Abstract
Background:
Diacylglycerol acyltransferases (DGATs) esterify sn-1,2-diacylglycerol with a long-chain fatty acyl-CoA, the last and rate-limiting step of triacylglycerol (TAG) biosynthesis in eukaryotic organisms. Understanding the roles of DGATs will help to create transgenic plants with value-added properties and provide information for therapeutic intervention for obesity and related diseases. At least 74 DGAT2 sequences from 61 organisms have been identified from the Genbank databases, but the expression of any DGAT2 as a partial or full-length protein in E. coli had not been reported. The objective of this study was to develop a procedure for expression and purification of recombinant DGAT2 (rDGAT2).
Results:
An expression plasmid was engineered to express tung tree DGAT2 fused to maltose binding protein (MBP) and poly-histidine (His) affinity tags. Immunoblotting showed that rDGAT2 was detected in the soluble, insoluble and membrane fractions of E. coli. The rDGAT2 in the soluble fraction of E. coli was partially purified by amylose resin, Ni-NTA beads, and tandem affinity chromatography. Multiple proteins co-purified with rDGAT2. Size exclusion chromatography estimated the size of the rDGAT2-enriched fraction to be approximately eight times the monomer size. Affinity-purified rDGAT2 fractions had a yellow tint and contained fatty acids. The rDGAT2 in the insoluble fraction of E. coli was partially solubilized by 7 detergents (Brij 35, CHAPS, NP-40, SDS, Triton X-100, Tween 20 and Tween 80) with SDS being the most effective. Recombinant DGAT2 was purified to near homogeneity by SDS solubilization and Ni-NTA affinity chromatography. Mass spectrometry identified rDGAT2 as a component in the bands corresponding to the monomer and dimer forms as observed by SDS-PAGE. Protein bands with monomer and dimer sizes were also observed in the microsomal membranes of Saccharomyces cerevisiae expressing hemagglutinin (HA)-tagged DGAT2.
Conclusions:
This study describes a reliable procedure for producing the first recombinant DGAT2 from any species using an E. coli expression system. The results suggest that recombinant DGAT2 is present as monomer and dimer forms on SDS-PAGE and associated with other proteins, lipids, and membranes.
Background
Diacylglycerol acyltransferases (DGATs) catalyze the last and rate-limiting step of triacylglycerol (TAG) biosynthesis in eukaryotic organisms [1]. DGATs esterify sn-1,2-diacylglycerol with a long-chain fatty acyl-CoA. Plants and animals deficient in DGATs accumulate less TAG [2–4]. Animals with reduced DGAT activity are resistant to diet-induced obesity [3, 5] and lack milk production [3]. Over-expression of the DGAT enzymes increases TAG content in plants [6–12], animals [13–16], and yeast [17]. Therefore, understanding the roles of DGATs will help to create transgenic plants with value-added properties and provide information for therapeutic intervention for obesity and related diseases.
DGAT genes have been isolated from many organisms [18]. At least two forms of DGATs are present in mammals [19, 20] and plants [21, 22]. DGATs are integral membrane proteins with more than 40% of the total amino acid residues being hydrophobic [18]. The two forms have similar properties and amino acid composition except that DGAT1s are approximately 20 kDa larger than DGAT2s [18]. DGAT1s and DGAT2s have 41 and 16 completely conserved amino acid residues, respectively, although only two residues are shared by all DGATs [18]. DGAT isoforms have non-redundant functions in TAG biosynthesis in species such as mice [4] and tung tree (Vernicia fordii) [22, 23]. Mice deficient in DGAT1 are viable, have modest decreases in TAG, and are resistant to diet-induced obesity [3, 24]. In contrast, mice deficient in DGAT2 have severe reduction of TAG and die shortly after birth [4]. The inability of DGAT1 to compensate for the deficiency in DGAT2 knockout mice indicates the nonredundant functions of each DGAT isoform in TAG biosynthesis during mammal development.
Substantial progress has been made in the isolation and identification DGAT2 genes. Database searches have identified more than 74 DGAT2s from 61 organisms including plants, animals, fungi, and human. Phylogenetic analysis separates DGAT2 subfamily sequences into three distinct clusters (Figure 1). However, only a few studies have been directed to understanding the role of DGAT2s [11, 25–28] because purification of these proteins from endogenous tissues is extremely difficult [29]. Study of the roles of DGAT2 enzymes has focused solely on overexpression of DGAT2 genes in transgenic organisms [7, 30, 31]. Purification of DGAT enzymes from endogenous tissues or after over-expression in transgenic species has been difficult due to the integral membrane nature of these proteins. In fact, the expression of DGAT2 from any organism as a full-length or part of the protein in E. coli has not been reported. We recently established a procedure for expressing recombinant DGAT1 in E. coli [32]. But because of the differences in the primary sequences and predicted membrane topologies of DGAT1 and DGAT2 subfamilies [18], it is also necessary to develop a reliable procedure for recombinant DGAT2 expression.
Figure 1. Phylogenetic analysis of DGAT2s.
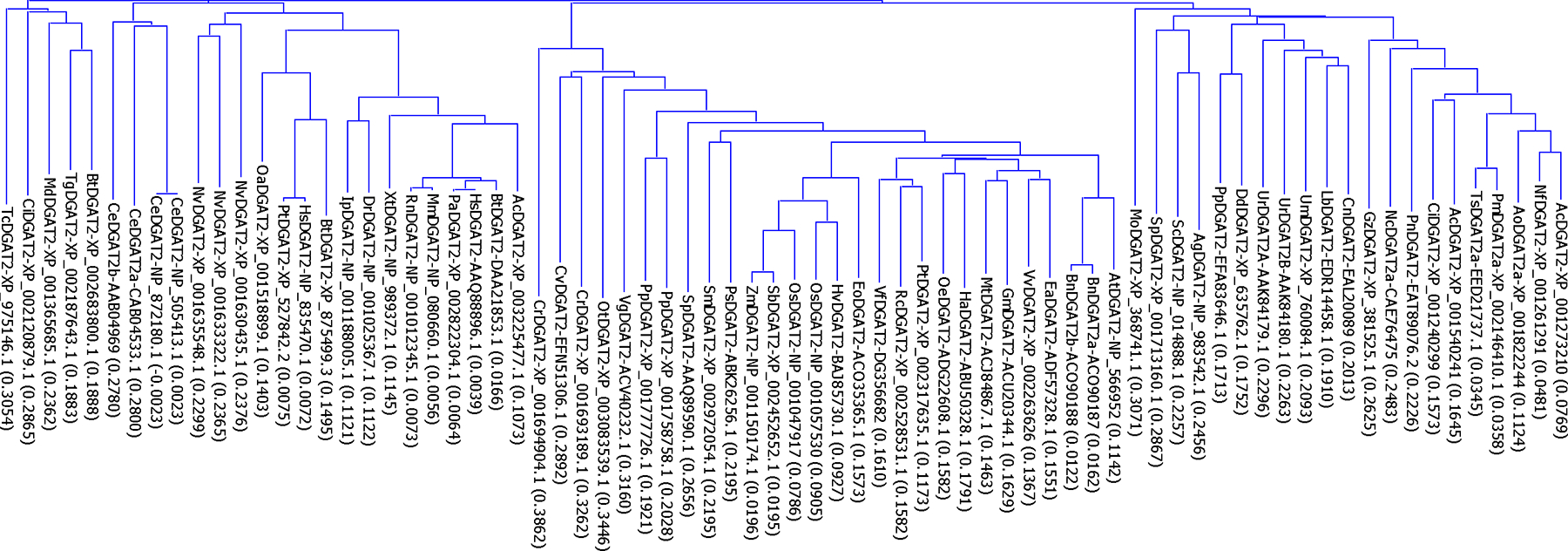
The evolutionary relationships among the 74 DGAT2s from 61 organisms were analyzed by phylogenetic analysis based on the Neighbor-Joining method of Saitou and Nei [54]. The name of each protein sequence consists of the initials of the organism followed by the assigned subfamily of DGATs in the databases and the GenBank accession number. The numbers in the parenthesis following DGAT names are the calculated distance values, which reflect the degree of divergence between all pairs of DGAT sequences analyzed. The abbreviations of the organisms are: Ac in AcDGAT1-EGC41804.1, Ajellomyces capsulatus; Ac in AcDGAT2-XP_003225477.1, Anolis carolinensis; Ac in AcDGAT2-XP_001273210, Aspergillus clavatus; Ag, Ashbya gossypii; Ao, Arthroderma otae; At, Arabidopsis thaliana; Bn, Brassica napus; Bt, Bos taurus; Ce, Caenorhabditis elegans; Ci in CiDGAT2-XP_002120879.1, Ciona intestinalis; Ci in CiDGAT2-XP_001240299, Coccidioides immitis; Cn, Cryptococcus neoformans: Cr, Chlamydomonas reinhardtii; Cv, Chlorella variabilis; Dd, Dictyostelium discoideum; Dr, Danio rerio; Ea, Euonymus alatus; Eo, Elaeis oleifera; Gm, Glycine max; Gz, Gibberella zeae; Ha, Helianthus annuus; Hs, Homo sapiens; Hv, Hordeum vulgare; Ip, Ictalurus punctatus; Lb, Laccaria bicolor; Md, Monodelphis domestica; Mm, Mus musculus; Mo, Magnaporthe oryzae; Mt, Medicago truncatula; Nc, Neurospora crassa; Nf, Neosartorya fischeri; Nv, Nematostella vectensis; Oa, Ovis aries; Oe, Olea europaea; Os, Oryza sativa; Ot, Ostreococcus tauri; Pa, Pongo abelii; Pm, Penicillium marneffei; Pn, Phaeosphaeria nodorum; Pp in PpDGAT2-EFA83646.1, Polysphondylium pallidum; Pp in PpDGAT2a-XP_001758758.1 and PpDGAT2b-XP_001777726.1, Physcomitrella patens; Ps, Picea sitchensis; Pt in PtDGAT2-XP_527842.2, Pan troglodytes; Pt in PtDGAT2-XP_002317635.1, Populus trichocarpa; Rc, Ricinus communis; Rn, Rattus norvegicus; Sb, Sorghum bicolor; Sc, Saccharomyces cerevisiae; Sm, Selaginella moellendorffii; Sp in SpDGAT2-AAQ89590.1, Spirodela polyrhiza; Sp in SpDGAT2-XP_001713160.1, Schizosaccharomyces pombe; Tc, Tribolium castaneum; Tg, Toxoplasma gondii; Ts, Talaromyces stipitatus; Um, Ustilago maydis; Ur, Umbelopsis ramanniana; Vf, Vernicia fordii; Vg, Vernonia galamensis; Vv, Vitis vinifera; Xt, Xenopus tropicalis; Zm, Zea mays. This analysis is updated with more DGAT2s from a previous analysis [32].
The objective of this study was to develop reliable procedures for the expression and purification of recombinant DGAT2 in E. coli. Tung tree DGAT2 was used as the model protein in the current study because it is thought to be the major form of DGAT responsible for the synthesis of tung oil in the seeds, which contains approximately 80% high-value eleostearic acid (18:3 _9cis,11trans,13trans) in the TAG fraction of tung tree seed oils [22, 33, 34].
Results
Construction of expression plasmids
Plasmid pMBP-DGAT2-His was constructed for the expression of recombinant protein MBP-DGAT2-His (rDGAT2) in E. coli. The rDGAT2 contained a MBP (maltose binding protein) at the amino terminus, a PreScission protease cleavage site, the full-length tung tree DGAT2 and 6 histidine residues (His) at the carboxyl terminus (Figure 2). In addition, plasmid pHA-DGAT2 was also designed to express the fulllength tung tree DGAT2 as a HA-tagged protein in yeast. The “Methods” section describes the details of the plasmid construction.
Figure 2. Plasmid map used for the construction of E. coli expression vector and primer sequences used for PCR-amplification of DGAT2 insert.
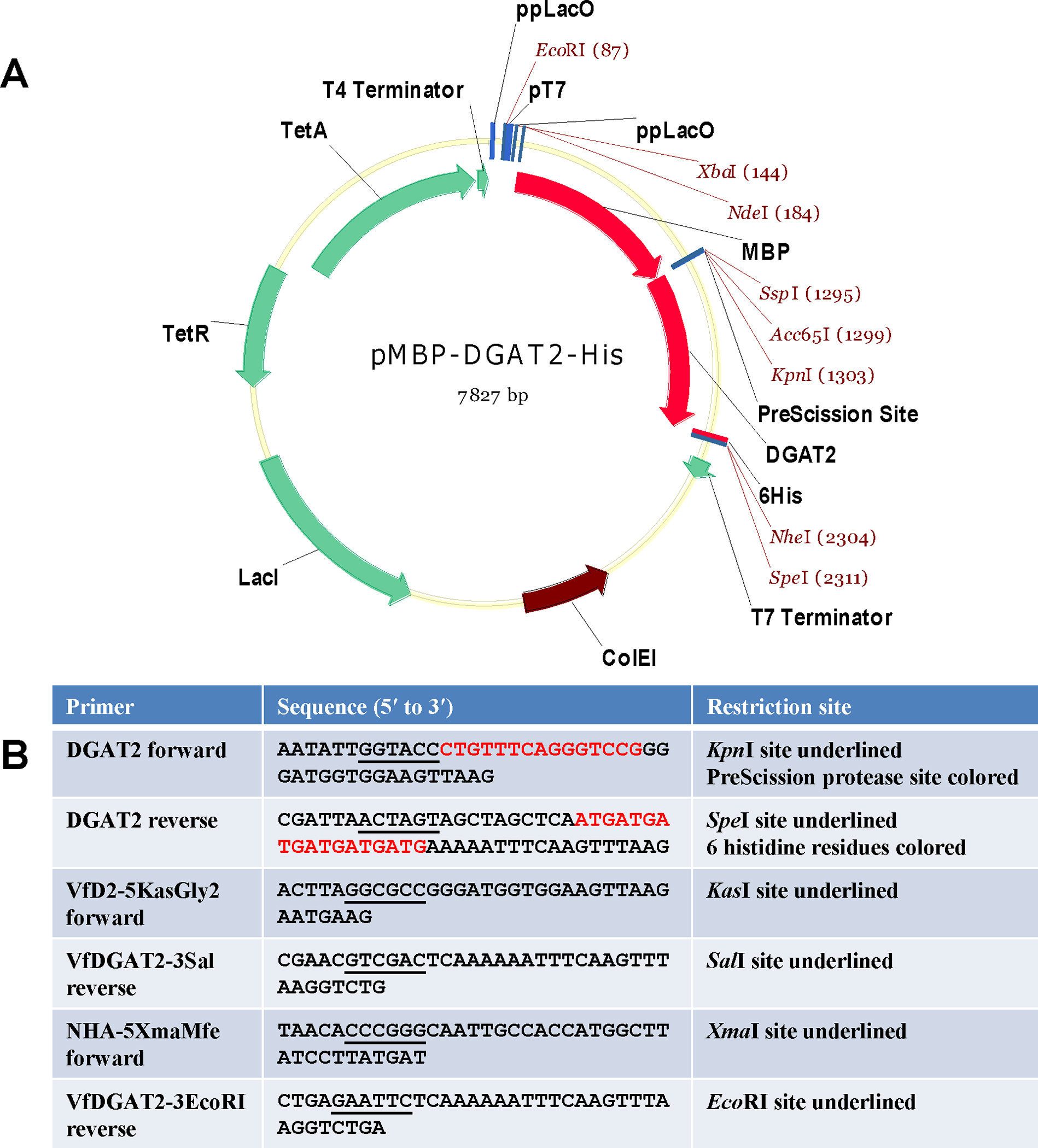
(A) Plasmid pMBP-hTTP [37] was used to express fulllengthDGAT2 in E. coli. Tung DGAT2 DNA was subcloned as described in the Materials and Methods section. (B) Primers for construction of E. coli and yeast expression plasmids. The sequences for restriction enzyme digestion sites are underlined. DGAT2 forward primer has a sequence coding for PreScission protease digestion site (5-CTGTTTCAGGGTCCG-3). DGAT2 reverse primer has a sequence coding for 6 histidine residues (5-ATGATGATGATGATGATG-3).
Expression of recombinant DGAT2 in E. coli
Plasmid pMBP-DGAT2-His was transformed into E. coli strain BL21(DE3). The expression of rDGAT2 was induced by IPTG. However, protein staining did not show a distinct band with a molecular mass corresponding to the full-length rDGAT2 (705 amino acid residues, 78.82 kDa) in the 10,000g supernatant from the un-induced or IPTG-induced cells (data not shown). The minimal expression level of rDGAT2 on staining gels was similar to that of rDGAT1 but in contrast to the massive induction of unfused MBP by IPTG under similar conditions [32]. The levels of rDGAT2 in the IPTG-induced cells were further evaluated by immunoblotting using anti-MBP-hTTP and anti-MBP-mTTP polyclonal antibodies, which were raised in rabbits against purified recombinant human and mouse TTP proteins fused to MBP. These antibodies were shown to cross-react with MBP and MBP fusion proteins with high specificity [35, 36]. Recombinant DGAT2 was also detected by commercial anti-MBP antibodies.
Immunoblotting showed that anti-MBP-hTTP antibodies cross-reacted with two major protein bands (at ~40 and 78 kDa) on immunoblots in samples from E. coli transformed with pMBP-DGAT2-His (Figure 3A). Similar results were obtained with anti-MBP-mTTP and anti-MBP antibodies (data not shown). The ~40 kDa was detected in both the control and all IPTG-induced samples (regardless of induction time), suggesting that this protein band was the endogenously expressed MBP protein from E. coli (Figure 3A). The higher molecular mass band in the IPTG-induced extracts corresponded to the predicted size of rDGAT2. The induction profile showed that the band corresponding to rDGAT2 was detectable after 30 min of IPTG induction and the greatest expression level was achieved after 1 h, but declined significantly after 6 h of IPTG induction (Figure 3A). The expression level of rDGAT2 was not significantly different when cells were grown at induction temperatures of 25 or 37°C or in cells cultured with or without 0.2% glucose (data not shown). Some minor protein bands were also detected by the antibodies in the IPTG-induced sample but not in the control extract (Figure 3A). These minor protein bands were probably degradation products of full-length rDGAT2.
Figure 3. Expression and identification of recombinant DGAT2 in E. coli.
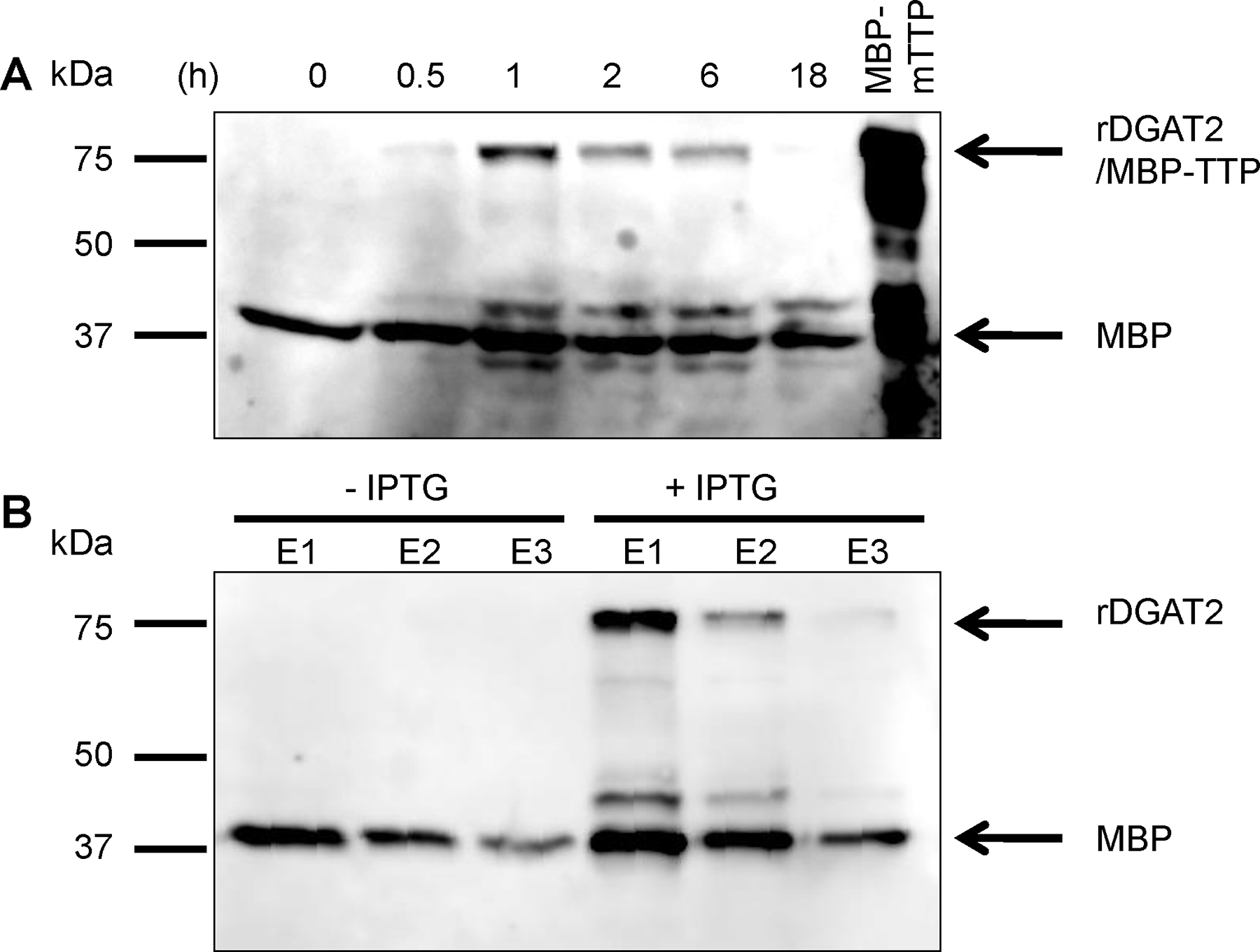
(A) Expression of rDGAT2 in E. coli. Plasmid pMBP-DGAT2-His was transformed into E. coli BL21(DE3). Protein expression was induced by IPTG for various times as indicated on the figure. DGAT2 fusion protein in the 10,000g supernatant was identified by polyclonal antibodies raised against MBP-hTTP fusion protein [35]. Partially purified MBP-mTTP [37] was used as a positive control for immunoblotting since this protein cross-reacts with the antibodies and has a similar molecular mass as rDGAT2. (B) Identification of rDGAT2 in E. coli. The extracts from cells induced with IPTG in panel A were pooled together and mixed with amylose resin affinity beads. Proteins bound to the beads were eluted by maltose solution and detected with anti-MBP-hTTP antibodies. The extracts from cells without IPTG induction were used as a negative control for the purification. E1, E2, and E3 represent three separate elutions with maltose solution. The lower molecular mass band was identified as endogenous MBP from E. coli. 15% SDSPAGE was used for the separation of proteins.
Confirmation of recombinant DGAT2 expressed in E. coli
To confirm that the larger molecular weight immunoreactive band shown on Figure 3A was rDGAT2, the 10,000g supernatants from E. coli treated with IPTG for 0.5–18 h in Figure 3A were pooled and purified by amylose resin affinity chromatography. Our previous studies have indicated that amylose resin effectively binds to MBP, MBP-hTTP, and MBP-mTTP [35, 37, 38]. Therefore, the amylose resin was chosen as an initial step to purify rDGAT2. As a control, the supernatants from cells without IPTG induction were also pooled separately and purified in the same way. Immunoblotting of the amylose resin affinity-purified samples (Figure 3B) were positive for the same two protein bands as seen in the IPTG induction time-course study (Figure 3A). The 78 kDa band was clearly present in the IPTG-induced extracts but was missing in the extract from cells without IPTG induction. The results also show that the majority of the larger molecular weight protein was eluted from the resin with the first maltose wash. The minor protein bands were also purified by the amylose resin, supporting the notion that these minor bands were degradation products of the full-length rDGAT2 that still contain the MBP domain.
Additional purification of rDGAT2 with amylose resin affinity beads was not efficient. The 10,000g supernatant was applied onto a MBPTrap HP column containing amylose resin. After extensive washing, the bound proteins were eluted with 20 mM maltose. However, only a small protein peak was eluted with maltose as observed from UV absorbance at 280 nm (Supplemental Figure 1). Immunoblotting confirmed that the eluted fraction contained rDGAT2 (Supplemental Figure 1), but the great majority of rDGAT2 did not bind to amylose resin and was recovered in the FPLC pass-through and unbound fractions (Supplemental Figure 1). Batch purification with amylose resin resulted in a higher recovery of rDGAT2 (Supplemental Figure 1), probably due to more extensive binding between the beads and rDGAT2 under batch purification. Like column purification, the great majority of rDGAT2 was still in the unbound fractions from the batch purification (data not shown). In addition, a significant amount of rDGAT2 was eluted from the MBPTrap HP column by 0.5 M NaOH (Supplemental Figure 1). Therefore, the proteins from the 10,000g pellet were solubilized using the alkali solution followed by neutralization with HCl. The neutralized protein solution was applied onto the same column. The rDGAT2 protein did not bind to the column and was recovered in the FPLC pass-through and unbound fractions (Supplemental Figure 2). Batch purification of the pass-through and unbound fraction confirmed that the great majority of rDGAT2 bound to amylose resin poorly because little rDGAT2 was eluted from amylose resin (Supplemental Figure 2). These results suggest that folding of rDGAT2 might have prevented rDGAT2 from binding to the amylose resin.
Localization of recombinant DGAT2 in E. coli
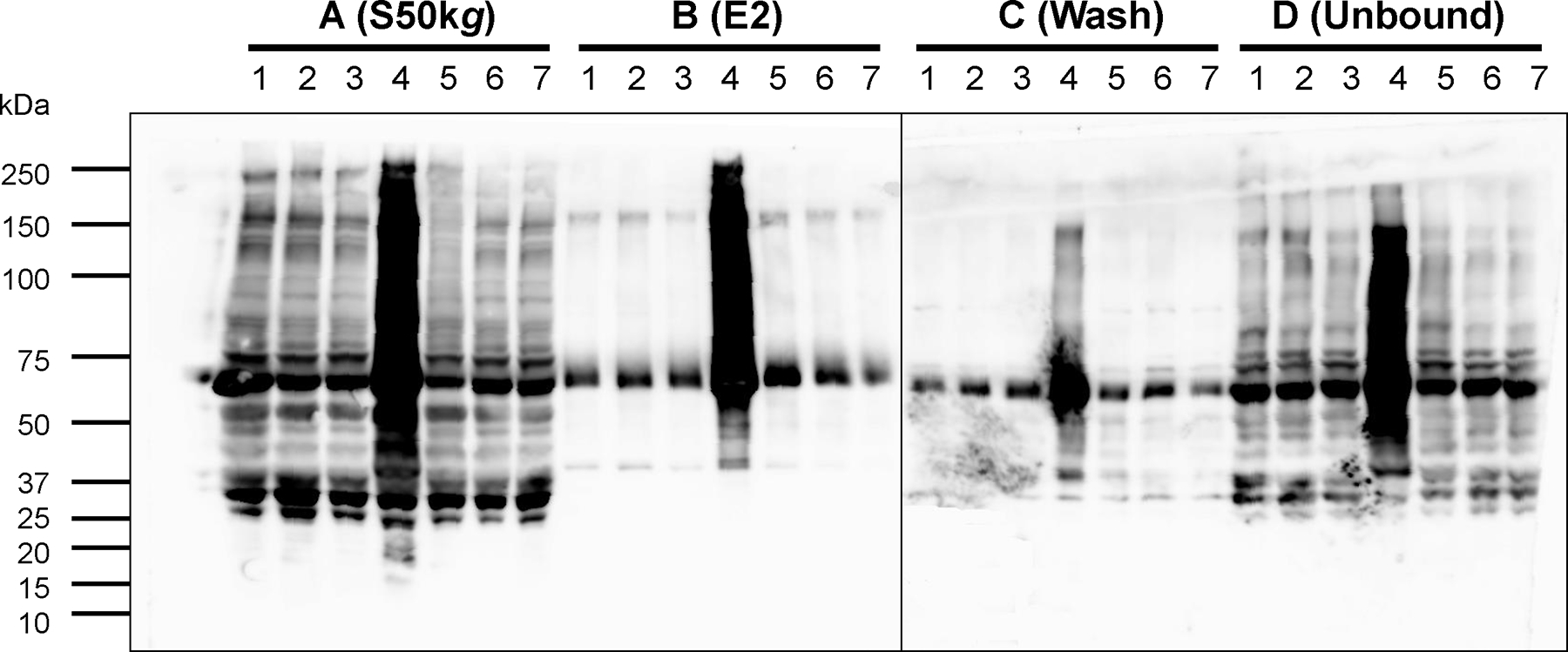
Cellular fractionation was performed to determine the localization of the over-expressed rDGAT2 in E. coli. The 10,000g supernatant, essentially free of inclusion bodies and protein aggregates [39, 40], was subjected to 50,000g centrifugation to generate the plasma membrane pellet [41]. The resultant supernatant was further centrifuged at 100,000g to generate the cytosolic fraction. Immunoblotting showed that large amounts of full-length rDGAT2 were detected in the membranes but very little was recovered in the 100,000g pellet (Figure 4). Amylose resin affinity purification of the cytosolic fraction showed a small fraction of rDGAT2 was detectable (Figure 4). Endogenous MBP was largely contained in the cytosolic fraction and enriched by amylose resin affinity purification, in agreement with the soluble nature of MBP (Figure 4).
Figure 4. Localization of recombinant DGAT2 in E. coli.
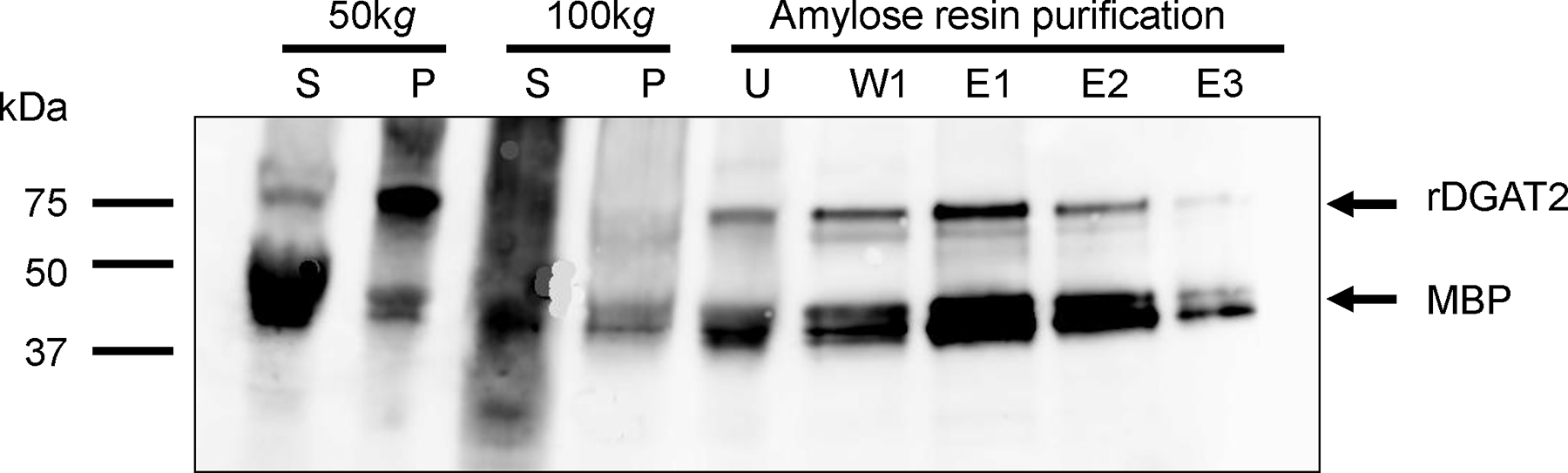
The 10,000g supernatant from over-expressed E. coli were sequentially centrifuged at 50,000g and 100,000g. The 100,000g supernatant was used for amylose resin affinity purification of rDGAT2. Proteins were separated by 4–20% SDS-PAGE. Proteins in the gel were transferred onto a nitrocellulose membrane for immunoblotting with anti-MBP-hTTP antibodies. S, supernatant (10 μL); P, membrane pellet (10 μL); U, unbound fraction (10 μL); W1, the first wash of the three washes (20 μL); E1, E2, and E3, three successive elutions with 20 mM maltose (20 μL). The full-length rDGAT2 and MBP are marked with arrows.
Purification of recombinant DGAT2 with tandem Ni-NTA and amylose resin affinity beads
Recombinant DGAT2 was engineered to contain 6 histidine residues at its carboxy terminus. Ni-NTA beads were used for attempted purification of the recombinant protein from the 10,000g supernatant of induced cells. The bound proteins were eluted with increasing imidazole concentrations ranging from 50 to 1000 mM. SDS-PAGE showed that the purified fractions contained a number of proteins as shown by Coomassie blue staining (Figure 5A-top, elutions 1–5), but the majority of the bound recombinant protein was eluted with 150–200 mM imidazole (Figure 5A-top, elutions 3–4). We concluded that rDGAT2 only partially bound to Ni-NTA beads because a significant amount of the full-length rDGAT2 was recovered in the unbound fraction (Figure 5A-bottom, lane U) and the washes (Figure 5A-bottom). As expected, the endogenous MBP protein did not bind to the Ni-NTA beads (Figure 5A-bottom, lane U vs. elutions 1–6). A protein band with twice the size of the full-length rDGAT2 was also observed in eluted fractions (Figure 5A-bottom, elutions 3–4). As described below, this band likely represents a homodimer of rDGAT2.
Figure 5. Purification of recombinant DGAT2 from E. coli with tandem Ni-NTA and amylose affinity beads.

(A) Ni-NTA purification. The 10,000g supernatant was mixed with Ni-NTA affinity beads. The beads were washed five times. Proteins bound to the beads were eluted by imidazole solution containing 50, 100, 150, 200, 250, 1000 mM (Elutions 1–6). Proteins were separated by 4–20% SDSPAGE and stained with Coomassie brilliant blue (A, top) or transferred onto nitrocellulose membranes for immunoblotting with anti-MBP-hTTP antibodies (A, bottom). The full-length rDGAT2, MBP, and the potential dimer of the full-length rDGAT2 are marked with arrows. H, homogenate, S2, 2,000g supernatant, S10, 10,000g supernatant, U, unbound fraction, B, Ni-NTA beads after 6 imidazole elutions. (B) The fractions purified by Ni-NTA affinity beads as shown in A were mixed with amylose resin affinity beads. The beads were washed five times. Proteins bound to the beads were eluted by amylase resin elution buffer containing 20 mM maltose (lanes 1–5) and three times with 0.5 M NaOH (lanes 6–8). Proteins were separated by 4–20% SDS-PAGE and stained with silver reagent (B, top) or transferred onto a nitrocellulose membrane for immunoblotting with anti-MBP-hTTP antibodies (B, bottom). The full length rDGAT2 and the potential dimer of the full-length rDGAT2 are marked with arrows. E4, proteins eluted with 200 mM imidazole solution from Ni-NTA beads (Figure 5).
The imidazole-eluted proteins (Figure 5A) were pooled and subjected to additional purification via amylose resin affinity chromatography. Silver staining of eluted fractions showed that the additional affinity purification step did not appear to improve rDGAT2 purity but resulted in significant reduction of the protein recovered from the eluted fractions compared to the proteins subjected to Ni-NTA purification alone (Figure 5B-top). Immunoblotting showed that a fraction of rDGAT2 was eluted in the first three elution steps, but most of the recombinant protein did not bind to the resin (Figure 5B-bottom, lane E4 vs. elutions 1–3). As in Figure 5A, an immunoreactive band was also observed in eluted fractions that correspond to a rDGAT2 dimer (Figure 5B-bottom).
Size estimation of recombinant DGAT2 with size exclusion chromatography
Fractions containing rDGAT2 from Ni-NTA affinity purification (Figure 5) were separated by size exclusion chromatography using a Superose 12 HR 10/30 column. The chromatogram for the size standards is shown in Figure 6A. Three protein peaks from the samples were observed on the size exclusion chromatogram (Figure 6B, insert panel). Protein fractions were analyzed by SDS–PAGE and immunoblotting. Recombinant DGAT2 was detected in a wide range of FPLC fractions (Figure 6C, insert panel). The largest amount of rDGAT2 was detected in FPLC fractions 14 and 15 (Figure 6C), which corresponded to the second protein peak on the chromatogram (Figure 6B) with elution volume similar to those of bovine thyroglobulin (670 kDa) (Figure 6A vs. 6B). A faint protein band with approximately twice the size of rDGAT2 was also detected in fractions from size exclusion chromatography (Figure 6C).
Figure 6. Size estimation of recombinant DGAT2 with size exclusion chromatography.
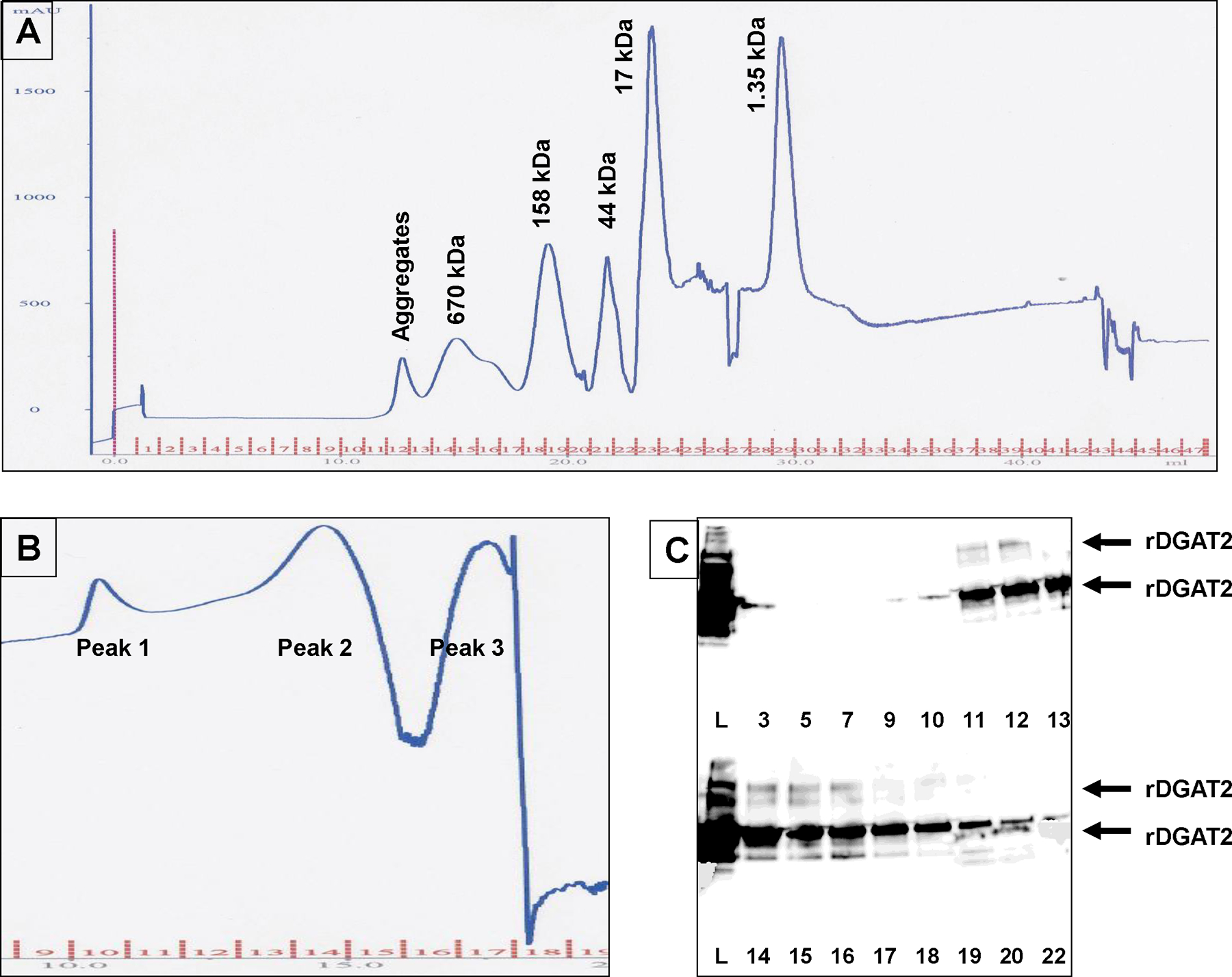
(A) The chromatogram for the protein size standards. The protein standards were vitamin B12, horse myoglobin, chicken ovalbumin, bovine γ-globulin, and bovine thyroglobulin. (B). Part of the chromatogram for rDGAT2. Recombinant DGAT2 fraction purified from Ni-NTA affinity beads was centrifuged at 10,000g before being loaded onto a Superose 12 HR 10/30 column and eluted with identical procedure for the size standards. (C). Immunoblotting detection of rDGAT2 using anti-MBP-hTTP serum (4–20% SDS-PAGE). The full-length rDGAT2 and the potential dimer of the protein are marked with arrows.
Solubilization of recombinant DGAT2 from insoluble fraction
Fractionation experiments showed that rDGAT2 could be largely recovered in the 30,000g pellet. Therefore, rDGAT2 from this pellet was solubilized by 7 detergents (Brij 35, CHAPS, NP-40, SDS, Triton X-100, Tween 20 and Tween 80) followed by centrifugation at 50,000g (Figure 7). Immunoblotting showed that rDGAT2 was only partially solubilized by these detergents at 0.5% final concentration and significant amounts of the protein were still detected in the 50,000g pellet following detergent solublization (data not shown). SDS was the most effective detergent for rDGAT2 solubilization (Figure 7A, lane 4). Affinity purification from the 50,000g supernatant with Ni-NTA beads confirmed this finding (Figure 7B, lane 4). Nonetheless, significant amounts of the recombinant protein still did not bind to the Ni-NTA beads and were recovered in the unbound fractions and the washes (Figure 7C & D).
Figure 7. Solubilization of recombinant DGAT2 by different detergents.
The insoluble fraction from 30,000g pellet of E. coli was solubilizd with 0.5% detergents at 4°C for 1 h followed by centrifugation at 50,000g for 30 min. The 50,000g supernatant was used for affinity purification with Ni-NTA beads. Various fractions were used for immunoblotting analysis with anti-MBP-hTTP antiserum. A, 50,000g supernatant, B, 200 mM imidazole elution, C, washes, D, unbound fractions. Lane 1, Brij 35, lane 2, CHAPS, lane 3, NP-40, lane 4, SDS, lane 5, Triton X-100, lane 6, Tween 20 and lane 7, Tween 80. Each lane was loaded with 20 μL of each fraction.
Purification of recombinant DGAT2 to near homogeneity from SDS-solubilized fraction
Since SDS was found to be an effective detergent, further solubilization was done with different concentrations of SDS (0.1, 0.3, 0.5, and 1% SDS) under different temperatures (4°C, 25°C, 37°C, and 80°C). Immunoblotting showed that 0.3% SDS treatment at 4°C effectively solubilized rDGAT2, but 0.5% and 1% SDS extracted more rDGAT2 protein at 25°C (Figure 8A & 8B). The putative dimer of fulllength rDGAT2 was also seen in the eluted fractions (Figure 8A & B). Silver staining showed that rDGAT2 was purified to near homogeneity by Ni-NTA affinity beads regardless of the varied SDS concentrations or solubilization temperatures (Figure 8C-F).
Figure 8. Purification of recombinant DGAT2 from solubilized fractions with Ni-NTA affinity chromatography.
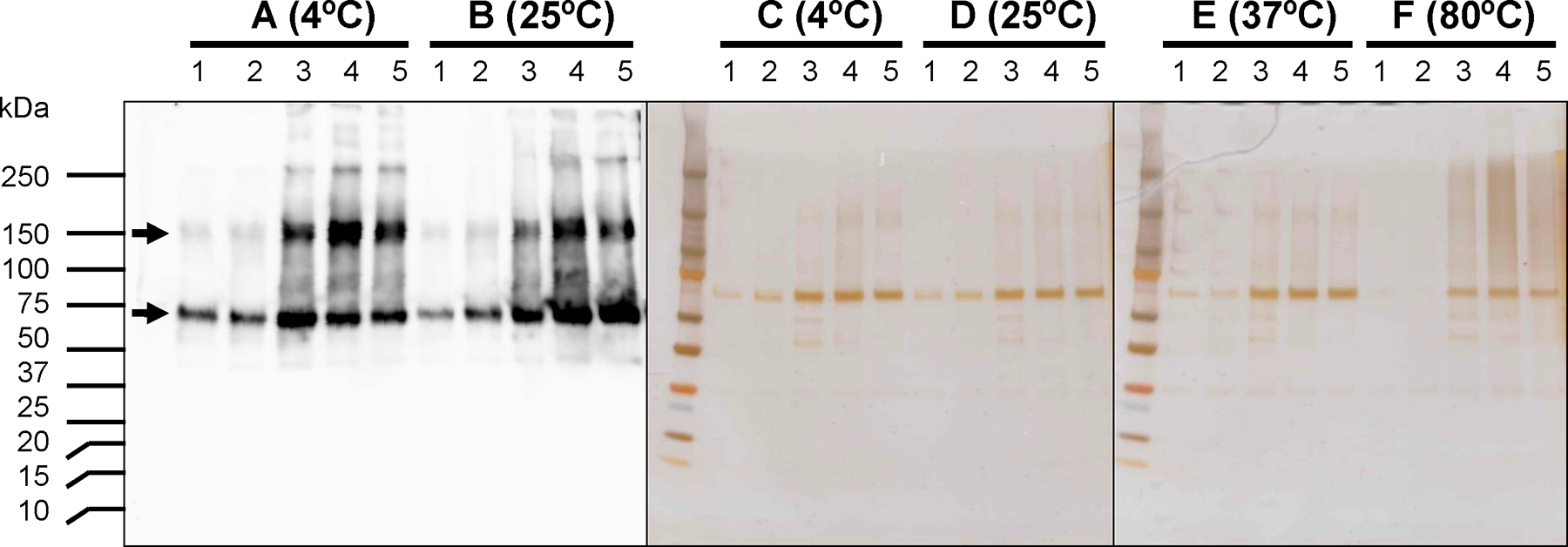
The insoluble fraction from 30,000g pellet of E. coli was solubilizd with various concentrations of SDS at different temperatures for 1 h followed by centrifugation at 50,000g for 30 min. The 50,000g supernatant was used for affinity purification with Ni-NTA beads. Various fractions were used for SDS-PAGE followed by staining with silver reagent and immunoblotting analysis with anti-MBP-hTTP antiserum. A, 4°C, B, 25°C, C, 37°C, D, 80°C, E, 37°C, F, 80°C. Lane 1, 0% SDS, lane 2, 0.1% SDS, lane 3, 0.3% SDS, lane 4, 0.5% SDS, lane 5, 1% SDS. Each lane was loaded with 10 μL of each fraction.
Identification of recombinant DGAT2 by mass spectrometry
Mass spectrometry was used to confirm the positive immunoblot band following SDS extraction and Ni-NTA affinity chromatography as containing rDGAT2. The protein bands corresponding to the monomer and dimer of rDGAT2 (Figure 9 A and B) were excised and digested with trypsin. The in-gel digested peptides were analyzed by LC/ESI/MS (liquid chromatography-electrospray ionization-mass spectrometry). Following LC/MS/MS analyses and database searching, rDGAT2 peptides were observed in both bands corresponding to monomeric and dimeric DGAT2. These MS/MS analyses correspond to 23% sequence coverage (Figure 9C) in the lower molecular weight band (monomer) and 12% sequence coverage in the higher molecular weight band (dimer). The MS/MS data of the triply charged ion of m/z 754.4 from the lower molecular weight band is shown in Figure 9D. These MS analyses provided further evidence that the purified proteins which cross-reacted with anti-MBP-TTP antibodies are likely rDGAT2.
Figure 9. Mass spectrometric analyses of protein bands corresponding to recombinant DGAT2.
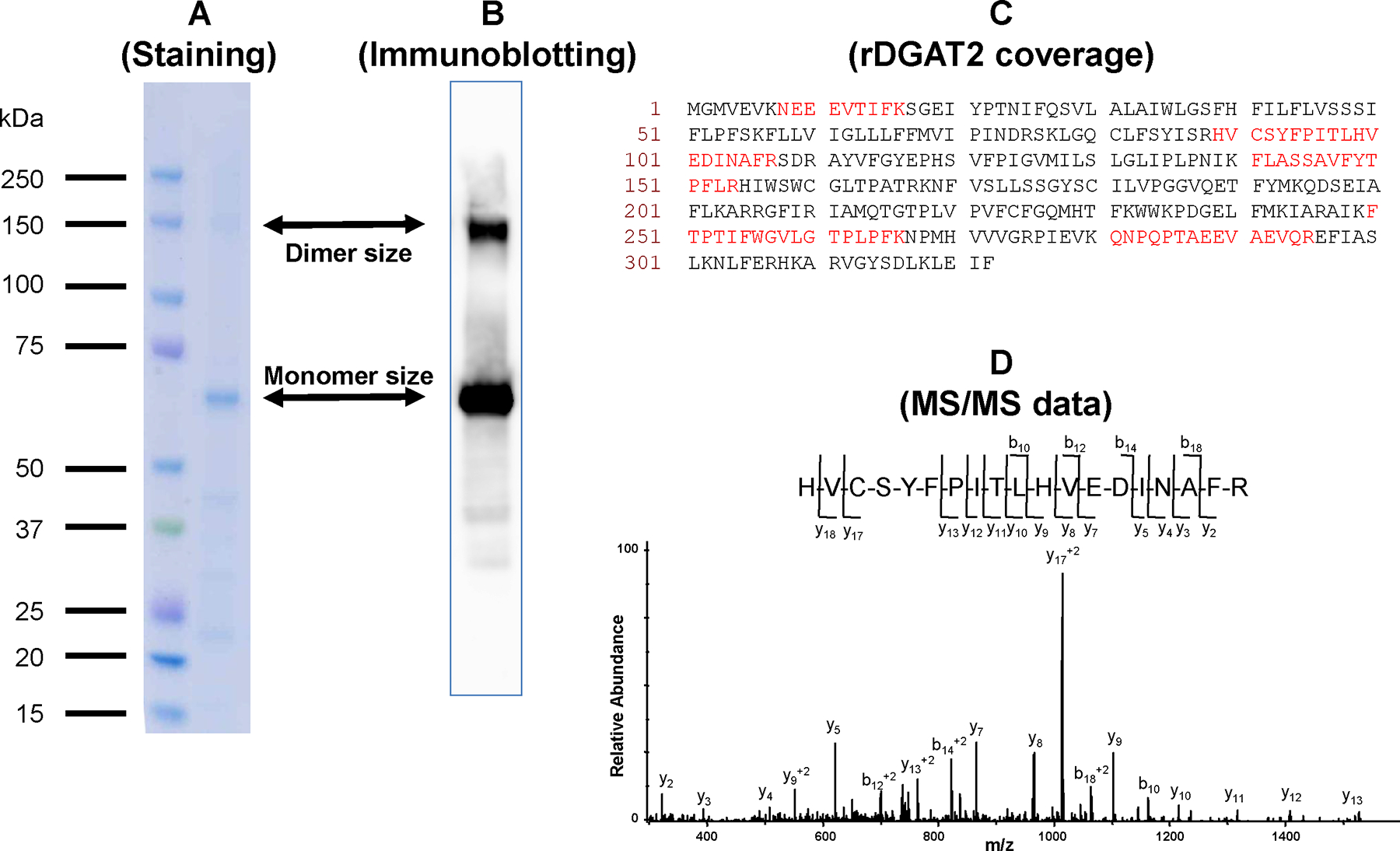
The proteins purified by Ni-NTA affinity chromatography following SDS extraction were separated by SDSPAGE and stained with Coomassie brilliant blue. The protein bands corresponding to the sizes of the monomer and dimer of rDGAT2 were excised and digested with trypsin. The in-gel digested peptides were analyzed by LC-ESI-MS. (A) The protein bands corresponding to monomer and dimer sizes on SDS-PAGE were used for MS analysis, (B) Immunoblotting identification of both protein bands cross30 reacted with anti-MBP-hTTP antibodies, (C) Sequence of rDGAT2 with amino acids observed by LC/MS/MS in the lower molecular weight band highlighted in red, (D) MS/MS data of the triply charged ion of m/z 754.4 corresponding in mass to amino acid residues 89–107 of rDGAT2.
Monomer and dimer forms of DGAT2 expressed in yeast cells
The above results suggest that rDGAT2 (MBP-DGAT2-His) forms dimers even after SDS solubilization. To test if this dimerization is DGAT2-specific, this protein was expressed as an N-terminal HA fusion protein in yeast cells, without the long MBP fusion partner. The homogenates of yeast cells transformed with either HA-DGAT2 plasmid or the control plasmid were sequentially centrifuged at 2,000g, 10,000g, and 100,000g resulting in the 100,000g microsomal membrane pellet [22, 42]. Immunoblotting showed that a protein band with a size similar to the predicted size of HA-DGAT2 was detected in the microsomal membranes of the yeast transformed with pHA-DGAT2, but not in the cytosol, nor in any of the fractions of yeast cells transformed with the control plasmid (Figure 10). Like those of the rDGAT2 from E. coli shown in Figures 5–6 and 8-9, an immunoreactive band of approximately 70 kDa (twice the size of the HA-DGAT2 monomer) was seen in the microsomal membranes but not in the cytosolic fraction, nor the control fractions (Figure 10). A 120-kDa band was detected in all samples, an indication of a non-specific protein that cross-reacted with the anti-HA antibody (Figure 10).
Figure 10. Monomer and dimer forms of DGAT2 in yeast microsomal membranes.
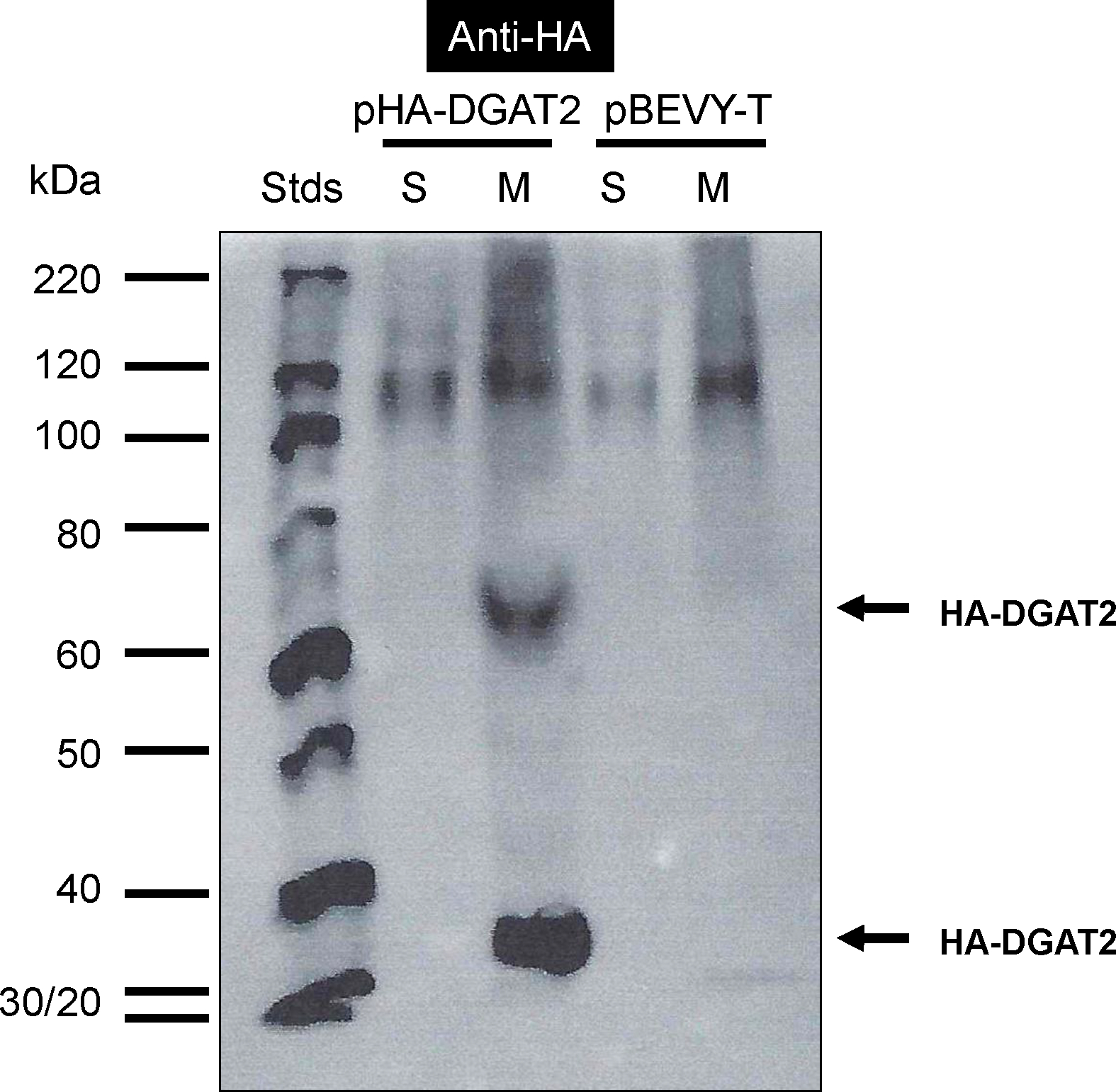
Yeast expression plasmids pHA-DGAT2 and empty pBEVY-T were transformed into the yeast Saccharomyces cerevisiae. Yeast cells were homogenized by mechanical methods using glass beads. The homogenate was sequentially centrifuged at 2,000g, 10,000g and 100,000g. Equal amounts of proteins in the 100,000g pellet and the supernatant (20 μg/lane) were separated by 10% SDS-PAGE. Proteins in the gels were transferred onto a nitrocellulose membrane for immunoblotting with anti-HA antibody. S, 100,000g supernatant (cytosol), M, 100,000g pellet (microsomal membranes). The full-length HA-DGAT2 and the potential dimer of the protein are marked with arrows.
Discussion
Diacylglycerol acyltransferases (DGATs) are responsible for the final and rate-limiting step of triacylglycerol (TAG) biosynthesis in eukaryotic organisms. Gain- and loss-of-function studies suggest that understanding the mechanisms of DGAT catalysis will help to generate transgenic plants with value added properties and provide key knowledge for therapeutic intervention for obesity and related diseases. Database search has identified at least 74 DGAT2 sequences from 61 organisms (Figure 1), but the expression of rDGAT2 as a full-length or partial protein from any organism had not been reported using E. coli expression system. We describe a reliable procedure for expressing full-length rDGAT2 in a bacterial expression system. In this study, the full-length DGAT2 was fused to MBP at the amino terminus and His-tag at the carboxyl terminus (Figure 2). The successful expression of the full-length rDGAT2 was probably due to the fusion to MBP, which was shown to increase the solubility of target proteins [43].
It is difficult to purify DGATs from any source, probably because these proteins are integral membrane proteins with approximately 40% amino acid residues being hydrophobic [18]. Although significant amounts of rDGAT2 were detected in the soluble fraction (Figure 3), most of rDGAT2 were associated with the plasma membranes in E. coli (Figure 4), which was in contrast to the localization of tung tree DGAT2 being exclusively in the microsomal membranes of yeast (Figure 10). In this study, rDGAT2 was engineered with double affinity tags for facilitating purification of the protein from E. coli. However, rDGAT2 was only partially purified from the soluble extract by either type of affinity beads or both kinds of beads in tandem (Figure 5). Solubilization of rDGAT2 from the membrane pellet facilitated affinity purification of rDGAT2 to high purity (Figures 7 & 8), which was identified by cross-reacting with antibodies and mass spectrometry (Figure 9). Attempts to measure the activity of rDGAT2 from Ni-NTA purified fractions, prior to or after SDS solubilization, using nonradioactive oleoyl-CoA and sn-1,2-diolein did not result in measurable TAG synthesis (as measured by thin layer chromatography, data not shown), probably due to the high threshold of detection inherent to the use of nonradioactive substrate. Further experiments are required to demonstrate the enzymatic activity of rDGAT2 using radioactive acyl-CoA.
Several experiments support the assignment of the full-length rDGAT2 as a monomer and dimer on immunoblots. First, MBP-TTP antibodies have been well-characterized in a number of previous publications which cross-react specifically with both MBP and TTP [35, 36]. Second, the detection of MBP by anti-MBP-TTP antibodies [35, 36] is identical to the commercial anti-MBP antibodies [37]. Third, the size of the full-length rDGAT2 on immunoblots corresponds to the calculated size of the protein. Finally, the identity of both bands as rDGAT2 was confirmed by mass spectrometry. Previous reports have shown that the N-terminal regions of DGAT1 from Brassica napus and Homo sapiens form dimers and other higher ordered multimers [44, 45]. Our data is the first to suggest that DGAT2 may also interact to form homopolymers. Further experiments are required to determine how the dimer of rDGAT2 persists under the otherwise strong denaturing conditions of SDS solubilization, reduction and heat treatment that occur during affinity chromatography and SDS-PAGE separation.
Recombinant DGAT2 is probably part of a large protein complex with itself and/or other proteins. First, size exclusion chromatography estimated the size of rDGAT2 peaked at approximately eight times its predicted monomer size (670 vs. 79 kDa) (Figure 6). Second, protein bands with twice the size of the monomer rDGAT2 were consistently detected by immunoblotting and confirmed by mass spectrometry (Figure 9). The dimerization of rDGAT2 protein is probably DGAT2-specific because the dimers were detected in protein samples induced from E. coli expressing MBP-DGAT2-His fusion protein (Figures 5–9) and yeast-expressing HA-DGAT2 (Figure 10). The dimer size of rDGAT2 protein was also consistently detected in fractions purified by affinity and size exclusion chromatography as well as Mono Q anion exchange chromatography (data not shown). Finally, the proteins coeluted from amylose resin and Ni-NTA affinity resins along with many other proteins (Figure 5), which are not seen when MBP was purified from amylose resin [35, 37]. The suggestion of rDGAT2 multimerlization described above is supported by the recently report with mouse DGAT2 [46]. The formation of complexes might be one of the reasons why MBP and His-tagged rDGAT2 did not effectively bind to amylose resin and Ni-NTA beads. However, the nature of the binding partners and their potential functional significance in TAG biosynthesis requires more detailed characterization.
Recombinant DGAT2 is associated with lipids and plasma membranes. The partially purified proteins eluted from affinity beads and size exclusion chromatography appeared yellow in color. Gas chromatography confirmed that rDGAT2 fraction purified by Ni-NTA affinity chromatography contained fatty acids (data not shown). This observation is in agreement with a previous report showing that DGAT2 is localized in lipid droplets of cultured mouse adipocytes (equivalent to oil bodies in plant seeds) [47]. This study demonstrates that rDGAT2 is also associated with E. coli plasma membranes because the majority of rDGAT2 was detected in the 50,000g plasma membranes instead of the 100,000g cytosol (Figure 4). The membrane association with rDGAT2 was also supported by the fact that most of rDGAT2 bound to amylose resin or Ni-NTA beads poorly, in contrast to those of MBP-TTP fusion proteins [37]. It is unlikely that the unbound rDGAT2 was associated with inclusion bodies or protein aggregates because they would be effectively removed by 10,000g centrifugation [39, 40] prior to affinity chromatography. The association of rDGAT2 with lipids and plasma membranes may explain why tandem affinity beads were not effective in purification of rDGAT2 to homogeneity.
Tung tree DGAT1 (526 residues, ~60 kDa) and DGAT2 (322 residues, ~37 kDa) have very limited similarity in primary amino acid sequence, but both are integral membrane proteins with similar properties [22]. These sequence similarities and differences are probably the main reasons for the observed similarities and differences in the expression and purification of rDGAT1 reported previously [32] and rDGAT2 reported here. The similarities of rDGAT1 and rDGAT2 expression and purification include: 1) expression levels of both rDGATs in E. coli are relatively low compared with those of MBP fusion partner; 2) the majority of both rDGATs are associated with the pellet/membranes of E. coli; 3) both rDGATs bind to amylose affinity resin poorly; 4) purification by Ni-NTA affinity chromatography results in of co-elution of multiple other proteins; 5) The semi-purified fractions appear yellow, and contain fatty acids; 6) SDS is the best reagent for solubilization of rDGATs; and 7) both monomer and dimer sizes of rDGATs are detected on immunoblots following SDS-PAGE separation.
A number of differences are observed between rDGAT1 and rDGAT2 expression and purification: 1) IPTG induction resulted in lower relative levels of soluble rDGAT1 compared to rDGAT2; 2) rDGAT1 is degraded much more extensively than rDGAT2; 3), rDGAT1 binds to Ni-NTA affinity beads more tightly than rDGAT2 because rDGAT1 is eluted with higher concentrations of imidazole than rDGAT2; and 4) Essentially pure fractions of rDGAT2 can be achieved by combining SDS solubilization and Ni-NTA affinity chromatography, which is not the case with rDGAT1.
Conclusions
This study describes a reliable procedure for producing the first recombinant DGAT2 from any species using an E. coli expression system. The results suggest that recombinant DGAT2 is present as monomer and dimer forms and associated with other proteins, lipids, and membranes. The ability to express and purify full-length DGAT2 in E. coli represents a step forward towards generating recombinant DGAT2 for further studies such as raising high titer antibodies and studying structure-function relationships.
Methods
Bacterial expression plasmid construction
Plasmid pMBP-DGAT2-His was designed to express the full-length Vernicia fordii type 2 diacylglycerol acyltransferase (DGAT2, GenBank Accession No. DQ356682 [22]) as a fusion protein in E. coli protein expression system. The recombinant protein MBP-DGAT2-His (rDGAT2) contained MBP (maltose binding protein) at the amino terminus and 6 histidine residues (His) at the carboxyl terminus. Plasmid pMBP-hTTP (Figure 2A) as the cloning vector was reported previously [37]. Plasmid pMBP-DGAT2-His was constructed by replacing the hTTP fragment in plasmid pMBP-hTTP with the PCR-amplified DGAT2 fragment at the KpnI and SpeI sites. Existing DGAT2 plasmid was used as the template for PCR-amplification of the DGAT2 DNA open reading frame [22]. DGAT2 forward primer contained DNA sequence for a KpnI/Asp718I restriction enzyme recognition site followed by a PreScission protease cleavage site (5-CTGTTTCAGGGTCCG-3) which codes for 5 amino acid residues (LFQGP) [37] between MBP and DGAT2 protein sequences (Figure 2B). DGAT2 reverse primer contained sequence for a His-tag (5’-ATGATGATGATGATGATG-3’) which codes for 6 histidine residues at the carboxyl terminus of DGAT2 (Figure 2B). The plasmid construction was confirmed by restriction enzyme digestion and DNA sequencing using the GenomeLab Dye Terminator Cycle Sequencing-Quick Start Kit and CEQ 8000 Genetic Analysis System (Beckman Coulter).
Yeast expression plasmid construction
Plasmid H80 (renamed here as pHA-DGAT2 for uniformity) was designed to express the full-length tung DGAT2 as a HA-tagged protein in yeast. The open reading frame for DGAT2 [22] was amplified with the forward primer VfD2–5KasGly2 and the reverse primer VfDGAT2–3Sal (Figure 2B) using Pfu Ultra HS polymerase master mix (Stratagene). The PCR-amplified DNA fragment was digested with restriction enzymes KasI and SalI and ligated into the shuttle vector B49 (unpublished), which was digested with the same two enzymes. Cloning into B49 fuses the 5’ end of the PCR product in-frame with DNA sequence that encodes a start methionine followed by a hemagglutinin (HA) epitope [48]. The 5’ end of the resulting construct codes for the peptide sequence MAYPYDVPDYAGSG, where the HA epitope sequence is underlined, and the last G denotes the second amino acid residue glycine of DGAT2 sequence (GenBank Accession No. DQ356682) [22]. The resulting HA-tagged shuttle plasmid, named B86, was used as a template for a new PCR amplification using the forward primer NHA-5XmaMfe and the reverse primer VfDGAT2–3EcoRI (Figure 2B), which provide recognition sites for XmaI upstream of the start methionine ATG codon and EcoRI downstream of the stop codon, respectively. The resulting product was digested with XmaI and EcoRI and ligated into similarly digested pBEVY-T (a bi-directional expression vector for yeast with constitutive promoters) [49] to create plasmid pHA-DGAT2. The suite of BEVY plasmids directs either constitutive or galactose-inducible protein production in Saccharomyces cerevisiae [49]. Bacterial colonies carrying properly assembled plasmids were identified by colony PCR followed by plasmid isolation and DNA sequencing using the GenomeLab Dye Terminator Cycle Sequencing-Quick Start Kit and CEQ 8000 Genetic Analysis System (Beckman Coulter).
Expression of recombinant DGAT2 in E. coli
Plasmid pMBP-DGAT2-His was transformed into the E. coli BL21(DE3) strain by electroporation. The optimum conditions for rDGAT2 expression were determined with several isopropylthio-β-galactoside (IPTG) induction conditions including various temperatures and induction times. The optimum conditions for rDGAT2 expression were as follows: a single colony was inoculated into Luria–Bertani (LB)-tetracycline (15 μg/mL) medium (LB-Tet) and grown overnight with shaking at 37°C. The overnight culture was inoculated at a 1:20 dilution into fresh medium and grown for about 2 h at 37°C to reach a cell density of about 0.6–1.0 at OD at 600 nm. IPTG was added to the culture medium (0.4 mM final concentration) and protein expression was induced at 37°C for 3 h. Cells were collected by centrifugation at 5,000g for 10 min and homogenized by sonication in homogenization buffer containing amylose resin wash buffer (20 mM Tris–HCl, pH 7.4, 200 mM NaCl, 10 mM β-mercaptoethanol, 1 mM EDTA) or nickel-nitrilotriacetic agarose (Ni-NTA) resin wash buffer (50 mM NaH2PO4, pH 7.4, 300 mM NaCl, 10 mM β-mercaptoethanol, 0.05% Tween-20), plus 0.2–1 mM phenylmethylsulfonyl fluoride (PMSF), and 1:100–1:500 dilution of the protease inhibitor cocktail (Sigma, P8340). The homogenate was centrifuged at 2,000g for 10 min to remove cell debris and the resulting supernatant was centrifuged at 10,000g for 10 min to remove inclusion bodies and protein aggregates [39, 40]. The supernatant and the pellet were used to evaluate the expression levels and solubility of rDGAT2. The expression of empty vector pMAL-c2X was reported previously [32].
Expression of recombinant DGAT2 in yeast
Plasmid pHA-DGAT2 and the control plasmid pBEVY-T were transformed into the yeast strain SCY1998 (dga1/iro1 double mutant) deficient in endogenous DGAT and PDAT activity [50]. Colonies were selected on synthetic dextrose minus tryptophan medium (SD-Trp, 2% dextrose, 0.67% yeast nitrogen base, 1x amino acids minus Trp). Single colonies were grown 45 h at 30°C, then back-diluted into 100 mL of synthetic galactose minus tryptophan medium (SG-Trp, 2% galactose, 0.67% yeast nitrogen base, 1x amino acids minus Trp) at a concentration of 0.2 OD at 600 nm units/mL. Yeast cells were harvested by centrifugation at 5,000g for 10 min after 16–24 h of growth.
Localization of recombinant DGAT2 in E. coli
E. coli cells were homogenized by sonication as described above. The homogenate was sequentially centrifuged at 2,000g for 10 min and 10,000g for 20 min. The 10,000g supernatant (2 mL) essentially free of inclusion bodies and protein aggregates [39, 40] was subjected to 50,000g centrifugation to generate the plasma membrane pellet [41]. The resultant 50,000g supernatant was further centrifuged at 100,000g to generate the cytosolic and microsomal membrane fractions. The 50,000g and 100,000g pellets were suspended in 0.2 mL membrane suspension buffer containing 10 mM Tris, pH 7.4, 1 mM EDTA, and 0.2 M sucrose. The 100,000g cytosol was used for batch purification with amylose resin affinity beads as described above with 3× 1 mL washes and 3× 0.2 mL elutions. Anti-MBP-hTTP antibodies were used to identify rDGAT2 in various fractions.
Localization of recombinant DGAT2 in yeast cells
Microsomal membranes and cytosol were isolated from yeast cells by differential centrifugation using a previously described procedure [42]. Briefly, yeast cells were homogenized by mechanical methods using glass beads (Sigma, G8772) in homogenization buffer (0.4 M sucrose, 50 mM NaH2PO4, pH 7.4, 1 mM EDTA, 1 mM PMSF, 10 mM DTT, and 1:100 dilution of the protease inhibitor cocktails P8340 from Sigma). The homogenate was sequentially centrifuged at 2,000g for 10 min, 10,000g for 20 min and 100,000g for 60 min. The 100,000g pelleted and the supernatant are commonly designated as microsomal membranes and the cytosol [22, 42]. The microsomal membranes were suspended in a membrane suspension buffer described above.
Batch purification of recombinant DGAT2 with Ni-NTA beads
Recombinant DGAT2 was partially purified from E. coli by batch method from the 10,000g supernatant using Ni-NTA beads according to a similar procedure [32]. The 10,000g supernatant was mixed with Ni-NTA Agarose (Qiagen). The mixtures were incubated at 4°C with rotation for 3 h followed by centrifugation at 1,000g for 5 min. The beads were washed five times each with 10 bead-volume of Ni-NTA resin wash buffer followed by rotation for 10 min and centrifugation. The bound proteins were eluted from the beads by gravity flow in a Bio-Rad mini-column with increasing concentrations of imidazole (Sigma) in Ni-NTA wash buffer.
Batch purification of recombinant DGAT2 with amylose resin
Recombinant DGAT2 was also partially purified from E. coli by batch method from the 10,000g supernatant using amylose resin affinity beads. [32] Briefly, the 10,000g supernatant was mixed with amylose resin (New England Biolabs). The mixtures were incubated at 4°C with rotation for 3 h followed by centrifugation at 1,000g for 5 min. The beads were washed five times each with 10 bead-volume of amylose resin wash buffer. The bound proteins were eluted from the beads by gravity flow in a Bio-Rad mini-column with 20 mM maltose (Sigma) in amylose resin wash buffer.
FPLC column purification of recombinant DGAT2 with amylose resin
Recombinant DGAT2 was subjected to purification with amylose resin affinity column using fast protein liquid chromatography (FPLC) (GE Healthcare Life Sciences) with similar procedures for the purification of MBP-TTP and MBP-ZFP36L1 fusion proteins [35, 37, 51]. The 10,000g supernatant or the partially purified rDGAT2 from Ni-NTA batch purification was loaded onto a MBPTrap HP column (GE Healthcare Life Sciences). The column was washed with 5 bed-volume of amylose resin wash buffer (20 mM Tris–HCl, pH 7.4, 200 mM NaCl, 10 mM β-mercaptoethanol, 1 mM EDTA) and then eluted with 10 bed-volume of amylose resin elution buffer (20 mM maltose in amylose resin wash buffer) followed by 0.5 M NaOH wash of the column. As a control for affinity purification, MBP was similarly purified from E. coli transformed with plasmid pMAL-c2x (New England Biolabs) by amylose resin affinity chromatography [32].
Size estimation of recombinant DGAT2 with size exclusion chromatography
Size-exclusion chromatography was used to estimate the molecular mass of rDGAT2 using a similar procedure [37]. rDGAT2 fraction purified from Ni-NTA affinity beads was centrifuged at 10,000g for 10 min before being loaded onto a Superose 12 HR 10/30 column (GE Healthcare Life Sciences). Proteins were eluted with amylose resin wash buffer (20 mM Tris–HCl, pH 7.4, 200 mM NaCl, 10 mM β-mercaptoethanol, 1 mM EDTA) and analyzed by SDS–PAGE and immunoblotting using anti-MBP-hTTP serum. Fractions containing rDGAT2 were pooled and concentrated by centrifugation at 2,000g using Amico Ultra-4 Centrifugal Filter Devices (Millipore). The protein size standards were run on the same column. The protein standards were vitamin B12 (1.35 kDa), horse myoglobin (17 kDa), chicken ovalbumin (44 kDa), bovine γ-globulin (158 kDa), and bovine thyroglobulin (670 kDa) (Bio-Rad Laboratories).
Purification of recombinant DGAT2 from detergent-solubilized E. coli pellets
Cellular fractionation studies indicated that DGAT2 fusion protein could be recovered from the 30,000g pellet. Therefore, the pellet was suspended in Ni-NTA sonication buffer and used for detergent solubilization at 4°C for 1 h with 0.5% of detergents (Brij 35, CHAPS, NP-40, SDS, Triton X-100, Tween 20 and Tween 80). After identifying SDS being the most effective detergent for the solubilization, the pellet was solubilized by 0.1, 0.3, 0.5, and 1% SDS under 4°C, 25°C, 37°C, and 80°C for 1 h. The solubilization mixtures were centrifuged at 50,000g for 30 min. The supernatant was used for recombinant DGAT2 purification with Ni-NTA affinity beads as described above.
Protein determination, SDS–PAGE, and immunoblotting
Protein concentrations were determined with the Bradford method using the Protein Assay Dye Reagent Concentrate (Bio-Rad Laboratories) following 0.5 M NaOH treatment of the samples [42]. Proteins were separated by 10%, 15%, or 4–20% SDS–PAGE and visualized by staining with Coomassie brilliant blue (Sigma) or silver staining reagent (Bio-Rad Laboratories) [35]. DGAT fusion proteins were detected by immunoblotting following previously described procedures using nitrocellulose membranes and SuperSignal West Pico Chemiluminescent Substrate (Pierce) [35, 42]. The primary antibodies were rabbit anti-MBP-hTTP antibodies [35] and anti-MBP-mTTP antibodies [36] and affinity-purified goat anti-HA antibody (Bethyl Laboratories). The secondary antibodies were affinity-purified goat anti-rabbit IgG (H+L) horseradish peroxidase conjugate (GAR-HRP) with human IgG absorbed (Bio-Rad Laboratory) and donkey anti-goat HRP (DAG-HRP) (Bethyl Laboratories).
In-gel tryptic digestion
The protein bands were manually excised from the gel, cut into small pieces, and transferred into a 96-well microtiter plate. Gel pieces were subjected to automatic tryptic digestion using an Investigator™ Progest protein digestion station (Genomic Solutions, Ann Arbor, MI) [37, 38]. Briefly, gel bands were sequentially washed twice with 25 mM ammonium bicarbonate buffer (pH 7) and acetonitrile, dehydrated, rehydrated with 25 μL of the enzyme solution, and digested at 37°C for 8 h. The enzyme solution used was sequencing grade modified trypsin (Promega Corp., Madison, WI) at a concentration of 0.01 mg/mL in 25 mM ammonium bicarbonate buffer (pH 7). Resulting tryptic peptides were extracted from the gel, lyopholized, and stored at −80°C. Prior to mass spectrometric analysis, the peptides were reconstituted in 40 μL of a 97:3 solution of water:acetonitrile (0.1% formic acid).
ESI Mass Spectrometry
For the nanoLC/ESI/MS/MS analyses, an Agilent XCT Ultra ion trap (Agilent Technologies, Inc., Santa Clara, CA) equipped with an HPLC-Chip Cube MS interface and an Agilent 1100 nanoLC system [52]. Injections of 20 μL from the peptide digests were made onto a 40 nL enrichment column followed by a 43 mm × 75 μm analytical column, packed with ZORBAX 300SB C18 particles. Peptides were separated and eluted using a linear gradient of 3–50% acetonitrile (0.1% formic acid) over 40 min, followed by a linear gradient of 50–95% acetonitrile over 7 min at a flow rate of 500 nL/min. The ion trap mass spectrometer was operated in the positive ion mode, standard enhanced mode using the following settings: capillary voltage, −2150 V; mass range, 300–1500; ICC smart target (number of ions in the trap prior to scan out), 100000 or 200 milliseconds of accumulation; and MS/MS fragmentation amplitude, 1.0 V. During the LC/MS/MS analyses, automated data dependent acquisition software was employed with the six most abundant ions (threshold requirement of 10000 counts) from each spectrum selected for MS/MS analysis.
Following the analyses, the MS/MS data were extracted and analyzed using Spectrum Mill MS Proteomics software (Agilent Technologies, Inc). To generate peak lists, the raw data files were processed using the Data Extractor function with the following parameters: deconvoluted ions of 300–6000 Da and a retention time of 10 to 60 min. MS scans with the same precursor m/z were merged based on a +/− 1.4 m/z window and a +/− 15 sec retention time window. Using the extracted data, searches were performed against the NCBI nonredundant protein database using the MS/MS search function. Parameters used for the searches included: precursor mass tolerance, +/− 1.5 Da; product mass tolerance, +/− 1 Da; enzyme specificity, trypsin, with maximum two missed cleavage sites; variable modifications, oxidized methionine and N-terminal pyroglutamic acid; and a minimum matched peak intensity, 80%. All MS/MS sequence assignments used for protein identifications were manually validated.
Supplementary Material
Supplemental Figure 1 Comparison of column and batch purification of rDGAT2 in the 10,000g supernatant from E. coli with amylose resin affinity beads. (A) FPLC chromatogram of column purification by MBPTrap HP column. The 10,000g supernatant was loaded onto a MBPTrap HP column. The column was washed extensively with amylose resin wash buffer. The bound proteins were eluted with amylose resin elution buffer containing 20 mM maltose and finally with 0.5 M NaOH. Protein peaks were identified by monitoring UV absorbance at 280 nm on the chromatogram. The three protein peaks corresponded to the unbound, wash, and maltose-eluted fractions. The chromatogram from NaOH elution is not shown. (B) Identification of rDGAT2 in the column fractions by immunoblotting. Proteins in the unbound, wash, maltose elution, and NaOH elution were separated by 10% SDS-PAGE. rDGAT2 and MBP were identified by anti-MBP-hTTP antibodies. The full-length rDGAT2 is marked with an arrow. P, 10,000g pellet, S, 10,000g supernatant. (C) Batch purification of rDGAT2 by amylose resin affinity beads. The 10,000g supernatant was mixed with amylose resin affinity beads. The beads were washed three times. Proteins bound to the beads were eluted by 20 mM maltose solution. rDGAT2 was detected by immunoblotting with anti-MBP-hTTP antibodies.
Supplemental Figure 2 Comparison of column and batch purification of rDGAT2 in the 10,000g pellet from E. coli with amylose resin affinity beads. The 10,000g pellet was solubilized with 0.5 M NaOH followed by neutralization with HCl. The neutralized protein solution was purified using procedures identical to those described in the legend for Figure 3. (A) FPLC chromatogram of column purification by the MBPTrap HP column. (B) Identification of rDGAT2 by immunoblotting in fractions of column purification by anti-MBP-hTTP antibodies. The full-length rDGAT2 is marked with an arrow. (C) Batch purification of rDGAT2 by amylose resin affinity beads. S, 10,000g supernatant, P, 10,000g pellet, U, unbound fraction.
Acknowledgements and Funding
This work was supported by USDA-ARS Quality and Utilization of Agricultural Products Research Program and, in part, by the Intramural Research Program of the NIH, National Institutes of Environmental Health Sciences (project ES050171). O.D. Howard Jr. is a graduate student from the Center for Genomics and Biotechnology and Department of Biology at Alcorn State University working on his thesis at USDA-ARS. The authors thank Ms. Katina Johnson of the Protein Microcharacterization Core Facility (NIH-NIEHS) for the mass spectrometry analyses, Drs. Kandan Sethumadhavan and Soheila J. Maleki for technical help and Drs. John M. Dyer, Thomas J. Caperna, Jeffrey W. Cary, Stephanie A. Boone, Kenneth B. Tomer, and Jeffrey F. Kuhn for helpful comments on the manuscript. A preliminary report was presented at the Plant Biology 2010 in Montreal, Canada, on July 31- August 4, 2010 [53]. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Lists of abbreviations
- DGAT
diacylglycerol acyltransferase
- ESI-MS
Electrospray ionization-mass spectrometry
- FPLC
fast protein liquid chromatography
- HA
hemagglutinin
- His
poly histidine
- IPTG
isopropylthio-β-galactoside
- MBP
maltose binding protein
- MS/MS
tandem mass spectrometry
- Ni-NTA
nickelnitrilotriacetic agarose
- PAGE
polyacrylamide gel electrophoresis
- rDGAT
recombinant diacylglycerol acyltransferase
- TAG
triacylglycerol
- TTP
tristetraprolin
Footnotes
Competing interests
The author declares that he has no competing interests.
References
- 1.Ichihara K, Takahashi T, Fujii S: Diacylglycerol acyltransferase in maturing safflower seeds: its influences on the fatty acid composition of triacylglycerol and on the rate of triacylglycerol synthesis. Biochim Biophys Acta 1988, 958:125–129. [DOI] [PubMed] [Google Scholar]
- 2.Zou J, Wei Y, Jako C, Kumar A, Selvaraj G, Taylor DC: The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J 1999, 19:645–653. [DOI] [PubMed] [Google Scholar]
- 3.Smith SJ, Cases S, Jensen DR, Chen HC, Sande E, Tow B, Sanan DA, Raber J, Eckel RH, Farese RV, Jr.: Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nat Genet 2000, 25:87–90. [DOI] [PubMed] [Google Scholar]
- 4.Stone SJ, Myers HM, Watkins SM, Brown BE, Feingold KR, Elias PM, Farese RV, Jr.: Lipopenia and skin barrier abnormalities in DGAT2-deficient mice. J Biol Chem 2004, 279:11767–11776. [DOI] [PubMed] [Google Scholar]
- 5.Chen HC, Rao M, Sajan MP, Standaert M, Kanoh Y, Miura A, Farese RV, Jr., Farese RV: Role of adipocyte-derived factors in enhancing insulin signaling in skeletal muscle and white adipose tissue of mice lacking Acyl CoA:diacylglycerol acyltransferase 1. Diabetes 2004, 53:1445–1451. [DOI] [PubMed] [Google Scholar]
- 6.Andrianov V, Borisjuk N, Pogrebnyak N, Brinker A, Dixon J, Spitsin S, Flynn J, Matyszczuk P, Andryszak K, Laurelli M, Golovkin M, Koprowski H: Tobacco as a production platform for biofuel: overexpression of Arabidopsis DGAT and LEC2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotechnol J 2010, 8:277–287. [DOI] [PubMed] [Google Scholar]
- 7.Burgal J, Shockey J, Lu C, Dyer J, Larson T, Graham I, Browse J: Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol J 2008, 6:819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durrett TP, McClosky DD, Tumaney AW, Elzinga DA, Ohlrogge J, Pollard M: A distinct DGAT with sn-3 acetyltransferase activity that synthesizes unusual, reduced-viscosity oils in Euonymus and transgenic seeds. Proc Natl Acad Sci U S A 2010, 107:9464–9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jako C, Kumar A, Wei Y, Zou J, Barton DL, Giblin EM, Covello PS, Taylor DC: Seed-specific overexpression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol 2001, 126:861–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lardizabal K, Effertz R, Levering C, Mai J, Pedroso MC, Jury T, Aasen E, Gruys K, Bennett K: Expression of Umbelopsis ramanniana DGAT2A in seed increases oil in soybean. Plant Physiol 2008, 148:89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J, Francis T, Mietkiewska E, Giblin EM, Barton DL, Zhang Y, Zhang M, Taylor DC: Cloning and characterization of an acyl-CoA-dependent diacylglycerol acyltransferase 1 (DGAT1) gene from Tropaeolum majus, and a study of the functional motifs of the DGAT protein using site-directed mutagenesis to modify enzyme activity and oil content. Plant Biotechnol J 2008, 6:799–818. [DOI] [PubMed] [Google Scholar]
- 12.Bouvier-Nave P, Benveniste P, Oelkers P, Sturley SL, Schaller H: Expression in yeast and tobacco of plant cDNAs encoding acyl CoA:diacylglycerol acyltransferase. Eur J Biochem 2000, 267:85–96. [DOI] [PubMed] [Google Scholar]
- 13.Liu L, Zhang Y, Chen N, Shi X, Tsang B, Yu YH: Upregulation of myocellular DGAT1 augments triglyceride synthesis in skeletal muscle and protects against fat-induced insulin resistance. J Clin Invest 2007, 117:1679–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roorda BD, Hesselink MK, Schaart G, Moonen-Kornips E, Martinez-Martinez P, Losen M, De Baets MH, Mensink RP, Schrauwen P: DGAT1 overexpression in muscle by in vivo DNA electroporation increases intramyocellular lipid content. J Lipid Res 2005, 46:230–236. [DOI] [PubMed] [Google Scholar]
- 15.Kamisaka Y, Kimura K, Uemura H, Shibakami M: Activation of diacylglycerol acyltransferase expressed in Saccharomyces cerevisiae: overexpression of Dga1p lacking the N-terminal region in the Deltasnf2 disruptant produces a significant increase in its enzyme activity. Appl Microbiol Biotechnol 2010, 88:105–115. [DOI] [PubMed] [Google Scholar]
- 16.Liu L, Shi X, Bharadwaj KG, Ikeda S, Yamashita H, Yagyu H, Schaffer JE, Yu YH, Goldberg IJ: DGAT1 expression increases heart triglyceride content but ameliorates lipotoxicity. J Biol Chem 2009, 284:36312–36323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamisaka Y, Tomita N, Kimura K, Kainou K, Uemura H: DGA1 (diacylglycerol acyltransferase gene) overexpression and leucine biosynthesis significantly increase lipid accumulation in the Deltasnf2 disruptant of Saccharomyces cerevisiae. Biochem J 2007, 408:61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao H: Structure-function analysis of diacylglycerol acyltransferase sequences from 70 organisms. BMC Research Notes 2011, 4:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cases S, Smith SJ, Zheng YW, Myers HM, Lear SR, Sande E, Novak S, Collins C, Welch CB, Lusis AJ, Erickson SK, Farese RV, Jr.: Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc Natl Acad Sci U S A 1998, 95:13018–13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cases S, Stone SJ, Zhou P, Yen E, Tow B, Lardizabal KD, Voelker T, Farese RV, Jr.: Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J Biol Chem 2001, 276:38870–38876. [DOI] [PubMed] [Google Scholar]
- 21.Lardizabal KD, Mai JT, Wagner NW, Wyrick A, Voelker T, Hawkins DJ: DGAT2 is a new diacylglycerol acyltransferase gene family: purification, cloning, and expression in insect cells of two polypeptides from Mortierella ramanniana with diacylglycerol acyltransferase activity. J Biol Chem 2001, 276:38862–38869. [DOI] [PubMed] [Google Scholar]
- 22.Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM: Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 2006, 18:2294–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Zhou G, Wang Y, Xu L: F-BOX and oleosin: Additional target genes for future metabolic engineering in tung trees? Ind Crops Prod 2010, 32:684–686. [Google Scholar]
- 24.Chen HC, Smith SJ, Ladha Z, Jensen DR, Ferreira LD, Pulawa LK, McGuire JG, Pitas RE, Eckel RH, Farese RV Jr.,.: Increased insulin and leptin sensitivity in mice lacking acyl CoA:diacylglycerol acyltransferase 1. J Clin Invest 2002, 109:1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Q, Siloto RM, Weselake RJ: Role of cysteine residues in thiol modification of acyl-CoA:diacylglycerol acyltransferase 2 from yeast. Biochemistry 2010, 49:3237–3245. [DOI] [PubMed] [Google Scholar]
- 26.Liu Q, Siloto RM, Snyder CL, Weselake RJ: Functional and Topological Analysis of Yeast Acyl-CoA:Diacylglycerol Acyltransferase 2, an Endoplasmic Reticulum Enzyme Essential for Triacylglycerol Biosynthesis. J Biol Chem 2011, 286:13115–13126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McFie PJ, Stone SL, Banman SL, Stone SJ: Topological orientation of acyl-CoA:diacylglycerol acyltransferase-1 (DGAT1) and identification of a putative active site histidine and the role of the n terminus in dimer/tetramer formation. J Biol Chem 2010, 285:37377–37387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stone SJ, Levin MC, Farese RV Jr., : Membrane topology and identification of key functional amino acid residues of murine acyl-CoA:diacylglycerol acyltransferase-2. J Biol Chem 2006, 281:40273–40282. [DOI] [PubMed] [Google Scholar]
- 29.Little D, Weselake R, Pomeroy K, Furukawa-Stoffer T, Bagu J: Solubilization and characterization of diacylglycerol acyltransferase from microspore-derived cultures of oilseed rape. Biochem J 1994, 304 ( Pt 3):951–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kroon JT, Wei W, Simon WJ, Slabas AR: Identification and functional expression of a type 2 acyl-CoA:diacylglycerol acyltransferase (DGAT2) in developing castor bean seeds which has high homology to the major triglyceride biosynthetic enzyme of fungi and animals. Phytochemistry 2006, 67:2541–2549. [DOI] [PubMed] [Google Scholar]
- 31.Li R, Yu K, Hatanaka T, Hildebrand DF: Vernonia DGATs increase accumulation of epoxy fatty acids in oil. Plant Biotechnol J 2010, 8:184–195. [DOI] [PubMed] [Google Scholar]
- 32.Cao H, Chapital DC, Shockey JM, Klasson KT: Expression of tung tree diacylglycerol acyltransferase 1 in E. coli. BMC Biotechnology 2011, 11:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abbott CE: Fruit-bud development in tire tung-oil tree. J Agric Res 1929, 38:679–695. [Google Scholar]
- 34.Dyer JM, Tang FQ, Chapital DC, Lax AR, Shepherd HS, Shih DS, Pepperman AB: Differential extraction of eleostearic acid-rich lipid-protein complexes in tung seeds. J Am Oil Chem Soc 1998, 75:1687–1690. [Google Scholar]
- 35.Cao H: Expression, purification, and biochemical characterization of the antiinflammatory tristetraprolin: a zinc-dependent mRNA binding protein affected by posttranslational modifications. Biochemistry 2004, 43:13724–13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao H, Tuttle JS, Blackshear PJ: Immunological characterization of tristetraprolin as a low abundance, inducible, stable cytosolic protein. J Biol Chem 2004, 279:21489–21499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao H, Dzineku F, Blackshear PJ: Expression and purification of recombinant tristetraprolin that can bind to tumor necrosis factor-alpha mRNA and serve as a substrate for mitogen-activated protein kinases. Arch Biochem Biophys 2003, 412:106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao H, Deterding LJ, Venable JD, Kennington EA, Yates JR, III, Tomer KB, Blackshear PJ: Identification of the anti-inflammatory protein tristetraprolin as a hyperphosphorylated protein by mass spectrometry and site-directed mutagenesis. Biochem J 2006, 394:285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massie HR, Zimm BH: Molecular weight of the DNA in the chromosomes of E. coli and B. subtilis. Proc Natl Acad Sci U S A 1965, 54:1636–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren Q, De RG, Kessler B, Witholt B: Recovery of active medium-chain-length-poly-3-hydroxyalkanoate polymerase from inactive inclusion bodies using ion-exchange resin. Biochem J 2000, 349:599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romanov GA, Spichal L, Lomin SN, Strnad M, Schmulling T: A live cell hormone-binding assay on transgenic bacteria expressing a eukaryotic receptor protein. Anal Biochem 2005, 347:129–134. [DOI] [PubMed] [Google Scholar]
- 42.Cao H, Sullivan TD, Boyer CD, Shannon JC: Bt1, a structural gene for the major 39–44 kDa amyloplast membrane polypeptides. Physiol Plant 1995, 95:176–186. [Google Scholar]
- 43.Kapust RB, Waugh DS: Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci 1999, 8:1668–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng D, Meegalla RL, He B, Cromley DA, Billheimer JT, Young PR: Human acyl-CoA:diacylglycerol acyltransferase is a tetrameric protein. Biochem J 2001, 359:707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weselake RJ, Madhavji M, Szarka SJ, Patterson NA, Wiehler WB, Nykiforuk CL, Burton TL, Boora PS, Mosimann SC, Foroud NA, Thibault BJ, Moloney MM, Laroche A, Furukawa-Stoffer TL: Acyl-CoAbinding and self-associating properties of a recombinant 13.3 kDa N-terminal fragment of diacylglycerol acyltransferase-1 from oilseed rape. BMC Biochem 2006, 7:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McFie PJ, Banman SL, Kary S, Stone SJ: Murine diacylglycerol acyltransferase-2 (DGAT2) can catalyze triacylglycerol synthesis and promote lipid droplet formation independent of its localization to the endoplasmic reticulum. J Biol Chem 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuerschner L, Moessinger C, Thiele C: Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic 2008, 9:338–352. [DOI] [PubMed] [Google Scholar]
- 48.Fritze CE, Anderson TR: Epitope tagging: general method for tracking recombinant proteins. Methods Enzymol 2000, 327:3–16. [DOI] [PubMed] [Google Scholar]
- 49.Miller CA III, Martinat MA, Hyman LE: Assessment of aryl hydrocarbon receptor complex interactions using pBEVY plasmids: expressionvectors with bi-directional promoters for use in Saccharomyces cerevisiae. Nucleic Acids Res 1998, 26:3577–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oelkers P, Cromley D, Padamsee M, Billheimer JT, Sturley SL: The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J Biol Chem 2002, 277:8877–8881. [DOI] [PubMed] [Google Scholar]
- 51.Cao H, Lin R, Ghosh S, Anderson RA, Urban JF, Jr.: Production and characterization of ZFP36L1 antiserum against recombinant protein from Escherichia coli. Biotechnol Prog 2008, 24:326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao H, Deterding LJ, Blackshear PJ: Phosphorylation site analysis of the anti-inflammatory and mRNA-destabilizing protein tristetraprolin [Review]. Expert Rev Proteomics 2007, 4:711–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao H, Chapital D, Mason C, Shockey J, Klasson KT: Expression of tung seed diacylglycerol acyltransferases (DGAT) in E. coli and yeast [abstract]. Plant Biology 2010. 2010:P02054. [Google Scholar]
- 54.Saitou N, Nei M: The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987, 4:406–425. [DOI] [PubMed] [Google Scholar]
- 55.Detweiler CD, Deterding LJ, Tomer KB, Chignell CF, Germolec D., Mason RP: Immunological identification of the heart myoglovin radical formed by hydrogen peroxide. Free Rad. Biol. Med. 2002, 33:364-369. [DOI] [PubMed] [Google Scholar]
- 56.Chatterjee S, Ehrenshaft M, Bhattacharjee S, Deterding LJ, Bonini MG, Corbett J, Kadiiska MB, Tomer KB, Mason RP: Immuno-spin trapping of a post-translational carboxypeptidase B1 radical formed by a dual role of xanthine oxidase and endothelial nitric oxide synthase in acute septic mice. Free Rad. Biol. Med. 2009, 46:454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 Comparison of column and batch purification of rDGAT2 in the 10,000g supernatant from E. coli with amylose resin affinity beads. (A) FPLC chromatogram of column purification by MBPTrap HP column. The 10,000g supernatant was loaded onto a MBPTrap HP column. The column was washed extensively with amylose resin wash buffer. The bound proteins were eluted with amylose resin elution buffer containing 20 mM maltose and finally with 0.5 M NaOH. Protein peaks were identified by monitoring UV absorbance at 280 nm on the chromatogram. The three protein peaks corresponded to the unbound, wash, and maltose-eluted fractions. The chromatogram from NaOH elution is not shown. (B) Identification of rDGAT2 in the column fractions by immunoblotting. Proteins in the unbound, wash, maltose elution, and NaOH elution were separated by 10% SDS-PAGE. rDGAT2 and MBP were identified by anti-MBP-hTTP antibodies. The full-length rDGAT2 is marked with an arrow. P, 10,000g pellet, S, 10,000g supernatant. (C) Batch purification of rDGAT2 by amylose resin affinity beads. The 10,000g supernatant was mixed with amylose resin affinity beads. The beads were washed three times. Proteins bound to the beads were eluted by 20 mM maltose solution. rDGAT2 was detected by immunoblotting with anti-MBP-hTTP antibodies.
Supplemental Figure 2 Comparison of column and batch purification of rDGAT2 in the 10,000g pellet from E. coli with amylose resin affinity beads. The 10,000g pellet was solubilized with 0.5 M NaOH followed by neutralization with HCl. The neutralized protein solution was purified using procedures identical to those described in the legend for Figure 3. (A) FPLC chromatogram of column purification by the MBPTrap HP column. (B) Identification of rDGAT2 by immunoblotting in fractions of column purification by anti-MBP-hTTP antibodies. The full-length rDGAT2 is marked with an arrow. (C) Batch purification of rDGAT2 by amylose resin affinity beads. S, 10,000g supernatant, P, 10,000g pellet, U, unbound fraction.


