Abstract
Arylsulphatases B (ASB) and A (ASA) are subject to a unique post-translational modification that is required for their function. The modification reaction, conversion of an active-site cysteine into a formylglycine, becomes saturated when these enzymes are overexpressed. We have removed the possibility of in vivo modification by expressing mutants of ASB and ASA in which the active-site cysteine is substituted with a serine. These mutants are expressed much more efficiently when compared with the native enzymes under identical conditions. The purified ASB mutant can then be converted into catalytically active ASB in vitro using vanadate and light.
Keywords: expression, mutagenesis, oxidation, sulphatase, toxicity, UV, vanadate
Abbreviations: ASA, arylsulphatase A; ASB, arylsulphatase B; CHO, Chinese-hamster ovary; DPBS, Dulbecco PBS; ER, endoplasmic reticulum; ERT, enzyme-replacement therapy; 4-MU, 4-methylumbelliferone; 4-MUS, 4-methylumbelliferyl sulphate; NPT, neomycin phosphotransferase; QPCR, quantitative PCR; RT, reverse transcriptase; SUMF1, sulphatase-modifying factor 1
INTRODUCTION
At least six lysosomal storage diseases are caused by deficiencies in a related family of mammalian sulphatase enzymes. ERT (enzyme-replacement therapy) provides a means of treating these disorders [1], but this approach presupposes an adequate supply of the recombinant enzyme.
The common biopharmaceutical manufacturing line CHO (Chinese-hamster ovary)-K1 has been used to establish clones expressing a number of sulphatases. Despite optimization of the expression vector, careful clonal selection and, in one case, dihydrofolate reductase amplification, high-yield cell lines have not been obtained [2–6]. Similarly, overexpression of ASB (arylsulphatase B) from recombinant retrovirus in deficient human fibroblasts has been reported to occur only at the expense of endogenous sulphatase expression levels [7]. Overexpression of ASA (arylsulphatase A) also causes a measurable decrease in endogenous sulphatases [8]. These results are consistent with a limit to the total sulphatase output of a mammalian cell.
A biosynthetic limitation specific to sulphatases has been described in the literature. All sulphatases share a unique post-translational modification [9]. A single active-site cysteine is converted into a hydrated formylglycine [10]. The converted residue is situated within a highly conserved peptide sequence that is required for the activation reaction to occur [11]. Conversion is strictly required for sulphatase activity and occurs shortly after translocation of nascent sulphatase into the ER (endoplasmic reticulum) of eukaryotes [12].
Cysteine conversion proceeds by sequential oxidation and hydrolysis reactions. The oxidation step is enzyme catalysed and, hence, saturable [13]. Since the cell normally expresses sulphatase enzymes at relatively low levels, it is not unexpected that the conversion step might be overwhelmed by large amounts of the recombinant enzyme. Saturation of the sulphatase activation process might be expected to have detrimental effects on enzyme yield and on the health of the overexpressing line.
Saturation of the sulphatase activation pathway in the ER will initially result in a failure to convert the active-site cysteine into formylglycine in newly translocated sulphatase. Failure to convert will result in an additional cysteine residue within the sulphatase sequence, which is not accounted for in the normal folding pathway. The introduction of additional cysteine residues into proteins can lead to misfolding as a result of inappropriate disulphide bonds [14–16]. The degree of misfolding probably depends on the particular sulphatase in question. Of note, the overexpression of iduronate-2-sulphatase and N-acetylgalactosamine-6-sulphatase results in secretion of large amounts of folded, unconverted enzyme, whereas the overexpression of ASB does not. In theory, the accumulation of misfolded sulphatase within the ER could stress the cell through the unfolded protein and the ER overload responses [17,18]. Subsequent retrotranslocation and accumulation of misfolded sulphatase in the cytoplasm could, in turn, tax the proteasome system and result in aggresomes, which are toxic to the cell [19]. These effects would select against cell lines that overexpress ASB.
The enzyme responsible for active-site cysteine conversion, SUMF1 (sulphatase-modifying factor 1), has recently been cloned and characterized [20,21]. Transient co-transfection of SUMF1 with iduronate-2-sulphatase and N-acetylgalactosamine-6-sulphatase has been reported to increase yields of active enzyme by 5- and 50-fold respectively. By relieving saturation of the sulphatase conversion step, co-expression of SUMF1 with sulphatase enzymes will probably remove the constraint that low sulphatase yield has had on efforts to treat sulphatase deficiencies by ERT [22] (it should be pointed out that the production of 1 mol of H2S for each mol of activated sulphatase might harm the cell under conditions where sulphatase-activation capacity is augmented).
We have sought alternative approaches to the overexpression of sulphatases. Previous studies have shown that sulphatase mutants in which a serine residue has been substituted for the active-site cysteine are expressed at levels that are at least comparable with those of the corresponding native enzymes [23–26]. A serine mutant might be expected in some cases to be expressed more efficiently than native enzyme since it will not be prone to misfolding as a result of inappropriate disulphide formation. For the same reason, and, again, depending on the specific sulphatase involved, overexpression of the serine mutant might be less detrimental to the host cell since cellular stress related to the accumulation of misfolded protein would be mitigated. Importantly, serine mutants of ASB and ASA have been shown to be virtually indistinguishable from their respective native counterparts by epitope analysis and X-ray diffraction [24,28,29]. The challenge is in finding a process for converting serine into formylglycine in secreted serine-mutant protein.
An in vitro sulphatase conversion reaction would ideally target a single amino acid, the active-site serine. The binding specificity of the sulphatase active site can be exploited to achieve this end. A wide variety of small oxo-anionic species bind and inhibit sulphatases, presumably by competing for the sulphate-binding site. Vanadate has been demonstrated to have a particularly high affinity [30,31]. The crystal structure of ASB complexed to vanadate has been solved: a single vanadate occupies the active-site sulphate-binding pocket [31]. Vanadate is a mimic of phosphate and sulphate, but vanadium has low-energy redox chemistry that phosphorus and sulphur do not have. In particular, vanadate (V5+) can undergo reduction to vanadyl (V4+) in the presence of suitable electron sources [32]. This reaction is facilitated by light. When vanadate is bound to protein, the electron sources for vanadate reduction are amino acid side chains. Specific side-chain oxidations mediated by vanadate and light have been described previously [33]. To date, these have all occurred in phosphate-binding proteins. In the best-characterized case, that of myosin, a serine residue within the ATP-binding pocket is converted into a formylglycine as a result of its proximity to vanadate [34–39].
We report that the serine mutants of ASB and ASA are expressed at higher levels than the native enzymes under identical conditions. We also report that vanadate and light can be used to convert the serine mutant of ASB (ASB C91S) into catalytically active ASB. The photoreaction is presumed to involve conversion of the active-site serine into a formylglycine in a manner analogous to that observed for the ATP-binding site of myosin. Vanadate-dependent conversion of sulphatase mutants represents a novel method of sulphatase production and an interesting example of the light-dependent activation of an enzyme.
MATERIALS AND METHODS
Materials
The BP15 rabbit and G192 sheep polyclonal anti-ASB antibodies were raised against native recombinant human enzymes. Rabbit polyclonal anti-ASA was a gift from Professor Arvan Fluharty (UCLA Mental Retardation Research Center, Los Angeles, CA, U.S.A.). Antibodies were purified over Protein G columns before use. PCR primers were synthesized by Qiagen. SybrGreen™ QPCR (quantitative PCR) reagents were purchased from Roche Biochemicals. Highly purified recombinant human ASB was manufactured at BioMarin Pharmaceutical.
Preparation of vanadate solutions
A 100 mM stock solution of sodium metavanadate (Sigma, S6383) was prepared as described previously [40].
ASB activity assays
Samples were diluted 1000-fold in assay buffer (50 mM NaOAc, pH 5.6). Aliquots of the dilutions (5 μl) were combined with 150 μl of 5 mM 4-MUS (4-methylumbelliferyl-sulphate substrate; Sigma) in assay buffer and incubated at 37 °C for 20 min in black Costar flat-bottom plates. Reactions were terminated with 150 μl of 350 mM glycine and 440 mM sodium carbonate (pH 10.7). Fluorescence was measured with a micro-plate fluorimeter (Molecular Devices, Sunnyvale, CA, U.S.A.) using an excitation wavelength of 366 nm and an emission wavelength of 446 nm. All samples were run in duplicate. One unit of enzyme activity is defined as 1 μmol of 4-MU (4-methylumbelliferone) produced per minute at 37 °C. For quantification of ASB using the activity assay described, a specific activity of 67 units/mg was used for ASB. Total protein was measured by Bradford assay (Bio-Rad Laboratories) using BSA as the standard.
Protein quantification
Concentrations of purified ASB and ASB C91S were calculated from A280 values and theoretical molar absorption coefficients were based on amino acid composition. The latter were obtained using ProtParam software (www.expasy.com). Both ASB and ASB C91S had theoretical molar absorption coefficients of 1.08×105 M−1·cm−1. Amounts of each protein were derived using a molecular mass of 56 kDa.
ASB ELISA
Nunc MaxiSorp™ ELISA plates were coated with sheep anti-ASB IgG (SH-G192; BioMarin Pharmaceutical) at 10 μg/ml in sodium carbonate buffer (pH 9.5). Excess antibody was removed by washing with Dulbecco's PBS containing 0.1% (v/v) Nonidet P40. Non-specific binding sites were blocked with the same buffer containing 2% (w/v) BSA. A series of ASB standards of known concentration were prepared by diluting a recombinant human ASB stock solution into the blocking buffer. Samples were similarly prepared at dilutions of 1:2, 1:5 and 1:10. Diluted standards and samples were added to coated wells in duplicate and incubated for 1 h at 37 °C. After incubation, wells were washed and the remaining bound antigen was labelled with rabbit anti-ASB IgG (BP15H; BioMarin Pharmaceutical), conjugated to horseradish peroxidase. After washing again to remove the unbound antibody, horseradish peroxidase activity was detected colorimetrically using a Bio-Rad TMB EIA substrate kit. Wells were read with a micro-plate spectrophotometer (Molecular Devices) and data analysed with SoftMax Pro software.
ASA Western blotting
For Western blotting, proteins were separated by reducing SDS/PAGE, electroblotted on to PVDF membranes, and blocked for 1 h at room temperature (22 °C) with blocking buffer containing 2% (w/v) dry milk powder in Tris-buffered saline (TBS: 20 mM Tris/HCl and 150 mM NaCl, pH 7.5) with 0.05% Nonidet P40. ASA antigen was detected with anti-(recombinant human ASA) antiserum diluted 25000-fold into blocking buffer. Bound primary antibodies were then detected with affinity-purified alkaline phosphatase-conjugated goat secondary antibody (Promega) diluted 5000-fold in blocking buffer. Blots were developed with Western Blue™ colorimetric substrate (Promega), rinsed with distilled water and scanned with a flat-bed scanner using Canvas software (Deneba; ACD Systems of America, Miami, FL, U.S.A.).
QPCR
RNA was extracted using Qiagen RNeasy™ reagents and reverse-transcribed with Superscript II+ (Invitrogen) and an oligo (T)25VN primer according to the manufacturer's instructions. cDNA was assayed using a Roche LightCycler™ with SybrGreen™ reagents. Primers specific for ASB (QASBF, 5′-AGACTTTGGCAGGGGGTAAT-3′; QASBR, 5′-CAGCCAGTCAGAGATGTGGA-3′), ASA (ASAF, 5′-CGCCCCTTCCTGTACTA-3′; ASAR, 5′-CAGGTCCATTGTCTGCAGTG-3′), NPT (neomycin phosphotransferase; MutSeqF, 5′-GCCGGCCACAGTCGATGAATCC-3′; MutSeqR, 5′-CTTGTCGATCAGGATGATCTGG-3′) and the hamster ribosomal S14 protein (CHOS14F, 5′-GAAACTATCTGCCGGGTGAC-3′; CHOS14R, 5′-TTTGATATGCAGGGCAGTGA-3′) were used. All samples were run in duplicate. The primers do not differentiate between native and mutant sequences. Quantification and melting curve analyses of the data were performed with LightCycler™ software (Roche).
ASB C91S expression construct
A human ASB cDNA was amplified from human liver cDNA (BD Biosciences) by high-stringency PCR using Stratagene PfuTurbo™ polymerase and the primers ASBF1 (5′-GCGATAGGTACCGCCATGGGTCCGCGCGGCGCGGCGAGC-3′) and ASBR2 (5′-GCGATACTCGAGCCCTGAAATCCTACATCCAAGGG-3′). The cDNA was sequenced and found to be identical with previous description (GenBank® accession number J05225). The cDNA was then reamplified with a different 5′-primer to decrease the GC bias at the 5′-end without changing the amino acid sequence (ASBatF, 5′-GCGATAGGTACCGCCATGGGTCCTAGAGGAGCTGCTTCTTTGCCTCGAGGACCCGGACCTCGGCGGCTGCTCCTC-3′). The engineered cDNA was digested with KpnI and XhoI and ligated into pcDNA3.1(+) (Invitrogen) to give pcDNA3.1-ASB. The 5′-modified version of ASB was used to make both the native and C91S proteins. To introduce the C91S mutation into the coding sequence of ASB, the pcDNA3.1-ASB plasmid was cut with AatII and the vector portion recircularized to remove the 5′-182 bp of the ASB cDNA. The deleted vector was then cut with PstI and the vector portion again recircularized to remove the 3′-760 bp of the cDNA. The truncated ASB construct was then cut a third time with EagI and XcmI, treated with alkaline phosphatase, gel-purified and ligated to a double-stranded oligonucleotide containing the C91S mutation, a T→A substitution. The mutated position is underlined (ASBmut3, 5′-PGGCCGGCGGGGTGCTCCTGGACAACTACTACACGCAGCCGCTGAGCACGCCGTCGCGGAGCCAGCTGC; ASBmut4, 5′-PCAGCTGGCTCCGCGACGGCGTGCTCAGCGGCTGCGTGTAGTAGTTGTCCAGGAGCACCCCGCC-3′). The mutation removes an ApaLI site from the ASB coding sequence. The remaining, mutagenized, portion of ASB, flanked by AatII and PstI sites was then used to reconstruct a full-length ASB C91S cDNA by ligation with KpnI–AatII and PstI–XbaI fragments from pcDNA3.1 (+)-ASB into pcDNA3.1 (+) to give pcDNA3.1-ASB C91S. The reconstructed cDNA was sequenced and found to be identical with the published sequence except for the engineered changes described.
ASA C69S expression construct
A human ASA cDNA was amplified from human liver cDNA (BD Biosciences) by high-stringency PCR using Stratagene PfuTurbo™ polymerase and the primers AryAF1 (5′-GCGATAGGTACCGCCATGGGGCACCGCGGTCCCTCCTC-3′) and AryAR (5′-GCGATACTCGAGTCAGGCATGGGGATCTGGGCAATG-3′). The amplified cDNA was cut with KpnI and XhoI for ligation into pcDNA3.1 (+). The cDNA was sequenced and found to be identical with that described previously (GenBank® accession number X52150). The ASA C69S was created in the same vector using the Promega Altered Sites II™ kit and a single mutagenic primer that introduces a T→A substitution. The mutated base is underlined (ASAser, 5′-PCGGCCGGTCAGGAGGGCGGCCCTAGAGGGTGTGCTCAGAGACACAGGCAC-3′).
Transfection of CHO-KI and selection of stably transfected pools of cells
The expression constructs, along with empty vector, were linearized with SalI and gel-purified. CHO-K1 cells were seeded at 2×105 cells/well in Corning Cell-Well™ 6-well polystyrene plates and allowed to grow overnight. Cells were transfected with Bio-Rad Cytofectene™ reagent according to the manufacturer's instructions. Cells were selected in UltraCHO™ (Cambrex, East Rutherford, NJ, U.S.A.) medium supplemented with 2.5% (w/v) dialysed fetal bovine serum, 2 mM glutamine, 50 μg/ml gentamicin, 2.5 μg/ml amphotericin and either 450 or 900 μg/ml G418. Resistant colonies were evident within a week. All transfections yielded hundreds of colonies. Mock transfection dishes had no colonies. After 12 days, resistant colonies for each of the construct types were pooled, expanded and frozen.
Experiments to compare expression of stably transfected pools of cells
Pooled cells stably transfected with each plasmid type and selection level were thawed and seeded in duplicate T-25 flasks at 1.2×104 cells/cm2 in UltraCHO™ medium supplemented with 2.5% dialysed fetal bovine serum, 2 mM glutamine, 50 μg/ml gentamicin, 2.5 μg/ml amphotericin and 450 or 900 μg/ml G418. After 24 h of incubation at 37 °C with 5% CO2, cell layers were rinsed with Hank's buffered salt solution and fed with seeding medium from which serum had been omitted. Flasks were then incubated for an additional 48 h. The supernatant was removed, clarified by filtration through a 0.2 μm polyethersulphone filter and stored at 4 °C. Cells were trypsinized and counted with a Coulter counter. Trypsinized cells were then pelleted at 500 g, washed with DPBS (Dulbecco PBS) and stored at –80 °C. Cell lysates were prepared by subjecting DPBS-resuspended cell pellets to three cycles of freeze–thaw using liquid nitrogen and a 37 °C water-bath.
Production of ASB C91S
The ASB C91S coding sequence was released from pcDNA3.1-ASB C91S by digesting with KpnI and XbaI and then ligated into a second vector, pBMPCINt1. The pBMPCINt1-ASB C91S construct was linearized with SalI, gel-purified and used to transfect CHO-S (Invitrogen) cells as described above. Cells were selected with 450 μg/ml G418. A total of 11 clones were picked and screened for expression of ASB C91S by ELISA as described above. Clone 7 had the highest volumetric productivity of ASB C91S (1 μg/ml) with an associated specific productivity of 3 pg·cell−1·day−1 under these conditions. Clone 7 was used for production of ASB C91S. Cells were seeded on to Cytopore 1™ beads in a 250 ml Corning spinner flask using Excell 302 medium (JRH Biosciences, Lenexa, KS, U.S.A.) supplemented with 2.5% dialysed fetal bovine serum, 2 mM glutamine, 50 μg/ml gentamicin, 2.5 μg/ml amphotericin and 450 μg/ml G418. Serum was gradually reduced to 0% over a 2-week period. The medium was harvested every 2 days, filtered through a 0.2 μm polyethersulphone sterile filter unit and stored at 4 °C.
Purification of ASB C91S
ASB C91S was purified on an anti-ASB (SH-G192; BioMarin Pharmaceutical) affinity column prepared by coupling the antibody with NHS-Sepharose beads (Bio-Rad Laboratories). Harvest medium was loaded on to the column after clarification by passage through a 0.2 μm polyethersulphone filter. Unbound proteins were removed by washing the column with DPBS. ASB C91S was eluted with 100 mM glycine/HCl (pH 2.25). Fractions were collected into tubes containing a Tris/HCl solution such that the pH of the eluate was raised immediately to 6.5. The purified material from the affinity column was buffer-exchanged and concentrated into 50 mM NaOAc (pH 6.5) with a Millipore Centricon Plus-20™ (8000 NMWL). Purified protein was quantified as described previously and stored at 4 °C.
Photochemical reactions
Photochemical reactions were performed in an Ace Glass Photochemical safety cabinet (7836-20). Two UVP Pen-Ray 5.5 W lamps (90-0012-02) were used as light sources. The lamps were powered by a UVP Photochemical UV power supply (99-0055-01) and housed in an Ace Glass jacketed immersion well (7892-45). Lamp temperature was held at 20 °C with a 1:1 mixture of Flag™ anti-freeze and tap water, which was pumped through the immersion well jacket at 16 litres/min. Coolant temperature was maintained with a recirculating chiller. Samples in microfuge tubes or microtitre plates were floated in a water bath in a 500 ml jacketed beaker. The beaker temperature was controlled with a second recirculating chiller. The lamp was positioned immediately below the samples. UVB radiation was removed with the anti-freeze mixture (which absorbs strongly at 280 nm) and with polystyrene plates. Irradiance was measured with an Oriel Goldilux™ Smartmeter Photometer using calibrated UVA (365 nm) and UVB (280 nm) probes. Samples were irradiated in Corning clear polystyrene 96-well plates or 1.5 ml polypropylene microfuge tubes (USA Scientific 1415-2600). Specific conditions for each experiment are described in the Figure legends.
RESULTS
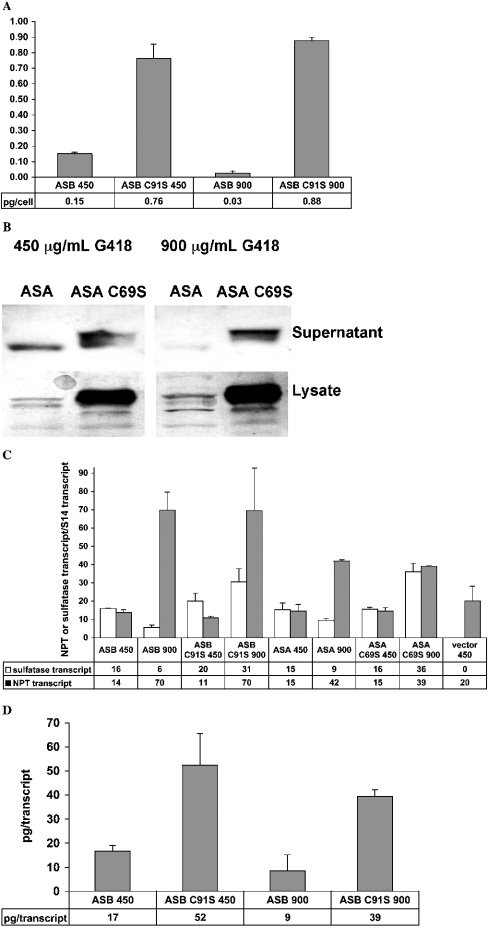
CHO cells expressing ASB, ASB C91S, ASA and ASA C69S were created to ascertain the level to which sulphatase activation affects the expression of sulphatase proteins. All sequences were cloned into identical vectors. All vectors contained the NPT gene in the same expression cassette to allow selection on G418. The sulphatase and NPT expression cassettes were proximate in the vectors, with ≤800 bp of sequence between them. Vectors expressing the native and mutant enzymes differed by a single base pair. Cells in replicate 6-well plates were transfected at the same time with identical amounts of plasmids using the same reagents. Transfected cells were selected at 450 and at 900 μg/ml G418, assuming that the latter level would enforce the requirement for high expression of NPT and the immediately adjacent sulphatase. Large pools of G418-resistant colonies were used for comparison of protein and transcript levels to eliminate clonal effects. Identical numbers of cells representing each stably transfected pool were used in the protein expression studies in duplicate flasks. Sulphatase protein in conditioned medium from each stably transfected pool was determined by ELISA for ASB and ASB C91S or by Western blot for ASA and ASA C69S. Protein levels were normalized to total cells in each flask as measured with a Coulter counter. Sulphatase and NPT transcript numbers in each pool of cells were quantified by RT (reverse transcriptase)–QPCR assay. Transcript levels were normalized to an internal control, the hamster ribosomal S14 protein transcript, also assayed by RT–QPCR [41]. Normalization of protein levels in conditioned medium to S14 transcript levels as a proxy for cell number did not change the relative relationships among the samples. These results indicate at least three differences in the expression behaviour of the native and serine-mutant sulphatases: (i) serine-mutant protein is present in conditioned medium at higher levels than native enzyme on both cell and transcript normalized bases; (ii) transcription of the native enzymes, but not the serine-mutant enzymes, becomes uncoupled from transcription of NPT with increasing G418 selection; and (iii) in a possibly related observation, serine-mutant transcript appears to be converted into secreted protein more efficiently than native transcript.
Protein expression of native and mutant sulphatases
When measuring the quantity of secreted sulphatase protein in conditioned medium by ELISA, it was evident that pools of stably transfected cells expressing ASB C91S protein did so at significantly higher levels than pools of cells expressing ASB (Figure 1A). At the 450 μg/ml G418 selection level, ASB C91S was present in the conditioned medium at normalized concentrations that were 5-fold greater than ASB. This difference became even more pronounced in cells selected at 900 μg/ml G418, where ASB C91S was present in conditioned medium at normalized concentrations that were 29-fold greater than ASB. Similarly, Western blotting with an anti-ASA antibody demonstrated that normalized ASA C69S levels in conditioned medium were significantly higher when compared with those of native ASA. The anti-ASA Western blot further revealed that the ASA C69S protein was present at much higher levels than ASA within the cell. A conservative estimate of Western blot band intensities for ASA and ASA C69S at a selection level of 900 μg/ml G418 suggests that there was at least 10-fold more ASA C69S in conditioned medium than native ASA expressed under identical conditions (Figure 1B). The difference in the amounts of intracellular ASA and ASA C69S appeared to be much greater than 10-fold. Strikingly, levels of secreted ASB (ELISA) and ASA (Western blot) both decreased on selection at higher G418 levels.
Figure 1. Quantification of sulphatase proteins and transcripts.
(A) Specific productivity of stably transfected pools of cells expressing either ASB or ASB C91S at 450 μg/ml G418 and 900 μg/ml G418. The medium was conditioned for 48 h before harvest. Samples were taken from duplicate T-25 flasks and clarified by centrifugation. Quantities of ASB and ASB C91S in the conditioned media were assayed in duplicate by ELISA as described in the Materials and methods section. Cell numbers were measured twice with a Coulter counter and averaged. The S.D. of total cells per well for all wells tested was 20% of the mean. Units for the y-axis are total pg of ASB or ASB C91S in the medium per cell at the end of the 48 h incubation period. Means±S.D. are shown. (B) Western blots of secreted (supernatant) and intracellular (lysate) protein preparations from pools of stably transfected CHO cells expressing ASA or ASA C69S. Equal amounts of protein, as determined by Bradford assay, were loaded in each well of an SDS/PAGE gel. Gels were blotted and probed with the anti-ASA antibody as described in the Materials and methods section. Samples from duplicate flasks of cells expressing ASA or ASA C69S were compared, with representative lanes shown. (C) Normalized NPT and sulphatase transcript levels obtained from pools of stably transfected cells selected at either 450 μg/ml G418 (450) or 900 μg/ml G418 (900) and expressing different sulphatase proteins. Cells were trypsinized, washed with DPBS and pelleted before isolation of RNA and reverse transcription as described in the Materials and methods section. QPCR was done in triplicate with means and S.D. shown. The hamster ribosomal S14 transcript was also quantified as an internal control for each RNA sample. NPT and sulphatase transcript levels between samples were normalized with the internal S14 controls. (D) Ratio of ASB or ASB C91S protein in conditioned medium per ASB or ASB C91S transcript within pools of stably transfected cells. Samples were taken from duplicate T-25 flasks. ASB proteins in the different samples were quantified in duplicate by ELISA and normalized to cell number measured twice with a Coulter counter as described in (A); transcript levels in total RNA isolated from each flask of cells were quantified by RT–QPCR and normalized to the ribosomal S14 internal standard as described in (C). RT–QPCRs were run in triplicate on each RNA sample. The units of the y-axis are pg of protein per transcript encoding the protein.
Transcription of native and mutant sulphatases
As expected, NPT transcript levels were higher in cells surviving at 900 μg/ml G418 when compared with that in cells surviving at 450 μg/ml G418. More surprising was the observation that native ASB and ASA transcript levels did not increase in parallel with the NPT transcripts (Figure 1C). In agreement with the levels of protein in the medium, transcript levels for the native enzymes actually decreased in cells surviving at 900 μg/ml G418. This result suggests that transcription of the native sulphatases and transcription of NPT become uncoupled in cells surviving at higher G418 concentrations. That this uncoupling would be required for cells to remain viable is indicative of some kind of toxicity associated with expression of native ASB and ASA. In contrast with the native transcripts, ASB C91S and ASA C69S transcript levels increased in parallel with NPT when G418 selection was increased from the 450 to 900 μg/ml. If it is toxicity that constrains increases in native sulphatase transcription with increasing G418 selection, then serine-mutant sulphatase transcription is not subject to the same effect.
Protein secretion efficiency
The quantity of secreted ASB and ASB C91S protein derived per transcript was calculated from the cell-normalized ASB and ASB C91S protein levels in conditioned medium and S14-normalized transcript levels in the cells expressing each protein. Significantly, more ASB C91S protein was derived from similar amounts of transcript relative to native ASB at both 450 and 900 μg/ml G418 (Figure 1D). This result is consistent with some native ASB being diverted from the secretory pathway, possibly as a result of misfolding and subsequent degradation. For ASA, the Western-blotting results demonstrate a prominent difference between intracellular levels of ASA and ASA C69S at the 450 μg/ml G418 selection level (Figure 1B), despite nearly identical levels of each transcript (Figure 1C).
Activation of ASB C91S with vanadate and light
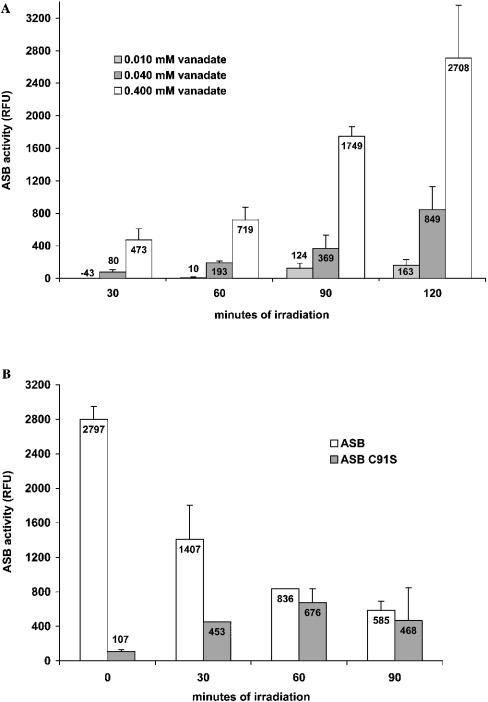
On the basis of the crystal structure of ASB complexed with vanadate, the near structural identity of ASB and ASB C91S [24] and the documented behaviour of myosin–vanadate complexes on irradiation, we reasoned that irradiation of solutions of ASB C91S and vanadate might convert the active-site serine of ASB C91S into a formylglycine and render the protein catalytically active. To test this hypothesis, affinity-purified ASB C91S was irradiated with UVA light in the presence of 2, 20, 40 and 400 μM sodium metavanadate in 50 mM NaOAc (pH 6.5) and 100 mM NaCl. Light below 300 nm was removed by placing appropriate absorptive materials between the light source and the samples (see the Materials and methods section). Treated samples were then assayed for activity against 4-MUS after dilution of the vanadate to concentrations below those necessary to inhibit the sulphatase. ASB C91S was converted into catalytically active enzyme by this procedure. Maximal activation was achieved at the highest vanadate concentration (Figure 2A). No activation was observed by irradiation in the absence of vanadate or with vanadate in the absence of irradiation. Increases in the amount of activated enzyme were observed through the last time point under the conditions used in this experiment. Activation was consistently observed over a number of experiments, with the results of a typical experiment shown.
Figure 2. Activation of ASB C91S with vanadate and light.
(A) The dependence of in vitro activation of ASB C91S on vanadate concentration and time of irradiation. For this experiment, ASB C91S (10 μl, 0.5 mg/ml) in 50 mM NaOAc (pH 5.6) and 100 mM NaCl was added to a 96-well Costar polystyrene round-bottom microtitre plate. Sodium metavanadate from a 100 mM stock was diluted 1000-, 100- and 25-fold in water. A 1 μl aliquot of each vanadate dilution was then added to wells containing ASB C91S to achieve final concentrations of 0.010, 0.040 and 0.400 μM vanadate. The plate was then irradiated for 120 min as described in the Materials and methods section, with the bottoms of the wells immersed in water maintained at 20 °C. Sample aliquots (2 μl) were placed on ice in the dark at 30, 60, 90 and 120 min and assayed as described in the Materials and methods section. The experiment was run with a single well per condition. Assays were run in duplicate. As a blank, the activity of ASB C91S that had not been treated with vanadate or light was subtracted from the activity of the test samples. Means are shown. Error bars represent half the distance between the minimum and maximum values. The y-axis represents RFU (relative fluorescence units) of 4-MU produced by the hydrolysis of 4-MUS. The negative activity value for the 0.010 mM vanadate reaction at 30 min of irradiation may have resulted from insufficient activation coupled with inhibition of residual arylsulphatase activity in the ASB C91S sample, despite dilution to reduce the vanadate concentration. The ASB C91S blank did not contain vanadate and would not have been subject to this inhibition. The effect was probably overcome by activation of ASB C91S with longer irradiation times. (B) In vitro activation of ASB C91S concomitant with inactivation of native ASB by vanadate and light. For this experiment, ASB C91S (50 μl, 0.24 mg/ml) and native ASB (50 μl, 0.25 mg/ml) in 50 mM NaOAc (pH 5.6) and 100 mM NaCl were placed in 1.5 ml polypropylene microcentrifuge tubes (USA Scientific). A 0.2 μl aliquot of a 2 mM sodium metavanadate solution in 20 mM Tris/HCl (pH 9.6) was added to each tube to give a final concentration of 0.008 mM vanadate. The tubes were then irradiated for 90 min as described in the Materials and methods section. Sample aliquots (5 μl) were placed on ice in the dark at 30, 60 and 90 min and assayed as described in the Materials and methods section. The experiment was run with a single well per condition. Assays were run in duplicate. As a blank, the activity of ASB C91S incubated with vanadate in the dark was subtracted from the activity of the test samples. Means are shown. Error bars represent half the distance between the minimum and maximum values. The y-axis represents RFU of 4-MU produced by the hydrolysis of 4-MUS. UVA irradiance was measured at 1, 15, 25 and 40 min during the course of the experiment with a probe fixed at the same distance from the source as the samples. Irradiance had a mean and S.D. over all measurements of 90±5 μW/cm at 365 nm.
The effect of the irradiation procedure on the activity of native ASB as well as the activation time course for the C91S mutant was then determined using a different experimental format (Figure 2B). ASB C91S reached a maximum level of activation at approx. 60 min using the specified conditions. Unfortunately, it was observed that the native enzyme was inactivated under the same conditions, indicating that further photo-oxidation may occur after initial conversion into formylglycine. Native ASB was not inactivated by irradiation in the absence of vanadate or with vanadate in the absence of irradiation. The effect of the activation procedure on native ASB was consistently observed over a number of experiments, with the results of a typical experiment shown. As a consequence of native ASB inactivation, the observed activity of a treated sample of ASB C91S at any given time will depend on the kinetics of oxidation to formylglycine as well as the kinetics of the subsequent reactions that inactivate the enzyme.
DISCUSSION
The results from these experiments with pools of stably transfected cells are consistent with enhanced protein secretion of serine-mutant sulphatases relative to native sulphatases. ASB C91S is present at significantly higher levels when compared with ASB in conditioned media under identical conditions. ASA C69S is present at significantly higher levels when compared with ASA in both conditioned media and within the cell under identical conditions. Reduced levels of the native enzymes may be a result of toxicity or of diminished efficiency of transit through the secretory pathway. It is conceivable that the former is a result of the latter, whereby blocked transit of the overexpressed native sulphatases leads to stresses on the cell that compromise the ability to survive under G418 selection. One explanation for this toxicity effect is that overexpression of the native sulphatases may lead to saturation of the SUMF1-mediated activation reaction in the ER. Saturation of this process would result in the persistence of a cysteine at position 91 of ASB and position 69 of ASA. The presence of an additional cysteine residue that is not accounted for in the normal folding pathway of the enzyme might, in turn, lead to misfolding as a result of inappropriate disulphide bond formation. Misfolded protein would not transit the remainder of the secretory pathway but instead be degraded either in the ER or in the cytoplasm. Problems in expression of native sulphatases pose a challenge to the development of these enzymes as therapeutics for treating associated lysosomal storage diseases by ERT. One potential means of answering this challenge may be co-expression of native sulphatases with SUMF1.
Another approach to overcoming limitations in sulphatase expression is to remove the requirement for in vivo activation of the enzymes entirely. We have demonstrated that inactive serine mutants of ASB and ASA can be expressed at much higher levels when compared with their native counterparts. The remaining task is to then convert the inactive mutant sulphatase into active enzyme. We have shown that this can be accomplished by oxidation with vanadate in the presence of UVA light. The structure of ASB complexed with vanadate is represented in the literature as the vanadate ester of a formylglycine hydroxy group [31]. A similar association between vanadate and a serine side chain can be envisaged to lead to light-catalysed two-electron transfer from the serine Cβ to vanadium. This reaction would result in the observed serine-mutant sulphatase conversion. The oxovanadium (V3+) generated as a by-product would be expected to com-proportionate in the presence of vanadate (V5+) to form 2 mol of vanadyl (V4+). Vanadyl slowly oxidizes back to vanadate in the presence of oxygen. The proximity of vanadate to the convertible residue leaves open the possibility of other redox mechanisms that do not require covalent association between vanadate and serine. In particular, hydrogen abstraction from Cβ by photoexcited vanadate might lead to di-radical intermediates that could resolve in a manner leading to conversion. Whereas the native form of ASB is inactivated by light and vanadate, we have not observed cleavage of the protein backbone as has been observed for other proteins. Cleavage at the oxidized serine in myosin is believed to occur by enolization of the resulting formylglycine through deprotonation of the backbone Cα. A second vanadate-dependent photo-oxidation step can then occur, leading to addition of molecular oxygen to the backbone Cα and eventual cleavage of the myosin peptide chain. Enolization of the aldehyde in the context of the sulphatase active site is less probable since the protein is expected to stabilize strongly the hydrated keto form of the side chain.
The sulphatase active site is highly conserved between both eukaryotic and prokaryotic sulphatases. For this reason, it is anticipated that light-dependent oxidation of serine mutants with vanadate will be applicable to other members of the sulphatase family. Improvements in the yield of active sulphatase would be required for this to be a practical method for sulphatase production. In particular, some form of intervention might be needed to protect the newly generated formylglycine residue from further reactions that destroy the catalytic activity of the enzyme. Intensive short-duration pulses of light might also be of benefit in this regard.
As a final note, this method represents an example of a light-activated signal amplification system in which the vanadate–sulphatase complex acts as the photo-detector and the activated sulphatase acts as the amplifier. The production of signal is self-limiting since the amplifier is inactivated under conditions of persistent stimulus.
Acknowledgments
We thank C. Cremo for helpful discussions on vanadate biochemistry and N. Zecherle for purification of the ASB C91S mutant.
References
- 1.Kakkis E. D., Muenzer J., Tiller G. E., Waber L., Belmont J., Passage M., Izykowski B., Phillips J., Doroshow R., Walot I., et al. Enzyme-replacement therapy in mucopolysaccharidosis I. N. Engl. J. Med. 2001;344:182–188. doi: 10.1056/NEJM200101183440304. [DOI] [PubMed] [Google Scholar]
- 2.Bielicki J., Fuller M., Guo X. H., Morris C. P., Hopewood J. J., Anson D. S. Expression, purification and characterization of recombinant human N-acetylgalactosamine-6-sulphatase. Biochem. J. 1995;311:333–339. doi: 10.1042/bj3110333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bielicki J., Hopwood J. J., Wilson P. J., Anson D. S. Recombinant human iduronate-2-sulphatase: correction of mucopolysaccharidosis-type II fibroblasts and characterization of the purified enzyme. Biochem. J. 1993;289:241–246. doi: 10.1042/bj2890241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bielicki J., Hopwood J. J., Melville E. L., Anson D. S. Recombinant human sulphamidase: expression, amplification, purification and characterization. Biochem. J. 1998;329:145–150. doi: 10.1042/bj3290145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Litjens T., Bielicki J., Anson D. S., Friderici K., Jones M. Z., Hopwood J. J. Expression, purification and characterization of recombinant caprine N-acetylglucosamine-6-sulphatase. Biochem. J. 1997;327:89–94. doi: 10.1042/bj3270089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webb E. Restricted integration into genomic DNA following transfection of Chinese-hamster ovary cells with N-acetylgalactosamine-4-sulphatase cDNA. Biotechnol. Appl. Biochem. 1996;23:205–207. [PubMed] [Google Scholar]
- 7.Anson D. S., Muller V., Bielicki J., Harper G. S., Hopwood J. J. Overexpression of N-acetylgalactosamine-4-sulphatase induces a multiple sulphatase deficiency in mucopolysaccharidosis-type-VI fibroblasts. Biochem. J. 1993;294:657–662. doi: 10.1042/bj2940657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohashi T., Matalon R., Barranger J. A., Eto Y. Overexpression of arylsulfatase A gene in fibroblasts from metachromatic leukodystrophy patients does not induce a new phenotype. Gene Ther. 1995;2:363–368. [PubMed] [Google Scholar]
- 9.Rommerskirch W., von Figura K. Multiple sulfatase deficiency: catalytically inactive sulfatases are expressed from retrovirally introduced sulfatase cDNAs. Proc. Natl. Acad. Sci. U.S.A. 1992;89:2561–2565. doi: 10.1073/pnas.89.7.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Figura K., Schmidt B., Selmer T., Dierks T. A novel protein modification generating an aldehyde group in sulfatases: its role in catalysis and disease. Bioessays. 1998;20:505–510. doi: 10.1002/(SICI)1521-1878(199806)20:6<505::AID-BIES9>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 11.Dierks T., Lecca M. R., Schlotterhose P., Schmidt B., von Figura K. Sequence determinants directing conversion of cysteine to formylglycine in eukaryotic sulfatases. EMBO J. 1999;18:2084–2091. doi: 10.1093/emboj/18.8.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dierks T., Lecca M. R., Schmidt B., von Figura K. Conversion of cysteine to formylglycine in eukaryotic sulfatases occurs by a common mechanism in the endoplasmic reticulum. FEBS Lett. 1998;423:61–65. doi: 10.1016/s0014-5793(98)00065-9. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt B., Selmer T., Ingendoh A., von Figura K. A novel amino acid modification in sulfatases that is defective in multiple sulfatase deficiency. Cell (Cambridge, Mass.) 1995;82:271–278. doi: 10.1016/0092-8674(95)90314-3. [DOI] [PubMed] [Google Scholar]
- 14.Wallis R., Cheng J. Y. Molecular defects in variant forms of mannose-binding protein associated with immunodeficiency. J. Immunol. 1999;163:4953–4959. [PubMed] [Google Scholar]
- 15.Simon P., Weiss F. U., Sahin-Toth M., Parry M., Nayler O., Lenfers B., Schnekenburger J., Mayerle J., Domschke W., Lerch M. M. Hereditary pancreatitis caused by a novel PRSS1 mutation (Arg-122→Cys) that alters autoactivation and autodegradation of cationic trypsinogen. J. Biol. Chem. 2002;277:5404–5410. doi: 10.1074/jbc.M108073200. [DOI] [PubMed] [Google Scholar]
- 16.Pomponio R. J., Norrgard K. J., Hymes J., Reynolds T. R., Buck G. A., Baumgartner R., Suormala T., Wolf B. Arg538 to Cys mutation in a CpG dinucleotide of the human biotinidase gene is the second most common cause of profound biotinidase deficiency in symptomatic children. Hum. Genet. 1997;99:506–512. doi: 10.1007/s004390050397. [DOI] [PubMed] [Google Scholar]
- 17.Cudna R. E., Dickson A. J. Endoplasmic reticulum signaling as a determinant of recombinant protein expression. Biotechnol. Bioeng. 2003;81:56–65. doi: 10.1002/bit.10445. [DOI] [PubMed] [Google Scholar]
- 18.Paschen W. Shutdown of translation: lethal or protective? Unfolded protein response versus apoptosis. J. Cereb. Blood Flow Metab. 2003;23:773–779. doi: 10.1097/01.WCB.0000075009.47474.F9. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Mata R., Gao Y. S., Sztul E. Hassles with taking out the garbage: aggravating aggresomes. Traffic. 2002;3:388–396. doi: 10.1034/j.1600-0854.2002.30602.x. [DOI] [PubMed] [Google Scholar]
- 20.Cosma M. P., Pepe S., Annunziata I., Newbold R. F., Grompe M., Parenti G., Ballabio A. The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell (Cambridge, Mass.) 2003;113:445–456. doi: 10.1016/s0092-8674(03)00348-9. [DOI] [PubMed] [Google Scholar]
- 21.Dierks T., Schmidt B., Borissenko L. V., Peng J., Preusser A., Mariappan M., von Figura K. Multiple sulfatase deficiency is caused by mutations in the gene encoding the human C(α)-formylglycine generating enzyme. Cell (Cambridge, Mass.) 2003;113:435–444. doi: 10.1016/s0092-8674(03)00347-7. [DOI] [PubMed] [Google Scholar]
- 22.Baenziger J. U. A major step on the road to understanding a unique posttranslational modification and its role in a genetic disease. Cell (Cambridge, Mass.) 2003;113:421–422. doi: 10.1016/s0092-8674(03)00354-4. [DOI] [PubMed] [Google Scholar]
- 23.Daniele A., Di Natale P. Heparan N-sulfatase: cysteine 70 plays a role in the enzyme catalysis and processing. FEBS Lett. 2001;505:445–448. doi: 10.1016/s0014-5793(01)02867-8. [DOI] [PubMed] [Google Scholar]
- 24.Brooks D. A., Robertson D. A., Bindloss C., Litjens T., Anson D. S., Peters C., Morris C. P., Hopwood J. J. Two site-directed mutations abrogate enzyme activity but have different effects on the conformation and cellular content of the N-acetylgalactosamine 4-sulphatase protein. Biochem. J. 1995;307:457–463. doi: 10.1042/bj3070457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Recksiek M., Selmer T., Dierks T., Schmidt B., von Figura K. Sulfatases, trapping of the sulfated enzyme intermediate by substituting the active site formylglycine. J. Biol. Chem. 1998;273:6096–6103. doi: 10.1074/jbc.273.11.6096. [DOI] [PubMed] [Google Scholar]
- 26.Millat G., Froissart R., Maire I., Bozon D. Characterization of iduronate sulphatase mutants affecting N-glycosylation sites and the cysteine-84 residue. Biochem. J. 1997;326:243–247. doi: 10.1042/bj3260243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reference deleted.
- 28.von Bulow R., Schmidt B., Dierks T., von Figura K., Uson I. Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis. J. Mol. Biol. 2001;305:269–277. doi: 10.1006/jmbi.2000.4297. [DOI] [PubMed] [Google Scholar]
- 29.Lukatela G., Krauss N., Theis K., Selmer T., Gieselmann V., von Figura K., Saenger W. Crystal structure of human arylsulfatase A: the aldehyde function and the metal ion at the active site suggest a novel mechanism for sulfate ester hydrolysis. Biochemistry. 1998;37:3654–3664. doi: 10.1021/bi9714924. [DOI] [PubMed] [Google Scholar]
- 30.Dibbelt L., Kuss E. Human placental sterylsulfatase. Interaction of the isolated enzyme with substrates, products, transition-state analogues, amino-acid modifiers and anion transport inhibitors. Biol. Chem. Hoppe. Seyler. 1991;372:173–185. doi: 10.1515/bchm3.1991.372.1.173. [DOI] [PubMed] [Google Scholar]
- 31.Bond C. S., Clements P. R., Ashby S. J., Collyer C. A., Harrop S. J., Hopwood J. J., Guss J. M. Structure of a human lysosomal sulfatase. Structure. 1997;5:277–289. doi: 10.1016/s0969-2126(97)00185-8. [DOI] [PubMed] [Google Scholar]
- 32.Sigel H., Sigel A. New York: Marcel Dekker; 1995. Vanadium and its Role in Life. [Google Scholar]
- 33.Muhlrad A., Ringel I. Use of vanadate-induced photocleavage for detecting phosphate binding sites in proteins. Met. Ions Biol. Syst. 1995;31:211–230. [PubMed] [Google Scholar]
- 34.Cremo C. R., Grammer J. C., Yount R. G. UV-induced vanadate-dependent modification and cleavage of skeletal myosin subfragment 1 heavy chain. 2. Oxidation of serine in the 23-kDa NH2-terminal tryptic peptide. Biochemistry. 1988;27:8415–8420. doi: 10.1021/bi00422a018. [DOI] [PubMed] [Google Scholar]
- 35.Cremo C. R., Grammer J. C., Yount R. G. Direct chemical evidence that serine 180 in the glycine-rich loop of myosin binds to ATP. J. Biol. Chem. 1989;264:6608–6611. [PubMed] [Google Scholar]
- 36.Cremo C. R., Long G. T., Grammer J. C. Photocleavage of myosin subfragment 1 by vanadate. Biochemistry. 1990;29:7982–7990. doi: 10.1021/bi00486a029. [DOI] [PubMed] [Google Scholar]
- 37.Cremo C. R., Grammer J. C., Yount R. G. Vanadate-mediated photocleavage of myosin. Methods Enzymol. 1991;196:442–449. doi: 10.1016/0076-6879(91)96038-s. [DOI] [PubMed] [Google Scholar]
- 38.Grammer J. C., Cremo C. R., Yount R. G. UV-induced vanadate-dependent modification and cleavage of skeletal myosin subfragment 1 heavy chain. 1. Evidence for active site modification. Biochemistry. 1988;27:8408–8415. doi: 10.1021/bi00422a017. [DOI] [PubMed] [Google Scholar]
- 39.Grammer J. C., Loo J. A., Edmonds C. G., Cremo C. R., Yount R. G. Chemistry and mechanism of vanadate-promoted photooxidative cleavage of myosin. Biochemistry. 1996;35:15582–15592. doi: 10.1021/bi961901g. [DOI] [PubMed] [Google Scholar]
- 40.Goodno C. C. Myosin active-site trapping with vanadate ion. Methods Enzymol. 1982;85:116–123. doi: 10.1016/0076-6879(82)85014-3. [DOI] [PubMed] [Google Scholar]
- 41.Cairns M. T., Church S., Johnston P. G., Phenix K. V., Marley J. J. Paraffin-embedded tissue as a source of RNA for gene expression analysis in oral malignancy. Oral Dis. 1997;3:157–161. doi: 10.1111/j.1601-0825.1997.tb00028.x. [DOI] [PubMed] [Google Scholar]