Abstract
Avian pathogenic Escherichia coli (APEC) cause avian colibacillosis and accurately distinguishing infectious isolates is critical for controlling its transmission. Multilocus sequence typing (MLST) is an accurate and efficient strain identification method for epidemiological surveillance. This research aimed to develop a fast and high-throughput workflow that simultaneously sequences the Achtman typing scheme's 7 housekeeping genes of multiple E. coli isolates using the Oxford Nanopore Technologies (ONT) platform for large-scale APEC study. E. coli strains were isolated from poultry farms, the housekeeping genes were amplified, and amplicons were sequenced on an R9.4 MinION flow cell using the Nanopore GridION sequencer (ONT, Oxford, UK) following the initial workflow (ONT-MLST). Moreover, the workflow was revised by introducing large-scale DNA extraction and multiplex PCR into the ONT-MLST workflow and applied to 242 new isolates, 18 isolates from the previous workflow, and 5 ATCC reference strains using Flongle flow cell on the Nanopore MinION Mk1C sequencer (ONT, Oxford, UK). Finally, the sequence type (ST) results of the 308 isolates collected from infected chickens and poultry farm environments were reported and analyzed. Data indicated that E. coli belonging to ST159, ST8578, and ST355 have the potential to infect multiple organs in broiler. In addition, zoonotic STs, ST69, ST10, ST38, and ST131, were detected from poultry farms. With the advantages of the high throughput of ONT, this study provides a rapid workflow for large-scale E. coli typing and identified frequently isolated sequence types related to APEC infection in poultry.
Key words: avian Escherichia coli, Oxford Nanopore, large-scale, high-throughput, field study
INTRODUCTION
Avian pathogenic Escherichia coli (APEC) is an extra-intestinal pathogenic E. coli that causes avian colibacillosis, which is one of the leading causes of mortality and morbidity in poultry (Fancher et al., 2020; Alber et al., 2021; Kathayat et al., 2021). Additionally, some subsets of APEC, O18:K1:H7 for example, has been detected in human samples, and constitutes a potential zoonotic risk (Moulin-Schouleur et al., 2006; Zhuge et al., 2021). Identifying and accurately distinguishing the strains of infectious pathogens is key to monitoring and controlling APEC transmission (Kobayashi et al., 2021).
Over the previous decades, numerous molecular approaches including PCR, Sanger sequencing, and Illumina sequencing have been actively utilized to identify isolates and localize disease outbreaks (Iguchi et al., 2015; Cole et al., 2019; Moore et al., 2021). Still, the lack of portability along with high labor and sequencing costs limited their usage in pathogen epidemiology. Multilocus sequence typing (MLST) is a nucleotide sequence-based approach utilized broadly for characterizing and subtyping E. coli strains (Adiri et al., 2003; Peng et al., 2019). The MLST based on Achtman typing scheme (Wirth et al., 2006) uses primer sets designed for PCR amplification of 7 conserved housekeeping genes using high-fidelity DNA polymerase, followed by sequencing of the PCR amplicons. Sequence information is then compared to the MLST database to obtain an allele number. Combining the 7 allele numbers determine the isolated sample's sequence type (ST). ST information can be used to trace the spread of infections caused by E. coli, monitor the emergence of new virulent strains, and identify outbreaks caused by a common source. In addition, E. coli ST can also support rapid genomic analysis of antibiotic resistance patterns, enabling the tracking of antibiotic-resistant strains and the development of more effective treatments (Brehony et al., 2019). Overall, E. coli MLST is an important tool for the study, control, and public health management of E. coli infections. Controlling the transmission of APEC demands comprehensive and timely genetic analyses to accurately map and predict its spread. A high-throughput MLST workflow is essential to promptly process a multitude of samples, allowing for rapid interventions in poultry populations and mitigating potential threats to the poultry industry.
Traditional MLST studies relied on Sanger sequencing to obtain the sequences of housekeeping genes. Although reliable, Sanger sequencing involves laborious bench work and is limited to sequencing a single molecule at a time. These limitations have led researchers to explore other sequencing alternatives. As the era moves to third-generation sequencing, the Oxford Nanopore Technologies (ONT) system emerges as a promising alternative to both Sanger and second-generation sequencing approaches, especially for high-throughput, long-read sequencing studies (Liao et al., 2022). The ONT system is capable of producing sequence reads up to 4 Mb in length and comes with the advantage of reduced costs (Enright and Spratt, 1999; Doumith et al., 2015); however, the primary concern with this method is its relatively high sequencing error rates. This poses a challenge for MLST analysis, as even a single nucleotide variation in the 7 housekeeping genes can lead to a distinct sequencing type.
To investigate the practical application of the ONT system in APEC research settings, we have refined and optimized a method for large-scale E. coli MLST sequencing (ONT-MLST). We evaluated the method for its accuracy in ST identification by comparison with the results obtained from Sanger MLST sequencing and Illumina whole genome sequencing. We then applied the ONT-MLST method to a large number of E. coli isolates to survey several broiler farms in Mississippi. The present study encompasses 2 primary objectives: firstly, to thoroughly investigate the accuracy and feasibility of employing the ONT for extensive MLST analysis; and secondly, to facilitate large-scale MLST study of APEC distribution and transmission in poultry.
MATERIALS AND METHODS
Bacterial Strains and DNA Extraction
The E. coli strains were isolated during 2018-2021 from broiler farms in north-central Mississippi. A total of 308 E. coli isolates were collected, including 263 disease-related strains collected from typical lesions of broilers showing colibacillosis, and 45 asymptomatic strains collected from litter, feces, and cloacal swab samples from broiler farms. Samples were cultured on MacConkey agar overnight at 37°C and selected bacterial colonies were further isolated on LB agar. For each isolate, 1.5 mL of LB broth cell culture from overnight incubation at 37°C was pelleted in a microcentrifuge tube and subjected to genomic DNA isolation.
The 308 isolates were split into 2 sets: the Beta set contained 66 isolates, and the Pro set contained 242 isolates. For the Pro set, an additional 18 isolates from the Beta set and 5 ATCC reference strains were also sequenced and served as sequencing references. The genomic DNA (gDNA) for the Beta set was extracted using the GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific, Waltham, MA) following the manufacturer's instructions. The Zymo Research Quick-DNA 96 Kit (Zymo Research, Irvine, CA) was used to extract the gDNA for the Pro set following the manufacturer's instructions. The quality of the extracted DNA was examined using 0.8% w/v agarose gel electrophoresis, and the DNA purity and concentrations were determined using a NanoDrop 1 spectrophotometer (Thermo Scientific, Wilmington, DE).
Primer Design and PCR Amplification
MLST was carried out using the Achtman typing scheme with 7 housekeeping genes: adenosine kinase (adk), fumarate hydratase (fumC), DNA gyrase subunit B (gyrB), isocitrate dehydrogenase (icd), malate dehydrogenase (mdh), purine-rich element binding protein A (purA), DNA recombination/repair protein (recA) (Wirth et al., 2006). We modified 3 primer sets (fumC, purA, and recA) using degenerate oligonucleotide primers to broaden the target specificity of amplification based on E. coli genomic similarity of our former whole-genome sequencing results (Jia et al., 2024). All primers, including updated primers, are listed in Table 1.
Table 1.
The information on primers used for 7 MLST housekeeping genes.
| Housekeeping gene |
Primer |
||||
|---|---|---|---|---|---|
| Target1 | Abbreviation | Orientation | Sequence (5′–3′) | Length (nt) | Amplicon size (bp) |
| adenylate kinase | adk | Forward | ATTCTGCTTGGCGCTCCGGG | 20 | 584 |
| Reverse | CCGTCAACTTTCGCGTATTT | 20 | |||
| fumarate hydratase | fumC | Forward | ACAGGTCGCMAGCGCTTCAA | 20 | 737 |
| Reverse | CTCARCGCTCGCTGGAGCATT | 21 | |||
| DNA gyrase subunit B | gyrB | Forward | TCGGCGACACGGATGACGGC | 20 | 880 |
| Reverse | ATCAGGCCTTCACGCGCATC | 20 | |||
| isocitrate dehydrogenase | icd | Forward | ATGGAAAGTAAAGTTGTTCCGGCACA | 26 | 878 |
| Reverse | GGACGCAGCAGGATCTGTT | 19 | |||
| malate dehydrogenase | mdh | Forward | ATGAAAGTCGCAGGCGCTGCTGGCGG | 26 | 932 |
| Reverse | TTAACGAACTCCTCGATATCTTTCTT | 26 | |||
| adenylosuccinate dehydrogenase | purA | Forward | GCCGCGCTGATGAAAGAGATG | 21 | 820 |
| Reverse | GCATACGGTAAGCCACRCAGA | 21 | |||
| DNA recombination/repair protein | recA | Forward | CGCATTCGCTTTACCYTGACC | 21 | 734 |
| Reverse | TCGTCGAAATCTACGGACCRGA | 22 | |||
The reference sequence was based on Escherichia coli str. K-12 substr. MG1655, complete genome (Accession No.: NC_000913.3) and avian pathogenetic E. coli whole-genome sequencing results conducted for this study.
For the Beta set, 7 housekeeping genes were individually amplified using 30 ng gDNA template, Phusion High-Fidelity PCR Master Mix (Thermo Fisher Scientific, Waltham, MA), and forward and reverse primers (Table 1) in a total volume of 15 µL with an annealing temperature set at 65℃. The yield and quality of each PCR amplicon were then assessed using agarose gel electrophoresis.
To decrease the number of PCR reactions needed, 2 multiplexed PCR assays were created and used with the Pro set: genes adk, fumC, and gyrB were multiplex amplified in a 25-µL reaction; genes purA, icd, mdh, and recA were multiplex amplified in a 35-µL reaction. In each reaction, 1 or 2 µL of gDNA without concentration normalization was used. Each reaction was run with an annealing temperature of 60℃. The detailed reaction components for each multiplex PCR amplification are shown in Table 2. Again, the yield and quality of the amplicons were assessed using agarose gel electrophoresis.
Table 2.
Optimized multiplex PCR reactions targeting for 7 housekeeping gene alleles used in E. coli MLST analysis.
| mpxPCR1 (3 genes) |
mpxPCR1 (4 genes) |
||
|---|---|---|---|
| Component | Volume (µL) | Component | Volume (µL) |
| Phusion High-Fidelity PCR Master Mix (2×) | 12.5 | Phusion High-Fidelity PCR Master Mix (2×) | 17.5 |
| adk Forward Primer (10 µM) | 0.25 | icd Forward Primer (10 µM) | 0.3 |
| adk Reverse Primer (10 µM) | 0.25 | icd Reverse Primer (10 µM) | 0.3 |
| fumC Forward Primer (10 µM) | 0.25 | mdh Forward Primer (10 µM) | 0.5 |
| fumC Reverse Primer (10 µM) | 0.25 | mdh Reverse Primer (10 µM) | 0.5 |
| gyrB Forward Primer (10 µM) | 0.25 | purA Forward Primer (10 µM) | 0.3 |
| gyrB Reverse Primer (10 µM) | 0.25 | purA Reverse Primer (10 µM) | 0.3 |
| H2O | 9.6 | recA Forward Primer (10 µM) | 0.5 |
| gDNA template | 1 | recA Reverse Primer (10 µM) | 0.5 |
| Total | 25 | H2O | 12.3 |
| gDNA template | 2 | ||
| Total | 35 | ||
Library Preparation and Sequencing
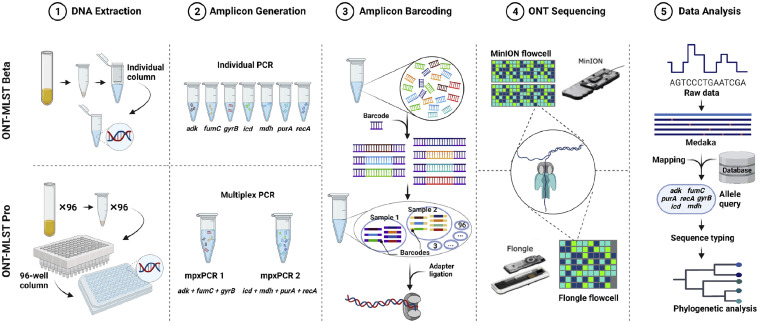
For both sets of isolates, the amplicons from the same isolate were pooled together and purified with AMPure XP beads (Beckman Coulter, Brea, CA). Nanopore barcoded amplicon libraries were prepared using the pooled and cleaned amplicons with the Ligation Sequencing Kit (SQK-LSK109; Oxford Nanopore Technologies, Oxford, UK) and Native Barcoding Expansion Kit (EXP-NBD196; Oxford Nanopore Technologies, Oxford, UK) according to the manufacturer's instructions. All 66 isolates of the Beta set were pooled and run on a single MinION R9.4 flow cell using the Nanopore GridION sequencer (Oxford Nanopore Technologies, Oxford, UK). The Pro isolate set was split into 4 pools and sequenced on 4 Flongle flow cells using the portable Nanopore MinION Mk1C sequencer (Oxfornd Nanopore Technologies, Oxford, UK). The generated reads were basecalled using Guppy (v4.3.4, ONT) with the high-accuracy base-calling model, and then the Medaka Consensus pipeline (v.1.3.2, ONT) was used with the first 4 thousand reads to determine the consensus sequences of each allele using the sequences of 7 housekeeping genes from the E. coli representative NC_002695.2 in the NCBI Ref-seq database. The allele profile numbers for the consensus sequences, along with the strain ST results, were identified using PubMLST (Jolley et al., 2018). An illustration of variations between both isolate sets is shown in Figure 1.
Figure 1.
Workflow of multilocus sequence typing (MLST) using Oxford Nanopore Technologies (ONT). A general workflow for the DNA extraction, amplicon generation, amplicon barcoding, ONT sequencing, and data analysis.
Validation
The Illumina whole genome sequencing for the 66 isolates in the Beta set was previously conducted in our lab (data not shown). SRST2 v0.2.0 (Inouye et al., 2014) was run with the raw Illumina data for each isolate to call the allele profile and ST number. The Illumina-based allele profile and ST number served as the standard for verifying the accuracy of ONT-MLST.
To test the effect of read depth on the accuracy of the allele and ST classification, the Nanopore reads from the Beta set were randomly subsampled and reclassified. In brief, seqtk (v1.4-r130-dirty) was used to subset each sample at 8 read depths (4,000, 1,000, 800, 400, 100, 80, 40, and 10). Each subset was run through the medaka consensus pipeline as previous description, the consensus sequences were aligned using blastn (v2.14.0+) to all known alleles in the Pub-MLST database to find the most likely allele profile. The allele profiles were then compared to all profiles available at Pub-MLST for exact ST matches. The subsets were compared with the SRST2 classification and graphed with R v4.2.2 (R Core Team, 2013) and the tidyverse package v2.0.0 (Wickham et al., 2019).
Additionally, Sanger sequencing was performed to validate the accuracy of both the Illumina and Nanopore sequencing, as Sanger is the traditional sequencing method for MLST studies. The PCR products of 5 isolates among the 18 samples that are in both the Beta and Pro sets were chosen for sequencing (Eurofins Genomics; https://eurofinsgenomics.com/en/home/). In brief, the PCR products of 7 housekeeping genes were purified using the GeneJET PCR Purification Kit (ThermoFisher Scientific, Waltham, MA, USA) and subsequently cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) according to manufacturers’ protocols. For each PCR product amplified from a gene-specific primer pair, 3 independent recombinant plasmid DNAs were sent to Eurofins Genomics (Louisville, KY, USA) for Automatic Sanger Sequencing analysis. Again, PubMLST was used to identify the allele profile and ST number of each strain from the sequences.
Allele Comparisons Among Called ST Types
For each unique ST found in the samples, a concatenated sequence was created using the allele sequences of the ST allele profile. These sequences were aligned using MAFFT v7.522 (Katoh and Standley, 2013) and those alignments were used to create a neighbor-joining tree with the R package ape v5.7-1 (Paradis and Schliep, 2018). The tree, allele sequence plot, and heatmap were visualized using R (v4.2.2) along with the Biostrings (v2.66.0), tidyverse (v2.0.0), ape (v5.7-1), tidytree (v0.4.5), ggtree (3.6.2), and aplot (0.2.1) packages (Yu, 2020; Yu, 2022; Yu et al., 2017).
RESULTS AND DISCUSSION
ONT is a Promising Strategy for MLST Analysis
An illustration of the methods for ONT-MLST using the Beta set is shown in Figure 1, upper part. Out of the 66 characterized isolates, 32 distinct sequence types were identified. The sequencing results summary is shown in Table 3. The allele assignments (Supplementary Table 1) for the Beta set were completely consistent with the 5 isolates sequenced using the traditional Sanger method and were largely consistent with those obtained by Illumina sequencing (459 out of 462 alleles matched; 99.35%). Discrepancies were observed for 3 alleles from 2 isolates, fumC and mdh in isolate 6897, and recA in isolate MS1696.
Table 3.
Summary of the ONT sequencing results from ONT-MLST Beta workflow.
| Item | Nanopore parameter |
|---|---|
| Run summary | |
| Reads generated | 11.91 M |
| Passed bases | 11.41 Gb |
| Failed bases | 1.08 Gb |
| Mean length (bp) | 868.05 |
| No. of reads per barcode | |
| Minimum | 87,292 |
| Maximum | 242,574 |
| Mean | 144,998.77 |
| SD | 30481.13 |
| Reads quality per barcode (Q score) | |
| Minimum | 11.414 |
| Maximum | 11.952 |
| Mean | 11.797 |
| SD | 0.103 |
To further investigate these inconsistencies, 10 independent replicates for each of the 3 genes were Sanger sequence using the method described above. The replicates of the 3 genes sequenced aligned to multiple alleles (3 in fumC, 2 in mdh and recA; Supplementary Figure 1) indicating that the original isolates were not singular E. coli strains. The findings underline the importance of attaining pure E. coli cultures to ensure accurate sequence typing.
The primary cause of these allele inconsistencies can be traced back to the preculture phase, deeming it a systematic error. Such errors can be potentially mitigated by employing rigorous culture processes and thorough colony screening. Overall, our findings confirm the capability of the ONT-MLST workflow to produce reliable sequencing data for E. coli sequence typing.
In traditional Sanger sequencing, each housekeeping gene of every isolate requires individual PCR reaction due to the necessity for pure sequencing material. This implies distinct reactions, gel electrophoresis, clean-up processes, and sequencing preparations for each gene. Conversely, the ONT sequencing strategy is more streamlined. During library preparation, the 7 amplicons from an isolate are combined and tagged with a specific barcode sequence (Liou et al., 2020). This system leverages barcoding for each isolate, allowing for pooling multiple PCR products into a singular tube, followed by one consolidated clean-up step. This not only enhances efficiency but also considerably reduces the labor intensity compared to Sanger sequencing.
A Large-Scale Workflow for MLST Study
To facilitate the large-scale MLST sequencing of the 265 isolates, we made modifications for Pro set to 3 pivotal steps: DNA extraction, PCR, and the usage of Nanopore flow cell type. This modified MLST workflow can be visualized in the lower section of Figure 1, labeled as "ONT-MLST Pro."
The use of 96-well DNA extraction plate simplified the process for large-scale E. coli research compared to individual DNA extraction column method. However, while this high-throughput method streamlines the DNA extraction process, it tends to produce DNA with lower yield and purity (Supplementary Table 2 and Supplementary Figure 2). The DNA concentration from high-throughput DNA extraction method ranged from 3.13 to 97.75 ng/µL, with the purity A260/A280 and A260/A230 ranging from 1.34 to 2.08 and from 0.07 to 2.20, respectively. Unlikely, spin-column method yield DNA ranged from 32.30 to 138.50 ng/µL, with the purity A260/A280 and A260/A230 ranging from 1.70 to 1.93 and from 1.59 to 2.62, respectively (data not shown). Such compromises on DNA purity could potentially impact the efficiency of subsequent PCR and sequencing stages.
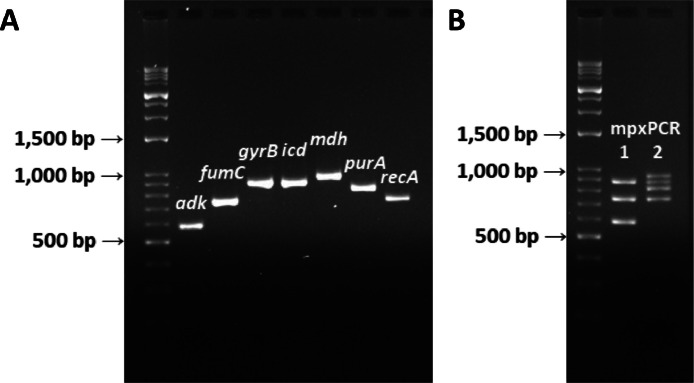
In the ONT-MLST Pro workflow, we finely optimized the efficiency of multiplex PCR reactions by grouping 7 housekeeping genes into 2 multiplex PCR reactions and adjusting the recipes of reagents in each multiplex PCR reaction (Table 2). The combination of genes in each multiplex PCR reaction was based on the amplicon size, primer efficiency, and the competition relations of the primers. The gel image of PCR products from both single PCR and multiplex PCR is shown in Figure 2. Seven housekeeping genes can be visualized on a 2% (w/v) agarose gel, and every DNA sample extracted using the 96-well extraction plate was successfully amplified (data not shown). While transitioning from individual to multiplex PCR, there was a significant reduction in reagent usage; specifically, 42% less Phusion High-Fidelity PCR Master Mix was needed (52.5 µL for individual PCR vs. 30 µL for multiplex PCR).
Figure 2.
Agarose gel image of the PCR products from single PCR (A) and multiplex PCR (B) of 1 E. coli isolate (MS1494). Primer ratio was adjusted based on efficiency. Ladder: VersaLadder (Gold Biotechnology, St. Louis, MO). Gel: 2 % (w/v) agarose gel.
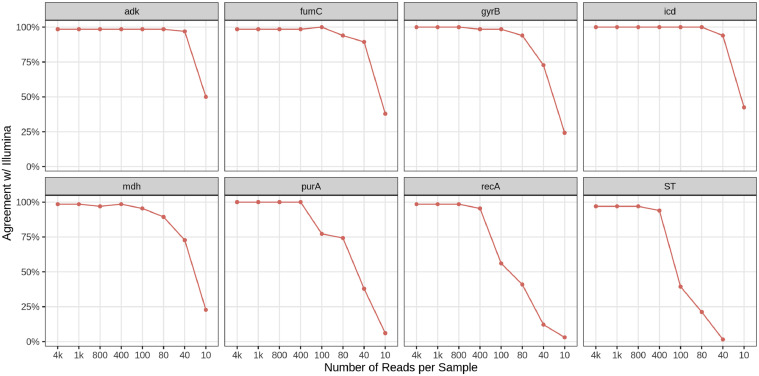
Sequencing the 66 isolates of the Beta set on a MinION flow cell produced approximately 145,000 reads per sample, or 20,714 reads per allele (Table 3). The read depth simulation results revealed that a total read depth of 400 sequences was adequate to accurately classify every allele in the ST identification for each isolate (Figure 3), excluding the 2 isolates that are likely not single strains. Due to the limited number of native barcodes available, the ONT-MLST protocol can process a maximum of 96 samples, which require a total of 38,400 (96 samples × 400 reads) reads for accurate classification. Based on the estimation, we switched MinION flow cell to Flongle flow cell. The actual sequencing results also supported the simulation results. Given the portable feature of Flongle flow cell and MinION Mk1C sequencer, the ONT-MLST workflow has the potential for evolution into an on-site sequencing protocol, which could eliminate the need for sample preservation, shipping procedures, and reduce associated time constraints and potential risks.
Figure 3.
Simulation of accuracy of the allele and ST calls for each subsample of the ONT compared to the SRST2 (Illumina) output. The sequencing data of 66 E. coli isolates from ONT-MLST Beta workflow and Illumina platform was used in this in silico simulation analysis.
In order to evaluate the performance of the updated ONT-MLST workflow on the downstream MLST analysis, we also included 16 randomly chosen isolates (Supplementary Table 1) from the Beta set. The results showed that these 16 isolates received the same ST results in the updated protocol, which indicated that downstream MLST results remained unaffected by the low DNA yield, DNA impurities, uneven PCR product distribution, and fewer sequence reads.
Field Application Reveals Disease-Related Sequence Types in Poultry
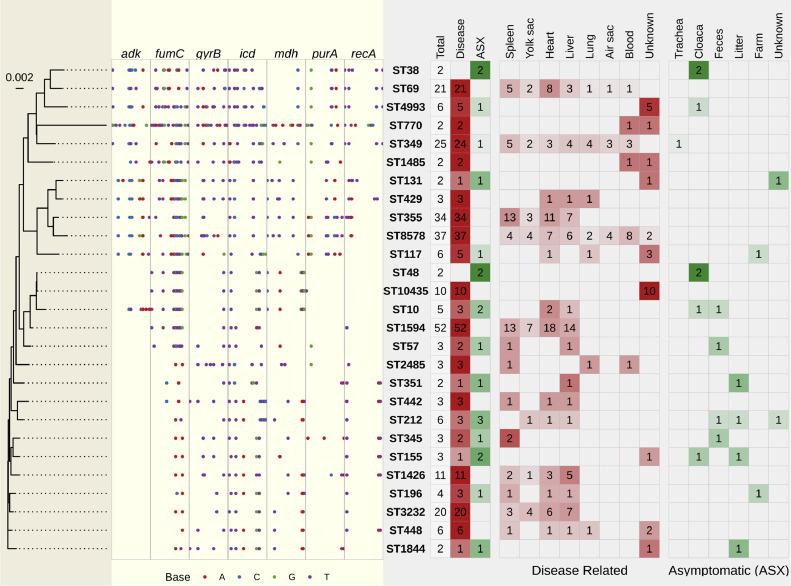
To conduct an extensive application of MLST in practical research related to the distribution and transmission of APEC in Mississippi, a total of 308 E. coli isolates collected from lesions of broilers showing the symptoms of colibacillosis and tissues of asymptomatic broilers and poultry environment were analyzed. A total of 27 STs were detected more than once, comprising 255 disease-related isolates and 20 asymptomatic isolates, accounting for 89.29% (275/308) of the total isolates. Figure 4 shows the SNP-based phylogenetic analysis, SNP variations among STs, and ST distribution of isolates for those of STs have more than one isolate.
Figure 4.
Phylogenetic analysis of 27 sequence types (ST) and the summary of source and count of isolates. Light wheat panel shows phylogenetic tree based on the concatenated sequences of the 7 housekeeping genes, the scale bar refers to a phylogenetic distance of 0.002; light yellow panel shows the sequence alignment of the 7 housekeeping genes, highlighting bases that are not the most common base in that position with red dots representing adenine (A), blue dots representing cytosine (C), green dots representing guanine (G), and purple dots representing thymine (T); and the light gray panel shows the sample source and isolate counts for each ST, with red squares indicating disease-related isolates and green squares indicating asymptomatic (ASX) isolates. The numbers in each square represent the isolate count for the corresponding category. The shade of each square represents the proportion (or frequency) of the total isolate count (isolate count/total isolates) for each sequence type (ST).
The most frequent sequence types among disease-related E. coli isolates were ST1594 (52 isolates), ST8578 (37 isolates), and ST355 (34 isolates) (Figure 4), accounting for 39.3% of all disease-related E. coli isolates. Of note, isolates with ST8578 were identified from 7 different organs of infected birds, including spleen, yolk sac, heart, liver, lung, air sac, and blood, highlighting its widespread distribution in the infected birds. Interestingly, despite the genetic similarities between ST8578 and ST355 (Figure 4), ST355 was not isolated from lung, air sac, or blood. Conversely, ST355 and ST1594 are genetically distinct based on the SNP analysis from the 7 housekeeping genes; however, these 2 STs exhibit similar infectious characteristics. This phylogenetic incongruence is common in E. coli because its genomic and phenotypic diversity (Leimbach et al., 2013), thus an accurate classification system, such as MLST, should be used to distinguish different sequence types. All 3 of these STs were commonly seen in spleen, yolk sac, heart, and liver of infected birds. Interestingly, these 3 STs were absent in asymptomatic isolates, possibly indicating a potent infectious capability but potentially limited spread inside poultry environments.
One of the objectives of this study was to investigate the APEC distribution and transmission in poultry farms of Mississippi. Among all disease-related isolates, twelve STs (ST131, ST117, ST4993, ST349, ST351, ST345, ST155, ST196, ST1844, ST212, ST57, and ST10) were also identified in asymptomatic chicken trachea, cloaca, feces, and litter, indicating a potential transmission of these STs via environment. Environmental disinfection is suggested to control the transmission of these twelve STs.
We found 3 specific STs (ST69, ST10, and ST38) that have also been previously reported as isolates from urine samples of patients diagnosed with urinary tract infection (Matsui et al., 2020), and ST131 has been reported as major cause of antimicrobial-resistant E. coli infections in the United States (Johnson et al., 2010), raising concerns about zoonotic transmission through poultry consumption.
Limitations
The MLST methodology is not exclusive to E. coli, the method can also be employed to identify other pathogens related to animal health or food safety (Ahmed et al., 2006; Salcedo et al., 2003) in the future. The high-throughput ONT-MLST workflow offers a rapid and cost-effective method for large-scale MLST studies. However, for studies involving a small number of samples, the traditional MLST method may still be the preferred choice.
CONCLUSION
In summary, this research has successfully established a workflow using Oxford Nanopore Technology (ONT) for identifying avian pathogenic Escherichia coli (APEC) strains through multilocus sequence typing (MLST). By simultaneously sequencing 7 housekeeping genes from multiple E. coli isolates, this approach proved invaluable for epidemiological surveillance and controlling APEC transmission. Notably, strains like ST1594, ST8578, and ST355 demonstrated the potential to infect multiple chicken organs and considered predominant STs in APEC isolates. The ONT-MLST workflow offers promise in revolutionizing large-scale epidemiological surveillance, setting the stage for its potential adoption as a standard lab protocol, thereby providing farms with vital data to guide treatments, vaccinations, and sanitation measures.
Acknowledgments
ACKNOWLEDGMENTS
This publication is a contribution of the project funded by USDA-ARS NACA 58-6064-2-014, USDA-ARS NACA 58-6066-0-064, USDA-ARS NACA 58-6064-9-014, USDA ARS CRIS project NE-1942/MIS-322430, and the Mississippi Agricultural and Forestry Experiment Station.
DISCLOSURES
The authors have declared no conflict of interest.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2024.104067.
Appendix. Supplementary materials
Supplementary Figure 1 Summary plot of DNA concentration and purity (based on A260/A280 and A260/A230 ratios) determined by Nanodrop 1 spectrophotometer (Thermo Scientific, Wilmington, DE). The genomic DNA extracted with a large-scale format using Zymo Research Quick-DNA 96 Kit (Zymo Research, Irvine, CA) exhibited a wide range display in both concentration and purity aspects of genomic DNA.
Supplementary Figure 2 The impact of impure isolate on Illumina and ONT-MLST sequencing outcomes, resulting in variations in allele numbers. The figure shows disparities in isolates 6897 (genes fumC and mdh) and MS1696 (gene recA) based on the comparison/alignment of single nucleotide polymorphisms (SNPs) of allele sequences retrieving from Illumina, ONT-MLST Beta, and Sanger sequencing methods. Total of 10 independent recombinant plasmid DNA samples from each gene allele were sequenced with Sanger platform. Sanger sequencing results were served as the “golden standard” to confirm the sequence of each allele. A: Adenine, red blocks; C: cytosine, blue blocks; G: guanine, green blocks; T: thymine, purple blocks.
REFERENCES
- Adiri R.S., Gophna U., Ron E.Z. Multilocus sequence typing (MLST) of Escherichia coli O78 strains. FEMS Microbiol. Lett. 2003;222:199–203. doi: 10.1016/S0378-1097(03)00295-7. [DOI] [PubMed] [Google Scholar]
- Ahmed N., Devi S.M., Valverde Mde L., Vijayachari P., Machang'u R.S., Ellis W.A., Hartskeerl R.A. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann. Clin. Microbiol. Antimicrob. 2006;5:28. doi: 10.1186/1476-0711-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber A., Stevens M.P., Vervelde L. The bird's immune response to avian pathogenic Escherichia coli. Avian Pathol. 2021;50:382–391. doi: 10.1080/03079457.2021.1873246. [DOI] [PubMed] [Google Scholar]
- Brehony C., McGrath E., Brennan W., Tuohy A., Whyte T., Brisse S., Maiden M., Jolley K., Morris D., Cormican M. An MLST approach to support tracking of plasmids carrying OXA-48-like carbapenemase. J. Antimicrob. Chemother. 2019;74:1856–1862. doi: 10.1093/jac/dkz136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole B.K., Ilikj M., McCloskey C.B., Chavez-Bueno S. Antibiotic resistance and molecular characterization of bacteremia Escherichia coli isolates from newborns in the United States. PLoS One. 2019;14 doi: 10.1371/journal.pone.0219352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumith M., Day M., Ciesielczuk H., Hope R., Underwood A., Reynolds R., Wain J., Livermore D.M., Woodford N. Rapid identification of major Escherichia coli sequence types causing urinary tract and bloodstream infections. J. Clin. Microbiol. 2015;53:160–166. doi: 10.1128/JCM.02562-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright M.C., Spratt B.G. Multilocus sequence typing. Trends Microbiol. 1999;7:482–487. doi: 10.1016/s0966-842x(99)01609-1. [DOI] [PubMed] [Google Scholar]
- Fancher C.A., Zhang L., Kiess A.S., Adhikari P.A., Dinh T.T.N., Sukumaran A.T. Avian pathogenic Escherichia coli and Clostridium perfringens: Challenges in no antibiotics ever broiler production and potential solutions. Microorganisms. 2020;8:1533. doi: 10.3390/microorganisms8101533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi A., Iyoda S., Seto K., Morita-Ishihara T., Scheutz F., Ohnishi M. Escherichia coli O-genotyping PCR: a comprehensive and practical platform for molecular o serogrouping. J. Clin. Microbiol. 2015;53:2427–2432. doi: 10.1128/JCM.00321-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Dashnow H., Raven L.A., Schultz M.B., Pope B.J., Tomita T., Zobel J., Holt K.E. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome. Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L., Arick M.A., Hsu C.Y., Peterson D.G., Evans J.D., Robinson K., Adhikari P., Zhang L. Complete genome sequences of two avian pathogenic Escherichia coli strains isolated from broilers exhibiting colibacillosis in Mississippi. Microbiol. Resour. Announc. 2024;13 doi: 10.1128/mra.01020-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.R., Johnston B., Clabots C., Kuskowski M.A., Castanheira M. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 2010;51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- Jolley K.A., Bray J.E., Maiden M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome. Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathayat D., Lokesh D., Ranjit S., Rajashekara G. Avian pathogenic Escherichia coli (APEC): An overview of virulence and pathogenesis factors, zoonotic potential, and control strategies. Pathogens. 2021;10:467. doi: 10.3390/pathogens10040467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecul. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Ikeda M., Okada Y., Higurashi Y., Okugawa S., Moriya K. Clinical and microbiological characteristics of recurrent Escherichia coli bacteremia. Microbiol. Spectr. 2021;9 doi: 10.1128/Spectrum.01399-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimbach A., Hacker J., Dobrindt U. E. coli as an all-rounder: the thin line between commensalism and pathogenicity. Curr. Top Microbiol. Immunol. 2013;358:3–32. doi: 10.1007/82_2012_303. [DOI] [PubMed] [Google Scholar]
- Liao Y.C., Wu H.C., Liou C.H., Lauderdale T.Y., Huang I.W., Lai J.F., Chen F.J. Rapid and routine molecular typing using multiplex polymerase chain reaction and MinION sequencer. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.875347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou C.H., Wu H.C., Liao Y.C., Yang Lauderdale T.L., Huang I.W., Chen F.J. NanoMLST: Accurate multilocus sequence typing using Oxford Nanopore Technologies MinION with a dual-barcode approach to multiplex large numbers of samples. Microb. Genom. 2020;6 doi: 10.1099/mgen.0.000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y., Hu Y., Rubin J., de Assis R.S., Suh J., Riley L.W. Multilocus sequence typing of Escherichia coli isolates from urinary tract infection patients and from fecal samples of healthy subjects in a college community. Microbiologyopen. 2020;9:1225–1233. doi: 10.1002/mbo3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.E.G., Paulin-Curlee G., Johnston B.D., Clabots C., DebRoy C., Johnson T.J., Weber B., Porter S., Armién A.G., Johnson J.R. Molecular characteristics, ecology, and zoonotic potential of Escherichia coli strains that cause hemorrhagic pneumonia in animals. Appl. Environ. Microbiol. 2021;87 doi: 10.1128/AEM.01471-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin-Schouleur M., Schouler C., Tailliez P., Kao M.R., Brée A., Germon P., Oswald E., Mainil J., Blanco M., Blanco J. Common virulence factors and genetic relationships between O18:K1:H7 Escherichia coli isolates of human and avian origin. J. Clin. Microbiol. 2006;44:3484–3492. doi: 10.1128/JCM.00548-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Schliep K. Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2018;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- Peng Z., Liang W., Hu Z., Li X., Guo R., Hua L., Tang X., Tan C., Chen H., Wang X., Wu B. O-serogroups, virulence genes, antimicrobial susceptibility, and MLST genotypes of Shiga toxin-producing Escherichia coli from swine and cattle in Central China. BMC Vet. Res. 2019;15:427. doi: 10.1186/s12917-019-2177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, R. 2013. R: A language and environment for statistical computing.
- Salcedo C., Arreaza L., Alcalá B., de la Fuente L., Vázquez J.A. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 2003;41:757–762. doi: 10.1128/JCM.41.2.757-762.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H., Averick M., Bryan J., Chang W., McGowan L.D.A., François R., Grolemund G., Hayes A., Henry L., Hester J. Welcome to the Tidyverse. J. Open Source Softw. 2019;4:1686. [Google Scholar]
- Wirth T., Falush D., Lan R., Colles F., Mensa P., Wieler L.H., Karch H., Reeves P.R., Maiden M.C., Ochman H., Achtman M. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. 2020. Aplot: Decorate a ‘ggplot'with associated information. R package Version 0.0 6.
- Yu G. CRC Press; Boca Raton, FL: 2022. Data integration, manipulation and visualization of phylogenetic trees. [Google Scholar]
- Yu G., Smith D.K., Zhu H., Guan Y., Lam T.T.Y. Ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017;8:28–36. [Google Scholar]
- Zhuge X., Zhou Z., Jiang M., Wang Z., Sun Y., Tang F., Xue F., Ren J., Dai J. Chicken-source Escherichia coli within phylogroup F shares virulence genotypes and is closely related to extraintestinal pathogenic E. coli causing human infections. Transbound. Emerg. Dis. 2021;68:880–895. doi: 10.1111/tbed.13755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Summary plot of DNA concentration and purity (based on A260/A280 and A260/A230 ratios) determined by Nanodrop 1 spectrophotometer (Thermo Scientific, Wilmington, DE). The genomic DNA extracted with a large-scale format using Zymo Research Quick-DNA 96 Kit (Zymo Research, Irvine, CA) exhibited a wide range display in both concentration and purity aspects of genomic DNA.
Supplementary Figure 2 The impact of impure isolate on Illumina and ONT-MLST sequencing outcomes, resulting in variations in allele numbers. The figure shows disparities in isolates 6897 (genes fumC and mdh) and MS1696 (gene recA) based on the comparison/alignment of single nucleotide polymorphisms (SNPs) of allele sequences retrieving from Illumina, ONT-MLST Beta, and Sanger sequencing methods. Total of 10 independent recombinant plasmid DNA samples from each gene allele were sequenced with Sanger platform. Sanger sequencing results were served as the “golden standard” to confirm the sequence of each allele. A: Adenine, red blocks; C: cytosine, blue blocks; G: guanine, green blocks; T: thymine, purple blocks.