Abstract
Glutathione S-transferases (GSTs) are dimeric proteins that play a major role in cellular detoxification. The GSTs in mosquito Anopheles dirus species B, an important malaria vector in South East Asia, are of interest because they can play an important role in insecticide resistance. In the present study, we characterized the Anopheles dirus (Ad)GST D3-3 which is an alternatively spliced product of the adgst1AS1 gene. The data from the crystal structure of GST D3-3 shows that Ile-52, Glu-64, Ser-65, Arg-66 and Met-101 interact directly with glutathione. To study the active-site function of these residues, alanine substitution site-directed mutagenesis was performed resulting in five mutants: I52A (Ile-52→Ala), E64A, S65A, R66A and M101A. Interestingly, the E64A mutant was expressed in Escherichia coli in inclusion bodies, suggesting that this residue is involved with the tertiary structure or folding property of this enzyme. However, the I52A, S65A, R66A and M101A mutants were purified by glutathione affinity chromatography and the enzyme activity characterized. On the basis of steady-state kinetics, difference spectroscopy, unfolding and refolding studies, it was concluded that these residues: (1) contribute to the affinity of the GSH-binding site (‘G-site’) for GSH, (2) influence GSH thiol ionization, (3) participate in kcat regulation by affecting the rate-limiting step of the reaction, and in the case of Ile-52 and Arg-66, influenced structural integrity and/or folding of the enzyme. The structural perturbations from these mutants are probably transmitted to the hydrophobic-substrate-binding site (‘H-site’) through changes in active site topology or through effects on GSH orientation. Therefore these active site residues appear to contribute to various steps in the catalytic mechanism, as well as having an influence on the packing of the protein.
Keywords: active site, Anopheles dirus, glutathione S-transferase, GSH-binding site, structure effect
Abbreviations: CDNB, 1-chloro-2,4-dinitrobenzene; FDNB, 1-fluoro-2,4-dinitrobenzene; G-site, GSH-binding site; GST, glutathione S-transferase; AdGST, Anopheles dirus GST; hGST, human GST; rGST, rat GST; H-site, hydrophobic-substrate-binding site; I52A, Ile-52→Ala substitution etc
INTRODUCTION
Glutathione S-transferases (GSTs, EC 2.5.1.18) are a widely distributed family of detoxifying dimeric enzymes found in most forms of life (e.g. vertebrates, plants, insects, yeasts and aerobic bacteria) [1,2]. GSTs catalyse the conjugation of hydrophobic substrates, such as drugs, herbicides and insecticides, with electrophilic centres to GSH [3,4]. The conjugation of GSH to such molecules increases their solubility and facilitates further metabolic processing [1,5–7]. In addition, these enzymes also carry out a range of other functions. They have peroxidase [5,8] and isomerase activity [9], they can inhibit the Jun N-terminal kinase (thus protecting cells against H2O2-induced cell death) [10], and they are able to non-catalytically bind a wide range of endogenous and exogenous ligands [11–13].
All cytosolic GSTs have the same basic protein folding, which comprises two domains. The N-terminal domain (domain I) adopts a α/β topology and provides the GSH-binding site (G-site) [3,14]. It is currently believed that the residues which contribute to binding glutathione involve a network of specific polar interactions between GSH and G-site residues that are either conserved or conservatively replaced between classes. The C-terminal domain (domain II) is an all-helical structure and provides the structural element for recognition of the broad range of hydrophobic co-substrate [H-site (hydrophobic-substrate-binding site)], which lies adjacent to the G-site [3,14]. It shows the greatest variability across the GST classes [15–20] and helps to define the substrate selectivity of the enzyme [3,14,21].
The enzyme catalysis of nucleophilic aromatic substitution reactions can be divided into several steps, involving substrate binding, activation of GSH by promoting and stabilizing the anionic thiol group, nucleophilic attack by the anionic glutathione to the hydrophobic substrate possessing an electrophilic centre, product formation and finally product release [22–25]. The function of several active site residues have been elucidated: Ser-9 is involved in the production and stabilization of the ionized GSH; Tyr-113 is involved in the formation and release of the GSH conjugate of Lucilia cuprina GST [24]; Cys-47 acts as a molecular switch for different conformations of human (h)GST P1-1 [26]; His-40, Lys-41 and Gln-53 play roles in GSH binding and the structural integrity of Zea mays GSTI [27]. However, there is still a need to determine the contribution of other important active site amino acids.
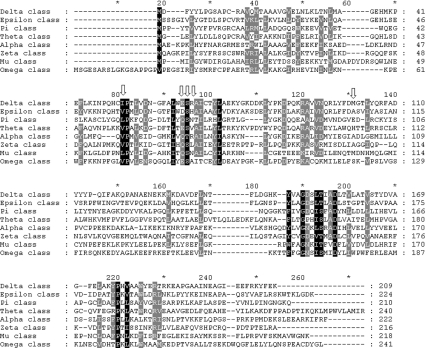
The Anopheles dirus adgst1AS1 gene is spliced to yield 4 different isoforms; D1, D2, D3 and D4 [28,29]. The present study focuses on AdGST D3, because it has high expression levels in Escherichia coli, displays high enzymic activity and the crystal structure is available [28,30]. The aim of this work is to characterize the function of the glutathione-binding residues in adGST D3-3. Five residues in the G-site which are shown in the crystal structure to directly interact with the glutathione were investigated. The residues Ile-52, Glu-64 and Ser-65 are conserved among the GST classes, whereas Arg-66 and Met-101 are variable across the GST classes (Figure 1). These residue positions were substituted with alanine and characterized for both structural and catalytic roles. The results showed that these active site residues make contributions not only to enzyme activity, but also to physical properties.
Figure 1. Amino acid alignments of several classes of GSTs.
Gaps introduced to maximize sequence similarity are shown by a horizontal dash. Black shadings represent 100% sequence similarity and gray shadings represent 80% sequence similarity. Vertical arrows indicate equivalent position of Ile-52, Glu-64, Ser-65, Arg-66 and Met-101 in Delta class. (GenBank® accession numbers are AF273039, NP_611323.1, A37378, NP_000844.1, NP_665683.1, O43708, NP_000552.1 and AH004172.1 respectively).
EXPERIMENTAL
Expression plasmid and site-directed mutagenesis
The plasmid pET3a-adgstD3, as previously described [28], was used to generate the active site residue mutants via PCR-based site-directed mutagenesis. The selected residues Ile-52, Glu-64, Ser-65, Arg-66 and Met-101, which interact directly with GSH, were substituted with alanine by using the mutagenic primers that have been designed according to the 5′ and 3′ sequence of the AdGST D3 wild-type gene (GenBank® accession number AF273039). Each mutant was randomly screened by restriction digestion analysis. Mutant plasmids could be distinguished from the template by digestion with the restriction enzyme corresponding to the restriction recognition site introduced by the mutagenic primers. The full-length GST coding sequence of the plasmids carrying the I52A, E64A, S65A, R66A and M101A mutations were verified by the dideoxy-chain-termination method.
Protein expression and purification
E. coli cells, containing wild-type and mutant plasmid pET3a-adgstD3, were grown in Luria–Bertani medium containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. The expression of GST was induced by the addition of 0.1 mM isopropyl 1-thio-β-galactopyranoside when D600 was 0.5. Following induction for 3 h, cells were harvested by centrifugation at 5000 g at 4 °C for 10 min. The soluble recombinant GST proteins were purified by GSTrap affinity chromatography (Amersham Pharmacia) or S-hexylglutathione agarose (Sigma) affinity chromatography in the case of low affinity towards the glutathione ligand [31]. Glycerol was then added to a final concentration of 50% (v/v) and the purified concentrated GSTs stored at −20 °C. After purification, the enzymes were homogeneous, as determined by SDS/PAGE. The protein concentration was determined by the Bradford method using BSA as a standard [32].
Kinetic studies
Steady-state kinetics were studied for wild-type and mutant enzymes at various concentrations of CDNB (1-chloro-2,4-dinitrobenzene) and GSH in 0.1 M phosphate buffer, pH 6.5. The reaction was monitored at 340 nm, ε=9600 M−1·cm−1. Apparent kinetic parameters, kcat, Km and kcat/Km were determined by fitting the collected data to a Michaelis–Menten equation by non-linear regression analysis using GraphPad Prism (GraphPad software, San Diego, CA, U.S.A.).
The pH dependence of kcat/KmCDNB was obtained by using 0.1 M sodium acetate buffers (from pH 5.0 to pH 5.5) and 0.1 M potassium phosphate buffer (from pH 6.0 to pH 8.5).
The second order kinetic constants at pH 6.5 for the spontaneous reaction of GSH with CDNB and FDNB (1-fluoro-2,4-dinitrobenzene), and the catalytic-centre activities (‘turnover numbers’) at pH 6.5 for adGST D3-3 with CDNB and FDNB as co-substrates, were obtained as described previously [33].
The effect of viscosity on kinetic parameters was assayed by using 0.1 M potassium phosphate buffer, pH 6.5, with various glycerol concentrations. Viscosity values (η) at 25 °C were calculated as described previously [34].
The specific activities of the enzymes were determined using a spectrophotometer with five different substrates: CDNB, 1,2-dichloro-4-nitrobenzene, ethacrynic acid, p-nitrophenethyl bromide and p-nitrobenzyl chloride as described previously [35,36].
Structural studies
One of the structural studies performed was to determine a half-life stability for the GST protein at 45 °C. The wild-type and mutant enzymes were incubated at 45 °C at a protein concentration of 1 mg/ml. The inactivation time courses were determined by taking suitable aliquots at the different time points for assay of remaining activity to calculate half-life of the enzyme [36].
Spectroscopic properties of the wild-type and mutant proteins were also studied. Intrinsic fluorescence emission spectra were measured at the excitation wavelength 295 nm, and the λmax and the fluorescence intensity of emission spectra were analysed at a protein concentration of 0.2 mg/ml [37].
A refolding experiment was also performed with the enzymes first being denatured in 4 M guanidinium chloride in renaturation buffer (0.2 M phosphate, 1 mM EDTA and 5 mM DTT, pH 7.0) at 25–27 °C for 30 min and then rapidly diluted (defining time 0) 1:40 into renaturation buffer. Therefore the final guanidinium chloride concentration was 0.1 M during refolding. Recovered activity was monitored as a function of time by taking appropriate aliquots of the renaturation mixture and immediately assaying for activity. Refolding rate constants were determined by non-linear regression analysis using a single exponential equation [37].
RESULTS AND DISCUSSION
Protein expression and purification
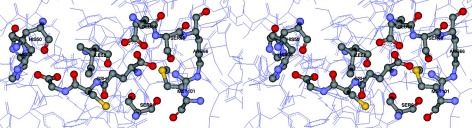
Five residues, shown by crystal structure to directly interact with glutathione, were studied to characterize their roles in catalysis and structure (Figure 2). These residues, Ile-52, Glu-64, Ser-65, Arg-66 and Met-101 in adGSTD3-3, were individually replaced with alanine by oligonucleotide-directed mutagenesis. The wild-type and mutant enzymes were expressed as soluble forms, except for I52A and E64A, which were insoluble at 37 °C. Decreasing the expression temperature to 25 °C allowed the I52A mutant protein to be expressed in a soluble form. However, the GST with the E64A substitution was still expressed as an insoluble form, even at 18 °C. Attempts at refolding the E64A protein were unsuccessful. This suggests that Glu-64 is a critical residue involved in the initial packing or folding of the protein to yield the final tertiary structure. The soluble GSTs were purified and gave a single band on SDS/PAGE. In each case, the unbound activity from the GSH affinity chromatography of the recombinant GSTs varied from 2–25% of the total activity, displaying a reduction in binding to the chromatography media.
Figure 2. Stereo view of the studied residues around the G-site of adGST D3-3.
Ile-52, Glu-64, Ser-65, Arg-66 and Met-101 interact directly with GSH.
Steady-state kinetics
Steady-state kinetics were performed with various concentrations of glutathione and CDNB as substrates. The reactions followed Michaelis–Menten kinetics, and the kinetic parameters kcat and Km were determined by non-linear regression analysis (Table 1). All of the mutations increased Km values for glutathione, especially for I52A and R66A, which had values 24- and 19-fold greater than wild-type, demonstrating a decreased affinity toward glutathione. However, the mutations yielded only slight differences in the affinity of binding towards the substrate CDNB when compared with the wild-type enzyme.
Table 1. Steady-state kinetic parameters and pKa values for the thiol group of GSH of wild-type and mutants of adGSTD3-3 for the CDNB conjugation reaction at pH 6.5 and 25 °C.
The enzyme activities were measured at varying concentrations of CDNB and GSH in 0.1 M phosphate buffer, pH 6.5. The pKa was obtained by using 0.1 M sodium acetate buffers (from pH 5.0 to pH 5.5) and 0.1 M potassium phosphate buffer (from pH 6.0 to pH 8.5).The reaction was monitored at 340 nm, ε=9600 M−1·cm−1.
| Enzyme | kcat (s−1) | KmGSH (mM) | KmCDNB (mM) | kcat/KmGSH (s−1/mM) | kcat/KmCDNB (s−1/mM) | pKa |
|---|---|---|---|---|---|---|
| Wild-type | 35.4 | 0.27±0.05 | 0.14±0.01 | 130.69 | 245.95 | 6.36±0.11 |
| I52A | 16.9 | 6.50±0.61 | 0.39±0.03 | 2.60 | 43.72 | 6.78±0.14 |
| S65A | 29.8 | 1.22±0.12 | 0.34±0.06 | 24.50 | 87.18 | 6.89±0.23 |
| R66A | 3.3 | 5.10±0.40 | 0.22±0.04 | 0.64 | 14.59 | 7.23±0.18 |
| M101A | 63.0 | 1.08±0.09 | 0.19±0.01 | 58.48 | 337.12 | 6.10±0.07 |
The differences in kcat value in the nucleophilic aromatic substitution reaction with CDNB observed for I52A and R66A decreased approx. 2- and 10-fold respectively, whereas the catalytic-centre activity for the M101A mutant is about 2 times greater than wild-type (Table 1). The question arose of whether the changes in kcat value were due to changes in the activation of the GSH substrate bound in binary complex with the enzyme. The pH dependence of the kinetic parameters in the binary complex was determined to give the pKa values in Table 1. The pKa of the R66A mutant was about 1 pH unit higher than that found for the wild-type. This increased pKa was also observed for a conserved hydroxy side-chain amino acid, Ser-9 in Delta class and tyrosine in Alpha, Mu and Pi classes, which play a important role in promoting and stabilizing anionic glutathione [3,23,24,38]. Several reports have shown that the glutamyl α-carboxylate of glutathione contributes to the ionization of the glutathione thiol group [39,40]. Arg-66 interacts directly with the glutamyl α-carboxylate of glutathione and appears to influence the ionization process of glutathione in Delta class GST. The involvement of positively charged residues in regulation of the electrostatic field also has been observed with other GSTs. For example, Arg-107 from hGST M2-2 [41] and Arg-15 hGST A1-1 [42] have been shown to contribute to active site ionization. For the other residues there are only slight differences in the pKa of glutathione for the I52A, S65A and M101A GSTs, suggesting these residues may contribute to glutathione orientation to yield a suitable position for conjugation with the electrophilic substrate.
Determining the rate-limiting step in the adGST-D3-3-catalysed reaction
Normally, the reaction of nucleophilic aromatic substitution proceeds via a σ-complex intermediate. Substitution of the chlorine leaving group in the CDNB molecule by a more electronegative fluorine increases the second order rate constant of the spontaneous reaction with glutathione about 50-fold, indicating that the rate-limiting step is the σ-complex formation [24]. Therefore the effect on the catalytic-centre activity of the fluorine substitution for the chlorine in CDNB was examined. The results show that the kcat does vary for the different GSTs (Table 2). This is most likely due to packing changes yielding a different architecture of the active site of the enzyme that responds differently to the smaller fluorine leaving group. However, this data indicates that the σ-complex formation is not the rate-limiting step. Therefore, the rate-limiting step in the enzymic reaction appears to be a physical step, rather than a chemical step.
Table 2. Effect of fluoride/chloride leaving group substitution on the rate of catalysis.
The catalytic-centre activities of the conjugation reaction catalyzing by adGST D3-3 enzymes of GSH with CDNB and FDNB as co-substrates were calculated at pH 6.5.
| Enzyme | kcatFDNB (s−1) | kcatCDNB (s−1) |
|---|---|---|
| Wild-type | 85.1 | 35.4 |
| I52A | 27.5 | 16.9 |
| S65A | 157.3 | 29.8 |
| R66A | 18.1 | 3.3 |
| M101A | 265.9 | 63.0 |
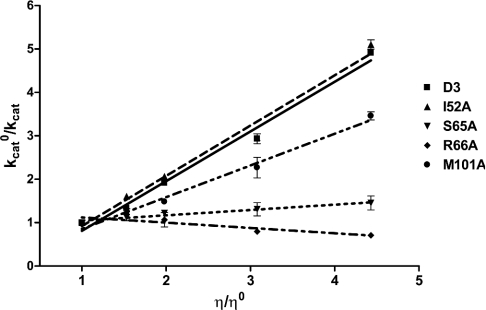
Next we examined the effect of viscosity on the kinetic parameters to determine the rate-limiting step of the catalytic reaction. A decrease of the rate constant by increasing the medium viscosity would reflect the influence of diffusion on catalysis [43]. It would indicate that the rate-limiting step of the reaction is related to the product release or the diffusion-controlled structural transition of the protein. A plot of the reciprocal of the relative catalytic constant (kcato/kcat) against the relative viscosity (η/ηo) should be linear. The slope should be equal to unity when the product release or structural transition is limited by a strictly diffusional barrier. If the slope approaches zero the chemistry or another non-diffusion barrier is rate-limiting. As shown in Figure 3 the inverse relative rate constant for the enzyme-catalysed reaction shows a linear dependence on the relative viscosity with a slope very close to unity for the GST D3-3 wild-type (1.14±0.01), similar to the I52A mutant GST (1.16±0.05). Diffusion-controlled motions of the protein have been reported to modulate the catalysis of other GST isoenzymes: a conformational change in the case of ternary complex formation of hGST P1-1 [33] and product release for rat (r)GST M1-1 [43], L. cuprina GST [24], rGST T2-2 [44] and Alpha-class GSTs [45]. In contrast the mutants S65A (0.12±0.09), R66A (−0.12±0.01) and M101A (0.73±0.08) exhibited kcat values with different degrees of viscosity dependence compared with the wild-type (Figure 3). In particular, the slopes of for the S65A and R66A mutants are very close to zero, indicating that the mutation changed the rate-limiting step of the enzyme from a physical to a non-physical step.
Figure 3. Viscosity effect on kinetic parameters of wild-type and mutant enzymes.
The effect of viscosity on kinetic parameters was assayed by using 0.1 M potassium phosphate buffer, pH 6.5, with various glycerol concentrations. Dependence of the reciprocal of the relative catalytic-centre activity (kcato/kcat) on the relative viscosity (η/ηo) for CDNB as co-substrate. D3, wild-type.
Substrate specificity
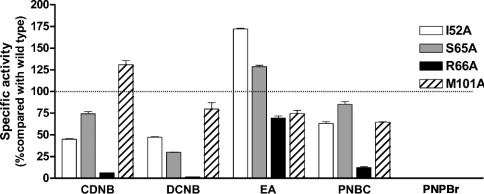
Substrate specificity determination revealed differences in the specificity or the interaction of the enzymes with several hydrophobic substrates (Figure 4). This data shows that mutations of the glutathione-binding residues changed the specificity toward various hydrophobic substrates. This suggests a rearrangement of the active site residues, resulting in changes in the topology of the active site pocket. Alternatively, the changes in the G-site residues may result in changes in orientation of glutathione within the active site which then affects the hydrophobic substrate binding. However, R66A showed a decrease in all substrate specificities, which is most likely due to the decreased contribution to GSH ionization, instead of the interaction with the hydrophobic substrates.
Figure 4. Substrate specific activity as a percentage change compared with the adGSTD3-3 (wild-type).
Five substrates: CDNB, 1,2-dichloro-4-nitrobenzene (DCNB), ethacrynic acid (EA), p-nitrophenethyl bromide (PNPBr) and p-nitrobenzyl chloride (PNBC) were used for enzyme activity assays.
Thermal stability
The wild-type enzyme was subjected to a heat inactivation assay, and it was observed that the GST activity began to decrease at 45 °C [46]. This temperature was used to determine half-life stabilities for the recombinant enzymes. The half-life corresponds to the time of incubation when there is 50% remaining activity. The I52A mutant enzyme was shown to be less stable than wild-type by approx. 5-fold (Table 3). However, the replacement at Arg-66 increased the stability of the mutant enzyme by approx. 60-fold. These data demonstrated that Ile-52 and Arg-66 are involved in structural stabilization of the enzyme. The changes in these two residues would appear to change the packing of the active site, which affects the overall structure of the enzyme.
Table 3. Thermal stability of wild-type and mutants of adGSTD3-3 at 45 °C.
The wild-type and mutant enzymes were incubated at 45 °C at the protein concentration of 1 mg/ml. The inactivation time courses were determined by taking suitable aliquots at the different time points for assay of remaining activity to calculate half-life of the enzyme.
| Enzyme | Half-life at 45 °C (min−1) |
|---|---|
| Wild-type | 2.33±0.12 |
| I52A | 0.42±0.01 |
| S65A | 1.70±0.08 |
| R66A | 145.73±10.47 |
| M101A | 5.72±0.71 |
Structural studies on wild-type and mutants
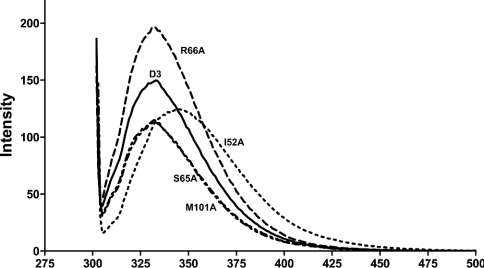
The intrinsic fluorescence spectra show differences between the wild-type, I52A and R66A mutations (Figure 5). Changes in amino acid side chains around tryptophan resulted in an increase in fluorescence intensity, and a red-shifted spectrum is observed as the protein unfolds to random coil [47]. The λmax values of wild-type and mutant enzymes were the same at 333±1 nm, except for I52A, which had a spectra red shifted to give a λmax at 346±2 nm. These data indicated that the tryptophan residue is more exposed to the solvent, suggesting that the packing of the tertiary structure of the I52A protein is looser. Although the λmax of the R66A enzyme was the same as the wild-type, the intensity of the spectra was increased about 1.6-fold, suggesting that the mutation at this residue causes a rearrangement of the amino acids around the tryptophan that decreased the quenching.
Figure 5. Intrinsic fluorescence spectra of AdGST D3-3 and mutants.
Intrinsic fluorescence emission spectra were measured at the excitation wavelength 295 nm, and the λmax and the fluorescence intensity of emission spectra were analysed at a protein concentration of 0.2 mg/ml.
Refolding experiment
The denaturant 4 M guanidinium chloride was sufficient to completely unfold the proteins, as shown by CD spectrum at 222 nm (results not shown). All of the data sets fitted to a single exponential equation for the refolding kinetics. In each case, 5–10 min of incubation in the refolding solution was sufficient to reach the maximum reactivation. In general the reactivation yields of the mutant enzymes were similar to the wild-type (Table 4). However, I52A mutant showed a reactivation velocity about 50-times greater than the wild-type GST, with recovery of about 95% of the activity. This suggests that Ile-52 plays a key role in the folding process.
Table 4. Refolding rate constants of adGST D3-3 variants and percentage recovered activity.
The enzymes were denatured in 4 M guanidinium chloride in renaturation buffer at 25–27 °C for 30 min and then rapidly diluted (defining time 0) 1:40 into renaturation buffer. Recovered activity was monitored as a function of time by withdrawal of appropriate aliquots of the renaturation mixture and immediately assaying for activity. Refolding rate constants were determined by non-linear regression analysis using a single exponential equation.
| Enzyme | kref (min−1) | Recovered activity (%) |
|---|---|---|
| Wild-type | 0.518±0.053 | 43.8 |
| I52A | 25.51±2.53 | 94 |
| S65A | 0.375±0.026 | 50.3 |
| R66A | 0.778±0.249 | 72.5 |
| M101A | 0.279±0.025 | 36.1 |
One possibility may be the side chain of this residue points into the protein molecule and interacts with Leu-6 and Leu-33 to form a small hydrophobic core, as the small side chain of alanine may fit more easily than the side chain of isoleucine for an increased rate in its function in the nucleation of the initial protein folding [36].
CONCLUSION
The present study investigated the glutathione-binding residues of AdGST D3-3 by site-directed mutagenesis. The residues in the G-site appear to impact upon both chemical and physical properties of the enzyme. For example, Ile-52 can affect several aspects of both catalytic and structural properties of the GST. As discussed above the Ile-52 forms a small hydrophobic core with Leu-6 and Leu-33, which, when replaced with the small side chain residue alanine, affected the interaction of the main chain polypeptide between β-sheets 3 and 4, resulting in looser packing of β-sheet 4. Trp-63 is located in β-sheet 4 and the changes at this position have yielded a red shift in intrinsic fluorescence spectra and changes in the half-life and the refolding rate constant of the enzyme. Demonstrating the structural impact of this residue position, Ile-52 is also involved in the initial binding of the glutathione substrate as shown by GSH affinity changes. Glu-64 had a major effect on structure, because the substitution at this residue caused the enzyme to fold improperly which caused the protein to express as insoluble form. The hydroxy group of Ser-65, which is conserved among the GSTs classes, has a function in glutathione binding and plays an important role in a non-physical step in the enzyme catalysis that is no longer performed by alanine substitution, as demonstrated by the viscosity experiment. Mutational studies of the equivalent amino acid in hGST A1-1 replaced Thr-68 by valine, which caused a shift in pH dependence of the enzyme-catalyzed reaction, suggesting a role in the ionization process of GSH [40]. However, we did not observe the same effect on the mutation in Ser-65 in Delta class which was also seen for Ser-67 in θ class [27]. As shown by the available crystal structure, although Ser-65 is an equivalent residue with the same functional group, the direction of the side chain is different. The hydroxy group of Thr-68 in hGST A1-1 appears to interact with the carboxylate of the glutamate residue of GSH, whereas Ser-65 in adGST D3-3 interacts with the amino group of Arg-66. Arg-66 is involved in the catalytic mechanism through a contribution to the glutathione ionization process, which is why changing this residue changes the rate-limiting step of the enzyme from a physical step to a non-physical step. Although the alignment of GSTs shows variation in this position, several classes of GST still have a conserved arginine residue located in the G-site, for example, Arg-15 in hGST A1-1 [42] and Arg-107 in hGST M2-2 [41]. It has been proposed the positive charge of arginine could provide a counter ion to promote ionization of the sulphydryl group of GSH or assist the function of the glutamate α-carboxylate of GSH in the GSH activation by acting as a proton acceptor in the catalytic mechanism [39–41]. Moreover, this amino acid position also influences structural maintenance of the protein. The R66A mutant enzyme is more stable than the wild-type enzyme, approx. 60-fold, and we also observed a 35% enhancement in emission intensity which would be a consequence of movements of the indole side chain. The tryptophan may be moving away from the potential quenching effect of Asp-47 and Gln-49 and changing the packing of the active site, as well as affecting the overall structure of the enzyme. The Met-101 position would appear to exert its influence through packing effects. It also may be involved in dimerization, because this residue is located at the subunit interface and interacts with several amino acids from the other subunit. These interaction and packing effect changes then impact upon the enzyme active site as shown by the changes in substrate specificity, as well as changes in kcat and Km. These effects have been observed previously for a subunit interface residue in a Pi class GST [48].
These experiments demonstrate that glutathione-binding residues contribute to various steps in the catalytic mechanism. Surprisingly, the present studies also implicated the active site residues as contributors to the overall structure with regard to their associations in the folding mechanism and structural stabilization. This contribution was also reflected by the change in the enzyme activity that resulted from an influence on the packing of the protein, in addition to a direct function in the catalytic mechanism.
Acknowledgments
This work was supported by the Thailand Research Fund and P.W. was supported by a Royal Golden Jubilee Scholarship.
References
- 1.Sheehan D., Meade G., Foley V. M., Dowd C. A. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001;360:1–16. doi: 10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ketterer B. A bird's eye view of the glutathione transferase field. Chem. Biol. Interact. 2001;138:27–42. doi: 10.1016/s0009-2797(01)00277-0. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong R. N. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem. Res. Toxicol. 1997;10:2–18. doi: 10.1021/tx960072x. [DOI] [PubMed] [Google Scholar]
- 4.Jakoby W. B., Habig W. H. In: Enzymatic Basis of Detoxication, vol 2. Jakoby W. B., editor. New York: Academic Press; 1980. pp. 63–94. [Google Scholar]
- 5.Mannervik B., Danielson U. H. Glutathione transferases – structure and catalytic activity. CRC Crit. Rev. Biochem. 1988;23:283–337. doi: 10.3109/10409238809088226. [DOI] [PubMed] [Google Scholar]
- 6.Hayes J. D., Pulford D. J. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 7.Eaton D. L., Bammler T. K. Concise review of the glutathione S-transferases and their significance to toxicology. Toxicol. Sci. 1999;49:156–164. doi: 10.1093/toxsci/49.2.156. [DOI] [PubMed] [Google Scholar]
- 8.Zhao T., Singhal S. S., Piper J. T., Cheng J., Pandya U., Clark-Wronski J., Awasthi S., Awasthi Y. C. The role of human glutathione S-transferases hGSTA1-1 and hGSTA2-2 in protection against oxidative stress. Arch. Biochem. Biophys. 1999;367:216–224. doi: 10.1006/abbi.1999.1277. [DOI] [PubMed] [Google Scholar]
- 9.Johansson A.-S., Mannervik B. Human glutathione transferase A3-3, a highly efficient catalyst of double-bond isomerization in the biosynthetic pathway of steroid hormones. J. Biol. Chem. 2001;276:32061–32065. doi: 10.1074/jbc.M104539200. [DOI] [PubMed] [Google Scholar]
- 10.Yin Z., Ivanov V. N., Habelhah H., Tew K., Ronai Z. Glutathione S-transferase p elicits protection against H2O2-induced cell death via coordinated regulation of stress kinases. Cancer Res. 2000;60:4053–4057. [PubMed] [Google Scholar]
- 11.Bhargava M. M., Listowsky I., Arias I. M. Ligandin. Bilirubin binding and glutathione S-transferase activity are independent processes. J. Biol. Chem. 1978;253:4112–4115. [PubMed] [Google Scholar]
- 12.Dulhunty A., Gage P., Curtis S., Chelvanayagam G., Board P. The glutathione transferase structural family includes a nuclear chloride channel and a ryanodine receptor calcium release channel modulator. J. Biol. Chem. 2001;276:3319–3323. doi: 10.1074/jbc.M007874200. [DOI] [PubMed] [Google Scholar]
- 13.Lo Bello M., Nuccetelli M., Caccuri A. M., Stella L., Parker M. W., Rossjohn J., McKinstry W. J., Mozzi A. F., Federici G., Polizio F., et al. Human glutathione transferase P1-1 and nitric oxide carriers: A new role for an old enzyme. J. Biol. Chem. 2001;276:42138–42145. doi: 10.1074/jbc.M102344200. [DOI] [PubMed] [Google Scholar]
- 14.Dirr H., Reinemer P., Huber R. X-ray crystal structures of cytosolic glutathione S-transferases. Implications for protein architecture, substrate recognition and catalytic function. Eur. J. Biochem. 1994;220:645–661. doi: 10.1111/j.1432-1033.1994.tb18666.x. [DOI] [PubMed] [Google Scholar]
- 15.Board P. G., Coggan M., Chelvanayagam G., Easteal S., Jermiin L. S., Schulte G. K., Danley D. E., Hoth L. R., Griffor M. C., Kamath A. V., et al. Identification, characterization, and crystal structure of the Omega class glutathione transferases. J. Biol. Chem. 2000;275:24798–24806. doi: 10.1074/jbc.M001706200. [DOI] [PubMed] [Google Scholar]
- 16.Ji X., Von Rosenvinge E. C., Johnson W. W., Tomarev S. I., Paitigorsky J., Armstrong R. N., Gilliland G. L. Three-dimensional structure, catalytic properties, and evolution of a sigma class glutathione transferase from squid, a progenitor of the lens S-crystallins of cephalopods. Biochemistry. 1995;34:5317–5328. doi: 10.1021/bi00016a003. [DOI] [PubMed] [Google Scholar]
- 17.Reinemer P., Dirr H. W., Ladenstein R., Huber R., Lo Bello M., Federici G., Parker M. W. Three-dimensional structure of class π glutathione S-transferase from human placenta in complex with S-hexylglutathione at 2.8 Å resolution. J. Mol. Biol. 1992;227:214–226. doi: 10.1016/0022-2836(92)90692-d. [DOI] [PubMed] [Google Scholar]
- 18.Reinemer P., Prade L., Hof P., Neuefeind T., Huber R., Zettl R., Palme K., Schell J., Koelln I., Barrunik H. D., Bieseler B. Three-dimenstional structure of glutathione S-transferase from Arabidopsis thaliana at 2.2 Å resolution: structural characterization of herbicide-conjugating plant glutathione S-transferases and a novel active site architecture. J. Mol. Biol. 1996;255:289–309. doi: 10.1006/jmbi.1996.0024. [DOI] [PubMed] [Google Scholar]
- 19.Sinning I., Kleywegt G. J., Cowan S. W., Reinemer P., Dirr H. W., Huber R., Gilliland G. L., Armstrong R. N., Ji X., Board P. G., et al. Structure determination and refinement of human Alpha class glutathione transferase A1-1, and a comparison with the Mu and Pi class enzymes. J. Mol. Biol. 1993;232:192–212. doi: 10.1006/jmbi.1993.1376. [DOI] [PubMed] [Google Scholar]
- 20.Wilce M. C. J., Board P. G., Feil S. C., Parker M. W. Crystal structure of a theta-class glutathione transferase. EMBO J. 1995;14:2133–2143. doi: 10.1002/j.1460-2075.1995.tb07207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilce M. C. J., Parker M. W. Structure and function of glutathione S-transferases. Biochim. Biophys. Acta. 1994;1205:1–18. doi: 10.1016/0167-4838(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 22.Armstrong R. N., Rife C., Wang Z. Structure, mechanism and evolution of thiol transferases. Chem. Biol. Interact. 2001;133:167–169. [Google Scholar]
- 23.Caccuri A. M., Ascenzi P., Antonini G., Parker M. W., Oakley A. J., Chiessi E., Nuccetelli M., Battistoni A., Bellizia A., Ricci G. Structural flexibility modulates the activity of human glutathione transferase P1-1. Influence of a poor co-substrate on dynamics and kinetics of human glutathione transferase. J. Biol. Chem. 1996;271:16193–16198. doi: 10.1074/jbc.271.27.16193. [DOI] [PubMed] [Google Scholar]
- 24.Caccuri A. M., Antonini G., Nicotra M., Battistoni A., Lo Bello M., Board P. G., Parker M. W., Ricci G. Catalytic mechanism and role of hydroxyl residues in the active site of theta class glutathione S-transferases. Investigation of Ser-9 and Tyr-113 in a glutathione S-transferase from the Australian sheep blowfly, Lucilia cuprina. J. Biol. Chem. 1997;272:29681–29686. doi: 10.1074/jbc.272.47.29681. [DOI] [PubMed] [Google Scholar]
- 25.Caccuri A. M., Lo Bello M., Nuccetelli M., Rossi P., Antonini G., Federici G., Ricci G. Proton release upon glutathione binding to glutathione transferase P1-1: kinetic analysis of a multistep glutathione binding process. Biochemistry. 1998;37:3028–3034. doi: 10.1021/bi971903g. [DOI] [PubMed] [Google Scholar]
- 26.Lo Bello M., Battistoni A., Mazzetti A. P., Board P. G., Muramatsu M., Federici G., Ricci G. Site-directed mutatgenesis of human glutathione transferase P1-1. Spectral, kinetic, and structural properties of Cys-47 and Lys-54 mutants. J. Biol. Chem. 1995;270:1249–1253. [PubMed] [Google Scholar]
- 27.Labrou N. E., Mello L. V., Clonis Y. D. Functional and structural roles of the glutathione-binding residues in maize (Zea mays) glutathione S-transferase I. Biochem. J. 2001;358:101–110. doi: 10.1042/0264-6021:3580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jirajaroenrat K., Pongjaroenkit S., Krittanai C., Prapanthadara L., Ketterman A. J. Heterologous expression and characterization of alternatively spliced glutathione S-transferases from a single Anopheles gene. Insect Biochem. Mol. Biol. 2001;31:867–875. doi: 10.1016/s0965-1748(01)00032-7. [DOI] [PubMed] [Google Scholar]
- 29.Pongjaroenkit S., Jirajaroenrat K., Boonchauy C., Chanama U., Leetachewa S., Prapanthadara L., Ketterman A. J. Genomic organization and putative promotors of highly conserved glutathione S-transferases originating by alternative splicing in Anopheles dirus. Insect Biochem. Mol. Biol. 2001;31:75–85. doi: 10.1016/s0965-1748(00)00107-7. [DOI] [PubMed] [Google Scholar]
- 30.Oakley A. J., Harnnoi T., Udomsinprasert R., Jirajaroenrat K., Ketterman A. J., Wilce M. C. J. The crystal structures of glutathione S-transferases isozymes 1-3 and 1-4 from Anopheles dirus species B. Prot. Sci. 2001;10:2176–2185. doi: 10.1110/ps.ps.21201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wongsantichon J., Harnnoi T., Ketterman A. J. A sensitive core region in the structure of glutathione S-transferases. Biochem. J. 2003;373:759–765. doi: 10.1042/BJ20030394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 33.Caccuri A. M., Ascenzi P., Lo Bello M., Federici G., Battistoni A., Mazzetti P., Ricci G. Are the steady state kinetics of glutathione transferase always dependent on the deprotonation of the bound glutathione? New insights in the kinetic mechanism of GST P1-1. Biochem. Biophys. Res. Commun. 1994;200:1428–1434. doi: 10.1006/bbrc.1994.1610. [DOI] [PubMed] [Google Scholar]
- 34.Wolf A. V., Brown M. G., Prentiss P. G. Handbook of Chemistry and Physics. Boca Raton, Florida: CRC Press, Inc.; 1985. Concentrative properties of aqueous solutions: conversion tables; pp. D219–D232. [Google Scholar]
- 35.Habig W. H., Pabst M. J., Jakoby W. B. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 1974;249:7130–7139. [PubMed] [Google Scholar]
- 36.Vararattanavech A., Ketterman A. Multiple roles of glutathione binding-site residues of glutathione S-transferase. Protein Pept. Lett. 2003;10:441–448. doi: 10.2174/0929866033478654. [DOI] [PubMed] [Google Scholar]
- 37.Stenberg G., Dragani B., Cocco R., Mannervik B., Aceto A. A conserved ‘hydrophobic staple motif’ plays a crucial role in the refolding of human glutathione transferase P1-1. J. Biol. Chem. 2000;275:10421–10428. doi: 10.1074/jbc.275.14.10421. [DOI] [PubMed] [Google Scholar]
- 38.Caccuri A. M., Antonini G., Board P. G., Parker M. W., Nicotra M., Lo Bello M., Federici G., Ricci G. Proton release on binding of glutathione to Alpha, Mu and Delta class glutathione transferases. Biochem. J. 1999;344:419–425. [PMC free article] [PubMed] [Google Scholar]
- 39.Gustafsson A., Pettersson P. L., Grehn L., Jemth P., Mannervik B. Role of the glutamyl α-carboxylate of the substrate glutathione in the catalytic mechanism of human glutathione transferase A1-1. Biochemistry. 2001;40:15835–15845. doi: 10.1021/bi010429i. [DOI] [PubMed] [Google Scholar]
- 40.Widersten M., Björnestedt R., Mannervik B. Involvement of the carboxyl groups of glutathione in the catalytic mechanism of human glutathione transferase A1-1. Biochemistry. 1996;35:7731–7742. doi: 10.1021/bi9601619. [DOI] [PubMed] [Google Scholar]
- 41.Patskovsky Y. V., Patskovska L. N., Listowsky I. The enhanced affinity for thiolate anion and activation of enzyme-bound glutathione is governed by an arginine residue of human Mu class glutathione S-transferases. J. Biol. Chem. 2000;275:3296–3304. doi: 10.1074/jbc.275.5.3296. [DOI] [PubMed] [Google Scholar]
- 42.Björnestedt R., Stenberg G., Widersten M., Board P. G., Sinning I., Jones T. A., Mannervik B. Functional significance of arginine 15 in the active site of human class alpha glutathione transferase A1-1. J. Mol. Biol. 1995;247:765–773. doi: 10.1016/s0022-2836(05)80154-8. [DOI] [PubMed] [Google Scholar]
- 43.Johnson W. W., Liu S., Ji X., Gilliland G. L., Armstrong R. N. Tyrosine 115 participates both in chemical and physical steps of the catalytic mechanism of a glutathione S-transferase. J. Biol. Chem. 1993;268:11508–11511. [PubMed] [Google Scholar]
- 44.Jemth P., Mannervik B. Fast product formation and slow product release are important features in a hysteretic reaction mechanism of glutathione transferase T2-2. Biochemistry. 1999;38:9982–9991. doi: 10.1021/bi983065b. [DOI] [PubMed] [Google Scholar]
- 45.Allardyce C. S., McDonagh P. D., Lian L.-Y., Wolf C. R., Roberts G. C. K. The role of tyrosine-9 and the C-terminal helix in the catalytic mechanism of Alpha-class glutathione S-transferases. Biochem. J. 1999;343:525–531. [PMC free article] [PubMed] [Google Scholar]
- 46.Wongtrakul J., Udomsinprasert R., Ketterman A. Non-active site residues Cys69 and Asp150 affected the enzymatic properties of glutathione S-transferase AdGSTD3-3. Insect Biochem. Mol. Biol. 2003;33:971–979. doi: 10.1016/s0965-1748(03)00103-6. [DOI] [PubMed] [Google Scholar]
- 47.Dirr H. W., Wallace L. A. Role of the C-terminal helix 9 in the stability and ligandin function of class a glutathione transferase A1-1. Biochemistry. 1999;38:15631–15640. doi: 10.1021/bi991179x. [DOI] [PubMed] [Google Scholar]
- 48.Stenberg G., Abdalla A.-M., Mannervik B. Tyrosine 50 at the subunit interface of dimeric human glutathione transferase P1-1 is a structural key residue for modulating protein stability and catalytic function. Biochem. Biophys. Res. Commun. 2000;271:59–63. doi: 10.1006/bbrc.2000.2579. [DOI] [PubMed] [Google Scholar]