Abstract
The BEVS (baculovirus expression vector system) is widely used for the production of proteins. However, engineered proteins frequently experience the problem of degradation, possibly due to the lytic nature of the conventional BEVS (herein referred to as L-BEVS). In the present study, a non-lytic BEVS (N-BEVS) was established by random mutagenesis of viral genomes. At 5 days post-infection, N-BEVS showed only 7% cell lysis, whereas L-BEVS showed 60% lysis of cells. The quality of protein expressed in both N- and L-BEVSs was examined further using a novel FRET (fluorescence resonance energy transfer)-based assay. To achieve this, we constructed a concatenated fusion protein comprising LUC (luciferase) sandwiched between EYFP (enhanced yellow fluorescent protein) and ECFP (enhanced cyan fluorescent protein). The distance separating the two fluorescent proteins in the fusion protein EYFP–LUC–ECFP (designated hereafter as the YLC construct) governs energy transfer between EYFP and ECFP. FRET efficiency thus reflects the compactness of LUC, indicating its folding status. We found more efficient FRET in N-BEVS compared with that obtained in L-BEVS, suggesting that more tightly folded LUC was produced in N-BEVS. YLC expression was also analysed by Western blotting, revealing significantly less protein degradation in N-BEVS than in L-BEVS, in which extensive degradation was observed. This FRET-based in vivo folding technology showed that YLC produced in N-BEVS is more compact, correlating with improved resistance to degradation. N-BEVS is thus a convenient alternative for L-BEVS for the production of proteins vulnerable to degradation using baculoviruses.
Keywords: baculovirus expression vector system, fluorescence resonance energy transfer, protein degradation, protein folding
Abbreviations: AcMNPV, Autographa californica multiple nucleopolyhedrovirus; BEVS, baculovirus expression vector system; CMV(m), cytomegalovirus (minimal); dpi, days post-infection; ECFP, enhanced cyan fluorescence protein; EGFP, enhanced green fluorescent protein; EYFP, enhanced yellow fluorescent protein; FRET, fluorescence resonance energy transfer; EFRET, efficiency of FRET; FIVF, FRET-based in vivo folding; (L/N-)BEVS, lytic/non-lytic BEVS; LUC, luciferase; MOI, multiplicity of infection; Sf21 cells, Spodoptera frugiperda IPLB-Sf21 cells; YLC construct, EYFP–LUC–ECFP fusion protein
INTRODUCTION
Baculoviruses are a diverse group of common insect pathogens that primarily infect the order Lepidoptera. These viruses contain circular double-stranded DNA genomes of 90–160 kb in length [1–3]. Since its introduction in 1983 [4], the BEVS (baculovirus expression vector system) has become one of the most popular protein expression systems used in industry and molecular biology laboratories. BEVSs have several advantages over other recombinant protein expression systems, including high protein yields, ease of use and safety. However, although BEVSs generally perform post-translational protein modifications similar to those of mammalian cells, leading to correct secretion and subunit assembly [5,6], some recombinant proteins are extensively degraded [7–10].
Since cells infected with baculovirus are eventually disrupted at the late stage of viral infection, leading to the loss of cellular homoeostasis and the release of proteases in conventional BEVS, this system is thus referred to as lytic BEVS (L-BEVS). In addition, cell lysis may leak significant amounts of the engineered proteins into the medium, which are difficult to recover. In order to overcome protein degradation in BEVSs, efforts have been made to inhibit protease activity, including addition of inhibitor cocktails to the culture medium and elimination of viral protease from the viral genome [7,11,12]. However, protease inhibitors may impair cell growth when included in culture medium, and they are expensive, particularly in large-scale expression systems. Moreover, the vulnerabilities of engineered proteins to proteolytic degradation differ, and the proteolytic activities in different insect cell lines differ as well. The optimal conditions for each case require careful and time-consuming determination.
Previously stably transfected insect cell lines have been used to eliminate baculovirus-mediated cell lysis [13,14]. However, stably transfected insect cell lines take a long time to become established. There are great variations between the yields of different cell clones, and thus laborious screening is required. Furthermore, it is not possible to use strong late or very-late promoters for high-yield protein production. Thus the creation of a non-lytic BEVS (N-BEVS) could be very useful, allowing the easy harvesting of proteins without significant protein degradation.
In the present study, N-BEVS was successfully established by random mutagenesis of the baculovirus genome. Cells infected with non-lytic mutants showed significantly less protein degradation than in L-BEVS, and yields of protein expressed in N-BEVS were comparable with those in L-BEVS. While testing the quality of proteins expressed in both non-lytic and conventional BEVS, we found no available assay to analyse compactness and integrity of proteins in living cells. Previous studies have shown that FRET (fluorescence resonance energy transfer) is an ideal technique to study protein folding in vitro [15–18], to serve as a calcium sensor [19], to assay the action of protease [20], and to assay protein–protein interactions or protein phosphorylation in cells [21,22]. Therefore we designed a novel FRET-based in vivo assay for the visualization and quantification of protein compactness in living cells. By using this tool, the compactness and integrity of the engineered proteins in living cells was readily examined. We demonstrated that the engineered protein produced in the newly developed N-BEVS was compactly folded, and experienced less degradation than that found in conventional L-BEVS.
EXPERIMENTAL
Cells and viruses
The Spodoptera frugiperda IPLB-Sf21 (Sf21) cell line was cultured as a monolayer in TNM-FH insect medium containing 8% (v/v) heat-inactivated fetal bovine serum [23,24]. It was used for the propagation and infection of wild-type and recombinant AcMNPV (Autographa californica multiple nucleopolyhedrovirus). All viral stocks were prepared and titres were determined according to the standard protocol described by O'Reilly et al. [25].
Construction of plasmids and viruses
Plasmid pABhcmEpL, which contains two promoters to drive the expression of two different foreign proteins, was constructed as follows. The coding sequence of EGFP (enhanced green fluorescent protein), derived from plasmid pEGFP-C1 (ClonTech) was inserted into a pBacPAK8 (ClonTech) transfer vector under the control of a CMVm (cytomegalovirus minimal) promoter, enhanced by an hr1 sequence [26]. The DNA-coding sequence of firefly LUC (luciferase) was inserted into the same transfer vector (pBacPAK8) under the control of a polyhedrin promoter. Plasmid pABhcmEpL was co-transfected with vAcRP23.Laz (Pharmingen), which is a linearized viral DNA of AcMNPV, into the Sf21 cell line, according to the standard protocol described by O'Reilly et al. [25], to produce a typical lytic recombinant virus, vABhcmEpL.
For the FRET-based assay, we constructed a tandem fusion protein consisting of a LUC sandwiched between EYFP (enhanced yellow fluorescent protein) and ECFP (enhanced cyan fluorescent protein). To generate the fusion gene, eyfp–luc–ecfp, a DNA fragment encoding LUC was amplified using PCR. The luc gene was then cloned into pECFP-N1 (ClonTech), resulting in a plasmid containing the luc–ecfp fusion sequence. The resulting plasmid, pcLC, was used as a template to amplify the luc–ecfp fusion gene by PCR. The PCR products were subsequently cloned into pEYFP-C1 (ClonTech). The resultant plasmid, pcYLC, contained the eyfp–luc–ecfp (ylc) tandem fusion gene and was driven by a CMV promoter (‘YLC’ represents the EYFP–LUC–ECFP fusion protein). To prepare the transfer plasmid for BEVS, ylc fragment was cut from pcYLC using NheI and NotI, and was ligated into the XbaI and NotI sites of pBacPAK8 (ClonTech). The resultant transfer plasmid, pABpYLC, contained the ylc sequence under the control of the polyhedrin promoter. The polyhedrin promoter of pABpYLC was then replaced with an hr1-sequence-enhanced CMVm promoter in order to obtain pABhcmYLC. Two additional transfer plasmids, pABpC and pABpY, were created by digesting pECFP-N1 with PstI and NotI, and by digesting pEYFP-C1 with NheI and KpnI, after which the fragments were cloned into the PstI/NotI and XbaI/KpnI sites of pBacPAK8 respectively (supplementary Figure 1; http://www.biochemJ.org/bj/381/bj3810695add.htm).
Recombinant baculoviruses were prepared by co-transfection of transfer plasmids and linearized AcMNPV viral DNA (BaculoGold; Pharmingen), into Sf21 cells using Lipofectin® (Invitrogen). For the generation of non-lytic vC4-derivative recombinant viruses, transfer plasmids were transfected into cells, followed by infection with the vC4 non-lytic baculovirus.
Mutagenesis and mutant screening
Sf21 cells (2×105) were infected with vABhcmEpL at an MOI (multiplicity of infection) of 1 and incubated at 26 °C or 33 °C in the presence of 5-bromodeoxyuridine at concentrations of 10, 30 and 40 μg/ml. Culture fluids were harvested at 5 dpi (days post-infection), and excess 5-bromodeoxyuridine was removed by dialysis against PBS [137 mM NaCl/29 mM KCl/4.3 mM Na2HPO4,7H2O/1.4 mM KH2PO4 (pH 7.2) containing 0.5% (w/v) BSA]. An aliquot of 25 μl of dialysed culture fluid containing mutated viruses was then used to infect Sf21 cells, which were then incubated at 26 °C for the screening of non-lytic baculovirus mutants.
Essentially, cells infected with non-lytic mutant viruses are morphologically indistinguishable from lytic virus-infected cells prior to lysis, and are thus difficult to identify. Distinction becomes possible by identifying that once cell lysis occurs, EGFP can freely leak out from the lytic baculovirus-infected cells. Therefore the retention of EGFP is an efficient indicator for cells infected with non-lytic baculovirus. Non-lytic mutant viruses were thus isolated by examining the retention of EGFP with fluorescence microscopy. At 5 and 8 dpi, EGFP expression cells were examined using a fluorocytometer to identify viral isolates with a high level of EGFP retention in the cells. Non-lytic clones were isolated and purified by three rounds of end-point dilution on 96-well plates.
In addition to retention of EGFP, lytic and non-lytic baculovirus-infected cells were also examined by viable/dead dye, EthD-1 (Molecular Probes), to distinguish between non-lytic and lytic cells. The ultrastructure of lytic and non-lytic cells was examined by electron microscopy, as described by us previously [23].
Characterization of YLC
LUC activity and Western blot analysis of YLC were performed as described by us previously [26]. To characterize the fluorescence properties of the engineered tandem fusion protein, YLC was purified using a HisBind kit (Novagen) and Sephacryl S-200 (Pharmacia) gel-filtration resin. The purity of YLC was checked by SDS/PAGE, and the concentration was determined using a Micro-BCA (bicinchoninic acid) kit (Pierce). Fluorescence emission spectra of YLC (4 μg/ml) were recorded in quartz cells in a spectrofluorophotometer (Aminco Bowmen Series 2; Spectronics Unicam). For the acquisition of fluorescence emission of YLC, the excitation wavelength was set to 430 nm, and the emission spectrum from 450–600 nm was recorded. For comparison, the fluorescence emissions of ECFP and EYFP were also measured. Supernatants of cell lysates from vABpLC- and vABpY-infected cells, containing ECFP and EYFP, were directly subjected to spectrofluorophotometry. Excitation wavelengths of 430 and 510 nm were used for ECFP and EYFP respectively. All spectra were corrected for buffer fluorescence.
Measurement of FRET in baculovirus-infected cells
Sf21 cells infected with various recombinant baculoviruses expressing engineered YLC fusion proteins were examined using laser-scanning confocal fluorescence microscopy (Pascal LSM; Zeiss), and the images were collected with a 40×objective lens (Plan-NEOFLUAR; numerical aperture 0.75). For detection of ECFP, cells were excited at 458 nm by an argon laser; fluorescence emissions were then collected through a dual-wavelength beam splitter at 458/514 nm and a long-pass emission filter at 475 nm. For detection of EYFP, the same beam splitter and long-pass emission filter at 530 nm were used. For FRET measurements, the infected cells were excited by a wavelength of 458 nm, and yellow fluorescence was collected using the 458/514 nm beam splitter and long-pass 530 nm emission filter. The original donor cyan fluorescence emission was separated by placing another 515 nm beam splitter and band-pass 475–525 nm emission filter after the 458/514 nm beam splitter, thereby allowing simultaneous detection of cyan and yellow fluorescence.
To measure FRET, a donor-quenching and acceptor-bleaching method was used. For the photobleaching experiment, the cyan image was collected first at an excitation wavelength of 458 nm, followed by photobleaching of the acceptor fluorescence (yellow) in a single cell with a 514 nm laser line, after which the second donor image was acquired. FRET efficiency was calculated from individual cells as one minus the ratio of the donor images before and after photobleaching of the acceptor, equivalent to 1−(intensity of the cyan emission before photobleaching/intensity of cyan emission after photobleaching), i.e. the fraction of cyan fluorescence emission transferred to the yellow fluorescence protein.
For FRET analysis in L-BEVS and N-BEVS, Sf21 cells were infected with various recombinant viruses, and sets of 40 individual virus-infected cells were analysed using FRET. FRET efficiencies were calculated as an average value from 40 cells examined in triplicate. To reduce overestimation of the FRET effect by intermolecular energy transfer derived from the potential overcrowding of ECFP and EYFP in baculovirus-infected cells, two recombinant baculoviruses bearing either ecfp or eyfp genes were co-infected into the same cells at an MOI of 10 to ensure viral co-infection in most of the same cells. The present study has shown that only limited FRET (3–8%) was detected by such co-infection, suggesting intermolecular energy transfer has relatively little effect on our results.
RESULTS
Isolation of non-lytic mutant of baculovirus
To screen for viral mutants with a minimal cell lytic phenotype, a recombinant baculovirus (vABhcmEpL), encoding an EGFP, was constructed (Figure 1A). After chemical mutagenesis of the parental virus vABhcmEpL, 41 mutants showing reduced cell lysis, as evidenced by the retention of EGFP in baculovirus-infected cells, were isolated out of a total of 4006 clones.
Figure 1. Development of non-lytic baculovirus mutants.
(A) The map of transfer vector pABhcmEpL used to generate parental recombinant baculovirus vABhcmEpL is shown. In this plasmid, EGFP is driven by a CMVm promoter and luc is driven by the polyhedrin promoter. The ‘backbone’ of this transfer vector is pBacPAK8 (ClonTech). p-pol, polyhedrin promoter; p-CMVm, CMVm promoter. (B) Fluorescence microscopic analyses of cells infected with non-lytic C4 virus (panels a1 and a2), and lytic parental vABhcmEpL virus (panels b1 and b2). Sf21 cells showed green fluorescence (panels a1 and b1) when infected with the non-lytic virus C4 and parental virus vABhcmEpL respectively. Dead cells were identified by EthD-1 (viable/dead dye; Molecular Probes) staining with red fluorescence (panels a2 and b2). Arrows indicate that the same living cells in panels b1 and b2 emit green fluorescence only, and arrowheads indicate that the same dead cells in panels b1 and b2 emit red fluorescence only. (C) A plot showing the daily variations in the percentage of cells retaining green fluorescence. Sf21 cells were infected with lytic virus vABhcmEpL (shown by the dotted lines) and non-lytic vC4 mutant virus (shown by the solid lines) at an MOI of 1 (open circles) and 10 (solid squares), and the percentage of cells expressing and retaining green fluorescence was recorded for 8 days using a fluorocytometer. (D) Electron micrographs of baculovirus-infected cells: left panel, non-lytic C4 infection; right panel, lytic parental vABhcmEpL viral infection. Arrowheads indicate virus particles, whereas arrows indicate disruptions in the cell membrane. The inset in the left-hand panel is a magnified region showing that intact endoplasmic reticulum structures are maintained in non-lytic cells. The bar represents 1 μm.
Clone C4, which showed the least cell lysis, was used to infect Sf21 cells and cell lysis was examined further (Figure 1B). At 5 dpi, approx. 90% of C4 virus-infected cells still contained EGFP, as indicated by green fluorescence (panel a1); in sharp contrast, less than 30% of cells infected with parental viruses showed green fluorescence (panel b1). The loss of EGFP in virus-infected cells resulted from a loss of cell membrane integrity. To confirm further the loss of cellular integrity, a red fluorescent dye (EthD-1; Molecular Probes) was employed to stain specifically the dead cells. Panel a2, which shows the same field of cells as panel a1, reveals that only a few red cells were found, indicating that most cells were viable; in sharp contrast, many red cells were identified when cells were infected with the parental lytic viruses in panel b2, which is the same field of cells as in panel b1. These experiments showed that only the viable cells (Figure 1B, panels b1 and b2, indicated by arrows) retained EGFP; and the EGFP always leaked out of the cells after cell death (Figure 1B, panels b1 and b2, indicated by arrowheads).
EGFP retention was also analysed by a fluorocytometer, revealing that fluorescence in lytically infected cells started to decline after 3 dpi, with less than 10% of cells remaining fluorescent at 8 dpi, whereas fluorescence in non-lytically infected cells did not drop significantly at 4 dpi, and 50–73% of cells remained fluorescent at 8 dpi (Figure 1C). Morphologies of parental virus- and C4-infected cells were analysed by electron microscopy. Electron micrographs show that cytoplasmic membranes of C4-infected cells were intact, with well-preserved subcellular structures (Figure 1D), including the endoplasmic reticulum, an important site for folding, modification and production of secretory and membrane proteins. In contrast, intensive damage to cells infected with parental viruses was observed. It is known that quality control of proteins in eukaryotic cells involves many chaperones, proteasomes and organelles [27]. With significantly less cell lysis, the C4-infected cells are more likely to retain these quality-control processes. Here, although a low percentage of cell lysis was still found in C4-infected cells at 4 dpi, the most convenient time for protein harvesting using BEVS (Pharmingen), the C4-based BEVS is hereafter referred to as an N-(non-lytic) BEVS.
Design and strategy for the detection of protein folding in vivo
To analyse the quality of engineered proteins expressed in N-BEVS and in conventional L-BEVS, we developed a novel in vivo protein folding assay system based on FRET. FRET is a non-radioactive, dipole–dipole coupling process whereby energy from an excited donor fluorophore is transferred to an acceptor fluorophore in close proximity. FRET is a distance-dependent energy transfer process, and remains effective and typically detectable at a distance less than 10 nm. Here, an EYFP and an ECFP were used to tag opposite ends of LUC, resulting in a FRET-competent dual fluorescent protein pair, the YLC construct (Figure 2). YLC could be expressed by recombinant baculovirus in cells, and the efficiency of FRET between ECFP (the donor) and EYFP (the acceptor) could then be measured to assay the compactness of central LUC in vivo, revealing the compactness of protein (LUC) folding and/or degradation.
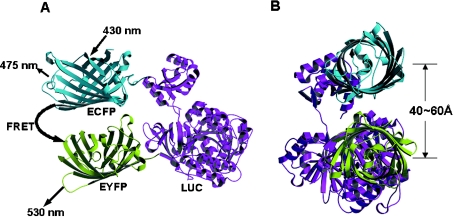
Figure 2. Schematic ribbon diagrams of a FRET-competent protein YLC.
Front view (A) and side view (B) of the YLC. EYFP (shown in yellow) and ECFP (shown in blue) were fused to the N- and C-termini of luciferase (LUC; shown in purple) respectively. The distance between ECFP (the donor) and EYFP (the acceptor) was estimated to be 5–6 nm, after the distance between the N- and C-termini of LUC (approx. 4 nm) and a β-can cylinder structure of fluorescent proteins (1.2 nm in radius and 4.2 nm in length) are taken into consideration [33,34]. Upon excitation of ECFP (the donor) at 430 nm, a sensitized yellow fluorescence emission (approx. 530 nm) from EYFP (the acceptor) signals the occurrence of FRET. This schematic structure was generated using MOLSCRIPT [35].
To begin with, we tested possible energy transfer in the YLC fusion protein by exciting this fusion protein at a wavelength of 430 nm. The fluorescence emission spectrum of purified YLC showed a cyan fluorescence emission (approx. 475 nm), and an even higher yellow fluorescence emission at approx. 530 nm was detected (Figure 3A). This indicated that FRET occurred between ECFP and EYFP in YLC. The energy transfer process was corroborated further by the fact that sensitized yellow emission was abolished upon the removal of ECFP from YLC by treatment with thrombin [Figure 3A; also see supplementary Figure 2 (http://www.biochemJ.org/bj/381/bj3810695add.htm)].
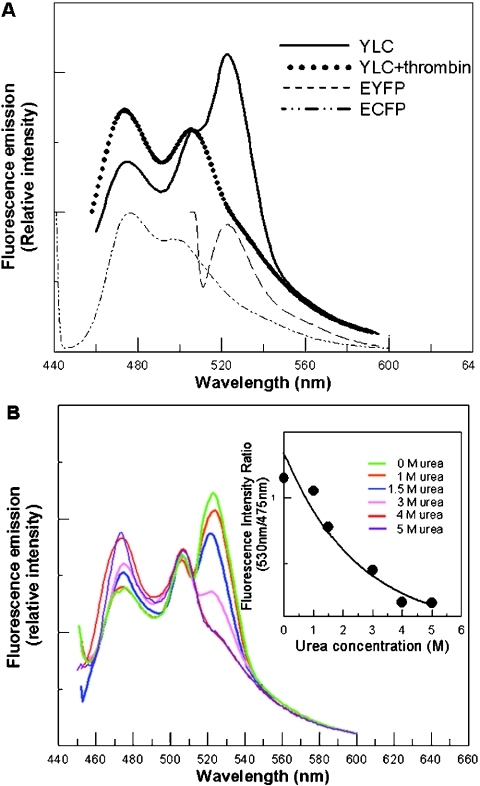
Figure 3. FRET and its correlation to the folding status of LUC in YLC.
(A) Fluorescence emission spectra of fusion proteins and individual fluorescent proteins. Upon excitation of YLC at a wavelength of 430 nm, cyan fluorescence occurred with a concomitant release of a greater emission of yellow fluorescence, whereas the sensitized yellow fluorescence was totally abolished upon the removal of EYFP by thrombin. A thrombin-cut sequence was engineered in-between EYFP and LUC, and the proper cleavage of YLC by thrombin was confirmed by Western blot analysis (supplemental Figure 2). ECFP and EYFP were produced by recombinant baculoviruses vABpC and vABpY (supplementary Figure 1) separately, to serve as controls for the emission spectra of single fluorescent proteins. (B) Correlation of FRET spectra with gradual unfolding of LUC in YLC. With the excitation of ECFP at a wavelength of 430 nm, sensitized fluorescence emission decreased concomitantly with an increasing urea concentration, from 0 to 5 M. The inset presents the decline in the fluorescence intensity ratio of yellow (530 nm) to cyan (475 nm). Since concentrations of urea less than 5 M (results not shown) have little or no effect on fluorescent proteins, the reduction in fluorescence emission at approx. 530 nm was attributed to the compromise in FRET concomitant with the loss of an ordered structure of LUC.
To study the effect of the compactness of LUC on the energy transfer between the ECFP/EYFP pair, we used urea to unfold LUC in a stepwise fashion. Upon titration of YLC with increasing amounts of urea (from 0 to 5 M), the 475 nm peak (ECFP emission) gradually increased, whereas the 530 nm peak (EYFP emission) gradually decreased to the background level (Figure 3B). In addition, the decrease in FRET was proportional to the decrease in LUC activity of YLC (supplementary Figure 3; http://www.biochemJ.org/bj/381/bj3810695add.htm). These results indicated that FRET could serve as an indicator for the monitoring of protein folding, revealing the compactness and integrity of target proteins.
The LUC expressed in N-BEVS is more compact than in L-BEVS
To compare the quality of YLC produced in N- and L-BEVSs, we constructed two lytic baculoviruses (vABpYLC and vABh-cmYLC) and two non-lytic baculoviruses (vC4pYLC and vC4hcmYLC), all of them encoding the YLC fusion protein. The engineered YLC proteins were driven either by a very late polyhedrin promoter [4] (denoted ‘p’ in vABpYLC and vC4pYLC) or by a synthetic early promoter [26] (denoted ‘hcm’ in vABhcmYLC and vC4hcmYLC), because cellular physiology may be different during early and very late stages of virus infection. Then, Sf21 cells were infected with these four different viruses and subjected to FRET measurement (Figure 4).
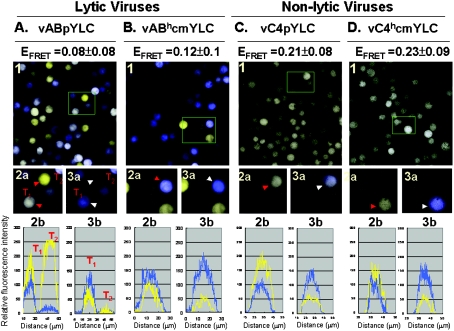
Figure 4. EFRET assays of cells infected with various recombinant baculoviruses.
The Figure shows Sf21 cells infected with lytic viruses (vABpYLC and vABhcmYLC; A and B) and non-lytic C4-based viruses (vC4pYLC and vC4hcmYLC; C and D) respectively. Panels 1 show fluorescence images of baculovirus-infected cells upon excitation at a wavelength of 458 nm. By simultaneously collecting cyan (donor) and yellow (acceptor) fluorescence, pseudo-colours were assigned to cells emitting cyan fluorescence (cyan) or yellow fluorescence (yellow) or both (pale yellow to white). Boxed regions were selected to illustrate cells before and after photobleaching at 514 nm, as shown in panels 2a and 3a respectively. Red arrowheads in panels 2a point to the cells before photobleaching, whereas white arrowheads in panels 3a mark cells which were first photobleached and then excited at 458 nm. Panels 2b and 3b are joint cyan (cyan traces) and yellow (yellow traces) fluorescence intensity profiles of the arrowhead-indicated cells in panels 2a and 3a respectively.
Similar to an excitation wavelength of 430 nm, a wavelength of 458 nm can also be properly used to excite cyan fluorescent protein. Owing to the fact that an excitation wavelength of 430 nm is not available in the confocal microscope, infected cells were excited at a convenient wavelength of 458 nm using a laser-scanning confocal fluorescence microscope (Pascal LSM; Zeiss), and the emission intensities of cyan (donor) and yellow (acceptor) fluorescence were simultaneously measured (Figure 4, panels 1, 2a and 2b). Then, the EYFP of YLC in individual cells was photobleached using energy of wavelength 514 nm to block energy transfer from the cyan donor to the yellow acceptor (Figure 4, panels 3a and 3b). After yellow elimination by photobleaching, the YLC constructs in the same cells were excited at 458 nm again. Without losing energy by transferring energy to the yellow acceptor, higher cyan emission was usually recorded in the second 475 nm acquisition (Figure 4, panels 3a and 3b). If such an enhancement of 475-nm acquisition does occur, the cells (white arrows) in panels 3a will become more bluish than those (red arrows) in panels 2a. EFRET (the efficiency of FRET) was then calculated from individual cells. The EFRET derived from the non-lytic system was significantly higher than that from the lytic system. Thus LUC expressed in the N-BEVS was in a more tightly packed conformation than that in the L-BEVS. As shown in Figure 3, it is reasonable to assume that a higher efficiency of FRET in Figure 4 is due to a better folding of the LUC centre.
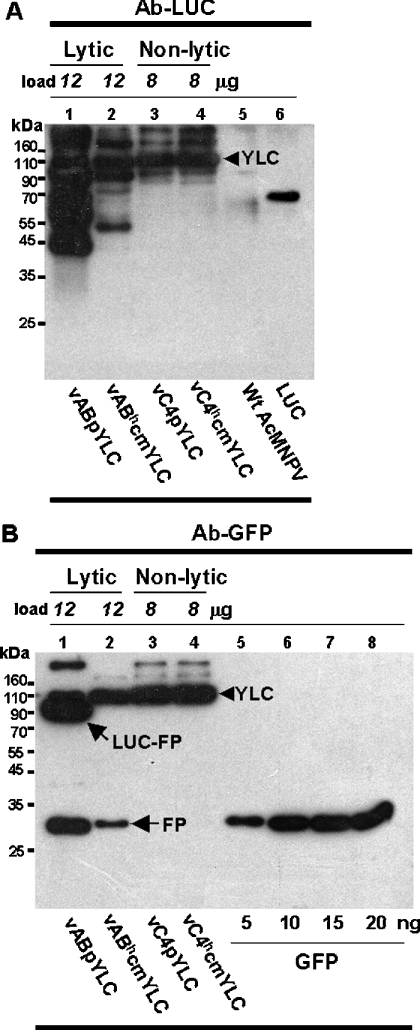
In theory, the emission of yellow fluorescence is a sign of proper FRET from cyan emission. Thus of particular interest were some yellow fluorescence-emitting cells (e.g. T2 cells in Figure 4A, panels 2a and 2b), because the cyan emission did not increase after the photobleaching of yellow fluorescence in these cells (T2 cells in Figure 4A, panels 3a and 3b). In fact, little cyan or yellow fluorescence was left in these lytic virus-infected cells after photobleaching (Figure 4A, panel 3b, the T2 trace), suggestive of truncated or incomplete YLC lacking ECFP being predominately generated in these cells. Using PCR analysis, we found that there was no deletion or truncation of the gene encoding YLC in the genome of lytic viruses (results not shown). Such protein truncation most likely resulted from partial protein degradation. Finally, YLC produced in both N- and L-BEVSs was studied further by Western blot analysis. Extensive degradation of YLC was obvious in L-BEVS (Figures 5A and 5B, lanes 1 and 2). In sharp contrast, little or no YLC degradation was observed in N-BEVS (Figures 5A and 5B, lanes 3 and 4).
Figure 5. Comparison of YLC degradation in N- and L-BEVSs.
Baculovirus-infected cell lysates were analysed by Western blotting. Antibodies against luciferase (Ab-LUC) and GFP (green fluorescent protein; Ab-GFP) were used to analyse the YLC proteins produced by lytic (vABpYLC and vABhcmYLC) or non-lytic (vC4pYLC and vC4hcmYLC) baculoviruses. Purified LUC (A, lane 6) and GFP (B, lanes 5–8) proteins (Sigma) were used as size and concentration standards. To normalize full-length YLC, different amounts of cell lysates were loaded. YLC, full-length YLC; LUC-FP, a fusion protein containing LUC and a copy of fluorescent protein; FP, fluorescent protein only. Numbers shown on the left of the panels represent the positions of molecular-mass markers.
DISCUSSION
In the present study, we have successfully established an N-BEVS with significantly less cell lysis than the L-BEVS. Cells infected with non-lytic baculoviruses showed intact morphology, whereas disrupted organelles were observed in cells infected with lytic baculoviruses. Our analyses using the FRET-based assay and Western blotting proved that the more intact the cells, the better the engineered proteins produced in terms of protein compactness. Table 1 summarizes the efficiency of FRET, the gross yields of YLC, the yields of full-length YLC, and the percentage of YLC degradation in both L- and N-BEVS, for a more detailed comparison. These comparisons clearly show that, although the gross yields of YLC were higher in L-BEVS than in N-BEVS, the majority of YLC was degraded in L-BEVS. If one compares the yields of full-length YLC, it is evident that much higher levels of full-length YLC were produced in N-BEVS than those produced by L-BEVS.
Table 1. Comparisons of YLC expression in the L-BEVS and N-BEVS.
| L-BEVS | N-BEVS | |||
|---|---|---|---|---|
| vABpYLC | vABhcmYLC | vC4pYLC | vC4hcmYLC | |
| FRET efficiency* | 0.08±0.08 | 0.12±0.1 | 0.21±0.08 | 0.23±0.09 |
| Gross YLC yield† | 412 | 109 | 322 | 365 |
| Full-length YLC | 97 | 89 | 322 | 365 |
| Degradation‡ (%) | 76 | 19 | n.d. | n.d. |
* The difference in FRET efficiency between early and very late promoters in lytic and non-lytic systems was insignificant (0.05<P<0.1), whereas the difference between the L-BEVS and N-BEVS was significant (P<0.05).
† YLC yield, consisting of both full-length and degraded YLC, was determined from Figure 5(B), expressed as μg/ml.
‡ Degradation (%) indicates the percentage of degraded fragments in all full-length and degraded YLC proteins. n.d., not detectable.
It is noteworthy that cells infected with non-lytic baculoviruses showed more intact subcellular structures compared with those cells infected with lytic viruses. It is possible that the functions of translational machinery may be affected detrimentally as cells start to become disrupted. Moreover, the efficiency of quality-control systems for translation may decrease as well. The preservation of intracellular structures and organelles, such as the endoplasmic reticulum and Golgi bodies, suggests that post-translational modifications are processed more efficiently in cells subjected to non-lytic baculovirus infection: this is especially important for the maturation of secretory proteins. Thus N-BEVS may be a promising tool for the expression of multidomain, secretory and membrane proteins, as well as proteins with complicated quaternary structures.
This FIVF (FRET-based in vivo folding) technology is very useful for measuring the overall compactness, determined by the folding and unfolding status, of target proteins. The status of the proteins expressed in BEVS is complicated, because other than folding and unfolding, protein degradation also occurs due to the infection of the virus. In Figure 3, we have shown that protein degradation also blocks FRET in the YLC protein, indicating that folding/unfolding and degradation of the target protein LUC can both be determined by the FIVF assay. Thus, to our knowledge, this is the first technology that can assess the folding status of proteins in living eukaryotic cells.
Protein folding plays an important and decisive role in protein function. In order to study the folding status of proteins, tedious protein purification and subsequent in vitro determination steps are currently routine practices [28]. To minimize possible artifacts derived from the non-biological in vitro environment, novel in vivo technologies for the study of protein folding and stability in Escherichia coli have been proposed and tested [29,30]. These studies rely on isotope labelling and subsequent protein analyses using either NMR spectroscopy or mass spectrometry after cell lysis. Since isotopes have to be labelled equally for all proteins in cells, overexpression of the target protein, such that it represents 25–30% of the total cellular protein content, is required to reduce huge background interference from all other cellular proteins. However, not all proteins can be expressed to high levels; in addition, too high a protein content in cells may interfere with correct protein folding, frequently resulting in the formation of inclusion bodies [31,32]. Moreover, the application of these methods has so far been limited to prokaryotic systems. The novel FIVF assay is a significant improvement on all the aforementioned technologies, in that it can be used to view and determine the folding status of a specific protein in a cellular environment containing thousands of other proteins.
In conclusion, we present two important and useful technological improvements in the present study. First, N-BEVS has many advantages over L-BEVS. (1) Proteins can be retained in and pelleted down with cells for simple, fast and efficient protein recovery. (2) Organelles and cellular machinery remain relatively intact for better protein modification without detectable degradation. (3) As opposed to the stable transfection of insect cells by which only early-type promoters can be used, stronger very-late promoters can be used in N-BEVS, due to the involvement of virus. With further improvements, the yields of the N-BEVS may be improved further. (4) Instead of low-efficiency stable transfection, protein production can be achieved by rapid virus infection.
Secondly, FIVF is a method that enables direct and rapid examination of the extent of protein folding in vivo among thousands of cellular background proteins. Owing to the high sensitivity of fluorescence, specific proteins can be studied without the need for high levels of expression by simply substituting the central protein (LUC) in YLC with essentially all other proteins. Thus, although this FRET-based technology is not designed for the study of detailed folding mechanisms, it is the first tool capable of real-time analysis of protein compactness and integrity in individual living cells in both eukaryotic and prokaryotic cells.
Acknowledgments
We thank S. P. Lee for her excellent work with electron microscopy. We also thank Mr Daniel Chamberlin and Kenrick J. Deen for suggesting revisions to the manuscript, and Dr Alexander Wlodawer, Dr Toshiya Endo, Dr Ching-Chung Wang, Dr James C.-K. Shen, and Dr Meng-Chao Yao for their comments. This work was supported by grants from the National Science Council (91-3112-P-001-030-Y and NSC92-2313-B-001-014) and Academia Sinica (IMB 3/15).
References
- 1.Smith G. E., Summer M. D. Restriction maps of five Autographa californica MNPV variants, Trihoplusia ni MNPV, and Galleria mellonella MNPV DNAs with endonucleases SmaI, KpnI, BamHI, SalI, XhoI, and EcoRI. J. Virol. 1979;30:828–838. doi: 10.1128/jvi.30.3.828-838.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Volkman L. E. Virus taxonomy: the classification and nomenclature of viruses. In: Murphy F. A., Fauquet C. M., Bishop D. H. L., Ghabrial S. A., Jarvis A. W., Martelli G. P., Mayo M. A., Summers M. D., editors. The Sixth Report of the ICTV. New York: Springer-Verlag Wien, Inc; 1995. pp. 104–113. [Google Scholar]
- 3.King L. A., Possee R. D. Advances in insect virology. Adv. Insect Physiol. 1994;25:2–73. [Google Scholar]
- 4.Smith G. E., Summers M. D., Fraser M. J. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol. Cell. Biol. 1983;3:2156–2165. doi: 10.1128/mcb.3.12.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarvis D. L., Summers M. D. Glycosylation and secretion of human tissue plasminogen activator in recombinant baculovirus-infected insect cells. Mol. Cell. Biol. 1989;9:214–223. doi: 10.1128/mcb.9.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumert T. F., Ito S., Wong D. T., Liang T. J. Hepatitis C virus structural proteins assemble into virus-like particles in insect cells. J. Virol. 1998;72:3827–3836. doi: 10.1128/jvi.72.5.3827-3836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pyle L. E., Barton P., Fujiwara Y., Mitchell A., Fidge N. Secretion of biologically active human proapolipoprotein A-I in a baculovirus-insect cell system: protection from degradation by protease inhibitors. J. Lipid Res. 1995;36:2355–2361. [PubMed] [Google Scholar]
- 8.Yamada K., Nakajima Y., Natori S. Production of recombinant sarcotoxin IA in Bombyx mori cells. Biochem. J. 1990;272:633–636. doi: 10.1042/bj2720633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz P. E., Martins P. C., Alves P. M., Peixoto C. C., Santos H., Moreira J., Carrondo M. J. T. Proteolytic activity in infected and noninfected insect cells: degradation of HIV-1 Pr55gag particles. Biotechnol. Bioeng. 1999;65:133–143. doi: 10.1002/(sici)1097-0290(19991020)65:2<133::aid-bit2>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 10.Ailor E., Betenbaugh M. J. Modifying secretion and post-translational processing in insect cells. Curr. Opin. Biotechnol. 1999;10:142–145. doi: 10.1016/s0958-1669(99)80024-x. [DOI] [PubMed] [Google Scholar]
- 11.Hom L. G., Volkman L. E. Preventing proteolytic artifacts in the baculovirus expression system. BioTechniques. 1998;25:18–20. doi: 10.2144/98251bm01. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki T., Kanaya T., Okazaki H., Katsuaki O., Usami A., Watanabe H., Kadono-Okuda K., Yamakawa M., Sato H., Mori H., Takahashi S., Oda K. Efficient protein production using a Bombyx mori nuclear polyhedrosis virus lacking the cysteine protease gene. J. Gen. Virol. 1997;78:3073–3080. doi: 10.1099/0022-1317-78-12-3073. [DOI] [PubMed] [Google Scholar]
- 13.Li E., Brown S. L., Dolman C. S., Brown G. B., Nemerow G. R. Production of functional antibodies generated in a nonlytic insect cell expression system. Protein Express. Purif. 2001;21:121–128. doi: 10.1006/prep.2000.1329. [DOI] [PubMed] [Google Scholar]
- 14.Farrell P. J., Lu M., Prevost J., Brown C., Behie L., Iatrou K. High-level expression of secreted glycoproteins in transformed lepidopteran insect cells using a novel expression vector. Biotechnol. Bioeng. 1998;60:656–663. doi: 10.1002/(sici)1097-0290(19981220)60:6<656::aid-bit2>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 15.Rhoades E., Gussakovsky E., Haran G. Watching proteins fold one molecule at a time. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3197–3202. doi: 10.1073/pnas.2628068100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lillo M. P., Szpikowska B. K., Mas M. T., Sutin J. D., Beechem J. M. Real-time measurement of multiple intramolecular distances during protein folding reactions: a multisite stopped-flow fluorescence energy-transfer study of yeast phosphoglycerate kinase. Biochemistry. 1997;36:11273–11281. doi: 10.1021/bi970789z. [DOI] [PubMed] [Google Scholar]
- 17.Schuler B., Lipman E. A., Eaton W. A. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Science. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 18.Nishimura C., Riley R., Eastman P., Fink A. L. Fluorescence energy transfer indicates similar transient and equilibrium intermediates in Staphylococcal nuclease folding. J. Mol. Biol. 2000;299:1133–1146. doi: 10.1006/jmbi.2000.3804. [DOI] [PubMed] [Google Scholar]
- 19.Miyawaki A., Llopis J., Heim R., McCaffery J. M., Adams J. A., Ikura M., Tsien R. Y. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature (London) 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 20.Kohl T., Heinze K. G., Kuhlemann R., Koltermann A., Schwille P. A protease assay for two-photon cross-correlation and FRET analysis based solely on fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12161–12166. doi: 10.1073/pnas.192433499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato M., Ozawa T., Inukai K., Asano T., Umezawa Y. Fluorescent indicators for imaging protein phosphorylation in single living cells. Nat. Biotechnol. 2002;20:287–294. doi: 10.1038/nbt0302-287. [DOI] [PubMed] [Google Scholar]
- 22.Nagai Y., Miyazaki M., Aoki R., Zama T., Inouye S., Hirose K., Iino M., Hagiwara M. A fluorescent indicator for visualizing cAMP-induced phosphorylation in vivo. Nat. Biotechnol. 2000;18:313–316. doi: 10.1038/73767. [DOI] [PubMed] [Google Scholar]
- 23.Lee J. C., Chen H. H., Chao Y. C. Persistent baculovirus infection results from deletion of the apoptotic suppressor gene p35. J. Virol. 1998;72:9157–9165. doi: 10.1128/jvi.72.11.9157-9165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin J. L., Lee J. C., Chen S. S., Wood H. A., Li M. L., Li C. F., Chao Y. C. Persistent Hz-1 virus infection in insect cells: evidence for insertion of viral DNA into host chromosomes and viral infection in a latent status. J. Virol. 1999;73:128–139. doi: 10.1128/jvi.73.1.128-139.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Reilly D. R., Miller L. K., Luckow V. A. New York: Oxford University Press; 1994. Baculovirus Expression Vectors: A Laboratory Manual. [Google Scholar]
- 26.Lo H. R., Chou C. C., Wu T. Y., Yuen J. P. Y., Chao Y. C. Novel baculovirus DNA elements strongly stimulate activities of exogenous and endogenous promoters. J. Biol. Chem. 2002;277:5256–5264. doi: 10.1074/jbc.M108895200. [DOI] [PubMed] [Google Scholar]
- 27.Wickner S., Maurizi M. R., Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 28.Righetti P. G., Verzola B. Folding/unfolding/refolding of proteins: present methodologies in comparison with capillary zone electrophoresis. Electrophoresis. 2001;22:2359–2374. doi: 10.1002/1522-2683(200107)22:12<2359::AID-ELPS2359>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 29.Ghaemmaghami S., Oas T. G. Quantitative protein stability measurement in vivo. Nat. Struct. Biol. 2001;10:879–882. doi: 10.1038/nsb1001-879. [DOI] [PubMed] [Google Scholar]
- 30.Serber Z., Votsch D. In-cell NMR spectroscopy. Biochemistry. 2001;40:14317–14323. doi: 10.1021/bi011751w. [DOI] [PubMed] [Google Scholar]
- 31.Clark E. D. B. Refolding of a recombinant proteins. Curr. Opin. Biotechnol. 1998;9:157–163. doi: 10.1016/s0958-1669(98)80109-2. [DOI] [PubMed] [Google Scholar]
- 32.Speed M. A., Wang D. I. C., King J. Specific aggregation of partially folded polypeptide chains: the molecular basis of inclusion body composition. Nat. Biotechnol. 1996;14:1283–1287. doi: 10.1038/nbt1096-1283. [DOI] [PubMed] [Google Scholar]
- 33.Yang F., Moss L. G., Phillips G. N. J. The molecular structure of green fluorescent protein. Nat. Struct. Biol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 34.Conti E., Franks N. P., Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996;4:287–298. doi: 10.1016/s0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- 35.Kraulis P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]