. [35] have provided evidence that human REV7 and REV1 form a stable heterodimer, and have shown that human REV7 does not affect the transferase activity or stability of REV1 in contrast with yeast REV3. Similar to human REV7, DmREV7 may have functions that are different from stimulation of the DNA synthetic activity of Dmpol ζ as observed in yeast.
As described briefly in the Introduction section, most of the spontaneous and DNA-damage-induced mutations in eukaryotes result from replication processes in which pol ζ plays major roles, and an important motive for studying pol ζ is the expectation that information about its activity will increase our understanding of the origins of cancer and the development of diversity in the immune system. Hitherto, the only available pol ζ protein was from yeast, a unicellular organism [23,29,32]. Now, we have succeeded in purifying pol ζ to near homogeneity through development of a REV1 protein-affinity isolation method. Using the directly purified native form, we determined for the first time the functions of pol ζ from a multicellular organism in vitro.
References
- 1.Ramadan K., Shevelev I. V., Maga G., Hübscher U. DNA polymerase λ from calf thymus preferentially replicates damaged DNA. J. Biol. Chem. 2002;277:18454–18458. doi: 10.1074/jbc.M200421200. [DOI] [PubMed] [Google Scholar]
- 2.Shimazaki N., Yoshida K., Kobayashi T., Toji S., Tamai K., Koiwai O. Over-expression of human DNA polymerase λ in E. coli and characterization of the recombinant enzyme. Genes Cells. 2002;7:639–651. doi: 10.1046/j.1365-2443.2002.00547.x. [DOI] [PubMed] [Google Scholar]
- 3.Eeken J. C., Romeijn R. J., de Jong A. W., Pastink A., Lohman P. H. Isolation and genetic characterisation of the Drosophila homologue of (SCE)REV3, encoding the catalytic subunit of DNA polymerase ζ. Mutant Res. 2001;485:237–253. doi: 10.1016/s0921-8777(01)00062-3. [DOI] [PubMed] [Google Scholar]
- 4.Lawrence C. W. Cellular roles of DNA polymerase ζ and Rev1 protein. DNA Repair. 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 5.Boyd J. B., Sakaguchi K., Harris P. V. Mus308 mutants of Drosophila exhibit hypersensitivity to DNA cross-linking agents and are defective in a deoxyribonuclease. Genetics. 1990;125:813–819. doi: 10.1093/genetics/125.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oshige M., Aoyagi N., Harris P. V., Burtis K. C., Sakaguchi K. A new DNA polymerase species from Drosophila melanogaster: a probable mus308 gene product. Mutat. Res. 1999;433:183–192. doi: 10.1016/s0921-8777(99)00005-1. [DOI] [PubMed] [Google Scholar]
- 7.Takata K., Ishikawa G., Hirose F., Sakaguchi K. Drosophila damage-specific DNA-binding protein 1 (D-DDB1) is controlled by the DRE/DREF system. Nucleic Acids Res. 2002;30:3795–3808. doi: 10.1093/nar/gkf490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlabach A., Fridlender B., Bolden A., Welssbach A. DNA-dependent DNA polymerases from HeLa cell nuclei II. Template and substrate utilization. Biochem. Biophys. Res. Commun. 1971;44:879–885. doi: 10.1016/0006-291x(71)90793-5. [DOI] [PubMed] [Google Scholar]
- 9.Richard F. M., Pia T., Sue C. Purification and characterisation of dRP-A: a single-stranded DNA binding protein from Drosophila melanogaster. FEBS Lett. 1994;342:139–144. doi: 10.1016/0014-5793(94)80488-5. [DOI] [PubMed] [Google Scholar]
- 10.Harris P. V., Boyd J. B. Pyrimidine dimers in Drosophila chromatin become increasingly accessible after irradiation. Mutat. Res. 1987;183:53–60. doi: 10.1016/0167-8817(87)90045-9. [DOI] [PubMed] [Google Scholar]
- 11.Kuroda K., Kagiyama-Takahashi R., Shinomiya T. Immunoaffinity purification and properties of Drosophila melanogaster DNA polymerase α–primase complex. J. Biochem. (Tokyo) 1990;108:926–933. doi: 10.1093/oxfordjournals.jbchem.a123316. [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa T., Uematsu N., Mizukoshi T., Iwai S., Iwasaki H., Masutani C., Hanaoka F., Ueda R., Ohmori H., Todo T. Mutagenic and nonmutagenic bypass of DNA lesions by Drosophila DNA polymerases dpolη and dpolι. J. Biol. Chem. 2001;276:15155–15163. doi: 10.1074/jbc.M009822200. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi M., Nishida Y., Moriuchi T., Hirose F., Hui C. C., Suzuki Y., Matsukage A. Drosophila proliferating cell nuclear antigen (cyclin) gene: structure, expression during development, and specific binding of homeodomain proteins to its 5′-flanking region. Mol. Cell. Biol. 1990;10:872–879. doi: 10.1128/mcb.10.3.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar A., Widen S. G., Williams K. R., Kedar P., Karpel R. L., Wilson S. H. Studies of the domain structure of mammalian DNA polymerase β. Identification of a discrete template binding domain. J. Biol. Chem. 1990;265:2124–2131. [PubMed] [Google Scholar]
- 15.Smith C. A., Taylor J.-S. Preparation and characterization of a set of deoxyoligonucleotide 49-mers containing site-specific cis-syn, trans-syn-I, (6-4), and Dewar photoproducts of thymidylyl(3′→5′)-thymidine. J. Biol. Chem. 1993;268:11143–11151. [PubMed] [Google Scholar]
- 16.Morioka H., Miura H., Kobayashi H., Koizumi T., Fujii K., Asano K., Matsunaga T., Nikaido O., Stewart J. D., Ohtsuka E. Antibodies specific for (6–4) DNA photoproducts: cloning, antibody modeling and construction of a single-chain Fv derivative. Biochim. Biophys. Acta. 1998;1385:17–32. doi: 10.1016/s0167-4838(98)00029-6. [DOI] [PubMed] [Google Scholar]
- 17.Aoyagi N., Matsuoka S., Furunobu A., Matsukage A., Sakaguchi K. Drosophila DNA polymerase δ. Purification and characterization. J. Biol. Chem. 1994;269:6045–6050. [PubMed] [Google Scholar]
- 18.Kunkel T. A. The mutational specificity of DNA polymerase-β during in vitro DNA synthesis. J. Biol. Chem. 1985;260:5787–5796. [PubMed] [Google Scholar]
- 19.Burgers P. M., Koonin E. V., Bruford E., Blanco L., Burtis K. C., Christman M. F., Copeland W. C., Friedberg E. C., Hanaoka F., Hinkle D. C., et al. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem. 2001;276:43487–43490. doi: 10.1074/jbc.R100056200. [DOI] [PubMed] [Google Scholar]
- 20.Sakaguchi K., Boyd J. B. Purification and characterization of a DNA polymerase β from Drosophila. J. Biol. Chem. 1995;260:10406–10411. [PubMed] [Google Scholar]
- 21.Aoyagi N., Ohshige M., Hirose F., Kuroda K., Matsukage A., Sakaguchi K. DNA polymerase ε from Drosophila melanogaster. Biochem. Biophys. Res. Commun. 1997;230:297–301. doi: 10.1006/bbrc.1996.5945. [DOI] [PubMed] [Google Scholar]
- 22.Murakumo Y., Ogura Y., Ishii H., Numata S., Ichihara M., Croce C. M., Fishel R., Takahashi M. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J. Biol. Chem. 2001;276:35644–35651. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 23.Nelson J. R., Lawrence C. W., Hinkle D. C. Thymine–thymine dimer bypass by yeast DNA polymerase ζ. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 24.Banks G. R., Boezi J. A., Lehman I. R. A high molecular weight DNA polymerase from Drosophila melanogaster embryos. Purification, structure, and partial characterization. J. Biol. Chem. 1979;254:9886–9892. [PubMed] [Google Scholar]
- 25.Wernette C. M., Kaguni L. S. A mitochondrial DNA polymerase from embryos of Drosophila melanogaster. Purification, subunit structure, and partial characterization. J. Biol. Chem. 1986;261:14764–14770. [PubMed] [Google Scholar]
- 26.Bamabara R. A., Uyemura D., Choi T. On the processive mechanism of Escherichia coli DNA polymerase I. Quantitative assessment of processivity. J. Biol. Chem. 1978;253:413–423. [PubMed] [Google Scholar]
- 27.Thomas D. C., Roberts J. D., Sabatino R. D., Myers T. W., Tan C. K., Downey K. M., So A. G., Bambara R. A., Kunkel T. A. Fidelity of mammalian DNA replication and replicative DNA polymerases. Biochemistry. 1991;30:11751–11759. doi: 10.1021/bi00115a003. [DOI] [PubMed] [Google Scholar]
- 28.Osheroff W. P., Jung H. K., Beard W. A., Wilson S. H., Kunkel T. A. The fidelity of DNA polymerase β during distributive and processive DNA synthesis. J. Biol. Chem. 1999;274:3642–3650. doi: 10.1074/jbc.274.6.3642. [DOI] [PubMed] [Google Scholar]
- 29.Johnson R. E., Washington M. T., Haracska L., Prakash S., Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature (London) 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 30.Johnson R. E., Prakash S., Prakash L. Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, Polη. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 31.Masutani C., Kusumoto R., Iwai S., Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson R. E., Haracska L., Prakash S., Prakash L. Role of DNA polymerase η in the bypass of a (6-4) TT photoproduct. Mol. Cell. Biol. 2001;21:3558–3563. doi: 10.1128/MCB.21.10.3558-3563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Todo T., Takemori H., Ryo H., Ihara M., Matsunaga T., Nikaido O., Sato K., Nomura T. A new photoreactivating enzyme that specifically repairs ultraviolet light-induced (6–4)photoproducts. Nature (London) 1993;361:371–374. doi: 10.1038/361371a0. [DOI] [PubMed] [Google Scholar]
- 34.Murakumo Y., Roth T., Ishii H., Rasio D., Numata S., Croce C. M., Fishel R. A human REV7 homolog that interacts with the polymerase ζ catalytic subunit hREV3 and the spindle assembly checkpoint protein hMAD2. J. Biol. Chem. 2000;275:4391–4397. doi: 10.1074/jbc.275.6.4391. [DOI] [PubMed] [Google Scholar]
- 35.Masuda Y., Ohmae M., Masuda K., Kamiya K. Structure and enzymatic properties of a stable complex of the human REV1 and REV7 proteins. J. Biol. Chem. 2003;278:12356–12360. doi: 10.1074/jbc.M211765200. [DOI] [PubMed] [Google Scholar]