Abstract
ABCG2 [also known as BCRP (breast cancer resistance protein) or MXR] is an ABC (ATP-binding cassette) protein shown to confer multidrug resistance. ABCG2 was initially identified in resistant breast carcinoma cells (MCF-7/AdrVp1000) selected with doxorubicin and verapamil. Later studies demonstrated the presence of a point mutation (Arg482 to Thr) in ABCG2 in MCF-7/AdrVp1000 cells. This mutation was shown to modulate the transport of Rh123 (rhodamine 123). In the present study, we have used a previously characterized photoreactive drug analogue of Rh123, IAARh123 (iodoaryl-azido-Rh123), to examine the effects of the Arg482→Thr mutation on Rh123 binding and transport by ABCG2. Our results show that both wild-type (ABCG2R482) and mutant (ABCG2T482) ABCG2 bound directly to IAARh123. Surprisingly, however, wild-type ABCG2R482, which does not transport Rh123, was more intensely photolabelled than mutant ABCG2T482. In addition, inhibition of IAARh123 photolabelling using various drug substrates of ABCG2 revealed some differences between wild-type and mutant ABCG2. For example, a molar excess of mitoxantrone was more effective at inhibiting IAARh123 labelling of wild-type than of mutant ABCG2, while excess cisplatin, taxol and methotrexate showed significant inhibition of IAARh123 binding to both wild-type and mutant ABCG2. Taken together, the results of this study provide the first demonstration of the direct binding of drugs to ABCG2.
Keywords: breast cancer resistance protein (BCRP), drug interactions, multidrug resistance, photoaffinity labelling, rhodamine 123
Abbreviations: ABC, ATP-binding cassette; BCRP, breast cancer resistance protein; IAARh123, iodo-arylazido-rhodamine 123; MDR, multidrug resistance; MEM, minimal essential medium; MRP1, multidrug resistance protein 1; P-gp1, P-glycoprotein; Rh123, rhodamine 123
INTRODUCTION
Members of the ABC (ATP-binding cassette) family of membrane proteins have been shown to confer MDR (multidrug resistance) on cancer cells and to protect diverse organisms against toxic drugs [1]. Both P-gp1 (P-glycoprotein) and MRP1 (MDR protein 1) extrude different anticancer drugs in an ATP-dependent manner [2,3]. Among the recently discovered genes with a potential role in drug resistance is that encoding ABCG2 [also known as BCRP (breast cancer resistance protein) or MXR] [4]. ABCG2 is a 655-amino-acid integral membrane protein with a large cytoplasmic domain including a single N-terminal nucleotide-binding domain, followed by six putative membrane-spanning domains. This topology, which is half that of P-gp1, is characteristic of the ABCG gene family that includes, among others, genes encoding the Drosophila white, brown and scarlet proteins, along with the human white homologue ABCG1. In addition, Allen et al. [5] have cloned the murine homologue Bcrp1 by overexpression in mouse fibroblast cells selected for resistance to doxorubicin, mitoxantrone or topotecan. Murine Bcrp1 has a topology similar to that of human ABCG2, with 81% identity, and also mediates drug resistance through energy-dependent efflux of drug substrates. ABCG2 and Bcrp1 have both been shown to be localized in plasma membranes using several polyclonal and monoclonal antibodies [6]. ABCG2 was found in different tissues in apical, liver and blood cells, indicating that ABCG2 overexpression may occur in many types of tumours [7].
Human breast cancer carcinoma MCF-7 cells selected for resistance to doxorubicin, in the presence of verapamil (an inhibitor of P-gp1), led to the selection of a multidrug-resistant sub-line (MCF-7/AdrVp) that exhibited cross-resistance to mitoxantrone and daunorubicin, but not to vinca alkaloids, paclitaxel and cisplatin [8]. Transfection studies using cDNA confirmed that ABCG2 confers resistance to drugs through ATP-dependent transport of ABCG2 substrates [4]. It was shown [9] that MCF-7/AdrVp1000 cells and a human colon carcinoma cell line selected for resistance to mitoxantrone, S1-M1-80, overexpress ABCG2 and efflux Rh123 (rhodamine 123) and doxorubicin in an ATP-dependent manner. However, other selected cell lines that also express high levels of ABCG2 did not efflux Rh123 or doxorubicin. Analysis of ABCG2 cDNA sequences from these cell lines revealed mutations at position 482: Arg482 to Thr in MCF-7/AdrVp1000 cells, and to Gly in S1-M1-81 cells. In addition, transfection studies using the cDNA of wild-type ABCG2R482, as well as the mutants ABCG2T482 and ABCG2G482, showed that only mutated ABCG2T/G482 could extrude Rh123 and doxorubicin out of the transfected cells [9]. The latter drug transport results were consistent with the resistance profile seen in cell lines selected for resistance to mitoxantrone. The direct interactions between Rh123 and ABCG2 are not known. Furthermore, Volk et al. [10] found that the Arg482→Thr and Arg482→Gly mutations in ABCG2 abolished methotrexate resistance compared with wild-type ABCG2, confirming that position 482 is crucial for drug recognition in ABCG2. In the present study, it was of interest to compare the interaction of Rh123 with and its transport by wild-type ABCG2R482 and mutant ABCG2T482 using the photoactive drug analogue of Rh123 [IAARh123 (iodo-arylazido-rhodamine 123)].
MATERIALS AND METHODS
Materials
The BXP-21 monoclonal antibody was purchased from Kamiya Biomedical Company (Seattle, WA, U.S.A.). 125I (100 mCi/ml) was purchased from Amersham Biomedical Inc. (Mississauga, Ontario, Canada). Protein A–Sepahrose was purchased from Pharmacia Inc. (Montreal, Quebec, Canada). Rh123, mitoxantrone, methotrexate and verapamil were purchased from Sigma-Aldrich (Oakville, Ontario, Canada). All other chemicals were of the highest commercial grade available.
Cell lines
The human breast cancer carcinoma MCF-7 cell line was subjected to a stepwise increase in mitoxantrone concentration. The resulting selected MCF-7/Mitox cell line could survive a mitoxantrone concentration of 78 nM. The resistant cell line was grown in α-MEM (minimal essential medium) plus 10% (v/v) fetal calf serum containing 78 nM mitoxantrone. The MCF-7/AdrVp1000 cells were also maintained in the same medium containing 100 ng/ml adriamycin and 10 μg/ml verapamil, as described previously [8].
Reverse transcription–PCR
Two primers were designed to obtain the ABCG2 gene from selected MCF-7/Mitox cells: sense 1206–1223, 5′-TGA TCT GCC TGC GTC AGC-3′; antisense 1663–1646, 5′-GTG AGT GTA GCC ATC CCA-3′. The expected PCR product was 457 bp in length. Random hexamers were used to prime the reverse transcription reaction, which was followed by 25 cycles of PCR as described elsewhere [11]. The cDNA was sequenced using double-strand sequencing to confirm the identity of the ABCG2 gene.
Plasma membrane preparations
MCF-7, MCF-7/AdrVp1000 and MCF-7/Mitox cells were grown in α-MEM containing 10% (v/v) fetal calf serum (Hyclone). Plasma membranes from the three cell lines were prepared as described previously [12]. Plasma membrane-enriched fractions were resuspended in NTE buffer [100 mM NaCl, 10 mM Tris/HCl, pH 7.4, and 10 mM EDTA in 30% (v/v) glycerol]. Protein concentrations were determined by the Lowry method [13]. Membranes were stored at −80 °C if not used immediately.
Photoaffinity labelling and immunoprecipitation
Total membranes from MCF-7, MCF-7/AdrVp1000 and MCF-7/Mitox cells were pre-incubated in the absence or presence of various concentrations of drugs prior to the addition of IAARh123 in 20 μl of labelling solution (5 mM Tris, pH 7.4, 250 mM sucrose with protease inhibitors: 2 μg/ml leupeptin, aprotenin and pepstatin and 2 mM PMSF) for 30 min in the dark. Membranes were then incubated for an additional 10 min on ice followed by UV irradiation at 254 nm for 10 min while on ice (Stratgene 1800 UV cross-linker; Stratagene, La Jolla, CA, U.S.A.) [14]. Following photoaffinity labelling, samples were mixed with 50 μl of buffer A (1% SDS and 0.05 M Tris, pH 7.4) and 200 μl of buffer B (1.25% Triton X-100, 190 mM NaCl and 0.05 M Tris, pH 7.4). Immunoprecipitation was carried out as described previously [12] using an ABCG2-specific monoclonal antibody, BXP-21 [15]. Immunoprecipitated proteins were resolved on SDS/PAGE using the Fairbanks gel system [16]. Gels were dried and exposed to Kodak X-AR film at −80 °C.
Accumulation of Rh123
MCF-7, MCF-7/AdrVp and MCF-7/Mitox cells, seeded in six-well plates (5×104 cells/well), were washed three times with PBS, pH 7.4, and incubated for 45 min at 37 °C with 50 mM glucose or 50 mM deoxyglucose and 15 mM sodium azide, or with 2 μM cyclosporine A. Rh123 was added to a final concentration of 1 μM, and cells were incubated for an additional 60 min. Cells were washed with ice-cold PBS and observed using a Nikon TE200 inverted microscope with a B-2A fluorescence filter.
Flow cytometry
MCF-7, MCF-7/AdrVp1000 and MCF-7/Mitox cells were grown in six-well plates in α-MEM, washed with PBS, and incubated with or without inhibitors of ATP synthesis (as indicated previously) for 30 min at 37 °C. Rh123 was added to each well at 1 μM final concentration with or without 2 μM cyclosporine A. The cells were left for 1 h at 37 °C, and then washed and suspended by trypsinzation. The cells were resuspended in cold PBS and subjected to cell sorting and analysis by FACscan using rhodamine excitation at λ=485 nm.
Cytotoxicity assay
MCF-7, MCF-7/Mito and MCF-7/AdrVp1000 cells were seeded at 1000–2000 cells per well in 24-well plates. Cells were allowed to grow overnight before the addition of Rh123 at increasing concentrations. Cells were grown for 6 days and then stained with 0.1% Methylene Blue in 50% (v/v) ethanol as described previously [17]. The staining of adherent cells was examined by incubating stained cells with 250 μl of 0.1% SDS in PBS at 37 °C for 1 h. The stained cells from each well were quantified at 630 nm using a Dynatech MR5000 plate reader. The effects of drugs on the viability of cells are expressed as means±S.D. of three independent experiments done in triplicate.
RESULTS
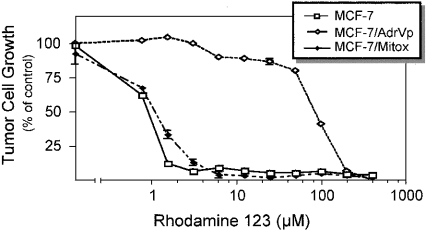
Rh123, a cationic fluorescent dye, has been shown to accumulate selectively in the mitochondria of eukaryotic cells [18]. P-gp1 and MRP1 have been shown to mediate the transport of Rh123 by direct binding [14,19]. Early studies have shown that the mutant ABCG2T482 could transport Rh123, while the wild-type ABCG2 could not [9]. In the present study, it was of interest to examine the effects of the Arg482→Thr point mutation on ABCG2 binding to, and transport of, Rh123. Figure 1 shows the growth of MCF-7, MCF-7/AdrVp1000 (expressing mutant ABCG2T482) and MCF-7/Mitox (expressing wild-type ABCG2R482) cells in the presence of increasing concentrations of Rh123. Figure 1 shows the IC50 of MCF-7/AdrVp1000 cells to be higher (∼100-fold) than that of MCF-7 or MCF-7/Mitox cells. These results are in agreement with earlier studies showing that Rh123 is a substrate for mutant ABCG2T482 or ABCG2G482, but not for the wild-type ABCG2 [9].
Figure 1. Effects of Rh123 on cell growth.
MCF-7, MCF-7/AdrVp1000 and MCF-7/Mitox cells were seeded at 1000–2000 cells/well in 24-well plates and grown in the absence or presence of increasing concentrations of Rh123 (0–400 μM). Cell viability was estimated 6 days later by staining the surviving cell colonies with 0.1% Methylene Blue in 50% (v/v) ethanol. Cell growth is expressed as a percentage of the control in the absence of drug. Each point is the mean±S.D. of three independent experiments.
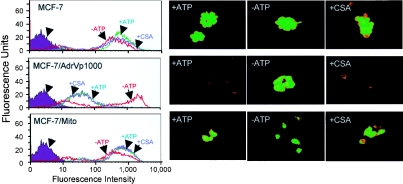
Next we determined if the differential sensitivity of MCF-7, MCF-7/Mito and MCF-7/AdrVp1000 cells to Rh123 is due to differences in Rh123 accumulation, and if it is ATP-dependent. Figure 2 (left panels) shows decreased Rh123 accumulation in cells expressing mutant ABCG2 (MCF-7/AdVp1000) in the presence of ATP. MCF-7 and MCF-7/Mitox cells expressing wild-type ABCG2 accumulated Rh123. The reduced accumulation of Rh123 in MCF-7/AdrVp1000 cells was reversed in the presence of inhibitors of ATP synthesis. This indicates that ATP is required for Rh123 efflux by mutant ABCG2T482. In addition, cyclosporine A, a P-gp1 modulator, did not reverse the efflux of Rh123 by mutant ABCG2. Figure 2 (right panels) shows the fluorescence of cells incubated with Rh123 under identical conditions as in Figure 2 (left panels). Together, the results in Figure 2 confirm early studies showing ATP-dependent efflux of Rh123 in tumour cells expressing mutant ABCG2T482, but not in those expressing wild-type ABCG2R482 [9,20].
Figure 2. Intracellular uptake of Rh123.
Cells (MCF-7, MCF-7/AdrVp1000 and MCF-7/Mitox) were grown in six-well plates, washed with PBS and incubated in the presence or absence of inhibitors of ATP synthesis or cyclosporine A (CSA), as described in the Materials and methods section. Cells were kept for 30 min at 37 °C before adding Rh123 to each well at a final concentration of 1 μM, with or without 2 μM cyclosporine A (as indicated in the Figure). Left panels: cells were washed and subjected to flow cytometry by FACscan using Rh123 excitation at λ=485 nm. Right panels: cells after incubation were viewed at ×400 magnification using a Nikon TE200 inverted microscope equipped with a fluorescence filter.
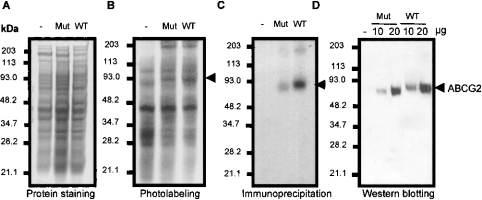
To determine whether mutation of Arg482 affects ABCG2 binding to Rh123, and hence whether only ABCG2T482 but not ABCG2R482 interacts directly with Rh123, drug binding of mutant ABCG2T482 and wild-type ABCG2R482 was examined using IAARh123. The results in Figure 3(B) (lanes 3 and 4) show the labelling of membranes from MCF-7, MCF-7/AdrVp1000 and MCF-7/Mitox cells with IAARh123. A ∼72 kDa protein in MCF-7/AdrVp1000 and MCF-7/Mitox cells, but not in MCF-7 cells, was photoaffinity labelled by IAARh123. To identify the 72 kDa protein, membranes of the three cell lines were photoaffinity labelled with IAARh123 and then immunoprecipitated with an ABCG2-specific monoclonal antibody, BXP-21. Figure 3(C) (lanes 2 and 3) shows a single polypeptide with an apparent molecular mass of 72 kDa that was not immunoprecipitated from MCF-7 membranes. Interestingly, membranes from MCF-7/Mitox cells expressing wild-type ABCG2 showed more intense photolabelling by IAARh123 than membranes from MCF-7/AdrVp1000 cells expressing the mutant ABCG2T482 (lane 3 compared with lane 2 in Figures 3B and 3C). To determine if the differences in ABCG2 photolabelling intensity were due to differences in ABCG2 expression in the three cell lines, Coomassie Blue staining was performed and the membranes were resolved on SDS/PAGE. Figure 3(A) (lanes 1–3) shows the similarity of protein expression levels in the three cell lines. To determine if ABCG2 is differentially expressed in MCF-7/AdrVp1000 and MCF-7/Mitox cells, membrane proteins from the two cell lines were resolved on SDS/PAGE and Western blotted using the monoclonal antibody BXP-21. Figure 3(D) shows the protein levels of ABCG2 expressed in MCF-7/AdrVp1000 and MCF-7/Mitox cells (lanes 2 and 3 compared with lanes 4 and 5). Both cell lines expressed similar amounts of ABCG2 protein, and the differences in photolabelling were due to the high affinity of IAARh123 for wild-type ABCG2 compared with mutant ABCG2T482.
Figure 3. Photoaffinity labelling of ABCG2 by IAARh123.
Total membranes prepared from MCF-7 (−), MCF-7/AdrVp1000 (Mut; expressing mutated ABCG2) and MCF-7/Mitox (WT; expressing wild-type ABCG2) cells were photoaffinity labelled with IAARh123 and resolved on SDS/PAGE (B). Panel (A) shows the Coomassie Blue staining of membrane proteins prepared from the indicated cell lines. (C) Photoaffinity-labelled membrane proteins from MCF-7, MCF-7/AdrVp1000 and MCF-7/Mitox cells immunoprecipitated with an ABCG2-specific monoclonal antibody (BXP-21). (D) Western blot of membranes (10 and 20 μg) prepared from the same cells using monoclonal antibody BXP-21. The position of ABCG2 is indicated by the arrowheads.
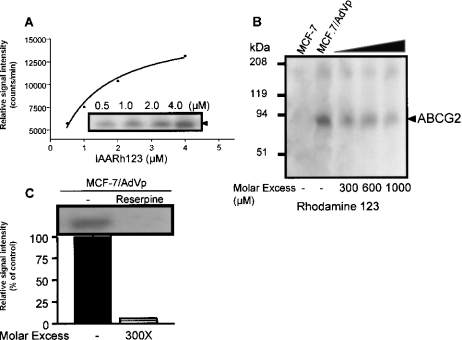
Given these unexpected results, it was important to determine the specificity of photolabelling of ABCG2 by IAARh123. Membranes from MCF-7/AdrVp1000 cells were photolabelled with increasing amounts of IAARh123 (0.5–4.0 μM). Figure 4(A) shows the intensity of mutant ABCG2 photoaffinity labelling with increasing IAARh123 concentrations. Photolabelling of mutant ABCG2 by IAARh123 was inhibited by increasing molar excesses of Rh123 added to the membranes prior to photolabelling (Figure 3B). A similar saturation curve and photolabelling inhibition by Rh123 were obtained with membranes containing wild-type ABCG2 (results not shown). To confirm that IAARh123 binds to a physiological drug binding site in ABCG2, Figure 4(C) shows the inhibition of photoaffinity labelling by reserpine, shown previously to reverse ABCG2-mediated MDR [21].
Figure 4. Specificity of ABCG2 photolabelling with IAARh123.
(A) Membrane proteins (50 μg) from MCF-7/AdrVp1000 cells were photolabelled with increasing concentrations (0–4 μM) of IAARh123. After SDS/PAGE, the bands corresponding to ABCG2 were excised and quantified. (B) Photolabelled ABCG2 from MCF-7/AdrVp1000 cells incubated in the absence or presence of a molar excess (0–1000-fold) of IAARh123. (C) Photolabelling of ABCG2 with IAARh123 in the absence or presence of reserpine at a 300× molar excess.
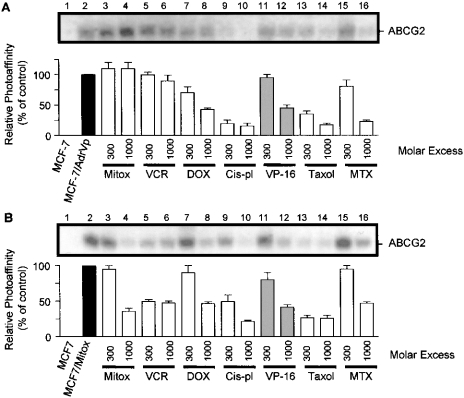
In addition to reserpine, it was important to determine if binding of IAARh123 to wild-type and mutant ABCG2 is affected by the presence of other substrates of ABCG2. Plasma membranes from MCF-7/AdrVp1000 and MCF-7/Mitox cells were photolabelled with IAARh123 in the presence of a molar excess of mitoxantrone, vincristine, doxorubicin, cisplatin, etoposide VP-16, taxol or methotrexate. The photolabelling of mutant ABCG2T482 was not inhibited by increasing the molar excess of mitoxantrone, a known substrate of ABCG2, and was in fact increased in the presence of a molar excess of mitoxantrone (Figure 5A, lanes 3 and 4). By contrast, photolabelling of wild-type ABCG2 was inhibited by increasing molar excess of mitoxantrone (Figure 5B, lanes 3 and 4). The different effects of mitoxantrone on ABCG2 photolabelling may be due to dissimilar binding affinities of the mutant as compared with wild-type ABCG2. Moreover, drugs that are not known to be substrates for ABCG2 have been shown to inhibit IAARh123 photolabelling of both mutant and wild-type ABCG2. Figures 5(A) and 5(B) (lanes 9–10 and 13–14) show that cisplatin and taxol significantly inhibited photolabelling of mutant ABCG2T482 and wild-type ABCG2R482 respectively. In addition, methotrexate, a substrate of wild-type ABCG2, inhibited photolabelling of both mutant and wild-type ABCG2 (lanes 15 and 16, Figures 5A and 5B). Similar inhibition of ABCG2 photoaffinity labelling occurred in the presence of increasing molar excess of vincristine, doxorubicin and etoposide VP-16 (Figures 5A and 5B). Consequently, these results show Rh123 to interact with mutant and wild-type ABCG2 at the same or overlapping site(s) as particular drugs such as cisplatin, taxol and methotrexate. However, other drugs such as vincristine, doxorubicin and etoposide VP-16 may interact with ABCG2 at different sites than Rh123. The inhibitory effect of mitoxantrone is different for mutant and wild-type ABCG2, probably as a result of the mutation of Arg482.
Figure 5. Effects of diverse drugs on the photoaffinity labelling of wild-type and mutant ABCG2 by IAARh123.
Membrane proteins from MCF-7, MCF-7/AdrVp1000 (A) or MCF-7/Mito (B) cells were photoaffinity labelled with IAARh123 in the absence or presence of a molar excess (300–1000-fold) of mitoxantrone (Mitox), vincristine (VCR), doxorubicin (DOX), cisplatin (Cis-pl), VP-16, taxol or methotrexate (MTX). The relative photolabelling signal of wild-type and mutant ABCG2 with IAARh123 in the presence of these drugs is also shown. The photoaffinity labelling results show a representative experiment that was repeated at least three times. The bar graphs show the relative decrease in the photoaffinity labelling of ABCG2 in the presence of increasing molar concentrations of the drugs averaged between three different independent experiments.
DISCUSSION
ABCG2 has been shown to confer resistance to mitoxantrone, daunorubicin and topotecan [7]. Surprisingly, MCF-7/AdrVp1000 cells and a mitoxantrone-selected S1-M1-80 colon cell line both contain a point mutation at amino acid 482 in ABCG2. In these mutants, the parental arginine is replaced by threonine or glycine respectively. This mutation results in greater resistance to anthracyclines and also enables transport of Rh123 [9]. In the present study, we have selected the MCF-7 cell line with mitoxantrone, causing the overexpression of wild-type ABCG2R482 (MCF-7/Mitox). Compared with the parental MCF-7 cell line, MCF-7/Mitox cells display drug resistance to mitoxantrone, doxorubicin and VP-16, but not to vinblastine (results not shown). We have also shown that the MCF-7/AdrVp1000 cell line, which expresses mutant ABCG2T482, is more resistant to Rh123 than are MCF-7/Mitox and parental MCF-7 cells (Figure 1). Although previous studies have proposed that the absence of drug transport in MCF-7/AdrVp1000 cells is due to a single point mutation in ABCG2, no direct evidence was provided. For example, it is plausible that the Arg482 to Thr change in the third transmembrane domain affects ABCG2 protein interactions (either homo- or hetero-protein interactions), which in turn leads to a gain in transport function. In addition, the transport of Rh123 in MCF-7/AdrVp1000 cells is ATP dependent, and can be inhibited by depleting ATP. Cyclosporine A, a P-gp1 modulator, did not inhibit Rh123 efflux. These observations are in agreement with earlier studies which indicated that the single mutation of amino acid Arg482 is essential for Rh123 transport by ABCG2 [4,9].
The latter finding, i.e. that a single point mutation completely abolishes the ability of ABCG2 to transport Rh123, persuaded us to examine how this point mutation affects the binding of Rh123 to ABCG2. Our results show that IAARh123 binding to ABCG2 is specific and is inhibited in the presence of a molar excess of Rh123 or reserpine (an inhibitor of ABCG2). Interestingly, while wild-type ABCG2 does not efflux Rh123, binding of IAARh123 is detected in greater quantities than with the mutant protein. It is still not clear if this difference is due to a change in the binding site affecting the affinity of Rh123 for ABCG2 or the transport of the drug by the protein. One possibility is that the slightly different chemical structure of IAARh123 may support binding to wild-type ABCG2, whereas the parent compound Rh123 cannot bind. Alternatively, it is possible that mutation at Arg482 could affect the oligomerization of ABCG2, which in turn affects the transport, but not the binding, of Rh123.
The specificity of the binding of IAARh123 to physiologically relevant site(s) in both mutant and wild-type ABCG2 was confirmed by competitive inhibition studies using molar excesses of different cytotoxic drugs. We found that mitoxantrone, a known substrate of ABCG2, was more effective at inhibiting the IAARh123 photolabelling of wild-type compared with mutant ABCG2. This may be contrasted with observations that the mutant protein is able to confer higher levels of resistance on mitoxantrone, compared with the wild-type. These studies, performed in ABCG2 transfectants, indicate that the gain of function that is observed with the mutant protein extends to mitoxantrone as well as Rh123, daunorubicin and Lysotracker Green [22], except that no transport of the latter three substrates is observed with the wild-type protein. However, the binding site for IAARh123 may be shifted in the mutant so that mitoxantrone is less effective in competition. This concept would be supported by the possibility that a different configuration in the third transmembrane domain results from the change from the positively charged, bulky amino acid arginine to the polar, smaller amino acid threonine. A similar effect is found with vincristine. It inhibits IAARh123 photolabelling of wild-type ABCG2 more than that of mutant ABCG2. Doxorubicin and VP-16 cause a mild inhibition of photolabelling of both wild-type and mutant ABCG2. Surprisingly, taxol and cisplatin caused a remarkable inhibition of the photolabelling by IAARh123 of both types of ABCG2. These two drugs have not been reported to be ABCG2 substrates [4,21]. This suggests the possibility that taxol and cisplatin both bind to ABCG2, but are not transported. The transport function of ABCG2 may require other physico-chemical changes in ABCG2, following drug binding. Indeed, our results are consistent with recent findings that drugs that inhibit the function of ABCG2 are not substrates for this protein [23]. In this latter study [23], it was demonstrated that HIV protease inhibitors affect ABCG2-associated MDR, but are not substrates of ABCG2. Consistent with this, it was reported that progesterone could inhibit P-gp1 photolabelling despite the fact that it was not consistently a substrate for P-gp1 [24]. On the other hand, methotrexate at high concentrations (1000-fold excess) inhibits the photolabelling of both mutant and wild-type ABCG2. However, an earlier study has shown that MCF-7/AdrVp1000 cells (containing ABCG2T482) displayed little or no cross-resistance to methotrexate, despite high resistance to mitoxantrone. Cells overexpressing wild-type ABCG2 demonstrate high cross-resistance to methotrexate [10]. This adds another important characteristic of the point mutant of ABCG2. Arg482 determines the drug specificity and affects the transport function of the protein, and impacts on photoaffinity labelling. Moreover, the drug competition studies provide more evidence that drug binding to the protein is only one step in drug transport.
In summary, we have shown for the first time that ABCG2 binds directly and specifically to a photoactive drug, IAARh123. This binding is more intense for wild-type ABCG2R482 than for mutant ABCG2T482. The competition of photolabelling by various drugs reveals differences in the drug binding and transport function of ABCG2.
Acknowledgments
We thank Ms Carole Damato for her careful editing of this manuscript before submission. This work was supported by grants from the Natural Sciences and Engineering Research Council (NSERC) of Canada to E.G.
References
- 1.Higgins C. F. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 2.Cole S. P., Bhardwaj G., Gerlach J. H., Mackie J. E., Grant C. E., Almquist K. C., Stewart A. J., Kurz E. U., Duncan A. M., Deeley R. G. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 3.Ling V. Multidrug resistance: molecular mechanisms and clinical relevance. Cancer Chemother. Pharmacol. 1997;40(Suppl.):S3–S8. doi: 10.1007/s002800051053. [DOI] [PubMed] [Google Scholar]
- 4.Doyle L. A., Yang W., Abruzzo L. V., Krogmann T., Gao Y., Rishi A. K., Ross D. D. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15665–15670. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen J. D., Brinkhuis R. F., Wijnholds J., Schinkel A. H. The mouse Bcrp1/Mxr/Abcp gene: amplification and overexpression in cell lines selected for resistance to topotecan, mitoxantrone, or doxorubicin. Cancer Res. 1999;59:4237–4241. [PubMed] [Google Scholar]
- 6.Scheffer G. L., Maliepaard M., Pijnenborg A. C., van Gastelen M. A., de Jong M. C., Schroeijers A. B., van der Kolk D. M., Allen J. D., Ross D. D., van der Valk P., et al. Breast cancer resistance protein is localized at the plasma membrane in mitoxantrone- and topotecan-resistant cell lines. Cancer Res. 2000;60:2589–2593. [PubMed] [Google Scholar]
- 7.Maliepaard M., van Gastelen M. A., de Jong L. A., Pluim D., van Waardenburg R. C., Ruevekamp-Helmers M. C., Floot B. G., Schellens J. H. Overexpression of the BCRP/MXR/ABCP gene in a topotecan-selected ovarian tumor cell line. Cancer Res. 1999;59:4559–4563. [PubMed] [Google Scholar]
- 8.Chen Y. N., Mickley L. A., Schwartz A. M., Acton E. M., Hwang J. L., Fojo A. T. Characterization of adriamycin-resistant human breast cancer cells which display overexpression of a novel resistance-related membrane protein. J. Biol. Chem. 1990;265:10073–10080. [PubMed] [Google Scholar]
- 9.Honjo Y., Hrycyna C. A., Yan Q. W., Medina-Perez W. Y., Robey R. W., van de Laar A., Litman T., Dean M., Bates S. E. Acquired mutations in the MXR/BCRP/ABCP gene alter substrate specificity in MXR/BCRP/ABCP-overexpressing cells. Cancer Res. 2001;61:6635–6639. [PubMed] [Google Scholar]
- 10.Volk E. L., Farley K. M., Wu Y., Li F., Robey R. W., Schneider E. Overexpression of wild-type breast cancer resistance protein mediates methotrexate resistance. Cancer Res. 2002;62:5035–5040. [PubMed] [Google Scholar]
- 11.Ross D. D., Doyle L. A., Schiffer C. A., Lee E. J., Grant C. E., Cole S. P., Deeley R. G., Yang W., Tong Y. Expression of multidrug resistance-associated protein (MRP) mRNA in blast cells from acute myeloid leukemia (AML) patients. Leukemia. 1996;10:48–55. [PubMed] [Google Scholar]
- 12.Georges E., Zhang J. T., Ling V. Modulation of ATP and drug binding by monoclonal antibodies against P-glycoprotein. J. Cell. Physiol. 1991;148:479–484. doi: 10.1002/jcp.1041480321. [DOI] [PubMed] [Google Scholar]
- 13.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 14.Nare B., Prichard R. K., Georges E. Characterization of rhodamine 123 binding to P-glycoprotein in human multidrug-resistant cells. Mol. Pharmacol. 1994;45:1145–1152. [PubMed] [Google Scholar]
- 15.Maliepaard M., Scheffer G. L., Faneyte I. F., van Gastelen M. A., Pijnenborg A. C., Schinkel A. H., van De Vijver M. J., Scheper R. J., Schellens J. H. Subcellular localization and distribution of the breast cancer resistance protein transporter in normal human tissues. Cancer Res. 2001;61:3458–3464. [PubMed] [Google Scholar]
- 16.Fairbanks G., Steck T. L., Wallach D. F. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971;10:2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- 17.Bradley G., Naik M., Ling V. P-glycoprotein expression in multidrug-resistant human ovarian carcinoma cell lines. Cancer Res. 1989;49:2790–2796. [PubMed] [Google Scholar]
- 18.Johnson L. V., Walsh M. L., Chen L. B. Localization of mitochondria in living cells with rhodamine 123. Proc. Natl. Acad. Sci. U.S.A. 1980;77:990–994. doi: 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daoud R., Kast C., Gros P., Georges E. Rhodamine 123 binds to multiple sites in the multidrug resistance protein (MRP1) Biochemistry. 2000;39:15344–15352. doi: 10.1021/bi0020574. [DOI] [PubMed] [Google Scholar]
- 20.Bates S. E., Robey R., Miyake K., Rao K., Ross D. D., Litman T. The role of half-transporters in multidrug resistance. J. Bioenerg. Biomembr. 2001;33:503–511. doi: 10.1023/a:1012879205914. [DOI] [PubMed] [Google Scholar]
- 21.Allen J. D., Jackson S. C., Schinkel A. H. A mutation hot spot in the Bcrp1 (Abcg2) multidrug transporter in mouse cell lines selected for Doxorubicin resistance. Cancer Res. 2002;62:2294–2299. [PubMed] [Google Scholar]
- 22.Robey R. W., Honjo Y., Morisaki K., Nadjem T. A., Runge S., Risbood M., Poruchynsky M. S., Bates S. E. Mutations at amino-acid 482 in the ABCG2 gene affect substrate and antagonist specificity. Br. J. Cancer. 2003;89:1971–1978. doi: 10.1038/sj.bjc.6601370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta A., Zhang Y., Unadkat J. D., Mao Q. HIV protease inhibitors are inhibitors but not substrates of the human breast cancer resistance protein (BCRP/ABCG2) J. Pharmacol. Exp. Ther. 2004;310:334–341. doi: 10.1124/jpet.104.065342. [DOI] [PubMed] [Google Scholar]
- 24.Yang C. P., DePinho S. G., Greenberger L. M., Arceci R. J., Horwitz S. B. Progesterone interacts with P-glycoprotein in multidrug-resistant cells and in the endometrium of gravid uterus. J. Biol. Chem. 1989;264:782–788. [PubMed] [Google Scholar]