Abstract
The platelet-derived growth factor receptor-β (PDGFR-β) has a number of conserved cysteine residues on its cytoplasmic domain. We have examined whether the cysteine residues play a role in the enzymic function of PDGFR-β. We found that N-ethylmaleimide, which selectively alkylates free thiol groups of cysteine residues, completely inhibited the kinase activity of PDGFR-β. We then identified, through site-directed mutagenesis, two conserved cysteine residues critical for the enzymic function of PDGFR-β. Cys to Ser mutations for either Cys-822, positioned in the catalytic loop, or Cys-940, located in the C-terminal kinase subdomain, significantly reduced the activities of autophosphorylation and phosphorylation towards exogenous substrates. The non-reducing gel analysis indicated that neither of these cysteine residues contributes to the kinase activity by disulphide-bond formation. In addition, the individual mutation of Cys-822 and Cys-940 had no effect on protein stability or the binding of substrates or ATP, implying that these cysteine residues are involved in enzyme catalysis. Finally, proteolytic cleavage assays showed that the mutation of Cys-940, but not Cys-822, induced a protein conformational change. Taken together, these results suggest that Cys-940 contributes to the catalytic activity of PDGFR-β by playing a structural role, whereas Cys-822 contributes through a different mechanism.
Keywords: catalysis, cysteine, kinase, mutagenesis, N-ethylmaleimide, platelet-derived growth factor (PDGF)
Abbreviations: CD, cytoplasmic domain; DMEM, Dulbecco's modified Eagle's medium; DTT, dithiothreitol; FBS, fetal bovine serum; GAP, GTPase-activating protein; GST, glutathione S-transferase; HEK-293T cells, human embryonic kidney 293T cells; MALDI–TOF-MS, matrix-assisted laser-desorption ionization–time-of-flight mass spectrometry; NEM, N-ethylmaleimide; PDGFR-β, platelet-derived growth factor receptor-β; PI3K, phosphoinositide 3-kinase; PLC-γ, phospholipase C-γ; PTP, protein tyrosine phosphatase; RTK, receptor tyrosine kinase; SH2 domain, Src homology 2 domain; WT, wild-type
INTRODUCTION
Platelet-derived growth factor receptor-β (PDGFR-β) belongs to a family of RTKs (receptor tyrosine kinases) and triggers the propagation of cellular signals by activating their intrinsic tyrosine kinase [1,2]. After PDGF binding, the receptors are brought together by dimerization and transphosphorylate each other on a number of tyrosine residues in the cytoplasmic domain. Phosphorylation of tyrosine residues, including Tyr-857 on the activation loop (A loop), leads to receptor kinase activation [3–5]. The phosphorylated tyrosine residues also serve as the binding sites for many SH2 domain (Src homology 2 domain)-containing signalling proteins such as PLC-γ1 (phospholipase C-γ1) [6–8], PI3K (phosphoinositide 3-kinase) [9–11] and RasGAP (GTPase-activating protein of Ras) [9,11]. Recruited SH2-domain proteins are often phosphorylated at tyrosine residues by the receptors to be activated for propagating their downstream signalling events [12].
The kinase domain of RTKs, present in the receptor's cytoplasmic portion, is well conserved in both its overall architecture and the mechanism of its activation. The structure of the kinase domain is divided into N- and C-terminal subdomains (N- and C-lobes respectively), and between these two lobes is located the deep cleft of the active site in which ATP is bound. The catalytic reaction of the kinase is initiated by binding of substrate peptide through the centrally located A loop, which has to be prestabilized by phosphorylation in an open and extended conformation. The hydroxy group of substrate tyrosine should then be in line with γ-phosphate of ATP for phosphotransfer reaction. Another loop structure known as the catalytic loop, which resides in close proximity to the active site, provides the critical catalytic residues whose precise spatial configuration is required for the in-line arrangement of the hydroxy group and γ-phosphate [13–15].
Previous studies have implicated cysteine residues in the enzymic function of several tyrosine kinases. The kinase activity of an epidermal growth factor receptor has been shown to be inhibited by thiol-modifying reagents such as NEM (N-ethylmaleimide); however, no evidence has been reported for the involvement of a cysteine residue in enzymic function [16]. Another RTK of oncogenic c-Ret was demonstrated to be superactivated by UV through disulphide-linked receptor dimerization [17]. The importance of cysteine residues in enzymic function has also been documented for non-RTKs such as the lymphocyte-specific tyrosine kinase, p56lck and v-Src kinase [18–20].
Li and Schlessinger [21] have reported that PDGFRs undergo disulphide-linked dimerization after ligand binding, and that these interchain disulphide bonds might be generated by some rearrangement of intrachain disulphide bonds. However, further studies including identification of the cysteine residues responsible for the receptor dimerization have not been reported.
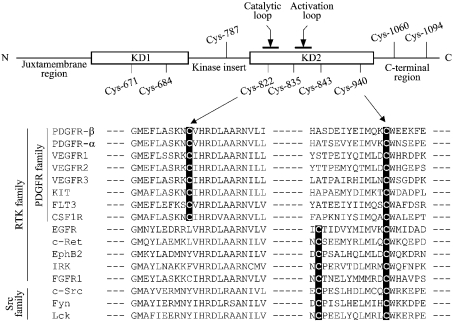
The CD (cytoplasmic domain) of PDGFR-β comprises the juxtamembrane region, the kinase domain that is separated by the sequence called kinase insert and the C-terminal region (Figure 1, top panel). In the CD of the human PDGFR-β, nine cysteine residues are present, with six scattered over the two separated kinase domains, one in the kinase insert and two in the C-terminal region. Cys-671 and Cys-684 are located in the N-terminal kinase domain (KD1) and Cys-822, Cys-835, Cys-843 and Cys-940 in the C-terminal kinase domain (KD2). Sequence alignment of various members of the tyrosine kinase family shows that Cys-940 is absolutely conserved throughout this family, with its C-terminal amino acid residue being invariably tryptophan (Figure 1, bottom panel). Cys-671, Cys-684, Cys-822 and Cys-843 are conserved only among the members of PDGFR family (Figure 1, bottom panel; results not shown). Interestingly, Cys-822 is positioned within the catalytic loop.
Figure 1. CD of the human PDGFR-β contains nine cysteine residues.
Top: CD of PDGFR-β (amino acids 557–1106) comprises a juxtamembrane region, two kinase domains (KD1 and KD2) separated by kinase insert and a C-terminal region. The positions of nine cysteine residues and the locations of the catalytic and activation loops are shown. The numbering of cysteine residues is based on the sequence of NP_002600. Bottom: sequence alignment of various human tyrosine kinases for the regions containing Cys-822 and Cys-940, which were identified in the present study to be critical for the enzymic function of PDGFR-β (see the Introduction section).
In the present study, we have examined whether these cysteine residues play roles in enzymic function of PDGFR-β. We identify two conserved cysteine residues, Cys-822 and Cys-940, to be critical for the kinase activity of PDGFR-β without affecting protein stability or the substrate or ATP binding. These cysteine residues do not contribute to PDGFR-β activity by disulphide-bond formation. Instead, the mutation of Cys-940, but not Cys-822, induces a protein conformational change of PDGFR-β.
MATERIALS AND METHODS
Materials
Horseradish peroxidase-linked rabbit IgG and mouse IgG were purchased from Amersham Biosciences. Ni2+-nitrilotriacetate resin was purchased from Qiagen. The polyclonal antibodies for PDGFR-β and PLC-γ, and site-specific phosphotyrosine antibodies were purchased from Santa Cruz Biotechnology. The monoclonal antibody for phosphotyrosine (4G10) and PDGF-BB were purchased from Upstate (Charlottesville, VA, U.S.A.). Unless specifically indicated, all other reagents were purchased from Sigma.
Expression and purification of PDGFR-β CD
His6-tagged human PDGFR-β CD was expressed and purified from Sf9 cells using Bac-to-Bac expression system from Life Technologies (Carlsbad, CA, U.S.A.). The cDNA fragment corresponding to the CD of human PDGFR-β (amino acids 557–1106) was PCR-amplified from the genomic DNA of HepG2 cells containing stably transfected WT (wild-type) human PDGFR-β cDNA [22]; upstream primer, TCCGGAATTCAGAAGAAGCCACGTTACGAG; and downstream primer, CTCGACAAGCTTCTACAGGAAGCTATCCTCTGC. The PCR product was cloned into EcoRI–HindIII sites of pFastBac HTb to generate pFastBac-PDGFR-β-CD and the sequence of the insert was verified by sequencing (ABI Prism 310). This plasmid was then cloned into the bacmid to generate a recombinant baculovirus. The baculovirus were infected into Sf9 cells at multiplicity of infection 4 for 72 h at 28 °C, and the cells were harvested and suspended in lysis buffer [20 mM Tris/HCl, pH 8.0, 100 mM KCl, 10% (v/v) glycerol, 2 mM 2-mercaptoethanol, 1 mM PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM 4-(2-aminoethyl)benzenesulphonyl fluoride, 10 μg/ml pepstatin and 1 mM Na3VO5] followed by homogenization after 1 cycle of freeze–thawing. His-tagged PDGFR-β CD was purified by Ni2+-nitrilotriacetate resin chromatography according to the manufacturer's instructions, and the proteins were dialysed and stored in the buffer containing 50 mM Tris/HCl (pH 8.0), 200 mM NaCl, 1 mM DTT (dithiothreitol) and 10% glycerol.
pFastBac-PDGFR-CD was used for site-directed mutagenesis (Quick-Xchange kit; Stratagene) to generate individual changes of the nine cysteine to serine residues. These plasmids were used for the generation of the cysteine mutant proteins of PDGFR-β CD using Sf9/baculovirus system as described above. One representative sequence of the oligonucleotides used for the site-directed mutagenesis is as follows: for C671S (Cys-671→Ser), sense: CAACCTGTTGGGGGCCTCTACCAAAGGAGGACCC; antisense: GGGTCCTCCTTTGGTAGAGGCCCCCAACAGGTTG. Complete sequence information is available on request.
In vitro kinase assays
In vitro kinase assays were performed in 50 μl volumes of reaction mixture containing 40 mM Hepes (pH 7.3), 10 mM MgCl2, 1 mM ATP (or 1 μCi [γ-32P]ATP; NEN Life Science, Boston, MA, U.S.A.), 0.1 mg/ml BSA, components contributed by the preparations of PDGFR-β CD and/or substrate proteins, 150 nM of PDGFR-β CD (unless otherwise indicated) and 1 μM of GST–PLC-γ (where GST stands for glutathione S-transferase) or GST–p85α (where indicated). Reactions were incubated at 30 °C for 20 min and subjected to 12% SDS/PAGE followed by either autoradiography (when [γ-32P]ATP was used) or Western-blot analysis (when unlabelled ATP was used). When NEM treatment is required, PDGFR-β was dialysed under anaerobic conditions in the same buffer as the above except DTT being omitted as described previously [23].
MALDI–TOF-MS (matrix-assisted laser-desorption ionization–time-of-flight mass spectrometry) analysis
PDGFR-β CD purified from baculovirus-infected Sf9 cells was incubated with 10 mM NEM at room temperature (20 °C) for 30 min, and was subjected to in-gel trypsin digestion and analysis with MALDI–TOF-MS (Voyager-DE STR; Applied Biosystems, Foster City, CA, U.S.A.). In brief, tryptic digest peptides were redissolved in a solution containing 0.1% trifluoroacetic acid and 50% (v/v) acetonitrile and mixed with an equal volume of matrix solution: 1 μl of this solution was spotted on to a stainless-steel MALDI target and analysed in delayed extraction and in positive-ion reflectron mode with an accelerating voltage of 20 kV, grid voltage at 64% and a delay time of 200 ns. For each spectrum, 200 laser shots were accumulated. The matrix used was 10 mg/ml α-cyano-4-hydroxycinnamic acid in 0.1% trifluoroacetic acid and 50% acetonitrile.
Construction of expression vectors for full-length PDGFR-β
The full-length human PDGFR-β cDNA was PCR-amplified from the HepG2 cells as described above and subcloned into the NheI–EcoRI sites of pcDNA3.1 (Invitrogen); upstream primer, AGCTGGCTAGCATGCGGCTTCCGGGTGCG; and downstream primer, CTCGACAAGCTTCTACAGGAAGCTATCCTCTGC. The cysteine mutants of full-length PDGFR-β were constructed by replacing approx. 1.5 kb of SacII–HindIII fragment of PDGFR-β cDNA with the corresponding sequence of pFastBac-PDGFR-β-CD containing Cys to Ser mutations.
Transfection and immunoprecipitation
HEK-293T cells (human embryonic kidney 293T cells) were cultured in DMEM (Dulbecco's modified Eagle's medium) with 10% (v/v) FBS (fetal bovine serum). Plasmid DNA (2 μg) was transfected into 5×105 cells on a 35 mm dish using CellFectin™ reagent (Invitrogen) according to the manufacturer's instructions. The transfected cells were incubated in DMEM with 10% FBS for 24 h and then further incubated for 16–20 h in DMEM containing 0.1% FBS before stimulation with 50 ng/ml PDGF-BB for 10 min. Cells were washed with ice-cold PBS and lysed in the lysis buffer (10 mM Tris/HCl, pH 7.4, 5 mM EDTA, 50 mM NaCl, 50 mM NaF, 1% Triton X-100, 10% glycerol, 10 μg/ml aprotinin, 1 mM PMSF, 10 μg/ml leupeptin, 10 μg/ml pepstatin A and 1 mM Na3VO4). Lysate was micro-centrifuged for 20 min and the soluble fraction was incubated with antibody against PDGFR-β at 4 °C for 24 h followed by incubation with 20 μl of Protein A plus agarose (Sigma) at 4 °C for 2 h. After an intensive wash with lysis buffer, the immunoprecipitates were subjected to SDS/PAGE and Western blot with ECL® detection system (Amersham Biosciences).
GST pull-down assays
In a 50 μl reaction, DTT-deprived PDGFR-β CD (150 nM) was incubated with 10 mM NEM in a buffer containing 40 mM Hepes (pH 7.3), 10 mM MgCl2 and 1 mM ATP at room temperature for 20 min. DTT was added to a final concentration of 10 mM and the reaction mixtures incubated at room temperature for 5 min, and then 1 μM of either GST–PLC-γ or GST–p85α was added and further incubated at 30 °C for 10 min. Glutathione-Sepharose 4B (20 μl; Amersham Biosciences) was added to the reactions and incubated for 30 min. The Sepharose beads were washed three times with ice-cold PBS and the bound proteins were subjected to SDS/PAGE and Western blot using PDGFR-β antibody.
Protease cleavage assays
PDGFR-β CD was mixed with buffer in 50 μl volumes to contain 37.5 mM Tris/HCl (pH 8.0), 150 mM NaCl, 1 mM DTT and 7.5% glycerol. After 10 μl of the mixture was taken (zero time reaction), trypsin (Sigma) was added to a final concentration ranging from 9.8 to 16.7 μg/ml and incubated at room temperature. A fraction (10 μl) of reaction mixture was taken and mixed with SDS loading buffer at the indicated time points, and subjected to 12% SDS/PAGE and Western blot with PDGFR-β antibody.
ATP filter-binding assays
Binding reactions of 50 μl volumes were set up to contain 40 mM Hepes (pH 7.3), 150 mM NaCl, 10 mM MgCl2, 1 mM DTT, 150 nM PDGFR-β CD, 1 μM GST–PLC-γ (where indicated) and 6.2 μCi [γ-35S]ATP (NEN Life Science). Reactions were incubated at room temperature for 30 min and then filtered through nitrocellulose membrane. After extensive washing, radioactivity retained on the membrane was measured in 1 ml of cocktail solution by liquid-scintillation counter.
RESULTS
PDGFR-β CD expressed and purified from Sf-9 cells is an active tyrosine kinase
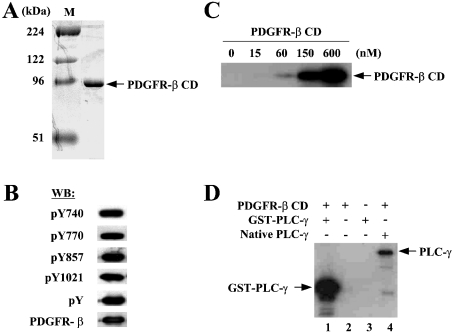
To study the role of the cysteine residues in the enzymic function of PDGFR-β, we decided to use a recombinant protein corresponding to the whole CD (amino acids 557–1106) of the human PDGFR-β. Ligand-independent dimerization and autophosphorylation have been observed when the full-length PDGF receptors were expressed at high densities in baculovirus-infected cells [24]. We tried to prepare the active enzyme by overexpressing PDGFR-β CD in Sf9 cells so as to be self-dimerized and autophosphorylated. Indeed, Western-blot analysis showed that purified PDGFR-β CD, more than 95% pure based on Coomassie-stained gel (Figure 2A), was phosphorylated at Tyr-857 on the A loop as well as other tyrosine residues known as the binding sites for SH2-containing signalling proteins such as PLC-γ, the p85α subunit of PI3K and GAP (Figure 2B). In vitro kinase assays showed that PDGFR-β CD possessed the tyrosine kinase activity. Incubation of increasing concentrations of PDGFR-β CD in the presence of 32P-γ-ATP resulted in a proportional increase in the levels of phosphorylation, indicative of transphosphorylation (Figure 2C). PDGFR-β CD was also capable of phosphorylating the exogenous substrates such as PLC-γ (Figure 2D) and the p85α subunit of PI3K (Figure 3C).
Figure 2. CD of PDGFR-β expressed and purified from Sf-9 cells is a functionally active enzyme.
(A) Coomassie-stained SDS/PAGE gel of purified CD of PDGFR-β is shown. The predicted mass of the protein is approx. 60 kDa but it migrates at approx. 95 kDa on SDS gel. The protein purity was estimated to be >95%. (B) Western-blot analysis was performed for the purified PDGFR-β CD with antibodies for the receptor or phosphotyrosine (pY, 4G10) or sequence-specific antibodies for phosphorylation at Tyr-1021 (binding site for PLC-γ), Tyr-857 (activation loop), Tyr-770 (binding site for GAP) or Tyr-740 (binding site for PI3K). (C) Autophosphorylation of PDGFR-β CD was demonstrated by in vitro kinase assays. Increasing concentrations of PDGFR-β CD were incubated under standard conditions containing [γ-32P]ATP and the reactions were subjected to SDS/PAGE followed by autoradiography. (D) The activity of PDGFR-β CD towards substrate phosphorylation was verified by in vitro kinase assays in which GST–PLC-γ (lane 1) or native PLC-γ (lane 4) was used as substrates in the presence of [γ-32P]ATP.
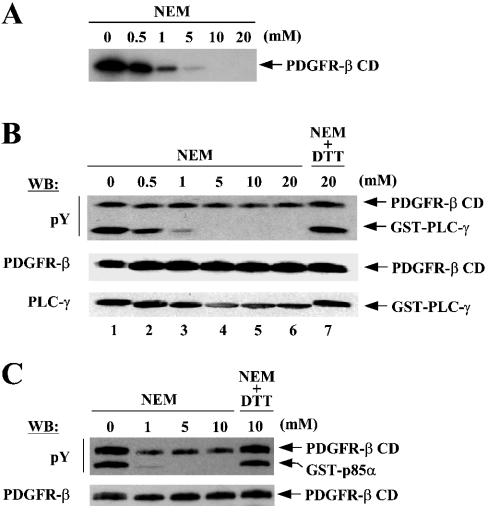
Figure 3. NEM inhibits the kinase activity of PDGFR-β.
Purified PDGFR-β CD, normally stored in a buffer containing 1 mM DTT, was dialysed in the buffer lacking DTT under anaerobic conditions before being subjected to in vitro kinase assays. PDGFR-β CD (150 nM) was incubated with increasing concentrations of NEM at 25 °C for 10 min and the reactions were subjected to kinase assays. (A) NEM-treated enzymes were further incubated under standard conditions containing [γ-32P]ATP and the reactions were subjected to SDS/PAGE followed by autoradiography. (B) NEM-treated enzymes were incubated with DTT to inactivate the remaining free NEM followed by further incubation with GST–PLC-γ under standard conditions except that 1 mM of unlabelled ATP were substituted for [γ-32P]ATP. The reactions were subjected to SDS/PAGE and Western blot, with the same membrane being sequentially probed by the indicated antibodies. Lane 7, as a control, is a treatment of enzyme with the mixture of equal amounts of NEM and DTT. (C) Similar experiments to the above were performed to see the NEM effects on the activity of PDGFR-β for the phosphorylation of another substrate, the p85α subunit of PI3K.
NEM inhibits the kinase activity of PDGFR-β CD
We examined whether cysteine residues are important for the enzymic function of PDGFR-β. PDGFR-β CD, normally stored in the buffer containing 1 mM DTT, was dialysed in the buffer lacking DTT under anaerobic conditions to remove DTT. The protein was incubated with NEM, which selectively alkylates free thiol groups of cysteine residues, and then subjected to kinase assays for autophosphorylation using [γ-32P]ATP. Treatment with NEM inhibited autophosphorylation in a dose-dependent fashion with the kinase activity being completely abolished at the concentration of 10 mM (Figure 3A).
We next examined whether NEM could also inhibit the activity of PDGFR-β for phosphorylating exogenous substrates. PDGFR-β CD was incubated with NEM as described above and DTT was added to the reactions to inactivate the remaining free NEM. The reaction mixture was then added to GST–PLC-γ (PLC-γ; amino acids 550–850) and incubated under standard conditions containing unlabelled ATP. The levels of tyrosine phosphorylation of both enzyme and substrate were determined by Western blot with phosphotyrosine antibody. NEM also inhibited PDGFR-β from phosphorylating the substrate GST–PLC-γ in a dose-dependent manner (Figure 3B, lanes 1–6). This inhibition was completely abolished when NEM was preincubated with DTT, indicating that the inhibition was attributed to NEM itself (Figure 3B, lane 7). In addition, the inhibitory effect of NEM was not due to a loss of enzyme or substrate as revealed by Western blot with antibodies for these proteins (Figure 3B, middle and lower panels). Similar experiments using the p85α subunit of PI3K as substrate further confirmed the NEM inhibition of PDGFR-β (Figure 3C). It should be noted that, since the steady-state levels of phosphorylation were measured in these assays, the levels of the phosphorylation on PDGFR-β CD were only partially decreased by NEM; NEM only blocks additional phosphorylation on the proteins that have already been highly phosphorylated after purification. Taken together, these results suggested that cysteine residues are critical for the kinase activity of PDGFR-β.
Identification of NEM-modified cysteine residues by MS
To determine which cysteine residues of PDGFR-β CD were modified by NEM treatment, we performed MS analysis. NEM-treated PDGFR-β CD was digested with trypsin and the tryptic digest peptides were subjected to MALDI–TOF-MS analysis. Expected monoisotopic masses for the peptides containing NEM-modified cysteine residues were detected for Cys-684, Cys-822, Cys-835, Cys-843 and Cys-940, indicating that the thiol groups of these cysteine residues were modified by NEM (Table 1). All these cysteine residues except Cys-843 appeared to be fully modified with NEM because the corresponding peptides with no NEM modification were minimally detected (results not shown). The expected masses for the NEM modification of Cys-671 and Cys-787 were not detected.
Table 1. MALDI–TOF-MS analysis for tryptic digest peptides containing NEM-modified cysteine residues of PDGFR-β CD.
PDGFR-β CD purified from baculovirus-infected Sf9 cells was treated with NEM, and subjected to tryptic digestion and MALDI–TOF-MS analysis. Expected and observed monoisotopic masses (MH+) of tryptic digest peptides containing NEM-modified cysteine residues are shown with the corresponding peptide sequences. Tryptic digestion does not generate short peptides containing Cys-1060 and Cys-1094. ND, not detected.
| Cysteine-containing peptides | Expected mass (MH+) | Observed mass (MH+) | |
|---|---|---|---|
| Cys-671 | (K)IMSHLGPHLNVVNLLGACTK(G) | 2242.24 | ND |
| Cys-684 | (K)GGPIYIITEYCR(Y) | 1509.79 | 1509.71 |
| Cys-787 | (K)YADIESSNYMAPYDNYVPSAPERTCR(A) | 3137.41 | ND |
| Cys-822 | (K)NCVHRDLAAR(N) | 1279.69 | 1279.65 |
| Cys-835 | (R)DLAARNVLICEGK(L) | 1526.85 | 1526.74 |
| Cys-843 | (K)ICDFGLAR(D) | 1019.55 | 1019.54 |
| Cys-940 | (K)CWEEK(F) | 819.39 | 819.33 |
Identification of two cysteine residues critical for the kinase activity of PDGFR-β
Having established that cysteine residues were critical for the activity of PDGFR-β, we attempted to determine which cysteine residues would be responsible. Nine cysteine residues present in the CD of PDGFR-β were individually changed to serine residues through site-directed mutagenesis, and these mutant proteins were expressed and purified from baculovirus-infected Sf9 cells. The expression levels and protein stability of these mutants in Sf9 cells were not distinguishable from those of WT protein (results not shown).
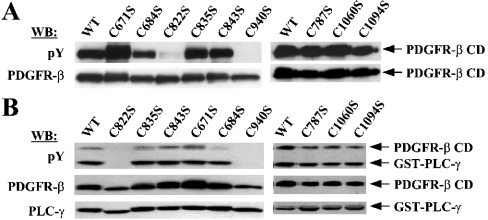
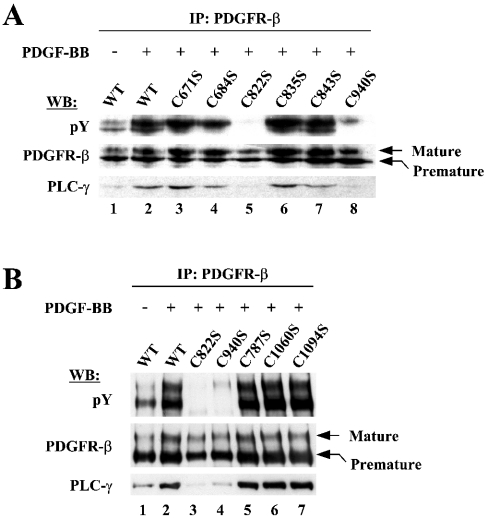
First, we checked the status of tyrosine phosphorylation of purified mutant proteins by Western blot. Strikingly, the mutants C822S and C940S, in which Cys-822 and Cys-940 were changed to Ser respectively showed barely detectable levels of tyrosine phosphorylation, whereas all the other mutants retained levels compatible with those of WT (Figure 4A). These two mutants, as could be expected from their extremely low levels of phosphorylation, were shown to be completely defective in the activity of phosphorylating exogenous substrates in in vitro kinase assays using GST–PLC-γ as a substrate (Figure 4B). These results demonstrated that, of the nine cysteine residues present in the CD, Cys-822 and Cys-940 are critical for the enzymic activity of PDGFR-β.
Figure 4. Cys-822 and Cys-940 were identified to be critical for the enzymic activity of PDGFR-β in vitro.
(A) Through site-directed mutagenesis, mutant PDGFR-β CDs were generated to contain individual Cys to Ser mutations for the nine cysteine residues. WT and mutant proteins, expressed and purified from Sf9 cells, were analysed for the levels of tyrosine phosphorylation by Western blot with the phosphotyrosine antibody. The same blot was reprobed with the receptor antibody to ensure equal protein loading. (B) The same set of proteins was examined with respect to the activity for the phosphorylation of substrate, GST–PLC-γ. The kinase reactions were performed as in Figure 3(B). Note that the order in which samples were loaded on the gel is different from that in (A).
Cys-822 and Cys-940 are required for the enzymic function of PDGFR-β in vivo
We next examined whether Cys-822 and Cys-940 would also critically function in the context of native form of PDGFR-β within the cells. A set of expression vectors (pcDNA3.1) containing the full-length PDGFR-β with individual Cys to Ser mutations was transfected into the HEK-293T cells, which were verified by Western blot to express no endogenous PDGFR-β (results not shown). PDGFR-β was then immunoprecipitated with receptor antibody and the immunocomplexes were subjected to Western blot. The expression levels and protein stability of these mutants in HEK-293T cells were not significantly different from those of WT (Figures 5A and 5B, middle panels).
Figure 5. Cys-822 and Cys-940 are critical for the enzymic activity of PDGFR-β in the context of full-length receptors in vivo.
(A) Expression vectors containing full-length PDGFR-β in the form of WT or Cys to Ser mutants for the cysteine residues within the kinase domain were transfected into the HEK-293T cells lacking endogenous PDGFR-β expression. Transfected cells were stimulated with 50 ng/ml PDGF-BB for 10 min after 24 h serum starvation (the control of the mock stimulation was included only for WT in this set of experiments). The receptors were immunoprecipitated from whole cell lysates and subjected to Western blot with the indicated antibodies. (B) Experiments, similar to those in (A) were performed for the mutations of the cysteine residues present outside the kinase domain.
After PDGF stimulation, the levels of tyrosine phosphorylation of WT PDGFR-β greatly increased, resulting in the enhancement of the binding of PLC-γ to the receptors (Figures 5A and 5B, lanes 1 and 2). In contrast, tyrosine phosphorylation was completely abolished for C822S and barely detectable for C940S receptors even in the presence of PDGF stimulation (Figure 5A, lanes 5 and 8; Figure 5B, lanes 3 and 4). Owing to the defect in tyrosine phosphorylation, these mutant receptors were severely compromised in their ability to bind PLC-γ (Figure 5A, lanes 5 and 8; Figure 5B, lanes 3 and 4). The rest of the mutant receptors showed similar levels of basal phosphorylation to those of the WT receptor in the absence of PDGF stimulation (results not shown). After PDGF stimulation, these receptors could be normally activated and support the PLC-γ binding at levels comparable with those of the WT receptor (Figure 5A, lanes 3, 4, 6 and 7; Figure 5B, lanes 5–7). These results clearly demonstrated that both Cys-822 and Cys-940 are critical for the enzymic function of PDGFR-β in vivo.
Cys-822 and Cys-940 do not contribute to PDGFR-β activity by disulphide-bond formation
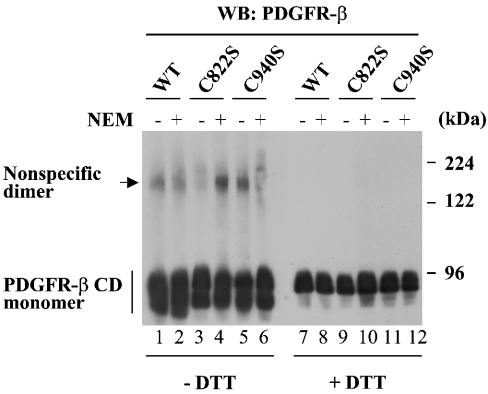
To understand the mechanisms by which Cys-822 and Cys-940 contribute to the enzymic function of PDGFR-β, first we examined whether these cysteine residues participate in the formation of disulphide bonds by non-reducing gel analysis as described previously [25]. A very small portion of WT PDGFR-β CD generated a species migrating as a dimer under non-denaturing conditions (without DTT), and this species disappeared in the presence of DTT (Figure 6, lanes 1, 2, 7 and 8). However, the formation of the dimer was also observed for C822S and C940S (Figure 6, lanes 3–6) as well as other Cys to Ser mutants (results not shown). Moreover, the dimer formation was still observed when WT protein was inactivated by pretreatment with NEM (lane 2). These results show that the activity of PDGFR-β is not dependent on this minor formation of the dimer. We believe that the observed dimers were generated by the formation of non-specific disulphide bonds probably through the oxidation during the boiling process of protein samples. Interestingly, all the PDGFR-β CD proteins showed multiple bands at the monomer position under non-denaturing conditions (lanes 1–8 and results not shown), indicative of existence of intrachain disulphide bonds as previously suggested [21].
Figure 6. Cys-822 and Cys-940 do not contribute to PDGFR-β activity by disulphide-bond formation.
Non-reducing gel analysis was performed as described previously [25] to see whether Cys-822 and/or Cys-940 participate in disulphide-bond formation. PDGFR-β CDs pretreated with (+) or without (−) NEM were dialysed with buffer to remove free NEM. The proteins were boiled in SDS sample buffer containing 40 mM NEM either in the absence or in the presence of DTT, and subjected to 5% SDS/PAGE and Western blot.
Neither Cys-822 nor Cys-940 is involved in substrate or ATP binding
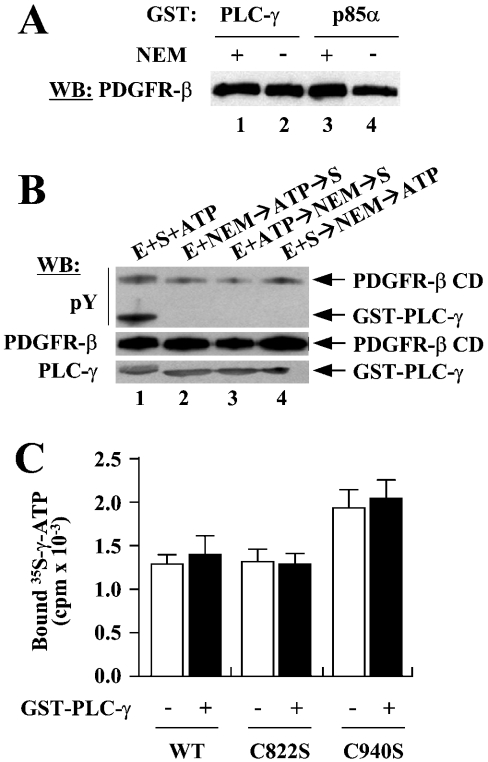
Next, we tested the possibility that Cys-822 and Cys-940 are involved in substrate or ATP binding. The C822S and C940S mutants essentially lack tyrosine phosphorylation and consequently do not support substrate binding. Therefore we asked whether NEM modification of Cys-822 and Cys-940 could affect the binding of substrates to PDGFR-β; NEM does not compromise the tyrosine phosphorylation that has already been established on the proteins (Figures 3B and 3C). GST pull-down experiments showed that NEM treatment did not affect the bindings of PLC-γ or p85α to PDGFR-β CD (Figure 7A), indicating that it is unlikely that NEM inhibits PDGFR-β by blocking substrate binding.
Figure 7. Neither Cys-822 nor Cys-940 is involved in substrate or ATP binding.
(A) GST pull-down assays were performed to see whether NEM blocks substrate binding. WT PDGFR-β CDs that had been incubated with (lanes 1 and 3) or without (lanes 2 and 4) NEM were subjected to pull-down assays using GST–PLC-γ or GST–p85α as bait. (B) To determine whether NEM blocks ATP binding, the effects of preoccupation of ATP on the NEM inhibition of PDGFR-β were examined. Lane 1, control reaction with no NEM treatment. Lane 2, normal reaction for the NEM inhibition of PDGFR-β; the enzyme was modified by NEM before the addition of ATP and substrate. Lane 3, PDGFR-β CD was incubated with ATP before NEM treatment and then the substrate was added to the reaction. Lane 4, PDGFR-β CD was incubated with substrate first, treated with NEM, and then ATP was added. (C) Filter-binding assays were performed to examine the effects of Cys to Ser mutations for Cys-822 or Cys-940 on ATP binding. WT, C822S or C940S were incubated with [γ-35S]ATP either with or without GST–PLC-γ, and the reactions were filtered through a nitrocellulose membrane. After intensive washing, the radioactivity retained with proteins was measured. A representative of three independent experiments is shown.
We then examined whether NEM blocks ATP binding to the enzyme. We rationalized that, if NEM blocks ATP binding, prior occupation with ATP before NEM modification should relieve the NEM inhibition of PDGFR-β to a certain extent. When PDGFR-β CD was incubated with ATP after NEM treatment, phosphorylation of PLC-γ was completely abolished as seen before (Figure 7B, lanes 1 and 2). Incubation of PDGFR-β CD with ATP before NEM treatment did not give rise to any detectable levels of the phosphorylation of PLC-γ, indicating that the NEM inhibition was not attributable to the blockage of ATP binding (Figure 7B, lane 3). NEM still showed its inhibitory effect when it was added to the reaction even after having permitted PLC-γ binding to PDGFR-β CD, again demonstrating that NEM has no effect on the enzyme–substrate interactions (Figure 7B, lane 4).
To examine directly whether Cys to Ser mutations for Cys-822 or Cys-940 could affect ATP binding, we performed filter-binding assays. WT, C822S or C940S of PDGFR-β CDs either in the absence or presence of PLC-γ was incubated with [γ-35S]ATP, a non-hydrolysable form of ATP, and the reactions were filtered through a nitrocellulose membrane. After intensive wash, the radioactivity retained by proteins on the membrane was measured. C822S showed a similar affinity for ATP when compared with WT, and C940S protein showed a little more binding of ATP when compared with WT (Figure 7C). Presence of PLC-γ in the binding reactions had no effect on the ATP binding to the proteins. These results demonstrated that neither Cys-822 nor Cys-940 plays a role in ATP binding.
Mutation of Cys-940, but not Cys-822, induces a conformational change of PDGFR-β CD
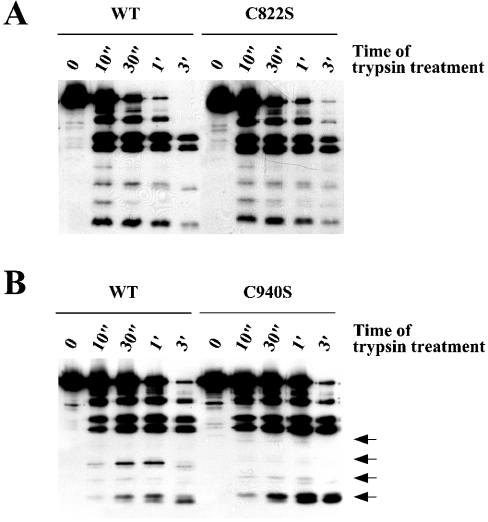
Finally, we tested the possibility of a structural role of Cys-822 and Cys-940. Alteration of a protein conformation is often reflected by changes of proteolytic cleavage pattern. PDGFR-β CD protein pairs of either WT/C822S or WT/C940S were subjected to partial proteolytic cleavage by trypsin, and the cleavage products were separated on SDS gel followed by Western blot with the receptor antibody. As shown in Figure 8, whereas the trypsin cleavage pattern of C822S was almost identical with that of WT, C940S showed a significantly different pattern from WT. These results suggest that mutation of Cys-940, but not Cys-822, induces a protein conformational change of PDGFR-β CD.
Figure 8. Mutation of Cys-940, but not Cys-822, induces a conformational change of PDGFR-β CD.
PDGFR-β CD (150 nM) was incubated with 16.7 μg/ml (A) and 9.8 μg/ml (B) of trypsin in 40 μl volumes of reaction buffer. A 10 μl aliquot of each reaction was removed and added to SDS sample buffer at the indicated time points (″, second; ′, minute). The first lane of each reaction was treated without trypsin. Each of the protein pairs was processed for trypsin digestion in parallel. Protein samples were separated by SDS (12% gel) followed by Western blot with the receptor antibody. Arrows on the right side of (B) indicate the bands of trypsin cleavage products, which are significantly different for the two proteins. A representative of four independent experiments is shown.
DISCUSSION
In the present study, we have evaluated a possible role for the conserved cysteine residues in the enzymic function of PDGFR-β. We clearly demonstrated that two conserved cysteine residues are critical for the kinase activity of PDGFR-β; both NEM modification and individual Cys to Ser mutation for Cys-822 and Cys-940 significantly reduce the activities of autophosphorylation and the phosphorylation of exogenous substrates. Cys-822, located on the catalytic loop at the enzyme active site, was chosen to demonstrate its critical role in tyrosine kinase activity for the first time in this study. The importance of Cys-940, located in the C-lobe of the kinase domain, has been documented for other tyrosine kinases, but our results suggest that the role of this cysteine in PDGFR-β activity is different from that of those kinases. Interestingly, Cys-822, unlike Cys-940, is only conserved among the members of the PDGFR family, implying that the function of this cysteine may be specific to this family of tyrosine kinases.
Cys-940 is absolutely conserved among the whole family of protein tyrosine kinases, and its critical role has been demonstrated for several tyrosine kinases. Mutation of either Cys-464 or Cys-475 (the Cys-940 equivalent) was shown to reduce significantly the enzymic activity of the lymphocyte-specific tyrosine kinase p56lck [18]. The equivalent cysteine in v-Src tyrosine kinase (Cys-498) has also been shown to be critical for its kinase activity and thereby the ability to cause cell transformation [19,20]. In addition, the equivalent cysteine of the oncogenic Ret tyrosine kinase (Cys-376) was shown to be important for UV-induced superactivation of this kinase [17]. Interestingly, the Cys-940 equivalents of p56lck and v-Src kinases contribute to protein stability, whereas that of the Ret kinase mediates the UV-induced dimerization via disulphide-bond formation [17–20]. However, Cys-940 of PDGFR-β neither plays a role in protein stability (Figures 4 and 5) nor mediates dimerization via disulphide-bond formation (Figure 6), implying that diverse mechanisms have evolved for the roles of Cys-940 in the enzymic function of tyrosine kinases.
Li and Schlessinger have shown that, on PDGF binding, PDGF receptors undergo covalent dimerization that is sensitive to the thiol-reducing and -alkylating agents such as 2-mercaptoethanol and NEM [21]. These results indicated that the dimerization of PDGF receptors is mediated by the formation of disulphide bonds. However, our results indicate that neither Cys-822 nor Cys-940 contributes to the enzymic activity of PDGFR-β by participating in the formation of disulphide bonds (Figure 6). We believe that the reported disulphide-linked dimerization of PDGF receptors is probably mediated by disulphide-bond formation between the receptors' extracellular domains.
MS data verified that both Cys-822 and Cys-940 were fully modified when PDGFR-β CD was treated with NEM. These results, together with the results from site-directed mutagenesis experiments, show that the NEM inhibition of PDGFR-β was actually attributable to the modification of those two cysteine residues. The thiol groups of cysteine residues, which are involved in enzymic function, often exist as reactive thiolate anions and are subjected to a redox regulation. For example, PTPs (protein tyrosine phosphatases) have a reactive cysteine in their active site and this cysteine can be readily oxidized by H2O2, resulting in enzyme inactivation [26,27]. In this regard, we examined whether Cys-822 and Cys-940 of PDGFR-β are susceptible to this type of oxidation, and found that the activity of PDGFR-β was not affected by H2O2 (results not shown). Previous studies indicate that H2O2 serves as an important signalling messenger in activating RTKs including PDGF receptors [28]. It is known that, on initial activation by PDGF binding, PDGFR-β transiently generates H2O2 and the increased concentrations of this H2O2 then lead to further enhancement of tyrosine phosphorylation for maximal receptor activation [28–30]. Inhibition of PTPs by H2O2 has been proposed to be mainly responsible for this positive feedback regulation [28]. Our observation that PDGFR-β is not susceptible to H2O2 inactivation is in keeping with the model for the indirect activation of this RTK via H2O2-mediated PTP inactivation.
How do Cys-822 and Cys-940 contribute to the enzymic function of PDGFR-β? Our results, showing that Cys-822 and Cys-940 are neither involved in substrate or ATP bindings nor mediate disulphide-linked dimerization, suggest that these cysteine residues play many roles in enzyme catalysis. To gain insights into how these cysteine residues could contribute to the catalytic mechanism, we studied the crystal structure of vascular endothelial growth factor receptor-2, another member of the PDGF receptor family, since the three-dimensional structure of PDGF receptor has not been determined. According to this structure, Cys-822 is predicted to exist at the active site ‘cleft’ so as to be positioned in close proximity to the three absolutely conserved amino acids, Asp-826, Asn-831 and Asp-844, whose precise spatial arrangement is essential for optimal phosphotransfer [31]. In contrast, Cys-940 is predicted to exist in an α-helix of the C-lobe, which is at a considerable distance located from the active site [31]. These predictions led us to hypothesize that Cys-822 plays a direct role in catalytic reaction (possibly phosphotransfer reaction), whereas Cys-940 makes an indirect contribution. Cys-940 seems to play a role in adopting the protein conformation which is optimal for catalytic reaction, since mutation of this cysteine leads to a conformational change of PDGFR-β (Figure 7B). More experiments will be required to substantiate this hypothesis.
Acknowledgments
We thank S.G. Rhee (National Institutes of Health, Bethesda, MD, U.S.A.) for the gift of antibodies for PLC-γ and tyrosine phosphorylated PLC-γ and Y.S. Bae (Ewha Womans University) for PLC-γ protein. We also thank A. Kazlauskas (Harvard Medical School, Boston, MA, U.S.A.) and J.R. Lee (Ewha Womans University) for providing the plasmids for GST–PLC-γ and GST–p85α respectively. This work was supported by the Korea Science and Engineering Foundation (KOSEF) grant to the Center for Cell Signaling Research (Ewha Womans University) and the Brain Korea 21 grant (to S.-R.L. and J.K.) and partially supported by the grant of Ewha-SK Corp. Drug Discovery Project.
References
- 1.Heldin C.-H., Östman A., Ronnstrand L. Signal transduction via platelet-derived growth factor receptors. Biochim. Biophy. Acta. 1997;1378:F79–F113. doi: 10.1016/s0304-419x(98)00015-8. [DOI] [PubMed] [Google Scholar]
- 2.Claesson-Welsh L. Platelet-derived growth factor receptor signals. J. Biol. Chem. 1994;269:32023–32026. [PubMed] [Google Scholar]
- 3.Baxter R. M., Secrist J. P., Vaillancourt R. R., Kazlauskas A. Full activation of the platelet-derived growth factor beta-receptor kinase involves multiple events. J. Biol. Chem. 1997;273:17050–17055. doi: 10.1074/jbc.273.27.17050. [DOI] [PubMed] [Google Scholar]
- 4.Kazlauskas A., Durden D. L., Cooper J. A. Functions of the major tyrosine phosphorylation site of the PDGF receptor β subunit. Cell Regul. 1991;2:413–425. doi: 10.1091/mbc.2.6.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fantl W. J., Escobedo J. A., Williams L. T. Mutations of the platelet-derived growth factor receptor that cause a loss of ligand-induced conformational change, subtle changes in kinase activity, and impaired ability to stimulate DNA synthesis. Mol. Cell. Biol. 1989;9:4473–4478. doi: 10.1128/mcb.9.10.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rönnstrand L., Mori S., Arvidsson A.-K., Eriksson A., Wernstedt C., Hellman U., Claesson-Welsh L., Heldin C.-H. Identifcation of two C-terminal autophosphorylation sites in the PDGF β-receptor: involvement in the interaction with phospholipase C-γ. EMBO J. 1992;11:3911–3919. doi: 10.1002/j.1460-2075.1992.tb05484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kashishian A., Cooper J. A. Phosphorylation sites at the C-terminus of the platelet-derived growth factor receptor bind phospholipase C-γ1. Mol. Biol. Cell. 1993;4:49–57. doi: 10.1091/mbc.4.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valius M., Bazenet C., Kazlauskas A. Tyrosines 1021 and 1009 are phosphorylation sites in the carboxy terminus of the platelet-derived growth factor receptor β subunit and are required for binding of phospholipase C-γ1 and a 64-kilodalton protein, respectively. Mol. Cell. Biol. 1993;13:133–143. doi: 10.1128/mcb.13.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fantl W. J., Escobedo J. A., Martin G. A., Turck C. W., del Rosario M., McCormick F., Williams L. T. Distinct phosphotyrosines on a growth factor receptor bind to specific molecules that mediate different signaling pathways. Cell (Cambridge, Mass.) 1992;69:413–423. doi: 10.1016/0092-8674(92)90444-h. [DOI] [PubMed] [Google Scholar]
- 10.Kazlauskas A., Kashishian A., Cooper J. A., Valius M. GTPase-activating protein and phosphatidylinositol 3-kinase bind to distinct regions of the platelet-derived growth factor receptor β subunit. Mol. Cell. Biol. 1992;12:2534–2544. doi: 10.1128/mcb.12.6.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kashishian A., Kazlauskas A., Cooper J. A. Phosphorylation sites in the PDGF receptor with different specifcities for binding GAP and PI3 kinase in vivo. EMBO J. 1992;11:1373–1382. doi: 10.1002/j.1460-2075.1992.tb05182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell (Cambridge, Mass.) 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 13.Huse M., Kuriyan J. The conformational plasticity of protein kinases. Cell (Cambridge, Mass.) 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 14.Hubbard S. R., Till J. H. Protein tyrosine kinase structure and function. Annu. Rev. Biochem. 2000;69:373–398. doi: 10.1146/annurev.biochem.69.1.373. [DOI] [PubMed] [Google Scholar]
- 15.Hubbard S. R., Mohammadi M., Schlessinger J. Autoregulatory mechanisms in protein-tyrosine kinases. J. Biol. Chem. 1998;273:11987–11990. doi: 10.1074/jbc.273.20.11987. [DOI] [PubMed] [Google Scholar]
- 16.Woltjer R. L., Staros J. V. Effects of sulfhydryl modification reagents on the kinase activity of the epidermal growth factor receptor. Biochemistry. 1997;36:9911–9916. doi: 10.1021/bi963007v. [DOI] [PubMed] [Google Scholar]
- 17.Kato M., Iwashita T., Takeda K., Akhand A. A., Liu W., Yoshihara M., Asai N., Suzuki H., Takahashi M., Nakashima I. Ultraviolet light induces redox reaction-mediated dimerization and superactivation of oncogenic ret tyrosine kinases. Mol. Biol. Cell. 2000;11:93–101. doi: 10.1091/mbc.11.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veillette A., Dumont D., Fournel M. Conserved cysteine residues are critical for the enzymatic function of the lymphocyte-specific tyrosine protein kinase p56lck. J. Biol. Chem. 1993;268:17547–17553. [PubMed] [Google Scholar]
- 19.Oo M. L., Senga T., Thant A. A., Amin R. R., Huang P., Mon N. N., Hamaguchi M. Cysteine residues in the C-terminal lobe of Src: their role in the suppression of the Src kinase. Oncogene. 2003;22:1411–1417. doi: 10.1038/sj.onc.1206286. [DOI] [PubMed] [Google Scholar]
- 20.Senga T., Miyazaki K., Machida K., Iwata H., Matsuda S., Nakashima I., Hamaguchi M. Clustered cysteine residues in the kinase domain of v-Src: critical role for protein stability, cell transformation and sensitivity to herbimycin A. Oncogene. 2000;19:273–279. doi: 10.1038/sj.onc.1203296. [DOI] [PubMed] [Google Scholar]
- 21.Li W., Schlessinger J. Platelet-derived growth factor (PDGF)-induced disulfide-linked dimerization of PDGF receptor in living cells. Mol. Cell. Biol. 1991;11:3756–3761. doi: 10.1128/mcb.11.7.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valius M., Kazlauskas A. Phospholipase C-gamma 1 and phosphatidylinositol 3 kinase are the downstream mediators of the PDGF receptor's mitogenic signal. Cell (Cambridge, Mass.) 1993;73:321–334. doi: 10.1016/0092-8674(93)90232-f. [DOI] [PubMed] [Google Scholar]
- 23.Lee S.-R., Kwon K. S., Kim S.-R., Rhee S. G. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J. Biol. Chem. 1998;273:15366–15372. doi: 10.1074/jbc.273.25.15366. [DOI] [PubMed] [Google Scholar]
- 24.Jensen R. A., Beeler J. F., Heidaran M. A., LaRochelle W. J. Characterization of baculovirus-expressed human α and β platelet-derived growth factor receptors. Biochemistry. 1992;31:10887–10892. doi: 10.1021/bi00159a032. [DOI] [PubMed] [Google Scholar]
- 25.Lee S. R., Yang K. S., Kwon J., Lee C., Jeong W., Rhee S. G. Reversible inactivation of the tumor suppressor PTEN by H2O2. J. Biol. Chem. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- 26.Xu D., Rovira I. I., Finkel T. Oxidants painting the cysteine chapel: redox regulation of PTPs. Dev. Cell. 2002;2:251–252. doi: 10.1016/s1534-5807(02)00132-6. [DOI] [PubMed] [Google Scholar]
- 27.Denu J. M., Dixon J. E. Protein tyrosine phosphatases: mechanisms of catalysis and regulation. Curr. Opin. Chem. Biol. 1998;2:633–641. doi: 10.1016/s1367-5931(98)80095-1. [DOI] [PubMed] [Google Scholar]
- 28.Rhee S. G., Bae Y. S., Lee S. R., Kwon J. Hydrogen peroxide: a key messenger that modulates protein phosphorylation through cysteine oxidation. Science STKE. 2000;53:PE1. doi: 10.1126/stke.2000.53.pe1. [DOI] [PubMed] [Google Scholar]
- 29.Bae Y. S., Sung J.-Y., Kim O. S., Kim Y.-J., Hur K. C., Kazlauskas A., Rhee S. G. Platelet-derived growth factor-induced H2O2 production requires the activation of phosphatidylinositol 3-kinase. J. Biol. Chem. 2000;275:10527–10531. doi: 10.1074/jbc.275.14.10527. [DOI] [PubMed] [Google Scholar]
- 30.Sundaresan M., Yu Z.-X., Ferrans V. J., Irani K., Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- 31.McTigue M. A., Wickersham J. A., Pinko C., Showalter R. E., Parast C. V., Tempczyk-Russell A., Gehring M. R., Mroczkowski B., Kan C.-C., Villafranca J. E., et al. Crystal structure of the kinase domain of human vascular endothelial growth factor receptor 2: a key enzyme in angiogenesis. Structure Fold. Des. 1999;7:319–339. doi: 10.1016/s0969-2126(99)80042-2. [DOI] [PubMed] [Google Scholar]