Abstract
The human splicing factor ASF/SF2 (alternative splicing factor/splicing factor 2) is modular in structure with two RNA-binding domains (RBD1 and RBD2) and a C-terminal domain rich in arginine–serine dipeptide repeats. ASF/SF2 is an essential splicing factor that also functions as an important regulator of alternative splicing. In adenovirus E1A (early region 1A) alternative pre-mRNA splicing, ASF/SF2 functions as a strong inducer of proximal 5′-splice-site selection, both in vitro and in vivo. In the present study, we tested the functional role of individual domains of ASF/SF2 in alternative splicing in vitro. We show that ASF/SF2-RBD2 is the critical domain controlling E1A alternative splicing. In fact, RBD2 alone is sufficient to mimic the activity of the full-length ASF/SF2 protein as an inducer of proximal 5′-splice-site selection in vitro. The RBD2 domain induces a switch to E1A-proximal 5′-splice-site usage by repressing distal 12 S splicing and simultaneously stimulates proximal 13 S splicing. In contrast, the ASF/SF2-RBD1 domain has a more general splicing enhancer phenotype and appears to stimulate preferentially cap-proximal 5′-splice-site selection. Furthermore, the SWQDLKD motif, which is conserved in all SR proteins (serine/arginine-rich proteins) containing two RBDs, and the ribonucleoprotein-1-type RNA recognition motif were both found to be necessary for the alternative splice-site-switching activity of ASF/SF2. The RNP-1 motif was necessary for efficient RNA binding, whereas the SWQDLKD motif most probably contributes by functioning as a surface-mediating critical protein–protein contact during spliceosome assembly.
Keywords: alternative splicing, alternative splicing factor/splicing factor 2 (ASF/SF2), early region 1A (E1A), RNA-binding domain (RBD), conserved RNA-binding motif 1 (RNP-1), serine/arginine-rich protein (SR protein)
Abbreviations: ASF/SF2, alternative splicing factor/splicing factor 2; E1A, early region 1A; RBD, RNA-binding domain; RNP, ribonucleoprotein; RNP-1 motif, conserved RNA-binding motif 1; RS domain, arginine- and serine-rich domain; hnRNP, heterogeneous nuclear ribonucleoprotein; snRNP, small nuclear ribonucleoprotein; SR protein, serine/arginine-rich protein
INTRODUCTION
The precise excision of introns from pre-mRNA is performed by the spliceosome, a macromolecular machine containing five small nRNAs and numerous proteins (reviewed in [1]). The spliceosome performs the two primary functions of splicing: it recognizes the splicing signals in the pre-mRNA and catalyses the removal of the intron and the joining of the exons. The mechanism of splice-site selection in constitutive and alternative splicing are closely connected, since components of the splicing machinery essential for constitutive splicing also have a role in the regulation of alternative splicing. For example, developmental or tissue-specific differences in the activity or the amount of general splicing factors and/or gene-specific splicing regulators can result in alternative splice-site usage. One such group of factors, which are often involved in regulated splicing, is the SR family of splicing factors [2].
SR proteins (serine/arginine-rich proteins), a group of highly conserved proteins in metazoans, are essential for constitutive splicing as well as regulation of alternative splicing. SR proteins have a characteristic structure and contain one or two N-terminal RNP (ribonucleoprotein)-type RBDs (RNA-binding domains) and a variable-length C-terminal RS domain (arginine- and serinerich domain; reviewed in [3]). SR proteins are modular in structure, with the RBDs making sequence-specific contact with the RNA [4,5] and the RS domain mediating protein–protein interactions that are supposed to be essential for the recruitment of other splicing components during spliceosome assembly [3,6–8]. SR proteins stimulate binding of U1 snRNP (small nuclear RNP) to the 5′-splice site, binding of U2AF (U2 snRNP auxiliary factor) to the 3′-splice site and recruitment of the U4/U6-U5 triple snRNP to form the pre-spliceosome [6,9,10]. SR proteins are, to a large extent, functionally redundant as activators of constitutive splicing. They also serve as regulators of alternative splicing. In general, increasing concentrations of these proteins tend to select the intron-proximal 5′-splice site in vitro [11,12] and in vivo [13–15]. However, individual SR proteins can sometimes have opposite effects on alternative splice-site selection, as for adenovirus E1A (early region 1A) pre-mRNA splicing (see below).
The adenovirus E1A transcription unit represents a natural pre-mRNA substrate that gives rise to three major mRNAs, namely 13, 12 and 9 S mRNAs, by alternative splicing using three 5′-splice sites and a common 3′-splice site [16]. E1A alternative splicing is subjected to a temporal regulation during a lytic infection. Thus the 9 S mRNA, which is almost undetectable at the early stages of infection, becomes the predominant E1A mRNA at later stages of infection (reviewed in [17]). The early-to-late shift in splice-site usage is probably determined by the intrinsic properties of the respective 5′-splice-site regions, with the 13 and 9 S 5′-splice sites being the strongest and weakest respectively. E1A alternative splicing has been shown to be highly sensitive to changes in various parameters, both in vitro and in vivo [18,19]. Therefore E1A has successfully been used as a model substrate to characterize the function of SR proteins as regulators of alternative RNA splicing. The activity of the whole family of SR proteins has been tested using the E1A unit as a model substrate, either in vitro and/or in transient co-transfection assays. The results show that many SR proteins have unique activities to activate one of the three 5′-splice sites in the E1A pre-mRNA. For example, ASF/SF2 (alternative splicing factor/splicing factor 2) and SC35 enhance proximal 13 S mRNA splicing [11,13,14,20–22], whereas SRp20 enhances 12 S mRNA splicing [14,23] and SRp54 enhances 9 S mRNA splicing [24]. SRp40 and 9G8 appear to stimulate both 13 and 12 S mRNA splicing [11,14,24,25]. The dose-dependent enhancement of the proximal 5′-splice-site selection by ASF/SF2 has been explained using an occupancy model where ASF/SF2 stimulates U1 snRNP recruitment to all the 5′-splice sites [26]. When all the sites are filled, splicing occurs between the 5′-splice sites closest to the 3′-splice site, i.e. the proximal 13 S 5′-splice site.
The function of the different ASF/SF2 domains in E1A alternative splicing has only been tested in vivo. The results show that the RS domain was dispensable for the 13 S 5′-splice-site-switching activity of ASF/SF2 [15,23]. Furthermore, ASF/SF2 mutant proteins lacking RBD1 or RBD2 are also active as regulators of E1A splicing [23,27]. A mutant protein lacking RBD1 had the same activity as wild-type ASF/SF2 and stimulated 13 S 5′-splice-site selection, whereas a mutant protein lacking RBD2 enhanced 12 S mRNA splicing.
In addition to functioning as splicing enhancer factors, SR proteins can also function as splicing repressor proteins (reviewed in [17]). This motivated us to investigate the function of ASF/SF2 domains in E1A alternative splicing. We have shown previously that ASF/SF2 and other SR proteins inhibit adenovirus IIIa pre-mRNA splicing by binding to the IIIa repressor element (3RE), which is located immediately upstream of the IIIa branch site [28]. In a follow-up study [29], we examined the functional role of the different domains of ASF/SF2 in the regulated splicing of the IIIa pre-mRNA in vitro. We showed that the second RBD (RBD2) of ASF/SF2 was both necessary and sufficient for ASF/SF2-mediated repression of IIIa pre-mRNA splicing. Furthermore, we showed that the SWQDLKD motif, conserved in all SR proteins containing RBD2, was essential for the ASF/SF2-RBD2-mediated repression of IIIa splicing.
In the present study, we have examined the contribution of different domains of ASF/SF2 to E1A alternative splicing in vitro. In contrast with previous results [30], we show that both RBDs of ASF/SF2 are active and capable of regulating E1A alternative splicing in vitro. However, the activity of the two domains was opposite on alternative 5′-splice-site usage. Thus RBD2 strongly promoted proximal 13 S 5′-splice-site usage and concomitantly inhibited distal 12 S 5′-splice-site selection, whereas RBD1 preferentially activated distal 12 S 5′-splice-site usage. Interestingly, the activity of RBD2 and RBD1 on E1A alternative splicing was also detected in the absence of splice-site competition. Thus RBD2 functioned as a repressor of 12 S 5′-splice-site usage and simultaneously activated 13 S 5′-splice-site selection. In contrast, RBD1 activated splicing of both the 12 and 13 S 5′-splice sites in the absence of competition, suggesting that RBD1 may preferentially activate the cap-proximal 5′-splice site in a pre-mRNA. Furthermore, the SWQDLKD motif, conserved in all SR proteins containing an RBD2, and RNP-1 (conserved RNA-binding motif 1) were both required for the alternative splice-site selection activity of ASF/SF2-RBD2. The RNP-1 motif was necessary for efficient RNA binding, whereas the SWQDLKD motif most probably contributes by functioning as a surface for protein–protein interactions between ASF/SF2-RBD2 and other components of the splicing machinery.
EXPERIMENTAL
Constructs and protein purification
Most plasmids expressing ASF/SF2 hybrid proteins have been described previously [29]. His–RBD1 and His–RBD2 were created from MS2–RBD1 and MS2–RBD2 by recloning the individual ASF/SF2-RBDs into the pET15b vector (Novagen). All the hybrid proteins were co-expressed in Escherichia coli BL21(DE3) together with SR protein kinase 1 [31] and purified under the conditions described previously [29]. The plasmid pML005SVR1-13S(mut) was generated by site-specific mutagenesis, changing four nucleotides at the E1A 13 S 5′-splice site (5′-CUACAGUAAGUGA-3′ mutated to 5′-CUACAugcAuUGA-3′ taken from [32]).
Synthesis of transcripts
The template for E1A pre-mRNA synthesis was generated by PCR amplification, using the upstream primer 5′-ATTAATACGACTCACTATAGGGTCCGGTTTCTATGCC-3′ and the downstream primer 5′-ACACAGGTGATGTCGGGCGTCTCAGGATAGCAGG-3′ on the plasmid pML005 SVR1 [33]. The truncated E1A pre-mRNA contained nucleotides 879–1336 from the adenovirus 2 E1A region [34]. The E1A 12 S pre-mRNA used in Figure 5 differs from the wild-type transcript in containing the four-nucleotide change that inactivates the 13 S 5′-splice site [32]. The template for E1A 13 S pre-mRNA synthesis was generated by PCR amplification, using the alternative upstream primer 5′-ATTAATACGACTCACTATAGGGACCCAGATATTATG-3′ together with the common downstream primer. The E1A 13 S pre-mRNA contained nucleotides 1052–1336 from the adenovirus 2 E1A region [34]. Capped and 32P-labelled E1A pre-mRNAs (CTP as the labelled nucleotide) were synthesized by in vitro run-off transcription from purified PCR product using T7 RNA polymerase, as described previously [35].
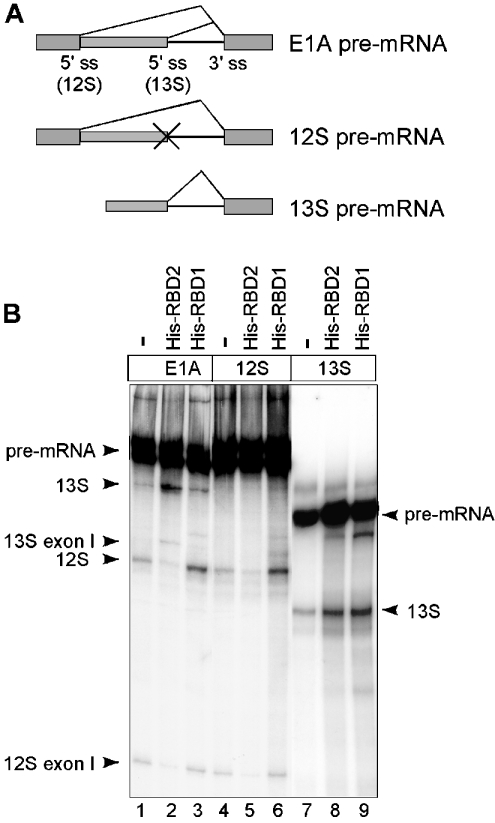
Figure 5. Effects of His–RBD1 and His–RBD2 on the activity of individual E1A 5′-splice sites.
(A) Schematic representation of the three E1A pre-mRNAs used to assay the activity of the ASF/SF2-RBDs on E1A 13 or 12 S 5′-splice-site activity, alone or in competition. (B) In vitro splicing reactions were performed under limiting splicing conditions in HeLa-NE using the indicated E1A pre-mRNAs and supplemented with the His–RBD1 or His–RBD2 proteins (150 pmol), as depicted in Figure 2(A). The products were resolved by gel electrophoresis and visualized by autoradiography. Positions of the pre-mRNA, free first exons and splicing products are indicated.
Templates for synthesis of the 115 nucleotide RNA fragments used in the UV cross-linking assay were generated by PCR amplification, using the upstream primer 5′-ATTAATACGACTCACTATAGGGACCCAGATATTATG-3′ and the downstream primer 5′-AATTACCACACCAAACCCACCACTCTATCACCC-3′ on pML005 SVR1 [33] or pML005SVRI-13S(mut) plasmid respectively. These short RNA fragments span the E1A 13 S 5′-splice site and contain nucleotides 1052–1166 from the adenovirus 2 E1A region [34]. Uncapped 32P-labelled transcripts (GTP and UTP as the labelled nucleotides) were prepared by in vitro run-off transcription from purified PCR product using T7 RNA polymerase.
In vitro splicing
HeLa nuclear extracts (HeLa-NE) and HeLa cytoplasmic extracts (HeLa-S100) were prepared as described previously [29,35]. Great care was taken to establish conditions that would generate low basal splicing but approximately equal usage of the 12 and 13 S 5′-splice sites. In our standard method, the in vitro splicing reactions contained 6% (v/v) HeLa-NE, 25% (v/v) HeLa-S100, 2.6% (w/v) poly(vinyl alcohol), 20 mM phosphocreatine, 2 mM ATP, 12% (v/v) glycerol, 12 mM Hepes (pH 7.9), 48 mM KCl, 2.1 mM MgCl2 and 15 fmol of 32P-labelled pre-mRNA in a total volume of 25 μl. Splicing reaction mixtures were incubated at 30 °C for 2 h. The reaction products were resolved on 8% denaturing polyacrylamide gels and visualized by autoradiography. Identities of spliced products and intermediates were assigned based on their size.
UV cross-linking
Approx. 30 fmol of 32P-labelled 115 nucleotide fragments encompassing the E1A 13 S wild-type or mutant 5′-splice site were incubated under splicing conditions (20 mM phosphocreatine/2 mM ATP/12% glycerol/12 mM Hepes, pH 7.9/48 mM KCl/2.1 mM MgCl2) with the indicated amounts of purified MS2–ASF/SF2 hybrid proteins in a total reaction volume of 12.5 μl, together with 2.5 μg of unlabelled competitor tRNA. Mixtures were incubated at 30 °C for 15 min and then irradiated with 254 nm UV light for 30 min on ice (output, 1200 μW/cm2; distance, 1 cm). RNA was digested with 50 μg of RNase A at 37 °C for 15 min, followed by 15 min incubation at 56 °C. Then, 4 μl of a 6×SDS gel loading buffer was added and the samples were resolved by SDS/PAGE (12% gel) under reducing conditions [36]. Labelled proteins were visualized by autoradiography.
RESULTS
ASF/SF2-RBD1 and -RBD2 have opposite effects on E1A alternative splice-site selection in vitro
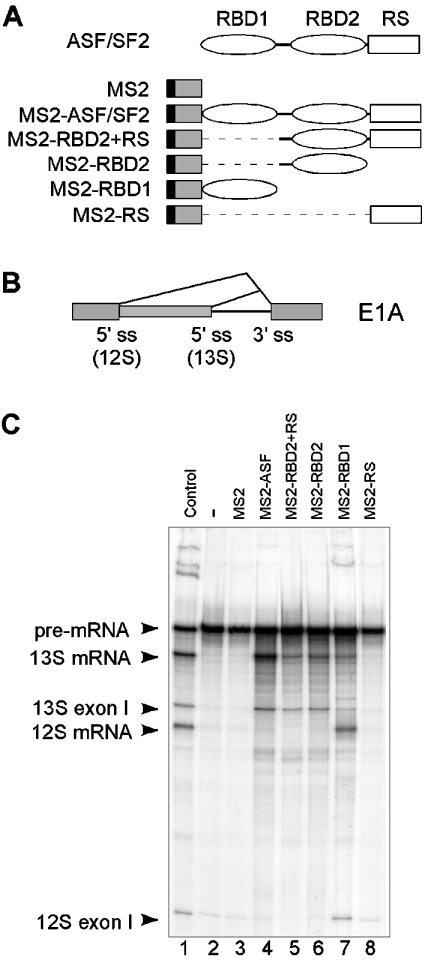
To investigate the role of individual domains of ASF/SF2 in alternative splicing, we analysed the activity of chimaeric ASF/SF2 proteins on adenovirus E1A pre-mRNA splicing in vitro. For the initial experiments, we used a series of MS2–ASF/SF2 chimaeric proteins (Figure 1A) that we have described previously [29]. The MS2–ASF/SF2 hybrid proteins were expressed and purified from E. coli as His-tagged proteins together with SR protein kinase 1. The co-expression of the MS2–ASF/SF2 fusion proteins with SR protein kinase 1 makes the proteins more soluble and more active as splicing regulators [29,31].
Figure 1. Role of structural domains of ASF/SF2 in the regulation of E1A alternative splicing.
(A) Structure of the MS2–ASF/SF2 hybrid proteins used to examine the domain requirements of ASF/SF2 in E1A alternative splicing. Hybrid proteins encode for the following ASF/SF2 sequences: MS2–ASF/SF2, amino acids 1–248; MS2–RBD2+RS, amino acids 92–248; MS2–RBD2, amino acids 92–197; MS2–RBD1, amino acids 1–92; and MS2–RS, amino acids 196–248. (B) Schematic representation of the E1A pre-mRNA showing the position of splice sites and the splicing events that generate the 13 and 12 S mRNAs respectively. (C) Alternative splicing enhancer activity of the MS2–ASF/SF2 hybrid proteins. In vitro splicing reactions were performed under limiting splicing conditions in HeLa-NE using the E1A pre-mRNA and the indicated MS2–ASF/SF2 fusion proteins (30 pmol). The products were resolved by gel electrophoresis and visualized by autoradiography. Positions of the pre-mRNA, free first exons and splicing products are indicated by arrowheads. Lane 1 shows a splicing reaction incubated under optimal splicing conditions.
The E1A transcription unit uses three 5′-splice sites and a common 3′-splice site to produce three major mRNAs: the 13, 12 and 9 S mRNAs [16]. For convenience, we used a shortened E1A pre-mRNA that contained only the 13 and 12 S 5′-splice sites (Figure 1B). As shown previously, E1A alternative splicing in HeLa-NE is strongly dependent on the reaction conditions [18]. Therefore great care was taken to mix HeLa-NE and HeLa-S100 extracts in proportions that ensured a low basal usage of both the 5′-splice sites (see the Experimental section). Importantly, it was essential to include a small amount of HeLa-NE, since S100 extracts do not contain SR proteins and, therefore, do not support constitutive splicing.
Although ASF/SF2 deletion proteins were created as MS2 fusion proteins, our initial experiments demonstrated that the effects of the MS2–ASF/SF2 hybrid proteins on E1A pre-mRNA splicing were not dependent on the presence of the MS2 operator sequence in the E1A pre-mRNA (results not shown). This finding is in line with previous results showing that ASF/SF2 proteins devoid of RBD1 or RBD2 are functional as regulators of E1A alternative splicing in transient transfection assays [23,27]. Therefore all the experiments presented here were performed using an E1A pre-mRNA substrate lacking an MS2 operator sequence.
In the present study, we evaluated the effects of the MS2–ASF/SF2 hybrid proteins on alternative splicing of E1A pre-mRNA in vitro. It is well established that ASF/SF2 promotes proximal 5′-splice-site utilization on the E1A pre-mRNA [20,21]. In agreement with this, addition of the wild-type MS2–ASF/SF2 protein efficiently promoted 13 S 5′-splice-site usage (Figure 1C, lane 4), whereas the same molar amount of the native MS2 protein had no effect on alternative splice-site selection (lane 3). Deletion of RBD1 caused a slight reduction in the general splicing enhancer activity of the protein (MS2–RBD2+RS; lane 5). However, this protein still stimulated proximal 5′-splice-site usage. Interestingly, the MS2–RBD2 hybrid protein promoted utilization of the proximal 5′-splice site as efficiently as the MS2–RBD2+RS fusion protein (lane 6), suggesting that the RS domain is not essential for this effect. This finding is in agreement with the observation that the MS2–RS fusion protein had no stimulatory effect on E1A splicing (lane 8). The MS2–RBD1 fusion protein activated both 12 and 13 S mRNA splicing (lane 7). However, the effect was most pronounced on distal 12 S mRNA splicing. Thus the net effect of MS2–RBD1 was a preferential stimulation of distal 12 S 5′-splice-site usage (see below).
The MS2 coat protein sequence is not required for the opposite effects of ASF/SF2-RBD1 and -RBD2 on E1A alternative splice-site usage
The MS2 coat protein is a dimeric protein [37]. Consequently, it cannot be excluded that the MS2–RBD1 and MS2–RBD2 proteins through an artificial dimerization mediated by the MS2 domain may mimic an ASF/SF2ΔRS protein variant, i.e. a protein with two RBDs. Also, the MS2 part of the chimaeric proteins may bind unspecifically to the RNA.
To determine whether the MS2 domain was required for the splice-site-switching activity of MS2–ASF/SF2 fusion proteins, we generated His–RBD1 and His–RBD2 (Figure 2A). These proteins are identical with MS2–RBD1 and MS2–RBD2 (Figure 1A), except that they lack the MS2 coat protein sequence. As shown in Figure 2, the histidine variants of RBD1 and RBD2 have the same activity as the corresponding MS2 fusion proteins: His–RBD1 inducing preferentially 12 S 5′-splice-site selection (Figure 2C) and His–RBD2 enhancing 13 S 5′-splice-site usage (Figure 2D). We conclude that ASF/SF2-RBD1 and -RBD2 function as inducers of splice-site switching in the absence of artificial dimer formation. Furthermore, this experiment showed that His–RBD2, in addition to activating 13 S 5′-splice-site selection, reduced 12 S mRNA splicing (Figure 2D). This effect was not unique to the His–RBD2 protein, since the MS2–RBD2 and MS2–RBD2+RS proteins also showed opposite effects on proximal and distal 5′-splice-site usage (Figures 1 and 3).
Figure 2. ASF/SF2-RBD1 and -RBD2 have different effects on E1A alternative 5′-splice-site selection.
(A) Schematic representation of the structure of His–RBD1 and His–RBD2 proteins. His–RBD1 contains amino acids 1–92 and His–RBD2 contains amino acids 92–197 from ASF/SF2. (B) Schematic representation of the E1A pre-mRNA showing the position of splice sites and the splicing events that generate the 13 and 12 S mRNAs respectively. (C, D) In vitro splicing reactions were performed under limiting splicing conditions in HeLa-NE using the E1A pre-mRNA and the indicated His–RBD fusion proteins (150 pmol). The products were resolved by gel electrophoresis and visualized by autoradiography. Positions of pre-mRNA, splicing products and free first exons are indicated by arrowheads.
Figure 3. The conserved SWQDLKD motif and the RNP-1-type RNA recognition motif are essential for the proximal 13 S 5′-splice-site enhancer activity of ASF/SF2-RBD2.
(A) Schematic representation of the structure of MS2–RBD2+RS mutant proteins. The amino acid sequence of RBD2 is shown on the top and the residues changed in each mutant protein are indicated. The conserved SWQDLKD motif is shown boxed in the sequence. (B) In vitro splicing reactions were performed under limiting splicing conditions in HeLa-NE using the E1A pre-mRNA and the indicated MS2–RBD2+RS hybrid proteins (24.5 pmol). The products were resolved by gel electrophoresis and visualized by autoradiography. Positions of pre-mRNA, splicing products and free first exons are indicated by arrowheads.
The α-helix 1 and RNP-1 RNA recognition motifs are necessary for the proximal splice-site-switching activity of ASF/SF2–RBD2
To characterize further the structural motifs in ASF/SF2–RBD2 required for the proximal 5′-splice-site-switching activity, we used a set of ASF/SF2 mutant proteins that we previously constructed to perturb specific motifs in RBD2 ([29] and Figure 3A). Although our results have shown that the RS domain is not required for the splice-site-switching activity of RBD2, we used the MS2–RBD2+RS hybrid protein as the parental protein for mutant construction because of its more robust character (see [29]). In an attempt to minimize structural and folding perturbations, these mutations were constructed by introducing conservative amino acid changes that were predicted to preserve the secondary structure of the various motifs mutated [29]. As shown in Figure 3, a mutation of the α-helix 2 did not have any negative effect on MS2–RBD2+RS both as an activator of proximal 13 S 5′-splice-site selection and as a repressor of 12 S 5′-splice-site usage (lane 3). In contrast, mutating either the RNP-1 motif or the conserved SWQDLKD motif in α-helix 1 completely abolished the activity of MS2–RBD2+RS as a regulator of E1A alternative splicing (lanes 4 and 5). In fact, supplementing these proteins with HeLa-NE appeared to have a slight repressive effect on basal E1A splicing.
Mutation of the α-helix 1 motif in ASF/SF2-RBD2 inhibits the splice-site-switching activity of RBD2 without impairing RNA binding
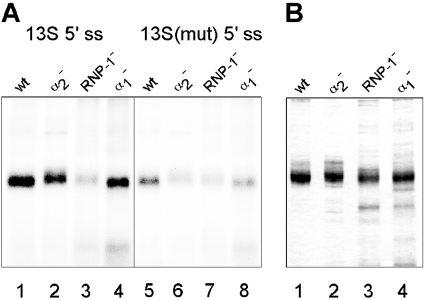
Previous studies have shown that purified recombinant ASF/SF2ΔRS can specifically bind to the E1A 13 S 5′-splice site [32]. To determine whether the failure of the RNP-1 and α-helix 1 mutant proteins to induce proximal 13 S 5′-splice-site selection correlated with a loss in specific RNA binding, we performed UV cross-linking experiments using a short RNA spanning the E1A 13 S 5′-splice site. In these experiments, the wild-type and mutant MS2–RBD2+RS proteins (depicted in Figure 3A) were cross-linked under splicing conditions to a 115-nucleotide 32P-labelled RNA fragment spanning the E1A 13 S 5′-splice site. As shown in Figure 4(A), a mutation affecting the RNP-1 motif (lane 3) resulted in a significant reduction in the RNA-binding capacity of the mutant protein. This result was expected since the RNP-1 motif is a part of the β-strands making contact with the RNA. Since this protein also failed to activate E1A splicing (Figure 3), the results suggest that the RNA-binding capacity of RBD2 is necessary for its function as a 13 S 5′-splice-site enhancer protein. Most interestingly, a mutation destroying the conserved SWQDLKD motif in α-helix 1 did not significantly reduce the RNA-binding capacity of the MS2–RBD2+RS protein (Figure 4, lane 4), but completely inactivated it as an E1A 13 S 5′-splice-site enhancer protein (Figure 3). This finding is noteworthy and suggests that the protein–protein interactions mediated by α-helix 1 may be essential for the splice-site-switching activity of ASF/SF2–RBD2. From this point, it is interesting to note that, in the predicted structure of ASF/SF2–RBD2, α-helix 1 is positioned opposite to the β-strands making contact with the RNA. As expected, the α-helix 2 mutant protein, which had wild-type splice-site-switching activity (Figure 3), was also efficient in RNA binding (lane 2). Figure 4(B) shows a Coomassie Blue-stained gel of the RBD2 mutant proteins used in the UV cross-linking experiment.
Figure 4. The RNP-1-type RNA recognition motif of ASF/SF2-RBD2, but not the SWQDLKD motif, is required for protein binding to the E1A 13 S 5′-splice site.
(A) RNA binding was tested by a UV cross-linking assay in which ASF/SF2-RBD2 mutant proteins (schematically depicted in Figure 3A) were incubated and cross-linked to a short 32P-labelled RNA, containing the E1A 13 S wild-type (lanes 1–4) or mutant 5′-splice site (lanes 5–8). The products were resolved on a 12% SDS/polyacrylamide gel and visualized by autoradiography. (B) Coomassie Blue-stained gel of the ASF/SF2-RBD2 mutant proteins used in the experiments shown in Figures 3 and 4.
To demonstrate that the ASF/SF2-RBD2 domain binds specifically to the E1A 13 S 5′-splice site, the UV cross-linking experiment was repeated using an RNA containing four mutations that were previously shown to be critical for ASF/SF2 binding to the 13 S 5′-splice site [32]. As shown in Figure 4(A), all the proteins showed a drastic reduction or complete abolishment of RNA binding (lanes 5–8). Collectively, these results suggest that the MS2–RBD2+RS fusion proteins interacted preferentially with the E1A 13 S 5′-splice site. It is interesting to note that the α-helix 2 mutant protein, which interacted almost as efficiently as the wild-type and α-helix 1 mutant proteins with the native 13 S 5′-splice site (Figure 4A, lane 2), was unable to interact with the mutated 13 S 5′-splice site (lane 6).
ASF/SF2-RBD2 is a repressor of E1A 12 S 5′-splice-site usage
To dissect further the activity of ASF/SF2 on E1A alternative splice-site usage, we tested the effect of the RBDs on splice-site activation of the 12 or 13 S 5′-splice sites, in the absence of competition. For this experiment, two additional pre-mRNAs were created (Figure 5A). The E1A 12 S pre-mRNA was generated by introducing four base changes that inactivate the E1A 13 S 5′-splice site in the E1A unit [32]. The E1A 13 S pre-mRNA was generated by synthesis of a shorter transcript encoding only the proximal E1A 13 S 5′-splice. As shown in Figure 5(B), addition of His–RBD2 to the tandem E1A pre-mRNA stimulated, as expected (Figure 2D), the proximal 13 S 5′-splice site and simultaneously repressed the distal 12 S 5′-splice-site usage (lane 2). Interestingly, on transcripts encoding the individual 5′-splice sites, His–RBD2 efficiently repressed E1A 12 S pre-mRNA splicing (lane 5), suggesting that this domain indeed has a splicing repressor phenotype. In addition, the His–RBD2 protein also had a slight stimulatory effect on E1A 13 S splicing (lane 8). In contrast, His–RBD1, which activates preferentially 12 S mRNA splicing in the tandem construct (lane 3), activated strongly both E1A 12 and 13 S splicing under non-competitive conditions (lanes 6 and 9). Taken together, these results suggest that the RBD1 domain has a more general splicing enhancer activity, possibly stimulating the cap-proximal 5′-splice site irrespective of its origin.
DISCUSSION
Previous studies have shown that the RS domain of ASF/SF2 is necessary for its activity as a constitutive splicing factor [30,38], but is not required for the activity of the protein as an inducer of alternative 5′-splice-site selection [30,38]. Thus an ASF/SF2ΔRS protein stimulates proximal 5′-splice-site selection, both in vitro and in vivo [15,30,38]. In the present study, we show that the ASF/SF2-RBD2 domain has a decisive and dominant role as a regulator of E1A alternative splicing. In fact, the ASF/SF2-RBD2 domain alone is sufficient to reproduce the activity of the full-length protein on E1A alternative splice-site activation in vitro under our experimental conditions. The specificity is identical although the activity is slightly lower compared with the full-length protein (Figure 1). Our results suggest that the RBD2 domain induces a shift towards 13 S splicing by possessing contrasting activities on the individual 12 and 13 S 5′-splice sites. Thus the shift in splice-site selection appears to result from a specific RBD2-mediated inhibition of 12 S 5′-splice-site selection combined with an enhancement of 13 S 5′-splice-site usage (Figure 5). In contrast, ASF/SF2-RBD1 had the opposite effect and promoted distal 12 S 5′-splice-site usage (Figure 1). On the individual 5′-splice sites, RBD1 stimulated both 12 and 13 S 5′-splice-site usage (Figure 5). Therefore it is quite probable that ASF/SF2-RBD1 has a more general activity as a splicing enhancer domain. The preferential activation of the 12 S 5′-splice site in the tandem construct (Figures 1 and 5) may be explained by a model where the ASF/SF2-RBD1 preferentially stimulates splicing to the cap-proximal 5′-splice site. Such a model would explain how RBD1 causes a shift towards 12 S splicing in the tandem construct while efficiently stimulating both the 12 and 13 S 5′-splice sites under non-competitive conditions.
The activity of ASF/SF2-RBD1 resembles the activity ascribed to the hnRNP (heterogeneous nuclear ribonucleoprotein) A1 protein. Thus hnRNP A1 has been shown to counteract the stimulatory activity of ASF/SF2 on 13 S mRNA splicing and to enhance cap-proximal 5′-splice-site selection [13,21,39]. An alignment of the RBDs of these two proteins indicates that ASF/SF2-RBD1 is more similar to the two RBDs of hnRNP A1 than to ASF/SF2-RBD2 [40].
The dose-dependent enhancement of proximal 5′-splice-site selection by ASF/SF2 has been explained using an occupancy model, where ASF/SF2 stimulates U1 snRNP recruitment to all the 5′-splice sites [26]. Results presented here support this model and suggest that the modular activities of the two RBDs of ASF/SF2 contribute differently to the overall E1A alternative splice-site-switching activity. Thus, in agreement with previous results [5,41], ASF/SF2-RBD2 appears to be the dominant regulator of E1A splice-site selection in the context of the wild-type protein, since the full-length protein has the same activity as the RBD2 domain (Figure 1). In fact, ASF/SF2-RBD2 alone causes an inhibition of 12 S mRNA splicing; i.e. a probable reduction in U1 snRNP occupancy at the 12 S 5′-splice site. However, since ASF/SF2-RBD1 has the opposite activity of stimulating cap-proximal 5′-splice-site occupancy, the wild-type ASF/SF2 protein may stimulate U1 snRNP recruitment to both the 5′-splice sites. As a result, when all the 5′-splice sites are filled, splicing occurs between the 5′-splice sites closest to the 3′-splice site [26].
The repressive function of the ASF/SF2-RBD2 domain on E1A 12 S 5′-splice-site splicing is reminiscent of our previous observation that ASF/SF2-RBD2 functions as a splicing repressor domain inhibiting IIIa 3′-splice-site usage [29]. However, in the present study, we show that the regulation of 5′-splice-site usage is more complex, with the RBD2 domain functioning both as an activator (13 S) and as a repressor (12 S) domain in splicing. Furthermore, the activity of ASF/SF2-RBD2 as a repressor of IIIa 3′-splice-site usage required an artificial tethering of the domain to the pre-mRNA [29], whereas this was not required for its function as a regulator of E1A 5′-splice-site usage. A possible explanation for the difference in results could be related to the fact that ASF/SF2 appears to bind directly to a 5′-splice site [6,32]. In contrast, ASF/SF2 and also SR proteins in general are believed to enhance or repress 3′-splice-site usage by binding to splicing enhancer or splicing repressor elements that can be located at some distance from the 3′-splice site (reviewed in [3]).
Mutating the RNP-1 motif completely inactivated the MS2–RBD2+RS protein as an E1A splicing enhancer protein (Figure 3). In our previous experiments on IIIa splicing, the same mutation had no effect on the activity of the protein as a 3′-splice-site repressor protein [29]. This result was as expected, since the activity of RBD2, as a regulator of IIIa 3′-splice-site usage, required the artificial tethering of the protein to the RNA via an MS2 operator sequence and, therefore, was not dependent on its own RNA-binding capacity. This difference in results suggests that restoring the RNA-binding capacity of the RNP-1 mutant protein, by introducing an MS2 operator at an appropriate position in the E1A pre-mRNA, would re-activate the MS2–RBD2+RS (RNP-1−) protein as a regulator of E1A splicing. We have tested this hypothesis by inserting an MS2 operator site at several positions in the E1A pre-mRNA. However, we were unable to restore the capacity of the RNP-1 mutant protein to function as an activator of E1A splicing (results not shown) at any of the tested positions. We showed previously that placing the highly structured MS2 operator too close to the IIIa 3′-splice site resulted in a general inhibition of splicing [29]. Because of this finding, we did not attempt to insert the MS2 operator directly at the 13 S 5′-splice site. We cannot therefore exclude that our experiments failed because the highly structured MS2 operator was not placed at the appropriate position to aid in the recruitment of U1 snRNP.
Mutating the SWQDLKD motif in ASF/SF2-RBD2 had no significant effect on the RNA-binding capacity of the mutant protein (Figure 4). This result was logical since α-helix 1 is predicted to be positioned opposite to the β1 and β3 strands making contact with the RNA in the predicted structure of RBD2. Therefore the complete loss of splicing enhancer activity of this protein suggests that this motif probably functions as an interaction surface mediating critical contacts between ASF/SF2 tethered to the pre-mRNA and other factors in the spliceosome. In this respect, it is interesting to note that this motif was also essential for the activity of the MS2–RBD2+RS protein to function as a repressor of adenovirus IIIa splicing [29]. Taken together, our results suggest that the SWQDLKD motif is a crucial domain mediating critical interactions between ASF/SF2 and the splicing machinery. Obviously, characterization of the partner proteins that interact with the α-helix 1 domain of ASF/SF2-RBD2 will be important to explain the multifaceted character of this domain on 5′- and 3′-splice-site usage.
Acknowledgments
We are grateful to Dr Martin Luetzelberger for fruitful discussions. This work was supported by the Swedish Cancer Society.
References
- 1.Hastings M. L., Krainer A. R. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 2001;13:302–309. doi: 10.1016/s0955-0674(00)00212-x. [DOI] [PubMed] [Google Scholar]
- 2.Caceres J. F., Kornblihtt A. R. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- 3.Graveley B. R. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tacke R., Manley J. L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandler S. D., Mayeda A., Yeakley J. M., Krainer A. R., Fu X. D. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3596–3601. doi: 10.1073/pnas.94.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohtz J. D., Jamison S. F., Will C. L., Zuo P., Luhrmann R., Garcia-Blanco M. A., Manley J. L. Protein–protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature (London) 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 7.Wu J. Y., Maniatis T. Specific interactions between proteins implicated in splice-site selection and regulated alternative splicing. Cell (Cambridge, Mass.) 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 8.Xiao S. H., Manley J. L. Phosphorylation of the ASF/SF2 RS domain affects both protein–protein and protein–RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 9.Zuo P., Maniatis T. The splicing factor U2AF35 mediates critical protein–protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]
- 10.Roscigno R. F., Garcia-Blanco M. A. SR proteins escort the U4/U6.U5 tri-snRNP to the spliceosome. RNA. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- 11.Zahler A. M., Neugebauer K. M., Lane W. S., Roth M. B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 12.Fu X. D., Mayeda A., Maniatis T., Krainer A. R. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice-site selection. Proc. Natl. Acad. Sci. U.S.A. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caceres J. F., Stamm S., Helfman D. M., Krainer A. R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 14.Screaton G. R., Caceres J. F., Mayeda A., Bell M. V., Plebanski M., Jackson D. G., Bell J. I., Krainer A. R. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J., Manley J. L. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- 16.Berk A. J., Sharp P. A. Structure of the adenovirus 2 early mRNAs. Cell (Cambridge, Mass.) 1978;14:695–711. doi: 10.1016/0092-8674(78)90252-0. [DOI] [PubMed] [Google Scholar]
- 17.Akusjärvi G., Stevenin J. Remodelling of the host cell RNA splicing machinery during an adenovirus infection. Curr. Top. Microbiol. Immunol. 2003;272:253–286. doi: 10.1007/978-3-662-05597-7_9. [DOI] [PubMed] [Google Scholar]
- 18.Schmitt P., Gattoni R., Keohavong P., Stevenin J. Alternative splicing of E1A transcripts of adenovirus requires appropriate ionic conditions in vitro. Cell (Cambridge, Mass.) 1987;50:31–39. doi: 10.1016/0092-8674(87)90659-3. [DOI] [PubMed] [Google Scholar]
- 19.Gattoni R., Chebli K., Himmelspach M., Stevenin J. Modulation of alternative splicing of adenoviral E1A transcripts: factors involved in the early-to-late transition. Genes Dev. 1991;5:1847–1858. doi: 10.1101/gad.5.10.1847. [DOI] [PubMed] [Google Scholar]
- 20.Harper J. E., Manley J. L. Multiple activities of the human splicing factor ASF. Gene Expr. 1992;2:19–29. [PMC free article] [PubMed] [Google Scholar]
- 21.Mayeda A., Krainer A. R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell (Cambridge, Mass.) 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 22.Himmelspach M., Cavaloc Y., Chebli K., Stevenin J., Gattoni R. Titration of serine/arginine (SR) splicing factors during adenoviral infection modulates E1A pre-mRNA alternative splicing. RNA. 1995;1:794–806. [PMC free article] [PubMed] [Google Scholar]
- 23.Caceres J. F., Misteli T., Screaton G. R., Spector D. L., Krainer A. R. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W. J., Wu J. Y. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol. Cell. Biol. 1996;16:5400–5408. doi: 10.1128/mcb.16.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cavaloc Y., Popielarz M., Fuchs J. P., Gattoni R., Stevenin J. Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eperon I. C., Makarova O. V., Mayeda A., Munroe S. H., Caceres J. F., Hayward D. G., Krainer A. R. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 2000;20:8303–8318. doi: 10.1128/mcb.20.22.8303-8318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Houven#x00A0;van Oordt W., Newton K., Screaton G. R., Caceres J. F. Role of SR protein modular domains in alternative splicing specificity in vivo. Nucleic Acids Res. 2000;28:4822–4831. doi: 10.1093/nar/28.24.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanopka A., Mühlemann O., Akusjärvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature (London) 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- 29.Dauksaite V., Akusjärvi G. Human splicing factor ASF/SF2 encodes for a repressor domain required for its inhibitory activity on pre-mRNA splicing. J. Biol. Chem. 2002;277:12579–12586. doi: 10.1074/jbc.M107867200. [DOI] [PubMed] [Google Scholar]
- 30.Caceres J. F., Krainer A. R. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yue B. G., Ajuh P., Akusjärvi G., Lamond A. I., Kreivi J. P. Functional coexpression of serine protein kinase SRPK1 and its substrate ASF/SF2 in Escherichia coli. Nucleic Acids Res. 2000;28:E14. doi: 10.1093/nar/28.5.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuo P., Manley J. L. The human splicing factor ASF/SF2 can specifically recognize pre-mRNA 5′ splice sites. Proc. Natl. Acad. Sci. U.S.A. 1994;91:3363–3367. doi: 10.1073/pnas.91.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bondesson M., Öhman K., Manervik M., Fan S., Akusjärvi G. Adenovirus E4 open reading frame 4 protein autoregulates E4 transcription by inhibiting E1A transactivation of the E4 promoter. J. Virol. 1996;70:3844–3851. doi: 10.1128/jvi.70.6.3844-3851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts R. J., Akusjärvi G., Aleström P., Gelinas R. E., Gingeras T. R., Sciaky D., Pettersson U. Vol. 8. Boston: Martin Nijhoff Publishing; 1986. A Consensus Sequence for the Adenovirus-2 Genome. [Google Scholar]
- 35.Mühlemann O., Akusjärvi G. Preparation of splicing competent nuclear extracts from adenovirus infected cells. In: Wold W. S. M., editor. Methods in Molecular Biology. Vol. 21. Totowa, NJ: Humana Press; 1998. pp. 203–216. [Google Scholar]
- 36.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. Boston: John Wiley & Sons; 1995. Current Protocols in Molecular Biology; pp. 10.2.4–10.2.16. [Google Scholar]
- 37.Valegard K., Murray J. B., Stockley P. G., Stonehouse N. J., Liljas L. Crystal structure of an RNA bacteriophage coat protein–operator complex. Nature (London) 1994;371:623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- 38.Zuo P., Manley J. L. Functional domains of the human splicing factor ASF/SF2. EMBO J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X., Bani M. R., Lu S. J., Rowan S., Ben-David Y., Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice-site selection in vivo. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Birney E., Kumar S., Krainer A. R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayeda A., Screaton G. R., Chandler S. D., Fu X. D., Krainer A. R. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol. Cell. Biol. 1999;19:1853–1863. doi: 10.1128/mcb.19.3.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]