Abstract
Angiotensin-converting enzyme (ACE) is a zinc metallopeptidase that plays a major role in blood homoeostasis and reproduction in mammals. In vertebrates, both transmembrane and soluble ACE, containing one or two homologous active sites, have been characterized. So far, several ACEs from invertebrates have been cloned, but only in insects. They are soluble and display a single active site. Using biochemical procedures, an ACE-like activity was detected in our model, the leech, Theromyzon tessulatum. Annelida is the most distant phylum in which an ACE activity has been observed. To gain more insight into the leech enzyme, we have developed a PCR approach to characterize its mRNA. The approx. 2 kb cDNA has been predicted to encode a 616-amino-acid soluble enzyme containing a single active site, named TtACE (T. tessulatum ACE). Surprisingly, its primary sequence shows greater similarity to vertebrates than to invertebrates. Stable in vitro expression of TtACE in transfected Chinese-hamster ovary cells revealed that the leech enzyme is a functional metalloprotease. As in mammals, this 79 kDa glycosylated enzyme functions as a dipeptidyl carboxypeptidase capable of hydrolysing angiotensin I to angiotensin II. However, a weak chloride inhibitory effect and acetylated N-acetyl-SDKP (Ac SDAcKP) hydrolysis reveal that TtACE activity resembles that of the N-domain of mammalian ACE. In situ hybridization shows that its cellular distribution is restricted to epithelial midgut cells. Although the precise roles and endogenous substrates of TtACE remain to be identified, characterization of this ancestral peptidase will help to clarify its physiological roles in non-insect invertebrate species.
Keywords: angiotensin-converting enzyme, cloning, enzymic activity, evolution, invertebrate, leech
Abbreviations: Ac, N-acetyl; ACE, angiotensin-converting enzyme; sACE, somatic ACE; tACE, testicular (germinal) ACE; TtACE, Theromyzon tessulatum ACE; Ang I, angiotensin I; CHO cell, Chinese-hamster ovary cell; DpaOH, N3(2,4-dinitrophenyl)L-2,3-diaminopropionyl-OH; HHL, Hippuryl-His-Leu; mAb, monoclonal antibody; Mca, (7-methoxycoumarin-4-yl)acetyl-; ORF, open reading frame; RT, reverse transcriptase; 3′-UTR, 3′-untranslated region
INTRODUCTION
Angiotensin-converting enzyme (dipeptidyl-peptidase A, kininase II; EC 3.4.15.1, dicarboxypeptidase 1, DCP1; referred to as ACE) is a dipeptidyl carboxypeptidase belonging to the M2-metalloprotease family. Within the renin–angiotensin system, this zincdependent metalloprotease converts the inactive decapeptide Ang I (angiotensin I) into the vasopressor octapeptide Ang II. ACE also inactivates bradykinin, a vasodilator peptide, and thus contributes to blood pressure increase in mammals (see [1,2] for reviews). In vertebrates, two separate isoforms have been characterized: somatic (sACE) and testicular (germinal) ACE (tACE) are encoded by a single gene from two alternate promoters. This gene arose from an ancestor gene possessing a unique active-sitecoding region [3]. sACE and tACE differ with regard to the number of their active sites: sACE presents two highly conserved gluzincin active sites, HEXXH, whereas tACE possesses only one, corresponding to the C-terminal domain of sACE. sACE N- and C-terminal active sites (referred to as N- and C-domains respectively) have distinct enzymic activities on endogenous and/or synthetic substrates. In addition, inhibitors of ACE can discriminate between the two domains. For example, it has been shown that the C-domain is responsible for the major part of Ang I hydrolysis by sACE [4]. In contrast, N-domain ACE cleaves more efficiently the haematoregulatory peptide N-acetyl-SDKP [5] and Ang 1–7 [6]. Both sACE and tACE present a C-terminal transmembrane domain, and are membrane-anchored isoforms that can be solubilized in the extracellular medium by a secretase [7–10]. This post-translational cleavage has been reported as ‘shedding’. It could influence ACE activity, since especially membrane-bound forms of ACE are catalytically active in vivo [11,12]. An ACE homologue, ACE2, has recently been characterized in humans [13,14] and mice [15]. Being membrane-bound, the single active site ACE2 functions as a monocarboxypeptidase and is insensitive to the classical ACE inhibitor captopril [13,16]. ACE2 competes with ACE for Ang I in humans and, furthermore, degrades Ang II to Ang 1–7. Therefore ACE2 (through the ACE/ACE2 ratio) is an essential regulator for functioning of the heart.
In invertebrates, ACE-related genes have only been cloned in insects so far. Interestingly, all these genes encode soluble, single active-site enzymes [17–19]. In the fruitfly, Drosophila melanogaster, six potential genes encoding ACE-like proteins have been identified in silico. Two zinc-binding-pattern-conserved cDNAs have been cloned, encoding homologous enzymes and named AnCE [20] and Race [17]. Both AnCE and Race show approx. 40% amino acid sequence identity with ACEs from vertebrates and share similar enzymic properties [21] while acting on a broad range of substrates. Moreover, these latter enzymes, which have not been reported to be involved in volaemia, were demonstrated to play a key role in development [13,22,23] and reproduction [24–26].
To our knowledge, no ACE-like protein has been characterized so far at the molecular level in non-insect invertebrates. In addition, no functional ACE is present in the genome of the nematode Caenorhabditis elegans. In this worm, Brooks et al. [27] identified genes encoding non-peptidase homologues among which ACN-1 (C. elegans ACE-like non-peptidase 1) has recently been demonstrated to play a crucial role in development despite being catalytically inactive. This raises relevant phylogenic questions about the appearance of ACE during the course of evolution and the situation encountered in more distant groups.
Our model, the freshwater-living leech, Theromyzon tessulatum (Annelida; Clitellata; Hirudinida; Hirudinea; Rhynchobdellida; Glossiphoniidae), is an aquatic-bird ectoparasite; its life cycle has been divided into four stages according to its three blood meals. The third and last meal induces drastic morphological as well as physiological modifications such as a significant weight increase and induction of sexual maturation. Based on a recent phylogenetic classification (see Tree Of Life Web Project at http://tolweb.org and [28] for references), this Lophotrochozoa leech is considered to be at least as distant from vertebrates as the Ecdysozoa C. elegans. However, biochemical procedures have shown previously that an active ACE-like enzyme is present in T. tessulatum [29,30], suggesting that this leech contains an ancestral ACE-like protease.
In the present study, we report the molecular cloning and biochemical characterization of TtACE (T. tessulatum ACE), an ACE-like enzyme in T. tessulatum. To our knowledge, TtACE is the first non-insect invertebrate functional ACE-like enzyme ever cloned. To gain more insight into the evolutionary steps of ACE and to question its putative physiological roles in the leech, enzymic assays were performed on recombinant TtACE expressed in CHO-K1 cells (where CHO cell stands for Chinese-hamster ovary cell), together with multiplex semi-quantitative RT (reverse transcriptase)–PCR studies and in situ hybridization.
EXPERIMENTAL
Animals
Mature specimens of the leech, T. tessulatum, reared under laboratory conditions by the method described by Malecha [31], were used in the present study. Leeches' developmental stages were assessed according to their three blood meals (stage 0, from birth to the first meal; stage 1, between the first and the second meals; stage 2, between the second and the third meals; and stage 3, after the third and last meal). Stage 3 is subdivided into several periods as follows: early stage 3 completes the leech's genital tract maturation; however, leeches naturally die after egg laying and hatching, which terminate stage 3.
Reverse transcription
Total RNA was isolated from tissues of stage 1, 2 and 3 leeches by the guanidium thiocyanate/phenol/chloroform method using TRIzol® according to the manufacturer's instructions (Gibco BRL, Cergy-Pontoise, France). Equivalent amounts of RNA (5 μg) were reverse-transcribed with 200 units of Superscript II RT (Invitrogen) according to the manufacturer's instructions, using dT adapter (5′-CGAGTCGACATCGATCGTTTTTTTTTTTTTTTTT-3′) as an antisense primer for the reaction.
Molecular characterization
The full-length TtACE cDNA sequence was obtained by primer walking on a PBM8-orientated stage 2 T. tessulatum plasmid cDNA library (Biomethodes, Evry, France). Degenerate sense and antisense primers (ACEinv-S1, 5′-GTSTGYCAYGCSWSYGCSTG-3′; ACEinv-AS1, 5′-GCYTCRTGRAASCCSGGRTT-3′) were designed within the highly conserved regions surrounding the active site, HEXXH, of ACE in numerous vertebrate as well as invertebrate species, i.e. VCHASAWDFY and GANPGFHEA. PCRs were performed on 50 ng of PBM8 cDNA library in a Mastercycler gradient thermal cycler (Eppendorf). Cycling parameters were 5 min, 94 °C; 30 cycles: 1 min, 94 °C; 1 min, 42 °C; 1 min 30 s, 72 °C; and 5 min, 72 °C. The expected 180 bp products (according to various known ACE sequences) were resolved on a 2% (w/v) agarose gel stained with ethidium bromide and subcloned into the pGEM-T easy vector (Promega, Charbonnières, France). They were sequenced using fluorescent dideoxyterminator (ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit) on an ABI prism 310 sequencer (both from Applied Biosystems) with pGEM-T easy-specific primers (F23, 5′-CCCAGTCACGACGTTGTAAAAGG-3′; R23, 5′-AGCGGATAACAATTTCACACAGG-3′).
Characterization of this fragment allowed us to design homologous primers, subsequently used in PCRs for the complete TtACE cDNA cloning. The 5′ part of TtACE cDNA sequence was obtained from successive PCRs upstream of the coding region (i.e. primer walking) using antisense homologous primers. These primers were designed from previously characterized PBM8-s1 (5′-CGCCGTTACAGATCCAAGCTCC-3′)/LACESAS1 (5′-AGGCCACGGAATTGAAGATCC-3′) PCR product sequences. The 3′ part of TtACE was achieved using LACESS1 (5′-TCCAAGTGGTGGCACAACAGG-3′) and dT adapter primers. The PCR conditions were similar to those described above, except the annealing parameters that were adapted to each pair of primers (data available on request). Products were subcloned and sequenced as described above.
Similar degenerate primers (ACEinv-S1 and ACEinv-AS1) were also used to assess the presence of an ACE-like enzyme in other annelid species (Erpobdella octoculata, Hirudo medicinalis and Nereis diversicolor) using the procedure described above. Nested PCRs under the same conditions were performed when required.
Cloning of full-length TtACE cDNA and expression in cultured mammalian cells
Construct
Full-length TtACE cDNA was amplified by PCR (94 °C, 3 min; 30 cycles: 94 °C, 1 min; 59.5 °C, 40 s; 72 °C, 2 min; and 72 °C, 5 min) from the PBM8 cDNA library (50 ng) by 5 units of Pfu DNA polymerase (Promega) using specific primers (LACEFLS3, 5′-GAATTGCTCGAGTCGACCCACGCGTCCG-3′; LACEFLAS4, 5′-AAGTGGTAGAAAAATTATGAAATGACATAAAATC-3′) based on the sequence characterized by the procedure described in the Molecular characterization section. These primers were designed from sequences upstream of the stop codon in position −15, and downstream of the stop codon within the 3′-UTR (3′-untranslated region) respectively. Products were nested with LACEFLS4 (5′-CGCGTCCGGATGATTTTTTAAATGTTTCG-3′) and LACEFLAS4 under the same PCR conditions, subcloned and sequenced as described above. Full-length TtACE cDNA was cloned into the EcoRV site of pcDNA3.1 plasmid (Invitrogen). The sequence of the obtained pTtACE construct was verified first by restriction profile and then by sequencing as described above, using pcDNA3 primers (sense, 5′-ACTACAGGGAGACCCAAGC-3′; antisense, 5′-AGTCGAGGCTGATCAGCG-3′) as well as internal TtACE-specific primers.
Expression
CHO-K1 cells were cultured on FB6 plates in Dulbecco's modified Eagle's medium (Gibco BRL) with serum until 85–90% confluent. Cells were stably transfected with 1 μg of pTtACE per well using LIPOFECTAMINE™ 2000 reagent (Invitrogen) according to the manufacturer's instructions, and were selected for resistance to Geneticin. Individual resistant colonies producing large amounts of TtACE were cloned by limiting dilution techniques as described previously [4].
Immunological characterization of recombinant TtACE
Secreted recombinant TtACE was obtained from the culture medium of transfected CHO cells grown for 2 days in serum-free medium (ULTRA CHO). After concentration, the recombinant enzyme was analysed by SDS/PAGE, followed by Western-blot analysis. To characterize the TtACE enzyme, a rabbit antiserum (TtAb1) was raised against a peptide corresponding to the sequence 403–417 (HEAIADIASLSVATP). The peptide and antisera were provided by Neosystem (Strasbourg, France).
Enzymic assays
ACE activity was measured using different substrates. Hip-His-Leu (HHL; 5 mM; Hip, Hippuryl) and AcSDAcKP (0.5 mM; where Ac is N-acetyl) were used as human C- and N-domain-specific substrates respectively [32,33]. Ang I (25 μM) was used as both a human C- and N-domain substrate and HHL-NH2 (5 mM) as an amidated substrate.
Assays were performed at 37 °C with 25 μl of TtACE culture medium for 30 or 60 min. The rate of hydrolysis of all substrates used was resolved and quantified by reverse HPLC (Waters, Milford, MA, U.S.A.). The specificity of the reaction was confirmed in the presence of 1 μM of the ACE inhibitor, lisinopril. The effect of chloride on HHL hydrolysis was determined under standard conditions with NaCl concentrations ranging from 0 to 1000 mM. Activity of TtACE was compared with that of the N- or C-domain of human ACE, as described previously [4].
Inhibition of Mca-ASDK-DpaOH hydrolysis by various ACE inhibitors
Inhibition studies were performed with Mca-ASDK-DpaOH [where Mca stands for (7-methoxycoumarin-4-yl)acetyl- and DpaOH stands for N3(2,4-dinitrophenyl)L-2,3-diaminopropionyl-OH] as a fluorescent ACE substrate, as described previously [34]. Assays were performed by recording fluorescence at 390 nm.
The potency of captopril and delaprilat for the inhibition of Mca-ASDK-DpaOH hydrolysis by recombinant TtACE was determined by establishing dose-dependent inhibition curves at equilibrium. The compounds were preincubated with TtACE for 1 h and reactions were initiated by the addition of 12 μM of substrate.
To compare the effects of these inhibitors, the same dose-dependent curves were studied with recombinant C- and N-sites of human ACE.
Inhibition of Mca-ASDK-DpaOH hydrolysis by ACE mAb (monoclonal antibody)
mAb i2H5 (a gift from Dr S. Danilov, Department of Chemistry, Moscow State University, Russia), an mAb raised against purified human lung ACE, recognizes epitopes in the N-terminal domain of ACE and inhibits the enzymic activity of the N-terminal active site only [35]. The inhibitory potency of mAb i2H5 towards the recombinant TtACE and C- and N-active sites of human ACE was determined by establishing dose-dependent inhibition curves.
Northern-blot analysis
Total RNA was extracted as described above, and was poly(A)+ (polyadenylated)-enriched using oligo(dT) resin-affinity chromatography. Poly(A)+ RNA (10 μg) from stage 2 leeches was subjected to Northern-blot analysis as described by Breton et al. [36]. A [32P]dCTP-radiolabelled 280-bp cDNA probe corresponding to nt 1426–1705 of TtACE cDNA sequence was generated with the Ready-to-go labelling kit (Amersham Biosciences). Nylon membranes were hybridized overnight at 42 °C and washed [final stringency conditions were 0.5×sodium citrate and 0.1% (w/v) SDS; 45 °C for 15 min]. Autoradiograms (Kodak) were visualized after exposure for 16 h at room temperature (20 °C).
Multiplex semi-quantitative PCR
Semi-quantification of TtACE expression was performed using the multiplex relatively quantitative RT–PCR validated method [37,38]. The constitutively expressed housekeeping gene, β-actin, was used as an internal standard (the sequence data for T. tessulatum is available at the GenBank® database under the accession no. CK640381). β-Actin primers (AcF, 5′-GATCTGGCATCACACCTTCTACAA-3′; AcR, 5′-GTTGAAGGTCTCGAACATGATCTG-3′) were kindly provided by Dr C. Lefebvre (Laboratoire de Neuroimmunologie des Annélides, University of Lille 1). cDNA from stage 1 (n=10), stage 2 (n=10) and stage 3 (n=8) leeches were obtained by the method described above. They were used as templates in 27-cycle PCRs. Previous experiments have allowed us to determine that 27 cycles correspond to the number of reaction steps required for the maximal exponential amplification phase of β-actin before reaching the plateau phase (results not shown). Each tube contained the following mixture: 50 ng of cDNA; 2 pmol each of the primers LACESS1 and LACESAS1, as well as AcF and AcR; 20 μM dNTPs; 1 unit of Taq DNA polymerase; MgCl2 in an appropriate buffer and water to a final volume of 50 μl. PCR cycling conditions were 94 °C, 3 min; 27 cycles: 94 °C, 40 s; 52 °C, 1 min; 72 °C, 1 min. Negative and positive controls were performed on water and 50 ng of pBM8 stage 2 cDNA library respectively. Products were resolved by 2% agarose-gel electrophoresis, stained with ethidium bromide as described above and digitally photographed under UV light (312 nm). Absorbance was analysed with the Claravision system v5.1 (Claravision Perfect Image software and Claravision Gel Analyst software; Claravision, Orsay, France). Statistical analysis was performed using unpaired Student's t test.
In situ hybridization
Cellular localization of TtACE mRNA synthesis was investigated by in situ hybridization on stage 2 and stage 3 leeches. Leeches were fixed overnight in 0.1 M phosphate buffer containing 4% (w/v) paraformaldehyde (pH 7.4). After dehydration, animals were embedded in paraplast and 7 μm sections were cut, mounted on poly(L-lysine)-coated slides and stored at 4 °C until use. The labelled, approx. 1 kb, cRNA probe was obtained from the library by PCR using the primers LACESS1 and dT adapter. The cDNA fragment (nt 1123–1848 and a part of the 3′-UTR) was cloned in pGEM-T easy (Promega) and sequenced as described above. [35S]UTP-labelled antisense and sense riboprobes were generated from linearized cDNA plasmids by in vitro transcription using SP6/T7 RNA-labelling kit (Roche). 35S-labelled riboprobes (100 ng or 1×106 c.p.m./slide) were hybridized as described previously [39]. Slides were observed under a Zeiss Axioscope microscope. The control consisted of replacing the antisense riboprobe with a sense riboprobe. RNAse control sections were obtained by adding a preincubation step with 10 mg/ml RNAse A before hybridization.
RESULTS
Molecular characterization
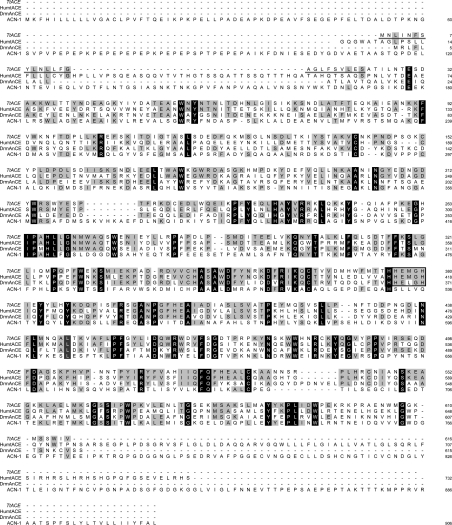
Using a PCR approach, we cloned an approx. 2 kb cDNA fragment presenting a 1848 bp ORF (open reading frame). This ORF possesses a transcription start (position 1), is delimited by two in-frame 5′- and 3′-stop codons (positions −15 and 1849 respectively) and a putative polyadenylation signal in the 3′-UTR. This ORF corresponds to the putative 616-amino-acid leech enzyme (GenBank® accession no. AY560004). It contains the zinc-binding motif HEXXH (positions 376–380) and the putative third zinc ligand, a glutamic residue located 24 amino acids downstream of the second histidine residue of this motif (position 403). Hydrophobicity predictions (SignalP V1.1, available at http://www.cbs.dtu.dk/services/SignalP) suggested post-translational cleavage of a signal peptide between residues 23 and 24 (SVLES↓ATIL). The deduced primary sequence revealed that, in agreement with what has been found in invertebrates, the enzyme did not seem to contain a C-terminal transmembrane domain. Moreover, the PROSCAN bioinformatics prediction tool program (available at http://cbs.dtu.dk/services/NetNGlyc/) emphasized the presence of six potential N-glycosylation sites. The in silico glycosylphosphatidylinositol-anchor-site prediction tool (big-PI Predictor, available at http://mendel.imp.univie.ac.at/gpi/cgi-bin/gpi_pred.cgi) [40] did not detect any pattern likely to undergo such a modification in the TtACE primary sequence. Taken together, these results suggested that TtACE might only exist under a soluble form. Subsequently, after the signal peptide cleavage and assuming that all potential sites had undergone N-glycosylation, the predicted molecular mass of mature TtACE was approx. 78 kDa. Interestingly, its amino acid sequence showed greater similarity to vertebrate than to invertebrate enzymes [53% identity with Gallus gallus ACE (GI: 1703072), 47 and 48% identity with Homo sapiens N- and C-domains respectively (GI: 37790804) and 39% identity with D. melanogaster AnCE (GI: 26006940) according to the BLAST server (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html)] [41]. Its primary sequence indicated that TtACE bore every functional residue at conserved positions for zinc ions (His376 and His380), chloride ions (Arg180, Tyr218, Trp478, Arg48 and Arg515) and lisinopril (His346, Ala347, Glu377, Lys504, His506, Tyr51 and Tyr517) binding when compared with human tACE [41,42] (Figure 1). Using the same approach, we cloned partial cDNA and found the existence of an ACE-like enzyme in all of the water-living annelid species examined, including non-parasitic (Erpobdella octoculata, Nereis diversicolor) as well as parasitic species (Hirudo medicinalis). The characterized sequences were all highly conserved in these species (i.e. more than 70% homology in nucleotides) whether parasitic or not (results not shown). It is noteworthy that, although all other known ACE sequences of the animal kingdom present a glutamic residue at position 6 after the last histidine of the zinc-binding motif HEXXH (according to the MEROPS database release 6.3, available at http://merops.sanger.ac.uk), a histidine residue was identified at this position in every annelid ACE sequence.
Figure 1. Amino acid sequence alignment between TtACE and ACEs of various species.
This alignment was obtained in silico using Clustal W software (available at http://www.ebi.ac.uk/clustalw/). HumtACE, H. sapiens C-domain ACE (GI: 113041); DrmAnCE, D. melanogaster AnCE (GI: 26006940); ACN-1, C. elegans ACE-like non-peptidase 1 (GI: 25151555). Amino acids are numbered on the right side of the first methionine residue. Dashes represent gaps. The residues identical between all sequences are shown in white within black boxes. The conserved amino acids with regard to TtACE are shaded grey. The M2 active site is boxed. TtACE putative signal peptide is underlined.
Biochemical characterization of recombinant TtACE
Expression of secreted TtACE in CHO cells
Culture medium of recombinant stable CHO cells was analysed by SDS/PAGE followed by Western-blot analysis. Recombinant TtACE migrated with an apparent molecular mass of 79 kDa (Figure 2).
Figure 2. Western-blot analysis of recombinant TtACE.
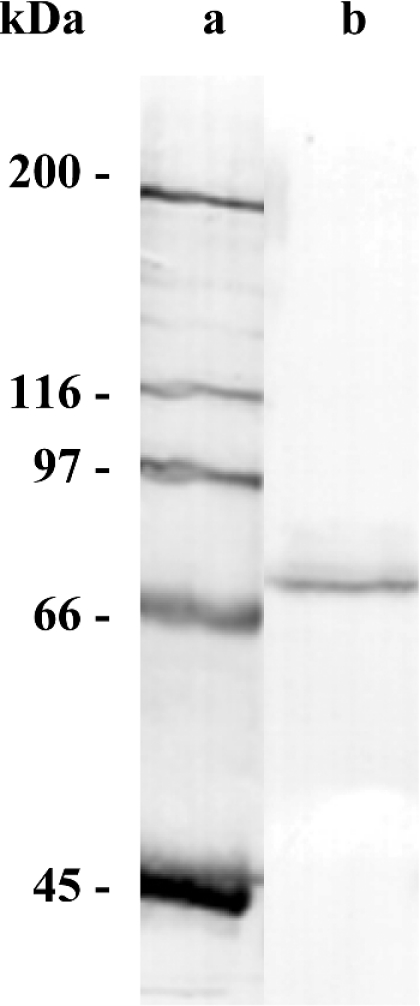
TtACE expressed in CHO cells was analysed by SDS/PAGE, followed by Western-blot analysis with the antiserum TtAb1. Lane a, molecular-mass standards; lane b, recombinant TtACE.
Enzymic properties of recombinant TtACE
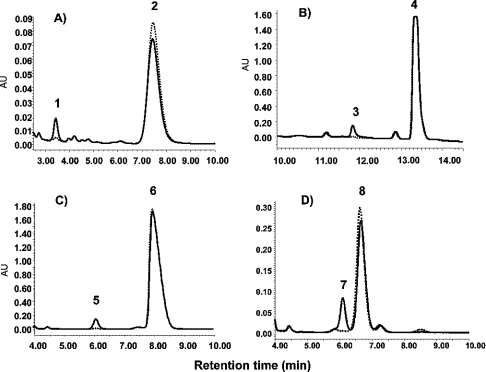
The enzymic activity of recombinant TtACE was studied using the mammalian ACE substrate Ang I and two synthetic substrates, specific to the N- and the C-domain active sites of ACE, namely AcSDAcKP and HHL respectively. Recombinant TtACE hydrolysed the three substrates, as shown by HPLC analysis (Figures 3A–3C). TtACE hydrolysed HHL-NH2, a substrate with a blocked C-terminus (Figure 3D). All these hydrolyses were fully inhibited by 1 μM lisinopril.
Figure 3. Hydrolysis of Ang I, AcSDAcKP, HHL and HHL-NH2 by TtACE.
Profiles are as follows: (A) Ang I (peak 2), Ang II (peak 1); (B) AcSDAcKP (peak 4), AcKP (peak 3); (C) HHL (peak 6), hippuric acid (peak 5); and (D) HHL-NH2 (peak 8), hippuric acid (peak 7). Inhibition of TtACE activity by 1 μM lisinopril is indicated (·········).
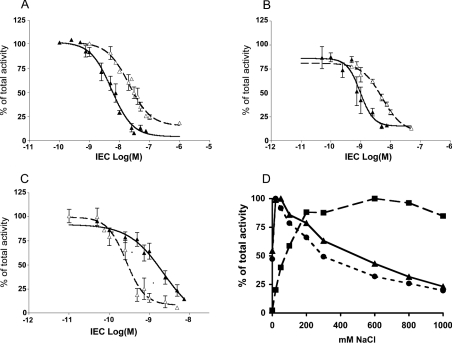
Since chloride dependence of HHL hydrolysis is a good index of the N- or C-domain property of the active sites of ACE, TtACE activity under different chloride concentrations was assessed (Figure 4D). TtACE maximal activity was observed at 50 mM Cl− concentration, similar to the single N-domain of human ACE.
Figure 4. Inhibitory effects of various ACE inhibitors on the hydrolysis of HHL by recombinant TtACE.
Effects of captopril (▴) and delaprilat (▵) on recombinant TtACE (A) and the N- (B) or C-terminal (C) functional active sites of human ACE. Results are expressed as the means for three experiments. (D) Effects of various chloride concentrations on the hydrolysis of HHL by recombinant TtACE (▴) and the N- and C-domains of human ACE (• and ▪ respectively). Recombinant TtACE from culture medium was dialysed against distilled water and various chloride concentrations were added to aliquots of the dialysed medium. Activity of ACE towards HHL was determined and compared with that of the single active-site N- and C-domains of ACE as described in [4].
Effect of two ACE inhibitors on TtACE activity
Inhibition of TtACE was studied using two mammalian ACE inhibitors selected for their ability to inhibit differentially either the N- or C-domain of human ACE, captopril and delaprilat respectively. TtACE was more potently inhibited by captopril (IC50=5.5 nM) than by delaprilat (IC50=20.7 nM), a result similar to that observed with the two human ACE domains (Figures 4A–4C). Finally, TtACE activity was inhibited by mAb i2H5 in a manner similar to human N-domain (result not shown).
mRNA expression
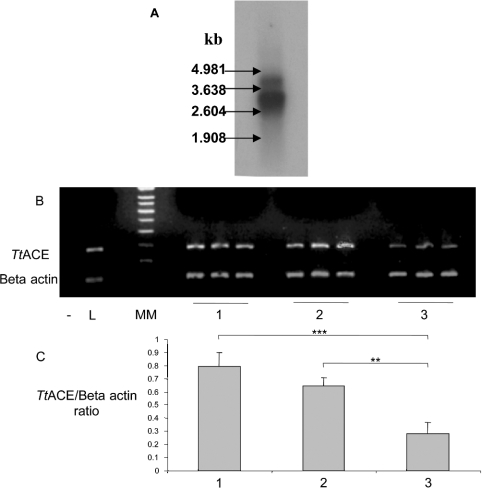
Results of Northern-blot analysis showed the existence of a major transcript of approx. 3 kb and a minor one migrating with an apparent molecular mass of 4.2 kb (Figure 5A). Performing multiplex semi-quantitative RT–PCR experiments using β-actin as an internal standard, we found variable amounts of ACE mRNA at all stages (stages 1–3) of the leech's life cycle (Figure 5B). TtACE mRNA was present from stage 1 and its expression level was similar at stages 1 and 2. However, a lower mRNA expression level was detected at stage 3, as assessed by fluorescence analysis of agarose gels (Figure 5C).
Figure 5. TtACE mRNA Northern-blot profile (A), multiplex semi-quantitative RT–PCR expression pattern during the life cycle of T. tessulatum (B) and the associated semi-quantification (C).
(A) kb, molecular-mass standards. (B) TtACE, 281 bp amplicon corresponding to TtACE; β-actin, 165 bp amplicon corresponding to β-actin. Lane –, negative control achieved using water instead of cDNA; lane L, positive control performed on a 50 ng pBM8 cDNA library; lane MM, 100 bp molecular-mass standards; lanes 1, 2 and 3, triplicate reactions on cDNA of stage 1, 2 and 3 animals respectively. The gel shown is representative of 28 experiments. (C) Semi-quantitative analysis of TtACE expression reported for β-actin expression. Agarose gels were digitally photographed with Claravision Perfect Image software v5.1 and analysed using the Claravision Gel Analyst software v5.1. The stage of the leech's life is indicated (stages 1–3) below the corresponding line. The TtACE/β-actin signal ratio is expressed in arbitrary units and values are the means±S.E.M. for n=10 (stage 1), n=10 (stage 2) and n=8 (stage 3) independent experiments. Statistical analysis was performed using unpaired Student's t test. **P<0.01; ***P<0.001, compared with stage 1 TtACE/β-actin ratio.
Cellular localization
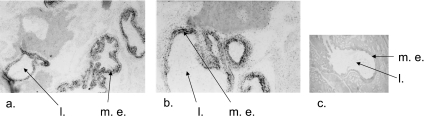
After in situ hybridization analysis of the whole body, TtACE mRNA labelling was exclusively observed in the epithelial cells of the midgut from stage 2 leeches (Figure 6). No labelling could be detected in stage 3 animals or in any other tissue, e.g. the reproductive tract.
Figure 6. Microphotographs of stage 2 T. tessulatum sections after in situ hybridization.
Radiolabelling is visible as black dots. (a) Antisense riboprobe, magnification ×20; (b) antisense riboprobe, magnification ×40; (c) sense riboprobe, magnification ×20; m.e., midgut epithelium; l., midgut lumen.
DISCUSSION
The present study brings molecular evidence for the existence of a functional ACE-like enzyme in non-insect invertebrates. It describes the first cloning of cDNA and the biochemical activity and distribution of an ACE-like enzyme in the leech, T. tessulatum, named TtACE. To date, only biochemical data about ACE-like activities in these species are available [29,43]. Their cDNA structure has never been identified so far. The 1848-bp cDNA ORF of TtACE encodes a putative 616-amino-acid protein of 79 kDa. This protein does not possess a membrane anchor, contains a unique M2 catalytic active site and shares a common structure with ACE from insects. However, its amino acid sequence shows a higher homology with ACE from vertebrates, particularly with ACE from chicken. Since the leech, T. tessulatum, is the only Lophotrochozoa species in which an active ACE-like enzyme has been characterized at the molecular level, TtACE may help us to address numerous questions that remain to be answered.
One relevant point concerns phylogenic and evolutive considerations, since C. elegans does not contain any functional ACE-like enzyme in its genome. Since C. elegans and arthropods belong to the Ecdysozoa group and T. tessulatum to the Lophotrochozoa, the present study brings evidence that appearance of a functional ACE-like protein occurs before or at the formation of the Bilateria (see Tree Of Life Web Project at http://tolweb.org and [28] for references). Interestingly, a non-peptidase ACE-like protein (ACN-1) has been identified in the C. elegans genome and has very recently been demonstrated to play a key role in development [27]. However, the reason why the C. elegans enzyme lost its protease activity remains to be elucidated. In any case, our results raise very interesting questions about the putative primitive and original functions of ACE-like enzymes in the animal kingdom. In view of the higher sequence homology between the isoforms from leeches and vertebrates than those identified in other non-annelid invertebrates, we speculated that this could be specific to the leech, T. tessulatum, an aquatic-bird parasite. We thus decided to look for the presence of a 180-bp ACE cDNA fragment surrounding the active site in non-parasitic annelid species. The existence of such a cDNA fragment, which shows a great sequence identity with TtACE even in non-parasitic annelid species, indicates that an active ACE-like protease is present in all the annelids studied, and its presence does not seem to be dependent on parasitic biology. Altogether, our results indicated that annelids contain a presumably active ACE-like protein that includes an active site, possessing a higher sequence identity with ACE from vertebrates than with insects' enzymes. Interestingly enough, all the annelid ACE-like enzymes characterized possessed a specific histidine residue located six amino acids downstream of the zinc-binding motif. This amino acid seems to be unique to annelids since it is replaced by a glutamic residue in all the biologically active ACE-like enzymes cloned so far. However, this conserved amino acid residue has not been demonstrated to play an important catalytic role in the active site of ACE as assessed by NMR and X-ray structural studies in human and Drosophila [42,44].
Biochemical studies of stably transfected CHO-K1 cells expressing TtACE showed that the cloned leech cDNA encoded a functional enzyme with a molecular mass of 79 kDa, as expected. Hydrolysis assays on various substrates showed that TtACE was capable of hydrolysing several mammalian ACE substrates, including a C-amidated substrate. In this respect, TtACE, similar to mammalian ACE, displayed an endopeptidase activity. It is interesting to note that the TtACE active-site characteristics were similar to the N-domain characteristics of ACE, such as (i) low chloride dependence on HHL hydrolysis, (ii) preferential inhibition by N-domain-type ACE inhibitor and (iii) preferential inhibition by an mAb specific to the N-domain. This shows that the N-domain characteristics of ACE are already present in a distant organism such as the leech.
To gain more insight into the physiological functions of TtACE, we compared its mRNA expression levels at different stages of the leech's life cycle. TtACE mRNA was detected at all the stages examined, but did not show any significant quantitative variation between stages 1 and 2 (i.e. after the first and the second blood meals respectively). In contrast, a remarkable decrease in the expression level of the TtACE gene occurred at stage 3 after the last blood meal. During the first two stages, the morphology of leeches was not markedly affected. Important diuretic mechanisms took place, leading to concentration of the host's blood cells. In contrast, the last blood meal induced drastic morphological and physiological changes such as a significant weight increase and induction of sexual maturation. Thus the expression level of TtACE was the lowest when digestion and ovogenesis were completed, suggesting a potential role of the ACE from leech in these functions.
Surprisingly, TtACE mRNA expression was not detected in the reproductive system, but seemed to be restricted to midgut epithelial cells. Although ACE mRNA has already been shown to be expressed in the locust and dipteran midgut, proteases from insects, in contrast with TtACE, exhibit a wide tissue distribution. The physiological role and the endogenous substrates of ACE from invertebrates in the midgut remain to be elucidated, although the high levels of ACE activity in midgut tissues of Diptera have led to speculations about a digestive role for ACE [19], which is consistent with our findings. As far as we know, the ACE from invertebrates is clearly known to control only developmental and reproductive functions [17–26]. For example, the AnCE gene, whose genetic invalidation leads to a lethal phenotype [17], is expressed in the spermatocytes and spermatids of Drosophila [24]. Interestingly, in the mosquito Anopheles stephensi, an ACE is induced by a blood meal, but accumulates secondarily in the developing ovary [26], suggesting that the reproductive function of the enzyme is under a dietary influence. Recently, using the grey fleshfly Neobellieria bullata as a model, it has been shown that ovarian ACE could be involved in the proteolysis of yolk proteins, suggesting that it may be a regulator of vitellogenic and developmental processes [45,46]. Our results did not support the role of TtACE in the latter processes, but indicated that the soluble enzyme could be secreted within the digestive tract and acts as a final degrading enzyme of the proteic nutrients. However, we cannot rule out that a minor mRNA expression of TtACE might have occurred outside the midgut tissues, but was not detected by in situ hybridization. Alternatively, as in Drosophila, T. tessulatum may express different ACE genes that could present distinct cellular distributions. Furthermore, Northern-blot analysis of TtACE displayed two bands, suggesting either alternative splicing of TtACE mRNA or expression of a close ACE-related gene. Nevertheless, further experiments will be needed to clarify this issue.
Comprehension of the physiological roles of TtACE will require the molecular characterization of its endogenous substrates and/or its potential inhibitors. Although the amount of biochemical and molecular data on ACE-like enzymes from invertebrates is increasing rapidly, little is known concerning their in vivo targets. Understanding these functions in invertebrates could provide new, interesting information on the roles of these ancestral peptidases.
Acknowledgments
We are grateful to A. Desmond and S. Ruault for their excellent technical assistance. We thank Professor C. Denorme (CUEPP, Université des Sciences et Technologies de Lille) for a critical reading of this paper. This study was partially supported by a grant from Conseil Régional du Nord-Pas de Calais (Lille cedex, France), by Institut National pour la Santé et la Recherche Médicale and by Centre National de la Recherche Scientifique (Paris, France).
References
- 1.Corvol P., Williams T. A., Soubrier F. Peptidyl dipeptidase A: angiotensin-I converting enzyme. In: Barrett A. J., editor. Methods in Enzymology. London: Academic Press; 1995. pp. 283–305. [DOI] [PubMed] [Google Scholar]
- 2.Turner A. J., Hooper N. M. The angiotensin-converting enzyme gene family: genomics and pharmacology. Trends Pharmacol. Sci. 2002;23:177–183. doi: 10.1016/s0165-6147(00)01994-5. [DOI] [PubMed] [Google Scholar]
- 3.Hubert C., Houot A. M., Corvol P., Soubrier F. Structure of the angiotensin I-converting enzyme gene. Two alternate promoters correspond to evolutionary steps of a duplicated gene. J. Biol. Chem. 1991;266:15377–15383. [PubMed] [Google Scholar]
- 4.Wei L., Alhenc-Gelas F., Corvol P., Clauser E. The two homologous domains of human angiotensin I-converting enzyme are both catalytically active. J. Biol. Chem. 1991;266:9002–9008. [PubMed] [Google Scholar]
- 5.Rousseau A., Michaud A., Chauvet M. T., Lenfant M., Corvol P. The hemoregulatory peptide N-acetyl-Ser-Asp-Lys-Pro is a natural and specific substrate of the N-terminal active site of human angiotensin-converting enzyme. J. Biol. Chem. 1995;270:3656–3661. doi: 10.1074/jbc.270.8.3656. [DOI] [PubMed] [Google Scholar]
- 6.Deddish P. A., Marcic B., Jackman H. L., Wang H. Z., Skidgel R. A., Erdos E. G. N-domain-specific substrate and C-domain inhibitors of angiotensin-converting enzyme: angiotensin-(1–7) and keto-ACE. Hypertension. 1998;31:912–917. doi: 10.1161/01.hyp.31.4.912. [DOI] [PubMed] [Google Scholar]
- 7.Eyries M., Michaud A., Deinum J., Agrapart M., Chomilier J., Kramers C., Soubrier F. Increased shedding of angiotensin-converting enzyme by a mutation identified in the stalk region. J. Biol. Chem. 2001;276:5525–5532. doi: 10.1074/jbc.M007706200. [DOI] [PubMed] [Google Scholar]
- 8.Oppong S. Y., Hooper N. M. Characterization of a secretase activity which releases angiotensin-converting enzyme from the membrane. Biochem. J. 1993;292:597–603. doi: 10.1042/bj2920597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parkin E. T., Trew A., Christie G., Faller A., Mayer R., Turner A. J., Hooper N. M. Structure-activity relationship of hydroxamate-based inhibitors on the secretases that cleave the amyloid precursor protein, angiotensin converting enzyme, CD23, and pro-tumor necrosis factor-α. Biochemistry. 2002;41:4972–4981. doi: 10.1021/bi015936e. [DOI] [PubMed] [Google Scholar]
- 10.Parvathy S., Oppong S. Y., Karran E. H., Buckle D. R., Turner A. J., Hooper N. M. Angiotensin-converting enzyme secretase is inhibited by zinc metalloprotease inhibitors and requires its substrate to be inserted in a lipid bilayer. Biochem. J. 1997;327:37–43. doi: 10.1042/bj3270037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esther C. R., Marino E. M., Howard T. E., Machaud A., Corvol P., Capecchi M. R., Bernstein K. E. The critical role of tissue angiotensin-converting enzyme as revealed by gene targeting in mice. J. Clin. Invest. 1997;99:2375–2385. doi: 10.1172/JCI119419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esther C. R., Jr, Howard T. E., Marino E. M., Goddard J. M., Capecchi M. R., Bernstein K. E. Mice lacking angiotensin-converting enzyme have low blood pressure, renal pathology, and reduced male fertility. Lab. Invest. 1996;74:953–965. [PubMed] [Google Scholar]
- 13.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ. Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 14.Tipnis S. R., Hooper N. M., Hyde R., Karran E., Christie G., Turner A. J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 15.Komatsu T., Suzuki Y., Imai J., Sugano S., Hida M., Tanigami A., Muroi S., Yamada Y., Hanaoka K. Molecular cloning, mRNA expression and chromosomal localization of mouse angiotensin-converting enzyme-related carboxypeptidase (mACE2) DNA Seq. 2002;13:217–220. doi: 10.1080/1042517021000021608. [DOI] [PubMed] [Google Scholar]
- 16.Turner A. J., Tipnis S. R., Guy J. L., Rice G., Hooper N. M. ACEH/ACE2 is a novel mammalian metallocarboxypeptidase and a homologue of angiotensin-converting enzyme insensitive to ACE inhibitors. Can. J. Physiol. Pharmacol. 2002;80:346–353. doi: 10.1139/y02-021. [DOI] [PubMed] [Google Scholar]
- 17.Tatei K., Cai H., Ip Y. T., Levine M. Race: a Drosophila homologue of the angiotensin converting enzyme. Mech. Dev. 1995;51:157–168. doi: 10.1016/0925-4773(95)00349-5. [DOI] [PubMed] [Google Scholar]
- 18.Taylor C. A., Coates D., Shirras A. D. The Acer gene of Drosophila codes for an angiotensin-converting enzyme homologue. Gene. 1996;181:191–197. doi: 10.1016/s0378-1119(96)00503-3. [DOI] [PubMed] [Google Scholar]
- 19.Wijffels G., Fitzgerald C., Gough J., Riding G., Elvin C., Kemp D., Willadsen P. Cloning and characterisation of angiotensin-converting enzyme from the dipteran species, Haematobia irritans exigua, and its expression in the maturing male reproductive system. Eur. J. Biochem. 1996;237:414–423. doi: 10.1111/j.1432-1033.1996.0414k.x. [DOI] [PubMed] [Google Scholar]
- 20.Cornell M. J., Williams T. A., Lamango N. S., Coates D., Corvol P., Soubrier F., Hoheisel J., Lehrach H., Isaac R. E. Cloning and expression of an evolutionary conserved single-domain angiotensin converting enzyme from Drosophila melanogaster. J. Biol. Chem. 1995;270:13613–13619. doi: 10.1074/jbc.270.23.13613. [DOI] [PubMed] [Google Scholar]
- 21.Coates D., Isaac R. E., Cotton J., Siviter R., Williams T. A., Shirras A., Corvol P., Dive V. Functional conservation of the active sites of human and Drosophila angiotensin I-converting enzyme. Biochemistry. 2000;39:8963–8969. doi: 10.1021/bi000593q. [DOI] [PubMed] [Google Scholar]
- 22.Crackower M. A., Sarao R., Oudit G. Y., Yagil C., Kozieradzki I., Scanga S. E., Oliveira-dos-Santos A. J., da Costa J., Zhang L., Pei Y., et al. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature (London) 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 23.Quan G. X., Mita K., Okano K., Shimada T., Ugajin N., Xia Z., Goto N., Kanke E., Kawasaki H. Isolation and expression of the ecdysteroid-inducible angiotensin-converting enzyme-related gene in wing discs of Bombyx mori. Insect Biochem. Mol. Biol. 2001;31:97–103. doi: 10.1016/s0965-1748(00)00112-0. [DOI] [PubMed] [Google Scholar]
- 24.Hurst D., Rylett C. M., Isaac R. E., Shirras A. D. The Drosophila angiotensin-converting enzyme homologue Ance is required for spermiogenesis. Dev. Biol. 2003;254:238–247. doi: 10.1016/s0012-1606(02)00082-9. [DOI] [PubMed] [Google Scholar]
- 25.Zhu W., Vandingenen A., Huybrechts R., Baggerman G., De Loof A., Poulos P., Velentza A., Breuer M. In vitro degradation of the Neb-Trypsin modulating oostatic factor (Neb-TMOF) in gut luminal content and hemolymph of the grey fleshfly, Neobellieria bullata. Insect Biochem. Mol. Biol. 2001;31:87–95. doi: 10.1016/s0965-1748(00)00111-9. [DOI] [PubMed] [Google Scholar]
- 26.Ekbote U., Coates D., Isaac R. E. A mosquito (Anopheles stephensi) angiotensin I-converting enzyme (ACE) is induced by a blood meal and accumulates in the developing ovary. FEBS Lett. 1999;455:219–222. doi: 10.1016/s0014-5793(99)00870-4. [DOI] [PubMed] [Google Scholar]
- 27.Brooks D. R., Appleford P. J., Murray L., Isaac R. E. An essential role in moulting and morphogenesis of Caenorhabditis elegans for ACN-1: a novel member of the angiotensin-converting enzyme family that lacks a metallopeptidase active site. J. Biol. Chem. 2003;278:53240–53246. doi: 10.1074/jbc.M308858200. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen C. 2nd edn. Oxford: Oxford University Press; 2001. Animal Evolution: Interrelationships of the Living Phyla. [Google Scholar]
- 29.Laurent V., Salzet M. Biochemical properties of the angiotensin-converting-like enzyme from the leech Theromyzon tessulatum. Peptides. 1996;17:737–745. doi: 10.1016/0196-9781(96)00074-5. [DOI] [PubMed] [Google Scholar]
- 30.Vandenbulcke F., Laurent V., Verger-Bocquet M., Stefano G. B., Salzet M. Biochemical identification and ganglionic localization of leech angiotensin-converting enzymes. Brain Res. Mol. Brain Res. 1997;49:229–237. doi: 10.1016/s0169-328x(97)00146-0. [DOI] [PubMed] [Google Scholar]
- 31.Malecha J. Osmoregulation in Hirudinea Rhynchobdellida Theromyzon tessulatum (OFM). Experimental localization of the secretory zone of a regulation factor of water balance. Gen. Comp. Endocrinol. 1983;49:344–351. doi: 10.1016/0016-6480(83)90198-3. [DOI] [PubMed] [Google Scholar]
- 32.Azizi M., Massien C., Michaud A., Corvol P. In vitro and in vivo inhibition of the 2 active sites of ACE by omapatrilat, a vasopeptidase inhibitor. Hypertension. 2000;35:1226–1231. doi: 10.1161/01.hyp.35.6.1226. [DOI] [PubMed] [Google Scholar]
- 33.Wei L., Alhenc-Gelas F., Soubrier F., Michaud A., Corvol P., Clauser E. Expression and characterization of recombinant human angiotensin I-converting enzyme. Evidence for a C-terminal transmembrane anchor and for a proteolytic processing of the secreted recombinant and plasma enzymes. J. Biol. Chem. 1991;266:5540–5546. [PubMed] [Google Scholar]
- 34.Dive V., Cotton J., Yiotakis A., Michaud A., Vassiliou S., Jiracek J., Vazeux G., Chauvet M. T., Cuniasse P., Corvol P. RXP 407, a phosphinic peptide, is a potent inhibitor of angiotensin I converting enzyme able to differentiate between its two active sites. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4330–4335. doi: 10.1073/pnas.96.8.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Danilov S., Jaspard E., Churakova T., Towbin H., Savoie F., Wei L., Alhenc-Gelas F. Structure-function analysis of angiotensin I-converting enzyme using monoclonal antibodies. Selective inhibition of the amino-terminal active site. J. Biol. Chem. 1994;269:26806–26814. [PubMed] [Google Scholar]
- 36.Breton C., Pechoux C., Morel G., Zingg H. H. Oxytocin receptor messenger ribonucleic acid: characterization, regulation, and cellular localization in the rat pituitary gland. Endocrinology. 1995;136:2928–2936. doi: 10.1210/endo.136.7.7540544. [DOI] [PubMed] [Google Scholar]
- 37.Wei Q., Bondy M. L., Mao L., Gaun Y., Cheng L., Cunningham J., Fan Y., Bruner J. M., Yung W. K., Levin V. A., et al. Reduced expression of mismatch repair genes measured by multiplex reverse transcription–polymerase chain reaction in human gliomas. Cancer Res. 1997;57:1673–1677. [PubMed] [Google Scholar]
- 38.Wong H., Anderson W. D., Cheng T., Riabowol K. T. Monitoring mRNA expression by polymerase chain reaction: the ‘primer-dropping’ method. Anal. Biochem. 1994;223:251–258. doi: 10.1006/abio.1994.1581. [DOI] [PubMed] [Google Scholar]
- 39.Mitta G., Vandenbulcke F., Noel T., Romestand B., Beauvillain J. C., Salzet M., Roch P. Differential distribution and defence involvement of antimicrobial peptides in mussel. J. Cell Sci. 2000;113:2759–2769. doi: 10.1242/jcs.113.15.2759. [DOI] [PubMed] [Google Scholar]
- 40.Eisenhaber B., Bork P., Eisenhaber F. Prediction of potential GPI-modification sites in proprotein sequences. J. Mol. Biol. 1999;292:741–758. doi: 10.1006/jmbi.1999.3069. [DOI] [PubMed] [Google Scholar]
- 41.Combet C., Blanchet C., Geourjon C., Deleage G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 42.Natesh R., Schwager S. L., Sturrock E. D., Acharya K. R. Crystal structure of the human angiotensin-converting enzyme-lisinopril complex. Nature (London) 2003;421:551–554. doi: 10.1038/nature01370. [DOI] [PubMed] [Google Scholar]
- 43.Kawamura T., Oda T., Muramatsu T. Purification and characterization of a dipeptidyl carboxypeptidase from the polychaete Neanthes virens resembling angiotensin I converting enzyme. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2000;126:29–37. doi: 10.1016/s0305-0491(00)00177-2. [DOI] [PubMed] [Google Scholar]
- 44.Kim H. M., Shin D. R., Yoo O. J., Lee H., Lee J. O. Crystal structure of Drosophila angiotensin I-converting enzyme bound to captopril and lisinopril. FEBS Lett. 2003;538:65–70. doi: 10.1016/s0014-5793(03)00128-5. [DOI] [PubMed] [Google Scholar]
- 45.Hens K., Vandingenen A., Macours N., Baggerman G., Karaoglanovic A. C., Schoofs L., De Loof A., Huybrechts R. Characterization of four substrates emphasizes kinetic similarity between insect and human C-domain angiotensin-converting enzyme. Eur. J. Biochem. 2002;269:3522–3530. doi: 10.1046/j.1432-1033.2002.03043.x. [DOI] [PubMed] [Google Scholar]
- 46.Vandingenen A., Hens K., Baggerman G., Macours N., Schoofs L., De Loof A., Huybrechts R. Isolation and characterization of an angiotensin converting enzyme substrate from vitellogenic ovaries of Neobellieria bullata. Peptides. 2002;23:1853–1863. doi: 10.1016/s0196-9781(02)00144-4. [DOI] [PubMed] [Google Scholar]